Abstract
Schizophrenia may result from a neurotransmission hypofunction of glutamatergic and N-methyl-d-aspartate (NMDA) receptors. Linkage disequilibrium mapping has identified several promising and novel positional candidates, including the G72/G30 and d-amino-acid oxidase (DAAO) genes. Since the first positive association report, many subsequent studies have attempted to replicate the association but the results have been mixed. To try to resolve this inconsistency and to elucidate the relationship between the important glutamate-related genes and schizophrenia, the current meta-analysis has combined samples involving 16 polymorphisms covering all published case-control and family-based association studies up to October 2005. The results suggest that there is weak evidence of association between the G72/G30 genes and schizophrenia.
SCHIZOPHRENIA is a serious psychiatric disease that affects up to 1% of the population worldwide (Cannon et al. 1998; Jablensky 2000). Studies have suggested that schizophrenia might result from a neurotransmission hypofunction of glutamatergic and N-methyl-d-aspartate (NMDA) receptors (Tsai and Coyle 2002; Harrison and Owen 2003; Hall et al. 2004; Owen et al. 2004; Rapoport et al. 2005). A number of studies have provided evidence of linkage scans and mapping to chromosome 13q22–q34 (Lin et al. 1997; Blouin et al. 1998; Shaw et al. 1998; Brzustowicz et al. 1999; Levinson et al. 2000). Chumakov et al. (2002) found robust evidence of genetic association within the 13q linkage region for schizophrenia. Two overlapping genes, G72 and G30, located at 13q34 and spanning a 65-kb segment, have been shown to be significantly associated with schizophrenia using both individual single nucleotide polymorphism (SNP) and haplotype analysis (Chumakov et al. 2002).
G72 has been hypothesized to produce protein PLG72, which is an agonist for the glycine-binding site of the NMDA glutamate receptors (Mothet et al. 2000). The postmortem analysis of schizophrenic patients has revealed overproduced G72 together with a lower NMDA glutamate receptor activity, which could result in glutamate-signaling hypofunction (Chumakov et al. 2002). Further expression and functional studies have also supported the role of G72 in the etiology of schizophrenia (Mothet et al. 2000; Chumakov et al. 2002; Tsai and Coyle 2002; O'Donovan et al. 2003; Shirts and Nimgaonkar 2004). G72 protein interacts with the gene for d-amino-acid oxidase (DAAO) on 12q24 to regulate glutaminergic signaling through the NMDA receptors pathway (Tsai and Coyle 2002), and variation within G72 could affect NMDA signaling through the functional pathway (Chumakov et al. 2002). The synergic effect on the risk for schizophrenia was higher than the effect of each individual gene due to the interaction between the proteins of G72 and DAAO (Chumakov et al. 2002). The story of G72/G30 is quite complex, as set out in an article by Detera-Wadleigh and McMahon (2006). G72/G30 is also believed to be associated with other psychiatric disorders, such as bipolar disorder (Hattori et al. 2003; Chen et al. 2004; Schumacher et al. 2004).
Chumakov et al. (2002) initially reported strong associations between schizophrenia and dozens of SNPs in the G72/G30 and DAAO genes. The associations have been replicated independently in several, but not in all, studies. The available evidence shows inconsistency with regard to which alleles show association with the disease. In an attempt to clarify this inconsistency and to measure the magnitude of the effect of the putative risk alleles, the current meta-analysis has combined all available case-control and family-based association studies dealing with the G72/G30 and DAAO genes and schizophrenia.
MATERIALS AND METHODS
Literature search:
The literature included in the analysis was selected using PubMed and focusing on the keywords “schizophrenia,” “G72,” “G30,” “d-amino-acid oxidase,” and “DAAO.” All references cited in these studies and published reviews were examined to identify additional work not indexed by MEDLINE. The analyzed data cover work from all English-language publications up to October 2005.
