Abstract
Post-transcriptional modifications of RNA are nearly ubiquitous in the principal RNAs involved in translation. However, in the case of rRNA the functional roles of modification are far less established than for tRNA, and are subject to less knowledge in terms of specific nucleoside identities and their sequence locations. Post-transcriptional modifications have been studied in the SSU rRNA from Thermotoga maritima (optimal growth 80°C), one of the most deeply branched organisms in the Eubacterial phylogenetic tree. A total of 10 different modified nucleosides were found, the greatest number reported for bacterial SSU rRNA, occupying a net of ∼14 sequence sites, compared with a similar number of sites recently reported for Thermus thermophilus and 11 for Escherichia coli. The relatively large number of modifications in Thermotoga offers modest support for the notion that thermophile rRNAs are more extensively modified than those from mesophiles. Seven of the Thermotoga modified sites are identical (location and identity) to those in E. coli. An unusual derivative of cytidine was found, designated N-330 (M r 330.117), and was sequenced to position 1404 in the decoding region of the rRNA. It was unexpectedly found to be identical to an earlier reported nucleoside of unknown structure at the same location in the SSU RNA of the archaeal mesophile Haloferax volcanii.
Keywords: post-transcriptional modification, Thermotoga maritima, Haloferax volcanii, small ribosomal subunit RNA, liquid chromatography–mass spectrometry, electrospray ionization
INTRODUCTION
Relatively little is known concerning the specific roles of post-transcriptional modifications in rRNA (summarized by Decatur and Fournier 2002), in contrast to tRNA, in which a number of modification functions are slowly being unraveled (Agris 2004). In the case of rRNA, the influence of nucleotide modification on ribosome assembly and maturation (Bachellerie and Cavaillé 1998; Maden 1998; Mason 1998) is perhaps the best documented (Vaughan et al. 1967; Cunningham et al. 1990, 1991; Rydén-Aulin et al. 1993; Sirum-Connolly and Mason 1993; Sirum-Connolly et al. 1995; Green and Noller 1996), but mechanistic details at the molecular level have been scant. There is strong circumstantial evidence in the archaeal thermophiles that ribose methylation at O2′ provides a significant mechanism for thermal stabilization of rRNA (Noon et al. 1998) and of archaeal tRNA (Kowalak et al. 1994; Noon et al. 2003), primarily through reduced flexibility resulting from strong thermodynamic enforcement of the C3′-endo sugar conformation (Kawai et al. 1992). Also, from the standpoint of function, selective methylation of rRNA nucleotides provides, in some instances, an effective mechanism for conferring resistance to antibiotics that target the bacterial ribosome (Douthwaite et al. 2005).
Many of the structures and sequence locations of rRNA modifications are relatively well conserved (Maden 1990; Rozenski and McCloskey 2005) and exhibit distinct differences among the three principal evolutionary domains, Archaea, Bacteria, and Eukarya. The complete modification map for SSU rRNA of the bacterial thermophile Thermus thermophilus was recently completed (Guymon et al. 2006) and found to exhibit a notably lower level of modification than that of the archaeal thermophile Sulfolobus solfataricus (Noon et al. 1998) growing in the same temperature range (70°C–75°C), and a pattern of modification (nucleoside structures and sequence locations) unexpectedly similar to that in the mesophile Escherichia coli. For a tabulation of E. coli rRNA modifications and leading references see http://medlib.med.utah.edu/RNAmods/.
We have examined the SSU rRNA modifications in a second bacterial thermophile, Thermotoga maritima (optimal growth at 80°C) (Huber et al. 1986), an organism placed by small subunit ribosomal RNA (SSU rRNA) phylogeny as one of the most deeply rooted organisms in the Eubacterial tree. Analysis of the complete Thermotoga genome sequence (Nelson et al. 1999) as well as earlier data (Aravind et al. 1998) led to conclusions that extensive lateral gene transfer between T. maritima and Archaea have occurred, reflected in the unusually high percentage (24%) of genes in T. maritima that are similar to those in Archaea. Examination of T. maritima SSU rRNA in the present study was intended in part to establish whether its modifications are to any extent archaeal in nature, and thus reflecting possible horizontal transfer of RNA modification enzymes. Further, this study serves to extend the knowledge base of bacterial SSU rRNA modification patterns (Rozenski and McCloskey 2005; Guymon et al. 2006) beyond E. coli and T. thermophilus, the only bacteria for which complete SSU modification maps are available, and beyond organisms for which RNase T1 catalog data were reported (Rozenski and McCloskey 2005) in which modifications were sometimes reported to occur, often without chemical identity of the modified nucleoside or firm knowledge of its sequence location in the 16S RNA molecule.
RESULTS
Modified nucleoside content of T. maritima 16S rRNA
Identities and approximate numbers of modified nucleosides, determined by LC/ESI-MS analysis of total nucleoside digests of 16S rRNA (Supplemental Fig. S1, at http://library.med.utah.edu/mccloskey) are: Ψ (3.8 residues), m3U (1.0), m4Cm (1.2), m5C (0.73), Cm (0.82), unknown nucleoside N-330 (UV molar absorptivity not known, so stoichiometry not estimated), m2G (2.3), m7G (1.1), Am (trace level to 1.3; see text), and m2 6A (2.1).
