Abstract
The accurate repair of DNA double-strand breaks is essential for cell survival and maintenance of genome integrity. Here we describe xlf1+, a gene in the fission yeast Schizosaccharomyces pombe that is required for repair of double-strand breaks by nonhomologous end joining during G1 phase of the cell cycle. Xlf1 is the ortholog of budding yeast Nej1 and human XLF/Cernunnos proteins.
DNA double-strand breaks (DSBs), which can arise from endogenous or exogenous sources, particularly ionizing radiation (IR), are considered among the most lethal and genome-destabilizing types of DNA damage (Pierce et al. 2001; Willers et al. 2004). Efficient and accurate repair of DSBs is essential to preserve genome integrity. The options for repair of DSBs depend on the circumstances in which the DNA damage is suffered (Paques and Haber 1999; van Gent et al. 2001; West 2003; Krogh and Symington 2004; Sancar et al. 2004). DSBs that arise in G2 phase of a haploid organism can be effectively repaired by homologous recombination (HR), an error-free mechanism that uses the undamaged sister chromatid as a template for repair of the broken chromosome. In contrast, when DSBs occur in G1 phase of a haploid cell, the sister chromatid is unavailable as a template for DNA repair. In this circumstance, the only option for repair of DSBs in unique DNA sequences is the mechanism of nonhomologous end joining (NHEJ). This error-prone mechanism of DNA repair, which was originally discovered in mammalian cells, has since been shown to be broadly conserved among eukaryotes, including the budding yeast Saccharomyces cerevisiae and the fission yeast Schizosaccharomyces pombe.
In mammalian cells, NHEJ depends on the Ku70/80 complex that is thought to function as an end-to-end synapsis factor, on DNA-dependent protein kinase (DNA–PK), on Artemis endonuclease, and on the XRCC–DNA ligase IV complex, which carries out the final step of NHEJ (Sancar et al. 2004; Weterings and van Gent 2004). NHEJ in S. cerevisiae requires Ku70/80, the Mre11–Rad50–Xrs2 (MRX) complex that is also essential for repair of DSBs by homologous recombination, DNA ligase IV in a complex with the XRCC4-like protein Lif1, and a protein known as Nej1 (Frank-Vaillant and Marcand 2001; Kegel et al. 2001; Valencia et al. 2001; Dudasova et al. 2004; Daley et al. 2005). The only known NHEJ factors in fission yeast are the Ku70/80 complex and DNA ligase IV (Manolis et al. 2001; Miyoshi et al. 2003; Prudden et al. 2003; Ferreira and Cooper 2004). Curiously, S. pombe does not have a homolog of XRCC4/Lif1, nor does it require the Mre11–Rad50–Nbs1 (MRN) complex for NHEJ (Manolis et al. 2001). Similarly, current evidence suggests that the MRN complex is not required for NHEJ in metazoan organisms (Di Virgilio and Gautier 2005). These data suggest that a core group of proteins are required for NHEJ in all species (Ku70/80 and DNA ligase IV), with a larger group of accessory factors (DNA–PK, Artemis, XRCC4/Lif1, and MRN/MRX) that are required for NHEJ in only a subset of organisms.
An exciting recent development in the field of DNA repair was the discovery of a novel human NHEJ factor known as XLF/Cernunnos and the realization that is an ortholog of S. cerevisiae Nej1 (Ahnesorg et al. 2006; Buck et al. 2006a; Callebaut et al. 2006; Sekiguchi and Ferguson 2006). Cernunnos patients display a set of phenotypes, including microcephaly and immune deficiency, which closely resemble the characteristics of patients defective for DNA ligase IV (O'Driscoll et al. 2001; Ben-Omran et al. 2005; Buck et al. 2006b). These data are consistent with the evidence that XLF/Cernunnos interacts with the XRCC4–DNA ligase IV complex, suggesting that XLF/Cernunnos is a third essential subunit of this complex. We had previously noted the existence of a potential Nej1/Xlf1/Cernunnos-like gene in the genome of fission yeast, as also recently reported by others (Callebaut et al. 2006). In view of the potential importance of this gene in NHEJ, and the apparently different functional requirements for NHEJ among S. cerevisiae, S. pombe, and humans, we decided to investigate the S. pombe homolog of XLF/Cernunnos/Nej1, hereafter called Xlf1. The aim of these studies was to determine if S. pombe Xlf1 is required for repair of DSBs by NHEJ.
