Abstract
Although H19 was the first imprinted noncoding transcript to be identified, the function of this conserved RNA has remained unclear. Here, we identify a 23-nucleotide microRNA derived from H19 that is endogenously expressed in human keratinocytes and neonatal mice and overexpressed in cells transfected with human or mouse H19 expression plasmids. These data demonstrate that H19 can function as a primary microRNA precursor and suggest that H19 expression results in the post-transcriptional downregulation of specific mRNAs during vertebrate development.
Keywords: microRNA, RNA interference, genomic imprinting, noncoding RNAs
INTRODUCTION
Genomic imprinting leads to the expression of only one copy of an autosomal gene, depending on the parental origin of the relevant chromosome (Pauler and Barlow 2006). Imprinted genes occur in clusters consisting of one or more protein coding genes and at least one noncoding RNA. One particularly well-studied, and highly conserved, imprinted vertebrate gene cluster includes the insulin-like growth factor 2 (Igf2) gene, which is paternally expressed, and the ∼2.3 kb H19 noncoding RNA, which is maternally expressed. Although the integrity of the regulatory elements that control H19 transcription is critical for the maintenance of Igf2 imprinting, the H19 RNA per se is not (Jones et al. 1998). Indeed, transgenic mice lacking the H19 transcript develop quite normally. However, H19 RNA does have some function(s), as H19 displays tumor-suppressor activity in culture (Hao et al. 1993; Juan et al. 2000) and H19 overexpression can affect prenatal viability (Brunkow and Tilghman 1991; Kono et al. 2002).
MicroRNAs (miRNAs) are a class of ∼22-nucleotide (nt)-long noncoding RNAs expressed by all metazoan eukaryotes (Bartel 2004). Cellular miRNAs are initially expressed as part of one arm of an ∼80-nt RNA hairpin that in turn forms part of a longer primary miRNA transcript transcribed by RNA polymerase II (Cullen 2004). Primary miRNA hairpins have a characteristic structure, consisting of an ∼30-base-pair (bp) imperfect stem, a large terminal loop, and flanking unstructured RNA sequences, which is necessary and sufficient for recognition by the nuclear RNase III enzyme Drosha acting in concert with its cofactor DGCR8 (Han et al. 2004; Zeng et al. 2005). Drosha then cleaves ∼22 bp down the stem to excise the ∼60-nt pre-miRNA intermediate (Fig. 1A). Because Drosha processing results in the cleavage of the primary miRNA into at least three RNA fragments, miRNAs are rarely found in the exons of coding transcripts. Instead, miRNA hairpins are generally located in introns or in the exons of noncoding RNAs (Cullen 2004). After nuclear export of the pre-miRNA intermediate, a second RNase III enzyme called Dicer cleaves at the stem:loop junction to generate the mature miRNA (Fig. 1A; Hutvagner et al. 2001; Ketting et al. 2001). The mature miRNA is then incorporated into the RNA-induced silencing complex (RISC), where it acts as a guide RNA to direct RISC to complementary mRNA species (Hammond et al. 2000; Martinez et al. 2002; Schwarz et al. 2002).
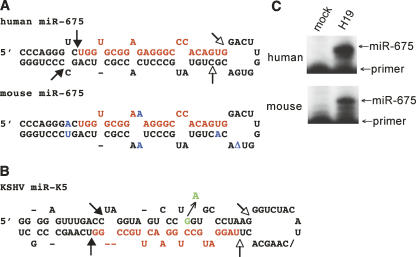
FIGURE 1.
Predicted structure and processing sites of the H19-derived primary miR-675 precursor. (A) Predicted RNA structures adopted by human and mouse pri-miR-675. Predicted Drosha processing sites are indicated by closed arrows and Dicer processing sites by open arrows. The predicted mature miRNA sequences are shown in red, including the miR-675 cDNA sequence cloned from human keratinocytes. The human and mouse pri-miR-675 precursors differ at six positions, indicated in blue, including a single nucleotide deletion in the terminal loop (shown as a delta). There is one difference in the predicted mature miRNA, a G to A change at position 10. This change lies outside the critical miRNA “seed” region (nucleotides 2–8) (Bartel 2004) and would continue to permit base pairing to a “U” residue in target mRNAs. (B) Predicted RNA structure adopted by the KSHV pri-miR-K5 hairpin. A previously described (Gottwein et al. 2006) natural polymorphism in the pri-miR-K5 precursor, which results in inefficient Drosha processing, is indicated in green. (C) Primer extension analysis to detect mature human or mouse miR-675. 293T cells were transfected with expression plasmids encoding similar ∼580-nt-long H19 cDNA sequences centered on the predicted pri-miR-675 hairpins (panel A). At 48 h after transfection, total RNA was harvested and subjected to primer extension analysis (Lee et al. 2003) using 18-nt primers fully complementary to the 3′ ends of the predicted human or mouse miR-675 sequence. The predicted 23-nt-long extension product was detected in each case. Mock-transfected 293T cells were used as a negative control.
