Abstract
Using cell lines and primary cells, it has been shown that translation control plays a key role regulating gene expression during physiological and pathological conditions. The relevance of this type of regulation in vivo (tissues, organs) remains to be elucidated, due to the lack of an efficient method for polysome-bound fractionation of solid tissue RNA samples. A simple and efficient method is described, in which tissue samples were pulverized in liquid nitrogen and lysed with NP40-lysis buffer in the presence of the RNAse inhibitors RNAsin and vanadyl-ribonucleoside complex. After cell lysis, the cytoplasmic extract was loaded into sucrose gradients, fractionated, and RNA prepared from each fraction. The obtained RNA was reverse transcribed with a low efficiency, a problem that was overcome by purifying polyA+ RNA. Aiming to use small quantities of solid tissue samples (10–20 mg/sample), polyA+ RNA purification was discarded, and the different components were individually screened for a negative effect on reverse transcription. The polysaccharide heparin, which is present as a nonspecific RNAse inhibitor, inhibits reverse transcriptase activity, and must be removed from RNA samples for an efficient reaction. Heparin was successfully removed by precipitation of the RNA with lithium chloride, as demonstrated by the reversal of the inhibition on RT-PCR reactions. In summary, we present a reliable method allowing us to prepare high-quality polysome-bound mRNA from small quantities of liquid-nitrogen–frozen solid tissue samples from both human and mouse origin, amenable for Northern blotting, RT-PCR reactions, and expression profiling analyses.
Keywords: polysome fractionation, tissue samples, sucrose gradients, RT-PCR, expression profiling
INTRODUCTION
Redistribution of mRNA between ribosome-free and polysome-bound fractions has been used to ascertain translational control of a given mRNA by Northern hybridization (Leibold and Munro 1988; Garcia-Sanz and Lenig 1996), and more recently in expression profiling analysis (Johannes et al. 1999; Zong et al. 1999; Mikulits et al. 2000). This can be done since translational control takes place, mainly at the initiation step (Mathews et al. 2000), and thus ribosome loading of a transcript is a robust indicator for translation efficiency. Indeed, genome-wide analyses of translationally regulated transcripts showed that 15%–40% of the regulated transcripts during physiological/pathological transitions are regulated at the translational level, indicating that translational control is a generalized mechanism of gene expression regulation (Pradet-Balade et al. 2001a). Examples of the major role of translational control stem from studies on T cell activation (Mikulits et al. 2000), epithelial-mesenchymal transition (Jechlinger et al. 2003), terminal erythroid differentiation (Blazquez-Domingo et al. 2005), and also from drug treatment (Grolleau et al. 2002), PDK1 signaling (Tominaga et al. 2005), or the early effects of the transcription factor Ras, in combination with AKT, which involves the differential recruitment of specific mRNAs to polysomes (Rajasekhar et al. 2003).
These data, which indicate the prominent role of translational control regulating gene expression, together with the lack of correlation between protein levels and levels of the corresponding mRNAs in expression profiling analyses (Tew et al. 1996; Gygi et al. 1999a,b), led us to propose that polysome-bound mRNA profiling more faithfully would represent cell phenotypes than the common profiling using total RNA, since polysome-bound profiling would allow us to identify, in addition to changes in transcription or mRNA stability (also detected by total mRNA expression profiling), changes in splicing efficiency, nucleo-cytoplasmic transport, and translation efficiency (Pradet-Balade et al. 2001a). This hypothesis, initially strengthened by the demonstration that polysome-bound profiling can detect changes observed in the proteome but undetectable by total RNA expression profiling (Grolleau et al. 2002), was subsequently confirmed by a large set of publications in which polysome-bound mRNA profiling allowed identifying translationally regulated genes. Indeed, this has been demonstrated in model systems as diverse as the response to Ras and AKT signaling (Rajasekhar et al. 2003), dehydration of Arabidopsis (Kawaguchi et al. 2004), hypoxia (Wouters et al. 2005), mating pheromone in yeast (MacKay et al. 2004), regulation of retinoic acid receptor in polymorphonuclear cells (Yost et al. 2004), inflammatory signals in neutrophils (Lindemann et al. 2004), analysis of breast cancer cell lines (Huber et al. 2004), osteosarcoma (Ju et al. 2003), erythroid differentiation (Joosten et al. 2004), or epithelial mesenchymal transition (Jung et al. 1997), which by now are represented by >50 publications.
Initially, the identification of translationally regulated genes was done by determining changes in the ratio between ribosome-free/polysome-bound signals for each gene (Johannes et al. 1999; Zong et al. 1999; Mikulits et al. 2000). The actual demonstration that the sum of signals from ribosome-free and polysome-bound fractions equals the signal from the total RNA for a the vast majority of genes (Pradet-Balade et al. 2001b) allowed us to ascertain that changes in translation efficiency correspond to changes in the ratio polysome-bound/total RNA, and thus to distinguish between changes in transcription/mRNA stability versus changes in translation control, splicing, or nucleo-cytoplasmic export.
Thus, once the relevance of translational control was established in tissue-cultured cells and primary cells, it became highly interesting to establish a methodology that would also allow reliable preparations of ribosome-free and polysome-bound mRNA from small solid tissue samples, amenable for RT-PCR, Northern blotting, and expression profiling. Here we describe such a protocol for isolation of polysome-bound mRNA from solid tissues that has been applied to different mouse and human samples (kidney, brain, mammary gland) to obtain high-quality RNA, suitable for polysome-bound profiling experiments. Furthermore, we tested the effects of the RNAse inhibitor vanadyl-ribonucleoside complex (VRC) on the quality of the data and describe an efficient method to avoid the inhibition of the reverse transcriptase due to the heparin used in polysome gradient fractionations as an unspecific RNAse inhibitor that allows us to directly use total RNA (instead of polyA+ RNA) in polysome-bound expression profiling experiments.