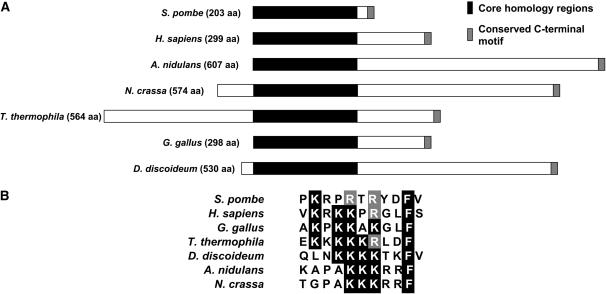
The S. pombe xlf1+ gene, whose systematic name is SPCC24B10.14c, is located on chromosome 3 (coordinates 924,212–924,936) between the snR94 and SPCTRNAARG.09 genes. The predicted mRNA contains one intron that, when spliced out, leaves a transcript that encodes a 203-amino-acid protein. The presence of this intron was confirmed by cloning xlf1+ from a cDNA library (our unpublished data). Putative homologs of xlf1+ exist in a diverse range of fungi and other organisms (Figure 1A) (Callebaut et al. 2006). Interestingly, the size and organization of Xlf1/Nej1 homologs varies widely throughout eukaryotic evolution. Mammalian homologs are ∼300 amino acids, homologs in filamentous fungi are ∼600 amino acids, and S. pombe Xlf1 is exceptional in being only 203 amino acids (Figure 1A). In fact, fission yeast Xlf1 consists almost entirely of an ∼175-amino-acid core homology region located in the conserved N-terminal region of most other Xlf1/Nej1 homologs (Figure 1A). The variable C-terminal regions of Xlf1/Nej1 homologs in filamentous fungi and other nonmetazoan species have no apparent sequence similarity to their metazoan counterparts. However, all of the homologs shown in Figure 1A, and homologs in most other species, share a conserved motif at their extreme termini (Figure 1B). This motif consists of a region rich in basic amino acids (R or K) followed by a phenylalanine residue at either the most C-terminal or the penultimate position (Figure 1B). The C-terminal cluster of basic amino acids was previously noted in metazoan XLF/Cernunnos proteins (Ahnesorg et al. 2006). It was speculated to be part of a nuclear localization signal (NLS) because a cluster of basic amino acids is typical of many NLS sequences (Ahnesorg et al. 2006). Alternatively, the highly conserved phenylalanine residue suggests that this region might be involved in a protein–protein interaction. Interestingly, this sequence is not found at the C terminus of S. cerevisiae Nej1.
Figure 1.—
Nej1/XLF/Cernurnnos homologs. (A) A representation of the Xlf1 homologs in S. pombe, Homo sapiens, Aspergillus nidulans, Neurospora crassa, Tetrahymena thermophila, Gallus gallus, and Dictyostelium discoideum. All of these proteins have a core homology domain of ∼175 amino acids that is ∼18% identical at the primary sequence level between any two of the proteins shown. They also all contain a conserved motif at their extreme C terminus. (B) Conserved C-terminal motif of Xlf1 homologs.
We performed a one-step deletion of the entire open reading frame of the xlf1+ gene, replacing it with the kanMx marker in a haploid strain. The correct replacement of xlf1+ by kanMx was confirmed by PCR analysis. The xlf1∷kanMx appeared to be fully viable. Genetic crosses confirmed that there were no suppressor mutations in the viable haploid xlf1∷kanMx cells. Unlike mutants defective in homologous recombination, such as nbs1Δ cells (Chahwan et al. 2003), the cells in xlf1Δ cultures were not elongated and there was no increase in dead cells relative to the wild-type control (our unpublished data).
