Abstract
We present a detailed analysis of linkage disequilibrium (LD) in the physical and genetic context of the barley gene Hv-eIF4E, which confers resistance to the barley yellow mosaic virus (BYMV) complex. Eighty-three SNPs distributed over 132 kb of Hv-eIF4E and six additional fragments genetically mapped to its flanking region were used to derive haplotypes from 131 accessions. Three haplogroups were recognized, discriminating between the alleles rym4 and rym5, which each encode for a spectrum of resistance to BYMV. With increasing map distance, haplotypes of susceptible genotypes displayed diverse patterns driven mainly by recombination, whereas haplotype diversity within the subgroups of resistant genotypes was limited. We conclude that the breakdown of LD within 1 cM of the resistance gene was generated mainly by susceptible genotypes. Despite the LD decay, a significant association between haplotype and resistance to BYMV was detected up to a distance of 5.5 cM from the resistance gene. The LD pattern and the haplotype structure of the target chromosomal region are the result of interplay between low recombination and recent breeding history.
THE ever-increasing availability of nucleotide sequence, and the concomitant improvement that this brings to our understanding of the organization of complex crop plant genomes, has created the opportunity to identify trait-related genes and to analyze their allelic diversity. The association between phenotype and genotype in diverse populations represents a powerful approach, but the applicability and design of such analyses depend critically on the extent and pattern of linkage disequilibrium (LD) present in the study population. Cultivated barley (Hordeum vulgare ssp. vulgare) has many of the hallmarks known to be associated with a high level of LD. Its effective recombination rate is dramatically reduced by its predominantly inbreeding habit, with an estimated outcrossing rate of ∼5% in winter barley and <0.5% in spring barley (Giles et al. 1974; Doll 1987; Abdel-Ghani et al. 2005). Domestication and intensive selection have introduced major bottlenecks in genetic variation, and these are thought to be largely responsible for the perceived narrowness of the modern gene pool (Badr et al. 2000; Russell et al. 2000; Matus and Hayes 2002). LD in modern spring barley, as estimated from a whole-genome survey, extends over distances of at least 10 cM, indicative of an extensive conservation of the genetic identity of barley chromosomes (Kraakman et al. 2004). However, estimates of genomewide LD conceal localized variation, which, as has been shown for a number of species, can be substantial and independent of the mating system (Gupta et al. 2005). In the self-pollinating species Arabidopsis thaliana LD varies from <10 kb in global (Tian et al. 2002) to 50–250 kb in local populations (Nordborg et al. 2002, 2005; Aranzana et al. 2005). Within a given set of maize (an outbreeding species) accessions, the extent of LD has been documented to vary widely around different genes and chromosomal regions (Remington et al. 2001). On chromosome 1, for example, it has been shown to decline within 0.1–0.2 kb (Tenaillon et al. 2001), while in a region under high selection pressure, it can extend up to 600 kb (Palaisa et al. 2004).
Little is known of the structure of LD in the physical and genetical vicinity of genes in cultivated barley (reviewed in Gupta et al. 2005). As the success of association studies in this major crop species depends critically on this knowledge, we have set out to evaluate LD in a well-characterized region of chromosome 3H, which harbors Hv-eIF4E, a gene encoding an important virus resistance. Two recessive alleles, rym4 and rym5, confer resistance to both soil-borne Bymoviruses, barley yellow mosaic virus (BaYMV) and barley mild mosaic virus (BaMMV) (Kanyuka et al. 2005; Stein et al. 2005). This combination of pathogens is commonly referred to as the barley yellow mosaic virus complex (BYMV). Carriers of rym4 are resistant to BaMMV, BaMMV-Sil, and BaYMV-1, but are susceptible to BaYMV-2, while those carrying rym5 are resistant to BaYMV-1, BaYMV-2, and BaMMV, but not to BaMMV-Sil (Kanyuka et al. 2004). Although at least seven independent loci conferring resistance to BYMV have been identified in barley to date (Ordon et al. 2005), European breeding has relied heavily on rym4 and rym5, both of which originate from single germplasm accessions. The Dalmatian landrace Ragusa is the source of rym4 (Huth 1985), while rym5 derives from the Chinese landrace Mokusekko 3 (Konishi et al. 1997; Graner et al. 1999; Friedt et al. 2000).
The cloning of Hv-eIF4E (Stein et al. 2005; Wicker et al. 2005) provides a focus for an LD analysis. Taking into consideration that breeding for resistance in Europe did not start until the 1980s, and that the Hv-eIF4E locus has been subjected to a high selection pressure over a short timescale, this case study reveals much of how genome structure and dynamics can be shaped by plant breeding.
MATERIALS AND METHODS
Plant material:
Included in the study were 127 cultivated barley cultivars and landraces originating from Europe, Asia, and America, along with four accessions of wild barley (ssp. spontaneum) collected in either Turkey or Israel. Geographical origins, pedigrees (where available), and phenotype with respect to reaction to BYMV are listed in supplemental Table S1 at http://www.genetics.org/supplemental/. Accessions carrying rym4 included Ragusa, 19 European cultivars, and four Asian landraces. With respect to rym5, five European and six Asian accessions, including the donor Mokusekko 3, were analyzed. All these accessions were tested in the field and by mechanical inoculation to verify their rym4/rym5 status (Götz and Friedt 1993; Ordon et al. 1993) (supplemental Table S1 at http://www.genetics.org/supplemental/). The remaining resistant accessions have not been tested for allelism to rym4 and rym5 (rym?; Table S1 at http://www.genetics.org/supplemental/). Nevertheless, their resistance is likely due to the presence of genes other than rym4/rym5, since their haplotypes differed markedly from those of the verified rym4 and rym5 accessions. In the context of this study, these genotypes are referred to as noncarriers of rym4/rym5 and were merged with the group of susceptible cultivars. Genomic DNA was extracted from a single plant per accession as described elsewhere (Graner et al. 1991).
Population structure:
Sixteen barley expressed-sequence-tag (EST)-derived simple sequence repeat markers (SSRs) (EST–SSRs) were selected to characterize population structure (supplemental Table S2 at http://www.genetics.org/supplemental/) (Thiel et al. 2003). The selection of appropriate markers was based both on map position (to obtain an even distribution across the genome) and on informativeness (high polymorphic information content value) (Weber 1990; Anderson et al. 1993). The markers were organized into multiplex sets comprising between three and six different primer pairs. Amplicons were generated with the Multiplex PCR kit (QIAGEN, Chatsworth, CA), using an amplification program of 95°/15 min, followed by 40 cycles of 94°/30 sec, 60°/30 sec, and 72°/15 sec, with a final extension step of 72°/10 min. PCR products were separated on 6% polyacrylamide gels using the ABI377 system (Applied Biosystems/Applera, Darmstadt, Germany), and profiles were analyzed with the software packages GeneScan 3.7.1 und Genotyper 3.7. The assessment of genetic population structure was performed within a Bayesian framework using a Markov chain Monte Carlo algorithm to sample from the joint posterior distribution of the subpopulation allele frequencies, and assignment of individuals to particular subgroups was effected with STRUCTURE version 2.1 (Pritchard et al. 2000a; http://pritch.bsd.uchicago.edu/structure.html). For a K setting of between 2 and 6, 10 independent simulations were performed, using the admixture model and a burn-in of 500,000 followed by 1,000,000 iterations.
