Abstract
The development of characteristic visceral asymmetries along the left–right (LR) axis in an initially bilaterally symmetrical embryo is an essential feature of vertebrate patterning. The allelic mouse mutations inversus viscerum (iv)1,2 and legless (lgl)3,4 produce LR inversion, or situs inversus, in half of live-born homozygotes. This suggests that the iv gene product drives correct LR determination, and in its absence this process is randomized2. These mutations provide tools for studying the development of LR-handed asymmetry and provide mouse models of human lateralization defects. At the molecular level, the normally LR asymmetric expression patterns of nodal5 and lefty6 are randomized in iv/iv embryos, suggesting that iv functions early in the genetic hierarchy of LR specification. Here we report the positional cloning of an axonemal dynein heavy-chain gene, left/right-dynein (lrd), that is mutated in both lgl and iv. lrd is expressed in the node of the embryo at embryonic day 7.5, consistent with its having a role in LR development7. Our findings indicate that dynein, a micro-tubule-based motor, is involved in the determination of LR-handed asymmetry and provide insight into the early molecular mechanisms of this process.
In humans, random development of left–right asymmetry is a feature of Kartagener’s syndrome, which is manifested by situs inversus, immotile cilia and the absence of ciliary dynein arms; this suggests that dynein function has a role in LR development8,9. Dyneins are a family of minus-end-directed microtubule-based motors which are traditionally classified as either cytoplasmic or axonemal. Cytoplasmic dyneins have been implicated in vesicular transport, nuclear migration, and spindle orientation10,11, whereas axonemal dyneins produce the motive forces that cause the sliding of adjacent microtubules in the axoneme10,11 and thus produce ciliary and flagellar movement. Dyneins function as large multi-subunit complexes containing up to three heavy chains, which include the force-producing motor domain, in addition to several intermediate and light chains. So far, 15 distinct dynein heavy-chain genes have been identified in vertebrates12–14. Two of these (Dnchc1 and Dnahc13) were mapped to the distal region of mouse chromosome 12, close to the previously identified map location of the iv and lgl mutations4,15.
We isolated and analysed overlapping YAC (yeast artificial chromosome) clones spanning the lgl transgene insertion site in this region of chromosome 12, which indicated that the lgl mutation includes a deletion of at least 600 kilobases (kb) (Fig. 1a). Our inability to isolate non-chimaeric YACs at one end of the contig or to identify the remaining mouse DNA–transgene junction suggests that this deletion may extend to the end of the chromosome. To determine whether a dynein gene is involved in the iv and lgl mutations, we screened the lgl YAC contig at low stringency with a DNA probe for the highly conserved first P-loop nucleotide-binding domain found in both cytoplasmic and axonemal dynein. This strategy identified a 2.1-kb genomic Bgl II fragment which cross-hybridizes to the dynein P-loop probe and is deleted in lgl DNA (see Supplementary information). Sequence analysis of this fragment revealed that it is distinct from Dnchc1 and Dnahc13 and includes two exons orthologous to a rat dynein-like gene, DLP11 (ref. 12). We cloned over 7 kb of coding sequence from this gene, including all four central P-loops, and sequence analysis reveals 67% identity and 81% similarity to the sea-urchin axonemal dynein-β heavy-chain gene, compared with only 31% identity and 54% similarity to the rat cytoplasmic dynein heavy-chain gene (Fig. 1b). In addition, the sequence immediately adjacent to the first P-loop (TETTKDL) defines this gene as an axonemal dynein isoform16. Recombination mapping using the same C57BL/6J X SI/Col-iv outcross/backcross panel initially used to determine the map position of iv (ref. 17) demonstrated no recombination between this gene, which we designate left/right-dynein (lrd), and iv out of 39 informative meioses, including the one animal that exhibited a recombination event between the distal end of IgH-V and iv17 (data not shown).
Figure 1.