In this report, we demonstrate that the imprinted H19 noncoding RNA can function as a primary miRNA transcript in both humans and mice. These data suggest that the biological role of H19, and potentially of some other imprinted noncoding RNAs, includes the formation of miRNAs that act to downregulate developmentally relevant mRNA species.
RESULTS AND DISCUSSION
During experiments designed to clone miRNAs expressed in a human keratinocyte cell line latently infected by human papillomavirus (Cai et al. 2006), we repeatedly cloned a 23-nt-long RNA derived from nucleotides 1014–1036 of the human H19 RNA. This sequence represented five out of 533 miRNA cDNA clones obtained from two independent cDNA libraries. Computer analysis predicts that this RNA sequence, termed miR-675, forms the upper part of one arm of an RNA hairpin that has the structure expected for a primary miRNA stem–loop (Fig. 1A). Sequence analysis showed that both the candidate miR-675 miRNA itself and the flanking RNA hairpin are evolutionarily conserved in rodents (Fig. 1A).
As noted above, miRNAs are processed from longer pri-miRNA precursors by sequential processing by the RNase III enzymes Drosha and Dicer (Cullen 2004). To examine whether H19 can indeed be processed in vivo to give rise to the mature 23-nt miRNA that we had cloned from human keratinocytes, we expressed similar ∼580-nt-long PCR-derived cDNA fragments, derived from either human or mouse H19 and centered on the predicted ∼73-nt-long primary miR-675 hairpin (Fig. 1A), in the cell line 293T, which does not normally express H19. Primer extension analysis revealed the readily detectable expression of the predicted 23-nt extension product in 293T cells transfected with either the human or the mouse H19 expression plasmid, but not in mock transfected cells (Fig. 1C). We therefore conclude that H19 can indeed be processed in vivo to give rise to the 23-nt-long miR-675 miRNA and that this ability is evolutionarily conserved in humans and mice.
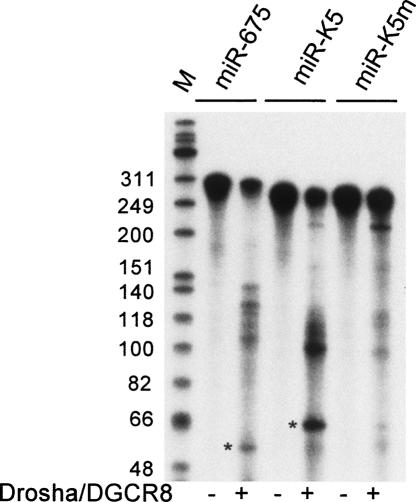
The first step in miRNA biogenesis is the cleavage of the primary miRNA precursor by the Drosha: DGCR8 dimer to release the pre-miRNA hairpin (Han et al. 2004). The in vitro incubation of a 295-nt-long, 32P-labeled human H19 transcript with recombinant Drosha:DGCR8 indeed resulted in the specific release of the predicted 57-nt-long pre-miR-675 hairpin (Fig. 2). As a control, we also performed in vitro Drosha:DGCR8 cleavage of a validated primary miRNA encoding the miR-K5 miRNA expressed by Kaposi's sarcoma associated herpesvirus (KSHV) (Fig. 1B; Gottwein et al. 2006). This again resulted in the release of the predicted pre-miRNA intermediate, which in this case is 62 nt in length (Fig. 2). This processing event is specific, as shown by the marked inhibition in miR-K5 processing seen with a naturally occurring sequence polymorphism (miR-K5m), which changes a single nucleotide in the miR-K5 hairpin (Fig. 1B) and inhibits Drosha:DGCR8 cleavage both in vitro and in vivo (Fig. 2; Gottwein et al. 2006).
FIGURE 2.
