Abstract
Chromosomes segregate using their kinetochores, the specialized protein structures that are assembled on centromeric DNA and mediate attachment to the mitotic spindle. Because centromeric sequences are not conserved, centromere identity is propagated by an epigenetic mechanism. All eukaryotes contain an essential histone H3 variant (CenH3) that localizes exclusively to centromeres. Because CenH3 is required for kinetochore assembly and is likely to be the epigenetic mark that specifies centromere identity, it is critical to elucidate the mechanisms that assemble and maintain CenH3 exclusively at centromeres. To learn more about the functions and regulation of CenH3, we isolated mutants in the budding yeast CenH3 that are lethal when overexpressed. These CenH3 mutants fall into three unique classes: (I) those that localize to euchromatin but do not alter kinetochore function, (II) those that localize to the centromere and disrupt kinetochore function, and (III) those that no longer target to the centromere but still disrupt chromosome segregation. We found that a class III mutant is specifically defective in the ability of sister kinetochores to biorient and attach to microtubules from opposite spindle poles, indicating that CenH3 mutants defective in kinetochore biorientation can be obtained.
ACCURATE chromosome segregation is critical for the maintenance of genomic stability and depends on the coordination of a number of events (for review, see Cleveland et al. 2003). During S-phase, replicated chromosomes (sister chromatids) become physically linked by a protein complex called cohesin, which holds sisters together while the chromosomes attach to the mitotic spindle (for review, see Nasmyth and Haering 2005). Chromosomes attach to microtubules via the kinetochore, the protein complex that assembles on centromeric DNA. Prior to segregation, sister kinetochores must biorient such that they bind to microtubules emanating from opposite spindle poles. Once kinetochores are bioriented, the pulling forces of the microtubules on linked sister kinetochores generate physical tension that draws the sister chromatids away from each other. At this time, cohesin is cleaved, allowing the sister chromatids to be pulled to opposite poles.
The fidelity of microtubule attachments to kinetochores is monitored by the spindle checkpoint, a conserved signal transduction system consisting of the Mad, Bub, and Mps1 proteins (for review, see Lew and Burke 2003). The spindle checkpoint prevents the premature segregation of improperly attached chromosomes by inhibiting the activity of the anaphase-promoting complex, a ubiquitin ligase that targets the anaphase inhibitor Pds1/securin for destruction. Experiments in several organisms suggest that the spindle checkpoint can be activated by the lack of kinetochore–microtubule attachments or defects in the tension exerted by microtubule-generated forces on kinetochores (for review, see Pinsky and Biggins 2005). In budding yeast, the Ipl1/aurora protein kinase distinguishes between attachment and tension defects because it is required only for spindle checkpoint activation when kinetochores do not come under tension (Biggins and Murray 2001). This appears to be due to its role in destabilizing inappropriate microtubule attachments to create unattached kinetochores that can activate the checkpoint (Pinsky et al. 2006b).
The best-characterized kinetochore is in budding yeast where >65 kinetochore proteins have been identified (for reviews, see Biggins and Walczak 2003; McAinsh et al. 2003). Most yeast kinetochore proteins are found in biochemically distinct subcomplexes called CBF3, CTF19/COMA, MTW1/Mis12, NDC80, and DAM1/DDD/DASH. Although the structure of the kinetochore is not known, dependency relationships and physical interactions subdivide the kinetochore into inner, central, and outer domains. The inner kinetochore contains the CBF3 complex that is required for the localization of all kinetochore proteins (He et al. 2001), as well as the DNA-binding proteins Mif2 and Cbf1 and the yeast centromeric histone H3 (CenH3). The central kinetochore contains the MTW1/Mis12, NDC80, and CTF19/COMA complexes. The outermost kinetochore complex appears to be the DAM1/DDD/DASH complex, which requires microtubules and many other kinetochore subcomplexes for its localization (Enquist-Newman et al. 2001; Cheeseman et al. 2002; Janke et al. 2002; Li et al. 2002; Scharfenberger et al. 2003).
Although kinetochore function is conserved, the size and sequence of centromeric DNA is highly variable among organisms. In multicellular eukaryotes, centromeres are characterized by megabases of repetitive DNA that lack strict sequence identity (for reviews, see Choo 2001; Cleveland et al. 2003). Budding yeast is the only organism with a defined centromeric DNA sequence (125 bp) that is sufficient to mediate kinetochore assembly (Fitzgerald-Hayes et al. 1982). Due to the lack of a conserved centromeric DNA sequence, centromere identity in all eukaryotes is thought to be propagated epigenetically.
A hallmark of all kinetochores is a conserved CenH3, which has been proposed to be the epigenetic mark that specifies centromere identity because it creates a specialized centromeric chromatin structure essential for chromosome segregation. CenH3 proteins contain a carboxy-terminal histone fold domain that is highly homologous to histone H3, but have divergent amino-terminal tails (for review, see Malik and Henikoff 2003). CenH3 can replace histone H3 in nucleosomes in vitro (Yoda et al. 2000; Black et al. 2004; Furuyama et al. 2006) and exists in centromeric nucleosomes in vivo (for review, see Kamakaka and Biggins 2005). Consistent with a role in specifying centromere identity, CenH3 proteins are essential for the localization of most kinetochore proteins (Blower and Karpen 2001; Oegema et al. 2001; Goshima et al. 2003; Collins et al. 2005), and ectopically localized CenH3 can lead to kinetochore formation (Heun et al. 2006).
Given the essential role for CenH3 in centromere identity and chromosome segregation, we hypothesized that CenH3 mutants that are defective in proper localization or function may be lethal when overexpressed for at least three different reasons. First, CenH3 mutants that incorporate into euchromatin when overexpressed may be lethal because they form ectopic kinetochores or interfere with other chromatin-based processes. These mutants could elucidate details about the exclusive targeting of CenH3 to the centromere. Second, CenH3 mutants that localize to the centromere could lead to kinetochore defects when overexpressed and may therefore reveal additional details about the kinetochore functions of CenH3 proteins. Finally, CenH3 mutants could be lethal when overexpressed because they do not localize to the centromere and therefore titrate one or more proteins important for segregation away from chromosomes. Studies of these mutants may identify critical CenH3-interacting proteins, elucidate details about the requirements of targeting CenH3 to centromeres, and reveal novel functions for CenH3 at the kinetochore. Consistent with our predictions, we report the isolation of three classes of CenH3 mutants that are lethal when overexpressed. Our subsequent characterization of a class III mutant that does not localize to the centromere revealed that it leads to a defect in kinetochore biorientation.
MATERIALS AND METHODS
Microbial techniques:
Media and microbial techniques were essentially as described (Sherman et al. 1974; Rose et al. 1990). For all inductions, 4% galactose (GAL) was used. Because all of the mutant strains exhibited a maximal large-budded arrest after 6 hr in GAL, we performed all GAL inductions for 6 hr. All experiments in which cells were released from a G1 arrest were carried out by α-factor (αF) arrest and release, using αF at 1 μg/ml. Nocodazole was used at 10 μg/ml. Analog-sensitive Ipl1 experiments were performed as described (Pinsky et al. 2006a). 1NA-PP1 was used at 50 μm. Yeast strains used in this study are listed in Table 1 and are derived from the W303 background. CEN8-GFP consists of 256 lactose operators integrated at QCR10 on chromosome VIII (2.1 kb from CEN8) and is visualized by expression of a GFP fusion to the lactose repressor (Straight et al. 1996). Epitope tagging by the PCR integration technique was according to Longtine et al. (1998), and it generated fusions that are functional at the permissive temperature. All primer sequences are available upon request. All experiments were carried out at 23° with the exception of the chromatin immunoprecipitation (ChIP) on microarray experiment, which was conducted at 30°.
TABLE 1.