RESULTS
Discrimination between actively translated and translationally silent mRNAs in the cell can be efficiently carried out using sucrose-gradient fractionation (polysome gradients), since this technique allows separation of free ribonucleoprotein particles (ribosome-free mRNA) from mRNAs bound to an increasing number of ribosomes (polysome-bound mRNA). Fractionation of cytoplasmic cell extracts in sucrose gradients has been restricted, for technical reasons, to cultured cell lines and primary lymphoid cells. Wishing to extend this type of analysis to solid tissues, we have modified the methodology used so far to allow an efficient preparation, after sucrose-gradient fractionation, of high-quality RNA using relatively small tissue samples (in the range of 10–20 mg), amenable for Northern blot, RT-PCR, and expression profiling analyses.
Polysome gradient fractionation from solid tissues
A series of changes had to be made to the method currently used to prepare sucrose gradients from cell lines to make it amenable for solid tissue samples, namely: (1) quick freezing of the sample in liquid nitrogen; (2) pulverization of the sample under liquid nitrogen; and (3) addition of VRC to the lysis buffer as a strong RNAse inhibitor, as RNAsin alone was not sufficiently effective in blocking RNAse activity in the samples. Since it was considered that this type of analysis might also be used for relatively small samples (i.e., biopsies), small amounts of tissue (in the range of 10–20 mg) were used.
Tissue samples of interest were transferred to siliconized microcentrifuge tubes and immediately frozen under liquid nitrogen. These samples were stored under liquid nitrogen or at −80°C until processing. The RNA extraction procedure begins with pulverization of the tissue under liquid nitrogen for which a mortar and a pestle, both RNAse free and previously cooled with liquid nitrogen, are used. The powder obtained is transferred to a microcentrifuge tube maintained for at least 5 min on dry ice, to minimize RNA degradation by avoiding thawing of the pulverized tissue. Afterward, the tissue powder is resuspended in lysis buffer (NP40 based) and the cells disrupted as quickly as possible by pipetting up and down 10 times, maintaining a ratio of tissue sample to buffer of 1 mL lysis buffer/10 mg tissue (Fig. 1). After lysis, intact nuclei are pelleted at 4°C, for 10 sec at 12,000g in a microcentrifuge. The supernatant containing the cytoplasmic extract is transferred to a fresh microcentrifuge tube containing 2× extraction buffer (see Materials and Methods), under conditions where ribosomes bound to mRNAs remain “frozen” there by the presence of cycloheximide. Cellular debris, including mitochondria, are removed by subsequent centrifugation at 12,000g for 5 min, and the supernatant is loaded onto a 15%–40% sucrose gradient (Müllner and Garcia-Sanz 1997b). During centrifugation, mRNAs distribute into the different fractions of the gradient, based on their ribosome load (Fig. 1; Müllner and Garcia-Sanz 1997b).
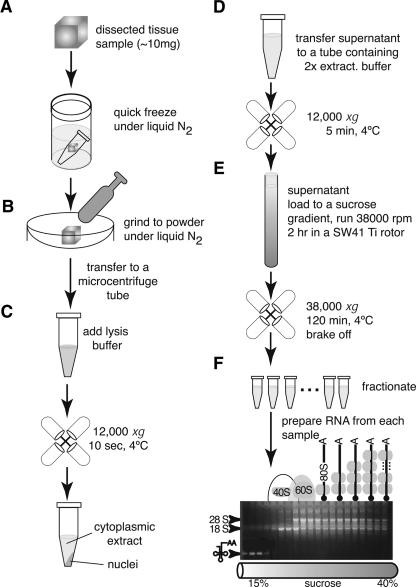
FIGURE 1.
Schematic representation of the protocol allowing isolation of polysome-bound mRNAs from small solid tissue samples. Dissected tissue samples (10–20 mg) are quickly frozen under liquid nitrogen (A). Frozen samples are crushed to powder in a mortar under liquid nitrogen (B). The powder is transferred to a microcentrifuge tube and lysed with NP40-lysis buffer in the presence of VRC (C). After nuclei removal, ribosomes are immobilized on mRNAs with cycloheximide (present in the 2× extraction buffer) (D); the cytoplasmic extract is loaded onto a 15%–40% sucrose gradient (E), spun for 2 h, and 20 fractions harvested. Each fraction is deproteinized and the RNA recovered after phenol-choloroform-isoamylalcohol extraction and subsequent ethanol precipitation (F). After fractionation, RNA fractionated on a sucrose gradient migrated on a Northern blot as shown. The top fractions are characterized by the presence of the tRNAs; next, fractions corresponding to a peak migration of the small 40S ribosomal subunit (revealed by the peak of 18S rRNA), followed by the peak migration of the large 60S ribosomal subunit (peak of 28S rRNA) contain tRNA. Fractions below contain mRNAs with one or more ribosomes attached to them.
This protocol was used to prepare polysome gradients from mouse mammary gland. Bioanalyzer profiles from these fractions demonstrate the quality of the fractionation as well as the RNA quality from each fraction (Fig. 2A). Subsequently, the RNA from each fraction was precipitated with 2 M LiCl, to remove contaminating heparin that would inhibit the reverse transcriptase (see Materials and Methods). The distribution in the gradient of several genes, including p53, p21, mdm2, and BAX, as well as of the eukaryotic translation initiation factor 1 α (eIF1α), was analyzed by quantitative RT-PCR on each fraction from the gradient. The data from Figure 2C show that a large fraction of the transcripts coding for the genotoxic-response genes p53, p21, and mdm2, and also the transcripts coding for eIF1α mRNA, are in the heavy sucrose fractions, most likely bound to a large amount of ribosomes (polysomes) (Fig. 2C).