H19 is processed by Drosha in vitro. A 295-nt-long human pri-miR-675 transcript, centered on the predicted miR-675 hairpin (Fig. 1A) was transcribed in vitro in the presence of α-32P CTP. Flag-tagged Drosha:DGCR8 was affinity purified from 293T cells cotransfected with Drosha–Flag and DGCR8–Flag expression plasmids (Gottwein et al. 2006). The 295-nt pri-miR-675 precursor is predicted to be processed by Drosha:DGCR8 to give a 57-nt-long pre-miRNA (indicated with an asterisk). Wild-type and mutant (m) versions of a previously described (Gottwein et al. 2006) pri-miR-K5 transcript served as controls. The pre-miRNA intermediate for pri-miR-K5 is predicted to be 62 nt long (asterisk). (M) DNA markers.
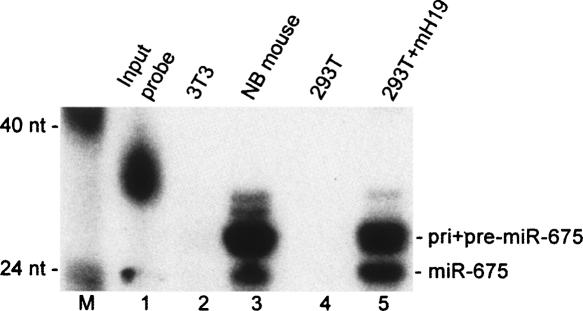
The imprinted H19 RNA is expressed at high levels during vertebrate embryo development but is downregulated in most tissues shortly after birth (Bartolomei et al. 1991). To further confirm that miR-675 is indeed expressed in vivo, we performed an RNase protection assay (RPA) that was designed to detect the mature, 23-nt-long murine miR-675 and also the pre- and pri-miRNA precursors, i.e., in the latter case the H19 transcript itself. Both the mature miR-675 RNA and the predicted precursors were readily detected in an RNA sample derived from a newborn mouse (Fig. 3, lane 3) but were not observed in RNA derived from murine 3T3 cells (Fig. 3, lane 2), which do not express detectable H19 RNA levels. Similarly, mature murine miR-675 was also readily detected by RPA in 293T cells transfected with the murine H19 expression plasmid analyzed in Figure 1C (Fig. 3, lane 5) but was not detected in mock-transfected 293T cells (Fig. 3, lane 4). We therefore conclude that the H19-derived miRNA miR-675 is indeed expressed during vertebrate development. We note that our data do not directly address whether miR-675 is itself imprinted, although our data showing that miR-675 can be processed out of the imprinted H19 RNA argue that this is very likely to be the case. Nevertheless, it remains formally possible that a portion of the observed endogenous miR-675 microRNAs are derived from an unknown, alternative transcript from the H19 locus that is distinct from H19 itself.
FIGURE 3.
Detection of mature miR-675 in newborn mice. This RPA analyzed 50 μg of total RNA derived from murine 3T3 cells (lane 2), a newborn (NB) mouse (lane 3), mock-transfected 293T cells (lane 4), or 293T cells transfected with a mouse H19 (mH19) expression plasmid (lane 5). The 31-nt-long single-stranded RPA probe used consisted of a 23-nt stretch complementary to the predicted mature mouse miR-675, a 5-nt 5′ extension complementary to part of the predicted terminal loop of the pre-miR-675 RNA hairpin (Fig. 1A), and finally a 3-nt nonspecific 5′ tag consisting of “GGG.” The probe was prepared by in vitro transcription, using T7 RNA polymerase, in the presence of α-32P-CTP. This RPA was performed using a miRVana miRNA detection kit (Ambion), as described by the manufacturer. (M) DNA markers.
Imprinted loci invariably contain at least one noncoding RNA, whose role, if any, has generally been unclear (Pauler and Barlow 2006). Here, we present evidence that the prototypic H19 noncoding RNA can be processed to release at least one miRNA that is conserved in humans and rodents. This processing event does not appear to be highly efficient, as full-length H19 transcripts can be readily detected during embryonic development (Bartolomei et al. 1991). We note that other noncoding pri-miRNA precursors, e.g., the “BIC” noncoding RNA that serves as the precursor for miR-155, are also readily detectable in vivo as full-length transcripts (Tam 2001). Moreover, miRNA processing itself has recently been shown to be developmentally regulated (Thomson et al. 2006; Obernosterer et al. 2006). Therefore, it is possible that the efficiency with which H19 is processed into miRNAs may vary significantly during different developmental stages and/or in different tissues. We note, however, that H19 processing by Drosha was readily detected in both transfected 293T cells (Fig. 1C) and in vitro (Fig. 2). Regardless, one would certainly predict that miR-675 excision would result in a decrease in full-length H19 expression, and it is possible that miR-675 may actually serve as a mechanism for the attenuation of H19 function in specific cells or tissues, assuming that full-length H19 indeed has a function.
