Abstract
RNase II is a 3′-5′ exoribonuclease that processively hydrolyzes single-stranded RNA generating 5′ mononucleotides. This enzyme contains a catalytic core that is surrounded by three RNA-binding domains. At its C terminus, there is a typical S1 domain that has been shown to be critical for RNA binding. The S1 domain is also present in the other major 3′-5′ exoribonucleases from Escherichia coli: RNase R and polynucleotide phosphorylase (PNPase). In this report, we examined the involvement of the S1 domain in the different abilities of these three enzymes to overcome RNA secondary structures during degradation. Hybrid proteins were constructed by replacing the S1 domain of RNase II for the S1 from RNase R and PNPase, and their exonucleolytic activity and RNA-binding ability were examined. The results revealed that both the S1 domains of RNase R and PNPase are able to partially reverse the drop of RNA-binding ability and exonucleolytic activity resulting from removal of the S1 domain of RNase II. Moreover, the S1 domains investigated are not equivalent. Furthermore, we demonstrate that S1 is neither responsible for the ability to overcome secondary structures during RNA degradation, nor is it related to the size of the final product generated by each enzyme. In addition, we show that the S1 domain from PNPase is able to induce the trimerization of the RNaseII–PNP hybrid protein, indicating that this domain can have a role in the biogenesis of multimers.
Keywords: S1 domain, RNase II, RNase R, PNPase, trimerization, RNA degradation
INTRODUCTION
Maturation, turnover, and quality control of RNA are performed by many different classes of ribonucleases. In Escherichia coli, several 3′-5′ exoribonucleases participate in all these processes (Régnier and Arraiano 2000; Arraiano and Maquat 2003). Exoribonuclease II (RNase II) is one of the main exoribonucleases in E. coli, and is responsible for 90% of the hydrolytic activity in crude extracts (Deutscher and Reuven 1991). This enzyme degrades RNA processively in the 3′-5′ direction, releasing 5′ mononucleotides. Its activity is sequence independent but sensitive to secondary structures, and poly(A) is the preferred substrate for this enzyme (Gupta et al. 1977; McLaren et al. 1991; Coburn and Mackie 1996; Marujo et al. 2000). RNase II expression is differentially regulated at the transcriptional and post-transcriptional levels (Zilhão et al. 1993, 1996a), and the protein can be regulated by the environmental conditions (Cairrão et al. 2001).
RNase II can be divided into several functionally distinct regions (Mian 1997; Amblar and Arraiano 2005; Amblar et al. 2006): an N-terminal segment (residues 1–103) containing a cold shock domain (CSD) (Graumann and Marahiel 1998) involved in RNA binding, a central region of ∼400 residues spanning the catalytic RNB domain (Mian 1997), and a C-terminal segment (residues 533–644) that corresponds to an S1 domain (Bycroft et al. 1997). Recently, the 3D structure of E. coli RNase II and its RNA complex have been determined (Frazão et al. 2006; McVey et al. 2006). Structural and functional analyses have established that the central region of RNase II is not only the catalytic core but also contains a third RNA-binding domain at its N-terminal region. This region (residues 85–156) has been identified as a second CSD since it has the typical oligosaccharide/oligonucleotide-binding fold (OB fold) and was shown to have RNA-binding ability (Amblar et al. 2006; Frazão et al. 2006).
The 3D structure of RNase II also showed that the residues from 557 to 636 at the C terminus correspond to an S1 domain with the typical S1-binding fold (Frazão et al. 2006), and biochemical studies have confirmed that this region is critical for the maintenance of stable RNA–protein complexes (Amblar et al. 2006). Furthermore, it has been recently reported that during RNase II cleavage the nucleotides at the 5′ end of the RNA substrate are situated in an anchor region in a deep cleft between the two CSD and the S1 domain (Frazão et al. 2006). S1 domains are RNA-binding modules that were originally identified in the ribosomal protein S1 (Boni et al. 1991). All of them display a similar OB fold that is shared by many other proteins of unrelated function (Murzin 1993; Arcus 2002; Theobald et al. 2003). The S1 domain is also present in many other ribonucleases such as RNase E, RNase G, polynucleotide phosphorylase (PNPase), and RNase R, and in all of them it seems to be mediating RNA recognition (Bycroft et al. 1997; Schubert et al. 2004; Amblar et al. 2006). In E. coli RNase E, the S1 domain is crucial for activity (McDowall et al. 1993), and it contains two distinct surface regions: one important for the ribonucleolytic activity, and the other important for autoregulation (Diwa et al. 2002). The role of this domain in PNPase is not yet clear. Previous studies have shown that mutations in the S1 domain of PNPase have a minimal effect on its activity and autoregulation (Jarrige et al. 2002), but indirectly facilitate the activity of the enzyme by promoting substrate binding and product release (Stickney et al. 2005).
In this work we have examined whether the S1 domain of three exoribonucleases from E. coli, RNase II, RNase R, and PNPase, can substitute for each other, and whether this domain is responsible for the differences in degradation activity of these enzymes. Through construction of chimeric proteins, we have demonstrated that the S1 domain from RNase II can be replaced by the S1 domain from different exoribonucleases, without abolishing the exonucleolytic activity of the enzyme. However, the S1 domains investigated are not equivalent. We have interpreted the data taking into account the ability of the enzymes to bind RNA and also regarding the degradation of different substrates. Interestingly, our results point to a possible involvement of the PNPase S1 domain in multimerization.
RESULTS AND DISCUSSION
Substitution of S1 domains in exoribonucleases
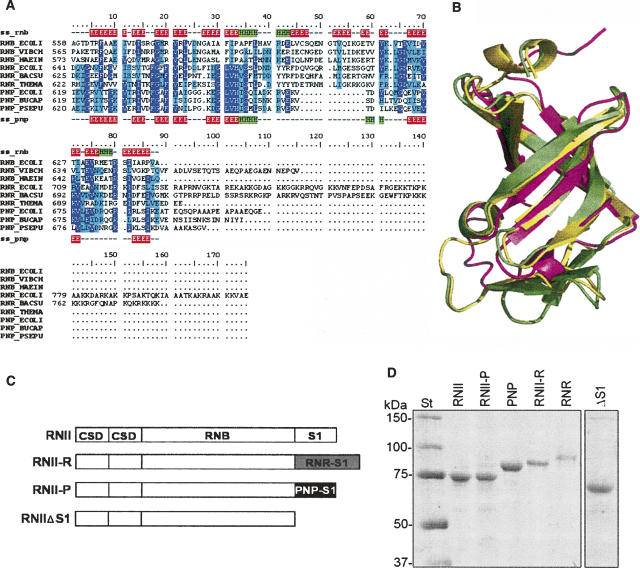
The S1 RNA-binding domain is largely present in RNA-binding proteins. The three main exoribonucleases of E. coli, RNase II, RNase R, and PNPase, present this domain at their C terminus. We have performed the structural alignment of the S1 domains of PNPase and RNase II as described in Materials and Methods. This alignment shows that both domains are similar in sequence (18% of sequence identity), and that they have the same secondary structural elements (Fig. 1A). We further extended this alignment, including the S1 domain of E. coli RNase R and the S1 domains of ribonucleases from other bacteria, which revealed several conserved residues. This multiple structural alignment has allowed us to construct a structure-based multiple sequence alignment used to extract evolutionary information related to specific residues for each family of proteins (Fig. 1A; Supplemental figures at http://novacripta.cbm.uam.es/bioweb/SuppMat/S1_domain_RNA07_supp.pdf). RNase R from E. coli and Bacillus subtilis show a larger C-terminal region containing a putative S1 domain, highly similar to those from RNase II and PNPase, and an extra lysine-rich region at the C terminus of unknown function. In the E. coli enzyme, the S1 domain of RNase R showed a 17% of sequence identity with the S1 of RNase II. Given the close homology, we were able to model the S1 domain of RNase R based on the structure of RNase II (Frazão et al. 2006). Superimposition of such a model with the structures of the S1 domains of PNPase and RNase II highlights a similar overall structure of the three S1 domains (Fig. 1B).