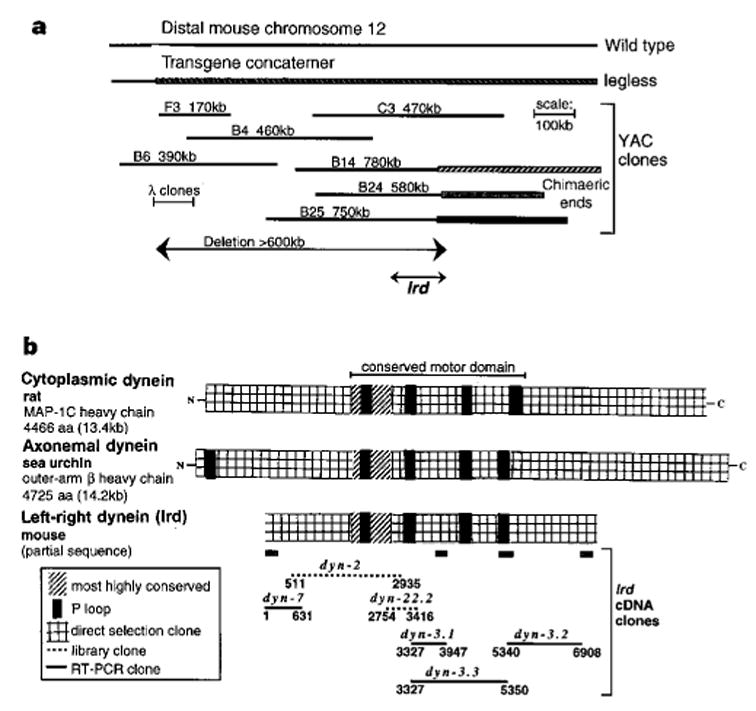
Identification of a dynein gene deleted in lgl. a, YAC contig spanning the mouse DNA/transgene junction in lgl. The location of the newly identified lrd gene (see text) is indicated. b, Homology of cloned lrd sequences with rat cytoplasmic and sea-urchin axonemal dynein heavy chains.
Comparison of lrd sequences from wild type and iv demonstrates a striking missense mutation in iv mice. Analysis of 7 kb of sequence obtained from cloned reverse transcription polymerase chain reaction (RT-PCR) products from wild-type (ICR outbred mice) and iv/iv homozygous mice revealed six silent base changes that did not alter coding potential, and are therefore probably strain polymorphisms. However, one difference was found that changes a glutamic acid to a lysine at a position corresponding to residue 2,249 of the sea-urchin sequence (Fig. 2a). This amino-acid substitution, resulting in a change from negative to positive charge, is located between the second and third P-loop motifs. This region of the protein is part of the highly conserved central portion of the heavy chain that constitutes the motor domain10 (Fig. 1b). The G → A base change in iv was confirmed by sequencing RT-PCR products from multiple RNA preparations and by sequencing the relevant exon from genomic DNAs of multiple iv and wild-type mice. The iv mutation arose in an outbred stock of mice that was subsequently crossed into the strains DBA, C3H and BALB/c; hence no parental strain is available for comparison. Thus, to establish that the mutation did not represent a strain polymorphism, we sequenced the genomic DNA encompassing this region of the lrd gene from eight different strains of laboratory mice (C57BL/6J, C3H, BALB/c, A/J, AKR/J, DBA, 129/J and SJL/J), the wild-derived strains TIRANO/Ei (Mus domesticus), SPRET/Ei (Mus spretus), and PANCEVO/Ei (Mus hortulanis), as well as rat (Rattus norvegicus). All contain the wild-type glutamic acid at this position. As shown in Fig. 2b, this region of the protein is highly conserved across widely divergent species and among both axonemal and cytoplasmic dynein heavy chains. The glutamic acid residue at this position is absolutely conserved among all known dynein heavy-chain sequences, which suggests that it is critical to the function of the protein. We conclude that this glutamic acid-to-lysine substitution is unique to iv mutants and therefore provides a molecular basis for the randomization of LR development seen in these mice.
