Abstract
The Saccharomyces cerevisiae non-Mendelian genetic element [PSI+] is the prion form of the translation termination factor Sup35p. The ability of [PSI+] to propagate efficiently has been shown previously to depend upon the action of protein chaperones. In this article we describe a genetic screen that identifies an array of mutants within the two major cytosolic Hsp70 chaperones of yeast, Ssa1p and Ssa2p, which impair the propagation of [PSI+]. All but one of the mutants was located within the ATPase domain of Hsp70, which highlights the important role of regulation of Hsp70–Ssa ATP hydrolysis in prion propagation. A subset of mutants is shown to alter Hsp70 function in a way that is distinct from that of previously characterized Hsp70 mutants that alter [PSI+] propagation and supports the importance of interdomain communication and Hsp70 interaction with nucleotide exchange factors in prion propagation. Analysis of the effects of Hsp70 mutants upon propagation of a second yeast prion [URE3] further classifies these mutants as having general or prion-specific inhibitory properties.
PRIONS are infectious proteins. They are a transmissible amyloid form of a cellular protein that replicates by converting the native protein into the same abnormal prion form. The term prion was first used to describe the nature of the scrapie infectious agent (Prusiner 1982). The conversion of the normal form of the mammalian prion protein PrPc to the infectious prion form PrPSc is associated with a number of transmissible spongiform encephalopathies (TSEs) in mammals (Collinge 2001). The discovery of proteins that can behave in a prion-like manner in yeast and filamentous fungi (Wickner 1994; Coustou et al. 1997) has expanded the prion theory from solely disease-causing agents into novel epigenetic phenomena that may carry out a functional biological role within the cell and raises the possibility that prions may be found in other species as well.
Saccharomyces cerevisiae has at least three proteins that meet the genetic criteria to be defined as a prion (Wickner 1994). Confirmed yeast prions include [PSI+], the prion form of Sup35p (Cox 1965), [URE3], the prion form of Ure2p (Aigle and Lacroute 1975), and [PIN+], the prion form of Rnq1p (Derkatch et al. 1997) but also possibly the prion form of a number of other prion-like proteins (Derkatch et al. 2001). The most widely studied yeast prion is [PSI+], which is the prion form of the translation termination factor Sup35p.
Sup35p can be subdivided into three distinct domains, an N-terminal domain of ∼130 amino acids that is required for prion formation and propagation, a highly charged middle domain, and a large C-terminal region that carries out essential functions in translation termination (reviewed in Tuite 2000). In [PSI+] cells a high proportion of Sup35p is present in the form of nonfunctional high molecular weight aggregates (Patino et al. 1996; Paushkin et al. 1997), which reduces the amount of translation termination release factor and therefore reduces the efficiency of translation termination. Thus, [PSI+] cells have high levels of nonsense suppression (Cox 1965). Natural variants of [PSI+] exist where a reduced amount of Sup35p can be found within these aggregates (Uptain et al. 2001) and such variants show a corresponding reduction in nonsense suppression. In contrast, the vast majority of Sup35p in [psi−] cells is present in a soluble functional form.
Another well-characterized yeast prion is [URE3]. Conversion of Ure2p into the infectious prion form causes a proportion or Ure2p to form high molecular weight amyloid aggregates that prevent the protein from carrying out its normal function in regulating nitrogen metabolism. Hence, [URE3] cells are capable of utilizing poor nitrogen sources while [ure-0] cells are not (Taylor and Wickner 2001; Lian et al. 2006).
Genetic screens have identified a number of protein chaperones and cochaperones as modulators of prion propagation in yeast (reviewed in Jones and Tuite 2005). The nonessential chaperone Hsp104p plays a critical role in propagation of all naturally occurring yeast prions (reviewed in True 2006). Hsp104p is a protein disaggregase that is required for recovery from exposure to stresses such as heat shock and works in concert with Hsp70 and Hsp40 in the refolding of aggregated proteins (Glover and Lindquist 1998). Deletion of Hsp104p prevents propagation of naturally occurring yeast prions while overexpression has been shown to efficiently cure [PSI+] but not [URE3] or [PIN+] (Chernoff et al. 1995; Derkatch et al. 1997; Moriyama et al. 2000), presumably due to differences in aggregated prion substrates.
Another major factor in efficient prion propagation is the cytosolic Hsp70 family. Hsp70's are a highly conserved family of chaperones that form the central part of a ubiquitous protein-folding system (Wegele et al. 2004). The yeast genome encodes at least 14 distinct Hsp70-related proteins that are located in various cellular compartments. Members of the Hsp70 family share the property of binding to short hydrophobic segments of partially folded or unfolded polypeptides, thereby preventing their aggregation. One subfamily of cytosolic Hsp70 that has been implicated in yeast prion propagation is the Hsp70–Ssa family. The Hsp70–Ssa family consists of four genes, SSA1–4, and the presence of at least one is required for vegetative growth (Werner-Washburne et al. 1989). Each Ssa protein is composed of an ATPase domain, a peptide-binding domain (PBD), and a C-terminal variable domain (James et al. 1997). SSA1 and SSA2 are constitutively expressed and under normal growth conditions ∼80% of Ssa protein is Ssa2p; expression of SSA3 and SSA4 is induced by stress conditions (Werner-Washburne et al. 1989).
The binding and hydrolysis of ATP dictates the interaction between Hsp70 and substrate. In the ATP-bound state the peptide substrate is rapidly exchanged between the peptide-binding pocket and the surrounding environment. Upon ATP hydrolysis the peptide-binding domain traps hydrophobic substrate, which is released upon nucleotide exchange from an ADP- to an ATP-bound state (reviewed in Mayer et al. 2001). The ATPase cycle of Hsp70 is influenced by cochaperones, including Hsp40's (e.g., Ydj1p in yeast) and tetratricopeptide repeat (TPR)-containing proteins (e.g., Sti1p in yeast) (reviewed in Jones and Tuite 2005). Efficient nucleotide exchange requires interactions with nucleotide exchange factors such as Fes1p (Kabani et al. 2002a,b) or Sse1p (Dragovic et al. 2006; Raviol et al. 2006).
Genetic and biochemical studies on Hsp70–Ssa proteins have provided insight into how cytosolic Hsp70 functions in yeast prion propagation. Overexpression of Hsp70–Ssa reduces the efficiency of curing of [PSI+] by Hsp104 overexpression (Newnam et al. 1999; Allen et al. 2005). Overexpression of Ssa1p also increases the de novo appearance of [PSI+] when Sup35p is overexpressed and Ssa1p appears to interact physically with Sup35p in vivo (Allen et al. 2005). Other studies have shown that overexpression of Ssa1p or the Hsp40 family members Ydj1p or Sis1p can efficiently cure natural and “artificial” variants of [PSI+] (Kushnirov et al. 2000; Krynduskin et al. 2002). Despite the high sequence identity between Ssa1p and Ssa2p (98% identity) they have been shown to have differing effects on propagation of [URE3] (Schwimmer and Masison 2002) and the presence of Ssa2p is required to maintain [URE3] (Roberts et al. 2004). Furthermore, different Hsp70–Ssa species have different effects upon the toxicity of amyloid formed by polyglutamine (Gokhale et al. 2005) and mutant α-synuclein (Flower et al. 2005).
