Abstract
S*S (Silver), S*N (wild type/gold), and S*AL (sex-linked imperfect albinism) form a series of alleles at the S (Silver) locus on chicken (Gallus gallus) chromosome Z. Similarly, sex-linked imperfect albinism (AL*A) is the bottom recessive allele at the orthologous AL locus in Japanese quail (Coturnix japonica). The solute carrier family 45, member 2, protein (SLC45A2), previously denoted membrane-associated transporter protein (MATP), has an important role in vesicle sorting in the melanocytes. Here we report five SLC45A2 mutations. The 106delT mutation in the chicken S*AL allele results in a frameshift and a premature stop codon and the corresponding mRNA appears to be degraded by nonsense-mediated mRNA decay. A splice-site mutation in the Japanese quail AL*A allele causes in-frame skipping of exon 4. Two independent missense mutations (Tyr277Cys and Leu347Met) were associated with the Silver allele in chicken. The functional significance of the former mutation, associated only with Silver in White Leghorn, is unclear. Ala72Asp was associated with the cinnamon allele (AL*C) in the Japanese quail. The most interesting feature concerning the SLC45A2 variants documented in this study is the specific inhibition of expression of red pheomelanin in Silver chickens. This phenotypic effect cannot be explained on the basis of the current, incomplete, understanding of SLC45A2 function. It is an enigma why recessive null mutations at this locus cause an almost complete absence of both eumelanin and pheomelanin whereas some missense mutations are dominant and cause a specific inhibition of pheomelanin production.
PIGMENTATION in birds and mammals is based on the synthesis of two different types of melanin, brown/black eumelanin and yellow/red pheomelanin. Tyrosinase is the rate-limiting enzyme of melanin biosynthesis, which takes place in melanosomes within melanocytes. Tyrosinase and the tyrosinase-related proteins Tyrp1 and Tyrp2 (Dct) are involved in the production of eumelanin. Only the presence of cysteine and some tyrosinase activity appear to be required for the production of pheomelanin. When tyrosinase is expressed at low levels, pheomelanin is produced by the addition of cysteine to dopaquinone (Kobayashi et al. 1995; Wakamatsu and Ito 2002; Kushimoto et al. 2003). Simplified, high tyrosinase activity is associated with synthesis of eumelanin whereas low activity results in the production of pheomelanin. The spherical pheomelanin premelanosomes are less organized than the rod-shaped eumelanin premelanosomes and contain less melanin (Brumbaugh 1968).
In birds, females are the heterogametic sex (ZW) and males the homogametic sex (ZZ). The sex-linked Silver locus controlling Silver (S*S) and wild type/gold (S*N) plumage color (Figure 1) in chicken (Gallus gallus) was described in 1912 by Sturtevant, who found a sex-linked factor (Silver) that inhibits red color pigmentation (Sturtevant 1912). Silver is incompletely dominant to wild type and its phenotypic expression is highly influenced by modifying genes. It can thus be difficult to identify Silver in some genetic backgrounds (Smyth 1990). Sex-linked imperfect albinism (S*AL) is the bottom recessive allele at this locus (Werret et al. 1959; Cole and Jeffers 1963) (Figure 1). S*AL birds have white plumage with a ghost pattern that depends on genetic background. The eyes are pink at hatching but darken with age except for the red pupils (Mueller and Hutt 1941; Hutt and Mueller 1943; Werret et al. 1959; Silversides and Crawford 1990).
Figure 1.—
Chickens expressing the wild type (S*N), Silver (S*S), and sex-linked imperfect albinism (S*AL) phenotypes.
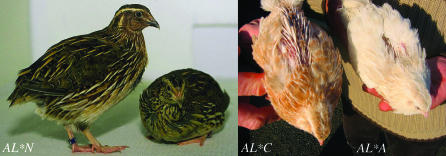
The recessive allele for sex-linked albinism in the Japanese quail (Coturnix japonica) was designated AL*A, while the wild-type allele was denoted AL*N (Lauber 1964; Minvielle et al. 2000) (Figure 2). By intergeneric crossing of male chickens (G. gallus) homozygous for S*S, S*N, or S*AL to female Japanese quail (C. japonica) hemizygous for AL*N or AL*A, it was found that S in chicken and AL in quail are orthologous. All five intergeneric hybrids from the crossing of albinos from the two Phasianidae species were albino (Silversides and Mérat 1991). Cinnamon (AL*C) is another allele at the AL locus in the Japanese quail (Figure 2). Both the AL*C and the AL*A mutations are caused by recessive alleles, and AL*C is dominant over AL*A (Truax and Johnson 1979; Cheng and Kimura 1990; Minvielle et al. 2000). Thus, the hierarchy of dominance for these alleles is AL*N > AL*C > AL*A. The plumage of wild-type Japanese quail is brown in variable shades (Figure 2). The quail albino chicks have bright pink eyes and yellow to white color. The adult birds have white plumage with buff ghost barring (Lauber 1964; Cheng and Kimura 1990). Cinnamon is phenotypically identical to the dark-eyed dilute (AL*D) allele (Cheng and Kimura 1990). The eyes of the AL*D chicks are red and have subnormal melanin pigmentation but darken with age. The AL*D mutation results in dilution of the brown pigment of the feathers but the plumage pattern is not affected (Cheng and Kimura 1990).
Figure 2.—
Japanese quails expressing the wild type (AL*N), cinnamon (AL*C), and sex-linked imperfect albinism (AL*A) phenotypes.
The gene encoding solute carrier family 45, member 2, protein (SLC45A2) is associated with pigmentation variation in several vertebrates; SLC45A2 was previously known as membrane-associated transporter protein (MATP). SLC45A2 mutations have been found in medaka (Fukamachi et al. 2001), humans (Newton et al. 2001), mouse (Newton et al. 2001; Du and Fisher 2002), and horse (Mariat et al. 2003). Oculocutaneous albinism type IV (OCA4) in humans is caused by mutations in SLC45A2 (Newton et al. 2001). SLC45A2 has 12 predicted transmembrane regions (Fukamachi et al. 2001) but the function of SLC45A2 is not fully understood. Mutations in SLC45A2 have been shown to disrupt tyrosinase processing and trafficking at the post-Golgi level (Kushimoto et al. 2003; Hearing 2005). SLC45A2 was first identified as an antigen in melanoma (AIM), AIM1 (Harada et al. 2001).
The sex-linked Silver locus in chicken is known to be located on the upper half of chicken chromosome Z, 2.4 cM from the slow-feathering (K) locus (Bitgood 1999), which is tightly linked to the chicken endogenous virus ev21 (Bacon et al. 1988). SLC45A2 is located in the vicinity of this viral insertion (within a 300-kb distance on chicken chromosome Z, http://www.genome.ucsc.edu). Here we show that mutations in SLC45A2 cause imperfect albinism both in chicken (S*AL) and in Japanese quail (AL*A) as well as the Silver (S*S) and cinnamon (AL*C) phenotypes in the two species.