Figure 2.
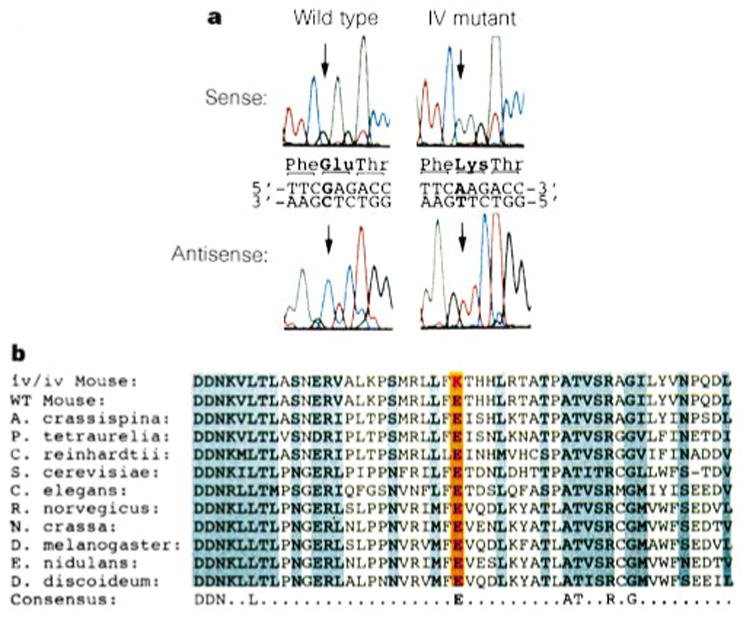
iv has a missense mutation in a highly conserved amino acid of Lrd. a, Raw sequence data. b, The E → K substitution is unique to iv and is in a very highly conserved amino-acid residue (orange). Amino acids that are identical or show only conservative changes across species are shaded. The consensus (bottom) shows residues that are identical among all known axonemal and cytoplasmic dynein sequences, including those not shown here. Amino-acid sequences in this region of Lrd were identical for all wild-type mouse strains analysed (12 in total). The rat orthologue also contains a glutamic acid at this position (data not shown).
The lgl phenotype, with hindlimb, brain and craniofacial malformations, as well as situs inversus3, is more severe than the iv phenotype. This could be the result of a more severe lrd mutation in lgl—a deletion versus the missense mutation found in iv—and/or the result of additional genes mutated in lgl mice.
Expression of lrd messenger RNA was examined by northern blot hybridization, RT-PCR and in situ hybridization. Northern blot hybridization demonstrates a low abundance lrd transcript of ~ 15 kb in adult brain (see Supplementary information). Expression was detected at a lower level in adult testes (data not shown). The transcript size appears similar in iv and wild-type RNA. RT-PCR showed that lrd mRNA is expressed as early as the blastocyst stage (embryonic day (E) 3.5) and is also expressed in embryonic stem (ES) cells, which are derived from the blastocyst inner cell mass (Fig. 3). Expression is also seen at E8, E9 and E16.5 (Fig. 3). No transcripts were detected in mouse embryo fibroblasts (MEFs; Fig. 3). In situ hybridization showed that lrd mRNA embryonic expression is highly restricted; at E7.5, before the time of asymmetric expression of nodal and lefty, lrd mRNA is detected only in the ventral cells of the node (Fig. 4a–c). We detected no expression in the heart at any stage. These results show that the onset of lrd mRNA expression precedes that of nodal and lefty, two of the earliest molecular markers of LR asymmetry, which are misexpressed in iv mutant embryos.5,6 Also, the presence of lrd transcripts in the node implicates this mammalian equivalent of Spemann’s organizer in the establishment of the LR axis. In the newborn and adult, expression of lrd mRNA was found in several types of ciliated epithelium, including the oviducts (Fig. 4d).
Figure 3.
