Abstract
The discovery of RNA interference (RNAi) has revolutionized biological research and has a huge potential for therapy. Since small double-stranded RNAs (dsRNAs) are required for various RNAi applications, there is a need for cost-effective methods for producing large quantities of high-quality dsRNA. We present two novel, flexible virus-based systems for the efficient production of dsRNA: (1) an in vitro system utilizing the combination of T7 RNA polymerase and RNA-dependent RNA polymerase (RdRP) of bacteriophage ϕ6 to generate dsRNA molecules of practically unlimited length, and (2) an in vivo RNA replication system based on carrier state bacterial cells containing the ϕ6 polymerase complex to produce virtually unlimited amounts of dsRNA of up to 4.0 kb. We show that pools of small interfering RNAs (siRNAs) derived from dsRNA produced by these systems significantly decreased the expression of a transgene (eGFP) in HeLa cells and blocked endogenous pro-apoptotic BAX expression and subsequent cell death in cultured sympathetic neurons.
Keywords: dsRNA, siRNA, RNA interference
INTRODUCTION
RNA silencing refers to a group of recently discovered mechanisms where dsRNA regulates eukaryotic gene expression in a sequence-specific manner (Dykxhoorn et al. 2003; Meister and Tuschl 2004). Various small RNAs are used routinely as powerful research tools, and their application as therapeutic reagents is being investigated. To date, RNA interference (RNAi) and related silencing mechanisms have been used to target expression of several cancer- and disease-related genes and to alleviate viral infections in cell cultures and in vivo models (Uprichard 2005; van Rij and Andino 2006).
Thus far, gene silencing experiments have been successfully carried out using sequence-specific siRNAs or short hairpin RNAs that have been either synthesized chemically or transcribed from template DNA using DNA-dependent RNA polymerases (Amarzguioui et al. 2005). However, since several factors affect the efficacy of an siRNA (Reynolds et al. 2004; Heale et al. 2005), identifying a functional sequence often is laborious and expensive. One way to circumvent this trial-and-error approach is to digest long dsRNAs into siRNA pools containing molecules of multiple sequences (Myers et al. 2003; Seyhan et al. 2005). In fact, in plants, fungi, and most invertebrates, full-length dsRNA molecules can be used to induce RNA silencing as such, enabling their potential use in the regulation of gene expression as well as a tool in functional genomics (Dykxhoorn et al. 2003; Meister and Tuschl 2004). Current methods for producing long dsRNAs rely on annealing two single-stranded RNA (ssRNA) strands. However, this may result in poor-quality dsRNA.
We developed two novel, flexible, virus-based systems for the efficient production of dsRNA: (1) an in vitro system utilizing the combination of T7 RNA polymerase and RNA-dependent RNA polymerase (RdRp) of bacteriophage ϕ6 (Makeyev and Bamford 2000a,b) to generate dsRNA molecules, and (2) an in vivo RNA replication system based on carrier state bacterial cells containing the ϕ6 polymerase complex (Onodera et al. 1992; Sun et al. 2004) to produce virtually unlimited amounts of dsRNA. Furthermore, we demonstrate the utility of this system by RNA knockdown experiments in two different cell types.
RESULTS AND DISCUSSION
We developed an alternative approach for generating functional siRNA pools for entire genes and small genomes. This in vitro dsRNA production method (Fig. 1A) combines the use of T7 RNA polymerase to synthesize ssRNA templates of heterologous cDNA and the RdRP of bacteriophage ϕ6, a dsRNA virus infecting Pseudomonas syringae cells. ϕ6 RdRP performs de novo initiation and produces full-length dsRNAs from given template ssRNAs. The enzyme is highly processive, replicating ssRNA molecules of practically unlimited length (Makeyev and Bamford 2000a,b; Butcher et al. 2001).
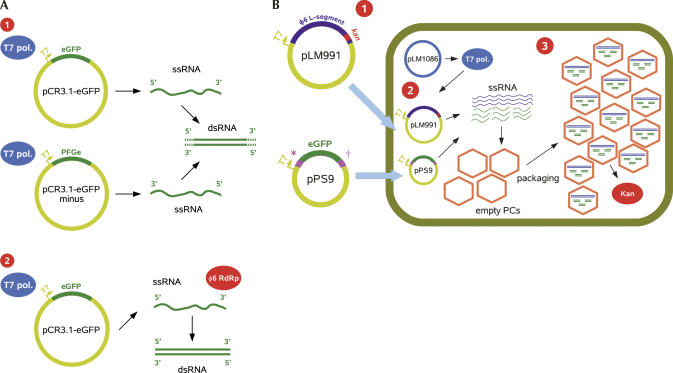
FIGURE 1.