MATERIALS AND METHODS
Animals:
A number of chicken pedigrees and breeds have been used in this study. A three-generation pedigree from an intercross between one red jungle fowl (RJF) male and three White Leghorn (WL) females has been generated for gene mapping (Schutz et al. 2002). From the F1 generation four males and 37 females were selected to generate an F2 generation, showing a wide diversity of plumage color (Kerje et al. 2003). Two other family materials were also used. The first segregated for Silver (White Buttercup, S*S vs. Brown Buttercup, S*N) and is an experimental cross, developed for the identification of plumage color genes. The origins of the cross are a broiler line and a brown layer line. The second segregated for Silver (S*S) and imperfect albinism (S*AL) and is a synthetic population (Mérat et al. 1986) currently raised at the INRA Génétique Factorielle Avicole (GFA) experimental unit in Nouzilly. The birds carrying S*AL were received from R. G. Somes in Connecticut in 1979. In the 1980s these birds were crossed with a synthetic French line carrying Silver. Rhode Island Red (S*N) has also been crossed into this population. A total of 50 animals exhibiting different genotypes at the S locus were sampled from this gene pool. DNA samples from different chicken breeds collected by the AvianDiv project (Hillel et al. 2003), red jungle fowl DNA from four different Scandinavian zoo populations (Håkansson and Jensen 2005), and White Leghorn DNA from the hypothyroid obese strain (OS) (Cole 1966) were also used.
Japanese quail DNA samples from wild type (AL*N), cinnamon (AL*C), and imperfect albinism (AL*A) raised at the INRA GFA experimental unit in Nouzilly were used for sequencing. Skin tissue samples used for RNA extractions were from AL*N and AL*A quails and heterozygous (S*S/S*AL or S*N/S*AL) male chicks.
Sequencing of genomic DNA:
Primers for amplification and sequencing of all SLC45A2 exons for both chicken and quail were designed using in silico predicted intron–exon boundaries and DNA sequences from the February 2004 chicken genome assembly (http://www.genome.ucsc.edu). In chicken, the gene was amplified in six parts, using the primer pairs ex1Fpcr/ex1Rpcr, ex2Fpcr/ex2Rpcr, ex3Fpcr/ex3Rpcr, ex4&5Fpcr/ex4&5Rpcr, ex6Fpcr/ex6Rpcr, and ex7Fpcr/ex7Rpcr; both the PCR primers and internal sequencing primers were used for direct sequencing (supplemental Table 1 at http://www.genetics.org/supplemental/). Up- and downstream regions of chicken SLC45A2 were sequenced using the primer pairs up10kbF/up10kbR, up20kbF/up20kbR, dwn9kbF/dwn9kbR, dwn20kbF/dwn20kbR, dwn30kbF/dwn30kbR, and dwn80kbF/dwn80kbR.
Due to the difficulties in amplifying quail DNA with chicken primers some additional primers were designed to amplify the quail exons. In quail, the gene was finally amplified in six parts, using the primer pairs ex1Fpcr/ex1Rpcr, ex2Fpcr/ex2Rseq, ex3Fseq/ex3Rpcr3, ex4&5Fpcr/ex4&5Rpcr, ex6Fpcr3/ex6Rpcr3, ex7Fpcr3/ex7Rpcr3, and ex7Fpcr5/ex7Rpcr5 (supplemental Table 1 at http://www.genetics.org/supplemental/).
All PCR reactions were carried out in a total volume of 10–20 μl and contained ∼50 ng genomic DNA, 1× PCR buffer II (Applied Biosystems, Foster City, CA), 2.5 mm MgCl2, 200 μm dNTPs, 0.75–1 unit AmpliTaq Gold DNA polymerase (Applied Biosystems), and 20 pmol of each primer. The PCRs were performed in an Applied Biosystems 2720 thermal cycler and started with 5 min at 94°, followed by a touchdown PCR program starting with denaturation at 94° for 30 sec, annealing for 30 sec, and elongation at 72° for 1 min/kb of PCR product. The annealing temperature started at 65° and was then decreased by 1°/cycle for 14 cycles, an additional 30 cycles was run on 51° constant annealing temperature, and the last cycle ended with 72° for 10 min.
PCR fragments were gel purified using the E.Z.N.A gel extraction kit (Omega Bio-tek, Doraville, GA) and sequenced directly using the DYEnamic ET dye terminator kit (MegaBACE) and the MegaBACE 1000 instrument (GE Healthcare Bio-Sciences, Uppsala, Sweden). Sequences were analyzed with Sequence Analysis software (GE Healthcare Bio-Sciences) and Sequencher 3.1.1 software (Gene Codes, Ann Arbor, MI).
cDNA sequencing plus 5′ and 3′ RACE:
The First-Strand cDNA Synthesis kit (GE Healthcare Bio-Sciences) was used for cDNA synthesis from 14-day-old whole-embryo chicken total RNA (Kerje et al. 2004). PCR amplifications were performed using the cDNAexon1F and cDNAexon7R primers (supplemental Table 1 at http://www.genetics.org/supplemental/) and the reactions were carried out as described above but with minor changes: 130 ng of cDNA per reaction and 35 cycles on 51° constant annealing temperature. The PCR fragment was cloned using the TOPO TA cloning kit (Invitrogen, Carlsbad, CA) and sequenced using the T7 and M13 universal primers. Primers for 5′ and 3′ RACE were designed on the basis of the obtained sequences.
5′ and 3′ RACE were performed on RACE-ready first-strand cDNA from broiler brain according to the Gene Racer kit protocol (Invitrogen). The SLC45A2-specific primers used in this experiment, together with the primers provided by the kit, were chRACE5′rev1, chRACE3′fwd2, and the nested primer chRACE3′fwd1 (supplemental Table 1 at http://www.genetics.org/supplemental/). The RACE-ready first-strand cDNA was diluted 1:9 and 1 μl was used for PCR amplification in a total volume of 20 μl with 1× PCR buffer II (Applied Biosystems), 2 mm MgCl2, 200 μm dNTPs, 1 unit AmpliTaq Gold DNA polymerase (Applied Biosystems), and 20 pmol of each primer. The PCR started with 5 min at 94°, followed by a touchdown PCR program starting with 94° for 30 sec, annealing for 30 sec, and elongation at 72° for 1 min. The annealing temperature started at 72° and was then decreased 1°/cycle for 9 cycles, an additional 40 cycles were run on 66° constant annealing temperature, and the last cycle ended with 72° for 10 min. The PCR fragments were cloned using the TOPO TA cloning kit (Invitrogen) and sequenced using the T7 and M13 universal primers.
Genotyping and linkage mapping:
Pyrosequencing with Pyro Gold chemistry was used to analyze chicken coding SNPs in exon 3 and exon 4 (Biotage, Uppsala, Sweden). A 113-bp fragment containing the SNP at position 902 A → G in exon 3 was amplified with the PYROex3Fseq and PYROex3Rbio primers and a 104-bp fragment containing the SNP at position 1111 C → A in exon 4 was amplified with PYROex4Fbio and PYROex4Rseq primers (supplemental Table 1 at http://www.genetics.org/supplemental/). PCR reactions were carried out as described above with minor changes. For the PYROex3Fseq/PYROex3Rbio the PCR started with 5 min at 94°, followed by 45 cycles of 94° for 30 sec, 51° for 30 sec, and 72° for 15 sec, and the last cycle ended with 72° for 10 min. For the PYROex4Fbio/PYROex4Rseq the regular touchdown program was used with 40 cycles at 51° constant annealing temperature. The PYROex3Fseq and PYROex4Rseq were also used as sequencing primers in their respective tests and were designed to anneal just prior the SNP of interest. SLC45A2 was mapped in relation to other markers genotyped in the RJF × WL pedigree using the TWOPOINT function in CRIMAP (Green et al. 1990). The BUILD and FLIPS functions were used to test the order of markers.