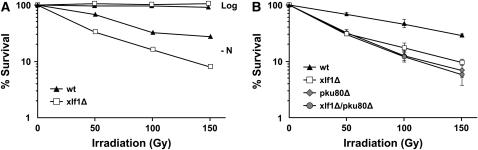
Fission yeast mutants defective in NHEJ exhibit little or no sensitivity to IR or other genotoxic agents that cause DSBs (Ferreira and Cooper 2004). This phenotype can be explained by the cell cycle distribution of S. pombe cells growing in optimum growth conditions. These cells have a long G2 phase and essentially no G1 phase; hence the vast majority of cells in an asynchronous culture have a 2C DNA content and thus are able to repair DSBs by homologous recombination. On the other hand, cells starved of nitrogen primarily arrest division in G1 phase; hence in these conditions NHEJ is needed for repair of DSBs (Ferreira and Cooper 2004). We therefore tested whether xlf1Δ cells were sensitive to IR when grown to log phase or in cultures starved of nitrogen. In log phase, the IR survival of xlf1Δ cells was equivalent to wild type (Figure 2A). In nitrogen-starved cells, the IR survival of wild-type cells was decreased relative to wild-type (and xlf1Δ cells) cells grown in rich media. This result is indicative of the relative inefficiency of NHEJ compared to HR in repairing DSBs. Significantly, in nitrogen-starved cells the IR survival of xlf1Δ cells was decreased relative to wild type (Figure 2A). From these results we conclude that Xlf1 is required for efficient repair of DSBs in G1 but not during G2 phase.
Figure 2.—
Xlf1 and Pku80 function in the same pathway required for survival of DSBs in G1 phase. (A) IR survival of wild type (PR110; leu1-32 ura4-D18) and xlf1Δ (SC4080; xlf1∷kanMx leu1-32 ura4-D18) strains. All incubations were performed at 30°. For log-phase cultures, cells were grown to midlog phase (OD600 = 0.5) in Edinburgh minimal media with leucine and uracil (EMM + LH) medium (Moreno et al. 1991). For nitrogen-starved cultures (−N), cells were prepared essentially as described (Ferreira and Cooper 2004). They were grown in EMM + LU to OD600 = 0.5, washed three times in EMM + LU without ammonium chloride, suspended in the same media at a cell concentration 1–5 × 106 cells/ml, incubated for 72 hr, and then readjusted to OD600 = 0.5 in the same media. Cells were irradiated with IR at 3.2 Gy/min using a 137Cs source at room temperature (∼26°) and then plated onto rich media (yeast extract, glucose, and supplements) plates at 30°. Colony numbers were counted 3 days after plating to determine cell survival rate. (B) The same experiment was repeated in triplicate with wild type (PR110), xlf1Δ (SC4080), pku80Δ (SC4082; pku80∷kanMx leu1-32 ura4-D18), and xlf1Δ pku80Δ (SC4084; xlf1∷kanMx pku80∷hygMx leu1-32 ura4-D18).
These findings suggested that Xlf1 was likely part of the NHEJ mechanism that repairs DSBs in G1 phase. To address this question more specifically, we performed genetic epistasis analysis with xlf1Δ and pku80Δ mutations. Previous studies showed that pKU80 is essential for NHEJ in fission yeast (Miyoshi et al. 2003). As observed in the previous experiment, neither of the single xlf1Δ and pku80Δ mutants were sensitive to IR when log-phase cultures were irradiated (data not shown). Likewise, the xlf1Δ pku80Δ double mutant was insensitive to IR in log-phase cultures (data not shown). However, when nitrogen-starved cells were irradiated, we observed that both the xlf1Δ and pku80Δ strains were more sensitive to IR compared to wild type, and their degree of IR sensitivity was approximately equal (Figure 2B). Importantly, the xlf1Δ pku80Δ double mutant was no more sensitive to IR than either of the single mutants (Figure 2B).