To assess the applicability of an association analysis within modern breeding material, association tests were performed in 51 European winter barley cultivars, comprising 20 rym4 carriers and 31 susceptible accessions. To infer the population structure of this sample, the estimated individual membership coefficient in each subgroup was accounted for with a structured population association test (STRAT) (Pritchard et al. 2000b). The test statistic is constructed by computing the likelihood ratio to test the null hypothesis that allele frequencies of subpopulations at the candidate locus are independent of phenotype. Phenotypic association was tested by treating single haplotypes as multiallelic marker loci, including only single polymorphic sites at which the minor allele occurred at a frequency of at least 0.05. Empirical P-values of the observed test statistic per marker locus and per polymorphic site were calculated using 100,000 permutations.
The variation in phenotype due to population structure was assessed by a multiple linear regression model (PLABSTAT version 2H; Utz 1993). The estimated membership fractions for each genotype in K clusters of K = 3 to K = 6 in the entire set and K = 2 for the subset of European winter cultivars were considered. The coefficient of determination from the regression (R2) was applied to quantify the effect on the trait of the genetic background structure.
SNP discovery and genotyping:
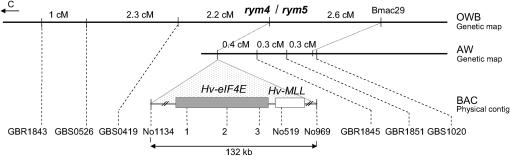
A number of mutations in Hv-eIF4E are known to confer resistance to BYMV. Thus the scan for polymorphic nucleotides within the set of 131 accessions was focused on an assembly of 12 genomic fragments within and surrounding Hv-eIF4E (Figure 1). The size of each fragment was between 175 and 1039 bp, encompassing in total 6.9 kb. This represents a genetic interval of ∼5.5 cM proximal to, and 0.9 cM distal to, Hv-eIF4E (Table 1). The LD structure in relation to physical distance was analyzed on the basis of six fragments spanning the ∼132-kb BAC contig AY661558 established during the cloning of Hv-eIF4E (Stein et al. 2005; Wicker et al. 2005). This included three genic fragments, covering all five exons (645 bp) and flanking intron regions (1840 bp), one fragment from the adjacent distally mapping gene Hv-MLL (marker No519; 1048 bp upstream), and one noncoding fragment mapping both distally (No969) and proximally (No1134) to the target. The proximal fragments GBR1843, GBS0526, and GBS0419 have been previously mapped in the Oregon Wolfe mapping population (OWB, Dom × Rec) (Costa et al. 2001), which consists of 94 F1-derived doubled haploid progeny (Stein et al. 2007). The distal fragments GBR1845, GBR1851, and GBS1020 were mapped in 115 segmental recombinant inbred lines of Alraune × W122.37, a population genetically equivalent to >4800 F2 progeny (AW) (Pellio et al. 2005). Because of a lack of polymorphism between the parents of both these mapping populations, it was not possible to combine all the markers onto a single map. Therefore, for the LD analysis, genetic map distances were considered separately for the OWB and AW populations.
Figure 1.—
Combined genetic and physical map of the region surrounding Hv-eIF4E on chromosome 3H. Genetic distances in centimorgans are indicated for markers flanking the locus in the OWB and the high-resolution AW maps. The arrow indicates the centromere (C). The schematic arrangement of marker fragments is depicted (not to scale) on the physical BAC contig (AY661558). Hv-MLL, barley MCT-1-like protein.
TABLE 1.
Summary of all sampled marker fragments
| Marker | Distance in cMa | Position on contig in bpb | Fragment size in bp | Polymorphic sites | Haplotypesc | Best BLASTx hit | Score | |
|---|---|---|---|---|---|---|---|---|
| GBR1843 | 5.5 | OWB | — | 347 | 4 | 4 | Unknown protein P0458E05.14 (Oryza sativa BAC05614) | 233 1.0E-60 |
| GBS0526 | 4.5 | OWB | — | 930 | 9 | 4 | Putative 60S ribosomal protein L38 (O. sativa BAC79676) | 132 1.0E-30 |
| GBS0419 | 2.2 | OWB | — | 371 | 4 | 3 | Putative β-fructofuranosidase (O. sativa BAC05626) | 354 9.0E-97 |
| No1134 | 0.02 | AW | 28,973 29,373 | 401 | 11 | 6 | None | AY661558 |
| Hv-eIF4E | 0 | — | 101,606 | 879 | 12 | 6 | Eukaryotic initiation factor of translation 4E (Hv-eIF4E) | AY661558 |
| fragment 1 | 102,484 | |||||||
| Hv-eIF4E | 0 | — | 103,313 | 1039 | 15 | 7 | Eukaryotic initiation factor of translation 4E (Hv-eIF4E) | AY661558 |
| fragment 2 | 104,351 | |||||||
| Hv-eIF4E | 0 | — | 106,146 | 565 | 12 | 6 | Eukaryotic initiation factor of translation 4E (Hv-eIF4E) | AY661558 |
| fragment 3 | 106,710 | |||||||
| No519 | 0 | — | 107,786 | 499 | 2 | 3 | Monocarboxylic acid transporter-like (Hv-MLL) | AY661558 |
| 108,284 | ||||||||
| No969 | 0 | — | 161,182 | 175 | 2 | 3 | None | AY661558 |
| 161,356 | ||||||||
| GBR1845 | 0.4 | AW | — | 265 | 5 | 4 | Putative cytochrome B5 (O. sativa BAB63673) | 110 8.0E-31 |
| GBR1851 | 0.7 | AW | — | 424 | 5 | 4 | Unknown protein P0518C01.5 (O. sativa BAB63668) | 75 8.0E-13 |
| GBS1020 | 1.0 | AW | — | 1011 | 2 | 3 | Putative cell cycle switch protein (O. sativa BAB63690) | 312 4.0E-89 |
Relative to ATG of Hv-eIF4E.
Start and end position of the corresponding fragment based on contig AY661558.
No. of haplotypes with a frequency >0.02 in the entire collection.
In preparation for sequencing, DNA fragments were amplified using fragment-specific PCR profiles, as given in supplemental Table S3 at http://www.genetics.org/supplemental/. PCR products were purified (MinElute 96 UF PCR purification, QIAGEN), and 50–100 ng of product was used as template for cycle sequencing with DYEnamic ET dye terminator chemistry (Amersham Bioscience, Freiburg, Germany) on a capillary automatic sequencing device (MegaBace 1000, Amersham). Alignments were compiled and analyzed using Sequencher v4.1 (Gene Codes, Ann Arbor, MI) and Bioedit v4.7.8 (Tom Hall, North Carolina State University; http://www.mbio.ncsu.edu/BioEdit/bioedit.html). The data set describing all genotyped polymorphic sites is available on request.
Statistical analyses:
For the purpose of statistical analyses, indels were regarded as single sites. Estimates for LD were obtained via the software package TASSEL (version 1.0.9, http://www.maizegenetics.net/bioinformatics/tasselindex.htm), applying the measurement r2 (squared correlation coefficient) (Hill and Robertson 1968). To estimate the strength of LD between haplotypes, we used the measure D′ (Lewontin 1964) modified for multiple alleles by calculating a weighted average of D′ value where the weights are the products of the corresponding allele frequencies (Hedrick 1987; Farnir et al. 2000). Significance of LD is determined by a two-sided Fisher's exact test for biallelic sites and by permutations (setting number 1000) for multiallelic sites. To preclude bias of low-frequency alleles on the LD calculation, polymorphic sites featuring allele(s) with a frequency of ≤0.05 were excluded. Values of r2 and D′ were either plotted as a function of the pairwise distance between the polymorphic sites or displayed as an LD matrix as generated by TASSEL.