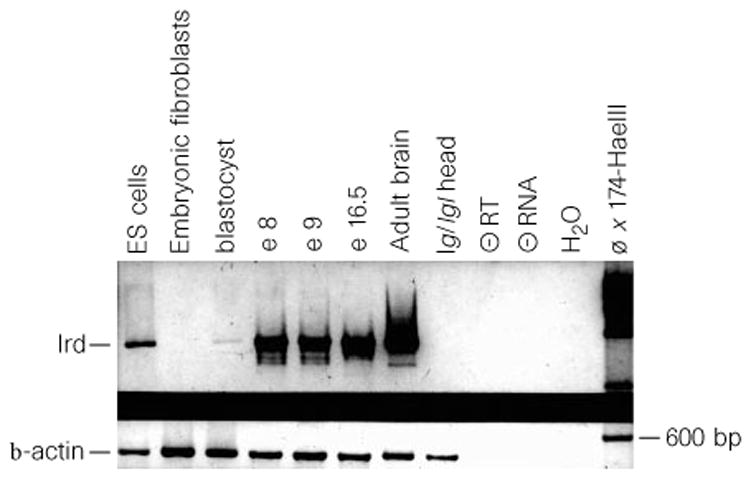
RT-PCR analysis of embryonic and adult expression. Primers used were specific for lrd (top) or control β -actin (bottom). Negative controls include: no reverse transcriptase (−RT), no tissue (medium control, −RNA), and no RNA (H2O).
Figure 4.
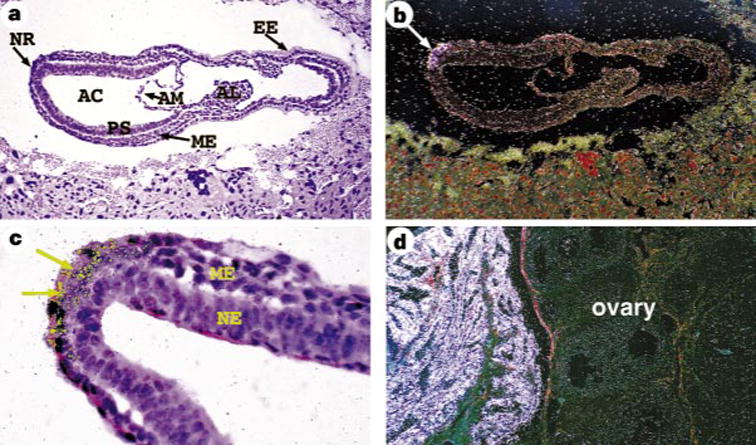
Expression of lrd localized by in situ hybridization. a–d, Hybridization to antisense lrd riboprobe; no signal was detected with a sense control probe. a, c, Bright-field image; b, d, dark-field images. a, b, Sagittal section of an E7.5 embryo demonstrating lrd expression in the node (arrow in b). c, Enlargement to show nodal localization of signal (arrows). d, lrd is expressed in the ciliated epithelium of the adult oviduct (left), but not in the ovary. NR, nodal region; AC, amniotic cavity; PS, primitive streak; AM, amnion; ME, mesoderm; AL, allantois; EE, extraembryonic ectoderm; NE, neural ectoderm. Computer-enhanced colour (yellow) was used to highlight the hybridization signal in c.
Expression of lrd mRNA in the embryo was observed primarily in non-ciliated cells, such as ES cells and blastocysts; it is unlikely that expression in the node is due to the presence of cilia, because cilia found in the node are immotile monocilia lacking dynein arms18. lrd mRNA expression was detected in many ciliated cells in the newborn and adult as well (Fig. 4d, and data not shown). As the sequence of lrd is more similar to axonemal than cytoplasmic dyneins, it is paradoxical that it is also found in cells lacking cilia; however, dyneins classified as axonemal have been found in non-ciliated cells19, suggesting that there is some blurring of the boundary between axonemal and cytoplasmic dyneins. We propose that lrd has dual functions: in the ciliated epithelium of adult structures it functions in ciliary beating, whereas embryonic expression indicates that mechanisms other than ciliary movement are involved in LR specification.