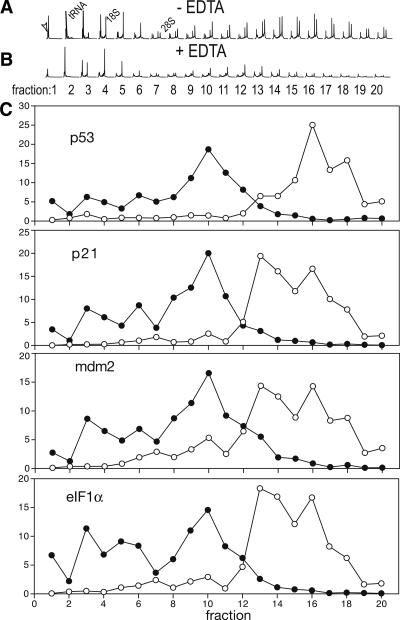
FIGURE 2.
Sucrose gradients from mammary tissue samples. Sucrose-gradient fractionation from mouse mammary gland tissue was carried out from samples of X-ray irradiated adult female mice. The processing of the samples was carried out as described in Figure 1. The quality of the sucrose fractionation and of the RNA in each fraction was determined with a Bioanalyzer (Agilent) (A), as well as in a control gradient in which the tissue was lysed in the presence of 25 mM EDTA (B) (EDTA release control), in which the presence of EDTA leads to the shift of mRNAs from the polysome-bound fraction toward lighter sucrose fractions (monosomes). The arrowhead indicates the peak corresponding to a marker RNA added to each sample, and the peaks corresponding to tRNA, 18S, and 28S RNAs are indicated. The RNA on each fraction was reverse transcribed and aliquots amplified with specific primers and the corresponding probes (UniversalProbe Library probes, Roche) by qPCR. For the amplification, genes implicated in DNA-damage responses (p53, p21, BAX, Mdm2) and eIF1α (see Materials and Methods for details) were used. The data are presented as percentage of the total amount of RNA on each fraction (C). (Empty circles) fractions of a polysome gradient prepared in the absence of EDTA; (filled circles) fractions of a polysome gradient prepared in the presence of EDTA.
To ascertain that indeed p53, p21, mdm2, and eIF1α transcripts were bound to polysomes, the lysis of the cells was carried out in the presence of the Ca++/Mg++ chelating agent EDTA (EDTA-release experiment), which leads to the redistribution of polysome-bound mRNAs toward fractions of lower density (monosomes), as previously described (Pastori and Schoenberg 1993; Beck and De Maio 1994; Ohashi et al. 2002). Indeed, the EDTA-release experiment demonstrates a redistribution of p53, p21, mdm2, and eIF1α mRNAs toward less-dense fractions (monosomes) (Fig. 2C), indicating that these transcripts are efficiently translated in the mouse mammary gland several hours after X-ray irradiation (Fig. 2C).
The same protocol was successfully used for sucrose-gradient fractionation of other mouse and human tissues (Fig. 3), where samples were analyzed by methylene blue staining of the corresponding Northern blots. Hybridization of the blots with a β-actin-specific cDNA probe revealed a distribution of the β-actin mRNA characteristic of each tissue (Fig. 3B).
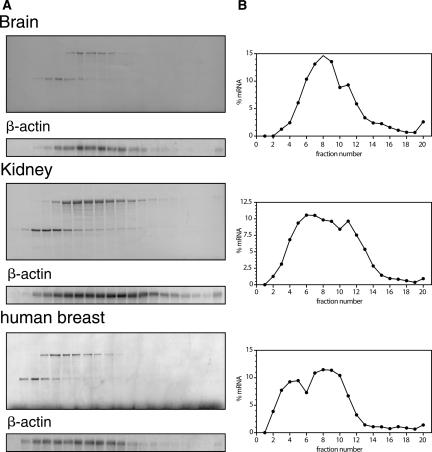
FIGURE 3.
Sucrose gradients from solid tissue samples. Sucrose-gradient fractionation of mRNAs from mouse brain and kidney as well as from human breast samples were carried out as described in Figure 1. Staining of the corresponding filters with methylene blue shows the fractionation of cellular RNA, according to its ribosomal load. The integrity of the mRNA was assessed after Northern hybridization with a β-actin cDNA probe (A). The hybridization signals of β-actin mRNA were quantified using a PhosphorImager, and the distribution of this mRNA in the sucrose gradients is depicted as percentage of the mRNA in each fraction (B).
Heparin inhibition of reverse transcriptase is fully reverted by lithium chloride
Initially we observed very low reverse-transcription yields from sucrose-fractionated RNA samples, as reflected by inefficient PCR amplifications and cRNA synthesis, implying that for profiling analysis to become feasible polyA+ RNA had to be used, requiring larger samples to quantitatively prepare polyA+ RNA. Since one of the aims of this work was to enable the use small samples (i.e., from biopsies), we analyzed the reasons for the inefficient reverse transcription reaction. Two lines of argument indicated that this was not due to the quality of the RNA obtained. First, polyA+ mRNA purified from these samples could be reverse transcribed with high efficiency. Second, analysis of polysome gradients gave an inhibition in the amplification not only of the mRNAs analyzed, but also cRNA amplified with the same primers in competitive RT-PCR reactions (J.A. Garcia-Sanz, unpubl.).
A literature search for inhibitors of the reverse transcriptase allowed identification of heparin as a possible inhibitor in the sucrose-gradient samples. Indeed, heparin, commonly used in sucrose-gradient fractionation (650 μg/mL) as an unspecific RNAse inhibitor (Ukita et al. 1962), in concentrations of 5 μg/mL inhibits 50% of the Superscript II reverse transcriptase activity (Gerard 1994). Thus, if only 1/100 of the original heparin concentration remained through the phenol-chloroform extraction and ethanol precipitation steps, reverse transcriptase activity should be at least partially inhibited.