For the clustering of haplotypes across the 132-kb contig and the assignment to haplogroups, neighbor-joining trees (Saitoh et al. 2004) were constructed on the basis of Kimura two-parameter distances and pairwise deletions of gaps, applying MEGA version 2.1 (Kumar et al. 1994). To test the robustness of derived tree topologies, 1000 bootstraps were performed. To estimate gene genealogies, a haplotype network of Hv-eIF4E was obtained by statistical parsimony (Templeton et al. 1992), using the program TCS version 1.18 (Clement et al. 2000). This program calculates the probability of parsimony for all pairwise differences until the probability exceeds 0.95.
To assess genomic variability across the entire target region and to measure and compare the diversity within and across the designated haplogroups, both nucleotide (pi) and haplotype (Hd) diversities were estimated using DnaSP (Version 3.51; Rozas and Rozas 1999; http://www.ub.es/dnasp/) software. For this purpose, indels were treated as single sites. Overall, 12 of the 127 H. vulgare genotypes were excluded from the analysis because of missing data.
RESULTS
Population structure:
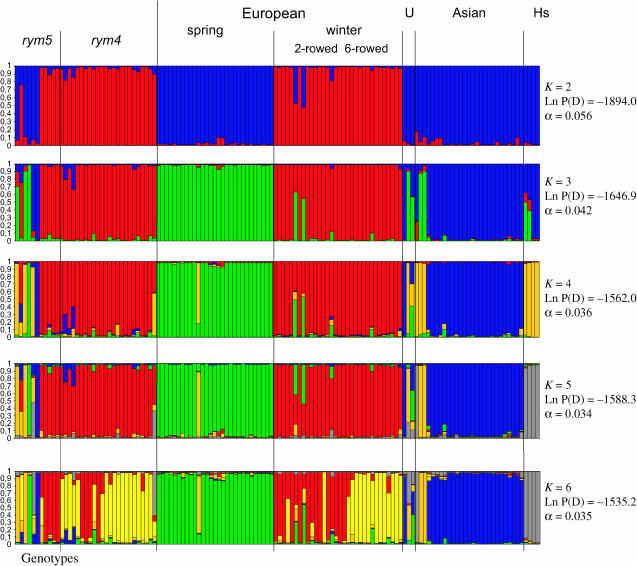
The 16 EST–SSR loci revealed 77 alleles, with 2–9 alleles/locus. The distribution of these alleles was in close accordance with the taxonomic and geographic subgroups defined in the germ-plasm collection, as illustrated by a Bayesian approach (Figure 2). However, it was difficult to fully determine the optimal number of subgroups, since the posterior probabilities for the number of clusters increased steadily. On the assumption of two subgroups (K = 2), the population was divided into winter and spring types. A stepwise increase to K = 6 led to the gradual separation of the Asian from the European accessions, of the two-row from the six-row spike types, and of the cultivated (vulgare) from the wild (spontaneum) types. Importantly, all rym4 carriers and all European rym5 carriers displayed allele frequencies similar to those of susceptible accessions belonging to different subgroups. Thus population structure could account for only 21% (K = 3) to 29% (K = 6) of the variation in BYMV resistance.
Figure 2.—
Estimated population structure based on 16 EST–SSRs. The population can be partitioned into K subpopulations, which are color labeled. A bar, whose colored segments represent the individual's estimated contribution to the individual subpopulations, represents each accession. Black vertical lines separate accessions of different resistance phenotype, origin, and growth habit. U, America; Hs, ssp. spontaneum.
Structure of LD:
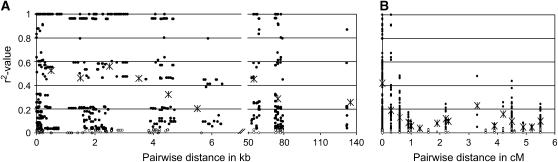
The 12 genomic fragments encompassing the target region were resequenced across the collection of 131 accessions. A total of 83 polymorphic sites, comprising 78 SNPs and five indels, were identified. Of the 83 sites, 5 were triallelic, and 4 of these, in addition to 14 of the biallelic sites, had an allele frequency of ≤0.05 and were excluded from the subsequent analysis. Across the entire collection, a considerable degree of LD was evident within the physical contig (Figure 3A), while at the genetic level, the r2 value dropped sharply to <0.3 within 1 cM of the target (Figure 3B). Despite the diminishing r2 value, a repeated increase of LD (r2 > 0.3) was observed at a larger distance. This was not an artifact of population stratification, as the allele distribution of the corresponding pairs of polymorphic sites did not associate with population structure.
Figure 3.—
LD structure at Hv-eIF4E. Plots show the pairwise LD measurement r2 related to (A) physical, and (B) genetic distances. The data consist of 67 polymorphic sites with a minor allele frequency >0.05 in the set of 131 accessions. Nonsignificant r2 values (P > 0.05) are indicated by open dots. Mean values of r2 are given by asterisks for (A) windows of 1 and 10 kb distance and (B) identical genetic distances.
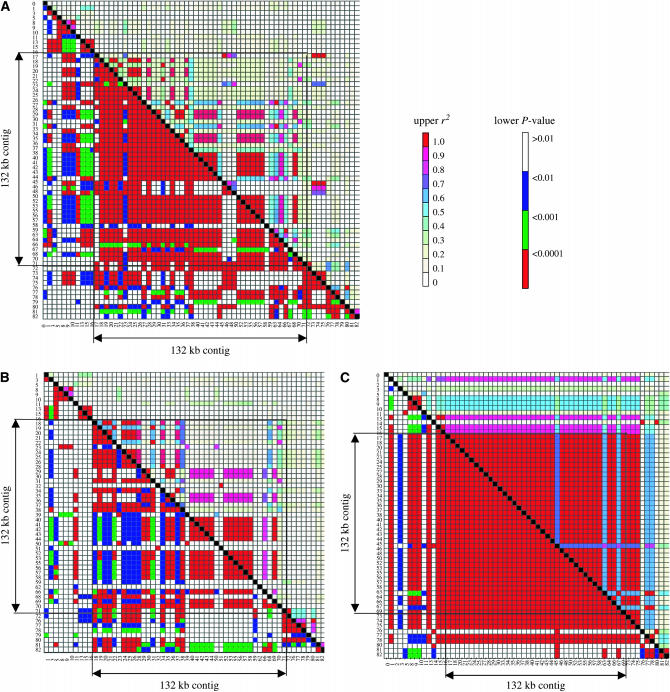
To compare the LD structure between rym4/rym5-resistant and -susceptible genotypes (including noncarriers of rym4/rym5), r2 was determined separately for these two subgroups (Figure 4). The extended structure of LD observed for the 132-kb contig was confirmed for all subgroups, but with varying strength. In the subgroup represented by the rym4 (n = 24) and the rym5 (n = 11) carriers, most pairs of polymorphic sites within the interval displayed complete LD (r2 = 1), while the susceptible subgroup (n = 96) showed fewer high r2 values. LD within the susceptible group fell, as it did for the entire set, within 1 cM of the target. On the other hand, the rym4/rym5 subgroup was characterized by a significantly inflated LD across the entire genetic region, demonstrating a high degree of conservation within resistant genotypes.
Figure 4.—
Strength and extent of LD in different germplasm sets. Polymorphic sites in the investigated interval with a minor allele frequency >5% were considered for a pairwise calculation of LD across (A) the entire collection of 131 accessions, (B) the non-rym4/rym5-resistant subpopulation of 96 accessions, and (C) the set of 35 rym4 or rym5 carriers. Each point in the LD matrix represents a comparison between a pair of polymorphic sites, with the r2 values displayed above the diagonal and the P-values for Fisher's exact test below. Points on the diagonal correspond to comparisons of each site with itself. Polymorphic sites located within the fragments of the physical contig are indicated. Color codes for r2 and P-values are given.