Production methods for long dsRNA. (A) In vitro dsRNA production systems. ➊ Existing method of generating dsRNA by ssRNA hybridization. cDNA of the desired gene, in this case egfp, was cloned in both orientations into a suitable vector under a T7 promoter. T7 RNA polymerase was used to synthesize both plus- and minus-stranded ssRNAs, which were annealed to yield dsRNA (dotted ends symbolize nonspecific annealing). ➋ New method of generating dsRNA by bacteriophage ϕ6 RdRP. cDNA of the desired gene was cloned into a suitable vector under a T7 promoter and ssRNA was synthesized by T7 RNA polymerase or the viral ssRNA was used directly. The RdRP of ϕ6 efficiently synthesizes an exact complementary minus strand beginning from the very 3′ end of the plus strand. (B) New in vivo dsRNA production system utilizing bacteriophage ϕ6. The diagram depicts the formation of a stable carrier state relationship between ϕ6 and P. syringae Cit7 (pLM1086) host cells. ➊ Plasmids containing cDNA of the ϕ6 Lkan segment (pLM991) and the SeGFP segment (egfp flanked by ϕ6 s-segment 5′-packaging (*) and 3′-replication (†) signals [pPS9]), placed under a T7 promoter, were electroporated into the host cells and maintained by kanamycin selection. ➋ The cells contain a plasmid (pLM1086) that constitutively expresses T7 RNA polymerase, transiently synthesizing ssRNA from the cDNA plasmids, which are nonreplicative in P. syringae. The ϕ6 Lkan segment contains the viral RdRP and other genes necessary for the formation of empty polymerase complexes (PCs), and a kanamycin resistance (kan) gene. Packaging begins with SeGFP ssRNA (the s-segment specific ssRNA is packaged first; Gottlieb et al. 1992), followed by l-segment-specific ssRNA. ➌ Upon packaging, an exact complementary strand is synthesized inside the PC particle by the viral RdRP. Packaged capsids contain on average three copies of the SeGFP segment and one copy of the ϕ6 Lkan segment.
The genome of bacteriophage ϕ6 consists of three segments (S 2948 base pairs [bp], M 4063 bp, and L 6374 bp). In each segment, the genes are clustered in functional groups flanked by 5′-packaging and 3′-replication signals. The L-segment encodes the components of the polymerase complex (Poranen and Bamford 1999). During the ϕ6 infection cycle, the empty polymerase complex (the procapsid, PC) packages the single-stranded genomic precursor molecules and converts them into dsRNA inside the procapsid. The cytoplasm is never exposed to dsRNA. The polymerase complexes transcribe mRNAs from dsRNA and export them to the cytoplasm for protein synthesis (Sun et al. 2004).
Recombinant ϕ6 RdRP is an excellent tool for small- to medium-scale in vitro dsRNA synthesis. To meet the need for large-scale dsRNA production, we utilized carrier state bacterial cells containing the ϕ6 polymerase complex (Poranen and Tuma 2004). Insertion of a reporter gene into a genomic segment of ϕ6 yields viable viruses that establish a carrier state relationship with the host cells (Onodera et al. 1992; Sun et al. 2004). An in vivo large-scale dsRNA production system was set up in nonpathogenic Pseudomonas syringae Cit7 cells (Wilson et al. 1999) containing a plasmid (pLM1086; Sun et al. 2004) constitutively expressing T7 RNA polymerase. In these cells, ϕ6-specific ssRNA and procapsid proteins are produced simultaneously in vivo by transfection with ColEI-based suicide plasmids containing cDNA copies of viral genome segments under control of the T7 promoter (Fig. 1B). The system is stable at the RNA level. Selection of stable carrier state cells is achieved by inserting a kanamycin resistance (kan) gene in the 3′ untranslated region of the L segment. Plasmids containing a T7 promoter and any heterologous cDNA, flanked by the viral packaging and replication signals, are transcribed and packaged into viral PC particles, followed by synthesis of the complementary RNA strand.
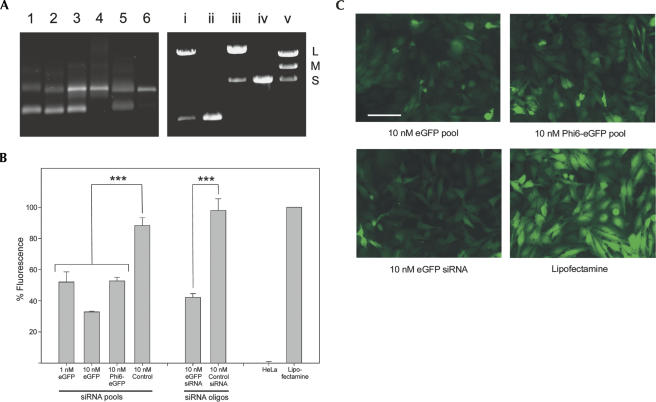
As proof of principle, we created siRNA pools of enhanced green fluorescent protein (eGFP) using both the in vitro and in vivo systems. In the in vitro approach, we used T7 RNA polymerase to synthesize eGFP-specific ssRNA from pCR3.1-eGFP. The ssRNA served as a template for ϕ6 RdRP, which produced high-quality dsRNA (Fig. 2A). This dsRNA was digested with recombinant Dicer, and the resulting siRNAs were purified by anion-exchange chromatography. After transfection into HeLa cells constitutively expressing eGFP, these siRNAs significantly and specifically decreased GFP fluorescence from ∼24 h after transfection until well beyond the day 3 time point depicted in Figure 2B,C. eGFP down-regulation was evident at 1 nM, but more prominent at 10 nM (Fig. 2B). Increasing the siRNA concentration to 25 nM did not decrease the fluorescence further (Supplemental Fig. 1). The eGFP pool was as effective as the commercial eGFP-siRNA oligonucleotide against a single site. No decrease in fluorescence was detected in cells transfected with a nonspecific control pool, control siRNA oligo, or the transfection reagent (Fig. 2B,C). Chromatographic purification was critical to remove residual long dsRNA that was toxic to the cells (data not shown; Sledz and Williams 2004).
FIGURE 2.