Yeast strains used in this study
| Strain | Genotype |
|---|---|
| RC65 | MATabar1 ura3-1 leu2-3,112 his3-11 trp1-1:pGAL-CSE4-353:TRP1 MCD1-HA6 can1-100 ade2-1 |
| RC66 | MATabar1 ura3-1 leu2-3,112 his3-11 trp1-1:pGAL-CSE4:TRP1 MCD1-HA6 can1-100 ade2-1 |
| SBY3 | MATabar1 ura3-1 leu2-3,112 his3-11 trp1-1 can1-100 ade2-1 |
| SBY617 | MATaCSE4:CSE4-myc12:URA3 bar1 ura3-1 leu2-3,112 his3-11 trp1-1 can1-100 ade2-1 |
| SBY818 | MATaPDS1-myc18:LEU2 bar1 ura3-1 leu2-3,112 his3-11:pCUP1-GFP12-LacI12:HIS3 trp1-1:256LacO:TRP1 lys2Δ can1-100 ade2-1 |
| SBY1071 | MATabar1 ura3-1:pGAL-myc13-CSE4:URA3 leu2-3,112 his3-11 trp1-1 can1-100 ade2-1 |
| SBY1737 | MATabar1 ura3-1 leu2-3,112 his3-11 trp1-1:pGAL-CSE4-350:TRP1 can1-100 ade2-1 |
| SBY1738 | MATabar1 ura3-1 leu2-3,112 his3-11 trp1-1:pGAL-CSE4-351:TRP1 can1-100 ade2-1 |
| SBY1739 | MATabar1 ura3-1 leu2-3,112 his3-11 trp1-1:pGAL-CSE4-352:TRP1 can1-100 ade2-1 |
| SBY1740 | MATabar1 ura3-1 leu2-3,112 his3-11 trp1-1:pGAL-CSE4-353:TRP1 can1-100 ade2-1 |
| SBY1741 | MATabar1 ura3-1 leu2-3,112 his3-11 trp1-1:pGAL-CSE4-356:TRP1 can1-100 ade2-1 |
| SBY1742 | MATabar1 ura3-1 leu2-3,112 his3-11 trp1-1:pGAL-CSE4-358:TRP1 can1-100 ade2-1 |
| SBY1743 | MATabar1 ura3-1 leu2-3,112 his3-11 trp1-1:pGAL-CSE4-359:TRP1 can1-100 ade2-1 |
| SBY1744 | MATabar1 ura3-1 leu2-3,112 his3-11 trp1-1:pGAL-CSE4-360:TRP1 can1-100 ade2-1 |
| SBY1745 | MATabar1 ura3-1 leu2-3,112 his3-11 trp1-1:pGAL-CSE4-361:TRP1 can1-100 ade2-1 |
| SBY1746 | MATabar1 ura3-1 leu2-3,112 his3-11 trp1-1:pGAL-CSE4-362:TRP1 can1-100 ade2-1 |
| SBY1775 | MATabar1 ura3-1 leu2-3,112 his3-11 trp1-1:pGAL-CSE4-354:TRP1 can1-100 ade2-1 |
| SBY1812 | MATabar1 ura3-1 leu2-3,112 his3-11 trp1-1:pGAL-CSE4:TRP1 can1-100 ade2-1 |
| SBY1893 | MATabar1 ura3-1 leu2-3,112 his3-11:pCUP1-GFP12-LacI12:HIS3 trp1-1:pGAL-CSE4-353:TRP1 can1-100 ade2-1 |
| SBY1894 | MATabar1 mad2ΔURA3 ura3-1 leu2-3,112 his3-11:pCUP1-GFP12-LacI12:HIS3 trp1-1:pGAL-CSE4-353:TRP1 can1-100 ade2-1 |
| SBY1949 | MATaura3-1:CSE4-myc12:URA3 bar1 leu2-3,112 his3-11 trp1-1:pGAL-CSE4-350:TRP1 can1-100 ade2-1 |
| SBY2640 | MATabar1 ura3-1 leu2-3,112 his3-11 trp1-1:pGAL-CSE4-352-myc13:TRP1:HIS3 can1-100 ade2-1 |
| SBY2567 | MATabar1 ura3-1 leu2-3,112 his3-11 trp1-1:pGAL-CSE4-353-myc13:TRP1:HIS3 can1-100 ade2-1 |
| SBY4278 | MATaMTW1-myc13:KanMX bar1 ura3-1 leu2-3,112 his3-11 trp1-1:pGAL-CSE4-353:TRP1 can1-100 ade2-1 |
| SBY4279 | MATaMIF2-myc13:KanMX bar1 ura3-1 leu2-3,112 his3-11 trp1-1:pGAL-CSE4-353:TRP1 can1-100 ade2-1 |
| SBY4280 | MATaCTF13-myc13:KanMX bar1 ura3-1 leu2-3,112 his3-11 trp1-1:pGAL-CSE4-353:TRP1 can1-100 ade2-1 |
| SBY4281 | MATaNDC80-myc13:KanMX bar1 ura3-1 leu2-3,112 his3-11 trp1-1:pGAL-CSE4-353:TRP1 can1-100 ade2-1 |
| SBY4282 | MATaCTF19-myc13:KanMX bar1 ura3-1 leu2-3,112 his3-11 trp1-1:pGAL-CSE4-353:TRP1 lys2Δ can1-100 ade2-1 |
| SBY4283 | MATaOKP1-myc13:KanMX bar1 ura3-1 leu2-3,112 his3-11 trp1-1:pGAL-CSE4-353:TRP1 can1-100 ade2-1 |
| SBY5014 | MATaPDS1-myc18:LEU2 bar1 ura3-1 leu2-3,112 his3-11 trp1-1:pGAL-CSE4-353:TRP1 can1-100 ade2-1 |
| SBY5050 | MATacdc20ΔLEU2 Chr8:CEN-LacO:TRP1 ura3-1 leu2-3,112 his3-11:pCUP1-GFP12-LacI12:HIS3 trp1-1:pGAL-CDC20:TRP1 can1-100 ade2-1 |
| SBY5210 | MATaMTW1-CFP3:URA3 DAM1-GFP3:URA3 cdc20ΔLEU2 ura3-1 leu2-3,112 his3-11 trp1-1:pGAL-CDC20:TRP1 can1-100 ade2-1 |
| SBY5223 | MATaMTW1-CFP3:URA3 DAM1-GFP3:URA3 ura3-1 leu2-3,112 his3-11 trp1-1:pGAL-CSE4-353:TRP1 can1-100 ade2-1 |
| SBY5313 | MATaChr8:CEN-LacO:TRP1 mad2ΔURA3 bar1 ura3-1:TUB1-CFP:URA3 leu2-3,112 his3-11:pCUP1-GFP12-LacI12:HIS3 trp1-1:pGAL-CSE4-353:TRP1 lys2Δ can1-100 ade2-1 (pRS315 LEU, CEN) |
| SBY5314 | MATaChr8:CEN-LacO:TRP1 mad2ΔURA3 bar1 ura3-1:TUB1-CFP:URA3 leu2-3,112 his3-11:pCUP1-GFP12-LacI12:HIS3 trp1-1 can1-100 ade2-1 (pRS315 LEU, CEN) |
| SBY5433 | MATaChr8:CEN-LacO:TRP1 bar1 ura3-1 leu2-3,112 his3-11:pCUP1-GFP12-LacI12:HIS3 trp1-1:pGAL-CSE4-358:TRP1 can1-100 ade2-1 |
| SBY5434 | MATaChr8:CEN-LacO:TRP1 bar1 ura3-1 leu2-3,112 his3-11:pCUP1-GFP12-LacI12:HIS3 trp1-1:pGAL-CSE4-359:TRP1 can1-100 ade2-1 |
| SBY5435 | MATaChr8:CEN-LacO:TRP1 bar1 ura3-1 leu2-3,112 his3-11:pCUP1-GFP12-LacI12:HIS3 trp1-1:pGAL-CSE4-361:TRP1 can1-100 ade2-1 |
| SBY5436 | MATaChr8:CEN-LacO:TRP1 bar1 ura3-1 leu2-3,112 his3-11:pCUP1-GFP12-LacI12:HIS3 trp1-1:pGAL-CSE4-362:TRP1 can1-100 ade2-1 |
| SBY5437 | MATaMTW1-GFP3:HIS3 SPC29-CFP:KanMX bar1 ura3-1 leu2-3,112 his3-11 trp1-1 can1-100 ade2-1 |
| SBY5446 | MATaMTW1-GFP3:HIS3 SPC29-CFP:KanMX bar1 ura3-1 leu2-3,112 his3-11 trp1-1:pGAL-CSE4-353:TRP1 can1-100 ade2-1 |
| SBY5455 | MATaipl1ΔKanMX:ipl1-as5:LEU2 PDS1-myc18:LEU2 bar1 ura3-1 leu2-3,112 his3-11:pCUP1-GFP12-LacI12:HIS3 trp1-1:pGAL-CSE4-353:TRP1 can1-100 ade2-1 |
| SBY5456 | MATaipl1ΔKanMX:ipl1-as5:LEU2 PDS1-myc18:LEU2 bar1 ura3-1 leu2-3,112 his3-11 trp1-1 can1-100 ade2-1 |
| SBY5621 | MATaChr8:CEN-LacO:TRP1 bar1 ura3-1 leu2-3,112 his3-11:pCUP1-GFP12-LacI12:HIS3 trp1-1:pGAL-CSE4-351:TRP1 can1-100 ade2-1 |
| SBY5622 | MATaChr8:CEN-LacO:TRP1 bar1 ura3-1 leu2-3,112 his3-11:pCUP1-GFP12-LacI12:HIS3 trp1-1:pGAL-CSE4-354:TRP1 can1-100 ade2-1 |
All strains were generated for this study and are isogenic with the W303 background.
Plasmid construction:
The pGAL-CSE4, 2μ plasmid used in the screen for CSE4 mutants (pSB465) was constructed by cloning 500 bp downstream of CSE4 into a pGAL-CSE4 TRP1 2μ plasmid (pSB462). Primers SB338 and SB339 that contain engineered SacII/SacI sites, respectively, were used to PCR amplify the downstream 500 bp of CSE4. The PCR product was digested with SacII and SacI and ligated into the same sites of pSB462 to create pSB465. The addition of the downstream 500 bp of noncoding sequence to the pGAL-CSE4 plasmid resulted in a mild increase in Cse4p levels in galactose media (K. A. Collins and S. Biggins, data not shown).