Haplotype structure:
Haplotypes were generated on the basis of the polymorphic sites, and their structure across the entire genetic interval was analyzed. Considering each individual fragment, between three and seven haplotypes achieved a frequency >2% (Table 1). LD strength between haplotypes of a single DNA fragment within subgroups of resistant (rym4 and rym5 carriers) and susceptible genotypes revealed remarkable differences (supplemental Figure S1 at http://www.genetics.org/supplemental/). Compared to the susceptible group, resistant accessions had higher r2 values across the entire region.
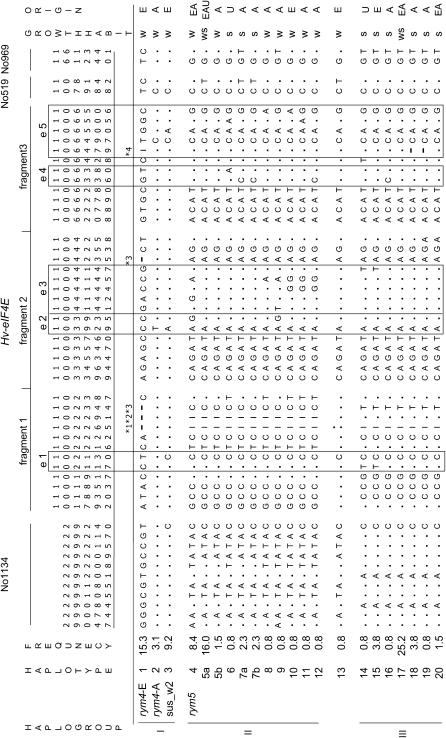
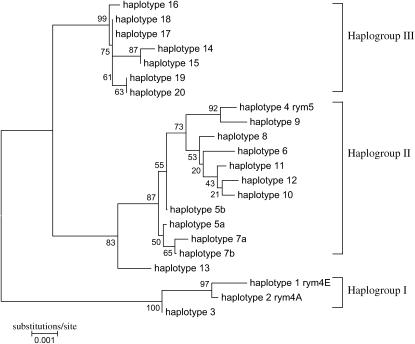
For the 132-kb contig, 22 haploytpes were identified on the basis of 54 polymorphic sites (Figure 5). The rym4 carriers included two haplotypes (haplotype 1 rym4-A and haplotype 2 rym4-E), which matched the two broad geographical origins of Asia (A) and Europe (E). All rym5 carriers shared the same haplotype (haplotype 4 rym5). Noncarriers of rym4/rym5 were represented by 19 haplotypes. More than half of them (10/19) were singletons while >67% of those genotypes belonged to one of the three major haplotypes 3, 5a, and 17. Using a neighbor-joining method, the haplotypes clustered into three major clades, hereafter referred to as haplogroups I–III (Figure 6). The formation of these haplogroups appeared to be independent of BYMV resistance (Figure 5). Thus, haplogroup I included three haplotypes comprising the European and Asian rym4 carriers (haplotypes 1 and 2) and a group of highly conserved susceptible two-row winter cultivars (haplotype 3). Only 6 of the 54 scored sites varied within this haplogroup. Haplogroup II contained rym5 carriers (haplotype 4) and geographically diverse accessions (haplotypes 5–12), which included both no, or as yet undetermined, resistance alleles/genes. A clear dimorphism between haplogroups I and II was obtained for half of the polymorphic sites of the contig. Dimorphic sites were located mainly in noncoding sequences, while 15 of the 16 polymorphic exon sites were shared between the two haplogroups. Only one cultivar (Posaune, haplotype 13) was intermediate between these groups, containing a signature of recombination both up- and downstream of the Hv-eIF4E exon 1. Haplogroup III (haplotypes 14–20) had a composite structure with at least three origins. The sequence data of No1134 are closely related to haplogroup I, with only two polymorphic sites present (Figure 5, position 29084n and 29201n). Moreover, variation within Hv-eIF4E fragment 1 resulted in a sequence, which is clearly distinct from that present in haplogroups I and II. The pattern of polymorphic sites downstream of Hv-eIF4E fragment 1, however, resembled haplogroup II. This apparent patchwork on either side of fragment 1 may indicate the occurrence of several historical recombination events within Hv-eIF4E. The overwhelming reduction in haplotype and nucleotide diversity within the haplogroups, as compared to what is present across the entire collection, validates the grouping (supplemental Table S4 at http://www.genetics.org/supplemental/). A separate analysis indicates that, within a given haplogroup, less diversity is present in the noncoding than in the coding sequence. Importantly, all sites within the coding sequence of Hv-eIF4E generate an amino acid exchange(s) in the protein sequence.
Figure 5.—
Polymorphic sites in the Hv-eIF4E contig. Polymorphic sites located in coding (e1, e2, etc.) and noncoding regions are designated according to their position in the AY661558 sequence. Dots indicate sites identical to haplotype 1 (rym4-E). For all indels, the starting point is given (*1, 108 bp; *2, 2 bp; *3, 1 bp; *4, 10 bp). Considering all 54 polymorphic sites, haplotypes 1–20 can be identified. Haplotypes 5 and 7 were separated into “a” and “b,” which, although sharing an identical haplotype for all three Hv-eIF4E genic fragments, differed at a site in the flanking region (haplotype 5 at No519; haplotype 7 at No969). Haplogroups I, II, and III are indicated on the left. Growth habit (w, winter; s, spring) and origin (E, Europe; A, Asia; U, America) are given for each haplotype.
Figure 6.—
Haplotype relationships within 132-kb interval. The midpoint-rooted neighbor-joining tree is based on the sequence of No1134, Hv-eIF4E fragments 1–3, No519, and No969 of the 22 haplotypes designated in Figure 5. The numbers on the branches indicate the frequency (%) with which a clade appeared in 1000 bootstrap samples. Branch lengths are proportional to the probable number of substitutions per site using Kimura two-parameter distances. Haplogroups I, II, and III are indicated.
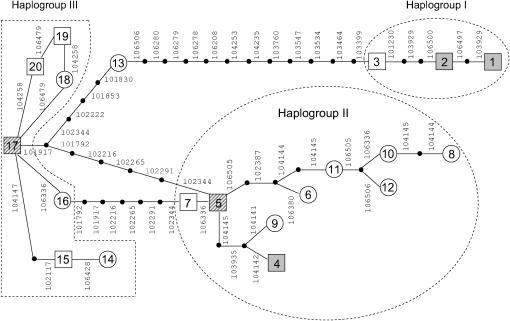
A haplotype network for Hv-eIF4E (Figure 7), constructed with a statistical parsimony algorithm, illustrates the relationships between the haplotypes between and within the haplogroups and emphasizes the genetic distance between the haplogroups I and II/III, with the presumed recombinant haplotype 13 representing the only link between them. Haplotypes 5 and 17 are central within the network and have generated a number of descendant haplotypes. Both are present in a significant number of accessions and are broadly distributed with respect to growth habit and origin. Thus they are probably the most ancient in the germplasm set. In contrast, on the basis of the termini of the network, the two resistant haplotypes, rym4-E and rym5, must have emerged rather recently.