(A, left panel) dsRNA produced by hybridization of complementary ssRNAs or by ϕ6 RdRP. (Lane 1) ssRNA from pCR3.1-eGFP, (lane 2) ssRNA from pCR3.1-eGFPminus, (lane 3) dsRNA generated by hybridization, (lane 4) dsRNA generated by hybridization after LiCl precipitation, (lane 5) dsRNA generated by ϕ6 RdRP from plus-stranded ssRNA, and (lane 6) dsRNA generated by ϕ6 RdRP from plus-stranded ssRNA after LiCl precipitation. (Right panel) Agarose gel analysis of dsRNA produced by the in vivo production system. (Lane i) LiCl precipitated dsRNA from eGFP carrier state cells, (lane ii) LiCl precipitated dsRNA from eGFP carrier state cells after HPLC, (lane iii) LiCl precipitated dsRNA from ϕ6 S-segment carrier state cells, (lane iv) LiCl precipitated dsRNA from ϕ6 S-segment carrier state cells after HPLC, and (lane v) wild-type ϕ6 dsRNA as size markers (L 6374 bp, M 4063 bp, S 2948 bp). (B) The effects of siRNAs on transgene expression. HeLa-eGFP cells were transfected with siRNA pools or siRNA oligonucleotides as indicated and fluorescence levels were analyzed by a VICTOR2 1420 multilabel counter 3 d later. The data (emission at 535 nm) are expressed as a percentage of mock-transfected (Lipofectamine) cells. Background values from medium were similar to those from parental HeLa cells and were subtracted. The averages of five independent experiments are shown ± S.E.M. *** = P < 0.001 as compared to 10 nM respective control values. The exact P values are given in Supplemental Table 2. (C) Microscopic images of typical HeLa-eGFP cultures transfected with indicated siRNAs. Scale bar represents 100 μm.
For the in vivo large-scale method, we cloned the egfp gene into plasmid pLM659 (containing cDNA of the ϕ6 S segment) and replaced all ϕ6-specific cDNA except the packaging and replication regions of the S segment. The resultant plasmid pPS9 encoding SeGFP was electroporated into the host cells together with pLM991 (L segment with kan gene; Sun et al. 2004) followed by kanamycin selection. Several colonies were screened for their dsRNA content. A stable ϕ6-eGFP carrier state cell line containing three copies of the eGFP segment and a single copy of the Lkan segment, as determined by densitometry analysis, was selected for dsRNA production. Packaging of multiple copies of reduced size genome segments in ϕ6 has been discussed previously (Mindich et al. 1995; Qiao et al. 1997; Poranen and Bamford 1999).
Approximately 1.6 mg dsRNA/g wet cells were produced (∼10% of the total RNA synthesized in the cell), both in batch cultures and in a 10 L fermenter, indicating a linear correlation between the dsRNA yield and the cell mass (Supplemental Table 1). Consequently, it is possible to produce kilogram quantities of dsRNA in industrial-scale bioreactors.
The in vivo method also produced high-quality dsRNA for gene silencing. Most ribosomal RNA (rRNA) was removed by lithium chloride (LiCl) precipitation (Fig. 2A). Anion-exchange chromatography removed residual rRNA and separated the two dsRNA segments. Our current technology separates ∼4.0 kb segments of interest from the 7.599 kb Lkan-segment, constituting the upper size limit of the insert. The presence of the dsRNA-encoding eGFP in the SeGFP segment and the RdRP in the Lkan segment were verified by RT-PCR (Supplemental Fig. 2).
To illustrate the activity of dsRNA extracted from ϕ6-eGFP carrier state cells in gene silencing, it was Dicer digested, purified, and transfected into HeLa-eGFP cells. This caused a significant decrease of fluorescence, comparable to the specific eGFP pool and the eGFP siRNA oligonucleotide (Fig. 2B,C). As an additional control, dsRNA of the genuine S segment of control carrier state cells was purified, Dicer digested, and introduced into HeLa-eGFP cells. This pool had no effect on fluorescence (Supplemental Fig. 1).
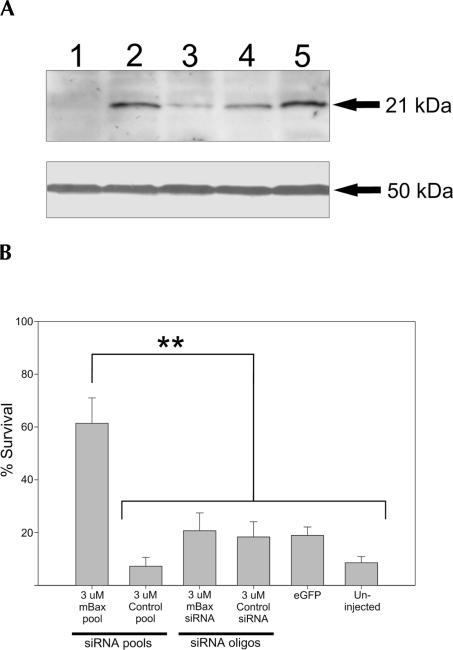
Although eGFP is an excellent silencing target for proof-of-principle experiments, the conditions for silencing an endogenous gene and a transgene may differ. To extend our analysis to endogenous targets, we used our in vitro method to generate a siRNA pool against Bax, which encodes a pro-apoptotic BCL-2 family protein (Deckwerth et al. 1996). The Bax pool transfected at 3 nM concentration strongly suppressed the levels of endogenous BAX protein in neuroblastoma Neuro2A-20 cells, whereas the control pool had no effect (Fig. 3A). At the same concentration, the commercial siRNA oligonucleotide against mouse Bax was less effective than the pool. We then tested the functional consequences of Bax silencing in apoptotic neurons. Cultured neonatal sympathetic neurons die apoptotically when deprived of nerve growth factor (NGF). This critically depends on the activation of BAX, as Bax-deficient neurons are completely resistant to NGF deprivation (Deckwerth et al. 1996). We microinjected our Bax pool into sympathetic neurons, removed NGF from the medium 2 d later, and counted the living injected neurons over a 3-d period after NGF deprivation. The Bax pool efficiently blocked the death of the neurons, whereas the control pool did not (Fig. 3B). The siRNA oligonucleotide was much less protective, most probably due to residual BAX protein expression. Its effect on the neurons did not differ notably from nonspecific controls.