FIGURE 1.
(A) A structure-based multiple sequence alignment of S1 domains from RNase II (RNB), RNase R (RNR), and PNPases (PNP) of several organisms (ECOLI: Escherichia coli; VIBCH: Vibrio cholerae; HAEIN: Haemophilus influenzae; BACSU: Bacillus subtilis; THEMA: Thermotoga maritima; BUCAP: Buchnera aphidicola ; PSEPU: Pseudomonas putida). Alignment is colored according to conservation: highly conserved residues are in purple, semiconserved residues are in cyan. Secondary structure elements of S1 domains from RNB_ECOLI (ss_rnb) and PNP_ECOLI (ss_pnp) are also indicated: β-sheets are in red and α-helices are in green. (B) A structural alignment of experimentally determined structures for the S1 domains of E. coli RNase II (yellow; PDB code: 2IX0) (Frazão et al. 2006) and PNPase (magenta; PDB code: 1SRO) (Bycroft et al. 1997) and a homology model generated for the S1 domain of E. coli RNase R (green). (C) A schematic representation of the domain organization of RNase II (RNII), RNase II-R (RNII-R), RNase II-P (RNII-P), and truncated protein RNase IIΔS1 (RNIIΔS1). Protein domains from RNase II (CSD, RNB, and S1) are represented as white rectangles, the S1 domain from RNase R is represented as a gray rectangle, and the S1 domain from PNPase as a black rectangle. (D). Purity of the enzymes was analyzed in a 10% SDS-PAGE; 0.5 μg of purified (His)6–RNase II (RNII), (His)6–RNase II-P (RNII-P), (His)6–PNPase (PNP), (His)6–RNase II-R (RNII-R), (His)6–RNase R (RNR), and (His)6–RNase IIΔS1 (ΔS1) were applied and visualized by Coomassie blue staining. The molecular weights of the standard proteins (St) are indicated on the left.
The three major exoribonucleases degrade RNA in the 3′ to 5′ direction in a processive and sequence-independent manner, but they show a different behavior against secondary structures. PNPase and RNase II degrade unstructured single-stranded RNAs and, although with certain differences, both of them are inhibited in vitro by the presence of secondary structures (Zilhão et al. 1996b; Spickler and Mackie 2000). In contrast, RNase R is able to degrade highly structured RNAs (Cheng and Deutscher 2002, 2004; Amblar et al. 2006; Andrade et al. 2006). Due to the special features shown by the C terminus of RNase R, we wanted to investigate whether this region could account for the capacity of RNase R to degrade folded RNAs. Therefore, we deleted the S1 domain of RNase II (ΔS1 mutant) and constructed two hybrid proteins by substituting the S1 domain with that of RNase R or PNPase. The C-terminal region of (His)6–RNase II (residues 529–644) was replaced by the C-terminal region of RNase R (residues 636–813) or PNPase (residues 616–711), thus obtaining the corresponding hybrid proteins (His)6–RNII-R and (His)6–RNII-P, respectively (Fig. 1C). In addition, genes encoding RNase R (rnr) and PNPase (pnp) from E. coli were cloned into the pET15 expression vector (Novagen) to obtain the N-terminal histidine-tagged fusion forms of each protein ([His]6–RNase R and [His]6–PNPase). The truncated form of (His)6–RNase II, lacking the S1 domain (residues 529–644), named ΔS1 (Amblar et al. 2006), was used as a control. All recombinant enzymes were overproduced in E. coli BL21(DE3) cells and remained soluble after 2 h of induction with IPTG. The corresponding proteins were purified as described in Materials and Methods, and the pure enzymes were analyzed by SDS-PAGE (Fig. 1D). Quantification of the gels revealed that in all cases, the purity of the proteins was higher than 65%.
The S1 domain from RNase II can be replaced by the S1 domain from different exoribonucleases
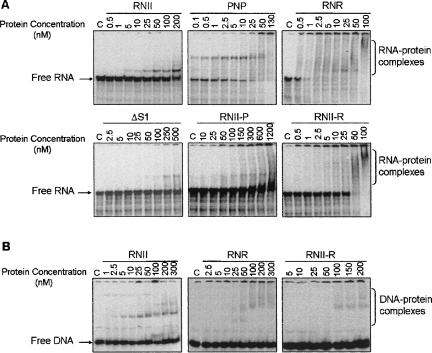
The effect of the S1 domain substitution was analyzed by testing the RNA-binding ability and exoribonucleotytic activity of the different constructs. The RNA-binding capability was tested using the in vitro transcribed mRNA malE–malF (McLaren et al. 1991). This transcript corresponds to the intergenic region of the malE–malF operon and consists of a 375 nucleotide (nt) RNA molecule containing two stem–loop structures. The band-shift experiments performed revealed that the three full-length polypeptides, RNase II, PNPase, and RNase R, were able to bind the malE–malF transcript, generating retardation bands that correspond to RNA–protein complexes (Fig. 2A). In addition, it seems that PNPase binds this substrate more efficiently than the other two enzymes, since stable RNA–protein complexes were detected with 0.1 nM of PNPase, the lowest protein concentration tested, while RNase II and RNase R required 5 nM or 2.5 nM, respectively. According to previous data (Amblar et al. 2006), the removal of the S1 domain drastically affected the RNA-binding ability of RNase II, and 250 nM of the truncated protein ΔS1 was necessary to detect retardation bands. However, if we added the C-terminal regions of PNPase or RNase R the RNA binding could be detected with 100 nM of RNII-P or 50 nM of RNII-R. Moreover, RNA binding with the two hybrid proteins appears like a smear of high molecular weight (MW) forms instead of clear bands, similar to RNase R. Such a smear could be due to a heterogeneous population of RNA–protein complexes.
FIGURE 2.
RNA- and DNA-binding ability of wild-type and hybrid proteins. malE–malF (2 fmol) (A) or 1 fmol ompA-3 DNA oligomer (B) were incubated under the conditions described in Materials and Methods. The enzyme concentration used is indicated in the figure. A control, “C,” reaction without enzyme added was performed in all experiments. Binding reactions were applied on a 5% nondenaturing PAA gel. The mobility of free and complexed substrates was detected using the PhosphorImager system from Molecular Dynamics.
It has been demonstrated that high concentrations of RNase II are able to degrade DNA, and that RNase R is much less efficient in degrading this substrate (Cheng and Deutscher 2002). In addition, RNase II activity on RNA is strongly inhibited by DNA oligomers (Cannistraro and Kennell 1994) while RNase R is not, suggesting a very poor ability of RNase R to bind DNA in comparison with RNase II (Cheng and Deutscher 2002). On this basis, and in order to investigate the role of the S1 domain in the ability of RNase II and RNase R to bind DNA, we tested the DNA-binding capability of these two proteins and the RNII-R hybrid enzyme using a 39-nt DNA oligomer (Andrade et al. 2006). The band-shift experiments revealed that the three proteins were able to bind the DNA oligomer but with very different efficiencies (Fig. 2B). RNase II was able to form stable DNA–protein complexes at only 5 nM of protein, revealing a similar binding ability with both DNA and RNA substrates. By contrast, RNase R required a 10-fold higher concentration of protein than RNase II to form DNA–protein complexes (50 nM of protein), and its ability to bind DNA was much lower than with RNA (>20-fold reduction). Therefore, RNase R has a very poor ability to bind DNA versus RNA, confirming that the low efficiency of RNase R on DNA degradation is due, at least partly, to a very poor binding. The RNII-R hybrid protein also showed a drastic reduction in DNA-binding ability versus RNA, since DNA–protein complexes were visible only at 100 nM of protein, while with this concentration, the whole RNA substrate was bound to the enzyme (Fig. 2A). Therefore, the replacement of the S1 domain of RNase II with the S1 of RNase R caused a decrease in DNA binding versus RNA. Our results demonstrate that the S1 from RNase II contributes to the binding of DNA, but this does not happen with the S1 domain from RNase R.