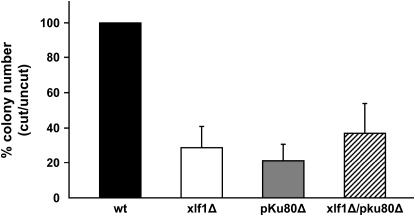
These results strongly indicated that Xlf1 is essential for the Ku80-dependent process of repair of DSBs by NHEJ. To specifically address this question, we performed linear plasmid ligation assays with the relevant strains. Intact plasmid pAL-SK+ or the same plasmid linearized with BamHI endonuclease was transformed into cells as previously described (Nakamura et al. 2004). The relative copy number of transformed colonies from cut vs. uncut plasmid was calculated for each strain and normalized relative to wild type. This analysis showed that there was a large deficiency in linear plasmid transformation efficiency in the xlf1Δ and pku80Δ strains relative to wild type (Figure 3). As seen for the IR survival assays, the defect of the xlf1Δ pku80Δ double mutant in the linear plasmid transformation efficiency was equivalent within statistical error to either of the single mutants (Figure 3), indicating that Xlf1 is essential for Ku80-dependent repair of DSBs by NHEJ.
Figure 3.—
Xlf1 is required for NHEJ. A plasmid repair assay to measure NHEJ efficiency was carried out for the indicated strains. Relative colony numbers obtained from transformation with cut vs. uncut plasmid are plotted. Intact plasmid pAL-SK+ or the same plasmid linearized with BamHI endonuclease were transformed into cells as previously described (Nakamura et al. 2004).
These genetic epistasis studies establish that Xlf1 is required for repair of IR-induced DSBs by the NHEJ pathway in fission yeast. Because xlf1Δ cells are insensitive to IR when irradiated in log-phase cultures, we further conclude that XLF1 is not required for repair of DSBs by homologous recombination. We have also tested whether log-phase xlf1Δ cells are sensitive to a variety of other genotoxic agents, such as hydroxyurea (HU), UV irradiation, methyl methanesulfonate, and camptothecin (CPT). These agents can damage DNA in a variety of ways. For example, HU causes replication forks to stall, while CPT cause replication fork breakage. Neither the xlf1Δ cells nor the pku80Δ mutant were abnormally sensitive to these DNA-damaging agents (our unpublished data), again pointing to a role of Xlf1 specifically in NHEJ. From these results we conclude that Xlf1 is an essential core component of NHEJ in fission yeast.
The discovery of the similarity between XLF and Nej1 suggests that the full battery of known budding-yeast NHEJ repair factors is present in metazoa. Intriguingly, fission yeasts seem to lack an ortholog of the Lif1/XRCC4 subunit. There is a slim possibility that a highly divergent ortholog remains to be identified, but given the high degree of similarity between the budding yeast and metazoan proteins, the inability to identify a sequence homolog might genuinely reflect a gene loss event from the fission yeast genomes. It has been suggested that in spite of the lack of any clear primary sequence similarity, XLF bears a high degree of resemblance to XRCC4 at the secondary structure level (Ahnesorg et al. 2006; Callebaut et al. 2006). It is therefore reasonable to speculate that S. pombe might rely on one protein, Xlf1, perhaps in a homodimeric state, to perform functions similar to those usually performed by an XRCC4-XLF heterodimer in other species. Such a scenario is not unprecedented. For instance, the two subunits of the heterodimeric structure-specific endonucleases Mus81-Eme1 and XPF-ERCC1 share a high level of predicted secondary structure similarity in the catalytic and DNA-binding domains. Interestingly, in Archea this family of heterodimeric nucleases is represented by a single protein, Hef, which homodimerizes to perform its catalytic functions (Nishino et al. 2005). Another possibility is that there exists a subset of breaks that are refractory to ligation in the absence of XRCC4. Conceivably, such reactions could be dependent on physiological contexts, such as a specific chromatin code around the site of the DNA break, that are species specific. For instance, the histone H3-K79 methyltransferase, Dot1, is present in budding and filamentous yeasts as well as in metazoa but absent from fission yeasts (Feng et al. 2002; Sanders et al. 2004). As such, the loss of a specific NHEJ-inhibitory factor could have mitigated the requirement for an XRCC4 ortholog in the fission yeasts, thereby leading to its subsequent loss. Future biochemical experiments should provide a more conclusive answer to these speculations.