The integrating plasmid that overexpresses CSE4 (pSB571) was constructed by cloning the KpnI/SacI fragment containing pGAL-CSE4 with the 500 bp downstream sequence from pSB465 into the same restriction sites of pRS304, a TRP1-integrating vector (Sikorski and Hieter 1989). Ten of the mutants were subcloned onto a TRP1-integrating plasmid by digesting the 2μ parent plasmids with KpnI/SacI and ligating the pGAL-CSE4 mutant fragments into pRS304 at the same sites (Sikorski and Hieter 1989). The following 2μ parent plasmids were used to generate the indicated integrating plasmids: pSB530 (pGAL-CSE4-350) to generate pSB543, pSB531 (pGAL-CSE4-351) to generate pSB544, pSB532 (pGAL-CSE4-352) to generate pSB545, pSB533 (pGAL-CSE4-353) to generate pSB544, pSB534 (pGAL-CSE4-354) to generate pSB556, pSB538 (pGAL-CSE4-358) to generate pSB554, pSB539 (pGAL-CSE4-359) to generate pSB547, pSB540 (pGAL-CSE4-360) to generate pSB548, pSB541 (pGAL-CSE4-361) to generate pSB555, and pSB542 (pGAL-CSE4-362) to generate pSB549. pSB536 (pGAL-CSE4-356) was digested with KpnI/PstI and cloned into the same sites of pRS304 to generate pSB536.
To generate integrating vectors to test for complementation of a CSE4 null strain, CSE4-353 and CSE4-354 were put under the control of the endogenous CSE4 promoter on URA3 integrating vectors. CSE4-353 was first cloned into a pGAL, URA3 vector by digesting pSB553 with XhoI/SacI and ligating the fragment into the same sites of pSB157 to create pSB973. The GAL1 promoter was then removed from pSB973 by digesting with KpnI/EcoRI and replaced with the CSE4 promoter (obtained from pSB887) using the same restriction sites to create pSB1079. The CSE4-354 plasmid was also constructed in two steps. First, pSB556 was digested with KpnI/EcoRI to remove the GAL1 promoter, and the CSE4 promoter (obtained from pSB887) was ligated using the same sites to create pSB1078. This plasmid was then digested with KpnI/SacI and ligated into the same sites of pRS306 (Sikorski and Hieter 1989) to generate pSB1080.
The isolation of pGAL-CSE4 mutants that are lethal when overexpressed:
Error-prone PCR was performed with primers SB340 and SB373 to amplify the CSE4 gene according to Castillo et al. (2002). Gap repair was used to recombine the plasmid backbone with the PCR product (Orr-Weaver et al. 1983). Specifically, pSB465 (pGAL-CSE4, 2μ, TRP1) was digested with EcoRI and SacII and then cotransformed with the PCR products into a wild-type yeast strain (SBY3). The PCR product and the gapped plasmid were titrated to determine the optimal concentrations for stimulating recombination in vivo. Transformants were selected on glucose (GLU) media without tryptophan and then replica plated to GLU and GAL media lacking tryptophan and glycerol plates. Glycerol plates were used to eliminate petite mutants from the screen (Wallis et al. 1972). Plates were incubated at 23° and scored for growth after 2 days. In total, ∼100,000 transformants were screened. Candidate mutants were colonies that grew on GLU and glycerol plates but died or showed reduced viability on GAL plates. To confirm that the lethal phenotype was dependent on the plasmid, the plasmids were isolated from candidate mutants by standard methods (Rose et al. 1990) and retested in SBY3. Thirteen mutants were reproducibly lethal on GAL plates. Mutants were then subcloned from 2μ plasmids to integrating plasmids and tested for viability on GAL media. Of the 13 mutants, 11 conferred a lethal phenotype when integrated at the TRP1 locus and were named CSE4-350-CSE4-354, CSE4-356, and CSE4-358-CSE4-362. Plasmids were sequenced with primer SB373.
Protein and immunological techniques:
Protein extracts were made and immunoblotted as described (Minshull et al. 1996). 9E10 antibodies (Covance, Princeton, NJ) that recognize the Myc tag were used at a 1:10,000 dilution. Anti-Tub1p antibodies that recognize tubulin were used at 1:1000 (Accurate Chemical & Scientific, Westbury, NY). Anti-rabbit Cse4p antibodies (Pinsky et al. 2003) were used at 1:500.
Chromatin immunoprecipitation experiments:
ChIP analysis was performed as described (Collins et al. 2005) with the exception that threefold serial dilutions of the crude lysates (input) and immunoprecipitated (IP) DNA are shown in all figures. Primers used to amplify HMR locus (315 bp) were SB208 and SB209 and primers for TEL6 (273 bp) were SB206 and SB207. Chromatin immunoprecipitation on microarray was according to Glynn et al. (2004) with the exception that a 12CA5 HA antibody (Roche Applied Science, Indianapolis) was used for the IP.
Microscopy and flow cytometry:
Flow cytometry was performed as described (Collins et al. 2005). All microscopy experiments were carried out on cells that were fixed (Biggins et al. 1999; Collins et al. 2005) and at least 200 cells were analyzed for all reported experiments. The Bernoulli formula was used to calculate the 99% confidence interval. Microscopy was performed with a Photometrics Cascade 512B camera (Photometrics, Tuscon, AZ) and acquired with MetaMorph software (Molecular Devices, Downington, PA). Indirect immunofluorescence was performed as described (Rose et al. 1990).
RESULTS
The isolation of pGAL-CSE4 mutants that are lethal when overexpressed:
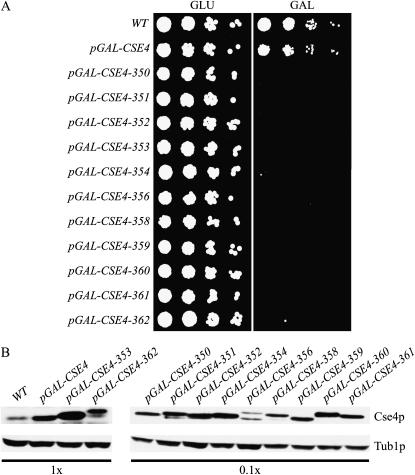
We hypothesized that mutants defective in the localization or functions of the budding yeast CenH3, Cse4p, would be lethal when overexpressed. Cells overexpressing the CSE4 gene do not have a detectable growth defect (Collins et al. 2004), so we performed error-prone PCR on CSE4 that was under the control of the inducible galactose promoter (pGAL-CSE4) on a high-copy plasmid (2μ) to maximize overexpression. The mutagenized plasmids were transformed into wild-type cells and selected on plates containing GLU to repress pGAL-CSE4 expression. Mutants were identified by isolating transformants that died when replica plated to GAL media. After screening >100,000 yeast transformants, we identified 11 pGAL-CSE4 mutants (pGAL-CSE4-350–354, -356, -358–362) that grow similarly to wild-type cells on GLU media but are lethal on GAL media (Figure 1A). Sequencing of the pGAL-CSE4 mutants and subsequent characterization studies revealed that each allele contains multiple mutations that are required for the lethal phenotype (supplemental Figure 1 at http://www.genetics.org/supplemental/; data not shown). We tested whether overexpression of the mutant proteins was required to generate the lethal phenotype by placing the CSE4-351, CSE4-353, and CSE4-354 genes under the control of the endogenous CSE4 promoter. We found that CSE4-351 and CSE4-353 could not support viability, while CSE4-354 cells were fully viable in the absence of the wild-type Cse4 protein. This indicates that at least one of the mutants requires overexpression to have a phenotypic consequence for the cell.
Figure 1.—
The isolation of pGAL-CSE4 mutants that are lethal when overexpressed. (A) Fivefold serial dilutions of wild-type (SBY3), pGAL-CSE4 (SBY1812), pGAL-CSE4-350 (SBY1737), pGAL-CSE4-351 (SBY1738), pGAL-CSE4-352 (SBY1739), pGAL-CSE4-353 (SBY1740), pGAL-CSE4-354 (SBY1775), pGAL-CSE4-356 (SBY1741), pGAL-CSE4-358 (SBY1742), pGAL-CSE4-359 (SBY1743), pGAL-CSE4-360 (SBY1744), pGAL-CSE4-361 (SBY1745), and pGAL-CSE4-362 (SBY1746) cells were incubated on GLU or GAL media at 23°. Although the overexpression of wild-type pGAL-CSE4 has no growth defect, overexpression of the pGAL-CSE4 mutants is lethal. (B) Most Cse4 mutant proteins are expressed at 10- to 100-fold higher levels than the wild-type Cse4 protein. Strains in A were induced with GAL for 6 hr and harvested for immunoblot analysis with anti-Cse4p and anti-Tub1p antibodies (loading control). To normalize for loading, 0.1× was loaded for the indicated Cse4p immunoblots.
We previously found that Cse4p is degraded by ubiquitin-mediated proteolysis (Collins et al. 2004), so we tested whether the mutations altered the steady-state levels of Cse4p. We found that 10 of the mutant strains had higher steady-state levels of Cse4p when induced with GAL (∼10- to 100-fold higher), while the levels of Cse4-362p were more similar to the overexpression of the wild-type Cse4 protein (Figure 1B). Due to the multiple mutations in each allele, there was variation in the mobility of the corresponding proteins. We confirmed that the levels of endogenous Cse4p were not altered by the overexpression of the mutant proteins by epitope tagging the mutant and endogenous proteins (K. A. Collins and S. Biggins, data not shown).