Further insight into how Hsp70–Ssa may influence prion propagation has come from characterization of the SSA1-21 mutant. SSA1-21 (L483W) is a dominant mutant of SSA1 that impairs propagation of [PSI+] but does not have any effect upon vegetative cell growth (Jung et al. 2000). Genetic analysis of SSA1-21 cells suggests that Ssa1-21p binds more avidly to prion aggregates and interferes with the production of propagons (Jung et al. 2000; Jones and Masison 2003). Analysis of prion aggregates in SSA1-21 cells suggests that the average size of aggregates is larger when compared to wild-type cells (Song et al. 2005). The ability of SSA1-21 to impair prion propagation is dependent on interaction with TPR cochaperones Sti1p and Cpr7p, and these effects appear independent of Hsp90 (Jones et al. 2004).
A previous study identified a number of SSA1 mutations located within the ATPase domain that impair [PSI+] propagation by a mechanism probably similar to that of the well-characterized SSA1-21 allele (Jones and Masison 2003). To further our understanding of how Hsp70–Ssa species are involved in yeast prion propagation we have carried out an extensive genetic screen to identify mutants in both the major cytosolic Ssa proteins, Ssa1p and Ssa2p, that impair [PSI+] propagation. All but one of these new Ssa mutants are located within the ATPase domain, which highlights the importance of the Ssa ATPase cycle in prion propagation. Location of mutated residues on the Hsp70 ATPase domain crystal structure supports a role for regulated interdomain communication in prion maintenance. Clustering of mutants within the ATPase domain in a region known to interact with the nucleotide exchange factor Fes1p indicates the importance of coordination of Hsp70 and its exchange factor(s) for efficient prion propagation. We also analyzed the effects of our new SSA mutants upon propagation of a second yeast prion [URE3], which has allowed the classification of SSA mutants into those having general effects or prion-specific effects. Furthermore, we demonstrate that a subset of these new SSA mutants appears to impair prion propagation by a mechanism that is distinct from that of previously well-characterized SSA1 mutants.
MATERIALS AND METHODS
Strains, plasmids, and genetic methods:
Strains used in this study were G400-1C (MATa ade2-1 SUQ5 kar1-1 his3 leu2 lys2 trp1 ura3 ssa1∷KanMX, ssa2∷HIS3, ssa3∷TRP1, ssa4∷URA3-1f/pRDW10 [PSI+]) (Jones and Masison 2003), G632 MATa ade2.1 SUQ5 kar1-1 his3 leu2 trp1 ura3 ssa1∷KanMX ssa2∷HIS3, which is equivalent to strain 1012 (Jones et al. 2004), and SB34 (Bach et al. 2003), which is a derivative of CC34 MATa, trp1-1, ade2-1, leu2-3,112, his3-11,15, URA2∷HIS3, [URE3] (Fernandez-Bellot et al. 2000), which has the ERG6 gene deleted with TRP1 and the DAL5 coding sequence replaced by ADE2. Plasmid pRDW10 and pJ120 have been described previously (Jones and Masison 2003) and are single-copy plasmids containing the SSA1 gene. Plasmid pGJ100 is the SSA2 coding sequence plus 500 bp of upstream and downstream sequence cloned into pRS315 (Sikorski and Hieter 1989). The SSA2 gene was amplified by PCR from strain 1001 (Jones et al. 2004) with oligonucleotide primers containing BamHI sites and subsequently cloned into the BamHI site of pRS315. All SSA1 mutants are derivatives of pJ120 and all SSA2 mutants are derivatives of pGJ100.
Monitoring of [PSI+] was carried out as described (Jones and Masison 2003). Briefly, the presence of [PSI+] (the nonfunctional aggregated form of Sup35p) and SUQ5 in our cells causes efficient translation readthrough of the ochre mutation in the ade2-1 allele. Nonsuppressed ade2-1 mutants are Ade− and are red when grown on medium containing limiting amounts of adenine due to the accumulation of a pigmented substrate of Ade2p. Partial suppression of ade2-1 by [PSI+] allows growth without adenine and eliminates the pigmentation (Cox 1965).
Monitoring of [URE3] again made use of the red/white selection based on the ADE2 gene. The strain SB34 has ADE2 under control of the DAL5 promoter. In [URE3] cells expression of the DAL5 promoter is high due to the action of Gln3p. In [ure-0] cells soluble Ure2p can interact with Gln3p and prevent transcription from the DAL5 promoter. Hence, when [URE3] is present the SB34 strain will grow on medium lacking adenine and is white on medium with limiting adenine. When [ure-0] this strain will not grow on medium lacking adenine and is red on medium with limiting adenine.
Generation of mutant SSA plasmid libraries:
Plasmids pJ120 and pGJ100 were subjected to treatment with hydroxylamine for 60 min (Schatz et al. 1988). For these plasmids this treatment results in mutation frequencies of ∼6%.
Isolation of SSA1 and SSA2 mutants that impair [PSI+] propagation:
SSA mutants were isolated using the plasmid shuffle technique. Strain G400-1C was transformed with either the SSA1 or the SSA2 mutagenized plasmid library. Transformed cells were selected on medium lacking leucine. Any red or dark-pink colonies were scored at this point as potential SSA mutants that could weaken [PSI+]. Transformation plates were replica plated onto medium containing limiting amounts of adenine and also 5-fluoroorotic acid (5-FOA), a chemical that selects against URA+ cells and hence against the presence of the pRDW10 plasmid. Colonies appearing red or dark pink at this stage were scored as potentially harboring a mutant SSA allele that cannot maintain [PSI+]. All potential SSA mutant-containing plasmids were isolated and retransformed back into G400-1C and analyzed for their effects upon [PSI+]. After retransformation the color phenotype of colonies was scored subjectively from 0 to 9, with 0 being white and 9 being red.
Assaying SSA mutant effects upon [URE3] propagation:
SB34 was grown to log phase growth under conditions that maintain [URE3] (medium lacking adenine). Cells were transformed with mutant SSA alleles and transformants were selected on medium lacking leucine. At this stage all cells (at least 100) were scored for color phenotype on the basis of being white, red, or sectored. Three white colonies were then selected and spread onto medium lacking both leucine and adenine, which will select for the SSA mutant plasmid and also for [URE3]. Cells were then restreaked onto medium lacking only leucine and scored for color phenotype on a subjective scale of 0–9, as described earlier.