It has been shown previously for RNA samples from heparinized clinical specimens that precipitation with 2 M lithium chloride (LiCl) is able to remove heparin (Jung et al. 1997). Thus, we tested if a similar treatment of polysome gradient samples would remove the inhibitory effect. For this purpose, RNA from a pool of polysome gradient fractions from Jurkat cells was divided in two parts: RNA from one half was precipitated with LiCl (see Materials and Methods), whereas the other half was ethanol precipitated (control) (Fig. 3A). Afterward an aliquot from each sample was used for quantitative RT-PCR using β-actin specific cDNA primers. Quantification of the results demonstrated an improvement (∼500-fold increase) in the amplified cDNA signal from the sample precipitated with LiCl (Fig. 3A).
To prove that indeed heparin had been removed by the LiCl precipitation, cytoplasmic RNA extracted from Jurkat cells prepared in the absence of heparin was spiked with an amount of heparin (650 μg/mL) equivalent to the concentration in polysome gradients. Thereafter, again, one aliquot of the RNA was precipitated with LiCl, whereas the other was precipitated with ethanol. Both RNA samples were analyzed using the same quantitative RT-PCR reaction as above. As additional controls, aliquots of Jurkat RNA without heparin were precipitated either with LiCl or ethanol. The results obtained demonstrate that, as expected, heparin at the concentration used fully inhibited amplification of β-actin mRNA. Notably, the LiCl precipitation was able to fully revert the inhibition of the reverse transcriptase activity since the amplification curve was similar to the one obtained in the aliquots devoid of heparin (Fig. 3B). LiCl precipitation itself did not have a noticeable effect on the amplification (Fig. 3B). Furthermore, comparison of the amplification profiles of similar amounts of cytoplasmic RNA (Fig. 3B) and polysome-fractionated RNA after LiCl precipitation (Fig. 3A) indicate that heparin is the major inhibitor of the reverse transcriptase present in the sucrose-gradient fractions.
The potent RNAse inhibitor VRC used for the sucrose fractionation (Gray 1974; Egberts et al. 1977; Berger and Birkenmeier 1979) does not affect reverse-transcriptase activity when used at 10 mM concentrations on polysome gradients (data not shown).
DISCUSSION
The method outlined above was designed to meet the need for the preparation of polysome-bound mRNA from small samples obtained from solid tissues, which will allow determining the role of translational control during physiological or pathological changes in these tissues or organs, even from biopsy samples. By using this protocol, sucrose-gradient fractionated RNA from three different tissues, two of mouse and one of human origin, were successfully prepared. The RNA obtained from these samples derived from brain, kidney, and mammary gland, was amenable to Northern blotting, RT-PCR, and expression profiling analysis (data not shown). One critical point of the method is that optimally small samples (10–20 mg each) must be quickly frozen under liquid nitrogen and then can be stored at −80°C for prolonged periods, which may be of interest for the use of human biopsy material. For processing of the samples, the first step is pulverization of tissue with a mortar and pestle. This step requires maintaining the tissue frozen: the mortar is filled with liquid nitrogen with the pestle inside. Once cold, the tissue is added and then pulverized until a fine powder is obtained, adding more liquid nitrogen when necessary. The powder is then transferred to a microcentrifuge tube precooled in dry ice, and NP40 lysis buffer containing RNAsin and VRC are added. Importantly, in our hands and with the tissue samples tested (in contrast to cell line derived material), RNAsin alone was clearly insufficient to prevent severe RNA degradation. Only the combination of RNAsin and VRC worked effectively. After lysis and removal of the nuclei and tissue debris, an equal volume of 2× extraction buffer, supplemented with cycloheximide, PMSF, and heparin, was added to the sample, which was subsequently centrifuged to remove mitochondria and membranous debris and then layered onto the 15%–40% sucrose gradients. It should be noted that with the equipment described the protocol cannot be scaled up further since there is a limitation in the volume of cytoplasmic extract (1–2 mL, corresponding to 10–20 mg of tissue) to be layered onto the gradient (10 mL).
Our data also show that purification of the initial RNA to obtain polyA+ RNA for reverse transcription/cRNA synthesis is not required. We can avoid problems with quantitative mRNA preparation for small samples if heparin is removed after preparation of the RNA by a LiCl precipitation step. This step is critical since the residual amount of heparin in the samples, even after phenol-choloroform-isoamylalcohol extraction and ethanol precipitation, inhibits reverse transcriptase activity ∼500-fold (Fig. 4A). LiCl precipitation is able to fully remove 650 μg/mL heparin, as demonstrated by the improved efficiency of RT-PCR amplification (see Fig. 4B). On the other hand, VCR, either alone or in combination with heparin, had any effect on reverse transcription after the usual purification steps (data not shown).
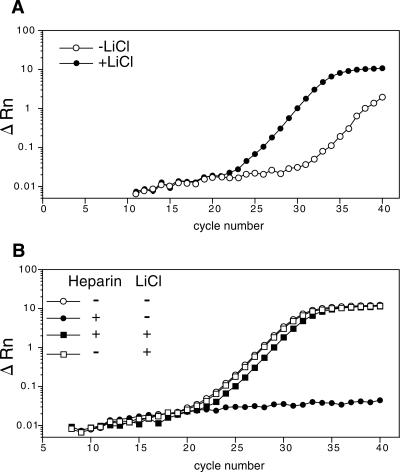
FIGURE 4.