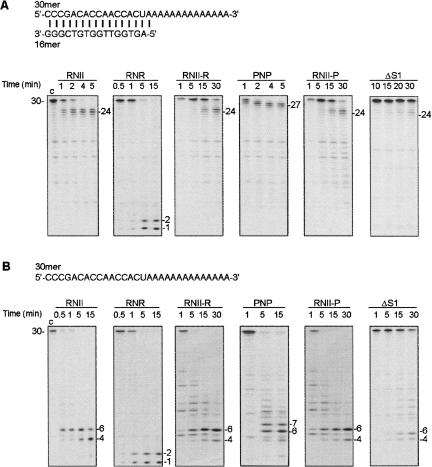
The involvement of the S1 domain in substrate specificity and exoribonucleolytic activity was also analyzed. We used a 30-nt oligomer of RNA in combination with the complementary 16-nt oligomer of DNA (see Materials and Methods) to generate two different RNA substrates: a double-stranded (ds) substrate with an additional 14-nt single-stranded extension at the 3′ end (16–30 ds) or a 30-nt single-stranded (ss) substrate (30 ss) (see Fig. 3). The choice of these substrates was based on previous reports that demonstrated their utility to detect different behaviors of RNases against double- and single-stranded RNAs (Cheng and Deutcher 2004; Amblar et al. 2006). The activity assays demonstrated that, according to previous results (Amblar et al. 2006), RNase II rapidly degraded the single-stranded region of the 16–30 ds substrate, rendering a 24-nt oligomer as a major product (Fig. 3A). By contrast, the smallest product generated by PNPase was a 27-nt oligomer, indicating that PNPase stalls before RNase II when encountering a double-stranded region. As expected, RNase R was able to degrade the 16–30 ds substrate to completion, confirming its ability to efficiently degrade an extensive region of double-stranded RNA as long as there is a 3′-single-stranded extension to allow the attachment of the enzyme (Cheng and Deutcher 2004). Elimination of the S1 domain in RNase II resulted in a drastic reduction of the exoribonucleolytic activity as revealed by the ΔS1 truncated protein, since degradation products were only detected after 30 min of reaction with 500 nM of protein. However, with RNase II, the same degradation products started to appear only after 1 min of reaction with only 0.5 nM of enzyme, and after 4 min of reaction the full-length substrate had disappeared. When we fused the S1 domain of RNase R or PNPase, the exoribonucleolytic activity was partially restored and the hybrid proteins showed a higher efficiency in RNA digestion than ΔS1 mutant protein. Degradation products were clearly visible with 60 nM of RNII-R and 200 nM of RNII-P after 15 min of incubation, and in 30 min the full-length substrate almost disappeared. Quantification of the percent of product released revealed that the activity of the hybrid proteins was low, but when compared to the ΔS1 mutant the RNII-P and the RNII-R proteins showed an 18- and 15-fold increase in activity, respectively. Similar results were obtained with the 30 ss substrate, where the hybrid proteins showed a higher efficiency in RNA degradation than the RNase IIΔS1 (Fig. 3B). From these results, we can conclude that the fusion of the C-terminal part of RNase R or the S1 domain of PNPase was able to compensate, at least in part, for the absence of the S1 domain in RNase II. Thus, the S1 domain of RNase II can be substituted by the S1 domain of the other two exoribonucleases, and the resulting chimeric proteins are still functional.
FIGURE 3.
Exoribonucleolytic activity of wild-type and hybrid proteins was assayed on a (A) double-stranded (16–30 ds) or (B) single-stranded substrate (30 ss). The enzyme concentrations used in A were: 0.5 nM of (His)6–RNase II (RNII), 0.5 nM of (His)6–RNase R (RNR), 60 nM of (His)6–RNase II-R (RNII-R), 0.5 nM of (His)6–PNPase (PNP), 200 nM of (His)6–RNase II-P (RNII-P), and 500 nM of (His)6–RNase IIΔS1 (ΔS1). The enzyme concentrations used in B were: 0.5 nM of (His)6–RNase II (RNII), 0.5 nM of (His)6–RNase R (RNR), 60 nM of (His)6–RNase II-R (RNII-R), 2 nM of (His)6–PNPase (PNP), 200 nM of (His)6–RNase II-P (RNII-P), and 50 nM of (His)6–RNase IIΔS1 (ΔS1). Reactions were performed as described in Materials and Methods and samples were withdrawn at the time points indicated in the figure. Reaction products were analyzed in a 20% PAA/7M urea gel and the bands detected by autoradiography. Substrate and product length are indicated. Control reactions, “c,” were incubated for 15 min with no enzyme added. The sequences of the oligoribonucleotides substrate used are depicted.
When using the 30 ss substrate the final products generated by the three wild-type (WT) enzymes, RNase II, PNPase, and RNase R, were different. Thus, according to previous reports, RNase II generated mainly a 4–6-nt fragment as the smallest product (Mitchell et al. 1997; LaCava et al. 2005; Amblar et al. 2006; Frazão et al. 2006), while PNPase and RNase R generated 6–7-nt and 1–2-nt fragments, respectively (Cheng and Deutcher 2004). The hybrid proteins and the ΔS1 truncated enzyme rendered the same products as RNase II, revealing that the S1 domain is not related to the size of the final products generated during degradation of a single-stranded substrate.
Activity assays using the 16–30 ds substrate revealed that the products generated with both hybrid proteins was the same as those generated by RNase II, and the major product observed with the three proteins corresponded to a 24-nt fragment (Fig. 3A). Therefore, both hybrid proteins behave like RNase II when encountering secondary structures in the RNA molecule. Given the differences detected in the C-terminal region of RNase R compared to RNase II and PNPase, a different behavior in degradation of double-stranded RNAs could be expected for the RNII-R hybrid polypeptide. However, the RNII-R hybrid protein was unable to degrade the double-stranded portion of the substrate and stalled at the same position as RNase II. Therefore, it seems that the ability of RNase R to overcome secondary structures does not reside either on its S1 domain, or on its C-terminal region.