Site-directed mutagenesis:
Mutant alleles were introduced into SSA coding sequence by using appropriate primers containing the required DNA base mismatch and using the Quickchange kit (Stratagene, La Jolla, CA) with PCR conditions as recommended by the manufacturer. All SSA alleles subjected to site-directed mutagenesis were fully sequenced to verify that only the desired mutation had been introduced.
Western analysis:
Western analysis was performed essentially as described in Jung et al. (2000) and Jones and Masison (2003). Hsp70 monoclonal antibody was purchased from Stressgen (Victoria, BC, Canada) (SPA 822) and Hsp104 polyclonal antibody was a gift from John Glover (University of Toronto).
RESULTS
Mutants of SSA1 and SSA2 that impair [PSI+] propagation:
Using the plasmid shuffle technique described in materials and methods we have isolated 25 new alleles of SSA1 and SSA2 that impair [PSI+] propagation (Table 1, Figure 1). Five mutants (A53V, T64I, R74K, G287D, and T295I) were isolated in both SSA1 and SSA2 (Table 1, columns 1 and 2). This strongly suggests that SSA1 and SSA2 are carrying out the same function in [PSI+] prion propagation and supports the suggestion that the SSA gene family provides a redundant activity in [PSI+] prion propagation (Allen et al. 2005). While all mutants greatly impair [PSI+] propagation when they are the sole Ssa source in the cell, only one (SSA2A176T) has any significant effect upon growth rate (Table 1, last column). This trend has been observed for virtually all previously characterized SSA1 mutants that alter [PSI+] propagation and supports the suggestion that either Ssa function in essential cellular pathways can be separated from Ssa function in prion propagation (Jung et al. 2000; Jones and Masison 2003) or [PSI+] is much more sensitive to perturbation in Ssa function compared to other cellular substrates.
TABLE 1.
Relative effects of SSA1 and SSA2 mutants on prion propagation and cell growth
| Mutationsa
|
Colorc in G400-1C
|
|||||
|---|---|---|---|---|---|---|
| In Ssa1p | In Ssa2p | E. coli DnaKb | Times isolated | Pre-FOA | Post-FOA | Generation time (% of wt)d |
| WT | WT | — | 0 | 0 | 100 | |
| A4T | I4 | 1 | 6 | 9 | 81 ± 8 | |
| D30N | A30 | 1 | 3 | 7 | 95 ± 1 | |
| G41D | A41 | 5 | 0–6 | 9 | 118 ± 2 | |
| G50D | G51 | 1 | 7 | 9 | 90 ± 9e | |
| A53V | A54 | 1 | 4 | 7 | 95 ± 4 | |
| A53V | 2 | 6 | 8 | 100 ± 5 | ||
| A53T | 4 | 7 | 8 | 105 ± 3 | ||
| T64I | T65 | 3 | 0–8 | 8 | 101 ± 2 | |
| T64I | 4 | 0–8 | 8 | 101 ± 4 | ||
| G73D | G74 | 3 | 5 | 9 | 101 ± 5 | |
| R74K | R75 | 1 | 5 | 7 | 94 ± 4 | |
| R74K | 1 | 6 | 7 | 77 ± 30 | ||
| S119F | A117 | 2 | 3 | 7 | 96 ± 4 | |
| A146T | A144 | 2 | 3 | 8 | 101 ± 3 | |
| A155V | A153 | 2 | 5 | 8 | 113 ± 10 | |
| A155T | 1 | 4 | 7 | 94 ± 4 | ||
| A176T | A174 | 1 | 4 | 8 | 131 ± 7 | |
| T223I | T225 | 8 | 2–7 | 8 | 94 ± 2 | |
| R259K | R261 | 2 | 5 | 9 | 100 ± 6 | |
| G287D | D289 | 4 | 7 | 9 | 102 ± 5 | |
| G287D | 5 | 7 | 9 | 101 ± 5 | ||
| T295I | T301 | 1 | 6 | 8 | 95 ± 3 | |
| T295I | 1 | 2 | 8 | 105 ± 5 | ||
| A297T | A303 | 1 | 4 | 8 | 107 ± 3 | |
| G481D | G482 | 1 | 2–6 | 9 | 98 ± 5 | |
Mutations isolated in a previous screen (Jones and Masison 2003) were also isolated in our screen: A17V, five times; R23H, one time; G32D, three times; and R34K, six times.
Corresponding amino acid in E. coli DnaK.
Color: 0, white, [PSI+]; 9, red, [psi−]; FOA, selection against presence of WT SSA1 URA3 plasmid.
Growth rates measured with SSA mutants as sole cytosolic Ssa species and in [psi−] variants.
G50D was the only allele to show a severe growth defect at 37°.
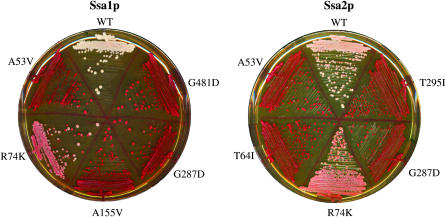
Figure 1.—
[PSI+] phenotypes of yeast cells expressing SSA1 or SSA2 mutant alleles as sole source of Ssa protein within the cell. Cells were streaked onto YPD and incubated at 30° for 2 days followed by a further 2 days at room temperature. Mutated residues are indicated. White color is indicative of the presence of [PSI+] prion, and pink and red colonies reflect a reduction in readthrough of the ade2-1 reporter gene and [psi−] cells, respectively.
The most striking observation about the mutants listed in Table 1 is that 24 of the 25 are located within the ATPase domain of Ssa (Table 1, columns 1 and 2). The one mutant located in the PBD is G481D, which is in very close proximity to the SSA1-21 allele (L483W) (Jung et al. 2000). Hence we suspect that G481D is behaving in a similar manner to L483W. The screen performed by Jones and Masison (2003) has previously alluded to the importance of the ATPase domain in prion propagation. In this more exhaustive screen of two Ssa species our findings support the importance of regulation of the Ssa ATPase cycle for propagation of [PSI+].
Pre-FOA treatment suggests that most SSA mutants show a degree of dominance with respect to [PSI+] propagation (Table 1, column 5). Only when plated onto FOA medium, thereby selecting for loss of the wild-type SSA1 plasmid, did all mutants develop colonies with a uniform color (Table 1, column 6). However, when placed in an environment containing all SSA genes the mutants have no or little effect on [PSI+] impairment (data not shown). The reason for this phenotypic difference is unknown but does not involve altered expression levels of Ssa3p or Ssa4p as these proteins are undetectable in these strains.