FIGURE 3.
(A) The effect of siRNAs on endogenous BAX protein (21 kDa) expression in Neuro2A-20 cells 3 d after transfection (upper panel), determined by Western blot analysis. (Lane 1) 3 nM mBax pool, (lane 2) 3 nM pLM659 control pool, (lane 3) 3 nM mBax siRNA oligo, (lane 4) 3 nM control siRNA oligo, (lane 5) Lipofectamine. The lanes contained equal amounts of total protein (β-tubulin, 50 kDa; lower panel). (B) The effect of siRNAs on apoptotic neurons. The neurons were microinjected with indicated siRNAs along with a plasmid for eGFP expression and deprived of NGF 2 d later. Fluorescent neurons living 3 d after NGF deprivation are expressed as the percentage of fluorescent neurons counted immediately after NGF deprivation. Neurons injected with eGFP plasmid alone serve as control for microinjection procedure. Shown are the averages of three independent experiments ± S.E.M. ** = P < 0.01. The exact P values are given in Supplemental Table 2.
We have shown here that medium and large quantities of dsRNA of a given sequence may be generated by novel in vitro or in vivo recombinant ϕ6 RdRP-based dsRNA production systems. The in vitro system offers an efficient and flexible method to produce high-quality dsRNA from an ssRNA molecule of practically unlimited length (such as an ssRNA virus gene or genome, even without prior knowledge of the sequence), eliminating the need for error prone hybridization. The in vivo system allows dsRNA to be produced in very large quantities for industrial-scale implementation. This approach requires some 300 bp of ϕ6-specific sequence at both ends of the desired gene. In the HeLa-eGFP cells, however, no disturbance of the RNAi activity was detected (Fig. 2). In fact, it has been proposed (Seyhan et al. 2005) that due to the mechanisms of Dicer activity, ∼200 bp of the nontargeted sequence at the ends of substrate dsRNA would enrich the targeted sequence in the resulting siRNA pool.
To our knowledge, this is the first description of an endogenous gene being successfully silenced in primary neurons by microinjecting siRNAs. The utilization of siRNA pools instead of individual siRNA oligonucleotides demonstrates the practical advantage of our new methodology. Although the Bax siRNA oligonucleotide significantly decreased the target protein level in cell cultures, gene silencing was not complete and consequently failed to protect primary neurons against apoptotic death (Fig. 3). With the Bax siRNA pool, which removed BAX much more efficiently and blocked the death, the laborious optimization of injection conditions due to a seemingly functional siRNA oligonucleotide was avoided.
For gene silencing, dsRNA must be Dicer digested for applications in mammalian systems, or it may be utilized unprocessed in invertebrates, plants, and fungi (Dykxhoorn et al. 2003). However, dsRNA and siRNAs have been shown to elicit various undesired cellular responses, including the activation of interferon-mediated pathways and off-targeting (Bridge et al. 2003; Jackson et al. 2003; Sledz and Williams 2004). The significance of these responses falls beyond the scope of this study. Several groups have elucidated the causes and consequences of the various adverse effects induced by different dsRNAs (Jackson and Linsley 2004; Cullen 2006). Some of these effects have been shown to be at least in part dependent on the cell type under investigation (Reynolds et al. 2006). As an example of a siRNA pool approach, it was recently reported that the efficient silencing of a target gene is feasible at least in certain mammalian cell lines by expressing long dsRNA intracellularly (Strat et al. 2006). An interferon response was not detected in this study, and the extent of off-targeting was modest.
The pools tested here were efficient at low concentrations, in particular the Bax pool, which was much more potent in silencing its target than the single siRNA oligonucleotide. Lowering siRNA concentrations has been shown to decrease off-targeting significantly (Jackson et al. 2003; Semizarov et al. 2003). Moreover, pooling several individual siRNA sequences against a single target has been proposed as an elementary control for silencing experiments (Jackson and Linsley 2004; Huppi et al. 2005). Since every siRNA has its own off-targets, pooling is thought to reduce the individual nonspecific contributions of each sequence, revealing the “true” RNAi phenotypes.
We envision RNA virus infections as the main therapeutic targets for RNAi. The novel methods described here provide an example of how large amounts of dsRNA required for efficient therapy could be synthesized cost effectively. When mucosal membranes are the route of infection, as is the case with influenza, HIV, and the common cold, delivery of siRNAs is feasible. The use of long target sites would reduce the generation of escape mutants, which are often detected when using single siRNAs (Westerhout et al. 2005; Wilson and Richardson 2005). Furthermore, interferon induction due to the controlled presence of long dsRNA molecules in combination with siRNAs might constitute a future direction of research.