The S1 of PNPase induces trimerization of the RNII–P hybrid protein
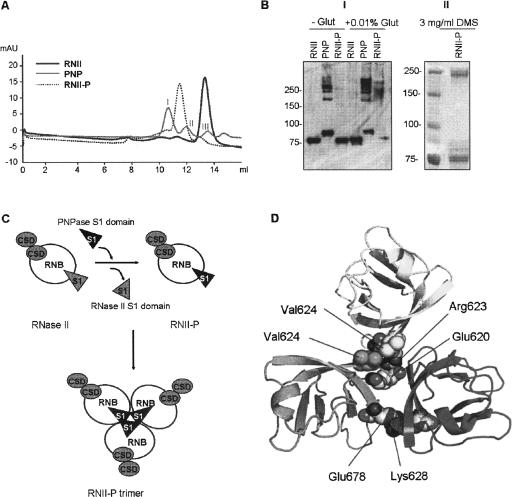
Recent data revealed that the role of the S1 domain is more complex than simply providing an RNA-binding surface. It has been demonstrated that the PNPase S1 domain is required for the functioning of the Yersinia type three secretion system (TTSS) during infection of macrophages, and that this function can be complemented by high copy expression of the S1 domains of E. coli PNPase, RNase R, and RNase II (Rosenzweig et al. 2005). Additionally, the S1 domain of E. coli RNase E dimerizes in vitro, and it has been postulated that its dimerization contributes to the formation of the tetrameric form in the native enzyme (Schubert et al. 2004). In E. coli PNPase the S1 domain has been shown to promote substrate recognition and product release, but it is not required for activity or autoregulation (Jarrige et al. 2002). Native PNPase is a trimeric enzyme, therefore, we wanted to investigate whether the RNII-P hybrid protein exists in a monomeric form, as the RNase II enzyme, or whether its quaternary structure corresponds to an oligomeric form such as PNPase. Initially, we undertook gel filtration analysis with the purified enzymes (Fig. 4A). RNase II revealed only one major peak that corresponded to an apparent molecular mass of ∼96 kDa, indicating that, as expected, RNase II adopts a monomeric form. PNPase presented one major peak that corresponded to an apparent molecular mass of 336 kDa (peak I), and two minor peaks of about 185 kDa (peak II) and 87 kDa (peak III), presumably corresponding to a trimer, a transient dimeric form, and a monomer, respectively. These results are in agreement with previous measurements performed on PNPase for which three bands corresponding to monomers, dimers, and trimers have been described (Portier 1975a,b). However, the RNII-P hybrid protein showed a major peak corresponding to an apparent molecular mass of 228 kDa, which is very similar to the real mass of a trimer (216.6 kDa). A smaller peak was also detected that corresponded approximately to 115 kDa, which could be either a monomer (72.2 kDa of real MW) or a transient dimer (144 kDa of real MW). Similar experiments were performed with RNase R and the hybrid RNII-R and, as expected, both of them were shown to be monomers (data not shown). The results obtained with RNase II, PNPase, and RNII-P were further confirmed by cross-linking experiments and light-scattering measurements. Upon mild treatment with glutaraldehyde, PNPase and RNII-P formed cross-linked oligomers, as revealed by Western blot analysis (Fig. 4B, panel I). PNPase showed an intermediate band slightly higher than 150 kDa, which was further converted to higher molecular weight bands, presumably corresponding to homotrimers, or even to larger oligomeric forms of PNPase (250 kDa or higher). Cross-linking of RNII-P hybrid protein revealed that, except for a small amount of protein that was in a monomeric form (around 75 kDa), most of it was an oligomer of about 250 kDa. In fact, at least three higher molecular weight bands were detected, two just under 250 kDa and one above it. These bands could represent trimers and tetramers, or even multiple cross-linked trimers. Similar results were obtained with dimethyl suberimidate as a cross-linker, which rendered two bands corresponding to monomers and trimers in SDS-PAGE (Fig. 4B, panel II). In contrast, and as expected, under the same conditions, the RNase II protein did not form oligomers and only the band corresponding to the monomer was detected after cross-linking. Light-scattering measurements revealed that the three proteins were stable and monodisperse in working conditions and confirmed the size of the native protein as well as of the aggregates present in solution. While RNase II exists in a monomeric form of about 100 kDa, both PNPase and RNII-P hybrid protein oligomerized as a trimer with a molecular mass of about 300 kDa. For instance, the RNase II sample showed 97% homogeneity and an average size of 8.56 nm, corresponding to a size of 101 kDa. PNPase and RNII-P samples showed 96% and 97% homogeneity, respectively. However, both samples presented an average size of about 14 nm (13.82 nm for PNPase and 13.64 nm for RNII–P) corresponding to a size of 300 kDa (more precisely, 310 kDa for PNPase and 300 kDa for RNII-P). These data indicated that, like PNPase, the RNII-P hybrid protein has a strong tendency to self-associate and adopts a predominantly trimeric form in solution. Therefore, the S1 domain of PNPase is able to induce trimerization of the hybrid RNII–P protein (Fig. 4C).
FIGURE 4.
(A) Gel filtration of (His)6–RNase II (RNII), (His)6–PNPase (PNP), and the hybrid protein (His)6–RNase II-P (RNII-P). (B) Panel I: Western blot of cross-linking experiments of (His)6–RNase II (RNII), (His)6–PNPase (PNP), and (His)6–RNase II-P (RNII-P). Samples of 0.5 μg of each protein were incubated in the absence (−) or in the presence (+) of 0.01% of gluteraldehyde and analyzed in a SDS-10% PAGE gel. Standard molecular weight is indicated on the left; Panel II: Western blot of cross-linking experiments of (His)6–RNase II-P (RNII-P). Ten micrograms of the protein were incubated in the presence of 3 mg/mL of dimethy suberimidate and analyzed in a SDS-10% PAGE gel. Standard molecular weight is indicated on the left. (C) Schematic representation of the model of RNase II trimerization induced by the S1 domain from PNPase. Catalytic RNB domain is represented as white circles, CSD domains are shown as gray circles, and S1 domains as triangles. (D) Detail of modeled PNPase S1 domain homotrimerization. Position of residues implicated in the two main S1 domain–S1 domain interactions in terms of stabilization of homotrimer are represented as spheres. Acidic chains of Glu 620 and Glu 678 of each monomer are faced to the side chains of positive residues Lys 628 and Arg 623, respectively, of the other two monomers. Contacts among side chains of all three Val 624 residues configure a local hydrophobic cluster. Only one representative of both interactions is depicted for clarity.
The crystal structure of PNPase revealed that contacts contributing to trimerization are located at the β-sheet trimerization interfaces at either end of the core domains, and in the loops around the central channel (Symmons et al. 2000). The S1 domain was not well ordered in the crystal but it seems to lie close to the upper surface of the trimer not involved in trimerization contacts. However, the fact that the S1 domain of E. coli PNPase causes trimerization in RNase II cannot be obviated and points to an intrinsic propensity of S1 domain to self-associate. A similar propensity has been reported for the S1 domain of RNase E, which is able to dimerize in solution (Schubert et al. 2004). On this basis a possible role of the S1 domain in promoting dimerization of RNase E has been proposed (Schubert et al. 2004). However, as occurs with PNPase, crystallographic studies on RNase E revealed that the S1 domain is not located at the dimer interface in the tetrameric enzyme (Callaghan et al. 2005). Therefore, the self-association tendency of the RNase E S1 domain could suggest a role in dimer biogenesis but not account for the stabilization on native RNase E. In light of the results obtained in this report, a similar function for the S1 domain of E. coli PNPase can be hypothesized.
Furthermore, taking into account only structural determinants (volume complementarities and electrostatics relationships) we were able to propose a model for a putative trimer of the S1 domain of PNPase (Fig. 4D). The model shows electrostatic contacts between Glu620–Arg623 and Lys628–Glu678 that can play an important role in trimerization. The alignment in Figure 1 shows that Glu620 is highly conserved in all PNPases but not in RNase II or RNase R. The Arg623 has a medium-high level of conservation in PNPase sequences, too, and the Lys628–Glu678 pair is almost completely conserved in all PNPases. On the contrary, in the RNase II protein this contact will be structurally substituted by a Glu–Glu repulsive pairing, thus preventing oligomerization. A hydrophobic cluster formed by the side chains of Val624 from the three S1 domains will contribute to the stability of the complex. The proposed model for trimerization is only theoretical, based on bioinformatics analysis of sequences and structures; but it presents a structurally possible solution to illustrate the experimentally observed formation of RNII-P trimers.
In this report we prove that the S1 domains from the three exoribonucleases of E. coli are interchangeable because the activity of ΔS1 has been recovered to a certain extent, but there is loss of activity when compared to the WT RNase II. This is probably due to the fact that each of the three S1 domains has been “customized” as each of the enzymes has evolved to perform a specific purpose in the cell. Thus, although the domains are not fully functionally interchangeable, they can be replaced one by each other, maintaining an evolutionarily conserved common activity. Moreover, the S1 domain does not influence the degradation properties of RNase II regarding substrate specificity and the length of final products. Our results also confirm that the S1 domain is more than simply an RNA-binding module, and as pointed out by previous observations, this domain may be involved in different processes such as protein multimerization.