Figure 7.—
The Hv-eIF4E haplotype network. Numbers correspond to the haplotype designations in Figure 5. Lines represent mutational changes and solid circles indicate intermediate haplotypes. The 95% confidence interval is 13 steps. Haplotypes shared between more than one genotype are indicated by squares. Shaded squares correspond to rym4-E (1), rym4-A (2), and rym5 (4) haplotypes, respectively. Haplotypes showing strong independence from growth habit and origin are marked with a hatched pattern. Haplogroups I, II, and III are indicated.
The genetic diversity within the European (n = 80) and Asian (n = 32) subgroups was compared across the physical contig. Despite the 2.5-fold excess of European material within the collection, the Asian accessions included a larger number of haplotypes. The more frequent occurrence of singletons in the latter set (seven vs. four) is largely responsible for its apparent wider diversity. With respect to common haplotypes (frequency within a subgroup ≥0.05), six were represented in the European set (95.5% of the total diversity) and seven in the Asian set (80.5% of the total diversity). Four of these common haplotypes were shared between the subgroups (haplotypes 4, 5a, 17, and 20; Figure 5). As a result, Hd and π-values were comparable between the European (Hd = 0.806 ± 0.022; pi = 0.00484 ± 0.00020) and Asian (Hd = 0.910 ± 0.025; pi = 0.00441 ± 0.00049) sets.
Association analysis:
Since BYMV resistance was most frequent among the European winter barleys, a test for association between the candidate locus and resistance was carried out for the subgroup of 51 European winter cultivars, of which 20 are rym4 carriers and the remainder are susceptible accessions. To ascertain the population structure within this group, a model-based clustering algorithm was applied. The average-likelihood values from 10 runs reached a maximum at K = 2 and fell for higher values (data not shown). The population explained 6% of the variation in BYMV resistance. Structured population association tests were carried out between BYMV resistance and haplotype at both the 12 marker fragments and at 55 of the 83 polymorphic sites (excluding those where the minimum allele frequency was <0.05). The association was significant (P < 0.01) for all nine loci mapping between No1134 and GBS1020, comprising the physical contig across Hv-eIF4E and the distal 1-cM region (Table 2). At six loci in this interval (No1134, Hv-eIF4E 1–3, No519, GBR1845M), rym4 carriers possessed haplotypes that were not observed in any susceptible accessions. Locus GBR1843, located 6.5 cM proximally to the physical contig, showed only a weak association (P < 0.05) with resistance. Over 56% of the SNPs were significantly associated with the resistant phenotype (P < 0.01), confirming the outcome of the haplotype test (supplemental Table S5 at http://www.genetics.org/supplemental/).
TABLE 2.
Structured population association tests between haplotype frequency at single-marker loci and rym4-encoded BYMV resistance
|
K = 1
|
K = 2
|
||||||
|---|---|---|---|---|---|---|---|
| Marker locus | Distance in cMa | χ2 | d.f. | TSb | P-value | TSb | P-value |
| GBR1834 | 5.5 | 11.40 | 4 | 6.41 | 0.0134* | 7.39 | 0.0427* |
| GBS0526 | 4.5 | 1.89 | 1 | 0.95 | 0.1670 | 1.55 | 0.2288 |
| GBS0419 | 2.2 | 7.39 | 1 | 3.99 | 0.0051** | 2.66 | 0.0812 |
| No1134 | 0.02 | 31.18 | 3 | 16.29 | <0.0001*** | 17.00 | <0.0001*** |
| Hv-eIF4E -1 | 0 | 31.34 | 3 | 16.44 | <0.0001*** | 16.92 | <0.0001*** |
| Hv-eIF4E -2 | 0 | 31.04 | 3 | 16.19 | <0.0001*** | 16.94 | <0.0001*** |
| Hv-eIF4E -3 | 0 | 31.04 | 3 | 16.19 | <0.0001*** | 16.94 | <0.0001*** |
| No519 | 0 | 31.00 | 2 | 16.00 | <0.0001*** | 17.27 | <0.0001*** |
| No969 | 0 | 7.64 | 2 | 4.04 | 0.0209* | 6.97 | 0.0066** |
| GBR1845 | 0.4 | 30.58 | 1 | 15.80 | <0.0001*** | 16.78 | <0.0001*** |
| GBR1851 | 0.7 | 20.93 | 2 | 10.84 | <0.0001*** | 11.79 | <0.0001*** |
| GBR1020 | 1.0 | 16.20 | 1 | 8.35 | <0.0001*** | 9.27 | <0.0001*** |
*P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001.
Relative to ATG of Hv-eIF4E.
Test statistic of STRAT (TS).
As a control, association tests were carried out between BYMV resistance and alleles of the evenly distributed SSR markers across the genome (supplemental Table S5 at http://www.genetics.org/supplemental/). Of 11 polymorphic SSR markers in the European winter barleys set, 10 were not associated with the phenotype and only one marker located on 6H showed a weak association (GBM1021; P = 0.035).
DISCUSSION
We have described a detailed evaluation of LD and haplotype patterns surrounding the gene Hv-eIF4E, which encodes a heavily utilized virus resistance in European barley.
Population structure:
Population structure has a major impact on patterns of LD and, consequently, on the outcome of association studies (Pritchard et al. 2000b). The diverse collection of cultivars and landraces from Europe, Asia, and America selected in this study could be clustered on the basis of growth habit, ear morphology, geographical origin, and subspecies and is similar in genetic breadth to collections used in other genomewide marker analyses in barley (Melchinger et al. 1994; Ordon et al. 1997; Thiel et al. 2003). Importantly, however, the population structure did not disturb the association between haplotype and BYMV resistance.
Linkage disequilibrium and genomic pattern:
In self-pollinating species such as A. thaliana and rice, LD has been observed to extend several kilobases beyond a target gene (Hagenblad and Nordborg 2002; Nordborg et al. 2002; Garris et al. 2003; Hagenblad et al. 2004; Olsen et al. 2004), and can even reach the centimorgan range (Nordborg et al. 2002; Zhu et al. 2003; Aranzana et al. 2005). Corresponding results apply at the Hv-eIF4E locus. The structure of the three conserved haplogroups in the physical vicinity of Hv-eIF4E reflected a high level of LD across the 132-kb interval. Similarly, a sustained level of LD has been revealed over a 212-kb stretch flanking the hardness locus in European elite barley cultivars (Caldwell et al. 2006). With respect to genetic distance, LD around Hv-eIF4E fell below the critical threshold of r2 = 0.3 within <1 cM. This result is inconsistent with a genomewide estimate for LD of up to 10 cM, as reported among modern spring barley cultivars (Kraakman et al. 2004). However, it has been conclusively established for both plants and humans that LD is highly variable, reflecting the combined influences of population structure, genomic region under consideration, and the number of polymorphic sites available (Akey et al. 2003; Ke et al. 2004).