Lithium chloride precipitation of the samples abrogates heparin inhibition of the reverse transcriptase. Quantitative RT-PCR amplification of β-actin mRNA from polysome-bound RNA (prepared with heparin), precipitated with either lithium chloride (+LiCl) or ethanol (−LiCl) (A). Aliquots of cytoplasmic Jurkat RNA (prepared in the absence of heparin) were mixed with heparin (650 μg/mL) and subsequently divided in two, one precipitated with lithium chloride (+heparin, +LiCl), or precipitated with ethanol (+heparin, −LiCl). Samples devoid of heparin and precipitated with either LiCl (−heparin, +LiCl) or with ethanol (−heparin, −LiCl) were used as controls. The quantitative amplification of the β-actin mRNA is illustrated for the different conditions (B).
In conclusion, we describe a protocol that allows the efficient preparation of ribosome-free and polysome-bound mRNAs from solid tissue samples. Furthermore, the lithium chloride precipitation of the RNA allows us to start from samples of smaller size, since this precipitation step efficiently depletes the samples of the reverse-transcriptase inhibitor heparin, and makes it unnecessary to quantitatively purify polyA+ RNA. We believe that this represents a significant improvement since, with the use of this protocol, it makes the fractionation of the RNA from solid tissues into ribosome-free and polysome-bound fractions amenable for analysis of biopsy samples by Northern blotting, RT-PCR, and expression profiling experiments, in which only the actively translated mRNAs from the cell are taken into account.
MATERIALS AND METHODS
Tissue samples
C57Bl/6 and BALB/c female mice, 6–12 wk old, were used to obtain tissue samples for the experiments described below. Euthanasia was carried out in a CO2 chamber for 5 min; mammary glands were obtained from a C56Bl/6 female, 16 h after whole body X-ray irradiation (20 Gy). Animals were housed and bred in our animal facility and in all experiments treated in accordance with the European Union and National Guide Lines and the Helsinki Declaration. Human primary breast cancer tissue samples were obtained through Drs. Carmen Rodriguez and Charles Theillet from the mammary tissue bank at the Pathology Department of the Val d'Aurelle Cancer Center (Montpellier, France).
Preparation of cytoplasmic RNA
Cytoplasmic RNA from Jurkat cells, a human leukemia cell line (ATCC #TIB152) (Schneider et al. 1977), was obtained following lysis of cells in NP40-lysis buffer (10 mM Tris-HCl at pH 8.0, 140 mM NaCl, 1.5 mM MgCl2, 0.5% Nonidet-P40, 20 mM dithiothreitol, 500 U/mL RNAsin [Blackburn et al. 1977], and 0.5% [w/v] deoxycholate), nuclei were pelleted at 12,000g for 10 sec, and the cytoplasmic extract digested with 10 μg/mL Proteinase K in the presence of 1% sodium dodecyl sulfate (SDS) and 10 mM EDTA (pH 8.0) for 30 min at 37°C as described (Müllner and Garcia-Sanz 1997c), after a modification from Favaloro et al. (1980).
Polysome gradient preparation
A piece of ∼20 mg solid tissue was pulverized under liquid nitrogen and the powder lysed in 1 mL of lysis buffer (10 mM Tris-HCl at pH 8.0, 150 mM NaCl, 5 mM MgCl2, 1% Nonidet-P40, 40 mM dithiothreitol, 500 U/mL RNAsin [Promega], 40 mM VRC [Berger and Birkenmeier 1979] [New England Bio Labs]) supplemented with 1% deoxycholate (Fluka). After lysing the cells by pipetting up and down several times, the nuclei were removed by centrifugation (12,000g, 10 sec, at 4°C), and the supernatant was supplemented with 500 μL of 2× extraction buffer (0.2 M Tris-HCl at pH 7.5, 0.3 M NaCl), 150 μg/mL cycloheximide, 650 μg/mL heparin, and 10 mM phenyl-methyl-sulfonyl fluoride, and centrifuged (12,000g, 5 min, at 4°C) to remove mitochondria and membranous debris. The supernatant was layered onto a 10 mL linear sucrose gradient (15%–40% sucrose [w/v], supplemented with 10 mM Tris-HCl at pH 7.5, 140 mM NaCl, 1.5 mM MgCl2, 10 mM dithiothreitol, 100 μg/mL cycloheximide, 0.5 mg/mL heparin) (Müllner and Garcia-Sanz 1997b) and centrifuged in a SW41Ti rotor (Beckman) for 120 min at 38,000 rpm at 4°C, with brake off. Fractions (20 μL × 550 μL) were collected and digested with 100 μg proteinase K in 1% SDS and 10 mM EDTA for 30 min at 37°C. RNAs were recovered by extraction with an equal volume of phenol-chloroform-isoamyl alcohol, followed by ethanol precipitation. As a control, an EDTA release experiment was made as described before, where the lysis buffer was supplemented with 25 mM EDTA (pH 8.0).
Northern blotting
After electrophoresis of RNA from the polysome gradients through denaturing 1.2% formaldehyde-agarose gels, RNA samples were transferred to nylon membranes (GeneScreen, NEN) and rRNA distribution visualized by methylene blue staining (Müllner and Garcia-Sanz 1997a). Northern blots were hybridized with a β-actin cDNA probe (the 1.1-kb PstI–PstI fragment of mouse β-actin cDNA, which hybridizes with both mouse and human samples) labeled with [α-32P]dCTP, using the random priming method. After washes, filters were exposed and quantified by a PhosphorImager (Molecular Dynamics). The background from each sample was subtracted using the local median algorithm of the ImageQuant program (version 1.2, Molecular Dynamics).