Phenotypic analyses divide the pGAL-CSE4 mutants into three classes:
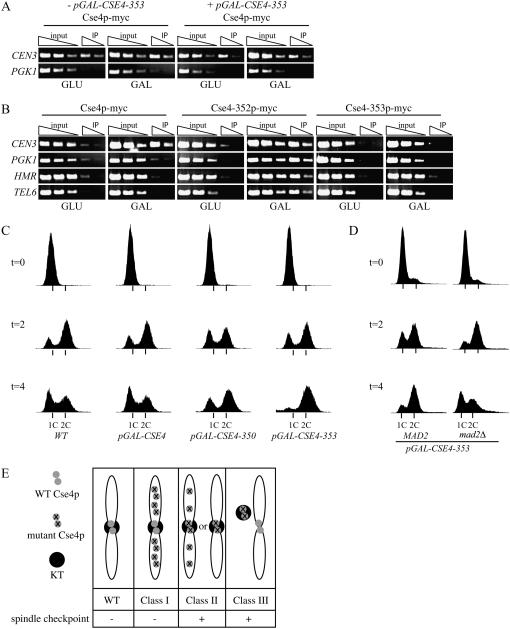
We hypothesized that the pGAL-CSE4 mutants could be categorized on the basis of the localization of the mutant and endogenous Cse4 proteins, as well as the phenotype due to overexpression of the mutant proteins. We therefore analyzed the localization of the endogenous wild-type Cse4 protein that was tagged with 13 myc epitopes (Cse4p-myc) by ChIP following overexpression of the mutant proteins. Cse4p-myc strains containing untagged pGAL-CSE4 mutants were grown in GLU or GAL for 6 hr and harvested for ChIP with anti-myc antibodies. Centromere localization was assayed by PCR with primers to CEN3, and PGK1 was used as a negative control locus (Westermann et al. 2003). In all cases, the endogenous Cse4p-myc protein was detected at the centromere but not at the control locus in the presence and absence of GAL (Figure 2A; Table 2). Therefore, when analyzed by the ChIP assay, the endogenous Cse4 protein still localized to the centromere when all of the mutants were overexpressed.
Figure 2.—
pGAL-CSE4 mutants can be divided into three classes on the basis of localization and spindle checkpoint activation. (A) The localization of wild-type Cse4p in cells overexpressing pGAL-CSE4-353. Cse4p-myc cells with (SBY1949) or without pGAL-CSE4-353 (SBY617) were induced with GAL for 6 hr and harvested for ChIP analysis with primers to CEN3 and PGK1. Threefold serial dilutions of input and IP are shown. The wild-type Cse4 protein still localizes to the centromere in cells overexpressing the Cse4-353 mutant protein. (B) Localization of the Cse4-352 and Cse4-353 mutant proteins. Cells containing pGAL-CSE4-myc (SBY1071), pGAL-CSE4-352-myc (SBY2640), or pGAL-CSE4-353-myc (SBY2567) were grown and processed for ChIP as in A. The Cse4-352 mutant protein localizes to CEN3, PGK1, HMR, and TEL6, while the Cse4-353 mutant protein does not localize to any of these loci. (C) FACS analysis of cells overexpressing pGAL-CSE4-350 and pGAL-CSE4-353. Wild-type (SBY3), pGAL-CSE4 (SBY1812), pGAL-CSE4-350 (SBY1737), and pGAL-CSE4-353 (SBY1740) cells were arrested in G1, released into GAL media, and harvested at the indicated times (hours) for FACS analysis. Wild-type and pGAL-CSE4 cells progress normally through the cell cycle, while cells overexpressing pGAL-CSE4-353 arrest in G2/M. Cells overexpressing pGAL-CSE4-350 show an increase in G2/M cells. (D) Cells overexpressing pGAL-CSE4-353 arrest due to spindle checkpoint activation. pGAL-CSE4-353 (SBY1893) and pGAL-CSE4-353 mad2Δ (SBY1894) cells were grown as in A and FACS was performed. (E) Schematic of the classes of pGAL-CSE4 mutants obtained. Class I mutants are defined by the localization of mutant Cse4p to euchromatin and by the absence of spindle checkpoint activation. Class II mutants are defined by Cse4p mutant localization to the centromere and activation of the spindle checkpoint. In this class, the endogenous Cse4 protein may or may not localize to the centromere and the mutant protein may or may not localize to euchromatin. Only two possibilities are shown. Class III mutants are defined by the absence of mutant Cse4p throughout the genome and activation of the spindle checkpoint.
TABLE 2.
Summary of the classes of pGAL-CSE4 mutants
| Class | Localization of endogenous Cse4p to CENa | Localization of mutant to CENa | Localization of mutant to euchromatina | Spindle checkpoint activationb | |
|---|---|---|---|---|---|
| Phenotype | I | + | ± | + | − |
| Phenotype | II | + | + | ± | + |
| Phenotype | III | + | − | ± | + |
| pGAL-CSE4-350 | I | + | ± | + | − |
| pGAL-CSE4-351 | I | + | − | + | − |
| pGAL-CSE4-352 | II | + | + | + | + |
| pGAL-CSE4-353 | III | + | − | − | + |
| pGAL-CSE4-354 | II | + | + | ± | + |
| pGAL-CSE4-356 | II | + | + | ± | + |
| pGAL-CSE4-358 | III | + | − | ± | + |
| pGAL-CSE4-359 | III | + | − | − | + |
| pGAL-CSE4-360 | II | + | + | ± | + |
| pGAL-CSE4-361 | III | + | − | ± | + |
| pGAL-CSE4-362 | III | + | − | − | + |
Summary of the localization of endogenous and mutant proteins is based on ChIP analyses. For pGAL-CSE4-351, ChIP on chip experiments indicated that Cse4-351p does not localize to the centromere in contrast to ChIP results.
Spindle checkpoint activation was determined by FACS analysis in the presence and absence of MAD2.
We next analyzed the localization of the mutant proteins that were epitope tagged with myc13 at the carboxy terminus and overexpressed from the galactose promoter. All mutants still conferred a lethal phenotype when epitope tagged (K. A. Collins and S. Biggins, data not shown). Cells containing the mutant proteins were grown in GLU or GAL for 6 hr and harvested for ChIP with anti-myc antibodies. The localization of the mutant proteins to CEN3, a silent mating-type locus (HMR), telomere VI (TEL6), and the PGK1 euchromatin control locus was analyzed by PCR (Collins et al. 2005) (Figure 2B; Table 2). Six of the mutant proteins (Cse4-350–352, -354, -356, and -360) localized to the centromere while the remaining five mutants did not (Cse4-353, -358 -359, -361 and -362). Strikingly, eight of the Cse4 mutant proteins could also be detected at the noncentromeric loci HMR, TEL6, and PGK1 by ChIP (Cse4-350-352, -354, -356, -358, -360 and -361) (Figure 2B; Table 2).
To further classify the mutants, we tested whether their overexpression caused a cell cycle arrest due to spindle checkpoint activation. Mutants that disrupt kinetochore function should arrest in metaphase in a spindle-checkpoint-dependent manner, whereas mutants that alter euchromatic processes would not activate the spindle checkpoint. We previously found that overexpression of the pGAL-CSE4-351 mutant causes cells to accumulate in M-phase in a checkpoint-independent manner (Collins et al. 2004). We therefore determined whether the remaining mutants arrest in M-phase by arresting cells in G1, releasing them into GAL media, and harvesting them for FACS and microscopy analyses. All of the mutants arrested as large-budded cells with a single mass of DNA and a 2C DNA content within 4 hr of GAL addition with the exception of pGAL-CSE4-350 (Figure 2C and K. A. Collins, and S. Biggins, data not shown). We tested whether the cells can recover from the arrest by returning the mutant cells to glucose after a 6-hr exposure to GAL. We found that CSE4-350, -351, -352, -356, -359, and -361 cannot form viable colonies when returned to glucose media, while CSE4-353, -354, -358, -360, and -362 form a reduced number of viable colonies (K. A. Collins and S. Biggins, data not shown). To determine if the mutants arrested in M-phase due to spindle checkpoint activation, we performed the same experiment in the absence of the spindle checkpoint gene, MAD2. In all cases, the mitotic arrest was dependent on the spindle checkpoint (Figure 2D; Table 2).
Taken together, our analyses of the localization of the endogenous and Cse4 mutant proteins, combined with the phenotypic data, allowed us to categorize the mutants into three classes (Figure 2E; Table 2). We defined class I mutants (pGAL-CSE4-350 and -351) as those in which the overexpressed mutant Cse4 protein localized to the centromere and euchromatin but did not lead to spindle checkpoint activation. These mutants are therefore unlikely to alter kinetochore function and likely to interfere with chromatin-based processes such as transcription and replication. The class II mutants (pGAL-CSE4-352, -354, -356, and -360) were categorized on the basis of the localization of the mutant protein to the centromere and activation of the spindle checkpoint. These data suggest that these mutants interfere with kinetochore function when overexpressed. The mutant class II proteins also localized to euchromatin, although they exhibited weaker euchromatic localization than the class I mutants. The class III mutants (pGAL-CSE4-353, -358, -359, -361, and -362) were classified as those in which the overexpressed Cse4 mutant protein did not localize to the centromere and the spindle checkpoint was activated. These mutants could therefore titrate away one or more proteins important for chromosome segregation away from the kinetochore. These mutant proteins failed to localize to euchromatin, with the exception of Cse4-358p and Cse4-361p, which exhibited slight euchromatic localization.