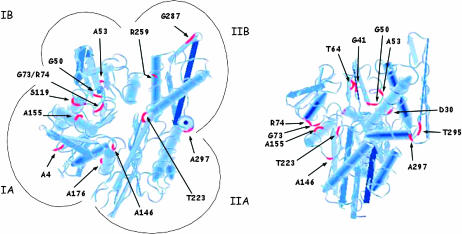
Given the high degree of homology between the ATPase domains of Hsp70 species across genera, it is reasonable to assume that the location of our new SSA mutants on the structure of the DnaK (Escherichia coli Hsp70) ATPase domain will provide insight into possible functional alterations. Figure 2 and Table 2 summarize the locations of the 18 ATPase domain SSA mutations within the ATPase domain of DnaK and also provide the corresponding residues for bovine Hsc70 to show further conservation.
Figure 2.—
Location of SSA mutant alleles on the ATPase domain of DnaK. A tubular diagram of the Protein Data Bank structure for 1DKG is shown from two different angles. Some mutants are shown on both structures to illustrate location more clearly.
TABLE 2.
Ssa ATPase domain mutations and location within Hsp70 ATPase crystal structure
| Mutation in Ssa | E. coli DnaK | bHsc70 | Location |
|---|---|---|---|
| A4T | I4 | A6 | IA: start β-strand |
| D30N | A30 | D32 | IA: loop |
| G41D | A41 | A43 | IB: middle β-strand |
| G50D | G51 | G52 | IB: end β-strand |
| A53V | A54 | A55 | IB: loop |
| T64I | T65 | T66 | IB: small β-strand |
| G73D | G74 | G75 | IB: loop |
| R74K | R75 | R76 | IB: loop |
| S119F | A117 | S121 | IA: start α-helix |
| A146T | A144 | A148 | IA′: loop |
| A155V | A153 | A157 | IA: middle α-helix |
| A176T | A174 | A178 | IA: middle α-helix |
| T223I | T225 | T226 | IIA: end β-strand |
| R259K | R261 | R262 | IIB: middle α-helix |
| G287D | D289 | G290 | IIB: top β-strand |
| T295I | T301 | T298 | IIB: end β-strand |
| A297T | A303 | A300 | IIB: start α-helix |
| G481D | G482 | G484 | Peptide-binding domain |
A striking observation from comparing the location of the ATPase domain mutants (Figure 2) is the clustering of residues both in primary and in tertiary structures. In addition to the clustering of specific residues in relation to each other, there is also a generalized clustering of residues to the broadly categorized IA, IB, and IIB region of the Hsp70 ATPase domain. There is only one mutated residue, T223, which is located within the IIA region (Table 2). We can see the clustering in three-dimensional space of G41 with T64 and also of G73 and R74 with A155.
Comparison of new SSA1 mutants to well-characterized SSA1 alleles that weaken [PSI+] propagation:
An array of SSA1 mutations have been reported previously that can suppress the prion-weakening effects of the SSA1-21 allele (Jones and Masison 2003). To assess whether our new SSA mutants may function in a similar manner to SSA1-21, we introduced three known SSA1-21 second-site suppressor mutations (A519T, E540K, and P636S) into five of our new SSA1 mutant alleles. We observed the effects upon [PSI+] propagation when these double SSA mutant alleles were the only SSA being expressed in the cell. As a control we used the A17V mutation of SSA1; as shown before, its prion-weakening effect is fully suppressed by the chosen suppressor mutations (Jones and Masison 2003).
The G73D mutation appears to function in a similar manner to the A17V allele in that G73D is fully suppressed by all three second-site suppressor mutations (Table 3). On the other hand the effect of G287D on [PSI+] propagation is unaltered by the presence of the second-site suppressors. This suggests that G287D is functioning differently than previously identified SSA1 mutants that alter [PSI+] propagation.
TABLE 3.
Effects of SSA1-21 known second-site suppressor mutations upon new prion-weakening SSA1 mutants
|
SSA1 mutation
|
||||||
|---|---|---|---|---|---|---|
| Second mutation | A17V | A53V | G73D | R259K | G287D | T295I |
| None | 9 | 7 | 9 | 9 | 9 | 8 |
| A519T | 0 | ND | 0 | ND | 9 | ND |
| E540K | 0a | 7b | 0a | 9b | 9a | 0a |
| P636S | 0 | ND | 0 | ND | 9 | ND |
Effects on colony color are scored subjectively as described in the Table 1 legend. ND, not determined.
Normal growth.
Poor growth.
The presence of E540K causes a loss of essential function for SSA1 and this loss of function can be overcome by A17V or L483W mutations (Jones and Masison 2003). All new SSA1 mutations tested in combination with E540K could suppress the growth defect, but not to the same extent as A17V or L483W (Table 3).
Hsp70 and Hsp104 abundance:
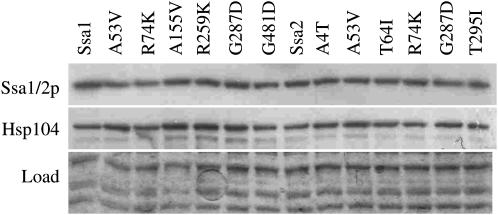
The propagation of [PSI+] can be influenced by the expression levels of Hsp70 and Hsp104 chaperones (Chernoff et al. 1995; Kushnirov et al. 2000). We therefore measured abundance of these chaperones in G400-1C expressing our new SSA mutants by Western blotting. In all of our mutants we found no difference in the abundance of Ssa or Hsp104 protein levels (Figure 3). This suggests that the effects upon [PSI+] propagation are not due to alterations in Ssa or Hsp104 protein abundance and suggest a more direct effect on prion propagation.
Figure 3.—
Abundance of Ssa1p or Ssa2p and Hsp104: Western blot to show chaperone levels in yeast cells harboring mutant alleles of SSA1 or SSA2 as sole Ssa protein in the cell. A representative sample of mutants is shown. Mutant alleles are as indicated. Identical aliquots were loaded onto two gels, one of which was probed with antibody to Hsp70, stripped, and probed with antibody to Hsp104. The second gel was stained with Coomassie to show equal loading of samples.
Different effects of SSA1 and SSA2 mutants upon propagation of [URE3]:
Overexpressed Ssa1p is capable of curing [URE3], whereas Ssa2p is not (Schwimmer and Masison 2002). The only known SSA allele to negatively affect [URE3] is ssa2-10 (Roberts et al. 2004). No SSA1 allele has been identified. It has therefore been suggested that Ssa1p and Ssa2p have different roles in propagation of the [URE3] prion. To investigate if our SSA mutants could alter the propagation of [URE3] we transformed yeast strain SB34 (Bach et al. 2003) with our mutants. This strain contains wild-type copies of all SSA genes and therefore any change in [URE3] propagation relies on a dominant effect from the mutant Ssa protein. In addition, we constructed plasmids carrying SSA1 and SSA2 alleles that harbored a mutation equivalent to ssa2-10 (P395L) to compare the effects of the ssa2-10 mutation with our new SSA alleles under the same experimental conditions.