MATERIALS AND METHODS
Cell lines
The ϕ6-based dsRNA production system was set up in the nonpathogenic Pseudomonas syringae Cit7 strain (a generous gift from Dr. Steven Lindow, Department of Plant and Microbial Biology, University of California, Berkeley, CA). Plasmid pLM1086, a derivative of the broad host range vector pRK290 that constitutively expresses T7 RNA polymerase, was isolated from P. syringae LM2691 (kindly provided by Professor Leonard Mindich, Department of Microbiology, The Public Health Research Institute, Newark, NJ). pLM1086 was electroporated (Sun et al. 2004) into P. syringae Cit7, yielding the host strain P. syringae Cit7 (pLM1086). The HeLa-eGFP cell line was generated by Dr. Yun-Fu Sun (Institute of Biotechnology, Helsinki, Finland). This HeLa cell line (ATCC) contains the pEGFP-C1 plasmid (Clontech Laboratories) and expresses egfp constitutively. Mouse neuroblastoma Neuro2A-20 cells are derived from Neuro2A cells and express the GFRα1 receptor (Lindahl et al. 2001). The cells were cultured in Dulbecco's modified Eagle's Medium (Sigma-Aldrich) or RPMI 1640 (ICN Biomedicals) containing 10% fetal bovine serum (HyClone Perbio Sciences) and 1% penicillin-streptomycin (GIBCO BRL, Invitrogen). G418-sulfate (Calbiochem) was also added to the culture medium (400 μg/mL for HeLa-eGFPs and 200 μg/mL for Neuro2A-20s).
Plasmids
pCR3.1-eGFP was constructed for the production of single-stranded egfp-RNA and contains an NheI/KpnI fragment of pEGFP-C1 inserted into pCR3.1 (Invitrogen). pCR3.1-eGFP minus has an NheI/KpnI fragment of pEGFP-C1 in the KpnI/XbaI sites of pCR3.1. For mBax-RNA production, a cDNA copy of Bax was cloned into pBluescript (Stratagene). pLM659 and pLM991 were generous gifts from Professor Leonard Mindich. pLM659 (Gottlieb et al. 1992) contains a cDNA copy of the 2948-bp S segment of the bacteriophage ϕ6 in pT7T319U. pLM991 (Sun et al. 2004) contains a 7599-bp cDNA copy of the L segment of ϕ6, including a kanamycin resistance gene insertion at the 3′ end. Plasmid pPS9 was derived from pLM659 and contains the egfp gene flanked by 5′ and 3′ regulatory sequences from the ϕ6S segment. pPS9 was constructed by amplifying the egfp gene by PCR from the pCR3.1-eGFP plasmid using primers 5′-CGGCCGGCCGAGCTCGAATTCGGTACCATATGTTTAAACATGGTGAGCAAGGGCGAGGAGCTG-3′ and 5′-CGGCCTGCAGATCTTAAGTTTAAACTTACTTGTACAGCTCGTCCATGCCGAGAGTG-3′ that contain restriction sites for EagI and PstI. The eGFP insert and pLM659 subsequently were digested with EagI and PstI and ligated together, yielding pPS9.
In vivo dsRNA production
Cultures of P. syringae Cit7 (pLM1086) cell transfected with pPS9 or pLM659 were incubated at 28°C with continuous stirring for 20 h (batch culture) or 24 h (fermentor). The cultures were chilled on ice for 15 min and the cells were pelleted at 8,000g for 8 min at 4°C. The pellet was suspended in 10 mL cold 18 Ω milli-Q water (Millipore)/g wet cells and the suspension was sonicated at 75% efficiency, cycle 0.5, for 1 min. The suspension was aliquotted into smaller batches, and an equal volume of TRIzol (Invitrogen) and one-fifth volume of chloroform were added. The mixture was centrifuged at 15,000g for 15 min at 4°C. The aqueous upper phase was recovered, one-half volume of isopropanol was added, and the mixture was consequently centrifuged at 15,000g for 10 min at 4°C. The pellet was washed once with 70% ethanol and resuspended in RNase-free 18 Ω milli-Q water. LiCl was added to a final concentration of 2 M to precipitate ssRNA. The samples were centrifuged at 15,000g for 30 min at 4°C. The supernatant was collected, LiCl was added to a final concentration of 4 M, after which the samples were centrifuged as above. The pellet was dissolved in RNase-free 18 Ω milli-Q water and precipitated again by adding 2.5 volumes of 96% ethanol. After incubation at −20°C for 30 min, dsRNA was recovered by centrifugation at 15,000g for 20 min at 4°C. The purified dsRNA pellet was dissolved in RNase-free 18 Ω milli-Q water and the concentration was determined by measuring absorption at 260 nm. The dsRNA segments were separated by anion-exchange high-performance liquid chromatography (HPLC) using a Waters Gen-Pak FAX column (Waters). dsRNA (90 μg) was injected onto the column in 25 mM Tris-HCl (pH 8.0), 1 mM EDTA. A linear gradient from 0 M NaCl to 1 M NaCl was applied (0.25 mL/min, 180 min), and fractions of 250 μL were collected. The segments eluted as separate peaks.
Reverse transcriptase polymerase chain reaction
Purified dsRNA originating from cells transfected with plasmids pPS9 or pLM659 was denatured and annealed to the appropriate forward primer. cDNA synthesis was performed using the Moloney Murine Leukemia Virus reverse transcriptase (Promega) according to the manufacturer's protocol. Residual RNA was removed by RNase H (Promega) and RNase III (Promega) treatment (37°C, 20 min). The reaction mixture was purified and the cDNA was amplified by standard PCR (Sambrook and Russell 2001) using a set of forward and reverse primers specific for certain genes or regulatory sequences. The reaction products were analyzed by electrophoresis in 0.8% agarose gels in TBE buffer (Sambrook and Russell 2001).