Cells overexpressing pGAL-CSE4-353 do not appear to alter the localization of kinetochore or cohesin proteins:
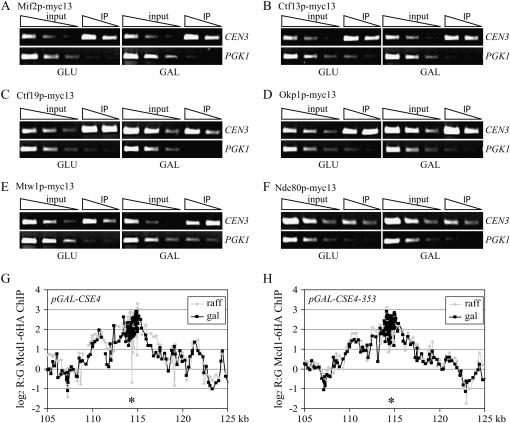
Because the class III mutants are the first reported examples of Cse4 mutant proteins that do not localize to the centromere, we further investigated the phenotypes associated with one of these mutants, pGAL-CSE4-353. In budding yeast, Cse4p is required for kinetochore assembly and the recruitment of pericentric cohesin to exogenous kinetochores (Tanaka et al. 1999; Collins et al. 2005). We therefore hypothesized that Cse4-353p overexpression is lethal because it titrates kinetochore or cohesin proteins away from the chromosome. To test this, we analyzed the localization of a representative member of each kinetochore subcomplex as well as a member of the cohesin complex in the presence and absence of Cse4-353p overexpression. First, we analyzed the localization of inner and central kinetochore proteins in cells overexpressing pGAL-CSE4-353 by ChIP. Our analysis included Mif2p, Ctf13p (of the CBF3 complex), Ctf19p and Okp1p (of the CTF19/COMA complex), Mtw1p (of the MTW1 complex), and Ndc80p (of the NDC80 complex). Cells expressing pGAL-CSE4-353 and a myc13 epitope-tagged kinetochore protein were grown in GLU or GAL media for 6 hr and harvested for ChIP analysis. We found that Mif2p (Figure 3A), Ctf13p (Figure 3B), Ctf19p (Figure 3C), Okp1p (Figure 3D), Mtw1p (Figure 3E), and Ndc80p (Figure 3F) localized to CEN3 in pGAL-CSE4-353 cells grown in either GLU or GAL, suggesting that the inner and central kinetochore complexes are intact. Because it is difficult to reliably analyze outer kinetochore protein localization by the ChIP technique, we performed fluorescence microscopy on cells expressing the outer kinetochore protein Dam1p fused to three copies of the green fluorescent protein (GFP; Dam1-GFP3). We found that Dam1-GFP3 always colocalized with the kinetochore protein Mtw1-CFP in cells overexpressing pGAL-CSE4-353 (supplemental Figure 2 at http://www.genetics.org/supplemental/). Because Dam1p requires other kinetochore subcomplexes for its kinetochore localization (Enquist-Newman et al. 2001; Cheeseman et al. 2002; Janke et al. 2002; Li et al. 2002; Scharfenberger et al. 2003), our data strongly suggest that the kinetochores are intact when Cse4-353p is overexpressed.
Figure 3.—
Kinetochore assembly and cohesin localization are normal in cells overexpressing pGAL-CSE4-353. (A–F) pGAL-CSE4-353 cells containing the indicated myc epitope-tagged kinetochore protein were grown in GLU or GAL media for 6 hr and harvested for ChIP analysis with primers to CEN3 and PGK1. Threefold serial dilutions of input and IP are shown. Mif2-myc13 (A, SBY4279), Ctf13-myc13 (B, SBY4280), Ctf19-myc13 (C, SBY4282), Okp1-myc13 (D, SBY4283), Mtw1-myc13 (E, SBY4278), and Ndc80-myc13 (F, SBY4281) all localize to kinetochores in the presence or absence of pGAL-CSE4-353 overexpression. (G and H) ChIP on microarray analysis of the cohesin Mcd1p in cells overexpressing pGAL-CSE4-353. Cells containing Mcd1-6HA that expressed pGAL-CSE4 (RC66) or pGAL-CSE4-353 (RC65) were grown in GAL and nocodazole and ChIP on chip was performed to localize Mcd1/Scc1 throughout the genome. The genomic region around CEN3 (*) is shown and indicates that there is no significant difference in the localization of Mcd1p when Cse4-353p is overexpressed.
Because kinetochore proteins did not appear to be mislocalized in the presence of high levels of Cse4-353p, we analyzed the localization of the Mcd1/Scc1 component of the cohesin complex using a highly sensitive chromatin IP on microarray approach (Glynn et al. 2004; Weber et al. 2004). Cells overexpressing pGAL-CSE4-353 arrest in metaphase, so we added nocodazole to the pGAL-CSE4 and pGAL-CSE4-353 cells containing Mcd1p-6HA in addition to GLU or GAL to compare metaphase-arrested populations of cells. Mcd1p-6HA was immunoprecipitated and the associated DNA was hybridized to microarrays to analyze the localization of cohesin throughout the genome. There was no significant change in the localization of Mcd1p in cells overexpressing Cse4p or Cse4-353p compared to wild-type cells (Figure 3, G and H; data not shown), indicating that Cse4-353p does not titrate cohesin away from the chromosomes.
Kinetochore clusters are farther apart in cells overexpressing pGAL-CSE4-353:
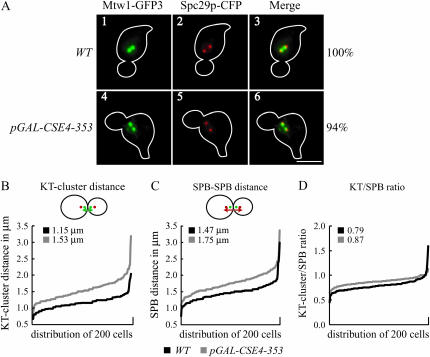
In budding yeast, kinetochores cluster into two foci when visualized by fluorescence microscopy due to the pulling forces of microtubules on linked sister chromatids (Goshima and Yanagida 2000; He et al. 2000; Tanaka et al. 2000; Pearson et al. 2001). During our microscopy analysis of Dam1-GFP3 localization, we noted that kinetochore clusters often appeared to be closer to the spindle poles in cells overexpressing pGAL-CSE4-353 compared to wild-type cells. We therefore further analyzed the position of kinetochores relative to the poles in cells overexpressing pGAL-CSE4-353 by localizing the kinetochores with a GFP fusion to the Mtw1 kinetochore protein (Mtw1-GFP3) and the spindle pole bodies (SPBs) with a cyan fluorescent protein (CFP) fusion to the SPB protein Spc29 (Spc29-CFP). Wild-type and cells overexpressing pGAL-CSE4-353 were arrested in G1 with the mating pheromone αF and released into GAL media. We analyzed the cells by microscopy every 10 min after release and found that >75% of wild-type and pGAL-CSE4-353 cells were in metaphase 100 min after release from G1. Consistent with our observations of cells containing Dam1-GFP3, the kinetochore clusters marked with Mtw1-GFP3 appeared farther apart in cells overexpressing Cse4-353p compared to wild-type cells (Figure 4A; compare panel 4 to panel 1).
Figure 4.—
Cells overexpressing pGAL-CSE4-353 have an increased distance between kinetochore clusters. Wild-type cells (SBY5437) and pGAL-CSE4-353 cells (SBY5446) expressing the kinetochore protein Mtw1-GFP3 and the SPB protein Spc29-CFP were arrested in G1 with αF, released into the cell cycle in GAL media, and analyzed 100 min after release from G1 by microscopy. (A) The Mtw1-GFP3 kinetochore clusters appear closer to the SPBs in pGAL-CSE4-353 cells compared to wild-type cells (compare panels 6 and 3). Bar, 5 μm. (B–D). Histograms of 200 cells of each strain indicate the following: (B) the distance between kinetochore clusters (KT-cluster distance), (C) the distance between SPBs (SPB–SPB distance), and (D) the ratio of the kinetochore cluster distance to SPB distance (KT/SPB normalized). The average of 200 cells is indicated. Cells overexpressing pGAL-CSE4-353 have a significantly greater distance between kinetochore clusters (P < 0.01).