Identification and sequence location of modified residues in T. maritima 16S rRNA
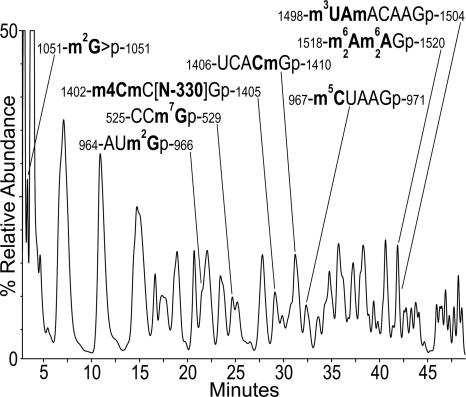
RNA modifications represented by changes in mass from the parent nucleotide were mapped. The locations of pseudouridines, which are mass silent, were not measured beyond the estimation of 3.8 net residues in the molecule. Chromatographic separation of RNase T1 digestion products of T. maritima SSU rRNA is shown in Figure 1, with the corresponding mass data for modified oligonucleotides given in Table 1. A section of the RNase U2 chromatogram in which modified oligonucleotides were eluted is shown in Supplemental Figure S2.
FIGURE 1.
HPLC separation of RNase T1 digestion products of T. maritima 16S rRNA, annotated to show modified oligonucleotides determined from mass shifts in conjunction with data in Table 1. UV absorbance detection at 260 nm.
TABLE 1.
Modified nucleosides in RNases T1 and U2 hydrolysis products from Thermotoga maritima 16S rRNA
To a certain extent the procedures used for identification and sequence placement of modified residues are analogous to those recently used and described in a study of T. thermophilus SSU rRNA (Guymon et al. 2006). Therefore, descriptions of results for seven modified residues in the present work can be found in the Supplemental Data section. These are: m7G-527 (Supplemental Fig. S3), m2G-966 and m5C-967 (Supplemental Fig. S4), m2G-1051 (Supplemental Table S2), m4Cm-1402 (Supplemental Fig. S5), Cm-1409 (Supplemental Figs. S6,S7), and 1518-m2 6Am2 6A-1519 placement. Results for N-330–1404, m3U-1498, and Am-1499 follow below.
N-330–1404
The presence of a novel structurally unknown nucleoside of M r 330 at position 1404 is indicated from the following four lines of evidence. (1) The sequencing spectrum of T1 oligonucleotide M r 1393 (Supplemental Fig. S5) can be assigned only if a unique nucleotide residue mass of 392 (N-330p minus H2O) is used in the third nucleotide position. (2) The base– (m/z 197) and N > p– (m/z 391) monomer fragment ion signals (Supplemental Table S1) were assigned from the data in Supplemental Figure S5 and time aligned as shown in Supplemental Figure S8. Ions corresponding to the modified residue m4Cm, whose presence is indicated from the total nucleoside analysis data in Supplemental Figure S1, were also time aligned. The exact coelution pattern of ions from N-330 and m4Cm is shown in Supplemental Figure S8, and supports the presence of both of those nucleosides in the T1 oligonucleotide M r 1393. The characteristic bacterial nucleoside m4Cm serves as an internal marker for position 1402 (Rozenski and McCloskey 2005) and correlates with the full oligonucleotide sequence derived by mass spectrometry in Supplemental Figure S5. (3) The presence of a base-modified unknown nucleoside of M r 330 (MH+, m/z 331; BH+ 2, m/z 199; λmax 270) eluting at 3.85 min in the total nucleoside digest of Thermotoga 16S rRNA, Supplemental Figure S1. (4) Modification of the universal C-1404 was also reported in several other bacterial SSU rRNAs (although not in E. coli) (Rozenski and McCloskey 2005), but not the identity of the modification.
m3U-1498 and Am-1499
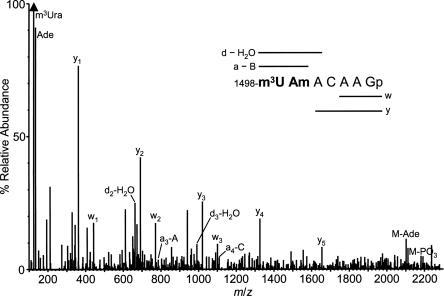
Assignment of the unusual tandem methylation sites 1498 and 1499 was made primarily through the T1 product M r 2318 (Table 1). The experimental mass 2318.43, using the corresponding gene sequence to calculate allowable RNA sequences, solely designates the unique sequence segment as shown in Table 1 corresponding to nucleotides 1498–1504 with two methyl groups. The sequence mass spectrum of this T1 fragment (Fig. 2) requires, through the presence of a clear y1–y5 ion series (McLuckey et al. 1992), that the methyl groups be confined to the first two nucleotides. The abundant methyluracil base ion m/z 125, in conjunction with the total nucleoside census (Supplemental Fig. S1) then implies m3U in position 1498 and Am, the only methylated A in the rRNA, at position 1499. (No methyladenine base ion, m/z 148, was observed). This methylation pattern was supported by the finding in the U2 digest of the 3′-truncated version of this sequence but having an extension of three nucleotides on the 5′ end, M r 1620 (see Table 1). However, the assignment of Am-1499 in these RNase products appeared to be at odds with the absence of the methylribose cyclic phosphate ion (m/z 225) in the spectrum, usually a strong indicator of presence of a ribose-methylated nucleotide (Phillips and McCloskey 1993). To address this point the product ion mass spectrum of chemically synthesized m3UAmACAAGp was acquired. It appeared essentially identical (data not shown) to the mass spectrum displayed in Figure 2, including the absence of ion m/z 225. The sequence of the Am-containing region is therefore assigned as shown in Table 1. The occurrence of Am is further supported by its presence in the total nucleoside digest (Fig. S1), with MH+ and BH+ 2 ions appearing at the expected retention time (Pomerantz and McCloskey 1990). Interestingly, a second culture of T. maritima showed an otherwise very similar overall modification data set, but with the 1495–1499 fragment occurring primarily as M r 1605.2 (see Table 1), i.e., with one fewer methyl group. We interpret this finding, taken in conjunction with variable amounts of Am found in nucleoside total digests from two different T. maritima cultures, as an indication that the level of Am-1499 is both substoichiometric and variable in the rRNA. To our knowledge, this is the first report of Am in RNA from bacteria; however, using LC/ESI-MS we have observed Am in unfractionated tRNA from E. coli MRE 600 (F. Qiu and J. McCloskey, unpubl., 1998).