In contrast to weak effects on [PSI+] when wild-type SSAs are present, the SSA mutants had major effects on [URE3] stability. As shown in Table 4, introduction of an extra copy of SSA1 or SSA2 did not have a dramatic effect upon [URE3] stability, although there was a reproducible tendency for the appearance of a small percentage of [ure-0] cells by an extra copy of SSA1. We also observed an effect on stability of [URE3] for both SSA1 and SSA2 carrying the P395L mutation. The SSA2 allele showed slightly more impact upon [URE3] strength than the SSA1 allele.
TABLE 4.
Relative effects of SSA1 and SSA2 mutants on [URE3] propagation
| Mutations
|
% white colonies | % red colonies | % sectored colonies | Color after restreak | |
|---|---|---|---|---|---|
| In Ssa1p | In Ssa2p | ||||
| WT | 92 | 7 | 1 | 0 | |
| WT | 100 | 0 | 0 | 0 | |
| A4T | 98 | 1 | 1 | 1 | |
| D30N | 31 | 54 | 15 | 1 | |
| G41D | 96 | 1 | 3 | 2 | |
| G50D | 99 | 1 | 0 | 1 | |
| A53V | 14 | 14 | 72 | 1 | |
| A53V | 2 | 76 | 22 | 5 | |
| A53T | 48 | 22 | 20 | 5 | |
| T64I | 97 | 0 | 3 | 1 | |
| T64I | 97 | 1 | 2 | 2 | |
| G73D | 76 | 7 | 17 | 2 | |
| R74K | 99 | 1 | 0 | 1 | |
| R74K | 96 | 3 | 1 | 1 | |
| S119F | 20 | 30 | 50 | 5 | |
| A146T | 9 | 33 | 58 | 6 | |
| A155V | 7 | 15 | 78 | 5 | |
| A155T | 12 | 30 | 58 | 4 | |
| A176T | 45 | 5 | 50 | 8 | |
| T223I | 90 | 5 | 5 | 1 | |
| R259K | 85 | 10 | 5 | 1 | |
| G287D | 99 | 1 | 1 | 3 | |
| G287D | 40 | 40 | 20 | 3 | |
| T295I | 90 | 5 | 5 | 2 | |
| T295I | 98 | 1 | 1 | 1 | |
| A297T | 33 | 34 | 33 | 1 | |
| P395La | 14 | 16 | 70 | 2 | |
| P395La | 17 | 29 | 54 | 4 | |
| G481D | 40 | 30 | 30 | 2 | |
Colony color was scored subjectively as for Table 1. Colony percentage is given after transformation of the SSA mutant into SB34 and color after restreak is for a colony from [URE3] selective medium, as described in materials and methods.
P395L is a SSA2 mutation isolated as weakening [URE3] propagation (Roberts et al. 2004) and was used to provide comparison with our new SSA mutants.
The most informative way to view the data in Table 4 is to combine the effect upon [URE3] strength after restreaking (Table 4, last column, i.e., reflected by colony color) with the initial effect upon [URE3] stability after transformation (Table 4, columns 3–5, i.e., reflected appearance of [ure-0] cells). It is clear that a number of mutants in SSA2 (A53V, A53T, A146T, and A176T) and SSA1 (S119F, A155V, and A155T) have a dramatic effect upon [URE3] propagation. The effect of these alleles on [URE3] propagation is greater than that of the previously identified P395L mutant in these experimental conditions. There are also a number of mutants in both SSA1 and SSA2, such as A4T and R74K, which behave similar to wild-type SSA under these experimental conditions.
The SSA mutants that showed the strongest effect upon [URE3] propagation (A146T and A176T) are SSA2 alleles. In the case of the A53V mutation, a greater effect upon [URE3] propagation is seen for the SSA2 version compared to the SSA1 version. However, other mutations found in both SSA1 and SSA2 (T64I, R74K, and T295I) behave in virtually the same manner with regard to [URE3] propagation.
Western blot analyses for Hsp70–Ssa and Hsp104p expression levels in SB34 strains carrying a copy of SSA mutants indicate that there is no correlation between expression levels of these chaperones and the ability to impair [URE3], thereby suggesting a more direct effect on prion propagation (data not shown).
Interestingly, we also found that cells harboring SSA1 or SSA2 carrying the P395L mutation as the sole SSA source show no effect whatsoever on propagation of [PSI+] (data not shown). This supports the view that there are differences in the way Ssa proteins influence the propagation of different prions.
DISCUSSION
We have identified 25 new mutations within the two major cytosolic Hsp70–Ssa molecular chaperones of S. cerevisiae that impair the propagation of the [PSI+] prion. Five of these mutants were identified in identical residues of both Ssa1p and Ssa2p. This provides clear evidence that the Hsp70–Ssa family is providing a redundant function with respect to the [PSI+] prion. All but one of the mutants were located within the ATPase domain, which further highlights the importance of the ATPase regulation of Hsp70–Ssa in its role in prion propagation. Only one mutant, SSA2A176T, had a major effect on growth rate. This supports previous observations that essential and prion-related functions of Hsp70–Ssa can be separated (Jung et al. 2000; Jones and Masison 2003) or that prion substrates are more sensitive to perturbation of Ssa function than other substrates.
We further classified our Ssa mutants by whether or not they impaired propagation of a second yeast prion [URE3]. Previous genetic studies have uncovered clear differences in how chaperones influence the propagation of different yeast prions. Hsp104p overexpression has been shown to efficiently cure [PSI+] but has no effect on [URE3] or [RNQ+] (Chernoff et al. 1995; Derkatch et al. 1997; Moriyama et al. 2000) and overexpression of Ssa1p but not Ssa2p can efficiently cure [URE3] (Schwimmer and Masison 2002). Only the ssa2-10 mutant allele has previously been shown to affect [URE3] propagation (Roberts et al. 2004). We identified some mutants in both Ssa1p and Ssa2p that are capable of impairing [URE3] propagation. The data clearly show that specific mutations in Hsp70–Ssa that impair one prion are not guaranteed to impair a different prion. These results are not too surprising as there is clearly a complex relationship between chaperones and amyloid propagation both in vivo and in vitro (Newnam et al. 1999; Kushnirov et al. 2000; Inoue et al. 2004; Shorter and Lindquist 2004; Flower et al. 2005; Gokhale et al. 2005; Krzewska and Melki 2006; Shorter and Lindquist 2006). Differences in how different chaperones alter amyloid propagation and how the same chaperone may alter propagation of different amyloid species probably arise from in vivo interactions of the chaperone with cochaperones and from the physical structure of the amyloid (reviewed in Jones and Tuite 2005).