To further quantify the difference in wild-type vs. pGAL-CSE4-353 cells, we measured the distance between the kinetochore clusters (KT-cluster distance) and the pole-to-pole distance (SPB–SPB distance) in 200 cells for each strain 100 min after release from G1. On average, the KT-cluster distance in cells overexpressing pGAL-CSE4-353 (1.53 μm) was significantly greater (P < 0.01) than that of the control cells (1.15 μm, Figure 4B). Similarly, the average SPB–SPB distance was significantly greater in cells overexpressing pGAL-CSE4-353 (1.75 μm) than in wild-type cells (1.47 μm, Figure 4C). Because both the KT-cluster distance and the SPB–SPB distance were greater in the mutant cells, we normalized the distances by dividing the KT-cluster distance by the SPB–SPB distance (KT/SPB ratio) for each cell and plotted the distribution of ratios (Figure 4D). A normalized ratio of 1.0 indicates that the KT-cluster distance is equal to the SPB–SPB distance, while a ratio <1 indicates that the KT clusters are closer together than the SPB–SPB distance. We found that the KT/SPB ratio in nearly all 200 cells overexpressing pGAL-CSE4-353 was significantly closer (P < 0.01) to 1.0 than that of the control cells (0.87 vs. 0.79), indicating that high levels of Cse4-353 cause the kinetochores to cluster closer to the spindle poles.
Cells overexpressing pGAL-CSE4-353 are defective in kinetochore biorientation:
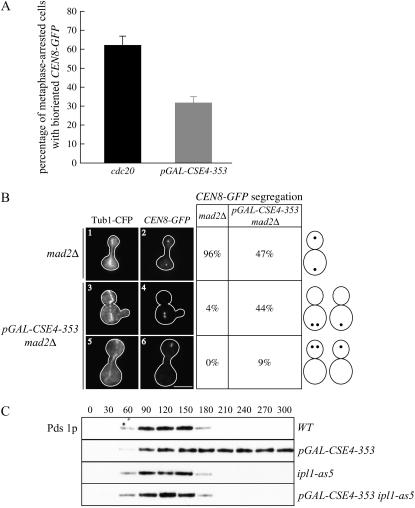
The distance between kinetochore clusters in metaphase cells is thought to result from the balance of microtubule-pulling forces on bioriented kinetochores that are opposed by the linkage between sister chromatids. We therefore considered the possibility that the increased KT-cluster distance in cells overexpressing pGAL-CSE4-353 could be due to the presence of mono-oriented kinetochore attachments. Mono-oriented sister kinetochores are attached to only one spindle pole, so they cannot resist the pulling forces of microtubules and therefore are pulled closer to that pole. In addition, this could lead to the spindle poles separating farther apart, another phenotype that we detected in cells overexpressing Cse4-353p. We therefore tested whether cells overexpressing pGAL-CSE4-353 have a biorientation defect by analyzing the separation of a single pair of sister kinetochores at metaphase by fluorescently marking CEN8-GFP (integrated 2.1 kb away from CEN8). As a control to compare metaphase-arrested populations, we depleted the anaphase-promoting complex component Cdc20. Cells containing two distinct CEN8-GFP signals were scored as bioriented, while cells with a single CEN8-GFP signal were considered mono-oriented. We found that 62% of the cdc20 cells contained bioriented CEN8 compared to only 32% of the pGAL-CSE4-353 mutant cells (Figure 5A), suggesting that Cse4-353p overexpression leads to a biorientation defect. Consistent with this, all of the other class III mutants were also defective in biorientation during the metaphase arrest, but a representative class I and class II mutant were not (supplemental Figure 3 at http://www.genetics.org/supplemental/).
Figure 5.—
Cells overexpressing pGAL-CSE4-353 are defective in kinetochore biorientation. (A) Analysis of biorientation in metaphase-arrested cells. pGAL-CDC20 (SBY5050) and pGAL-CSE4-353 cells (SBY5104) containing CEN8-GFP were grown in GLU or GAL media, respectively, for 6 hr. Cells with one CEN8 focus were scored as mono-oriented and cells with two distinct CEN8 foci were scored as bioriented. (B) Analysis of the segregation of a pair of sister chromatids in cells overexpressing pGAL-CSE4-353. Mad2Δ cells with (SBY5313) or without pGAL-CSE4-353 (SBY5314) that contain CEN8-GFP and Tub1-CFP were arrested in G1, released into GAL media, and scored for CEN8 segregation after 150 min. Bar, 5 μm. Representative pictures of sister-chromatid segregation and spindle elongation are shown at t = 150 min as well as quantification of the classes of phenotypes scored. Although spindles elongate in mad2Δ cells overexpressing pGAL-CSE4-353, 53% of the cells segregate both sisters to the same pole. (C) Cells overexpressing pGAL-CSE4-353 require the Ipl1/aurora protein kinase to activate the spindle checkpoint. Wild-type (SBY818), pGAL-CSE4-353 (SBY5014), ipl1-as5 (SBY5456), and pGAL-CSE4-353 ipl1-as5 (SBY5455) cells that contain Pds1-myc18 were arrested in G1 with αF and released into GAL media. The inhibitor 1NA-PP1 was added 80 min after release when small budded cells were first visible. Cells were harvested for immunoblot every 30 min and analyzed for Pds1p and Tub1p levels (data not shown).
To determine whether the biorientation defect results in lethal chromosome missegregation at anaphase, we analyzed the segregation of chromosome VIII in cells overexpressing pGAL-CSE4-353 that were allowed to progress through the cell cycle by deleting the MAD2 spindle checkpoint gene. Mad2Δ and pGAL-CSE4-353 mad2Δ cells containing CEN8-GFP and Tub1-CFP were arrested in G1 and released into GAL media. A total of 150 min after release, cells that had elongated their spindles as assayed by Tub1-CFP staining were scored for the segregation of CEN8-GFP. Although 96% of mad2Δ cells segregated CEN8-GFP to opposite poles, only 47% of pGAL-CSE4-353 cells properly segregated chromosome VIII (Figure 5B). In the 53% of cells in which both copies of CEN8-GFP segregated to the same pole, chromosome VIII segregated to the mother cell the majority of the time. Consistent with this observation, 48% of the pGAL-CSE4-353 mad2Δ cells appeared to have more DNA in the mother cell than in the daughter cell.
Although our data suggested that cells overexpressing pGAL-CSE4-353 contain mono-oriented chromosomes, it was possible that the lack of biorientation in these cells was due to a defect in microtubule attachment to kinetochores that could not be distinguished by these assays. We therefore tested whether the spindle-checkpoint-dependent arrest induced by Cse4-353p overexpression depends on the Ipl1/aurora protein kinase, which is required for the checkpoint when kinetochores are mono-oriented but not when they are unattached (Biggins and Murray 2001; Pinsky et al. 2006b). To control Ipl1 activity, we used the ipl1-as5 allele that can be inhibited by the small molecule 1NA-PP1 (Pinsky et al. 2006b). Wild-type, ipl1-as5, pGAL-CSE4-353, and pGAL-CSE4-353 ipl1-as5 cells were arrested in G1 and released into the cell cycle in GAL media containing 1NA-PP1. We analyzed the anaphase inhibitor Pds1 by immunoblotting and found that Pds1 levels cycle in wild-type and ipl1-as5 cells and are stabilized in cells overexpressing pGAL-CSE4-353, as expected (Figure 5C). Because Pds1 levels cycle in the pGAL-CSE4-353 ipl1-as5 cells, Ipl1/aurora activity is required for cells overexpressing Cse4-353p to activate the checkpoint. These data suggest that the kinetochores in pGAL-CSE4-353 cells are competent in attaching to microtubules, but that they make inappropriate mono-oriented attachments instead of proper bioriented attachments.
DISCUSSION
To further investigate the localization and functions of the budding yeast CenH3, we isolated and characterized 11 pGAL-CSE4 mutants that are lethal when overexpressed. We placed these mutants into three classes on the basis of the localization of the mutant proteins and spindle checkpoint activation. As we initially hypothesized, we were able to identify Cse4 mutant proteins that can localize to euchromatin when overexpressed, as well as mutants that do not localize to the centromere. Our previous characterization of Cse4-351, a class I mutant protein that localizes to euchromatin when overexpressed, revealed that ubiquitin-mediated proteolysis contributes to the exclusive localization of Cse4 by degrading excess protein that does not bind to the centromere (Collins et al. 2004). In this article, we analyzed the Cse4-353p mutant that does not localize to chromatin when overexpressed and found that it leads to a defect in kinetochore biorientation but not in kinetochore assembly. Our data raise the possibility that Cse4p has a role in biorientation in addition to specifying centromere identity.
The mechanism of the lethality of the pGAL-CSE4 mutants:
We identified three classes of pGAL-CSE4 mutants that are lethal when overexpressed. Because Cse4p is degraded by ubiquitin-mediated proteolysis (Collins et al. 2004), we expected that many of the mutants might be resistant to degradation. Consistent with this, we found that 10 of the 11 Cse4 mutant proteins have high steady-state levels and at least four are more stable than wild-type Cse4p (Collins et al. 2004; K. A. Collins and S. Biggins, data not shown). Although most of the mutant proteins are overexpressed at higher levels than wild-type Cse4p, they do not lead to the same mutant phenotype. This is probably due to the multiple, different mutations that are present in each allele. We attempted to identify the minimal number of mutations required for lethality and found that multiple mutations are necessary (this study; Collins et al. 2004). It is likely that many of the lethal phenotypes are due to a combination of increased protein stability and multiple mutations that alter function.