Confirmation of nonsense-mediated mRNA decay in imperfect albino chickens:
Total RNA was isolated from skin of chicks heterozygous for the S*AL deletion using the RNeasy fibrous tissue mini kit (QIAGEN, Valencia, CA). The tissues had been stored in RNAlater (Ambion, Austin, TX) at −80° prior to extraction. The RNA concentration and purity were checked using a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE). The RNA was treated with DNase according to the instructions for the DNA-free kit (Ambion) before cDNA synthesis using the First-Strand cDNA Synthesis kit (GE Healthcare Bio-Sciences). The chSALtestF/chSALtestR2 primers (supplemental Table 1 at http://www.genetics.org/supplemental/) were used to amplify and sequence the cDNA from chicks heterozygous for S*AL to test if the 106delT allele results in a downregulation of the mRNA due to nonsense-mediated mRNA decay (NMD). The reactions were carried out as described above with minor changes: 1 μl of the cDNA in a 20-μl reaction and 40 cycles at 51° constant annealing temperature. From the same individuals genomic DNA was extracted using standard methods and the ex1Fpcr/ex1Rpcr primers were used for amplification and sequencing as described above.
Expression analysis of the mutation causing imperfect albinism in Japanese quail:
Total RNA was isolated and cDNA synthesized from quail skin as described above for chicken skin. The test was designed to check if the G → T mutation at the splice acceptor site in the imperfect albinism allele at the intron3/exon 4 border results in an in-frame exon skipping. The AlQex3cDNAfwd/AlQex5cDNArev primers (supplemental Table 1 at http://www.genetics.org/supplemental/) were used for amplification of quail cDNA. The reactions were carried out as described above for the NMD test.
RESULTS
Assignment of SLC45A2 to the chicken linkage map:
The avian leucosis virus sequence (X54094) associated with the slow-feathering (K) locus was used as bait to search the May 2006 chicken genome assembly (http://www.genome.ucsc.edu) to find the chromosomal region harboring the Silver locus. A hit was found on chromosome Z around position 10.0 Mb. SLC45A2 is located ∼300 kb apart from the retroviral insertion at position ∼9.7 Mb, consistent with a map distance of a few centimorgans between K and Silver (Bacon et al. 1988). SLC45A2 was therefore identified as an obvious positional candidate gene for Silver. A 902 A → G SNP was found by direct sequencing of exon 3 and used to map SLC45A2 in relation to other markers genotyped in our RJF × WL intercross. Multipoint analysis revealed the following map order and map distances: ADL0022–(12.4 cM)–SLC45A2–(16.1 cM)–MCW0053. The order of loci was supported by a LOD score of 29.4 compared to the second most probable order.
Sequence analysis of chicken SLC45A2:
Ab initio predictions of transcripts from multiexon genes have been shown to be very error prone. GENSCAN, one of the best programs, has a sensitivity and specificity of ∼90% for detecting exons (Burge and Karlin 1997). Therefore, we decided to experimentally verify the exon composition of the full-length SLC45A2 transcript. The cDNA sequence obtained from a 14-day-old whole chicken embryo and broiler brain (deposited in GenBank with accession no. DQ900684) differed to a large extent from the one predicted by GENSCAN regarding exon/intron organization. A considerable part of exon 3 was missing and the last part of the sequence was erroneously predicted with an extra exon instead of the stop codon in exon 7. However, there is a predicted transcript XP429218 (http://www.ncbi.nlm.nih.gov/entrez) that correctly predicts exons 1–7 of SLC45A2 but also adds an exon 8 that we could not verify experimentally.
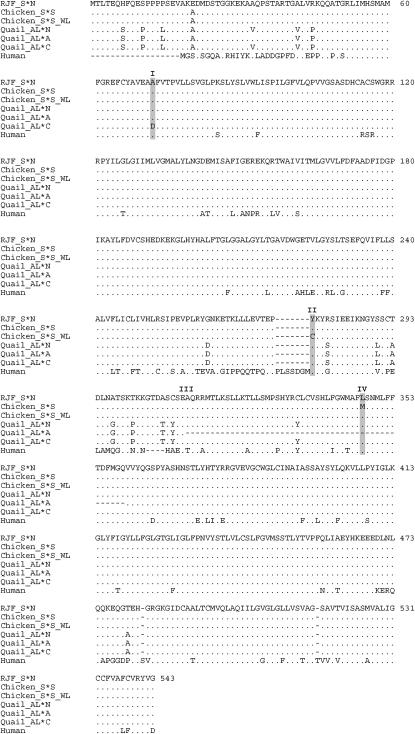
All seven exons, including the splice sites and adjacent intronic parts, of SLC45A2 were sequenced from genomic DNA samples representing different Silver alleles to evaluate SLC45A2 as a candidate gene for this locus. The analysis involved four red jungle fowl, eight Silver birds, one imperfect albino, and a domestic fowl assumed to carry the S*N allele. The S*S allele was associated with a C → A transition in exon 4 (Leu347Met) affecting transmembrane region 7 (Figures 3 and 4). This mutation was associated with Silver in all chicken breeds carrying this allele (the Silver, Fayoumi, Yurlov Crower, Rhode White, Friesian Fowl, and White Buttercup populations), except White Leghorn. An A → G transition in exon 3 (Tyr277Cys) affecting a loop region was found in the White Leghorn lines also assumed to carry Silver (Figures 3 and 4).
Figure 3.—
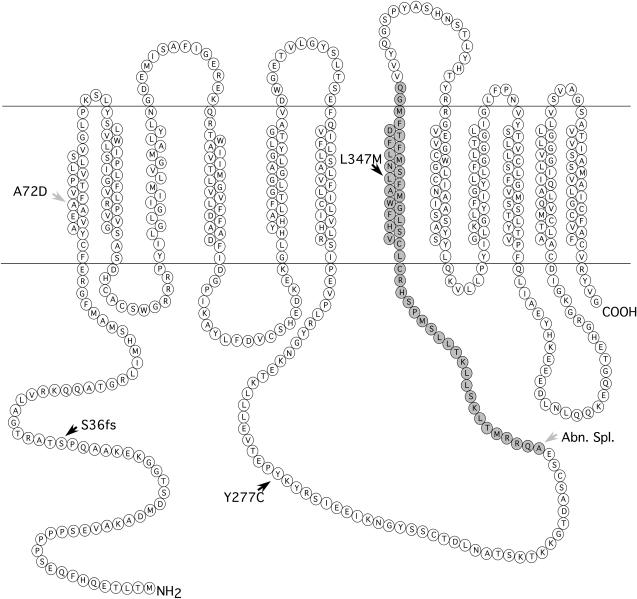
Membrane topology prediction of the SLC45A2 protein using TMHMM (v. 2.0; http://www.cbs.dtu.dk/services/TMHMM-2.0/). The locations of the frameshift mutation (S36fs) associated with imperfect albino (S*AL) and the two missense mutations Y277C and L347M associated with Silver (S*S) in chicken are indicated by solid arrowheads. The A72D mutation associated with cinnamon (AL*C) in Japanese quail is marked with a shaded arrowhead. The missing amino acids in the SLC45A2 protein encoded by sex-linked imperfect albinism (AL*A) in Japanese quail are shaded. Abn.Spl., aberrant splicing.