MATERIALS AND METHODS
Materials
Restriction enzymes, T4 DNA ligase, and T4 polynucleotide kinase were purchased from New England Biolabs; T7 RNA polymerase was obtained from Promega; and Pfu DNA polymerase from Fermentas. Unlabeled oligonucleotide primers were synthesized by Thermolab and Sigma Genosis.
Strains and plasmids
The E. coli strains used were JM109, F′ (traD36 proA+B+ lacIq Δ[lacZ]M15/ Δ[lac-proAB] glnV44 e 14- gyrA96 recA1 relA1 endA1 thi hsdR17 (Yanish- Perron et al. 1985), for cloning experiments, and BL21(DE3) (F− rB − mB − gal ompT [int∷PlacUV5 T7 gen1 imm21 nin5]) (Studier and Moffatt 1986) for expression and purification of enzymes. The plasmid used for in vitro transcription reactions was pCH77 (McLaren et al. 1991).
Construction of plasmids expressing RNase R, PNPase, and hybrid proteins
The pFCT6.1 plasmid (Cairrão et al. 2001) was used for expression of E. coli histidine-tagged RNase II protein. This plasmid contains the rnb gene cloned into pET-15b vector (Novagen) under the control of φ10 promoter, allowing the expression of the (His)6-tagged RNase II fusion protein. Similar constructions were performed for the expression of RNase R and PNPase from E. coli. The rnr and pnp genes from E. coli encoding RNase R and PNPase proteins, respectively, were cloned into pET-15b vector (Novagen), thus obtaining the corresponding six-histidine fusion proteins at the N-terminal end ([His]6-RNase R and [His]6-PNPase). The rnr gene was obtained from the E. coli MG1693 strain containing the pHM102 plasmid (Cairrão et al. 2003). This plasmid consists on a 5.3-kb chromosomal fragment from E. coli, which includes the rnr gene, cloned into a pUC19 vector. The pnp gene was obtained from the E. coli strain IBPC5321 containing the pB15.6 plasmid (Zilhão et al. 1996a). Both rnr and pnp genes were amplified from pHM102 and pB15.6 plasmids, respectively, using the RNR-forward (5′-CGGAGGAACCCATATGTCACAAGATCCTTT-3′) and RNR-reverse (5′-GATGCACTACGTGGATCCGGCCTACATGAT-3′) primers for the rnr gene, and PNP-forward (5′-GAAAGAGAAAGGATATTCATATGCTTAATC-3′) and PNP-reverse (5′-TGAATGAACGTCCGGATCCCGGTTGCTAAC-3′) primers for the pnp gene. The forward (RNR- and PNP) and reverse (RNR- and PNP) primers contained NdeI and BamHI sites (underlined), respectively, which were used for cloning into pET-15b vector, obtaining plasmids pABA–RNR and pABA–PNP.
Hybrid proteins were constructed by substituting the C-terminal region of (His)6–RNase II by the C-terminal part of (His)6–RNase R or (His)6–PNPase, thus obtaining the corresponding hybrid proteins: (His)6–RNII-R and (His)6–RNII-P, respectively. For this purpose, the SpeI restriction site was first introduced in the rnb gene of the pFCT6.1 plasmid at the 934 nt position by mutagenic PCR (Higuchi 1990) using: 5′-TTCGATATTATCTTCACTAGTGCCATCAGCGGAGA-3′, and 5′-TCTCCGCTGATGGCACTAGTGAAGATAATATCGAA-3′ as mutagenic primers (the SpeI restriction site is underlined), and RNB2019 (5′-AACCTATAAAAATAGGCGTATCACGAGGCC-3′) and RNB1028 (5′-GTGACTGGCAGCCTGAAAGTGAAGCAATTG-3′) as amplifying primers. The PCR product was further cloned in the pFCT6.1 plasmid, thus obtaining the pFCTSpe934 plasmid. The sequences corresponding to residues 636–813 of RNase R and residues 616–711 of PNPase were amplified using SPE-R (5′-GGCTGACTGGCTGACTAGTGACTTCATGCT-3′ and HIN-R 5′-CGCATTTTGTCAGCAAGCTTACCCTCTTCT-3′) primers for RNase R, and SPE-P (5′-CGAAGAGATCACTAGTGAAATCGAAGTGGG-3′ and HIN-P 5′-CAGCGGAGGGCAAATGGCAAGCTTACT-3′) primers for PNPase, containing SpeI (SPE-R and SPE-P) and HindIII (HIN-R and HIN-P) restriction sites (underlined). The SpeI–HindIII 1128-nt fragment of the rnb gene from the pFCTSpe934 plasmid was substituted by the corresponding fragments from rnr and pnp, obtaining the pABAII-R and pABAII-P, respectively.
Overexpression and purification of proteins
All plasmids constructed were transformed into a BL21(DE3) E. coli strain to allow the expression of the recombinant proteins upon IPTG induction. Cells were grown at 37°C in LB medium supplemented with 150 μg/mL amplicillin to an A600 of 0.45 and then induced by addition of 1 mM IPTG. Samples were withdrawn at different induction times and crude extracts were prepared as previously described (Amblar et al. 2001) in order to analyze the total protein content. The solubility of each protein during induction was tested by separation of the soluble and insoluble protein fraction as previously described (Amblar and López 1998), followed by fractionation in SDS/PAGE.
Purification of (His)6–RNase II, (His)6–RNase R, (His)6–RNase IIΔS1, and the hybrid (His)6–RNII-R and (His)6–RNII-P proteins was performed by histidine affinity chromatography using HiTrap Chelating HP columns (Amersham Biosciences) and AKTA HLPC system (Amersham Biosciences) following the protocol previously described (Amblar et al. 2006). Briefly, cells from 100 mL of IPTG induced cultures were harvested by centrifugation and lysated by mechanical disruption using a French Press. The clarified extracts were added to a HiTrap Chelating Sepharose 1 mL column equilibrated in buffer A (20 mM Tris–HCl, 0.5 M NaCl, pH8) plus 20 mM imidazol and 2 mM β-mercaptoethanol. Protein elution was achieved by a continuous imidazol gradient (from 20 mM to 500 mM) in buffer A. The fractions containing the purified protein were pooled together and loaded into a desalting 5 mL column (Amersham Biosciences) to change the buffer. For (His)6–RNase II, (His)6–RNase IIΔS1, and hybrid proteins the column was equilibrated with buffer B containing 20 mM Tris–HCl pH8, 100 mM KCl, and 2 mM β-mercaptoethanol. For (His)6–RNase R the desalting column was equilibrated with buffer C containing 20 mM Tris–HCl pH8, 300 mM KCl, and 2 mM β-mercaptoethanol. In the case of the (His)6–PNPase protein, cell lysis was performed as described above but the Tris component of the buffer was substituted by 20 mM of Na2HPO4. Clarified extract was added to a HiTrap Chelating Sepharose 1-mL column equilibrated in the same phosphate buffer plus 20 mM imidazol and 2 mM β-mercaptoethanol. After a washing step with 70 mM imidazol, protein was one-step eluted with 0.5 M imidazol. Fractions containing the purified protein were pooled together and loaded into the same desalting 5-mL column as described above, equilibrated with buffer D containing 30 mM Tris–HCl pH8, 60 mM KCl, 10 mM NaH2PO4, 1 mM MgCl2, and 2 mM DTT. Eluted proteins were concentrated by centrifugation at 15°C with Amicon Ultra Centrifugal Filter Devices (Millipore). Protein concentration was determined by spectrophotometry and 50% (v/v) glycerol was added to the final fractions prior to storage at −20°C.
In vitro transcription of RNAs
The malE–malF RNA molecule was obtained by in vitro transcription using the pCH77 plasmid linearized with EcoRI (McLaren et al. 1991) as a template. The transcription reaction was performed using the Riboprobe kit from Promega following the instructions given by the manufacturers, in a 20 μL volume, containing 20 μCi of [α-32P]-rUTP (Amersham Biosciences). Radioactively labeled RNA transcript was purified on a 6% PAA/7M urea gel as previously described (Conrad et al. 1998).