As a component of the dynein motor complex, lrd is an excellent candidate for involvement in LR determination, based on previous studies of dynein function and models of the development of handed asymmetry. As a microtubule-based motor, dynein might provide an initial LR bias to the embryo by asymmetric movement along a microtubule scaffolding which is oriented relative to the anterior–posterior and dorsal–ventral axes. Organized microtubule arrays are required for axis development in several model organisms20–22. In Xenopus, for example, disruption of microtubule arrays in early embryos prevents cortical rotation and results in diminished dorsal–anterior development and randomized LR cardiac asymmetries22. Additionally, RNA encoding Vgl, a protein that can regulate Xenopus LR development23, is asymmetrically localized to the vegetal cortex of the embryo through association with microtubules24. These observations demonstrate the importance of microtubules in axis determination and the asymmetric localization of putative morphogens. Furthermore, the expression of lrd in the node is consistent with a role in LR specification, because several of the molecules implicated in LR development, such as nodal, HNF-3β and shh, are also expressed there. A model of LR development25 proposed the existence of handed molecules that are oriented with respect to the anterior–posterior and dorsal–ventral axes of the early embryo; these molecules would establish LR asymmetry by directional movements of morphogens. Based on our results showing that an axonemal dynein is mutated in iv, the handed molecules may be microtubules and dynein the molecular motor required to generate LR asymmetry.
Methods
Mice
The lgl mutation has been described.3 SI/Col-iv/iv mice were obtained from Jackson Labs. For embryo isolation, noon on the day of finding a copulation plug (after overnight breeding) was considered as embryonic day 0.5.
YAC cloning
F3, B4 and B6 YACs (Fig. 1a) were isolated as described26. B14, B24 and B25 YACs were isolated from the Princeton YAC library screened with B4 YAC sequences (5′ -GAAGAGCAGTCGGTGCTTTTACCAG-3′ and 5′ CAGACAGTTGTGAGCCACTGGATATA-3′ ), and C3 was isolated from the Baylor mouse YAC library using the same primers. YAC end-specific fragments were isolated as described26, and interspecific backcross chromosome mapping (Jackson Labs Backcross DNA Panel Map Service) was used to show that B14, B24 and B25 are chimaeric; the right end of C3 contains repetitive sequences and was therefore not mapped. Probes from the left end of F3, the right end of B4, and all non-repetitive probes isolated from C3 are deleted in lgl/lgl DNA.
lrd gene isolation
A 270-bp dynein P-loop probe was cloned from mouse brain cDNA with primers (5′ -AACCTTTGATTTCCAGGC-3′ and 5′ -ATGGT-GATGAAGATAGCC-3′ ) based on conserved sequences flanking the first P-loop of the human and rat cytoplasmic dynein genes (GenBank accession numbers L23958 and L08505). This probe hybridized at reduced stringency (62° ) to a 2.1-kb Bgl II fragment in C3, B14, B24 and B25 YAC DNAs. This cross-hybridizing fragment was cloned from a C3 YAC plasmid library and was used to screen adult mouse brain and testis cDNA libraries (Clontech) to isolate dyn-2 and dyn-22.2 cDNA clones.
The C3 YAC was used to clone additional cDNAs by direct cDNA selection as described27, with the following modifications. cDNA prepared from wild-type adult brain mRNA using oligo(dT) and random primers and the Marathon cDNA-amplification kit (Clontech) was digested with AluI, HaeIII or RsaI before linker addition and PCR amplification. cDNA was preblocked with 2 μ g Cot1 DNA (Gibco BRL) and 8 μ g lgl/lgl DNA to duplex repetitive and non-deleted sequences before hybridization. After two rounds of hybridization, selected cDNAs were subcloned into the pT7blue vector (Novagen).