LiCl precipitation of RNA
Cytoplasmic RNA or pools of fractions containing polysome-bound mRNA, before being reverse transcribed, when indicated, were precipitated with 2 M LiCl on ice at 4°C overnight (Jung et al. 1997). After centrifugation (12,000g, 15 min at 4°C), pellets were washed twice with 70% ETOH (prestored at −20°C), air-dried, and resuspended in appropriate volumes of RNAse-free water. For control purposes (−LiCl), RNAs were precipitated with 0.3 M NaAc, 2.5 vol. 100% ETOH, and 1 μL glycogen (20 mg/mL, Roche). After centrifugation (12,000g, 15 min at 4°C), these pellets also were resuspended in RNAse-free water.
Quantitative PCR
RNA (5 μg) was reverse-transcribed using random hexamers and 100 U Superscript II RT (Invitrogen). Real-time PCR was performed on an ABI Prism 7700 (Applied Biosystems), using SYBR Green PCR Core Reagents (Applied Biosystems). The final MgCl2 concentration was 1.5 mM, and primers were used at 0.4 μM. Reactions were incubated 2 min at 50°C, 10 min at 95°C, followed by 40 cycles of 15 sec at 95°C, and 90 sec at 67°C. Specific primers were designed with PrimerExpress software (Applied Biosystems); amplified sequences spanned different exons. Differences in cDNA load were corrected for the amount of amplified 28S rRNA. mRNA expression levels are shown as ΔRn signal in each cycle.
RT-qPCR from each fraction from a polysome gradient was done using the Transcriptor First Strand cDNA Synthesis Kit (Roche), by random hexamers priming at 50°C for 1 h, following the manufacturer's instructions. Since the RNA amount on each fraction was different, in order to avoid efficiency differences of the reverse transcriptase, the RNA amounts were equalized by adding an appropriate amount of luciferase RNA to each fraction (negative control), giving a final amount of 1 μg of RNA on each sample. After synthesis of the cDNA first strand, qPCR was performed on an ABI Prism 7900 (Applied Biosystems), using the FastStart Taqman Probe Master (Rox) (Roche) and with sets of primers and Universal ProbeLibrary probes (Roche) designed online with ProbeFinder version 2.20 (Roche). Probes specific for eIF1α (forward primer: 5′-CGCTGCGTTTTGGTCACTA, reverse primer: 5′-CCTGGCTTCTGAGAGCTACAA, and Universal ProbeLibrary probe #63, which gives a 93-nt amplicon); Mdm2 (forward primer: 5′-TGGAGTCCCGAGTTTCTCTG, reverse primer: 5′-AGCCACTAAATTTCTGTAGATCATTG, and Universal ProbeLibrary probe #99, which gives a 70-nt amplicon); kinase inhibitor 1 (p21) (forward primer: 5′-TCCACAGCGATATCCAGACA, reverse primer: 5′-GGACATCACCAGGATTGGAC, and Universal ProbeLibrary probe #21, which leads to a 60-nt amplicon); tumor antigen p53 (forward primer: 5′-ACGCTTCTCCGAAGACTGG, reverse primer: 5′-AGGGAGCTCGAGGCTGATA, and Universal ProbeLibrary probe #25, which leads to a 67-nt amplicon); and Bcl2-associated X protein (BAX) (forward primer: 5′-CGGCGAATTGGAGATGAA, reverse primer: 5′-GTGTCCACGTCAGCAATCAT, and Universal ProbeLibrary probe #56, which leads to a 65-nt amplicon). Each sample was amplified with 1 cycle at 95°C for 10 min (to activate the polymerase) and 40 cycles at (95°C for 15 sec and 60°C for 1 min).
ACKNOWLEDGMENTS
We thank Drs. Carmen Rodriguez and Charles Theillet (INSERM, Montpellier) for providing tissue samples and Dr. Angel Zaballos (CNB, Madrid) for his help with the quantitative PCR reaction. The work in the authors’ laboratories was supported by grants from the U.S. Army Medical Research and Acquisition Activity (Breast Cancer Program) Contract number DAMD17-02-1-0339 (to J.A.G.-S.) and from the Spanish Ministry of Education and Science, contract SAF2003-00519 (to J.A.G.-S.), and the Austrian “Fonds zur Förderung der wissenschaftlichen Forschung, FWF” (to E.W.M.). R.V. is on leave from the Department of Conservative Odontology, Dentistry School, University of Chile. We also acknowledge the Unidad de Genomica, Parque Científico de Madrid, and Universidad Complutense de Madrid.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.79407.