FIGURE 2.
Product ion mass spectrum from RNase T1 fragment M r 2318, showing m3U-1498 adjacent to Am-1499. For clarity, only peaks used for sequence assignment are annotated. Sequence coverage from the four sequence ion series (a–B, d–H2O, w, and y) (McLuckey et al. 1992) is shown by horizontal bars.
3′-Terminal RNase T1 fragment from T. maritima 16S rRNA
The terminal fragment is a 15-mer, expected to contain the anti-Shine–Dalgarno mRNA-binding sequence. Because of the recent unusual finding of adjacent pseudouridines at 1540 and 1541 in the 3′-tail of T. thermophilus (Guymon et al. 2006), the Thermotoga 3′-terminal fragment was subjected to a Ψ detection protocol (Pomerantz and McCloskey 2005) using LC/ESI-MS/MS. No Ψ was detected. The terminal fragment was mass measured as 4591.7 (4591.60 calc.), and MS-sequenced as AUCACCUCCUUUCUAOH (Supplemental Fig. S9), thus revealing the molecular terminus as AOH-1545 (E. coli numbering) in the processed 16S molecule.
Presence of nucleoside N-330 in Haloferax volcanii 16S rRNA
The discovery of a new modified nucleoside in the Thermotoga SSU at the same position previously described as containing a modified cytidine in H. volcanii 16S rRNA (Gupta et al. 1983) prompted a revisit of our previously published study of the Haloferax SSU RNA modifications (Kowalak et al. 2000). When the ion chromatograms for the diagnostic MH+ and BH+ 2 ions for N-330 nucleoside (m/z 331 and 199, respectively) were extracted from our Haloferax total nucleoside digest data, their presence was confirmed by the appearance of these two ions in a peak exactly coeluting at 3.6 min (Fig. 3). If N-330 is, in fact, the “C*” nucleotide previously reported, the calculated mass of the predicted Haloferax N-330-containing T1 oligonucleotide (CCC*Gp) would be M r 1365.7. Reconstructed ion chromatograms were therefore generated using the M− and M2− ions (m/z 1364.7 and m/z 681.8, respectively), and for the B− ion for the base of N-330 (m/z 197). As shown in Supplemental Figure S10, each of the three ions forms a peak eluting at 16.0 min, indicating N-330 to be the modified C* nucleotide found by Woese and colleagues (Gupta et al. 1983).
FIGURE 3.
Demonstration of presence of modified nucleoside N-330 in a total nucleoside digest of H. volcanii 16S rRNA (from previously acquired data) (Kowalak et al. 2000), detected from reconstructed ion chromatograms (RIC) for characteristic positively charged ions. (A) UV absorbance at 260 nm. (B) RIC for the MH+ ion of N-330 (m/z 331). (C) RIC for the protonated base fragment ion of N-330 (m/z 199).