RT-PCR using TaqStart Ab (Clontech) and Expand long-range PCR (Boehringer-Mannheim) was used to clone cDNAs connecting the library and direct selection cDNA sequences and to clone cDNAs from iv/iv mutant adult-brain RNA.
Chromosome mapping
Recombination mapping was done using the outcross/backcross panel previously described17. An 800-bp BstXI restriction fragment probe from dyn-2 cDNA detected a polymorphism between SI/Col and C57BL/6J DNA digested with PstI, allowing the C57BL/6J portion of the backcross panel to be used.
Mutation analysis
The region of lrd encompassing the mutation in iv was amplified using the following primers: dyn3.6 (5′ -ATGATAA-CAAGGTGCTGACCC-3′ ), dyn3.7 (5′ CAAAATGCCAGCTCTGGAGACC-3′ ), dyn 3.6A (5′ -GCCAGCAATGAACGAGTGGCCCTCAAACCT-3′ ), and dyn 3.7A (5′ -AGCTCTGGAAACCGTGGCTGGTGTGGCTGT-3′ ). Primers dyn3.6 and dyn3.7 were used for RT-PCR to clone fragments from wild-type (Outbred ICR mice, Harlan) and iv/iv adult brain RNA. Three different RT-PCR clones per genotype, representing two different RNA samples, were analysed. For confirmation, these primers were used to isolate PCR products from genomic DNA of wild-type mouse strains DBA, 129/J, C57BL/6J, C3H, SJL/J and Mus spretus, as well as iv/iv (SI/Col), for direct sequencing with primers dyn3.7 and dyn3.8 (genomic-specific: 5′ -TGAGTCTACCTGTGGCT-GTCC-3′ ). Additionally, primers dyn3.6A and dyn3.7A were used to clone PCR fragments from SI/Col-iv/iv and the wild-type strains (C57BL/6J, C3H, BALB/c, A/J, AKR/J, SPRET/Ei (Mus spretus), PANCEVO/Ei (Mus hortulanus), TIRANO/Ei (Mus domesticus) and rat (Rattus norvegicus). At least two independent clones per DNA were sequenced. DNAs were obtained from the Jackson Labs.
GenBank accession numbers for sequences shown in Fig. 2b: Anthocidaris crassispina (sea urchin) ciliary dynein-β heavy chain (D01021), Paramecium tetraurelia outer arm dynein-β heavy chain (U19464), Chlamydomonas reinhardtii dynein-β heavy chain (U02963), Saccharomyces cerevisiae cytoplasmic dynein heavy chain (Z21877), Caenorhabditis elegans dynein (Z75536), Rattus norvegicus cytoplasmic dynein heavy chain (L08505), Neurospora crassa cytoplasmic dynein heavy chain (P45443), Drosophila melanogaster cytoplasmic dynein heavy chain (P37276), Emericella nidulans cytoplasmic dynein (P45444), and Dictyostelium discoideum cytoplasmic dynein heavy chain (A44357).
RT-PCR
cDNA was prepared using SuperScript (Gibco BRL). PCR was done using Amplitaq Gold (Perkin-Elmer) and primers dyn3.3 (5′ -AGAGCTT-CATCCTCAAAGTTGTCC-3′ ) and dyn3.5 (5′ -CAACTGGCCACATACG-GATTC-3′ ) (94 ° C for 30 s, 65 ° C for 30 s, 72 ° C for 30 s; 42 cycles).
In situ hybridization
Serial-section in situ hybridization was performed as described28 with a cDNA probe from a non-conserved region of lrd (the most 3′ 694 nucleotides of the sequence indicated in Fig. 1b); this probe does not cross-hybridize with other dynein genes by Southern blot analysis.