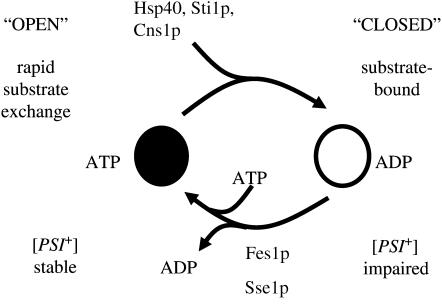
Previous genetic studies have suggested that promoting the substrate-bound ADP form of Hsp70–Ssa can impair prion propagation (Figure 4) (Jones and Masison 2003; Jones et al. 2004). While direct biochemical evidence is lacking to confirm this hypothesis, strong indirect evidence exists in the form of biochemical analysis of equivalent DnaK mutations (Mayer et al. 2001) and also the effect of Hsp40 and TPR cochaperones on ATPase stimulation of Ssa1p (Cyr et al. 1992; Wegele et al. 2003; Hainzl et al. 2004). Promoting the interaction of Hsp70–Ssa with the prion substrate may disrupt the finely tuned ATPase hydrolysis cycle and thus prevent prion propagation by not releasing substrate for prion conversion. Deregulation of the Hsp70–Ssa ATPase cycle can be achieved by interfering with interdomain communication or by altering interaction with cochaperones that influence ATP hydrolysis.
Figure 4.—
ATPase cycle of Ssa1p. The highly coordinated hydrolysis of ATP is used to switch between the open and closed forms of Hsp70 chaperone. The ATPase cycle is influenced by the action of Hsp40, TPR cochaperones, and nucleotide exchange factors. Genetic and biochemical data suggest that promotion of the ADP closed form of Hsp70 promotes impairment of prion propagation.
NMR studies (Revington et al. 2005) have recently identified a number of residues in bacterial Hsp70 that are important in communication between the ATPase domain and the PBD. Comparing our new SSA mutants to the data of Revington et al. (2005), we can infer that A53, G73, R74, A146, A176, and T223 may all play an important role in interdomain communication in Ssa proteins. Mutations at these residues may therefore disrupt the communication between the ATPase and the PBD of Ssa1p and Ssa2p and in turn alter [PSI+] propagation.
Vogel et al. (2006) have proposed an allosteric regulation of Hsp70 conformation by a proline switch. In DnaK, residues K70, P143, R151, and E171 play a role in controlling conformational changes in the substrate-binding domain and ATP hydrolysis. A significant number of our new Ssa mutants (G73D, R74K, A146T, A155V, and A176T) are located in regions around the corresponding residues in Ssa1p and Ssa2p, which further suggests that they may alter the interdomain communication control mechanism. We did not isolate mutants at these specific residues. However, these are essential residues for this interdomain communication mechanism and alteration would inactivate Hsp70 function. The nature of our genetic screen in identifying Hsp70 mutants requires maintenance of essential functions, ensuring that we would never succeed in isolating mutations at these essential sites.
Recently a crystal structure of bovine Hsc70 lacking the C terminus of the protein was solved (Jiang et al. 2005). This structure has allowed the identification of residues that form molecular interactions at the interface between the ATPase and substrate binding domains of Hsc70. A number of our new Hsp70 mutants (A146T, A155V, A176T, and T223I) are located in regions close to residues that are predicted to be important in forming productive interactions at the interface of ATPase and the PBD of Ssa proteins. This further adds weight to the prediction that a significant number of our Ssa mutants are affecting interdomain communication.
Altering the interaction between Hsp70–Ssa and its nucleotide exchange factors Fes1p or Sse1p will affect the finely tuned ATP hydrolysis cycle. A number of mutants (R259K, G287D, T295I, and A297T) are located within the IIB region that has been implicated as the interaction site between mammalian Hsp70 and the mammalian Fes1p homolog HspBP1 (Shomura et al. 2005). The IIB mutants may therefore be altered in their interaction with the S. cerevisiae nucleotide exchange factor Fes1p in a way that alters [PSI+] propagation. Fes1p has already been implicated in playing a role in [PSI+] propagation (Jones et al. 2004). When these mutant alleles are the sole SSA source in the cell [PSI+] propagation is severely impaired and this impairment is much stronger than is observed for a fes1Δ strain (Jones et al. 2004). This clear difference suggests that the SSA mutants do not cause a simple reduction or loss of interaction with this nucleotide exchange factor but probably affect the interaction in a more complex manner. Recently the yeast Hsp70/Hsp110 family member Sse1p has been identified as a second nucleotide exchange factor for Hsp70–Ssa (Dragovic et al. 2006; Raviol et al. 2006). It is conceivable that both exchange factors interact with Hsp70–Ssa through the IIB region and that our mutants are altering interaction with both or either of the nucleotide exchange factors in a complex manner. Similar to fes1Δ a deletion of SSE1 in our genetic backgrounds has a minor effect upon [PSI+] propagation (H. M. Loovers and G. W. Jones, unpublished data). If our Ssa mutants within the IIB region are promoting or increasing the time period Ssa stays bound to the prion-like substrate this may explain why these mutants impair prion propagation (Figure 4).
Hsp70–Ssa is an abundant protein chaperone that has the potential to interact with a wide variety of substrates and influence protein folding. It was recently reported that mature functional Sup35p is readily converted into the prion form (Satpute-Krishnan and Serio 2005). Sup35p is often found in complex with many other cellular proteins. It is possible that Hsp70–Ssa may play a role in [PSI+] propagation by participating in the process of remodeling protein complexes that contain Sup35p and during this process provide potential Sup35p prion substrate. Ssa1p and Ssa2p are involved in remodeling of the Rad9–Rad53 complex in response to DNA damage (Gilbert et al. 2003) and this remodeling function could extend to many as yet unidentified cellular complexes within yeast.
Further genetic and biochemical analysis of these new Ssa mutants will aid in understanding the precise role of Hsp70–Ssa in amyloid prevention, formation, and propagation within the yeast cell. Given the extent of conservation between yeast and mammalian cytosolic Hsp70's, what we discover in the yeast system may well be directly applicable in the mammalian cell.
Acknowledgments
We are indebted to Dan Masison for strains and plasmids. We thank Marc Blondel for strain SB34 and John Glover for Hsp104p antibody. We thank Sarah Perrett and Cathy Jones for critical reading of the manuscript. We thank David O'Connor for general help and advice and Peter Mowlds for initial experiments to identify some mutant alleles. This work was supported by a grant from the Irish Health Research Board (RP/04/227) and by a Marie Curie International Reintegration grant (516449) awarded to G.W.J.