DISCUSSION
Modification identities and levels in T. maritima SSU RNA
The finding of 10 different modified nucleosides at a net occupancy level of ∼14 sequence sites marks T. maritima, and similarly T. thermophilus (Guymon et al. 2006), as the most extensively modified bacterial SSU RNAs presently known. Examination of a number of catalogued RNase T1 modification maps, although by their nature less complete than the full modification maps from E. coli (leading citations in ref. Bakin et al. 1994; see also http://medlib.med.utah.edu/RNAmods), and T. thermophilus (Guymon et al. 2006) SSU RNAs, suggests that the levels and identities of modifications are perhaps narrower and more conserved in Bacteria than in the Archaea and Eukarya. For example, modification levels in Archaeal 16S RNA range from five residues in H. volcanii (Gupta et al. 1983) to ∼38 in S. solfataricus (Noon et al. 1998). Modification at ∼8–11 SSU RNA sequence sites appears to be most common in bacteria (E. coli has 11) while the elevation by about 30% of modification levels in the bacterial thermophiles (versus mesophiles) supports the conclusion that post-transcriptional modification in general serves to support structural stabilization of RNA (Sampson and Uhlenbeck 1988; Derrick and Horowitz 1993; Kowalak et al. 1994). The presence of 11 modifications was recently reported for T. thermophilus 23S rRNA (Mengel-Jorgensen et al. 2006), compared with ∼23 in E. coli LSU RNA. This relative concentration is significantly lower than expected, based on our finding of 14 sites in the Thermus 16S RNA. However, the methodology used (Mengel-Jorgensen et al. 2006) was not designed to provide a complete census of modifications that required placement. As in the case of T. thermophilus (Guymon et al. 2006), rRNA modification levels in Thermotoga are characteristically much lower than in the Archaeal thermophiles growing at about the same temperature (Noon et al. 1998), with considerably less reliance on ribose O-2′ methylation as a major means of structural stabilization. The finding of a net 3.8 residues of Ψ in Thermotoga SSU RNA is notable in that Ψ has been shown to play a strong, although often overlooked role in RNA stabilization (Davis 1995, 1998), and the level in Thermotoga appears to be the highest concentration of Ψ reported in bacterial SSU RNAs (Ofengand and Rudd 2000).
The availability of three bacterial SSU RNA modification maps, E. coli (Brosius et al. 1978), Thermus (Guymon et al. 2006), and now Thermotoga, when coupled with information from T1 catalogs (Rozenski and McCloskey 2005) permits an estimation of the most highly conserved modification sites that are unique to bacteria, even though most of the T1 catalog data identify sites but not chemical structures of modifications. In bacteria these sites are C*-967 in h31 and U*-1498 in the center of the decoding region in h44. An exception to the bacterial uniqueness of a modified C-967 appears to occur in the archaeon Thermoproteus tenax (Woese et al. 1984). The position 967 modification is reported to be m5C in all four cases in which the nucleoside structure has been established (E. coli, Thermus, Thermotoga, and Proteus). The adjacent modification m2G-966, implicated in P-site tRNA binding (von Ahsen and Noller 1995; Korostelev et al. 2006), represents an essentially universal SSU modification site in all phylogenetic domains (Rozenski and McCloskey 2005) and is represented by an interesting diversity of modified nucleoside structures in Archaea and Eukarya (Kowalak et al. 2000). The identity of U*-1498 has been established specifically as m3U in the same four organisms. The unusual dimethylcytidine m4Cm-1402 is unique to bacterial rRNA (Limbach et al. 1994), but this site appears to be modified in fewer than half of the reported cases (Rozenski and McCloskey 2005). In T. thermophilus the m4Cm residue serves to stabilize the third tRNA nucleotide by H binding of its phosphate to the 4-methylamino moiety (Korostelev et al. 2006).
Modifications at the functional center of the ribosome
As deduced in earlier literature, modification sites in bacterial SSU RNAs tend to occur, in three-dimensional space, near the decoding center of the RNA (Brimacombe et al. 1993; Decatur and Fournier 2002). Four of the 16S modifications (m2G-966, m5C-1400, m4Cm-1402, and m3U-1498) were determined by X-ray crystallography to support interaction between 16S RNA and the P-site codon and anticodon stem–loop (Korostelev et al. 2006). These observations reflect the net importance of modification to efficient ribosomal function, as has been stated (Decatur and Fournier 2002). The high level of modification in the upper portion of helix 44 in SSU RNA occurs at the interface with the LSU RNA, forming a cavity in which translation occurs. These modifications in h44 include six methyl groups each in Thermotoga and Thermus, opposite an analogous concentration of modifications on the 23S side of the Thermus LSU (Mengel-Jorgensen et al. 2006). Mengel-Jorgensen et al. (2006) have concluded that the occurrence of modifications in the 23S RNA of Thermus principally at the RNA–RNA interface suggests that they play a role in modulating the RNA–RNA interface contact. Their conclusions are supported by the locations of modifications in 16S RNA from Thermotoga (present work), Thermus (Guymon et al. 2006), and a series of studies on E. coli (Brimacombe et al. 1993; Decatur and Fournier 2002), showing that the ribosomal subunit interface is intimately associated with post-transcriptional modifications.
Modified nucleoside N-330–1404 in the decoding region of the RNA
Unknown N-330 is remarkable in two ways: first, in terms of its structural properties as inferred thus far; second, its unexpected occurrence in another phylogenetic domain, at the same location in the SSU RNA of the archaeal mesophile H. volcanii. Interestingly, C-1404 has been reported as modified but with unknown structure in the RNase T1 SSU maps of five bacteria (Rozenski and McCloskey 2005), and two archaea, H. volcanii (Gupta et al. 1983) and S. solfataricus (Woese et al. 1984). C-1404 pairs with essentially universal G-1497 (Cannone et al. 2002) near the top of h44. N-330 is implied to be a derivative of cytidine by the corresponding gene sequence (Gupta et al. 1983). The molecular mass of 330 Da is unique among all known modified nucleosides in RNA (see http://medlib.med.utah.edu/RNAmods) and suggests its structure to be more complex than any of the nine known modified cytidines in rRNA (Rozenski and McCloskey 2005). The accurate molecular mass of N-330 was measured as 330.117 ± 0.002 using a Micromass Q-Tof mass spectrometer. This value is consistent with an elemental composition of C12H18N4O7 (calc. 330.1175), thus requiring a side chain having one N atom, with three Ns accounted for by the cytosine heterocycle. Unfortunately, the exact chemical structure of N-330-1404 cannot be further pursued due to laboratory closure and retirement of the corresponding author.