Inclusion criteria:
Eligible studies had to meet all of the following criteria: (1) the studies were published in peer-reviewed journals and were independent studies using original data; (2) the studies provided sufficient data to calculate the odds ratio (OR) with confidence interval and P-value; (3) the studies investigated one or more of the 16 polymorphisms using either case-control or family-based approaches; (4) the studies described the genotyping primers, equipment, and protocols used or provided reference to them; (5) the studies diagnosed schizophrenia patients according to the International Classification of Diseases, Diagnostic and Statistical Manual, or Chinese Classification of Mental Disorders systems; and (6) the studies used healthy individuals as controls. Authors were contacted in cases where there were questions regarding their studies.
Quality assessments:
For association studies with inconsistent results based on the same polymorphisms, the methodological quality needed to be assessed using appropriate criteria to limit the risk of introducing bias into meta-analyses or systematic reviews. The classification method known as the “extended-quality score” (Li et al. 2006; Li and He 2006) was used to assess the quality of association studies. The extended-quality score categorizes studies as “high,” “medium,” or “poor” quality.
Statistical analyses:
Any study that contained data from different ethnic populations (or genders) was considered effectively as several individual studies. Data from the case-control studies were summarized in two-by-two tables and transmission disequilibrium test (TDT) studies were summarized in two-by-one tables. From each table, a log-OR and its sampling variance were calculated (Cho et al. 2005). Cochran's chi-square-based Q-statistic test was used to assess possible heterogeneity among the individual studies. Heterogeneity Q-tests were also performed to test for differences in OR between design types (case control vs. family based). A test for funnel plot asymmetry, described by Egger et al. (1997), was used to assess evidence for publication bias. ORs were pooled using the method of DerSimonian and Laird (1986), and 95% C.I.'s were constructed using Woolf's method. The significance of the overall OR was determined by the Z-test. For the sensitivity analysis, each study in turn was removed from the total, and the remaining were reanalyzed. This procedure was used to ensure that no individual study was entirely responsible for a finding. The type I error rate was set at 0.05. P-values are two tailed. An R-project program was used to depict the degree of differences and trend of association of risk allele frequency from controls to patients. If the vector had the same direction, this indicated the same kind of association, and vice versa.
Haplotype construction, counting, and linkage disequilibrium (LD) block defining were performed on 30 Centre d'Etude du Polymorphisme Humain trios (Utah residents) using Haploview software (http://www.hapmap.org). The multiallelic D′ was computed by performing a series of pairwise D′ calculations using each haplotype in turn as an allele, with all other haplotypes at the locus serving as the other allele. This was then repeated for each haplotype at each locus and averaged by haplotype frequency. Maximum-likelihood haplotype blocks were calculated using an EM algorithm.
Electronic database information:
Accession numbers and URLs for data in this article are as follows: for G72/G30 and DAAO, Online Mendelian Inheritance in Man at http://www.ncbi.nlm.nih.gov/Omim; genotype data for G72/G30 and DAAO, http://www.hapmap.org/; and genome data for G72/G30 and DAAO, http://genome.ucsc.edu/.
RESULTS
The combined search yielded 49 references. After discarding overlapping references and those that clearly did not meet the criteria, 14 studies were retained. These studies were then filtered to ensure conformity with the inclusion criteria. For the G72/G30 genes, two studies (Hattori et al. 2003; Chen et al. 2004) were excluded because they were concerned with bipolar affective disorders rather than schizophrenia; for the DAAO gene, one study (Chumakov et al. 2002) was excluded on the grounds of insufficient data. Finally, 11 studies, consisting of 6 case-control (1292 cases and 1392 controls) (Chumakov et al. 2002; Korostishevsky et al. 2004; Schumacher et al. 2004; Wang et al. 2004) and 3 TDT studies (Addington et al. 2004; Mulle et al. 2005; Zou et al. 2005) for the G72/G30 genes, and 2 case-control studies (846 cases and 836 controls) (Liu et al. 2004; Schumacher et al. 2004) for the DAAO gene, met our criteria for inclusion. The 11 studies included 2138 cases, 2228 controls, and 463 parent–offspring trios. They all fell into the category of medium-to-high quality, with no study falling into the “poor” category.