The drastic decay of LD at the genetic level, as it was observed in this study, was mainly attributable to the susceptible accessions. While in both rym4- and rym5-resistant groups considerable haplotype conservation persisted, the haplotypes of susceptible accessions revealed a high level of recombination in the regions flanking the 132-kb fragment. Various forces can contribute to haplotype conservation, including (a) prior genetic bottlenecks resulting in a low effective population size, (b) introgression of rym4 and rym5 from a restricted number of sources, (c) a very recent history of intensive selection for resistance against BYMV, (d) a lack of recombination in the target region, and (e) gene function. While it is difficult to demonstrate a specific bottleneck affecting the accessions investigated in this study, the effect of severe domestication-related bottlenecks on haploytpe diversity between modern barley cultivars and wild ssp. spontaneum has been repeatedly described (Badr et al. 2000; Matus and Hayes 2002; Piffanelli et al. 2004). A comparison between the diversity at Hv-eIF4E within wild and cultivated barley should provide more information regarding the history of this locus. Generally, modern breeding has narrowed the genetic base by altering allele frequencies. With the increased use of ssp. spontaneum as a donor for resistance genes, novel alleles have been introgressed into the gene pool of cultivated barley. The typical outcome of such introgressions has been analyzed elsewhere (Ivandic et al. 1998; Pillen et al. 2004). While it initially generates a spike in genetic diversity, strong selection for the exotic allele will gradually erode the frequencies of the “old” alleles, even leading to their complete loss (Russell et al. 2000; Collins et al. 2001; Bundock and Henry 2004). Such a scenario is likely to have occurred during breeding for resistance to BYMV. The analysis of pedigree and molecular marker data provides evidence that there was only a single source each of rym4 and rym5 in European elite germ plasm (Huth 1985; Graner and Bauer 1993; Friedt et al. 2000). Thus, strong selection for resistance resulted in an enrichment of the corresponding alleles in European winter barley. A similar situation applies to the selection of yellow endosperm in maize, which started in the early 20th century, and has been traced to two independent introgression events. As a result, genetic variation in the vicinity of the target locus Y1 is very low (Palaisa et al. 2003, 2004).
The short timescale over which intensive selection for BYMV resistance has operated has provided as yet only limited opportunities for recombination around the resistance gene. However, targeted selection for rym4/rym5 is probably not the only reason for the observed LD, as the pedigree triplets (parent1, parent2, offspring) formed by the susceptible cultivars Ursel, Ultra, Villa, and Volla, which are all represented in the collection, also showed no recombination across the entire region genotyped. Since susceptible alleles of Hv-eIF4E are not likely to be subjected to selection pressure, the observed pattern is more probably an outcome of related pedigrees and a limited number of meiotic events during the breeding process.
A low recombination rate in the target region has possibly also governed the size of the introgression segment and the LD pattern. In this regard, a comparison of genetic and physical distances in the region of Hv-eIF4E revealed marked differences in recombinational activity and, in particular, a reduction proximal to the gene and an increase distal to it (Stein et al. 2005). Except for marker fragment No1134, which borders an interval, characterized by a low ratio of physical-to-genetic distance (0.8–2.3 Mb/cM), the contig is located in a region with a high ratio (30–50 Mb/cM). The low recombination frequencies are consistent with the large proportion of transposable element sequence present in the 439.7-kb BAC contig, which contains only one genic island of 10 kb harboring Hv-eIF4E and Hv-MLL (Wicker et al. 2005). Meiotic recombination in eukaryotes is confined mainly to genes (Thuriaux 1977; Civardi et al. 1994; Dooner and MartinezFerez 1997), and studies in maize have confirmed that genes located close to retrotransposons have less recombinational activity than those present within gene clusters (Fu et al. 2002). A possible additional factor contributing to LD relates to selection pressure on genes located on either side of the BAC contig. The maintenance of LD across several genes has been demonstrated in maize, although it was largely restricted to gene-rich regions (Palaisa et al. 2004).
Further investigations are clearly needed to determine whether the genomic patterns observed can be attributed to a single factor such as limited sample size, limited effective population size, the small number of generations since introgression, or low recombination, or whether it is the result of a combination of some or all of these forces.
Haplotype network for the Hv-eIF4E locus:
On the basis of the derived network model, it can be suggested that the resistant and susceptible alleles have descended relatively recently from a common ancestor. The separation between haplogroups I and II lends strength to the notion that both rym4 and rym5 have an independent evolutionary history. Evidence for a geographic pattern has been provided for several loci in wild barley (Morrell et al. 2003, 2005), but the strong dimorphism associated with rym4 and rym5 did not correspond to any geographic subdivision. The fact that the germplasm has been subjected to breeding, material exchange, and subsequent introgression events of course may obscure such a division. Thus, to gain a better understanding of the evolution of the haplogroups, comprehensive allele genotyping within a set of geographically widely distributed landraces and wild barleys will be required.
Surprisingly, the strong clustering of haplotypes identified across Hv-eIF4E was exclusively attributable to polymorphic sites in noncoding sequence, while the genetic variation within each group was due mainly to amino acid replacement mutations in the exons. Why the dimorphism among the haplogroups is due largely to noncoding polymorphic sites remains unexplained. In particular, the haplotype diversity within haplogroup II is mostly attributable to rare polymorphic sites present in single Asian genotypes (haplotypes 8–12). Ongoing tests for allelism will show whether these genotypes carry new resistance alleles at the Hv-eIF4E locus, or whether the resistant phenotype is conferred by an independent locus.
Consequences for association studies:
The maintenance of the resistant haplotype blocks, resulting from recent, nonrecombined introgression event(s), has resulted in the formation of significant associations between resistance and haplotypes up to a distance of at least 1 cM from the resistance gene. Such a coarse level of resolution is inadequate for map-based gene cloning. On the other hand, it does imply that a fairly low marker density is sufficient to detect associations between a target region and resistance. If this is typical for other genes within breeding germplasm, the prospects are good for identifying chromosomal segments associated with traits of interest. Further resolution may be achievable by using populations that have been intermated over many generations, thereby promoting the breakdown of linkage blocks. This situation is not common in breeding material and is more likely to be found in landraces and wild populations. For spp. spontaneum, levels of LD comparable to that of the outbreeding species maize have been reported (Lin et al. 2002; Morrell et al. 2005; Caldwell et al. 2006), and these should be sufficient to provide the genetic resolution necessary to identify the functional polymorphism associated with the trait variation.
Acknowledgments
We thank M. Koornneef for his critical review of the manuscript, S. Wiedemann and S. Jakob for helpful discussions, and K. Trnka for technical assistance. We also acknowledge the linguistic advice of http://www.smartenglish.co.uk in preparing this article.