Activity assays
Exoribonucleolytic activity was assayed using oligoribonucleotides as a substrate. The 30 mer oligoribonucleotide (5′-CCCGACACCAACCACUAAAAAAAAAAAAAA-3′), was labeled at its 5′ end with [γ-32ATP] and T4 polynucleotide kinase. The RNA oligomer was then purified using Microcon YM-3 Centrifugal Filter Devices (Millipore) to remove the nonincorporated nucleotide. When indicated, the 30 mer oligoribonucleotide was hybridized to the complementary 16 mer oligodeoxiribonucletide (5′-AGTGGTTGGTGTCGGG-3′), thus obtaining the corresponding 16–30 ds substrate. The hybridization was performed in a 1:1 (mol:mol) ratio in the Tris component of the activity buffer by 5 min of incubation at 68°C followed by 45 min at room temperature. The exoribonucleolytic reactions were carried out in a final volume of 10 μL containing 30 nM of substrate. For (His)6–RNase II, (His)6–RNase IIΔS1, and hybrid proteins the activity buffer contained 20 mM Tris–HCl pH8, 100 mM KCl, 1 mM MgCl2, and 1 mM DTT (Amblar and Arraiano 2005). For (His)6–RNase R and (His)6–PNPase the activity buffers used were the optimal buffers previously described for each protein, i.e., 20 mM Tris–HCl pH8, 300 mM KCl, 1 mM MgCl2, and 1 mM DTT for RNase R (Cheng and Deutscher 2002) and 50 mM Tris–HCl pH8, 60 mM KCl, 1 mM MgCl2, 10 mM Na2HPO4 pH8, and 2 mM DTT for PNPase (Spickler and Mackie 2000; Regonesi et al. 2004). The amount of each enzyme added to the reaction was adjusted to obtain linear conditions and is indicated in Figure 3. Reactions were started by the addition of the enzyme and incubated at 37°C. Samples were withdrawn at the time points indicated in Figure 3, and the reaction was stopped by adding formamide-containing dye supplemented with 10 mM EDTA. Reaction products were resolved in a 20% polyacrylamide/7 M urea and analyzed by autoradiography. Exoribonucleolytic activity was quantified by measuring the relative intensity of the product bands. Therefore, exoribonucleolytic activity was estimated as the percentage of product released by 1 nM of enzyme in 1 min at 37°C.
Electrophoretic mobility shift assay (EMSA)
EMSAs with malE–malF mRNA was performed as described previously (Amblar and Arraiano 2005). Mixtures containing an increasing concentration of each enzyme were incubated for 10 min at 37°C and analyzed in a 5% nondenaturing polyacrylamide gel as previously described. In the EMSAs performed with a DNA oligomer, the ompA-3 oligonucleotide (5′-TAATACGACTCACTATAGGAAAAAAAACCCCGCAGCAGC-3′) (Andrade et al. 2006) was labeled at its 5′ end using [γ-32ATP] and T4 polynucleotide kinase. The substrate was then purified with Microcon YM-3 Centrifugal Filter Devices (Millipore) to remove the nonincorporated nucleotide. Binding reactions were performed in 10 μL of volume containing 20 mM Tris–HCl pH8, 100 mM KCl, 1 mM DTT, 0.5 μg/μL BSA, and 10 mM EDTA, 1 fmol of ompA-3 substrate, and increasing concentration of enzyme. Mixtures were incubated for 10 min at 37°C and analyzed in a 5% nondenaturing polyacrylamide gel as previously described (Amblar and Arraiano 2005). The RNA– and DNA–protein complexes were detected by using the PhosphorImager system from Molecular Dynamics.
Gel filtration chromatography
Purified proteins were subjected to gel filtration on a Sephacryl S-200 HR column (Amersham Biosciences) preequilibrated with 20 mM Tris–HCl pH 8, 100 mM KCl, and 2 mM β-mercaptoethanol. Elution was performed at a flow rate of 0.5 mL/min in the same buffer. Apparent molecular masses were assessed based on elution volumes of suitable markers (catalase 232 kDa, bovine serum albumin 67 kDa, ovoalbumin 45 kDa, chemotripsine 25 kDa, and RNase A 15 kDa).
Cross-linking experiments
Chemical cross-linking reactions were carried out based on previous reports (Regonesi et al. 2004; Schubert et al. 2004). For the first cross-linking experiment 0.5 μg of purified protein was incubated in 15 μL of 20 mM phosphate buffer pH7, with 50 mM NaCl and 0.01% (v/v) glutaraldehyde at 37°C for 10 min. Reactions were quenched with 1 μL of 1 M Tris–HCl pH8 and stored on ice.
For the second cross-linking confirmation experiment 10 μg of the RNII–PNP enzyme was incubated with 3 mg/mL of dimethyl suberimidate at 4°C in 0.2 M triethanolamine hydrochloride (pH 8.5) for a period of 16 h. All the protein samples were subsequently boiled for 15 min prior to separation on a SDS-8% PAGE gel. In the experiments performed with gluteraldehyde as the cross-linker the gel was further subjected to Western blot analysis using a 1:3000 dilution of Anti-His antibody (Amersham Biosciences) and the detection was carried out using the ECL system (Amersham Biosciences).
Dynamic light scattering
The oligomerization state of (His)6–RNase II, (His)6–RNase II-P, and (His)6–PNPase was analyzed using a Zetasizer Nano from Malvern Instruments Ltd. Measurements were performed in the presence of 20 mM Tris–HCl pH8 and 100 mM KCl, for RNase II and RNII-P hybrid protein, and 50 mM Tris–HCl pH8, 60 mM KCl, for PNPase.
Molecular modeling
Multiple structure-based amino acid sequence alignments of carboxyterminal domains of the RNase II, RNaseR, and PNPase families of proteins, and the crystallographic coordinates of S1 domains of RNase II (Protein Data Bank [PDB] entries: 2IX0 and 2IX1; Frazão et al. 2006; McVey et al. 2006) and PNPase (PDB entry: 1SRO; Bycroft et al. 1997) were performed using ClustalW (Thompson et al. 1994) and T-COFFEE (Guex and Peitsch 1997; Notredame et al. 2000) algorithms for sequence comparison and the DeepView package for structure fitting (Guex and Peitsch 1997). Based on the obtained alignment, the structural model for the S1 domain of E. coli RNase R was constructed using homology modeling procedures implemented in the SWISS-MODEL server (Guex and Peitsch 1997; Schwede et al. 2003) facilities at http://www.expasy.ch/swissmod/SWISS-MODEL.html, and their structural quality was checked using the WHAT-CHECK routines (Vriend 1990; Hooft et al. 1996) from the WHAT IF program (Vriend 1990) from the same server. Finally, in order to optimize geometries, release local constraints, and correct possible bad contacts, the modeled structures were energy minimized with the implementation of the GROMOS 43B1 force field in the program DeepView (Guex and Peitsch 1997) using 500 steps of steepest descent minimization followed by 500 steps of conjugate-gradient minimization.
The structural model for the molecular interaction between E. coli PNPase S1 domains was built using the computational methods for protein–protein rigid docking implemented in the program Hex (Ritchie and Kemp 2000). For all the docking models generated, molecular mechanics energy minimization over the force field implemented in Hex was finally performed. From the overall models, after discarding the nonrealistic ones, the best complexes in terms of highest steric and electrostatic correlation between the pair of protein structures were selected. Structure plots were generated using PyMOL (DeLano 2002).
SUPPLEMENTAL DATA
Supplemental material can be found at http://novacripta.cbm.uam.es/bioweb/SuppMat/S1_domain_RNA07_supp.pdf.