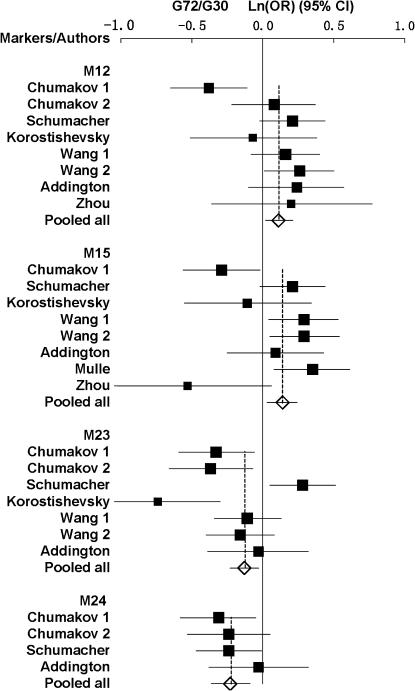
For M12 of the G72/G30 genes, all studies showed a pooled P-value of 0.02 [overall OR = 1.12 (1.02, 1.24)] with evidence of heterogeneity between studies (P = 0.003) (Table 1). There was no evidence of heterogeneity between design types (case control vs. TDT) (P > 0.05) (supplemental Table 1 at http://www.genetics.org/supplemental/).
TABLE 1.
The overall results of all polymorphisms of the G72/G30 genes
| Markers | OR (95% C.I.) | P(Z) | P(Q) |
|---|---|---|---|
| M12 (A/G)a (8)b | 1.12 (1.02, 1.24) | 0.0225 | 0.0031 |
| M13 (A/C) (4) | 1.06 (0.9, 1.24) | 0.4794 | 0.3857 |
| M14 (A/G) (8) | 0.99 (0.89, 1.1) | 0.8909 | 0.1388 |
| M15 (A/G) (8) | 1.15 (1.04, 1.27) | 0.0086 | 0.0030 |
| M16 (A/G) (3) | 1.12 (0.92, 1.35) | 0.2521 | 0.7874 |
| M18 (A/C) (3) | 0.95 (0.79, 1.13) | 0.5571 | 0.5008 |
| M19 (A/G) (4) | 0.98 (0.83, 1.16) | 0.8451 | 0.2428 |
| M20 (A/G) (3) | 1.03 (0.86, 1.23) | 0.7663 | 0.7641 |
| M21 (C/T) (4) | 1.14 (0.97, 1.34) | 0.1090 | 0.7115 |
| M22 (A/G) (7) | 1.04 (0.93, 1.17) | 0.5096 | 0.0151 |
| M23 (C/T) (7) | 0.88 (0.79, 0.97) | 0.0136 | 0.0004 |
| M24 (A/T) (4) | 0.8 (0.69, 0.91) | 0.0010 | 0.6421 |
| rs1935062 (A/C) (4) | 0.91 (0.79, 1.04) | 0.1611 | 0.3605 |
P(Z), Z-test used to determine the significance of the overall OR. P-values < 0.05 are in italics. P(Q), the Cochran's χ2-based Q-statistic test was used to assess the heterogeneity. P(T), T-test was used to evaluate the significance of publication bias. No P(T) < 0.05 (not shown). The NCBI rs ID for the M-SNPs (Chumakov et al. 2002) are the following: M12 (rs3916965), M13 (rs3916966), M14 (rs3916967), M15 (rs2391191), M16 (rs3918341), M18 (rs947267), M19 (rs778294), M20 (rs3916970), M21 (rs3916971), M22 (rs778293), M23 (rs3918342), and M24 (rs1421292).
The first allele is the risk allele.
The number of studies included are indicated in parentheses.
For M15, all studies showed a significant P-value of 0.0086 [overall OR = 1.15 (1.04, 1.27)] with evidence of heterogeneity (P = 0.003) (Table 1). There was no evidence of heterogeneity between design types (supplemental Table 1 at http://www.genetics.org/supplemental/). For M23, all studies showed a pooled P-value of 0.01 [overall OR = 0.88 (0.79, 0.97)] also with evidence of heterogeneity (P = 0.0004) (Table 1). For M24, all studies showed a significant P-value of 0.001 [overall OR = 0.8 (0.69, 0.91)], and there was no evidence of heterogeneity (P = 0.64) (Table 1). The forest plots for the four SNPs are shown in Figure 1.