Finally, we note that an independent study of S. pombe Xlf1 appeared while this article was under review (Hentges et al. 2006). In agreement with the findings reported here, Hentges et al. (2006) found that Xlf1 is required for efficient transformation of S. pombe with a linear plasmid and that it is also required for maximum survival of spores exposed to ionizing radiation. It also found that Xlf1 multimerizes in vitro and binds to DNA.
Acknowledgments
We thank the members of the Russell lab for support, advice, and encouragement. This research was funded by National Institutes of Health grant CA77325 awarded to P.R.
References
- Ahnesorg, P., P. Smith and S. P. Jackson, 2006. XLF interacts with the XRCC4-DNA ligase IV complex to promote DNA nonhomologous end-joining. Cell 124: 301–313. [DOI] [PubMed] [Google Scholar]
- Ben-Omran, T. I., K. Cerosaletti, P. Concannon, S. Weitzman and M. M. Nezarati, 2005. A patient with mutations in DNA ligase IV: clinical features and overlap with Nijmegen breakage syndrome. Am. J. Med. Genet. 137: 283–287. [DOI] [PubMed] [Google Scholar]
- Buck, D., L. Malivert, R. de Chasseval, A. Barraud, M. C. Fondaneche et al., 2006. a Cernunnos, a novel nonhomologous end-joining factor, is mutated in human immunodeficiency with microcephaly. Cell 124: 287–299. [DOI] [PubMed] [Google Scholar]
- Buck, D., D. Moshous, R. de Chasseval, Y. Ma, F. le Deist et al., 2006. b Severe combined immunodeficiency and microcephaly in siblings with hypomorphic mutations in DNA ligase IV. Eur. J. Immunol. 36: 224–235. [DOI] [PubMed] [Google Scholar]
- Callebaut, I., L. Malivert, A. Fischer, J. P. Mornon, P. Revy et al., 2006. Cernunnos interacts with the XRCC4 × DNA-ligase IV complex and is homologous to the yeast nonhomologous end-joining factor Nej1. J. Biol. Chem. 281: 13857–13860. [DOI] [PubMed] [Google Scholar]
- Chahwan, C., T. M. Nakamura, S. Sivakumar, P. Russell and N. Rhind, 2003. The fission yeast Rad32 (Mre11)-Rad50-Nbs1 complex is required for the S-phase DNA damage checkpoint. Mol. Cell. Biol. 23: 6564–6573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daley, J. M., P. L. Palmbos, D. Wu and T. E. Wilson, 2005. Nonhomologous end joining in yeast. Annu. Rev. Genet. 39: 431–451. [DOI] [PubMed] [Google Scholar]
- Di Virgilio, M., and J. Gautier, 2005. Repair of double-strand breaks by nonhomologous end joining in the absence of Mre11. J. Cell Biol. 171: 765–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudasova, Z., A. Dudas and M. Chovanec, 2004. Non-homologous end-joining factors of Saccharomyces cerevisiae. FEMS Microbiol. Rev. 28: 581–601. [DOI] [PubMed] [Google Scholar]
- Feng, Q., H. Wang, H. H. Ng, H. Erdjument-Bromage, P. Tempst et al., 2002. Methylation of H3-lysine 79 is mediated by a new family of HMTases without a SET domain. Curr. Biol. 12: 1052–1058. [DOI] [PubMed] [Google Scholar]
- Ferreira, M. G., and J. P. Cooper, 2004. Two modes of DNA double-strand break repair are reciprocally regulated through the fission yeast cell cycle. Genes Dev. 18: 2249–2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank-Vaillant, M., and S. Marcand, 2001. NHEJ regulation by mating type is exercised through a novel protein, Lif2p, essential to the ligase IV pathway. Genes Dev. 15: 3005–3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentges, P., P. Ahnesorg, R. S. Pitcher, C. K. Bruce, B. Kysela et al., 2006. Evolutionary and functional conservation of the DNA non-homologous end-joining protein, XLF/Cernunnos. J. Biol. Chem. 281: 37517–37526. [DOI] [PubMed] [Google Scholar]