While this report was being prepared, Mineno et al. (2006) reported the use of “massively parallel signature sequencing” technology to compile an exhaustive list of all the miRNAs expressed in the mouse embryo. This effort identified 195 novel murine miRNAs, one of which, miR-675, is identical to the murine miR-675 miRNA proposed in Figure 1A except that it is shorter by 1 nt at the 3′ end. While Mineno et al. (2006) neither mentioned nor discussed the fact that miR-675 is predicted to derive from the imprinted H19 RNA, this exhaustive sequencing effort does further confirm the hypothesis that murine H19 can indeed function as a primary miRNA precursor in vivo.
While the role(s) of miR-675 in mammalian development remains to be established, the parental conflict hypothesis (Moore and Haig 1991) suggests that imprinted, maternally expressed genes, such as H19, will generally act to suppress excessive embryonic growth. It is therefore possible that miR-675 functions to post-transcriptionally downregulate mRNAs, some of which may be paternally expressed, that encode growth factors or growth factor receptors or their downstream targets. Moreover, these data raise the possibility that some other imprinted noncoding RNAs might also give rise to miRNAs. Indeed, one other example of an imprinted miRNA locus located in the Dlk1–Gtl2 cluster has already been reported (Seitz et al. 2003). In cases where an imprinted noncoding RNA lies directly anti-sense to a protein-coding mRNA showing the opposite imprinting pattern (Pauler and Barlow 2006), it is also possible that imprinted noncoding RNA might give rise to an miRNA that could directly inhibit the expression of this mRNA via an RNA interference mechanism, as has indeed been recently demonstrated for miRNAs expressed from the imprinted Dlk1–Gtl2 gene cluster (Davis et al. 2005).
ACKNOWLEDGMENTS
This research was funded by NIH grant GM071408. We thank Marc Caron for the gift of reagents used in this research.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.351707.
REFERENCES
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Bartolomei, M.S., Zemel, S., Tilghman, S.M. Parental imprinting of the mouse H19 gene. Nature. 1991;351:153–155. doi: 10.1038/351153a0. [DOI] [PubMed] [Google Scholar]
- Brunkow, M.E., Tilghman, S.M. Ectopic expression of the H19 gene in mice causes prenatal lethality. Genes & Dev. 1991;5:1092–1101. doi: 10.1101/gad.5.6.1092. [DOI] [PubMed] [Google Scholar]
- Cai, X., Li, G., Laimins, L.A., Cullen, B.R. Human papillomavirus genotype 31 does not express detectable microRNA levels during either latent or productive virus replication. J. Virol. 2006;80:10890–10893. doi: 10.1128/JVI.01175-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen, B.R. Transcription and processing of human microRNA precursors. Mol. Cell. 2004;16:861–865. doi: 10.1016/j.molcel.2004.12.002. [DOI] [PubMed] [Google Scholar]
- Davis, E., Caiment, F., Tordoir, X., Cavaillé, J., Ferguson-Smith, A., Cockett, N., Georges, M., Charlier, C. RNAi-mediated allelic trans-interaction at the imprinted Rtl1/Peg11 locus. Curr. Biol. 2005;15:743–749. doi: 10.1016/j.cub.2005.02.060. [DOI] [PubMed] [Google Scholar]
- Gottwein, E., Cai, X., Cullen, B.R. A novel assay for viral microRNA function identifies a single nucleotide polymorphism that affects Drosha processing. J. Virol. 2006;80:5321–5326. doi: 10.1128/JVI.02734-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond, S.M., Bernstein, E., Beach, D., Hannon, G.J. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature. 2000;404:293–295. doi: 10.1038/35005107. [DOI] [PubMed] [Google Scholar]
- Han, J., Lee, Y., Yeom, K.-H., Kim, Y.-K., Jin, H., Kim, V.N. The Drosha-DGCR8 complex in primary microRNA processing. Genes & Dev. 2004;18:3016–3027. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao, Y., Crenshaw, T., Moulton, T., Newcomb, E., Tycko, B. Tumour-suppressor activity of H19 RNA. Nature. 1993;365:764–767. doi: 10.1038/365764a0. [DOI] [PubMed] [Google Scholar]