Figure 4.—
Multiple-protein alignment of SLC45A2 using chicken, quail, and human sequences. (I) The quail A72D mutation associated with cinnamon. (II) The chicken Y277C mutation associated with Silver in White Leghorn. (III) The beginning of the region that is missing in sex-linked imperfect albinism in Japanese quail. (IV) The chicken L347M mutation associated with Silver. A dot indicates identity to the master sequence and a dash indicates insertion/deletion differences between the sequences.
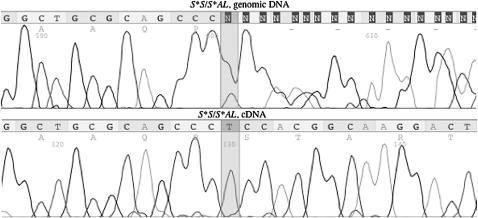
A 1-bp deletion (106delT) was revealed in S*AL, resulting in a frameshift at codon 36 and thereby a stop codon in exon 1 (Figure 3). Since a BglI site was created by the 106del mutation, a PCR–RFLP analysis was set up using BglI digestion. The test revealed a complete concordance between the presence of the 106del mutation and the imperfect albino genotype among 44 chickens, 31 albino (28 hemizygous females, 3 homozygous males) and 13 heterozygous carriers. Furthermore, the 106del mutation was not found in 6 nonalbino females, showing either the gold or the silver phenotype. A comparison of genomic DNA and cDNA from S*AL heterozygotes strongly suggested that NMD leads to degradation of the S*AL transcript (Figure 5). Direct sequencing of genomic DNA confirmed that the two tested birds were heterozygous for the S*AL deletion. Sequencing of cDNA from the same animals revealed only transcripts from the wild-type allele.
Figure 5.—
Sequence traces for part of chicken SLC45A2 exon 1 indicating nonsense-mediated mRNA decay (NMD) of the S*AL transcript. The top sequence shows sequenced genomic DNA of a chicken heterozygous for 106delT; position 106 is heterozygous T/delT and the two sequences are out of frame after this position. The bottom sequence shows the cDNA sequence from the same individual; only the wild-type sequence is apparent.
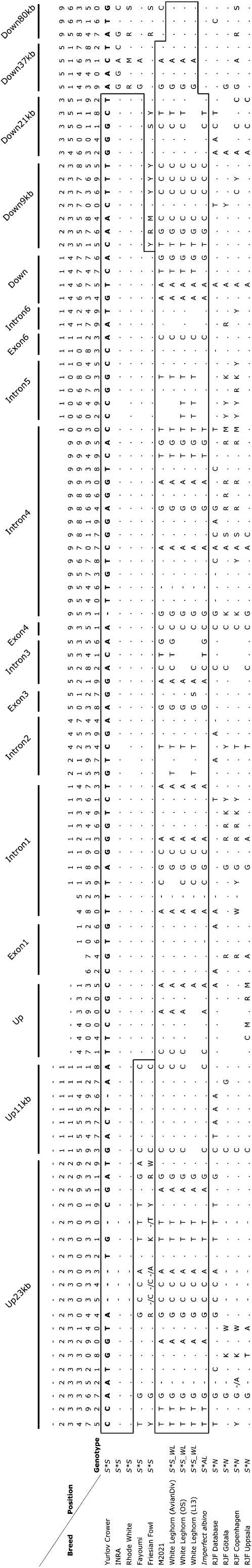
By additional sequencing of the up- and downstream regions of SLC45A2 across populations with different genotypes we could document that all chicken breeds carrying the L347M mutation had a minimum shared haplotype <35 kb, including the entire SLC45A2 coding sequence (Figure 6). The minimum shared haplotype among three different White Leghorn lines was even larger and extended for the entire region (>100 kb) covered in this study (Figure 6). It is worth noting that one sequenced bird (M2021) from the Nutreco experimental cross and the sequenced imperfect albino (S*AL) shared the same haplotype as White Leghorns for the entire SLC45A2 gene but not for the downstream region (Figure 6).
Figure 6.—
Alignment of nucleotide sequence polymorphisms in SLC45A2 and flanking regions among chickens with different genotypes at the Silver locus. All seven exons (1959 bp) were sequenced. Totals of 4081 bp of intronic sequence adjacent to the exons and 4128 bp of up- and downstream sequences were analyzed. The Yurlow Crower is used as the master sequence, dots indicate identity to the master sequence, dashes indicate insertion/deletion differences between the sequences, and open spaces indicate no information. Heterozygote positions are denoted as follows: R, A/G; Y, C/T; K, G/T; M, A/C; S, G/C; and W, A/T. The marked areas illustrate shared haplotypes between breeds. All sequences originate from breeds included in Table 1 except RJF Database and RJF Uppsala. RJF Database is the publicly available genome sequence of the chicken genome and RJF Uppsala is the breed used in our red jungle fowl/White Leghorn intercross.
Sequence analysis of Japanese quail SLC45A2:
SLC45A2 in quail was sequenced using genomic DNA and chicken-specific primers. A mutation was found in the imperfect albino (AL*A) birds, causing a G → T transversion at the splice acceptor site just preceding exon 4. Sequencing of cDNA prepared from skin samples from these birds confirmed that exon 4 is not present in the transcript (Figure 3). Another mutation was found in exon 1 of the cinnamon (AL*C) birds, causing a transition from C → A at nt 287 (Ala72Asp) (Figures 3 and 4).
A sequence comparison between the wild-type quail vs. the public red jungle fowl chicken genome sequence revealed a sequence identity in exons of 96.3%.
Association analysis across chicken breeds:
A number of chicken breeds with or without the Silver phenotype and including four captive red jungle fowl populations were screened for the two missense mutations Y277C and L347M (Table 1). The L347M mutation showed an almost complete association with Silver across all populations presumed to carry this allele except the White Leghorns and it was not found in birds with the wild-type allele at this locus. We are convinced that the few discrepancies concerning the L347M mutation are due to phenotyping errors. Two White Buttercup (S*S/S*N or S*S/W) and five Brown Buttercup (S*N/S*N or S*N/W) birds showed discrepant SLC45A2 genotypes (Table 1). These are most likely phenotyping errors since the scoring was done using 1-day-old chicks. At that age it can be difficult to unequivocally distinguish the phenotypes.
TABLE 1.