References
- Aigle, M., and F. Lacroute, 1975. Genetical aspects of [URE3], a non-mitochondrial, cytoplasmically inherited mutation in yeast. Mol. Gen. Genet. 136: 327–335. [DOI] [PubMed] [Google Scholar]
- Allen, K. D., R. D. Wegrzyn, T. A. Chernova, S. Muller, G. P. Newnam et al., 2005. Hsp70 chaperones as modulators of prion life cycle: novel effects of Ssa and Ssb on the Saccharomyces cerevisiae prion [PSI+]. Genetics 169: 1227–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bach, S., N. Talarek, T. Andrieu, J. M. Vierfond, Y. Mettey et al., 2003. Isolation of drugs active against mammalian prions using a yeast-based screening assay. Nat. Biotechnol. 21: 1075–1081. [DOI] [PubMed] [Google Scholar]
- Chernoff, Y. O., S. L. Lindquist, B. Ono, S. G. Inge-Vechtomov and S. W. Liebman, 1995. Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor [psi+]. Science 268: 880–884. [DOI] [PubMed] [Google Scholar]
- Collinge, J., 2001. Prion diseases of humans and animals: their causes and molecular basis. Annu. Rev. Neurosci. 24: 519–550. [DOI] [PubMed] [Google Scholar]
- Coustou, V., C. Deleu, S. Saupe and J. Begueret, 1997. The protein product of the het-s heterokaryon incompatibility gene of the fungus Podospora anserina behaves as a prion analog. Proc. Natl. Acad. Sci. USA 94: 9773–9778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox, B. S., 1965. [PSI+], a cytoplasmic suppressor of super-suppressors in yeast. Heredity 20: 505–521. [Google Scholar]
- Cyr, D. M., X. Lu and M. G. Douglas, 1992. Regulation of Hsp70 function by a eukaryotic DnaJ homolog. J. Biol. Chem. 267: 20927–20931. [PubMed] [Google Scholar]
- Derkatch, I. L., M. E. Bradley, P. Zhou, Y. O. Chernoff and S. W. Liebman, 1997. Genetic and environmental factors affecting the de novo appearance of the [PSI+] prion in Saccharomyces cerevisiae. Genetics 147: 507–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derkatch, I. L., M. E. Bradley, J. Y. Hong and S. W. Liebman, 2001. Prions affect the appearance of other prions: the story of [PIN(+)]. Cell 106: 171–182. [DOI] [PubMed] [Google Scholar]
- Dragovic, Z., S. A. Broadley, Y. Shomura, A. Bracher and F. U. Hartl, 2006. Molecular chaperones of the Hsp110 family act as nucleotide exchange factors of Hsp70s. EMBO J. 25: 2519–2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Bellot, E., E. Guillemet and C. Cullin, 2000. The yeast prion [URE3] can be greatly induced by a functional mutated URE2 allele. EMBO J. 19: 3215–3222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flower, T. R., L. S. Chesnokova, C. A. Froelich, C. Dixon and S. N. Witt, 2005. Heat shock prevents alpha-synuclein-induced apoptosis in a yeast model of Parkinson's disease. J. Mol. Biol. 351: 1081–1100. [DOI] [PubMed] [Google Scholar]
- Gilbert, C. S., M. van den Bosch, C. M. Green, J. E. Vialard, M. Grenon et al., 2003. The budding yeast Rad9 checkpoint complex: chaperone proteins are required for its function. EMBO Rep. 4: 953–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover, J. R., and S. Lindquist, 1998. Hsp104, Hsp70, and Hsp40: a novel chaperone system that rescues previously aggregated proteins. Cell 94: 73–82. [DOI] [PubMed] [Google Scholar]
- Gokhale, K. C., G. P. Newnam, M. Y. Sherman and Y. O. Chernoff, 2005. Modulation of prion-dependent polyglutamine aggregation and toxicity by chaperone proteins in the yeast model. J. Biol. Chem. 280: 22809–22818. [DOI] [PubMed] [Google Scholar]
- Hainzl, O., H. Wegele, K. Richter and J. Buchner, 2004. Cns1 is an activator of the Ssa1 ATPase activity. J. Biol. Chem. 279: 23267–23273. [DOI] [PubMed] [Google Scholar]
- Inoue, Y., H. Taguchi, A. Kishimoto and M. Yoshida, 2004. Hsp104 binds to yeast Sup35 prion fiber but needs other factor(s) to sever it. J. Biol. Chem. 279: 52319–52323. [DOI] [PubMed] [Google Scholar]
- James, P., C. Pfund and E. A. Craig, 1997. Functional specificity among Hsp70 molecular chaperones. Science 275: 387–389. [DOI] [PubMed] [Google Scholar]
- Jiang, J., K. Prasad, E. M. Lafer and R. Sousa, 2005. Structural basis of interdomain communication in the Hsc70 chaperone. Mol. Cell 20: 513–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, G. W., and D. C. Masison, 2003. Saccharomyces cerevisiae Hsp70 mutations affect [PSI+] prion propagation and cell growth differently and implicate Hsp40 and tetratricopeptide repeat cochaperones in impairment of [PSI+]. Genetics 163: 495–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, G. W., and M. F. Tuite, 2005. Chaperonin prions: The cellular machinery for propagating an infectious protein? BioEssays 27: 823–832. [DOI] [PubMed] [Google Scholar]
- Jones, G., Y. Song, S. Chung and D. C. Masison, 2004. Propagation of Saccharomyces cerevisiae [PSI+] prion is impaired by factors that regulate Hsp70 substrate binding. Mol. Cell. Biol. 24: 3928–3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung, G., G. Jones, R. D. Wegrzyn and D. C. Masison, 2000. A role for cytosolic hsp70 in yeast [PSI+] prion propagation and [PSI+] as a cellular stress. Genetics 156: 559–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabani, M., J. M. Beckerich and J. L. Brodsky, 2002. a Nucleotide exchange factor for the yeast Hsp70 molecular chaperone Ssa1p. Mol. Cell. Biol. 22: 4677–4689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabani, M., C. McLellan, D. A. Raynes, V. Guerriero and J. L. Brodsky, 2002. b HspBP1, a homologue of the yeast Fes1 and Sls1 proteins, is an Hsc70 nucleotide exchange factor. FEBS Lett. 531: 339–342. [DOI] [PubMed] [Google Scholar]
- Kryndushkin, D. S., V. N. Smirnov, M. D. Ter-Avanesyan and V. V. Kushnirov, 2002. Increased expression of Hsp40 chaperones, transcripional factors, and ribosomal protein Rpp0 can cure yeast prions. J. Biol. Chem. 277: 23702–23708. [DOI] [PubMed] [Google Scholar]