- Hutvagner, G., McLachlan, J., Pasquinelli, A.E., Balint, E., Tuschl, T., Zamore, P.D. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293:834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- Jones, B.K., Levorse, J.M., Tilghman, S.M. Igf2 imprinting does not require its own DNA methylation or H19 RNA. Genes & Dev. 1998;12:2200–2207. doi: 10.1101/gad.12.14.2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juan, V., Crain, C., Wilson, C. Evidence for evolutionarily conserved secondary structure in the H19 tumor suppressor RNA. Nucleic Acids Res. 2000;28:1221–1227. doi: 10.1093/nar/28.5.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketting, R.F., Fischer, S.E., Bernstein, E., Sijen, T., Hannon, G.J., Plasterk, R.H. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans . Genes & Dev. 2001;15:2654–2659. doi: 10.1101/gad.927801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kono, T., Sotomaru, Y., Katsuzawa, Y., Dandolo, L. Mouse parthenogenetic embryos with monoallelic H19 expression can develop to day 17.5 of gestation. Dev. Biol. 2002;243:294–300. doi: 10.1006/dbio.2001.0561. [DOI] [PubMed] [Google Scholar]
- Lee, M.-T.M., Coburn, G.A., McClure, M.O., Cullen, B.R. Inhibition of human immunodeficiency virus type 1 replication in primary macrophages by using Tat- or CCR5-specific small interfering RNAs expressed from a lentivirus vector. J. Virol. 2003;77:11964–11972. doi: 10.1128/JVI.77.22.11964-11972.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez, J., Patkaniowska, A., Urlaub, H., Lührmann, R., Tuschl, T. Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell. 2002;110:563–574. doi: 10.1016/s0092-8674(02)00908-x. [DOI] [PubMed] [Google Scholar]
- Mineno, J., Okamoto, S., Ando, T., Sato, M., Chono, H., Izu, H., Takayama, M., Asada, K., Mirochnitchenko, O., Inouye, M., et al. The expression profile of microRNAs in mouse embryos. Nucleic Acids Res. 2006;34:1765–1771. doi: 10.1093/nar/gkl096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore, T., Haig, D. Genomic imprinting in mammalian development: A parental tug-of-war. Trends Genet. 1991;7:45–49. doi: 10.1016/0168-9525(91)90230-N. [DOI] [PubMed] [Google Scholar]
- Obernosterer, G., Leuschner, P.J.F., Alenius, M., Martinez, J. Post-transcriptional regulation of microRNA expression. RNA. 2006;12:1161–1167. doi: 10.1261/rna.2322506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauler, F.M., Barlow, D.P. Imprinting mechanisms—It only takes two. Genes & Dev. 2006;20:1203–1206. doi: 10.1101/gad.1437306. [DOI] [PubMed] [Google Scholar]
- Schwarz, D.S., Hutvágner, G., Haley, B., Zamore, P.D. Evidence that siRNAs function as guides, not primers, in the Drosophila and human RNAi pathways. Mol. Cell. 2002;10:537–548. doi: 10.1016/s1097-2765(02)00651-2. [DOI] [PubMed] [Google Scholar]
- Seitz, H., Youngson, N., Lin, S.P., Dalbert, S., Paulsen, M., Bachellerie, J.P., Ferguson-Smith, A.C., Cavaille, J. Imprinted microRNA genes transcribed antisense to a reciprocally imprinted retrotransposon-like gene. Nat. Genet. 2003;34:261–262. doi: 10.1038/ng1171. [DOI] [PubMed] [Google Scholar]
- Tam, W. Identification and characterization of human BIC, a gene on chromosome 21 that encodes a noncoding RNA. Gene. 2001;274:157–167. doi: 10.1016/s0378-1119(01)00612-6. [DOI] [PubMed] [Google Scholar]
- Thomson, J.M., Newman, M., Parker, J.S., Morin-Kensicki, E.M., Wright, T., Hammond, S.M. Extensive post-transcriptional regulation of microRNAs and its implications for cancer. Genes & Dev. 2006;20:2202–2207. doi: 10.1101/gad.1444406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng, Y., Yi, R., Cullen, B.R. Recognition and cleavage of primary microRNA precursors by the nuclear processing enzyme Drosha. EMBO J. 2005;24:138–148. doi: 10.1038/sj.emboj.7600491. [DOI] [PMC free article] [PubMed] [Google Scholar]