Acknowledgments
We thank R. B. Vallee, M. A. Ghee and K. T. Vaughn for their comments and for providing a mouse cytoplasmic dynein cDNA probe; J. McGrath for assistance in embryo isolation; M. Lee for help with chromosome mapping; J. M. Corrales for help with cDNA cloning; K. Saalfeld, P. Groen and L. Artmeyer for in situ hybridization; A. Emley for photographic assistance; S. Bell for help with the expression studies; A. Horwich for comments and discussion; and the staff at the Keck Biotechnology Center at Yale University and at the University of Cincinnati DNA Core Facility for DNA sequencing. This work was supported by grants from the NIH (S.S.P. and D.M.S.), the American Heart Association, Ohio Division (D.P.W.), and The March of Dimes (M.B.)
References
- 1.Hummel KP, Chapman DB. Visceral inversion and associated anomalies in the mouse. J Hered. 1959;50:9–13. [Google Scholar]
- 2.Layton WM. Random determination of a developmental process. J Hered. 1976;67:336–338. doi: 10.1093/oxfordjournals.jhered.a108749. [DOI] [PubMed] [Google Scholar]
- 3.McNeish JD, et al. Phenotypic characterization of the transgenic mouse insertional mutation Legless. J Exp Zool. 1990;253:151–162. doi: 10.1002/jez.1402530205. [DOI] [PubMed] [Google Scholar]
- 4.Singh G, et al. legless insertional mutation: morphological, molecular, and genetic characterization. Genes Dev. 1991;5:2245–2255. doi: 10.1101/gad.5.12a.2245. [DOI] [PubMed] [Google Scholar]
- 5.Lowe LA, et al. Conserved left-right asymmetry of nodal expression and alterations in murine situs inversus. Nature. 1996;381:158–161. doi: 10.1038/381158a0. [DOI] [PubMed] [Google Scholar]
- 6.Meno C, et al. Left–right asymmetric expression of the TGFβ -family member lefty in mouse embryos. Nature. 1996;381:151–155. doi: 10.1038/381151a0. [DOI] [PubMed] [Google Scholar]
- 7.Lohr JL, Danos MC, Yost HJ. Left-right asymmetry of a nodal-related gene is regulated by dorsoanterior midline structures during Xenopus development. Development. 1997;124:1465–1472. doi: 10.1242/dev.124.8.1465. [DOI] [PubMed] [Google Scholar]
- 8.Afzelius BA. A human syndrome caused by immotile cilia. Science. 1976;193:317–319. doi: 10.1126/science.1084576. [DOI] [PubMed] [Google Scholar]
- 9.Afzelius BA. Situs inversus and ciliary abnormalities: What is the connection? Int J Dev Biol. 1995;39:839–844. [PubMed] [Google Scholar]
- 10.Holzbaur ELF, Vallee RB. Dyneins: molecular structure and cellular function. Annu Rev Cell Biol. 1994;10:339–372. doi: 10.1146/annurev.cb.10.110194.002011. [DOI] [PubMed] [Google Scholar]
- 11.Asai DJ. Multi-dynein hypothesis. Cell Motil Cytoskel. 1995;32:129–132. doi: 10.1002/cm.970320212. [DOI] [PubMed] [Google Scholar]
- 12.Tanaka Y, Zhang Z, Hirokawa N. Identification and molecular evolution of new dynein-like protein sequences in rat brain. J Cell Sci. 1995;108:1883–1893. doi: 10.1242/jcs.108.5.1883. [DOI] [PubMed] [Google Scholar]
- 13.Vaughan KT, et al. Multiple mouse chromosomal loci for dynein-based motility. Genomics. 1996;36:29–38. doi: 10.1006/geno.1996.0422. [DOI] [PubMed] [Google Scholar]
- 14.Andrews KL, Nettesheim P, Asai DJ, Ostrowski LE. Identification of seven rat axonemal dynein heavy chain genes: expression during ciliated cell differentiation. Mol Biol Cell. 1996;7:71–79. doi: 10.1091/mbc.7.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McGrath J, Horwich AL, Brueckner M. Duplication/deficiency mapping of situs inversus viscerum (iv), a gene that determines left–right asymmetry in the mouse. Genomics. 1992;14:643–648. doi: 10.1016/s0888-7543(05)80163-6. [DOI] [PubMed] [Google Scholar]