The two class I mutants that we identified (pGAL-CSE4-350 and -351) are resistant to proteolysis and are characterized by the localization of the mutant Cse4 protein in noncentromeric regions of the genome (this study; Collins et al. 2004), similar to the overexpression of wild-type CenH3 in multicellular eukaryotes (Henikoff et al. 2000; Ahmad and Henikoff 2001; Heun et al. 2006). Because these mutants did not alter the localization of the endogenous Cse4 protein nor activate the spindle checkpoint, it is unlikely that the lethality is related to altered kinetochore function. We considered the hypothesis that the overexpression of the class I mutants leads to lethality because they interfere with proteasome activity such that other substrates cannot be degraded. However, we were unable to detect a defect in nuclear proteasome function (K. A. Collins and S. Biggins, data not shown). We also did not detect replication defects due to aberrant localization of these Cse4 mutant proteins to euchromatin when analyzed by FACS analysis, so we currently favor the possibility that the class I mutants are lethal because they interfere with transcription. In the future, microarray experiments will be required to analyze the effects of class I mutants on transcription.
The four class II mutants (pGAL-CSE4-352, -354, -356, and -360) are characterized by the localization of the mutant proteins to the centromere and subsequent spindle checkpoint activation. However, these four mutant proteins also localized to euchromatin, making the nature of the lethality unclear. Because these mutants localize to the centromere and activate the spindle checkpoint, it is likely that they disrupt kinetochore assembly and function. Future experiments will be required to identify the nature of the kinetochore defects that occur when the class II mutants are overexpressed.
The five class III mutants are characterized by failure of the mutant protein to localize to the centromere or to any other chromosomal locus when overexpressed. These mutant proteins may have lost the ability to form nucleosomes or to interact with a CenH3 assembly factor. Because these mutants also activate the spindle checkpoint, we hypothesized that the lethality was due to the titration of one or more proteins away from the kinetochore when the mutant Cse4 proteins were overexpressed. Consistent with this hypothesis, we found that Cse4-353p is nuclear (K. A. Collins and S. Biggins, data not shown). However, we were unable to identify any protein that had an altered localization when Cse4-353p was overexpressed, despite multiple approaches to addressing this issue. For example, we analyzed the localization of a representative kinetochore protein from each subcomplex, but we were not able to detect altered kinetochore localization of any of the proteins that we tested. In addition, we directly analyzed the physical interactions between Cse4-353p and the kinetochore proteins Mif2 and Mtw1 that are known to associate with Cse4p (Pinsky et al. 2003; Westermann et al. 2003), but we did not detect an enhanced interaction between Cse4-353p and these proteins (K. A. Collins and S. Biggins, data not shown). Because Cse4p is required for cohesin localization to an exogenous centromere (Tanaka et al. 1999) and because the kinetochore acts as an enhancer of pericentric cohesin binding (Weber et al. 2004), we also analyzed the localization of the Mcd1/Scc1 cohesin protein. However, similar to our analysis of kinetochore proteins, we did not detect altered localization of cohesin. It is therefore possible that we have not yet assayed the protein that is titrated away or that there are differences in the localization of kinetochore proteins among cells that we cannot identify because our assays monitor the entire population of cells. Alternatively, it is possible that Cse4-353p does not titrate proteins away from the kinetochore. The most likely candidate protein that class III mutants might titrate is histone H4 (Glowczewski et al. 2000; Westermann et al. 2003); however, the overexpression of H4 only mildly suppressed the lethality of class III mutants (K. A. Collins and S. Biggins, data not shown). It will therefore take additional studies to determine whether the mechanism of lethality involves titrating away one or more proteins that regulate chromosome segregation.
Cells overexpressing pGAL-CSE4-353 have biorientation defects:
Our characterization of cells overexpressing a class III mutant, pGAL-CSE4-353, revealed a specific defect in kinetochore biorientation. We found that the majority of kinetochores clustered closer to the poles in pGAL-CSE4-353 cells compared to wild-type cells, suggesting the presence of mono-oriented chromosomes that could not resist microtubule-pulling forces. Consistent with this, there was a significant decrease in the biorientation of a single centromere, CEN8, in pGAL-CSE4-353 cells relative to control metaphase-arrested cells. A number of observations strongly suggest that pGAL-CSE4-353 cells are specifically defective in kinetochore biorientation and not in the ability of microtubules to attach to kinetochores. First, the CEN8 kinetochores clustered near the pole when they were mono-oriented during metaphase, suggesting that they were attached to microtubules. Second, chromosome VIII segregated to the pole when cells overexpressing pGAL-CSE4-353 were allowed to enter anaphase. If these chromosomes were not attached to microtubules, they would have remained distal to the pole instead of being pulled to the pole. Finally, cells overexpressing pGAL-CSE4-353 required the Ipl1/aurora protein kinase to activate the spindle checkpoint. Because Ipl1/aurora is not required for checkpoint activation when there are unattached kinetochores (Biggins and Murray 2001), our data strongly suggest that pGAL-CSE4-353 cells make attachments to kinetochores, although they are mono-oriented instead of bioriented.
Our isolation of the Cse4-353p mutant that appears competent in kinetochore assembly but defective in biorientation may have revealed an independent role for Cse4p in kinetochore biorientation. Although it is possible that the Cse4-353 mutant is a gain-of-function mutant that disrupts kinetochore biorientation without affecting kinetochore assembly, we favor the hypothesis that the endogenous Cse4 protein functions in biorientation. There are a number of ways in which centromeric chromatin could facilitate biorientation that could be independent from a role in assembling the kinetochore. For example, the proper centromeric chromatin structure could be required to sterically orient sister kinetochores toward opposite poles. Alternatively, centromeric chromatin may allow microtubules to generate the appropriate tension on the kinetochore and thus stabilize proper bioriented attachments (Nicklas and Ward 1994; King and Nicklas 2000). In the future, isolating recessive cse4 alleles that are functional in kinetochore assembly but defective in segregation may allow us to determine whether the endogenous Cse4 protein regulates biorientation in addition to kinetochore assembly.
Summary:
Our characterization of pGAL-CSE4 mutants that are lethal when overexpressed has determined that Cse4p is regulated by proteolysis and that it may also function in kinetochore biorientation. Future studies of these mutants may also reveal additional details about the functions of Cse4p, the requirements for Cse4p targeting to centromeres, and the mechanisms that exclude Cse4p from euchromatin. In addition, the isolation of pGAL-CSE4 mutants that are lethal when overexpressed highlights the importance of maintaining proper Cse4p levels and functions for genomic integrity.
Acknowledgments
We are especially grateful to Suzanne Furuyama for generating pSB887, and we also thank the Gottschling lab, Orna Cohen-Fix, and Sue Jaspersen for reagents. We are grateful to Bungo Akiyoshi, Chris Breed, Suzanne Furuyama, Rich Gardner, Chitra Kotwaliwale, Harmit Malik, Paul Megee, Ben Pinsky, Prerana Ranjitkar, Toshi Tsukiyama, and Danielle Vermaak for discussions and helpful comments on the manuscript. J.L.G. and R.C. were supported by funds from the Stowers Institute. K.A.C. was supported by a Viral Oncology Training grant. S.B. was supported by a National Institutes of Health grant and a Beckman Young Investigator Award. S.B. is a Scholar of the Leukemia and Lymphoma Society.