Distribution of the Y277C and L347M mutations in chicken SLC45A2 across populations
| Y277C
|
L347M
|
||||||
|---|---|---|---|---|---|---|---|
| Breed (origin) | S allele | A/A, A/W (wt) | G/– (S*S_WL) | Total | C/C, C/W (wt) | A/– (S*S) | Total |
| Fayoumi (INRA, France) | S*S | 5 | 0 | 5 | 0 | 5 | 5 |
| Yurlov Crower (INRA, France) | S*S | 5 | 0 | 5 | 0 | 5 | 5 |
| Rhode White (AvianDiv) | S*S | 8 | 0 | 8 | 0 | 8 | 8 |
| Friesian Fowl (AvianDiv) | S*S | 2 | 0 | 2 | 0 | 2 | 2 |
| White Buttercup (Nutreco, Holland) | S*S | 9 | 5 | 14 | 2 | 104 | 106 |
| White Leghorn (AvianDiv) | S*S_WL | 0 | 8 | 8 | 8 | 0 | 8 |
| White Leghorn (SLU13, Sweden) | S*S_WL | 0 | 15 | 15 | 16 | 0 | 16 |
| White Leghorn (OS, Sweden) | S*S_WL | 0 | 6 | 6 | 6 | 0 | 6 |
| Brown Buttercup (Nutreco, Holland) | S*N | 0 | 21 | 21 | 116 | 5 | 121 |
| Godollo Nhx (AvianDiv) | S*N | 1 | 7 | 8 | 8 | 0 | 8 |
| Green-Legged Partridge (AvianDiv) | S*N | 0 | 8 | 8 | 8 | 0 | 8 |
| Rhode Island Red (AvianDiv) | S*N | 1 | 7 | 8 | 8 | 0 | 8 |
| Ukrainian Bearded (AvianDiv) | S*N | 0 | 8 | 8 | 7 | 0 | 7 |
| Friesian Fowl (AvianDiv) | S*N | 4 | 1 | 5 | 6 | 0 | 6 |
| Red jungle fowl (Ebeltoft, Denmark) | S*N | 2 | 1 | 3 | 5 | 0 | 5 |
| Red jungle fowl (Frösö, Sweden) | S*N | 3 | 0 | 3 | 3 | 0 | 3 |
| Red jungle fowl (Götala, Sweden) | S*N | 5 | 0 | 5 | 5 | 0 | 5 |
| Red jungle fowl (Copenhagen) | S*N | 2 | 0 | 2 | 3 | 0 | 3 |
| Total | 47 | 87 | 134 | 201 | 129 | 330 | |
White Leghorns are presumed to carry the Silver allele. The tested White Leghorns representing three different lines did not carry the L347M mutation but were homo- or hemizygous for the Y277C mutation. However, the data across populations did not provide an unequivocal support for this being a causative mutation since it was also common in domestic fowl assumed to be non-Silver, including the Rhode Island Red, which apparently expresses red pigmentation. This missense mutation was uncommon in the red jungle fowl, and only 1 of 13 birds carried this mutation (Table 1).
Weak association between Y277C and plumage color in a red jungle fowl/White Leghorn intercross:
The extensive variation in plumage color in our intercross between the red jungle fowl and White Leghorn is expected to be controlled by at least four loci: Dominant white, Extension, Silver, and Barred. We have previously demonstrated that Dominant white in White Leghorns is due to a 9-bp insertion in the coding sequence of PMEL17 (Kerje et al. 2004) and Extended black represents a missense mutation in the gene for the melanocortin 1-receptor (MC1R) (Kerje et al. 2003). Since the single red jungle fowl male was homozygous for the SLC45A2 277Y allele and the three White Leghorn founder females were hemizygous for 277C we could utilize the data on plumage color from ∼800 F2 progeny to investigate a possible association between this mutation and plumage color. No significant association was recognized in males (data not shown) while a weak, but significant, association was revealed in females (Table 2). The difference between sexes was expected since 277C was present only in the heterozygous condition among males, due to the experimental design. It is well known that Silver is not fully expressed in heterozygotes (Smyth 1990). The observed association in females was to a large extent consistent with the expected effect of Silver, but the effect was not fully penetrant. Most birds expressing the reddish Cream phenotype were heterozygous for Dominant white, which primarily inhibits expression of black eumelanin, and the majority of these birds carried the wild-type SLC45A2 allele (277Y), which should allow full expression of red pigmentation. However, as many as 13 of 41 birds expressed cream color despite being hemizygous for 277C (Table 2). Similarly, a fraction of the birds expressing the wild-type color pattern lacked yellow/red pigmentation and were denoted grayish wild type. Twelve of 16 birds were hemizygous for 277C, consistent with the expected effect of Silver, but the remaining 4 were hemizygous for the wild type, and hence the association was not complete. A small number of birds were classified as gray and they were, somewhat unexpectedly, all hemizygous for the wild-type SLC45A2 allele. This could be due to the segregation of another feather color mutation, the Blue mutation, which is known to be present in White Leghorns (Smyth 1990). Heterozygous carriers of this mutation are blue–gray when they also carry the dominant black allele at the E/MC1R locus.
TABLE 2.
Association between the SLC45A2 Y277C missense mutation and variation in plumage color among female F2 progeny in a red jungle fowl/White Leghorn intercross
|
SLC45A2 genotype
|
Chi square (d.f. = 1) | |||
|---|---|---|---|---|
| Phenotype | 277Y/W | 277C/W | Total | |
| Whitea | 79 | 84 | 163 | 0.1 |
| Creama | 28 | 13 | 41 | 5.5* |
| White with black spotsa | 16 | 10 | 26 | 1.4 |
| Black with white spots | 6 | 2 | 8 | 2.0 |
| Gray | 5 | 0 | 5 | 5.0* |
| Black | 9 | 6 | 15 | 0.6 |
| Wild type, grayish | 4 | 12 | 16 | 4.0* |
| Wild type, normal | 4 | 6 | 10 | 0.4 |
| Barred, gray | 10 | 10 | 20 | 0 |
| Barred, yellow | 2 | 1 | 3 | 0.3 |
P < 0.05.
These birds were all homozygous or heterozygous for the Dominant white (I) allele, whereas all other classes were homozygous wild type (i/i).
The incomplete association between SLC45A2 and plumage color in this intercross may reflect incomplete penetrance because other genetic factors are suppressing the expression of the SLC45A2 mutation. Another explanation is that the effect is due to a linked locus showing a fairly high recombination rate with SLC45A2. We tested the latter hypothesis by carrying out the same association analysis using the flanking markers ADL0022 and MCW0053 located 12.4 and 16.1 cM from SLC45A2, respectively. However, none of these markers showed a stronger association to variation in plumage color. This result suggests that the association is due to SLC45A2 or another locus in the near vicinity showing an incomplete penetrance in regard to plumage color.
DISCUSSION
This article demonstrates that SLC45A2 is the causative gene for the sex-linked Silver locus in chicken and sex-linked imperfect albinism in Japanese quail. Previous studies using intergeneric crosses demonstrated that matings between albino chicken and quails (S*AL and AL*A, respectively) produce only albino progeny and these alleles must thus be due to mutations in a homologous gene (Silversides and Mérat 1991). Our observation of disrupting mutations associated with both albino alleles, a frameshift mutation at codon 36 in chicken, and a splice-site mutation leading to exon skipping in the quail therefore provides conclusive evidence that SLC45A2 is underlying these loci.