- 16.Asai DJ, et al. The dynein genes of Paramecium tetraurelia: sequencies adjacent to the catalytic P-loop identify cytoplasmic and axonemal heavy chain isoforms. J Cell Sci. 1994;107:839–847. doi: 10.1242/jcs.107.4.839. [DOI] [PubMed] [Google Scholar]
- 17.Brueckner M, D’Eustachio P, Horwich AL. Linkage mapping of a mouse gene, iv, that controls left–right asymmetry of the heart and viscera. Proc Natl Acad Sci USA. 1989;86:5035–5038. doi: 10.1073/pnas.86.13.5035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bellomo D, Lander A, Harragan I, Brown NA. Cell proliferation in mammalian gastrulation: the ventral node and notochord are relatively quiescent. Dev Dyn. 1996;205:471–485. doi: 10.1002/(SICI)1097-0177(199604)205:4<471::AID-AJA10>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 19.Vaisberg EA, Grissom PM, McIntosh JR. Mammalian cells express three distinct dynein heavy chains that are localized to different cytoplasmic organelles. J Cell Biol. 1996;133:831–841. doi: 10.1083/jcb.133.4.831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Theurkauf WE. Microtubules and cytoplasm organization during Drosophila oogenesis. Dev Biol. 1994;165:352–360. doi: 10.1006/dbio.1994.1258. [DOI] [PubMed] [Google Scholar]
- 21.Jesuthasan S, Strahle U. Dynamic microtubules and specification of the zebrafish embryonic axis. Curr Biol. 1996;7:31–42. doi: 10.1016/s0960-9822(06)00025-x. [DOI] [PubMed] [Google Scholar]
- 22.Danos MC, Yost HJ. Linkage of cardiac left-right asymmetry and dorsal–anterior development in Xenopus. Development. 1995;121:1467–1474. doi: 10.1242/dev.121.5.1467. [DOI] [PubMed] [Google Scholar]
- 23.Hyatt BA, Lohr JL, Yost HJ. Initiation of vertebrate left–right axis formation by maternal Vg1. Nature. 1996;384:62–65. doi: 10.1038/384062a0. [DOI] [PubMed] [Google Scholar]
- 24.Elisha Z, Havin L, Ringel I, Yisraeli JK. Vg1 RNA binding protein mediates the association of Vg1 RNA with microtubules in Xenopus oocytes. EMBO J. 1995;14:5109–5114. doi: 10.1002/j.1460-2075.1995.tb00193.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brown NA, McCarthy A, Wolpert L. Development of handed body asymmetry in mammals. Ciba Found Symp. 1991;162:182–201. doi: 10.1002/9780470514160.ch11. [DOI] [PubMed] [Google Scholar]
- 26.Supp DM, Witte DP, Branford WW, Smith EP, Potter SS. Sp4, a member of the Sp1-family of zinc finger transcription factors, is required for normal murine growth, viability, and male fertility. Dev Biol. 1996;176:284–299. doi: 10.1006/dbio.1996.0134. [DOI] [PubMed] [Google Scholar]
- 27.Segre JA, Nemhauser JL, Taylor BA, Nadeau JH, Lander ES. Positional cloning of the nude locus: genetic, physical, and transcription maps of the region and mutations in the mouse and rat. Genomics. 1995;28:549–559. doi: 10.1006/geno.1995.1187. [DOI] [PubMed] [Google Scholar]
- 28.Kern MJ, Witte DP, Valerius MT, Aronow BJ, Potter SS. A novel murine homeobox gene isolated by a tissue specific PCR cloning strategy. Nucleic Acids Res. 1992;20:5189–5195. doi: 10.1093/nar/20.19.5189. [DOI] [PMC free article] [PubMed] [Google Scholar]