It is interesting to consider the possibility that the structures and distributions of post-transcriptionally modified nucleosides—the specific products of a sizable array of RNA modification enzymes—could, in some instances, be indicators of past horizontal transfer of DNA coding for modification enzymes. This notion is supported by the sharp lines of phylogenetic demarcation of some modified nucleoside families (Limbach et al. 1994; Motorin and Grosjean 1998). The possibility of lateral transfer was earlier raised (McCloskey et al. 2001) in light of the strong clustering of the (otherwise bacterial) anticodon nucleoside mnm5s2U in tRNAs from the Methanococci, a lineage of methanogenic marine archaea. Interestingly, nucleoside N-330 is the only rRNA nucleoside occurring in Thermotoga that is shared only by bacteria and archaea, all others (with the exception of m4Cm-1402, which is uniquely bacterial) being found in all three phylogenetic domains. Nucleoside N-330 might therefore be, in part, the product of RNA modification enzyme genes transferred horizontally between Haloferax and Thermotoga. This speculation is based on the report of extensive lateral transfer between Thermotoga and archaea (Aravind et al. 1998; Logsdon and Faguy 1999), such that almost one-quarter of the Thermotoga genome (24%) was found to be archaeal in nature (Nelson et al. 1999). Work in the area of RNA modification enzymes (Leung et al. 1998), however, while extensive in some notable cases, such as the yeast tRNA modification enzymes (Johansson and Byström 2005) and the pseudouridine synthases (Ofengand and Fournier 1998), by its nature generally lags behind knowledge of modified nucleoside structures (now numbering about 106; see http://medlib.med.utah.edu/RNAmods) and their phylogenetic distributions. Pursuit of this possibility that N-330 is a product of lateral gene transfer will require knowledge of the chemical structure of N-330, and also of the sequences of specific enzymes responsible for its formation.
MATERIALS AND METHODS
Thermotoga maritima MSB8 cells were grown at 80°C at The University of Georgia Bioexpression and Fermentation Facility, under the supervision of T.E. Davies, and RNA from this source was acquired in two ways. First, purified 16S rRNA was obtained as a gift from V. Ramakrishnan (Department of Biochemistry, University of Utah; present address MRC Laboratory of Molecular Biology, Cambridge, U.K.). Second, T. maritima 30S ribosomal subunits were obtained from the same source and 16S RNA was extracted from the 30S ribosomes with TRI Reagent (Chomczynski and Sacchi 1987) following the manufacturer's protocol. 16S rRNAs (typically, 100 μg; 200 pmol) were digested totally to nucleosides with nuclease P1, phosphodiesterase I, and BAP (Crain 1990). Aliquots of RNA (5–100 pmol) were digested with 1000 units of RNase T1 (Ambion) in 10 mM Tris+ 1 mM EDTA, pH 7 for 30 min at 37°C or 45 min at 55°C. Alternatively, the RNA was digested with RNase U2 (Industrial Research Ltd.) in 20 mM diammonium citrate (DAC), pH 5, 1 mM EDTA, or in 20 mM DAC, pH 5, 1 mM EDTA, 8 M urea, with 5–10 units RNase U2 for 15 min at 60°C, then an additional 5–10 units of enzyme was added and digestion continued for 15 min. Details of the mass spectrometry methods used in this study can be found in the Supplemental Data section.
SUPPLEMENTAL DATA
The following supplemental material can be found at http://library.med.utah.edu/mccloskey: LC/ESI-MS analysis of nucleosides in Thermotoga 16S rRNA; partial chromatogram from LC/ESI-MS analysis of RNase U2 digest of Thermotoga 16S rRNA; assignments for monomer ions used in Table 1; data and discussion for placements of m7G, m2G (two each), m5C, m6 2 A (two each), and Cm; additional comments on nucleoside N-330; sequence mass spectrum of T1 oligonucleotide M r 4591.7; mass spectrometry Materials and Methods.
ACKNOWLEDGMENTS
This work was supported by NIH Grant R01 GM29812 (J.A.M.). We are indebted to V. Ramakrishnan for T. maritima ribosomes and rRNA, and to M. Tarry for preparative work in the V. Ramakrishnan laboratory. We thank H. F. Noller for a copy of his 70S ribosome-tRNA manuscript prior to publication, and F. Kirpekar for a preprint of his paper on T. thermophilus 23S rRNA modifications.
Footnotes
Abbreviations: LC/ESI-MS, combined liquid chromatography–electrospray ionization mass spectrometry; Ψ, pseudouridine; m3U, 3-methyluridine; Cm, 2′-O-methylcytidine; m4Cm, N 4,O-2′-dimethylcytidine; m5C, 5-methylcytidine; m2G, N 2-methylguanosine; m7G, 7-methylguanosine; Am, 2′-O-methyladenosine; m2 6A, N 6,N 6-dimethyladenosine; MS/MS, tandem mass spectrometry; M3−, molecular ion having three negative charges, corresponding to loss of three protons from the neutral molecule M, i.e., (M − 3H)3− (analogous definitions for other charge states); MH+, protonated molecule ion; BH+ 2, protonated base ion in which B is the base fragment attached to ribose and BH corresponds to the neutral base; CID, collision-induced dissociation.