- Krzewska, J., and R. Melki, 2006. Molecular chaperones and the assembly of the prion Sup35p, an in vitro study. EMBO J. 25: 822–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushnirov, V. V., D. S. Kryndushkin, M. Boguta, V. N. Smirnov and M. D. Ter-Avanesyan, 2000. Chaperones that cure yeast artificial [PSI+] and their prion-specific effects. Curr. Biol. 10: 1443–1446. [DOI] [PubMed] [Google Scholar]
- Lian, H. Y., Y. Jiang, H. Zhang, G. W. Jones and S. Perrett, 2006. The yeast prion protein Ure2: structure, function and folding. Biochim. Biophys. Acta 1764: 535–545. [DOI] [PubMed] [Google Scholar]
- Mayer, M. P., D. Brehmer, C. S. Gassler and B. Bukau, 2001. Hsp70 chaperone machines. Adv. Protein Chem. 59: 1–44. [DOI] [PubMed] [Google Scholar]
- Moriyama, H., H. K. Edskes and R. B. Wickner, 2000. [URE3] prion propagation in Saccharomyces cerevisiae: requirement for chaperone Hsp104 and curing by overexpressed chaperone Ydj1p. Mol. Cell. Biol. 20: 8916–8922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newnam, G. P., R. D. Wegrzyn, S. L. Lindquist and Y. O. Chernoff, 1999. Antagonistic interactions between yeast chaperones Hsp104 and Hsp70 in prion curing. Mol. Cell. Biol. 19: 1325–1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patino, M. M., J. J. Liu, J. R. Glover and S. Lindquist, 1996. Support for the prion hypothesis for inheritance of a phenotypic trait in yeast. Science 273: 622–626. [DOI] [PubMed] [Google Scholar]
- Paushkin, S. V., V. V. Kushnirov, V. N. Smirnov and M. D. Ter-Avanesyan, 1997. Interaction between yeast Sup45p (eRF1) and Sup35p (eRF3) polypeptide chain release factors: implications for prion-dependent regulation. Mol. Cell. Biol. 17: 2798–2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prusiner, S. B., 1982. Novel proteinaceous infectious particles cause scrapie. Science 216: 136–144. [DOI] [PubMed] [Google Scholar]
- Raviol, H., H. Sadlish, F. Rodriguez, M. P. Mayer and B. Bukau, 2006. Chaperone network in the yeast cytosol: Hsp110 is revealed as an Hsp70 nucleotide exchange factor. EMBO J. 25: 2510–2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revington, M., Y. Zhang, G. N. Yip, A. V. Kurochkin and E. R. Zuiderweg, 2005. NMR investigations of allosteric processes in a two-domain Thermus thermophilus Hsp70 molecular chaperone. J. Mol. Biol. 349: 163–183. [DOI] [PubMed] [Google Scholar]
- Roberts, B. T., H. Moriyama and R. B. Wickner, 2004. [URE3] prion propagation is abolished by a mutation of the primary cytosolic Hsp70 of budding yeast. Yeast 21: 107–117. [DOI] [PubMed] [Google Scholar]
- Satpute-Krishnan, P., and T. R. Serio, 2005. Prion protein remodelling confers an immediate phenotypic switch. Nature 437: 262–265. [DOI] [PubMed] [Google Scholar]
- Schatz, P. J., F. Solomon and D. Botstein, 1988. Isolation and characterization of conditional-lethal mutations in the TUB1 alpha-tubulin gene of the yeast Saccharomyces cerevisiae. Genetics 120: 681–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwimmer, C., and D. C. Masison, 2002. Antagonistic interactions between yeast [PSI(+)] and [URE3] prions and curing of [URE3] by Hsp70 protein chaperone Ssa1p but not by Ssa2p. Mol. Cell. Biol. 22: 3590–3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shomura, Y., Z. Dragovic, H. C. Chang, N. Tzvetkov, J. C. Young et al., 2005. Regulation of Hsp70 function by HspBP1: structural analysis reveals an alternate mechanism for Hsp70 nucleotide exchange. Mol. Cell 17: 367–379. [DOI] [PubMed] [Google Scholar]
- Shorter, J., and S. Lindquist, 2004. Hsp104 catalyzes formation and elimination of self-replicating Sup35 prion conformers. Science 304: 1793–1797. [DOI] [PubMed] [Google Scholar]
- Shorter, J., and S. Lindquist, 2006. Destruction or potentiation of different prions catalyzed by similar Hsp104 remodeling activities. Mol. Cell 23: 425–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski, R. S., and P. Hieter, 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122: 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, Y., Y. X. Wu, G. Jung, Y. Tutar, E. Eisenberg et al., 2005. Role for Hsp70 chaperone in Saccharomyces cerevisiae prion seed replication. Eukaryot. Cell 4: 289–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, K. L., and R. B. Wickner, 2001. [URE3] and [PSI]: prions of Saccharomyces cerevisiae. Contrib. Microbiol. 7: 21–31. [DOI] [PubMed] [Google Scholar]
- True, H. L., 2006. The battle of the fold: chaperones take on prions. Trends Genet. 22: 110–117. [DOI] [PubMed] [Google Scholar]
- Tuite, M. F., 2000. Yeast prions and their prion-folding domain. Cell 100: 289–292. [DOI] [PubMed] [Google Scholar]
- Uptain, S. M., G. J. Sawicki, B. Caughey and S. Lindquist, 2001. Strains of [PSI(+)] are distinguished by their efficiencies of prion-mediated conformational conversion. EMBO J. 20: 6236–6245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel, M., B. Bukau and M. P. Mayer, 2006. Allosteric regulation of Hsp70 chaperones by a proline switch. Mol. Cell 21: 359–367. [DOI] [PubMed] [Google Scholar]
- Wegele, H., M. Haslbeck, J. Reinstein and J. Buchner, 2003. Sti1 is a novel activator of the Ssa proteins. J. Biol. Chem. 278: 25970–25976. [DOI] [PubMed] [Google Scholar]
- Wegele, H., L. Muller and J. Buchner, 2004. Hsp70 and Hsp90—a relay team for protein folding. Rev. Physiol. Biochem. Pharmacol. 151: 1–44. [DOI] [PubMed] [Google Scholar]
- Werner-Washburne, M., J. Becker, J. Kosic-Smithers and E. A. Craig, 1989. Yeast Hsp70 RNA levels vary in response to the physiological status of the cell. J. Bacteriol. 171: 2680–2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner, R. B., 1994. [URE3] as an altered URE2 protein: evidence for a prion analog in Saccharomyces cerevisiae. Science 264: 566–569. [DOI] [PubMed] [Google Scholar]