SLC45A2 transcription is thought to be indirectly regulated by the melanocyte-specific transcription factor MITF (Du and Fisher 2002) and SLC45A2 ESTs have not been found in chicken brain cDNA libraries (http://www.genome.ucsc.edu). Surprisingly, we could amplify SLC45A2 5′ and 3′ cDNA sequences from a RACE-ready first-strand brain cDNA library. However, it is possible that SLC45A2 is expressed at a low level in brain and that we could amplify this sequence due to the high sensitivity of the RT–PCR method.
Here we describe five different SLC45A2 mutations associated with variation in plumage color in chicken and Japanese quail. Two of them are apparent null mutations causing sex-linked imperfect albinism. The deletion of 1 bp (106delT) in chicken S*AL exon 1 results in a frameshift and a premature stop codon and the transcript is apparently degraded by nonsense-mediated mRNA decay (Maquat 2005). Melanocytes from S*AL birds, in situ and in culture, have been found to have morphologically strange melanosomal/lysosomal organelles that are positive for both tyrosinase and acid phosphatase activities, and numerous morphologically normal premelanosomes were found to be lacking both enzymes (Boissy et al. 1987). The conclusion was that in the S*AL melanocyte, tyrosinase is not efficiently shuttled to the premelanosome. Since the normal function of SLC45A2 is believed to be in directing tyrosinase to these premelanosomes (Costin et al. 2003), our finding of a loss-of-function mutation in S*AL individuals is fully consistent with the observed phenotype of the S*AL melanocyte.
The splice-site mutation in quail albinos results in an in-frame skipping of exon 4 (Figure 3) and 47 amino acids are missing in the mature protein, including transmembrane region 7. This also leads to a phase shift for the location of the remaining loop regions in relation to different sides of the membrane. Thus, it is not surprising that this mutation causes imperfect albinism in the quail. Previous studies on this mutant have revealed that the mutant melanocytes show tyrosinase activity in the Golgi–endoplasmic reticulum–lysosome (GERL) region and in the Golgi vesicles, but not in the eumelanosomes. The same study also concluded that the AL gene may affect the transport of tyrosinase from the GERL or Golgi vesicles to the melanosomes (Yamamoto et al. 1987), once again fully consistent with the known function of SLC45A2 (Costin et al. 2003). Sex-linked imperfect albinism has been found in several other avian species such as turkey, budgerigar, and canary (Hutt 1949). Furthermore, sex-linked imperfect albinos have appeared a number of times in different populations of chicken and quail, suggesting that there are many more SLC45A2 mutations to be found in avian species.
The mutation (Ala72Asp) associated with cinnamon in Japanese quail was found in a highly conserved transmembrane region (Figures 3 and 4). This allele gives a more severe phenotype than Silver in chicken. The eyes of the chicks are red and have subnormal melanin pigmentation that darkens with age as in the imperfect albino birds (Cheng and Kimura 1990). The mutation results in a dilution of brown pigment in the feathers but the plumage pattern is not affected; the same ghost patterning is seen in imperfect albinos. The Ala72Asp mutation is a nonconservative amino acid substitution that may well explain the strong phenotypic effect of this missense mutation.
Two different missense mutations were associated with Silver in chicken. One of these (Leu347Met) was located in a highly conserved transmembrane region (Figures 3 and 4) and showed a complete association with the presence of Silver across breeds (Fayoumi, Yurlov Crower, Rhode White, Friesian Fowl, and White Buttercup) with the exception of White Leghorn. We therefore postulate that this is a causative mutation for the Silver phenotype among chickens. Seven birds with conflicting genotypes were observed in a family material but we are convinced that these are due to phenotyping errors since the phenotyping was performed using 1-day-old chicks.
We could not obtain conclusive evidence that the Tyr277Cys mutation associated with Silver in White Leghorns is causative. This mutation was found in a number of birds that were not presumed to carry Silver (Godollo Nhx, Green-legged Partridge, Rhode Island Red, Ukrainian Bearded, birds from the Nutreco experimental cross, and one red jungle fowl), suggesting that this could rather be a linked polymorphism. Furthermore, the mutation occurs in a poorly conserved region of the protein, but it is a nonconservative substitution at a site that is conserved between chicken and humans. The following observations strongly suggest that White Leghorns carry an SLC45A2 mutation: (i) all White Leghorns from three different lines were homozygous for the same SLC45A2 haplotype; (ii) the size of this shared haplotype among different White Leghorn lines was >100 kb, strongly indicative of a selective sweep at this locus; and (iii) the SLC45A2 segregation shows a weak, but significant, association to variation in plumage color in our red jungle fowl/White Leghorn intercross. Interestingly, the expression of red pigment most likely due to a leaky Silver allele has been observed as a problem in White Leghorn lines that can be corrected by selecting against the phenomenon (R. Okimoto, personal communication). This supports the notion that the Silver allele in the White Leghorn is not fully penetrant and its expression is influenced by the genetic background. Therefore, we consider Tyr277Cys as a candidate mutation for Silver in White Leghorns but it is also possible that Silver in White Leghorns is caused by a regulatory mutation located within the +100-kb haplotype shared by different strains of White Leghorn.
There has been strong selection at the Silver locus in chicken since the presence of Silver has been required for many breed-specific plumage patterns like the white color in White Leghorns. All breeds included in this study carrying Silver at present are expected to be fixed for this allele. Thus, the data presented in Figure 6 are fully consistent with this as the five divergent breeds carrying the Silver allele associated with the SLC45A2 347M mutation are homozygous for the same haplotype across SLC45A2. Similarly, the three different lines of White Leghorns are all homozygous for another haplotype carrying the 277C mutation. In sharp contrast, an examination of the sequence data from three red jungle fowls representing different Scandinavian zoo populations plus the genome sequence based on a single red jungle fowl bird reveals as many as 31 polymorphic sites in SLC45A2 (Figure 6). This marked difference in the degree of polymorphism between populations is a hallmark of a selective sweep for a favorable mutation (Maynard-Smith and Haigh 1974; Andersson and Georges 2004). The minimum shared haplotype among all chicken breeds carrying the 347M allele, firmly associated with Silver, was in the range 15–35 kb. This suggests that a marker density of one SNP/10 kb would have been sufficient to identify the Silver locus by a genomewide association analysis across breeds. The minimum shared haplotype among different White Leghorn lines, presumed to carry a second Silver allele, was even larger (>100 kb). This illustrates that the SNP density required for a genomewide association analysis will depend on the size affected by a selective sweep and the genetic distance among populations carrying the same mutation.
The most interesting feature of the SLC45A2 mutations described here is the specific inhibition of the expression of red pheomelanin in birds carrying Silver. This phenotypic effect is well documented in previous ultrastructural characterization of melanosomes from Silver and non-Silver birds (Brumbaugh 1971). A dilution of red pigment with no or only a minor effect on black pigment is also observed in horses heterozygous for a D153N mutation in SLC45A2 (Mariat et al. 2003). Horses homozygous for this mutation, though, have blue eyes and very little coat pigmentation as a result of a considerable dilution of both red and black pigments. In contrast, Silver in chicken has no effect on black pigment even in the homozygous condition. The specific inhibition of the production of pheomelanin that occurs in chickens carrying the Silver allele and in horses that are heterozygous for the D153N mutation cannot be explained by the current understanding of SLC45A2 function.