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.361607.
REFERENCES
- Agris, P.F. Decoding the genome: A modified view. Nucleic Acids Res. 2004;32:223–238. doi: 10.1093/nar/gkh185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind, L., Tatusov, R.L., Wolf, Y.I., Walker, D.R., Koonin, E.V. Evidence for massive gene exchange between archaeal and bacterial hyperthermophiles. Trends Genet. 1998;14:442–444. doi: 10.1016/s0168-9525(98)01553-4. [DOI] [PubMed] [Google Scholar]
- Bachellerie, J.-P., Cavaillé, J. Small nucleolar RNAs guide the ribose methylations of eukaryotic rRNAs. In: Grosjean H., Benne R., editors. Modification and editing of RNA. ASM Press; Washington, DC: 1998. pp. 255–272. [Google Scholar]
- Bakin, A., Kowalak, J.A., McCloskey, J.A., Ofengand, J. A single pseudouridine residue in E. coli 16S RNA is located at position 516. Nucleic Acids Res. 1994;22:3681–3684. doi: 10.1093/nar/22.18.3681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brimacombe, R., Mitchell, P., Osswald, M., Stade, K., Bochkariov, D. Clustering of modified nucleotides at the functional center of the ribosome. FASEB J. 1993;7:161–167. doi: 10.1096/fasebj.7.1.8422963. [DOI] [PubMed] [Google Scholar]
- Brosius, J., Palmer, M.L., Kennedy, P.J., Noller, H.F. Complete nucleotide sequence of a 16S ribosomal RNA gene from Escherichia coli . Proc. Natl. Acad. Sci. 1978;75:4801–4805. doi: 10.1073/pnas.75.10.4801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannone, J.J., Subramanian, S., Schnare, M.N., Collett, J.R., D'Souza, L.M., Du, Y., Feng, B., Lin, N., Madabusi, L.V., Muller, K.M., et al. The comparative RNA web (CRW) site: An online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics. 2002;3:2. doi: 10.1186/1471-2105-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski, P., Sacchi, N. Single-step method of RNA isolation by acid guanidinium thiocyanate–phenol–chloroform extraction. Anal. Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Crain, P.F. Preparation and enzymatic hydrolysis of RNA and DNA for mass spectrometry. Methods Enzymol. 1990;193:782–790. doi: 10.1016/0076-6879(90)93450-y. [DOI] [PubMed] [Google Scholar]
- Cunningham, P.R., Weitzmann, C.J., Nègre, D., Sinning, J.G., Frick, V., Nurse, K., Ofengand, J. In vitro analysis of the role of rRNA in protein synthesis: Site-specific mutation and methylation. In: Hill W.E., et al., editors. The ribosome: Structure, function, and evolution. ASM Press; Washington, DC: 1990. pp. 243–252. [Google Scholar]
- Cunningham, P.R., Richard, R.B., Weitzmann, C.J., Nurse, K., Ofengand, J. The absence of modified nucleotides affects both in vitro assembly and in vitro function of the 30S-ribosomal subunit of Escherichia coli . Biochimie. 1991;73:789–796. doi: 10.1016/0300-9084(91)90058-9. [DOI] [PubMed] [Google Scholar]
- Davis, D.R. Stabilization of RNA stacking by pseudouridine. Nucleic Acids Res. 1995;23:5020–5026. doi: 10.1093/nar/23.24.5020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis, D.R. Biophysical and conformational properties of modified nucleosides in RNA (nuclear magnetic resonance studies) In: Grosjean H., Benne R., editors. Modification and editing of RNA. ASM Press; Washington, DC: 1998. pp. 85–102. [Google Scholar]
- Decatur, W.A., Fournier, M.J. rRNA modifications and ribosome function. Trends Biochem. Sci. 2002;27:344–351. doi: 10.1016/s0968-0004(02)02109-6. [DOI] [PubMed] [Google Scholar]
- Derrick, W.B., Horowitz, J. Probing structural differences between native and in vitro transcribed Escherichia coli valine transfer RNA: Evidence for stable base modification-dependent conformers. Nucleic Acids Res. 1993;21:4948–4953. doi: 10.1093/nar/21.21.4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douthwaite, S., Fourmy, D., Yoshizawa, S. Nucleotide methylations in rRNA that confer resistance to ribosome-targeting antibiotics. In: Grosjean H., editor. Fine-tuning of RNA functions by modification and editing. Springer-Verlag; Berlin: 2005. pp. 285–307. [Google Scholar]
- Green, R., Noller, H.F. In vitro complementation analysis localizes 23S rRNA posttranscriptional modifications that are required for Escherichia coli 50S ribosomal subunit assembly and function. RNA. 1996;2:1011–1021. [PMC free article] [PubMed] [Google Scholar]