Densitometry analysis
In vivo produced dsRNA and wild-type ϕ6 dsRNA were subjected to agarose gel electrophoresis. Following electrophoresis, the digital image of the gel was captured using a CCD camera (Uvitech Gel Documentation and Analysis System). Image analysis (densitometry) was performed using TINA software (Raytest Isotopenmeßgeräte GmbH), comparing the optical density/base pair of the segment of interest to that of the L segment.
In vitro dsRNA production
Linearized and purified pCR3.1-eGFP, pLM659, and pBluescript-mBax were used as templates for T7 RNA polymerase. The resulting ssRNAs were treated with DNaseI (Promega), purified by chloroform extraction and LiCl-ethanol precipitation, and used as templates for ϕ6 polymerase. The ϕ6 RdRP reaction was performed essentially as described (Yang et al. 2001). Briefly, the reaction mixture contained 50 mM HEPES-KOH (pH 7.8), 20 mM ammonium acetate, 6% (w/v) polyethylene glycol 4K, 5 mM MgCl2, 2 mM MnCl2, 0.1 mM EDTA, 0.1% Triton X-100, 1 mM of each NTP, 1 U/μL RNAsin (Promega), and 2 U ϕ6 RdRp/μg ssRNA. The dsRNA was separated from ssRNA by sequential 2 M and 4 M LiCl-ethanol precipitation.
siRNAs
dsRNA molecules produced either in vitro or in vivo were digested with recombinant Dicer (Stratagene) for 18 h at 37°C. Subsequently, the reaction mixture was desalted using a NAP-5 or NAP-10 column (Amersham Biosciences, GE Healthcare), injected onto Waters Gen-Pak FAX, and eluted as above, but using a 120-min linear NaCl gradient. siRNA molecules eluted at ∼400 mM NaCl as a separate peak from undigested dsRNA. The siRNA fractions were combined, desalted as described above, and concentrated using a SpeedVac concentrator (Thermo Savant). The siRNA concentration was determined visually by agarose gel analysis in 4% NuSieve GTG Agarose (Cambrex) by comparing band intensity to known standards. The eGFP siRNA oligonucleotide with target sequence 5′-GCGACGTAAACGGCCACAAGTTC-3′ (Dharmacon accession number U76561) and nonspecific control siRNA oligonucleotides with target sequences 5′-CAAUCACCGACCACCGGAC-3′ and 5′-CGTACGCGGAATACTTCGA-3′ (5′-Cy3 labeled sense strand) were purchased from Dharmacon. The siRNA oligonucleotide against mBax (sc-29213) and a nonspecific control siRNA (sc-37007) were purchased from Santa Cruz Biotechnology.
Transfections
HeLa-eGFP, HeLa, or Neuro2A-20 cells were plated on 24-well plates (BD Biosciences) or 35-mm dishes (BD Biosciences), some containing 13-mm coverslips (Menzel Gläser), and transfected on the following day with Lipofectamine2000 (Invitrogen) essentially according to the manufacturer's instructions. G418-sulfate was omitted from the medium at this point. For HeLa-eGFP cells, transfection efficiency was ∼90%, as assessed by fluorescence microscopy. To prevent cell overgrowth, the concentration of FBS was lowered to 4% at 24 h after transfection. For fluorescence measurements, phenol red also was omitted from the culture medium.
Quantification of eGFP expression
The fluorescence levels of transfected cells on 24-well plates were measured daily after transfection using a VICTOR2 1420 multilabel counter (Wallac, PerkinElmer Life and Analytical Sciences) for 1 sec at 490 nm excitation and 535 nm emission (Cormack et al. 1996). The experiment was repeated independently five times (see below for statistical analysis). The cells growing on coverslips were fixed 3 d after transfection by incubating 30 min in 4% paraformaldehyde, washed with phosphate buffered saline (PBS) and mQ, and mounted on object glasses with Mowiol (Calbiochem). The samples were analyzed microscopically with an Olympus AX70 microscope using a 20× objective, and images were captured with an F-View camera and AnalySIS software (Soft Imaging System). For illustration, the images were similarly pseudocolored and levels linearly adjusted using Adobe Photoshop.
Western blotting
Neuro 2A-20 cells were lysed in 20 mM Tris-HCl (pH 7.5), 150 mM NaCl, 5 mM EDTA, 1% Nonidet P-40, 1% Triton X-100 3 d after transfection. The proteins were separated by 15% SDS-PAGE and transferred to a nitrocellulose filter. They were probed with monoclonal anti-BAX 6A7 antibodies (BD Pharmingen) and with polyclonal neuronal class III B-tubulin PRB-435P antibodies (Nordic BioSite) for loading control.
Neuronal culture and microinjections
Neonatal mouse sympathetic neurons were cultured and microinjected essentially as described (Sun et al. 2001). Briefly, the neurons were cultured in Neurobasal medium (Invitrogen) with B27 supplement (Invitrogen) and 30 ng/mL of 2.5S NGF (Promega) for 5–6 d. Then 3 μM of the siRNA pools or commercial siRNA oligos (Santa Cruz) were microinjected into the nuclei together with pEGFP-C1 plasmid. The cultures were maintained with NGF for 2 d to allow degradation of preexisting BAX protein. Apoptosis was then triggered by washing with NGF-free medium and adding function-blocking anti-NGF MAB5260Z antibodies (Chemicon). The living injected neurons, identified by coinjected eGFP, were counted 3 d later and expressed as a percentage of initial injected neurons counted immediately after NGF deprivation. The experiment was repeated independently three times (see below for statistical analysis).
Statistical analyses
To analyze the fluorescence levels of transfected HeLa-eGFP cells (n = 5) and the survival of microinjected neurons (n = 3), the means of independent experiments were compared with one-way ANOVA followed by a post hoc Tukey test using the SPSS program. The exact P values are given in Supplemental Table 2.