The amino acid cysteine is essential for the synthesis of red pigment whereas very little tyrosinase activity is required. It is therefore tempting to speculate that one of the functions of SLC45A2 is to transport cysteine into the melanosome and that this function is disrupted by the mutations associated with Silver. In previous studies it has been suggested that Silver may block the incorporation of sulfhydryls in pheomelanin since ultrastructural studies showed that sparsely melanized pheomelanosomes were coincident with histochemically sulfhydryl-negative melanocytes (Brumbaugh 1971). The subtle gray (sut) mouse has a mutation in the Slc7a11 gene encoding the cystine/glutamate exchanger xCT (Chintala et al. 2005). xCT has 12 predicted transmembrane regions and the cystine taken up by the cell via xCT is rapidly reduced to cysteine (Sato et al. 1999). An abnormal accumulation of tyrosinase (possibly in the trans-Golgi network) was found in the sut melanocyte, showing an anomalous trafficking of tyrosinase in the absence of cysteine. This abnormality does not seem to have any affect on eumelanin production (Chintala et al. 2005). SLC45A2 is also a 12-transmembrane transport protein but it is still unclear which molecules it transports (Fukamachi et al. 2001). The phenotypic effects of Silver in chicken and the D153N mutation in horse suggest that SLC45A2 may be crucial for the intracellular transport or melanosome content of cysteine after the xCT exchanger has allowed cystine to enter the melanocytes. This cysteine could be essential for the transport vesicles that direct tyrosinase from the trans-Golgi network to the pheomelanosome, and therefore mutations in Slc7a11 and some SLC45A2 mutations are affecting only pheomelanin production. There are also other mutations found to affect pheomelanin production. The gray-lethal (gl) mouse mutation in the GL/OSTM1 gene results from stalled pheomelanin migration due to clustered pheomelanin granules (Chalhoub et al. 2003), and the γ-glutamyl transpeptidase (GGT) knockout mice lack cysteine as a result of the nonworking cleavage of glutathione (Lieberman et al. 1996). However, if SLC45A2 has an important role for the incorporation of sulfhydryls into pheomelanin it cannot be its sole function since a defect in cystine/cysteine transport should not affect eumelanogenesis, whereas SLC45A2 null mutations almost completely abolish the production of both eumelanin and pheomelanin. It is an enigma why recessive null mutations cause an almost complete absence of both eumelanin and pheomelanin, whereas some missense mutations are dominant and cause a specific inhibition of the expression of red pheomelanin. Therefore, further experimental work based on the missense mutations in chicken and horse may shed light on SLC45A2 function.
Acknowledgments
The 14-day-old whole-embryo chicken total RNA used for cDNA synthesis and the RACE-ready first-strand cDNA from broiler brain were kindly provided by S. Kerje and P. Wahlberg. S. Kerje also contributed to discussion regarding the manuscript. We thank U. Gustafson for sequencing, J. Håkansson for the red jungle fowl samples, A.-S. Sahlqvist for the OS samples, D. Gourichon for quail and chicken samples, and L. W. Hillier for sharing information on the assembly of the chicken Z chromosome. We also thank D. Gourichon and G. Coquerelle for maintaining a gene pool of feather-color mutations and providing the tissue samples of S*AL embryo chicks and the pictures and the blood samples of the Fayoumi breed (AvianDiv samples). Sincere appreciation is also due to all the original providers of the DNA samples for AvianDiv populations: G. Virag, Small Animal Research Institute, Godollo, Hungary (Godollo Nhx); K. Cywa-Benko, Animal Research Institute, Cracow, Poland (Green-Legged Partridge); Y. Bondarenko, Research Institute, Kharkov, Ukrainia (Ukrainian Bearded); M. Protais, ISA breeders, France (Rhode Island Red and Rhode White); R. Crooijmans, WUR, The Netherlands (Friesian Fowl); and I. Moiseyeva, Research Institute, Moscow, Russia (Yurlov Crower). This work was supported by the Foundation for Strategic Research and the Swedish Research Council for Environment, Agricultural Sciences and Spatial Planning.
References
- Andersson, L., and M. Georges, 2004. Domestic-animal genomics: deciphering the genetics of complex traits. Nat. Rev. Genet. 5: 202–212. [DOI] [PubMed] [Google Scholar]
- Bacon, L. D., E. Smith, L. B. Crittenden and G. B. Havenstein, 1988. Association of the slow feathering (K) and an endogenous viral (ev21) gene on the Z chromosome of chickens. Poult. Sci. 67: 191–197. [DOI] [PubMed] [Google Scholar]
- Bitgood, J. J., 1999. Linkage relationships of the Z-linked silver, slow feathering, and pop-eye loci. Poult. Sci. 78: 1100–1101. [DOI] [PubMed] [Google Scholar]
- Boissy, R. E., G. E. Moellmann and R. Halaban, 1987. Tyrosinase and acid phosphatase activities in melanocytes from avian albinos. J. Invest. Dermatol. 88: 292–300. [DOI] [PubMed] [Google Scholar]
- Brumbaugh, J. A., 1968. Ultrastructural differences between forming eumelanin and pheomelanin as revealed by the pink-eye mutation in the fowl. Dev. Biol. 18: 375–390. [DOI] [PubMed] [Google Scholar]
- Brumbaugh, J. A., 1971. The ultrastructural effects of the I and S loci upon black-red melanin differentiation in the fowl. Dev. Biol. 24: 392–412. [DOI] [PubMed] [Google Scholar]
- Burge, C., and S. Karlin, 1997. Prediction of complete gene structures in human genomic DNA. J. Mol. Biol. 268: 78–94. [DOI] [PubMed] [Google Scholar]
- Chalhoub, N., N. Benachenhou, V. Rajapurohitam, M. Pata, M. Ferron et al., 2003. Grey-lethal mutation induces severe malignant autosomal recessive osteopetrosis in mouse and human. Nat. Med. 9: 399–406. [DOI] [PubMed] [Google Scholar]
- Cheng, K. M., and M. Kimura, 1990. Mutations and major variants in Japanese quail, pp. 333–362 in Poultry Breeding and Genetics, edited by R. D. Crawford. Elsevier, Amsterdam.
- Chintala, S., W. Li, M. L. Lamoreux, S. Ito, K. Wakamatsu et al., 2005. Slc7a11 gene controls production of pheomelanin pigment and proliferation of cultured cells. Proc. Natl. Acad. Sci. USA 102: 10964–10969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole, R. K., 1966. Hereditary hypothyroidism in the domestic fowl. Genetics 53: 1021–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole, R. K., and T. K. Jeffers, 1963. Allelism of silver, gold, and imperfect albinism in the fowl. Nature 200: 1238–1239. [Google Scholar]
- Costin, G. E., J. C. Valencia, W. D. Vieira, M. L. Lamoreux and V. J. Hearing, 2003. Tyrosinase processing and intracellular trafficking is disrupted in mouse primary melanocytes carrying the underwhite (uw) mutation. A model for oculocutaneous albinism (OCA) type 4. J. Cell Sci. 116: 3203–3212. [DOI] [PubMed] [Google Scholar]
- Du, J., and D. E. Fisher, 2002. Identification of Aim-1 as the underwhite mouse mutant and its transcriptional regulation by MITF. J. Biol. Chem. 277: 402–406. [DOI] [PubMed] [Google Scholar]
- Fukamachi, S., A. Shimada and A. Shima, 2001. Mutations in the gene encoding B, a novel transporter protein, reduce melanin content in medaka. Nat. Genet. 28: 381–385. [DOI] [PubMed] [Google Scholar]
- Green, P., K. Falls and S. Crook, 1990. Documentation for CRIMAP, Ver 2.4. Washington University School of Medicine, St. Louis.