- Gupta, R., Lanter, J.M., Woese, C.R. Sequence of the 16S ribosomal RNA from Halobacterium volcanii, an archaebacterium. Science. 1983;221:656–659. doi: 10.1126/science.221.4611.656. [DOI] [PubMed] [Google Scholar]
- Guymon, R., Pomerantz, S.C., Crain, P.F., McCloskey, J.A. The influence of phylogeny on posttranscriptional modification of rRNA in thermophilic prokaryotes: The complete modification map of 16S rRNA of Thermus thermophilus . Biochemistry. 2006;45:4888–4899. doi: 10.1021/bi052579p. [DOI] [PubMed] [Google Scholar]
- Huber, R., Langworthy, T.A., König, H., Thomm, M., Woese, C.R., Sleytr, U.B., Stetter, K.O. Thermotoga maritima sp. nov. represents a new genus of unique extremely thermophilic eubacteria growing up to 90°C. Arch. Microbiol. 1986;144:324–333. [Google Scholar]
- Johansson, M.J.O., Byström, A.S. Transfer RNA modifications and modifying enzymes in Saccharomyces cerevisiae . In: Grosjean H., editor. Fine-tuning of RNA functions by modification and editing. Springer-Verlag; Berlin: 2005. pp. 87–120. [Google Scholar]
- Kawai, G., Yamamoto, Y., Kamimura, T., Masegi, T., Sekine, M., Hata, T., Iimori, T., Watanabe, T., Miyazawa, T., Yokoyama, S. Conformational rigidity of specific pyrimidine residues in tRNA arises from posttranscriptional modifications that enhance steric interaction between the base and the 2′-hydroxyl group. Biochemistry. 1992;31:1040–1046. doi: 10.1021/bi00119a012. [DOI] [PubMed] [Google Scholar]
- Korostelev, A., Trakhanov, S., Laurberg, M., Noller, H.F. Crystal structure of a 70s ribosome-tRNA complex reveals functional interactions and rearrangements. Cell. 2006;126:1065–1077. doi: 10.1016/j.cell.2006.08.032. [DOI] [PubMed] [Google Scholar]
- Kowalak, J.A., Dalluge, J.J., McCloskey, J.A., Stetter, K.O. Role of posttranscriptional modification in stabilization of transfer RNA from hyperthermophiles. Biochemistry. 1994;33:7869–7876. doi: 10.1021/bi00191a014. [DOI] [PubMed] [Google Scholar]
- Kowalak, J., Bruenger, E., Crain, P.F., McCloskey, J.A. Identities and phylogenetic comparisons of post-transcriptional modifications in 16S ribosomal RNA from Haloferax volcanii . J. Biol. Chem. 2000;275:24484–24489. doi: 10.1074/jbc.M002153200. [DOI] [PubMed] [Google Scholar]
- Leung, H.-C.E., Hagervall, T.G., Björk, G.R., Winkler, M.E. Genetic locations and database accession numbers of RNA-modifying and -editing enzymes. In: Grosjean H., Benne R., editors. Modification and editing of RNA. ASM Press; Washington, DC: 1998. pp. 561–565. [Google Scholar]
- Limbach, P.A., Crain, P.F., McCloskey, J.A. Summary: The modified nucleosides of RNA. Nucleic Acids Res. 1994;22:2183–2196. doi: 10.1093/nar/22.12.2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logsdon, J.M., Faguy, D.M. Evolutionary genomics: Thermotoga heats up lateral gene transfer. Curr. Biol. 1999;9:R747–R751. doi: 10.1016/s0960-9822(99)80474-6. [DOI] [PubMed] [Google Scholar]
- Maden, B.E.H. The numerous modified nucleosides in eukaryotic ribosomal RNA. Prog. Nucleic Acid Res. Mol. Biol. 1990;39:241–303. doi: 10.1016/s0079-6603(08)60629-7. [DOI] [PubMed] [Google Scholar]
- Maden, B.E.H. Intracellular locations of RNA-modifying enzymes. In: Grosjean H., Benne R., editors. Modification and editing of RNA. ASM Press; Washington, DC: 1998. pp. 421–440. [Google Scholar]
- Mason, T.L. Functional aspects of the three modified nucleotides in yeast mitochondrial large-subunit rRNA. In: Grosjean H., Benne R., editors. Modification and editing of RNA. ASM Press; Washington, DC: 1998. pp. 273–280. [Google Scholar]
- McCloskey, J.A., Graham, D.E., Zhou, S., Crain, P.F., Ibba, M., Konisky, J., Söll, D., Olsen, G.J. Post-transcriptional modification in archaeal tRNAs: Identities and phylogenetic relations of nucleotides from mesophilic and hyperthermophilic. Methanococcales. Nucleic Acids Res. 2001;29:4699–4706. doi: 10.1093/nar/29.22.4699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLuckey, S.A., Van Berkel, G.J., Glish, G.L. Tandem mass spectrometry of small, multiply charged oligonucleotides. J. Am. Soc. Mass Spectrom. 1992;3:60–70. doi: 10.1016/1044-0305(92)85019-G. [DOI] [PubMed] [Google Scholar]
- Mengel-Jorgensen, J., Jensen, S.S., Rasmussen, A., Poehlsgaard, J., Kirpekar, F. Modifications in Thermus thermophilus 23S ribosomal RNA are centered in regions of RNA–RNA contact. J. Biol. Chem. 2006;281:22108–22117. doi: 10.1074/jbc.M600377200. [DOI] [PubMed] [Google Scholar]