ACKNOWLEDGMENTS
We thank David Guerra Aragão for the light-scattering measurements at ESRF (European Synchrotron Radiation Facility), France. The authors also thank M. Schubert for critical reading of this manuscript and J. Andrade and S. Viegas for helpful discussions. M.A. was a recipient of a Post Doctoral fellowship and A.B. was a recipient of a Ph.D. fellowship, both funded by FCT-Fundação para a Ciência e a Tecnologia, Portugal. The work at the ITQB was supported by FCT-Fundação para a Ciência e Tecnologia, Portugal. The work at the CBMSO was supported by “Ministerio de Educación y Ciencia” Grant SAF2004-06843 and by an institutional grant of “Fundación Ramón Areces.”
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.220407.
REFERENCES
- Amblar, M., Arraiano, C.M. A single mutation in Escherichia coli ribonuclease II inactivates the enzyme without affecting RNA binding. FEBS J. 2005;272:363–374. doi: 10.1111/j.1742-4658.2004.04477.x. [DOI] [PubMed] [Google Scholar]
- Amblar, M., López, P. Purification and properties of the 5′-3′ exonuclease D190→A mutant of DNA polymerase I from Streptococcus pneumoniae . Eur. J. Biochem. 1998;252:124–132. doi: 10.1046/j.1432-1327.1998.2520124.x. [DOI] [PubMed] [Google Scholar]
- Amblar, M., Lacoba, M.G.D., Corrales, M.A., López, P. Biochemical analysis of point mutations in the 5′-3′ exonuclease of DNA polymerase I of Streptococcus pneumoniae. Functional and structural implications. J. Biol. Chem. 2001;276:19172–19181. doi: 10.1074/jbc.M008678200. [DOI] [PubMed] [Google Scholar]
- Amblar, M., Barbas, A., Fialho, A.M., Arraiano, C.M. Characterization of the functional domains of Escherichia coli RNase II. J. Mol. Biol. 2006;360:921. doi: 10.1016/j.jmb.2006.05.043. [DOI] [PubMed] [Google Scholar]
- Andrade, J.M., Cairrão, F., Arraiano, C.M. RNase R affects gene expression in stationary phase: Regulation of ompA. Mol. Microbiol. 2006;60:219–228. doi: 10.1111/j.1365-2958.2006.05092.x. [DOI] [PubMed] [Google Scholar]
- Arcus, V. OB-fold domains: A snapshot of the evolution of sequence, structure and function. Curr. Opin. Struct. Biol. 2002;12:794. doi: 10.1016/s0959-440x(02)00392-5. [DOI] [PubMed] [Google Scholar]
- Arraiano, C.M., Maquat, L.E. Post-transcriptional control of gene expression: Effectors of mRNA decay. Mol. Microbiol. 2003;49:267–276. doi: 10.1046/j.1365-2958.2003.03545.x. [DOI] [PubMed] [Google Scholar]
- Boni, I.V., Isaeva, D.M., Musychenko, M.L., Tzareva, N.V. Ribosome-messenger recognition: mRNA target sites for ribosomal protein S1. Nucleic Acids Res. 1991;19:155–162. doi: 10.1093/nar/19.1.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bycroft, M., Hubbard, T.J., Proctor, M., Freund, S.M., Murzin, A.G. The solution structure of the S1 RNA binding domain: A member of an ancient nucleic acid-binding fold. Cell. 1997;88:235–242. doi: 10.1016/s0092-8674(00)81844-9. [DOI] [PubMed] [Google Scholar]
- Cairrão, F., Chora, A., Zilhão, R., Carpousis, J., Arraiano, C.M. RNase II levels change according to the growth conditions: Characterization of gmr, a new Escherichia coli gene involved in the modulation of RNase II. Mol. Microbiol. 2001;276:19172–19181. doi: 10.1046/j.1365-2958.2001.02342.x. [DOI] [PubMed] [Google Scholar]
- Cairrão, F., Cruz, A., Mori, H., Arraiano, C.M. Cold shock induction of RNase R and its role in the maturation of the quality control mediator SsrA/tmRNA. Mol. Microbiol. 2003;50:1349–1360. doi: 10.1046/j.1365-2958.2003.03766.x. [DOI] [PubMed] [Google Scholar]
- Callaghan, A.J., Marcaida, M.J., Stead, J.A., McDowall, K.J., Scott, W.G., Luisi, B.F. Structure of Escherichia coli RNase E catalytic domain and implications for RNA turnover. Nature. 2005;437:1187. doi: 10.1038/nature04084. [DOI] [PubMed] [Google Scholar]
- Cannistraro, V.J., Kennell, D. The processive reaction mechanism of ribonuclease II. J. Mol. Biol. 1994;243:930–943. doi: 10.1006/jmbi.1994.1693. [DOI] [PubMed] [Google Scholar]
- Cheng, Z.-F., Deutscher, M.P. Purification and characterization of the Escherichia coli exoribonuclease RNase R. Comparison with RNase II. J. Biol. Chem. 2002;277:21624–21629. doi: 10.1074/jbc.M202942200. [DOI] [PubMed] [Google Scholar]
- Cheng, Z.-F., Deutcher, M.P. An important role for RNase R in mRNA decay. Mol. Cell. 2004;17:313–318. doi: 10.1016/j.molcel.2004.11.048. [DOI] [PubMed] [Google Scholar]
- Coburn, G.A., Mackie, G.A. Differential sensitivities of portions of the mRNA for ribosomal protein S20 to 3′-exonucleases dependent on oligoadenylation and RNA secondary structure. J. Biol. Chem. 1996;271:15776–15781. doi: 10.1074/jbc.271.26.15776. [DOI] [PubMed] [Google Scholar]
- Conrad, C., Rauhut, R., Klug, G. Different cleavage specificities of RNases III from Rhodobacter capsulatus and Escherichia coli . Nucleic Acids Res. 1998;26:4446–4453. doi: 10.1093/nar/26.19.4446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLano, W.L. The PyMOL molecular graphics system. DeLano Scientific; San Carlos, CA: 2002. [Google Scholar]
- Deutscher, M.P., Reuven, N.B. Enzymatic basis for hydrolytic versus phosphorolytic mRNA degradation in Escherichia coli and Bacillus subtilis . Proc. Natl. Acad. Sci. 1991;88:3277–3280. doi: 10.1073/pnas.88.8.3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diwa, A.A., Jiang, X., Schapira, M., Belasco, J.G. Two distinct regions on the surface of an RNA-binding domain are crucial for RNase E function. Mol. Microbiol. 2002;46:959–969. doi: 10.1046/j.1365-2958.2002.03231.x. [DOI] [PubMed] [Google Scholar]
- Frazão, C., McVey, C.E., Amblar, M., Barbas, A., Vonrhein, C., Arraiano, C.M., Carrondo, M.A. Unravelling the dynamics of RNA degradation by ribonuclease II and its RNA-bound complex. Nature. 2006;443:110. doi: 10.1038/nature05080. [DOI] [PubMed] [Google Scholar]
- Graumann, P.L., Marahiel, M.A. A superfamily of proteins that contain the cold-shock domain. Trends Biochem. Sci. 1998;23:286–290. doi: 10.1016/s0968-0004(98)01255-9. [DOI] [PubMed] [Google Scholar]
- Guex, N., Peitsch, M.C. SWISS-MODEL and the Swiss-PdbViewer: An environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- Gupta, R.S., Kasai, T., Schlessinger, D. Purification and some novel properties of Escherichia coli RNase II. J. Biol. Chem. 1977;252:8945–8949. [PubMed] [Google Scholar]
- Higuchi, R. Recombinant PCR. In: Innis M.A., et al., editors. PCR protocols. A guide to methods and applications. Academic Press; San Diego, CA: 1990. pp. 177–183. [Google Scholar]
- Hooft, R.W., Vriend, G., Sander, C., Abola, E.E. Errors in protein structures. Nature. 1996;381:272. doi: 10.1038/381272a0. [DOI] [PubMed] [Google Scholar]
- Jarrige, A.-C., Brechemier-Baey, D., Mathy, N., Duche, O., Portier, C. Mutational analysis of polynucleotide phosphorylase from Escherichia coli . J. Mol. Biol. 2002;321:397. doi: 10.1016/s0022-2836(02)00645-9. [DOI] [PubMed] [Google Scholar]