REFERENCES
- Beck, S.C., De Maio, A. Stabilization of protein synthesis in thermotolerant cells during heat shock: Association of heat shock protein-72 with ribosomal subunits of polysomes. J. Biol. Chem. 1994;269:21803–21811. [PubMed] [Google Scholar]
- Berger, S.L., Birkenmeier, C.S. Inhibition of intractable nucleases with ribonucleoside–vanadyl complexes: Isolation of messenger ribonucleic acid from resting lymphocytes. Biochemistry. 1979;18:5143–5149. doi: 10.1021/bi00590a018. [DOI] [PubMed] [Google Scholar]
- Blackburn, P., Wilson, G., Moore, S. Ribonuclease inhibitor from human placenta. Purification and properties. J. Biol. Chem. 1977;252:5904–5910. [PubMed] [Google Scholar]
- Blazquez-Domingo, M., Grech, G., von Lindern, M. Translation initiation factor 4E inhibits differentiation of erythroid progenitors. Mol. Cell. Biol. 2005;25:8496–8506. doi: 10.1128/MCB.25.19.8496-8506.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egberts, E., Hackett, P.B., Traub, P. Inhibition of ribonucleases by ribonucleotides and transition state analogs in cell-free extracts from Ehrlich ascites tumor cells. Hoppe Seylers Z. Physiol. Chem. 1977;358:475–490. doi: 10.1515/bchm2.1977.358.1.475. [DOI] [PubMed] [Google Scholar]
- Favaloro, J., Treisman, R., Kamen, R. Transcription maps of polyoma virus-specific RNA: Analysis by two-dimensional nuclease S1 gel mapping. Methods Enzymol. 1980;65:718–749. doi: 10.1016/s0076-6879(80)65070-8. [DOI] [PubMed] [Google Scholar]
- Garcia-Sanz, J.A., Lenig, D. Translational control of interleukin 2 messenger RNA as a molecular mechanism of T cell anergy. J. Exp. Med. 1996;184:159–164. doi: 10.1084/jem.184.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerard, G.F. Inhibition of SuperScriptTM II reveste transcriptase by common laboratory chemicals. Focus. 1994;16:102–103. [Google Scholar]
- Gray, J.C. The inhibition of ribonuclease activity and the isolation of polysomes from leaves of the French bean, Phaseolus vulgaris. Arch. Biochem. Biophys. 1974;163:343–348. doi: 10.1016/0003-9861(74)90485-8. [DOI] [PubMed] [Google Scholar]
- Grolleau, A., Bowman, J., Pradet-Balade, B., Puravs, E., Hanash, S., Garcia-Sanz, J.A., Beretta, L. Global and specific translational control by rapamycin in T cells uncovered by microarrays and proteomics. J. Biol. Chem. 2002;277:22175–22184. doi: 10.1074/jbc.M202014200. [DOI] [PubMed] [Google Scholar]
- Gygi, S.P., Rist, B., Gerber, S.A., Turecek, F., Gelb, M.H., Aebersold, R. Quantitative analysis of complex protein mixtures using isotope-coded affinity tags. Nat. Biotechnol. 1999a;17:994–999. doi: 10.1038/13690. [DOI] [PubMed] [Google Scholar]
- Gygi, S.P., Rochon, Y., Franza, B.R., Aebersold, R. Correlation between protein and mRNA abundance in yeast. Mol. Cell. Biol. 1999b;19:1720–1730. doi: 10.1128/mcb.19.3.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber, M., Bahr, I., Kratzschmar, J.R., Becker, A., Muller, E.C., Donner, P., Pohlenz, H.D., Schneider, M.R., Sommer, A. Comparison of proteomic and genomic analyses of the human breast cancer cell line T47D and the antiestrogen-resistant derivative T47D-r. Mol. Cell. Proteomics. 2004;3:43–55. doi: 10.1074/mcp.M300047-MCP200. [DOI] [PubMed] [Google Scholar]
- Jechlinger, M., Grunert, S., Tamir, I.H., Janda, E., Ludemann, S., Waerner, T., Seither, P., Weith, A., Beug, H., Kraut, N. Expression profiling of epithelial plasticity in tumor progression. Oncogene. 2003;22:7155–7169. doi: 10.1038/sj.onc.1206887. [DOI] [PubMed] [Google Scholar]
- Johannes, G., Carter, M.S., Eisen, M.B., Brown, P.O., Sarnow, P. Identification of eukaryotic mRNAs that are translated at reduced cap binding complex eIF4F concentrations using a cDNA microarray. Proc. Natl. Acad. Sci. 1999;96:13118–13123. doi: 10.1073/pnas.96.23.13118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joosten, M., Blazquez-Domingo, M., Lindeboom, F., Boulme, F., Van Hoven-Beijen, A., Habermann, B., Lowenberg, B., Beug, H., Mullner, E.W., Delwel, R., et al. Translational control of putative protooncogene Nm23-M2 by cytokines via phosphoinositide 3-kinase signaling. J. Biol. Chem. 2004;279:38169–38176. doi: 10.1074/jbc.M401283200. [DOI] [PubMed] [Google Scholar]
- Ju, J.F., Huang, C.L., Minskoff, S.A., Mayotte, J.E., Taillon, B.E., Simons, J.F. Simultaneous gene expression analysis of steady-state and actively translated mRNA populations from osteosarcoma MG-63 cells in response to IL-1 alpha via an open expression analysis platform. Nucleic Acids Res. 2003;31:5157–5166. doi: 10.1093/nar/gkg702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung, R., Lubcke, C., Wagener, C., Neumaier, M. Reversal of RT-PCR inhibition observed in heparinized clinical specimens. Biotechniques. 1997;23:24–28. doi: 10.2144/97231bm03. [DOI] [PubMed] [Google Scholar]
- Kawaguchi, R., Girke, T., Bray, E.A., Bailey-Serres, J. Differential mRNA translation contributes to gene regulation under non-stress and dehydration stress conditions in Arabidopsis thaliana . Plant J. 2004;38:823–839. doi: 10.1111/j.1365-313X.2004.02090.x. [DOI] [PubMed] [Google Scholar]