SUPPLEMENTAL DATA
Supplemental Data files can be found at http://blogit.helsinki.fi/bamford/supplements.htm.
ACKNOWLEDGMENTS
The authors thank Riitta Tarkiainen, Congjun Zheng, Petri Papponen, and Sampo Vehma for their excellent technical assistance. This work was supported by the Academy of Finland Finnish Centre of Excellence Programme 2006-2011 (grants 1213467, 1213992 to D.H.B.), the Sigrid Jusélius Foundation, Biocentrum Helsinki, and Academy of Finland Systems Biology Programme grant 1105237 (to M.S.).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.348307.
REFERENCES
- Amarzguioui, M., Rossi, J.J., Kim, D. Approaches for chemically synthesized siRNA and vector-mediated RNAi. FEBS Lett. 2005;579:5974–5981. doi: 10.1016/j.febslet.2005.08.070. [DOI] [PubMed] [Google Scholar]
- Bridge, A.J., Pebernard, S., Ducraux, A., Nicoulaz, A.-L., Iggo, R. Induction of an interferon response by RNAi vectors in mammalian cells. Nat. Genet. 2003;34:263–264. doi: 10.1038/ng1173. [DOI] [PubMed] [Google Scholar]
- Butcher, S.J., Grimes, J.M., Makeyev, E.V., Bamford, D.H., Stuart, D.I. A mechanism for initiating RNA-dependent RNA polymerization. Nature. 2001;410:235–240. doi: 10.1038/35065653. [DOI] [PubMed] [Google Scholar]
- Cormack, B.P., Valdivia, R.H., Falkow, S. FACS-optimized mutants of the green fluorescent protein (GFP) Gene. 1996;173:33–38. doi: 10.1016/0378-1119(95)00685-0. [DOI] [PubMed] [Google Scholar]
- Cullen, B.R. Enhancing and confirming the specificity of RNAi experiments. Nat. Methods. 2006;3:677–681. doi: 10.1038/nmeth913. [DOI] [PubMed] [Google Scholar]
- Deckwerth, T.L., Elliot, J.L., Knudson, C.M., Johnson E.M., Jr, Snider, W.D., Korsmeyer, S.J. BAX is required for neuronal death after trophic factor deprivation and during development. Neuron. 1996;17:401–411. doi: 10.1016/s0896-6273(00)80173-7. [DOI] [PubMed] [Google Scholar]
- Dykxhoorn, D.M., Novina, C.D., Sharp, P.A. Killing the messenger: Short RNAs that silence gene expression. Nat. Rev. Mol. Cell Biol. 2003;4:457–467. doi: 10.1038/nrm1129. [DOI] [PubMed] [Google Scholar]
- Gottlieb, P., Strassman, J., Qiao, X., Frilander, M., Frucht, A., Mindich, L. In vitro packaging and replication of individual genomic segments of bacteriophage ϕ6 RNA. J. Virol. 1992;66:2611–2616. doi: 10.1128/jvi.66.5.2611-2616.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heale, B.S.E., Soifer, H.S., Bowers, C., Rossi, J.J. siRNA target site secondary structure predictions using local stable substructures. Nucleic Acids Res. 2005;33:e30. doi: 10.1093/nar/gni026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huppi, K., Martin, S.E., Caplen, N.J. Defining and assaying RNAi in mammalian cells. Mol. Cell. 2005;17:1–10. doi: 10.1016/j.molcel.2004.12.017. [DOI] [PubMed] [Google Scholar]
- Jackson, A.L., Linsley, P.S. Noise amidst the silence: Off-target effects of siRNAs? Trends Genet. 2004;20:521–524. doi: 10.1016/j.tig.2004.08.006. [DOI] [PubMed] [Google Scholar]
- Jackson, A.L., Bartz, S.R., Schelter, J., Kobayashi, S.V., Burchard, J., Mao, M., Li, B., Cavet, G., Linsley, P.S. Expression profiling reveals off-target gene regulation by RNAi. Nat. Biotechnol. 2003;21:635–637. doi: 10.1038/nbt831. [DOI] [PubMed] [Google Scholar]
- Lindahl, M., Poteryaev, D., Yu, L., Arumäe, U., Timmusk, T., Bongarzone, I., Aiello, A., Pierotti, M.A., Airaksinen, M.S., Saarma, M. Human glial cell line-derived neurotrophic factor receptor α4 is the receptor for persephin and is predominantly expressed in normal and malignant thyroid medullary cells. J. Biol. Chem. 2001;276:9344–9351. doi: 10.1074/jbc.M008279200. [DOI] [PubMed] [Google Scholar]
- Makeyev, E.V., Bamford, D.H. Replicase activity of purified recombinant protein P2 of double-stranded RNA bacteriophage ϕ6. EMBO J. 2000a;19:124–133. doi: 10.1093/emboj/19.1.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makeyev, E.V., Bamford, D.H. The polymerase subunit of a dsRNA virus plays a central role in the regulation of viral RNA metabolism. EMBO J. 2000b;19:6275–6284. doi: 10.1093/emboj/19.22.6275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister, G., Tuschl, T. Mechanisms of gene silencing by double-stranded RNA. Nature. 2004;431:343–349. doi: 10.1038/nature02873. [DOI] [PubMed] [Google Scholar]
- Mindich, L., Qiao, X., Qiao, J. Packaging of multiple copies of reduced-size genomic segments by bacteriophage ϕ6. Virology. 1995;212:213–217. doi: 10.1006/viro.1995.1470. [DOI] [PubMed] [Google Scholar]
- Myers, J.W., Jones, J.T., Meyer, T., Ferrell J.E., Jr Recombinant Dicer efficiently converts large dsRNAs into siRNAs suitable for gene silencing. Nat. Biotechnol. 2003;21:324–328. doi: 10.1038/nbt792. [DOI] [PubMed] [Google Scholar]