- LaCava, J., Houseley, J., Saveanu, C., Petfalski, E., Thompson, E., Jacquier, A., Tollervey, D. RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell. 2005;121:713–724. doi: 10.1016/j.cell.2005.04.029. [DOI] [PubMed] [Google Scholar]
- Marujo, P.E., Hajnsdorf, E., Le Derout, J., Andrade, R., Arraiano, C.M., Régnier, P. RNases II removes the oligo(A) tails that destabilizes the rpsO mRNA of E. coli . RNA. 2000;6:1185–1193. doi: 10.1017/s135583820000073x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDowall, K., Hernandez, R.G., Lin-Chao, S., Cohen, S.N. The ams-1 and rne-3071 temperature-sensitive mutations in the ams gene are in close proximity to each other and cause substitutions within a domain that resembles a product of the Escherichia coli mre locus. J. Bacteriol. 1993;175:4245–4249. doi: 10.1128/jb.175.13.4245-4249.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaren, R.S., Newbury, S.F., Dance, G.S.C., Causton, H.C., Higgins, C.F. mRNA degradation by processive 3′-5′ exoribonucleases in vitro and the implications for prokaryotic mRNA decay in vivo. J. Mol. Biol. 1991;221:81–95. [PubMed] [Google Scholar]
- McVey, C.E., Amblar, M., Barbas, A., Cairrão, F., Coelho, R., Romão, C., Arraiano, C.M., Carrondo, M.A., Frazão, C. Expression, purification, crystallization and preliminary diffraction data characterization of Escherichia coli ribonuclease II (RNase II) Acta Crystallog., Sect. F: Struct. Biol. Cryst. Commun. 2006;62:684–687. doi: 10.1107/S1744309106021506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mian, I.S. Comparative sequence analysis of ribonucleases HII, III, II, PH and D. Nucleic Acids Res. 1997;25:3187–3195. doi: 10.1093/nar/25.16.3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell, P., Petfalski, E., Shevchenko, A., Mann, M., Tollervey, D. The exosome: A conserved eukaryotic RNA processing complex containing multiple 3′→ 5′ exoribonucleases. Cell. 1997;91:457–466. doi: 10.1016/s0092-8674(00)80432-8. [DOI] [PubMed] [Google Scholar]
- Murzin, A.G. OB(oligonucleotide/oligosaccharaide binding)-fold: Commom structural and functional for non-homologous sequences. EMBO J. 1993;12:861–867. doi: 10.1002/j.1460-2075.1993.tb05726.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notredame, C., Higgins, D.G., Heringa, J. T-coffee: A novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 2000;302:205. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- Portier, C. Quaternary structure of Escherichia coli polynucleotide phosphorylase: New evidence for a trimeric structure. FEBS Lett. 1975a;50:79–81. doi: 10.1016/0014-5793(75)81045-3. [DOI] [PubMed] [Google Scholar]
- Portier, C. Quaternary structure of polynucleotide phosphorylase from Escherichia coli: Evidence of a complex between two types of polypeptide chains. Eur. J. Biochem. 1975b;55:573–582. doi: 10.1111/j.1432-1033.1975.tb02194.x. [DOI] [PubMed] [Google Scholar]
- Régnier, P., Arraiano, C.M. Degradation of mRNA in bacteria: Emergence of ubiquitous features. Bioessays. 2000;22:235–244. doi: 10.1002/(SICI)1521-1878(200003)22:3<235::AID-BIES5>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Regonesi, M.E., Briani, F., Ghetta, A., Zangrossi, S., Ghisotti, D., Tortora, P., Deho, G. A mutation in polynucleotide phosphorylase from Escherichia coli impairing RNA binding and degradosome stability. Nucleic Acids Res. 2004;32:1006–1017. doi: 10.1093/nar/gkh268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie, D.W., Kemp, G.J. Protein docking using spherical polar Fourier correlations. Proteins: Struct. Funct. Genet. 2000;39:178–194. [PubMed] [Google Scholar]
- Rosenzweig, J.A., Weltman, G., Plano, G.V., Schesser, K. Modulation of yersinia type three secretion system by the S1 domain of polynucleotide phosphorylase. J. Biol. Chem. 2005;280:156–163. doi: 10.1074/jbc.M405662200. [DOI] [PubMed] [Google Scholar]
- Schubert, M., Edge, R.E., Lario, P., Cook, M.A., Strynadka, N.C., Mackie, G.A., McIntosh, L.P. Structural characterization of the RNase E S1 domain and identification of its oligonucleotide-binding and dimerization interfaces. J. Mol. Biol. 2004;341:37–54. doi: 10.1016/j.jmb.2004.05.061. [DOI] [PubMed] [Google Scholar]
- Schwede, T., Kopp, J., Guex, N., Peitsch, M.C. Swiss-Model: An automated protein homology-modeling server. Nucleic Acids Res. 2003;31:3381–3385. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spickler, C., Mackie, A. Action of RNases II and polynucleotide phosphorylase against RNAs containing stem–loops of defined structure. J. Bacteriol. 2000;182:2422–2427. doi: 10.1128/jb.182.9.2422-2427.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stickney, L.M., Hankins, J.S., Miao, X., Mackie, G.A. Function of the conserved S1 and KH domains in polynucleotide phosphorylase. J. Bacteriol. 2005;187:7214–7221. doi: 10.1128/JB.187.21.7214-7221.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studier, F.W., Moffatt, B.A. Selective expression of cloned genes directed by T7 RNA polymease. J. Mol. Biol. 1986;189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- Symmons, M.F., Jones, G.H., Luisi, B.F. A duplicated fold is the structural basis for polynucleotide phosphorylase catalytic activity, processivity, and regulation. Struct. Fold. Des. 2000;8:1215–1226. doi: 10.1016/s0969-2126(00)00521-9. [DOI] [PubMed] [Google Scholar]
- Theobald, D.L., Mitton-Fry, R.M., Wuttke, D.S. Nucleic acid recognition by OB-fold proteins. Annu. Rev. Biophys. Biomol. Struct. 2003;32:115–133. doi: 10.1146/annurev.biophys.32.110601.142506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, J.D., Higgins, D.G., Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vriend, G. WHAT IF: A molecular modelling and drug design program. J. Mol. Graph. 1990;8:52–56. doi: 10.1016/0263-7855(90)80070-v. [DOI] [PubMed] [Google Scholar]
- Yanish-Perron, C., Vieira, J., Messing, J. Improved M13 phage cloning vectors and host strains: Nucleotide sequence of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Zilhão, R., Camelo, L., Arraiano, C.M. DNA sequencing and expression of the gene rnb encoding Escherichia coli ribonuclease II. Mol. Microbiol. 1993;8:43–51. doi: 10.1111/j.1365-2958.1993.tb01201.x. [DOI] [PubMed] [Google Scholar]
- Zilhão, R., Cairrão, F., Régnier, P., Arraiano, C.M. PNPase modulates RNase II expression in Escherichia coli: Implications for mRNA decay and cell metabolism. Mol. Microbiol. 1996a;20:1033–1042. doi: 10.1111/j.1365-2958.1996.tb02544.x. [DOI] [PubMed] [Google Scholar]
- Zilhão, R., Plumbridge, J., Hajnsdorf, E., Régnier, P., Arraiano, C.M. Escherichia coli RNase II: Characterization of the promoters involved in the transcription of rnb . Microbiology. 1996b;142:367–375. doi: 10.1099/13500872-142-2-367. [DOI] [PubMed] [Google Scholar]