- Leibold, E.A., Munro, H.N. Cytoplasmic protein binds in vitro to a highly conserved sequence in the 5′-untranslated region of ferritin heavy- and light-subunit mRNAs. Proc. Natl. Acad. Sci. 1988;85:2171–2175. doi: 10.1073/pnas.85.7.2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindemann, S.W., Yost, C.C., Denis, M.M., McIntyre, T.M., Weyrich, A.S., Zimmerman, G.A. Neutrophils alter the inflammatory milieu by signal-dependent translation of constitutive messenger RNAs. Proc. Natl. Acad. Sci. 2004;101:7076–7081. doi: 10.1073/pnas.0401901101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKay, V.L., Li, X.H., Flory, M.R., Turcott, E., Law, G.L., Serikawa, K.A., Xu, X.L., Lee, H., Goodlett, D.R., Aebersold, R., et al. Gene expression analyzed by high-resolution state array analysis and quantitative proteomics—Response of yeast to mating pheromone. Mol. Cell. Proteomics. 2004;3:478–489. doi: 10.1074/mcp.M300129-MCP200. [DOI] [PubMed] [Google Scholar]
- Mathews, M.B., Sonenberg, N., Hershey, J.W.B. Origins and principles of translational control. In: Sonenberg N., et al., editors. Translational control of gene expression. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2000. pp. 1–33. [Google Scholar]
- Mikulits, W., Pradet-Balade, B., Habermann, B., Beug, H., Garcia-Sanz, J.A., Mullner, E.W. Isolation of translationally controlled mRNAs by differential screening. FASEB J. 2000;14:1641–1652. doi: 10.1096/fj.14.11.1641. [DOI] [PubMed] [Google Scholar]
- Müllner, E.W., Garcia-Sanz, J.A. Analysis of RNA expression by Northern blotting. In: Lefkovits I., editor. Immunology methods manual. Academic Press; London: 1997a. pp. 407–424. [Google Scholar]
- Müllner, E.W., Garcia-Sanz, J.A. Polysome gradients. In: Lefkovits I., editor. Manual of immunological methods. Academic Press; London: 1997b. pp. 457–462. [Google Scholar]
- Müllner, E.W., Garcia-Sanz, J.A. Preparation of RNA. In: Lefkovits I., editor. Manual of immunological methods. Academic Press; London: 1997c. pp. 389–406. [Google Scholar]
- Ohashi, S., Koike, K., Omori, A., Ichinose, S., Ohara, S., Kobayashi, S., Sato, T.A., Anzai, K. Identification of mRNA/protein (mRNP) complexes containing Puralpha, mStaufen, fragile X protein, and myosin Va and their association with rough endoplasmic reticulum equipped with a kinesin motor. J. Biol. Chem. 2002;277:37804–37810. doi: 10.1074/jbc.M203608200. [DOI] [PubMed] [Google Scholar]
- Pastori, R.L., Schoenberg, D.R. The nuclease that selectively degrades albumin mRNA in vitro associates with Xenopus liver polysomes through the 80S ribosome complex. Arch. Biochem. Biophys. 1993;305:313–319. doi: 10.1006/abbi.1993.1428. [DOI] [PubMed] [Google Scholar]
- Pradet-Balade, B., Boulme, F., Beug, H., Mullner, E.W., Garcia-Sanz, J.A. Translation control: Bridging the gap between genomics and proteomics? Trends Biochem. Sci. 2001a;26:225–229. doi: 10.1016/s0968-0004(00)01776-x. [DOI] [PubMed] [Google Scholar]
- Pradet-Balade, B., Boulme, F., Mullner, E.W., Garcia-Sanz, J.A. Reliability of mRNA profiling: Verification for samples with different complexities. Biotechniques. 2001b;30:1352–1357. doi: 10.2144/01306rr03. [DOI] [PubMed] [Google Scholar]
- Rajasekhar, V.K., Viale, A., Socci, N.D., Wiedmann, M., Hu, X., Holland, E.C. Oncogenic Ras and Akt signaling contribute to glioblastoma formation by differential recruitment of existing mRNAs to polysomes. Mol. Cell. 2003;12:889–901. doi: 10.1016/s1097-2765(03)00395-2. [DOI] [PubMed] [Google Scholar]
- Schneider, U., Schwenk, H.U., Bornkamm, G. Characterization of EBV-genome negative “null” and “T” cell lines derived from children with acute lymphoblastic leukemia and leukemic transformed non-Hodgkin lymphoma. Int. J. Cancer. 1977;19:621–626. doi: 10.1002/ijc.2910190505. [DOI] [PubMed] [Google Scholar]
- Tew, K.D., Monks, A., Barone, L., Rosser, D., Akerman, G., Montali, J.A., Wheatley, J.B., Schmidt D.E., Jr Glutathione-associated enzymes in the human cell lines of the National Cancer Institute Drug Screening Program. Mol. Pharmacol. 1996;50:149–159. [PubMed] [Google Scholar]
- Tominaga, Y., Tamguney, T., Kolesnichenko, M., Bilanges, B., Stokoe, D. Translational deregulation in PDK-1−/− embryonic stem cells. Mol. Cell. Biol. 2005;25:8465–8475. doi: 10.1128/MCB.25.19.8465-8475.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ukita, T., Terao, T., Irie, M. Inhibition of pancreatic ribonuclease-I activity by heparin. J. Biochem. 1962;52:455–457. doi: 10.1093/oxfordjournals.jbchem.a127644. [DOI] [PubMed] [Google Scholar]
- Wouters, B.G., van den Beucken, T., Magagnin, M.G., Koritzinsky, M., Fels, D., Koumenis, C. Control of the hypoxic response through regulation of mRNA translation. Semin. Cell Dev. Biol. 2005;16:487–501. doi: 10.1016/j.semcdb.2005.03.009. [DOI] [PubMed] [Google Scholar]
- Yost, C.C., Denis, M.M., Lindemann, S., Rubner, F.J., Marathe, G.K., Buerke, M., McIntyre, T.M., Weyrich, A.S., Zimmerman, G.A. Activated polymorphonuclear leukocytes rapidly synthesize retinoic acid receptor-α: A mechanism for translational control of transcriptional events. J. Exp. Med. 2004;200:671–680. doi: 10.1084/jem.20040224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zong, Q., Schummer, M., Hood, L., Morris, D.R. Messenger RNA translation state: The second dimension of high-throughput expression screening. Proc. Natl. Acad. Sci. 1999;96:10632–10636. doi: 10.1073/pnas.96.19.10632. [DOI] [PMC free article] [PubMed] [Google Scholar]