- Kegel, A., J. O. Sjostrand and S. U. Astrom, 2001. Nej1p, a cell type-specific regulator of nonhomologous end joining in yeast. Curr. Biol. 11: 1611–1617. [DOI] [PubMed] [Google Scholar]
- Krogh, B. O., and L. S. Symington, 2004. Recombination proteins in yeast. Annu. Rev. Genet. 38: 233–271. [DOI] [PubMed] [Google Scholar]
- Manolis, K. G., E. R. Nimmo, E. Hartsuiker, A. M. Carr, P. A. Jeggo et al., 2001. Novel functional requirements for non-homologous DNA end joining in Schizosaccharomyces pombe. EMBO J. 20: 210–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi, T., M. Sadaie, J. Kanoh and F. Ishikawa, 2003. Telomeric DNA ends are essential for the localization of Ku at telomeres in fission yeast. J. Biol. Chem. 278: 1924–1931. [DOI] [PubMed] [Google Scholar]
- Moreno, S., A. Klar and P. Nurse, 1991. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194: 795–823. [DOI] [PubMed] [Google Scholar]
- Nakamura, T. M., L. L. Du, C. Redon and P. Russell, 2004. Histone H2A phosphorylation controls Crb2 recruitment at DNA breaks, maintains checkpoint arrest, and influences DNA repair in fission yeast. Mol. Cell. Biol. 24: 6215–6230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishino, T., K. Komori, Y. Ishino and K. Morikawa, 2005. Structural and functional analyses of an archaeal XPF/Rad1/Mus81 nuclease: asymmetric DNA binding and cleavage mechanisms. Structure 13: 1183–1192. [DOI] [PubMed] [Google Scholar]
- O'Driscoll, M., K. M. Cerosaletti, P. M. Girard, Y. Dai, M. Stumm et al., 2001. DNA ligase IV mutations identified in patients exhibiting developmental delay and immunodeficiency. Mol. Cell 8: 1175–1185. [DOI] [PubMed] [Google Scholar]
- Paques, F., and J. E. Haber, 1999. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 63: 349–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce, A. J., J. M. Stark, F. D. Araujo, M. E. Moynahan, M. Berwick et al., 2001. Double-strand breaks and tumorigenesis. Trends Cell Biol. 11: S52–S59. [DOI] [PubMed] [Google Scholar]
- Prudden, J., J. S. Evans, S. P. Hussey, B. Deans, P. O'Neill et al., 2003. Pathway utilization in response to a site-specific DNA double-strand break in fission yeast. EMBO J. 22: 1419–1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancar, A., L. A. Lindsey-Boltz, K. Unsal-Kacmaz and S. Linn, 2004. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu. Rev. Biochem. 73: 39–85. [DOI] [PubMed] [Google Scholar]
- Sanders, S. L., M. Portoso, J. Mata, J. Bahler, R. C. Allshire et al., 2004. Methylation of histone H4 lysine 20 controls recruitment of Crb2 to sites of DNA damage. Cell 119: 603–614. [DOI] [PubMed] [Google Scholar]
- Sekiguchi, J. M., and D. O. Ferguson, 2006. DNA double-strand break repair: a relentless hunt uncovers new prey. Cell 124: 260–262. [DOI] [PubMed] [Google Scholar]
- Valencia, M., M. Bentele, M. B. Vaze, G. Herrmann, E. Kraus et al., 2001. NEJ1 controls non-homologous end joining in Saccharomyces cerevisiae. Nature 414: 666–669. [DOI] [PubMed] [Google Scholar]
- van Gent, D. C., J. H. Hoeijmakers and R. Kanaar, 2001. Chromosomal stability and the DNA double-stranded break connection. Nat. Rev. Genet. 2: 196–206. [DOI] [PubMed] [Google Scholar]
- West, S. C., 2003. Molecular views of recombination proteins and their control. Nat. Rev. Mol. Cell Biol. 4: 435–445. [DOI] [PubMed] [Google Scholar]
- Weterings, E., and D. C. van Gent, 2004. The mechanism of non-homologous end-joining: a synopsis of synapsis. DNA Rep. 3: 1425–1435. [DOI] [PubMed] [Google Scholar]
- Willers, H., J. Dahm-Daphi and S. N. Powell, 2004. Repair of radiation damage to DNA. Br. J. Cancer 90: 1297–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]