References
- Ahmad, K., and S. Henikoff, 2001. Centromeres are specialized replication domains in heterochromatin. J. Cell Biol. 153: 101–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggins, S., and A. W. Murray, 2001. The budding yeast protein kinase Ipl1/Aurora allows the absence of tension to activate the spindle checkpoint. Genes Dev. 15: 3118–3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggins, S., and C. E. Walczak, 2003. Captivating capture: how microtubules attach to kinetochores. Curr. Biol. 13: R449–R460. [DOI] [PubMed] [Google Scholar]
- Biggins, S., F. F. Severin, N. Bhalla, I. Sassoon, A. A. Hyman et al., 1999. The conserved protein kinase Ipl1 regulates microtubule binding to kinetochores in budding yeast. Genes Dev. 13: 532–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black, B. E., D. R. Foltz, S. Chakravarthy, K. Luger, V. L. Woods, Jr. et al., 2004. Structural determinants for generating centromeric chromatin. Nature 430: 578–582. [DOI] [PubMed] [Google Scholar]
- Blower, M. D., and G. H. Karpen, 2001. The role of Drosophila CID in kinetochore formation, cell-cycle progression and heterochromatin interactions. Nat. Cell Biol. 3: 730–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo, A. R., J. B. Meehl, G. Morgan, A. Schutz-Geschwender and M. Winey, 2002. The yeast protein kinase Mps1p is required for assembly of the integral spindle pole body component Spc42p. J. Cell Biol. 156: 453–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman, I. M., S. Anderson, M. Jwa, E. M. Green, J. Kang et al., 2002. Phospho-regulation of kinetochore-microtubule attachments by the aurora kinase Ipl1p. Cell 111: 163–172. [DOI] [PubMed] [Google Scholar]
- Choo, K. H., 2001. Domain organization at the centromere and neocentromere. Dev. Cell 1: 165–177. [DOI] [PubMed] [Google Scholar]
- Cleveland, D. W., Y. Mao and K. F. Sullivan, 2003. Centromeres and kinetochores: from epigenetics to mitotic checkpoint signaling. Cell 112: 407–421. [DOI] [PubMed] [Google Scholar]
- Collins, K. A., S. Furuyama and S. Biggins, 2004. Proteolysis contributes to the exclusive centromere localization of the yeast Cse4/CENP-A histone H3 variant. Curr. Biol. 14: 1968–1972. [DOI] [PubMed] [Google Scholar]
- Collins, K. A., A. R. Castillo, S. Y. Tatsutani and S. Biggins, 2005. De novo kinetochore assembly requires the centromeric histone H3 variant. Mol. Biol. Cell 16: 5649–5660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enquist-Newman, M., I. M. Cheeseman, D. Van Goor, D. G. Drubin, P. B. Meluh et al., 2001. Dad1p, third component of the Duo1p/Dam1p complex involved in kinetochore function and mitotic spindle integrity. Mol. Biol. Cell 12: 2601–2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald-Hayes, M., L. Clarke and J. Carbon, 1982. Nucleotide sequence comparisons and functional analysis of yeast centromere DNAs. Cell 29: 235–244. [DOI] [PubMed] [Google Scholar]
- Furuyama, T., Y. Dalal and S. Henikoff, 2006. Chaperone-mediated assembly of centromeric chromatin in vitro. Proc. Natl. Acad. Sci. USA 103: 6172–6177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glowczewski, L., P. Yang, T. Kalashnikova, M. S. Santisteban and M. M. Smith, 2000. Histone-histone interactions and centromere function. Mol. Cell. Biol. 20: 5700–5711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glynn, E. F., P. C. Megee, H. G. Yu, C. Mistrot, E. Unal et al., 2004. Genome-wide mapping of the cohesin complex in the yeast Saccharomyces cerevisiae. PLoS Biol. 2: E259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima, G., and M. Yanagida, 2000. Establishing biorientation occurs with precocious separation of the sister kinetochores, but not the arms, in the early spindle of budding yeast. Cell 100: 619–633. [DOI] [PubMed] [Google Scholar]
- Goshima, G., T. Kiyomitsu, K. Yoda and M. Yanagida, 2003. Human centromere chromatin protein hMis12, essential for equal segregation, is independent of CENP-A loading pathway. J. Cell Biol. 160: 25–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, X., S. Asthana and P. K. Sorger, 2000. Transient sister chromatid separation and elastic deformation of chromosomes during mitosis in budding yeast. Cell 101: 763–775. [DOI] [PubMed] [Google Scholar]
- He, X., D. R. Rines, C. W. Espelin and P. K. Sorger, 2001. Molecular analysis of kinetochore-microtubule attachment in budding yeast. Cell 106: 195–206. [DOI] [PubMed] [Google Scholar]
- Henikoff, S., K. Ahmad, J. S. Platero and B. van Steensel, 2000. Heterochromatic deposition of centromeric histone H3-like proteins. Proc. Natl. Acad. Sci. USA 97: 716–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heun, P., S. Erhardt, M. D. Blower, S. Weiss, A. D. Skora et al., 2006. Mislocalization of the Drosophila centromere-specific histone CID promotes formation of functional ectopic kinetochores. Dev. Cell 10: 303–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke, C., J. Ortiz, T. U. Tanaka, J. Lechner and E. Schiebel, 2002. Four new subunits of the Dam1-Duo1 complex reveal novel functions in sister kinetochore biorientation. EMBO J. 21: 181–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamakaka, R. T., and S. Biggins, 2005. Histone variants: Deviants? Genes Dev. 19: 295–310. [DOI] [PubMed] [Google Scholar]
- King, J. M., and R. B. Nicklas, 2000. Tension on chromosomes increases the number of kinetochore microtubules but only within limits. J. Cell Sci. 113 (Pt. 21): 3815–3823. [DOI] [PubMed]
- Lew, D. J., and D. J. Burke, 2003. The spindle assembly and spindle position checkpoints. Annu. Rev. Genet. 37: 251–282. [DOI] [PubMed] [Google Scholar]
- Li, Y., J. Bachant, A. A. Alcasabas, Y. Wang, J. Qin et al., 2002. The mitotic spindle is required for loading of the DASH complex onto the kinetochore. Genes Dev. 16: 183–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine, M. S., A. McKenzie, III, D. J. Demarini, N. G. Shah, A. Wach et al., 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14: 953–961. [DOI] [PubMed] [Google Scholar]
- Malik, H. S., and S. Henikoff, 2003. Phylogenomics of the nucleosome. Nat. Struct. Biol. 10: 882–891. [DOI] [PubMed] [Google Scholar]
- McAinsh, A. D., J. D. Tytell and P. K. Sorger, 2003. Structure, function, and regulation of budding yeast kinetochores. Annu. Rev. Cell Dev. Biol. 19: 519–539. [DOI] [PubMed] [Google Scholar]
- Minshull, J., A. Straight, A. Rudner, A. Dernburg, A. Belmont et al., 1996. Protein phosphatase 2A regulates MPF activity and sister chromatid cohesion in budding yeast. Curr. Biol. 6: 1609–1620. [DOI] [PubMed] [Google Scholar]
- Nasmyth, K., and C. H. Haering, 2005. The structure and function of SMC and kleisin complexes. Annu. Rev. Biochem. 74: 595–648. [DOI] [PubMed] [Google Scholar]
- Nicklas, R. B., and S. C. Ward, 1994. Elements of error correction in mitosis: microtubule capture, release, and tension. J. Cell Biol. 126: 1241–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oegema, K., A. Desai, S. Rybina, M. Kirkham and A. A. Hyman, 2001. Functional analysis of kinetochore assembly in Caenorhabditis elegans. J. Cell Biol. 153: 1209–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr-Weaver, T. L., J. W. Szostak and R. J. Rothstein, 1983. Genetic applications of yeast transformation with linear and gapped plasmids. Methods Enyzmol. 101: 228–245. [DOI] [PubMed] [Google Scholar]
- Pearson, C. G., P. S. Maddox, E. D. Salmon and K. Bloom, 2001. Budding yeast chromosome structure and dynamics during mitosis. J. Cell Biol. 152: 1255–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinsky, B. A., and S. Biggins, 2005. The spindle checkpoint: tension vs. attachment. Trends Cell Biol. 15 (9): 486–493. [DOI] [PubMed] [Google Scholar]
- Pinsky, B. A., S. Y. Tatsutani, K. A. Collins and S. Biggins, 2003. An Mtw1 complex promotes kinetochore biorientation that is monitored by the Ipl1/Aurora protein kinase. Dev. Cell 5: 735–745. [DOI] [PubMed] [Google Scholar]
- Pinsky, B. A., C. V. Kotwaliwale, S. Y. Tatsutani, C. A. Breed and S. Biggins, 2006. a Glc7/protein phosphatase-1 regulatory subunits can oppose the Ipl1/Aurora protein kinase by redistributing Glc7. Mol. Cell. Biol. 26: 2648–2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinsky, B. A., C. Kung, K. M. Shokat and S. Biggins, 2006. b The Ipl1-Aurora protein kinase activates the spindle checkpoint by creating unattached kinetochores. Nat. Cell Biol. 8: 78–83. [DOI] [PubMed] [Google Scholar]
- Rose, M. D., F. Winston and P. Heiter, 1990. Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Scharfenberger, M., J. Ortiz, N. Grau, C. Janke, E. Schiebel et al., 2003. Nsl1p is essential for the establishment of bipolarity and the localization of the Dam-Duo complex. EMBO J. 22: 6584–6597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman, F., G. Fink and C. Lawrence, 1974. Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Sikorski, R. S., and P. Hieter, 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122: 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straight, A. F., A. S. Belmont, C. C. Robinett and A. W. Murray, 1996. GFP tagging of budding yeast chromosomes reveals that protein-protein interactions can mediate sister chromatid cohesion. Curr. Biol. 6: 1599–1608. [DOI] [PubMed] [Google Scholar]
- Tanaka, T., M. P. Cosma, K. Wirth and K. Nasmyth, 1999. Identification of cohesin association sites at centromeres and along chromosome arms. Cell 98: 847–858. [DOI] [PubMed] [Google Scholar]
- Tanaka, T., J. Fuchs, J. Loidl and K. Nasmyth, 2000. Cohesin ensures bipolar attachment of microtubules to sister centromeres and resists their precocious separation. Nat. Cell Biol. 2: 492–499. [DOI] [PubMed] [Google Scholar]
- Wallis, O. C., P. Ottolenghi and P. A. Whittaker, 1972. Induction of petite mutants in yeast by starvation in glycerol. Biochem. J. 127: 46P–47P. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber, S. A., J. L. Gerton, J. E. Polancic, J. L. DeRisi, D. Koshland et al., 2004. The kinetochore is an enhancer of pericentric cohesin binding. PLoS Biol. 2: E260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermann, S., I. M. Cheeseman, S. Anderson, J. R. Yates, III, D. G. Drubin et al., 2003. Architecture of the budding yeast kinetochore reveals a conserved molecular core. J. Cell Biol. 163: 215–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoda, K., S. Ando, S. Morishita, K. Houmura, K. Hashimoto et al., 2000. Human centromere protein A (CENP-A) can replace histone H3 in nucleosome reconstitution in vitro. Proc. Natl. Acad. Sci. USA 97: 7266–7271. [DOI] [PMC free article] [PubMed] [Google Scholar]