- Motorin, Y., Grosjean, H. Appendix 1: Chemical structures and classification of posttranscriptionally modified nucleosides in RNA. In: Grosjean H., Benne R., editors. Modification and editing of RNA. ASM Press; Washington, DC: 1998. pp. 543–549. [Google Scholar]
- Nelson, K.E., Clayton, R.A., Gill, S.R., Gwinn, M.L., Dodson, R.J., Haft, D.H., Hickey, E.K., Peterson, J.D., Nelson, W.C., Ketchum, K.A., et al. Evidence for lateral gene transfer between Archaea and Bacteria from genome sequence of Thermotoga maritima . Nature. 1999;399:323–329. doi: 10.1038/20601. [DOI] [PubMed] [Google Scholar]
- Noon, K.R., Bruenger, E., McCloskey, J.A. Posttranscriptional modifications in 16S and 23S rRNAs of the archaeal hyperthermophile Sulfolobus solfataricus . J. Bacteriol. 1998;180:2883–2888. doi: 10.1128/jb.180.11.2883-2888.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noon, K.R., Guymon, R., Crain, P.F., McCloskey, J.A., Thomm, M., Lim, M., Cavicchioli, R. Influence of temperature on tRNA modification in Archaea: Methanococcoides burtonii (Topt 23°C) and Stetteria hydrogenophila (Topt 95°C) J. Bacteriol. 2003;185:5483–5490. doi: 10.1128/JB.185.18.5483-5490.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ofengand, J., Fournier, M.J. The pseudouridine residues of rRNA: Number, location, biosynthesis, and function. In: Grosjean H., Benne R., editors. Modification and editing of RNA. ASM Press; Washington, DC: 1998. pp. 229–253. [Google Scholar]
- Ofengand, J., Rudd, K.E. Bacterial, archaeal, and organellar rRNA pseudouridines and methylated nucleosides and their enzymes. In: Garrett R.A., et al., editors. The ribosome: Structure, function, antibiotics, and cellular interactions. ASM Press; Washington, DC: 2000. pp. 175–189. [Google Scholar]
- Phillips, D.R., McCloskey, J.A. A comprehensive study of the low energy collision-induced dissociation of dinucleoside monophosphates. Int. J. Mass Spectrom. Ion Process. 1993;128:61–82. [Google Scholar]
- Pomerantz, S.C., McCloskey, J.A. Analysis of RNA hydrolyzates by LC/MS. Methods Enzymol. 1990;193:796–824. doi: 10.1016/0076-6879(90)93452-q. [DOI] [PubMed] [Google Scholar]
- Pomerantz, S.C., McCloskey, J.A. Detection of the common RNA nucleoside pseudouridine in mixtures of oligonucleotides by mass spectrometry. Anal. Chem. 2005;77:4687–4697. doi: 10.1021/ac058023p. [DOI] [PubMed] [Google Scholar]
- Rozenski, J., McCloskey, J.A. The small subunit rRNA modification database. Nucleic Acids Res. 2005;33:D135–D138. doi: 10.1093/nar/gki015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rydén-Aulin, M., Shaoping, Z., Kylsten, P., Isaksson, L.A. Ribosome activity and modification of 16S RNA are influenced by deletion of ribosomal protein S20. Mol. Microbiol. 1993;7:983–992. doi: 10.1111/j.1365-2958.1993.tb01190.x. [DOI] [PubMed] [Google Scholar]
- Sampson, J.R., Uhlenbeck, O.C. Biochemical and physical characterization of an unmodified yeast phenylalanine transfer RNA transcribed in vitro. Proc. Natl. Acad. Sci. 1988;85:1033–1037. doi: 10.1073/pnas.85.4.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirum-Connolly, K., Mason, T.L. Functional requirement of a site-specific ribose methylation in ribosomal RNA. Science. 1993;262:1886–1889. doi: 10.1126/science.8266080. [DOI] [PubMed] [Google Scholar]
- Sirum-Connolly, K., Peltier, J.M., Crain, P.F., McCloskey, J.A., Mason, T.L. Implications of a functional large ribosomal RNA with only three modified nucleotides. Biochimie. 1995;77:30–39. doi: 10.1016/0300-9084(96)88101-6. [DOI] [PubMed] [Google Scholar]
- Vaughan, M.H., Soeiro, R., Warner, J.R., Darnell, J.E. Role of RNA methylation in rRNA maturation. Proc. Natl. Acad. Sci. 1967;58:1527–1534. doi: 10.1073/pnas.58.4.1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Ahsen, U., Noller, H.F. Identification of bases in 16S rRNA essential for tRNA binding at the 30S ribosomal P site. Science. 1995;267:234–237. doi: 10.1126/science.7528943. [DOI] [PubMed] [Google Scholar]
- Woese, C.R., Gupta, R., Hahn, C.M., Zillig, W., Tu, J. The phylogenetic relationships of three sulfur dependent archaebacteria. Syst. Appl. Microbiol. 1984;5:97–105. doi: 10.1016/s0723-2020(84)80054-5. [DOI] [PubMed] [Google Scholar]