- Håkansson, J., and P. Jensen, 2005. Behavioural and morphological variation between captive populations of red junglefowl (Gallus gallus)—possible implications for conservation. Biol. Conserv. 122: 431–439. [Google Scholar]
- Harada, M., Y. F. Li, M. El-Gamil, S. A. Rosenberg and P. F. Robbins, 2001. Use of an in vitro immunoselected tumor line to identify shared melanoma antigens recognized by HLA-A*0201-restricted T cells. Cancer Res. 61: 1089–1094. [PubMed] [Google Scholar]
- Hearing, V. J., 2005. Biogenesis of pigment granules: a sensitive way to regulate melanocyte function. J. Dermatol. Sci. 37: 3–14. [DOI] [PubMed] [Google Scholar]
- Hillel, J., M. A. Groenen, M. Tixier-Boichard, A. B. Korol, L. David et al., 2003. Biodiversity of 52 chicken populations assessed by microsatellite typing of DNA pools. Genet. Sel. Evol. 35: 533–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutt, F. B., 1949. Genetics of the Fowl. Norton Creek Press, Blodgett, OR.
- Hutt, F. B., and C. D. Mueller, 1943. Independent identical mutations to albinism in the sex chromosome of the fowl. Am. Nat. 77: 181–184. [Google Scholar]
- Kerje, S., J. Lind, K. Schutz, P. Jensen and L. Andersson, 2003. Melanocortin 1-receptor (MC1R) mutations are associated with plumage colour in chicken. Anim. Genet. 34: 241–248. [DOI] [PubMed] [Google Scholar]
- Kerje, S., P. Sharma, U. Gunnarsson, H. Kim, S. Bagchi et al., 2004. The Dominant white, Dun and Smoky color variants in chicken are associated with insertion/deletion polymorphisms in the PMEL17 gene. Genetics 168: 1507–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi, T., W. D. Vieira, B. Potterf, C. Sakai, G. Imokawa et al., 1995. Modulation of melanogenic protein expression during the switch from eu- to pheomelanogenesis. J. Cell Sci. 108: 2301–2309. [DOI] [PubMed] [Google Scholar]
- Kushimoto, T., J. C. Valencia, G. E. Costin, K. Toyofuku, H. Watabe et al., 2003. The Seiji memorial lecture: the melanosome: an ideal model to study cellular differentiation. Pigment Cell Res. 16: 237–244. [DOI] [PubMed] [Google Scholar]
- Lauber, J. K., 1964. Sex-linked albinism in the Japanese quail. Science 146: 948–950. [DOI] [PubMed] [Google Scholar]
- Lieberman, M. W., A. L. Wiseman, Z. Z. Shi, B. Z. Carter, R. Barrios et al., 1996. Growth retardation and cysteine deficiency in gamma-glutamyl transpeptidase-deficient mice. Proc. Natl. Acad. Sci. USA 93: 7923–7926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maquat, L. E., 2005. Nonsense-mediated mRNA decay in mammals. J. Cell Sci. 118: 1773–1776. [DOI] [PubMed] [Google Scholar]
- Mariat, D., S. Taourit and G. Guerin, 2003. A mutation in the MATP gene causes the cream coat colour in the horse. Genet. Sel. Evol. 35: 119–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maynard-Smith, J., and J. Haigh, 1974. The hitch-hiking effect of a favourable gene. Genet. Res. 23: 23–35. [PubMed] [Google Scholar]
- Mérat, P., A. Bordas and G. Coquerelle, 1986. Caractéristiques de croissance, ponte et efficacité alimentaire associées au gène sal (albinos lié au sexe) chez la poule domestique. Genet. Sel. Evol. 18: 343–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minvielle, F., S. Ito, M. Inoue-Murayama, M. Mizutani and N. Wakasugi, 2000. Genetic analyses of plumage color mutations on the Z chromosome of Japanese quail. J. Hered. 91: 499–501. [DOI] [PubMed] [Google Scholar]
- Mueller, C. D., and F. B. Hutt, 1941. Genetics of the fowl: 12 - sex-linked, imperfect albinism. J. Hered. 32: 71–80. [Google Scholar]
- Newton, J. M., O. Cohen-Barak, N. Hagiwara, J. M. Gardner, M. T. Davisson et al., 2001. Mutations in the human orthologue of the mouse underwhite gene (uw) underlie a new form of oculocutaneous albinism, OCA4. Am. J. Hum. Genet. 69: 981–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato, H., M. Tamba, T. Ishii and S. Bannai, 1999. Cloning and expression of a plasma membrane cystine/glutamate exchange transporter composed of two distinct proteins. J. Biol. Chem. 274: 11455–11458. [DOI] [PubMed] [Google Scholar]
- Schutz, K., S. Kerje, O. Carlborg, L. Jacobsson, L. Andersson et al., 2002. QTL analysis of a red junglefowl x White Leghorn intercross reveals trade-off in resource allocation between behavior and production traits. Behav. Genet. 32: 423–433. [DOI] [PubMed] [Google Scholar]
- Silversides, F. G., and R. D. Crawford, 1990. Genetic aspects of a new mutation (sal-s) to sex-linked imperfect albinism in chickens. Genet. Sel. Evol. 22: 447–455. [Google Scholar]
- Silversides, F. G., and P. Mérat, 1991. Homology of the s+ locus in the chicken with Al+ in the Japanese quail. J. Hered. 82: 245–247. [Google Scholar]
- Smyth, J. R., Jr., 1990. Genetics of plumage, skin and eye pigmentation in chickens, pp. 109–167 in Poultry Breeding and Genetics, edited by R. D. Crawford. Elsevier, Amsterdam.
- Sturtevant, A. H., 1912. An experiment dealing with sex-linkage in fowls. J. Exp. Zool. 12: 499–518. [Google Scholar]
- Truax, R. E., and W. A. Johnson, 1979. Genetics of plumage color mutants in Japanese quail. Poult. Sci. 58: 1–9. [Google Scholar]
- Wakamatsu, K., and S. Ito, 2002. Advanced chemical methods in melanin determination. Pigment Cell Res. 15: 174–183. [DOI] [PubMed] [Google Scholar]
- Werret, W. F., A. J. Candy, J. O. King and P. M. Sheppard, 1959. Semi-albino: a third sex-linked allelomorph of silver and gold in the fowl. Nature 184(Suppl. 7): 480. [DOI] [PubMed] [Google Scholar]
- Yamamoto, H., K. Ito, S. Ishiguro and T. Takeuchi, 1987. Gene controlling a differentiation step in the quail melanocyte. Dev. Genet. 8: 179–185. [DOI] [PubMed] [Google Scholar]