References
- Abdel-Ghani, A. H., H. K. Parzies, S. Ceccarelli, S. Grando and H. H. Geiger, 2005. Estimation of quantitative genetic parameters for outcrossing-related traits in barley. Crop Sci. 45: 98–105. [Google Scholar]
- Akey, J. M., K. Zhang, M. M. Xiong and L. Jin, 2003. The effect of single nucleotide polymorphism identification strategies on estimates of linkage disequilibrium. Mol. Biol. Evol. 20: 232–242. [DOI] [PubMed] [Google Scholar]
- Anderson, J. A., G. A. Churchill, J. E. Autrique, S. D. Tanksley and M. E. Sorrells, 1993. Optimizing parental selection for genetic-linkage maps. Genome 36: 181–186. [DOI] [PubMed] [Google Scholar]
- Aranzana, M. J., S. Kim, K. Zhao, E. Bakker, M. Horton et al., 2005. Genome-wide association mapping in Arabidopsis identifies previously known flowering time and pathogen resistance genes. PLoS Genet. 1: e60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badr, A., K. Muller, R. Schafer-Pregl, H. El Rabey, S. Effgen et al., 2000. On the origin and domestication history of barley (Hordeum vulgare). Mol. Biol. Evol. 17: 499–510. [DOI] [PubMed] [Google Scholar]
- Bundock, P. C., and R. J. Henry, 2004. Single nucleotide polymorphism, haplotype diversity and recombination in the Isa gene of barley. Theor. Appl. Genet. 109: 543–551. [DOI] [PubMed] [Google Scholar]
- Caldwell, K. S., J. R. Russell, P. Langridge and W. Powell, 2006. Extreme population-dependent linkage disequilibrium detected in an inbreeding plant species, Hordeum vulgare. Genetics 172: 557–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Civardi, L., Y. J. Xia, K. J. Edwards, P. S. Schnable and B. J. Nikolau, 1994. The relationship between genetic and physical distances in the cloned A1-Sh2 interval of the Zea mays L. genome. Proc. Natl. Acad. Sci. USA 91: 8268–8272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clement, M., D. Posada and K. A. Crandall, 2000. TCS: a computer program to estimate gene genealogies. Mol. Ecol. 9: 1657–1659. [DOI] [PubMed] [Google Scholar]
- Collins, N. C., T. Lahaye, C. Peterhansel, A. Freialdenhoven, M. Corbitt et al., 2001. Sequence haplotypes revealed by sequence-tagged site fine mapping of the Ror1 gene in the centromeric region of barley chromosome 1H. Plant Physiol. 125: 1236–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa, J. M., A. Corey, P. M. Hayes, C. Jobet, A. Kleinhofs et al., 2001. Molecular mapping of the Oregon Wolfe barleys: a phenotypically polymorphic doubled-haploid population. Theor. Appl. Genet. 103: 415–424. [Google Scholar]
- Doll, H., 1987. Outcrossing rates in autumn and spring-sown barley. Plant Breed. 98: 339–341. [Google Scholar]
- Dooner, H. K., and I. M. Martinezferez, 1997. Recombination occurs uniformly within the bronze gene, a meiotic recombination hotspot in the maize genome. Plant Cell 9: 1633–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farnir, F., W. Coppieters, J. J. Arranz, P. Berzi, N. Cambisano et al., 2000. Extensive genome-wide linkage disequilibrium in cattle. Genome Res. 10: 220–227. [DOI] [PubMed] [Google Scholar]
- Friedt, W., K. Werner and F. Ordon, 2000. Genetic progress as reflected in highly successful and productive modern barley cultivars, pp. 271–279 in Proceedings of the 8th International Barley Genetics Symposium, Adelaide, Australia.
- Fu, H. H., Z. W. Zheng and H. K. Dooner, 2002. Recombination rates between adjacent genic and retrotransposon regions in maize vary by 2 orders of magnitude. Proc. Natl. Acad. Sci. USA 99: 1082–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garris, A. J., S. R. McCouch and S. Kresovich, 2003. Population structure and its effect on haplotype diversity and linkage disequilibrium surrounding the xa5 locus of rice (Oryza sativa L.). Genetics 165: 759–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giles, R. J., G. McConnell and J. L. Fyfe, 1974. Frequency of natural cross-fertilization in a composite cross of barley grown in Scotland. J. Agric. Sci. 83: 447–450. [Google Scholar]
- Götz, R., and W. Friedt, 1993. Resistance to the barley yellow mosaic-virus complex: differential genotypic reactions and genetics of BaMMV-resistance of barley (Hordeum vulgare L.). Plant Breed. 111: 125–131. [Google Scholar]
- Graner, A., and E. Bauer, 1993. Rflp mapping of the ym4 virus-resistance gene in barley. Theor. Appl. Genet. 86: 689–693. [DOI] [PubMed] [Google Scholar]
- Graner, A., A. Jahoor, J. Schondelmaier, H. Siedler, K. Pillen et al., 1991. Construction of an Rflp map of barley. Theor. Appl. Genet. 83: 250–256. [DOI] [PubMed] [Google Scholar]
- Graner, A., S. Streng, A. Kellermann, A. Schiemann, E. Bauer et al., 1999. Molecular mapping and genetic fine-structure of the rym5 locus encoding resistance to different strains of the barley yellow mosaic virus complex. Theor. Appl. Genet. 98: 285–290. [Google Scholar]
- Gupta, P. K., S. Rustgi and P. L. Kulwal, 2005. Linkage disequilibrium and association studies in higher plants: present status and future prospects. Plant Mol. Biol. 57: 461–485. [DOI] [PubMed] [Google Scholar]
- Hagenblad, J., and M. Nordborg, 2002. Sequence variation and haplotype structure surrounding the flowering time locus FRI in Arabidopsis thaliana. Genetics 161: 289–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagenblad, J., C. L. Tang, J. Molitor, J. Werner, K. Zhao et al., 2004. Haplotype structure and phenotypic associations in the chromosomal regions surrounding two Arabidopsis thaliana flowering time loci. Genetics 168: 1627–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedrick, P. W., 1987. Gametic disequilibrium measures: proceed with caution. Genetics 117: 331–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill, W. G., and A. Robertson, 1968. Linkage disequilibrium in finite population. Theor. Appl. Genet. 33: 54–78. [DOI] [PubMed] [Google Scholar]
- Huth, W., 1985. Versuche zur Virusdiagnose und Resistenzträgererstellung in Gerste gegen barley yellow mosaic virus. Vorträge für Pflanzenzüchtung 9: 107–120. [Google Scholar]
- Ivandic, V., U. Walther and A. Graner, 1998. Molecular mapping of a new gene in wild barley conferring complete resistance to leaf rust (Puccinia hordei Otth). Theor. Appl. Genet. 97: 1235–1239. [Google Scholar]
- Kanyuka, K., G. McGrann, K. Alhudaib, D. Hariri and M. J. Adams, 2004. Biological and sequence analysis of a novel European isolate of barley mild mosaic virus that overcomes the barley rym5 resistance gene. Arch. Virol. 149: 1469–1480. [DOI] [PubMed] [Google Scholar]
- Kanyuka, K., A. Druka, D. G. Caldwell, A. Tymon, N. Mccallum et al., 2005. Evidence that the recessive bymovirus resistance locus rym4 in barley corresponds to the eukaryotic translation initiation factor 4E gene. Mol. Plant Pathol. 6: 449–458. [DOI] [PubMed] [Google Scholar]
- Ke, X. Y., S. Hunt, W. Tapper, R. Lawrence, G. Stavrides et al., 2004. The impact of SNP density on fine-scale patterns of linkage disequilibrium. Hum. Mol. Genet. 13: 577–588. [DOI] [PubMed] [Google Scholar]
- Konishi, T., T. Ban, Y. Iida and R. Yoshimi, 1997. Genetic analysis of disease resistance to all strains of BaYMV in a Chinese barley landrace, Mokusekko 3. Theor. Appl. Genet. 94: 871–877. [Google Scholar]
- Kraakman, A. T. W., R. E. Niks, P. Van Den Berg, P. Stam and F. A. Van Eeuwijk, 2004. Linkage disequilibrium mapping of yield and yield stability in modern spring barley cultivars. Genetics 168: 435–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, S., K. Tamura and M. Nei, 1994. MEGA: molecular evolutionary genetics analysis software for microcomputers. Comput. Appl. Biosci. 10: 189–191. [DOI] [PubMed] [Google Scholar]
- Lewontin, R. C., 1964. The interaction of selection and linkage. I. General considerations; heterotic models. Genetics 49: 49–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, J.-Z., P. L. Morrell and M. T. Clegg, 2002. The influence of linkage and inbreeding on patterns of nucleotide sequence diversity at duplicate alcohol dehydrogenase loci in wild barley (Hordeum vulgare ssp. spontaneum). Genetics 162: 2007–2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matus, I. A., and P. M. Hayes, 2002. Genetic diversity in three groups of barley germplasm assessed by simple sequence repeats. Genome 45: 1095–1106. [DOI] [PubMed] [Google Scholar]
- Melchinger, A. E., A. Graner, M. Singh and M. M. Messmer, 1994. Relationships among European barley germplasm. 1. Genetic diversity among winter and spring cultivars revealed by Rflps. Crop Sci. 34: 1191–1199. [Google Scholar]
- Morrell, P. L., K. E. Lundy and M. T. Clegg, 2003. Distinct geographic patterns of genetic diversity are maintained in wild barley (Hordeum vulgare ssp spontaneum) despite migration. Proc. Natl. Acad. Sci. USA 100: 10812–10817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrell, P. L., D. M. Toleno, K. E. Lundy and M. T. Clegg, 2005. Low levels of linkage disequilibrium in wild barley (Hordeum vulgare ssp spontaneum) despite high rates of self-fertilization. Proc. Natl. Acad. Sci. USA 102: 2442–2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordborg, M., J. O. Borevitz, J. Bergelson, C. C. Berry, J. Chory et al., 2002. The extent of linkage disequilibrium in Arabidopsis thaliana. Nat. Genet. 30: 190–193. [DOI] [PubMed] [Google Scholar]
- Nordborg, M., T. T. Hu, Y. Ishino, J. Jhaveri, C. Toomajian et al., 2005. The pattern of polymorphism in Arabidopsis thaliana. PloS Biol. 3: 1289–1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen, K. M., S. S. Halldorsdottir, J. R. Stinchcombe, C. Weinig, J. Schmitt et al., 2004. Linkage disequilibrium mapping of Arabidopsis CRY2 flowering time alleles. Genetics 167: 1361–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ordon, F., R. Götz and W. Friedt, 1993. Genetic stocks resistant to barley yellow mosaic viruses (BaMMV, BaYMV, BaYMV-2) in Germany. Barley Genet. Newsl. 22: 46–49. [Google Scholar]
- Ordon, F., A. Schiemann and W. Friedt, 1997. Assessment of the genetic relatedness of barley accessions (Hordeum vulgare sl) resistant to soil-borne mosaic-inducing viruses (BaMMV, BaYMV, BaYMV-2) using RAPDs. Theor. Appl. Genet. 94: 325–330. [Google Scholar]
- Ordon, F., J. Ahlemeyer, K. Werner, W. Köhler and W. Friedt, 2005. Molecular assessment of genetic diversity in winter barley and its use in breeding. Euphytica 146: 21–28.