- Onodera, S., Olkkonen, V.M., Gottlieb, P., Strassman, J., Qiao, X., Bamford, D.H., Mindich, L. Construction of a transducing virus from double-stranded RNA bacteriophage ϕ6: Establishment of carrier states in host cells. J. Virol. 1992;66:190–196. doi: 10.1128/jvi.66.1.190-196.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poranen, M.M., Bamford, D.H. Packaging and replication regulation revealed by chimeric genome segments of double-stranded RNA bacteriophage ϕ6. RNA. 1999;5:446–454. doi: 10.1017/s1355838299981876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poranen, M.M., Tuma, R. Self assembly of double-stranded RNA bacteriophages. Virus Res. 2004;101:93–100. doi: 10.1016/j.virusres.2003.12.009. [DOI] [PubMed] [Google Scholar]
- Qiao, X., Qiao, J., Mindich, L. Stoichiometric packaging of the three genomic segments of double-stranded RNA bacteriophage ϕ6. Proc. Natl. Acad. Sci. 1997;94:4074–4079. doi: 10.1073/pnas.94.8.4074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds, A., Leake, D., Boese, Q., Scaringe, S., Marshall, W.S., Khvorova, A. Rational siRNA design for RNA interference. Nat. Biotechnol. 2004;22:326–330. doi: 10.1038/nbt936. [DOI] [PubMed] [Google Scholar]
- Reynolds, A., Anderson, E.M., Vermeulen, A., Fedorov, Y., Robinson, K., Leake, D., Karpilow, J., Marshall, W.S., Khvorova, A. Induction of the interferon response by siRNA is cell type- and duplex length-dependent. RNA. 2006;12:988–993. doi: 10.1261/rna.2340906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J., Russell, D.W. Molecular cloning: A laboratory manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, New York: 2001. [Google Scholar]
- Semizarov, D., Frost, L., Sarthy, A., Kroeger, P., Halbert, D.N., Fesik, S.W. Specificity of short interfering RNA determined through gene expression signatures. Proc. Natl. Acad. Sci. 2003;100:6347–6352. doi: 10.1073/pnas.1131959100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seyhan, A.A., Vlassov, A.V., Ilves, H., Egry, L., Kaspar, R.L., Kazakov, S.A., Johnston, B.H. Complete, gene-specific siRNA libraries: Production and expression in mammalian cells. RNA. 2005;11:837–846. doi: 10.1261/rna.7285805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sledz, C.A., Williams, B.R.G. RNA interference and double-stranded-RNA-activated pathways. Biochem. Soc. Trans. 2004;32:952–956. doi: 10.1042/BST0320952. [DOI] [PubMed] [Google Scholar]
- Strat, A., Gao, L., Utsuki, T., Cheng, B., Nuthalapaty, S., Mathis, J.M., Odaka, Y., Giordano, T. Specific and nontoxic silencing in mammalian cells with expressed long dsRNAs. Nucleic Acids Res. 2006;34:3803–3810. doi: 10.1093/nar/gkl532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, Y.F., Yu, L.Y., Saarma, M., Timmusk, T., Arumäe, U. Neuron-specific Bcl-2 homology 3 domain-only splice variant of Bak is anti-apoptotic in neurons, but pro-apoptotic in non-neuronal cells. J. Biol. Chem. 2001;276:16240–16247. doi: 10.1074/jbc.M010419200. [DOI] [PubMed] [Google Scholar]
- Sun, Y., Qiao, X., Mindich, L. Construction of carrier state viruses with partial genomes of the segmented dsRNA bacteriophages. Virology. 2004;319:274–279. doi: 10.1016/j.virol.2003.10.022. [DOI] [PubMed] [Google Scholar]
- Uprichard, S.L. The therapeutic potential of RNA interference. FEBS Lett. 2005;579:5996–6007. doi: 10.1016/j.febslet.2005.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rij, R.P., Andino, R. The silent treatment: RNAi as a defense against virus infection in mammals. Trends Biotechnol. 2006;24:186–193. doi: 10.1016/j.tibtech.2006.02.006. [DOI] [PubMed] [Google Scholar]
- Westerhout, E.M., Ooms, M., Vink, M., Das, A.T., Berkhout, B. HIV-1 can escape from RNA interference by evolving an alternative structure in its RNA genome. Nucleic Acids Res. 2005;33:796–804. doi: 10.1093/nar/gki220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, J.A., Richardson, C.D. Hepatitis C virus replicons escape RNA interference induced by a short interfering RNA directed against the NS5b coding region. J. Virol. 2005;79:7050–7058. doi: 10.1128/JVI.79.11.7050-7058.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, M., Hirano, S.S., Lindow, S.E. Location and survival of leaf-associated bacteria in relation to pathogenicity and potential for growth within the leaf. Appl. Environ. Microbiol. 1999;65:1435–1443. doi: 10.1128/aem.65.4.1435-1443.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, H., Makeyev, E.V., Bamford, D.H. Comparison of polymerase subunits from double-stranded RNA bacteriophages. J. Virol. 2001;75:11088–11095. doi: 10.1128/JVI.75.22.11088-11095.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]