- Palaisa, K. A., M. Morgante, M. Williams and A. Rafalski, 2003. Contrasting effects of selection on sequence diversity and linkage disequilibrium at two phytogene synthase loci. Plant Cell 15: 1795–1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palaisa, K., M. Morgante, S. Tingey and A. Rafalski, 2004. Long-range patterns of diversity and linkage disequilibrium surrounding the maize Y1 gene are indicative of an asymmetric selective sweep. Proc. Natl. Acad. Sci. USA 101: 9885–9890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellio, B., S. Streng, E. Bauer, N. Stein, D. Perovic et al., 2005. High-resolution mapping of the Rym4/Rym5 locus conferring resistance to the barley yellow mosaic virus complex (BaMMV, BaYMV, BaYMV-2) in barley (Hordeum vulgare ssp vulgare L.). Theor. Appl. Genet. 110: 283–293. [DOI] [PubMed] [Google Scholar]
- Piffanelli, P., L. Ramsay, R. Waugh, A. Benabdelmouna, A. D'Hont et al., 2004. A barley cultivation-associated polymorphism conveys resistance to powdery mildew. Nature 430: 887–891. [DOI] [PubMed] [Google Scholar]
- Pillen, K., A. Zacharias and J. Leon, 2004. Comparative AB-QTL analysis in barley using a single exotic donor of Hordeum vulgare ssp spontaneum. Theor. Appl. Genet. 108: 1591–1601. [DOI] [PubMed] [Google Scholar]
- Pritchard, J. K., M. Stephens and P. Donnelly, 2000. a Inference of population structure using multilocus genotype data. Genetics 155: 945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard, J. K., M. Stephens, N. A. Rosenberg and P. Donnelly, 2000. b Association mapping in structured populations. Am. J. Hum. Genet. 67: 170–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remington, D. L., J. M. Thornsberry, Y. Matsuoka, L. M. Wilson, S. R. Whitt et al., 2001. Structure of linkage disequilibrium and phenotypic associations in the maize genome. Proc. Natl. Acad. Sci. USA 98: 11479–11484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozas, J., and R. Rozas, 1999. DnaSP version 3: an integrated program for molecular population genetics and molecular evolution analysis. Bioinformatics 15: 174–175. [DOI] [PubMed] [Google Scholar]
- Russell, J. R., R. P. Ellis, W. T. B. Thomas, R. Waugh, J. Provan et al., 2000. A retrospective analysis of spring barley germplasm development from ‘foundation genotypes’ to currently successful cultivars. Mol. Breed. 6: 553–568. [Google Scholar]
- Saitoh, K., K. Onishi, I. Mikami, K. Thidar and Y. Sano, 2004. Allelic diversification at the C (OsC1) locus of wild and cultivated rice: nucleotide changes associated with phenotypes. Genetics 168: 997–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein, N., D. Perovic, J. Kumlehn, B. Pellio, S. Stracke et al., 2005. The eukaryotic translation initiation factor 4E confers multiallelic recessive Bymovirus resistance in Hordeum vulgare (L.). Plant J. 42: 912–922. [DOI] [PubMed] [Google Scholar]
- Stein, N., M. Prasad, U. Scholz, T. Thiel, H. Zhang et al., 2007. A 1,000-loci transcript map of the barley genome: new anchoring points for integrative grass genomics. Theor. Appl. Genet. (in press). [DOI] [PubMed]
- Templeton, A. R., K. A. Crandall and C. F. Sing, 1992. A cladistic analysis of phenotypic associations with haplotypes inferred from restriction endonuclease mapping and DNA sequence data. III. Cladogram estimation. Genetics 132: 619–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenaillon, M. I., M. C. Sawkins, A. D. Long, R. L. Gaut, J. F. Doebley et al., 2001. Patterns of DNA sequence polymorphism along chromosome 1 of maize (Zea mays ssp mays L.). Proc. Natl. Acad. Sci. USA 98: 9161–9166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel, T., W. Michalek, R. K. Varshney and A. Graner, 2003. Exploiting EST databases for the development and characterization of gene-derived SSR-markers in barley (Hordeum vulgare L.). Theor. Appl. Genet. 106: 411–422. [DOI] [PubMed] [Google Scholar]
- Thuriaux, P., 1977. Is recombination confined to structural genes on the eukaryotic genome? Nature 268: 460–462. [DOI] [PubMed] [Google Scholar]
- Tian, D. C., H. Araki, E. Stahl, J. Bergelson and M. Kreitman, 2002. Signature of balancing selection in Arabidopsis. Proc. Natl. Acad. Sci. USA 99: 11525–11530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utz, H. F., 1993. PLABSTAT Version 2H. Institute of Plant Breeding, Seed Science, and Population Genetics, University of Hohenheim, Stuttgart, Germany.
- Weber, J. L., 1990. Informativeness of human (Dc-Da)N.(Dg-Dt)N polymorphisms. Genomics 7: 524–530. [DOI] [PubMed] [Google Scholar]
- Wicker, T., W. Zimmermann, D. Perovic, A. H. Paterson, M. Ganal et al., 2005. A detailed look at 7 million years of genome evolution in a 439 kb contiguous sequence at the barley Hv-eIF4E locus: recombination, rearrangements and repeats. Plant J. 41: 184–194. [DOI] [PubMed] [Google Scholar]
- Zhu, Y. L., Q. J. Song, D. L. Hyten, C. P. Van Tassell, L. K. Matukumalli et al., 2003. Single-nucleotide polymorphisms in soybean. Genetics 163: 1123–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]