Figure 1.—
Forest plots of ln(OR) with 95% C.I. for the associated polymorphisms of the G72/G30 genes. Solid squares indicate the ln(OR), with the size of the square inversely proportional to its variance, and horizontal lines represent the 95% C.I.'s. The pooled results are indicated by the open diamond.
No statistically significant association was found in nine other SNPs (Table 1), and we found no significant association in the genotypic analyses for these SNPs whether the genotypes were combined with risk alleles or nonrisk alleles (Table 2).
TABLE 2.
The overall results of genotypic analysis
| Markers | Combinations | OR (95% C.I.) | P(Z) | P(Q) |
|---|---|---|---|---|
| M12 (A/G) | (11+12)/22 | 1.19 (0.89, 1.61) | 0.2406 | 0.1207 |
| 11/(12+22) | 1.38 (0.91, 2.08) | 0.1260 | 0.7559 | |
| M15 (A/G) | (11+12)/22 | 0.98 (0.48, 2) | 0.9577 | 0.0436 |
| 11/(12+22) | 1.35 (0.89, 2.04) | 0.1586 | 0.7418 | |
| M19 (A/G) | (11+12)/22 | 0.79 (0.59, 1.05) | 0.0985 | 0.5385 |
| 11/(12+22) | 0.95 (0.55, 1.65) | 0.8614 | 0.4701 | |
| M23 (C/T) | (11+12)/22 | 0.61 (0.12, 3.08) | 0.5527 | 0.00003 |
| 11/(12+22) | 1.16 (0.84, 1.61) | 0.3702 | 0.0524 | |
| MDAAO-4-5-6 | (11+12)/22 | 1.48 (1.09, 2.00) | 0.012 | 0.7617 |
| 11/(12+22) | 1.42 (1.17, 1.71) | 0.0003 | 0.7621 |
1 represents the first allele, and 2 represents the second allele. The NCBI rs ID for the MDAAO-SNPs are the following: MDAAO-4 (rs2111902), MDAAO-5 (rs3918346), and MDAAO-6 (rs3741775).
As for the DAAO gene, there was no evidence of association in the allelic analysis for any of the three polymorphisms MDAAO-4, -5, or -6 (P > 0.05) (not shown) although positive evidence of haplotypes for the three SNPs was reported (P < 0.00001) (Liu et al. 2004). However, when the genotype data of the three SNPs were combined, the pooled P-value was 0.0003 [overall OR = 1.42 (1.17, 1.71)] (Table 2). No publication bias was found for allelic and genotypic analyses with regard to any polymorphism [no P(T) < 0.05].
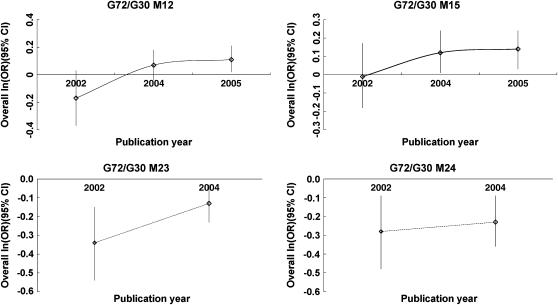
For M24, the results showed consistency, with the largest P-value being 0.015. However, for M12, M15, and M23, evidence of sensitivity was found, with the largest P-value > 0.05 (supplemental Table 2 at http://www.genetics.org/supplemental/). The retrospective asymptote lines of the analysis based on the publication year showed that a cumulative synthesis of M12 and M15 tended to be stable after 2004, in line with the meta-analysis. However, more replications are suggested for M23 and M24 due to instability of asymptotic slopes (Figure 2).
Figure 2.—
Retrospective analysis of associated polymorphisms was based on publication years since 2002.
The funnel plots and trend of allele frequency by R project are shown for G72/G30 as supplemental Figures 1 and 2 at http://www.genetics.org/supplemental/. Lack of space precluded the inclusion of the results of individual studies but these are available on request.
DISCUSSION
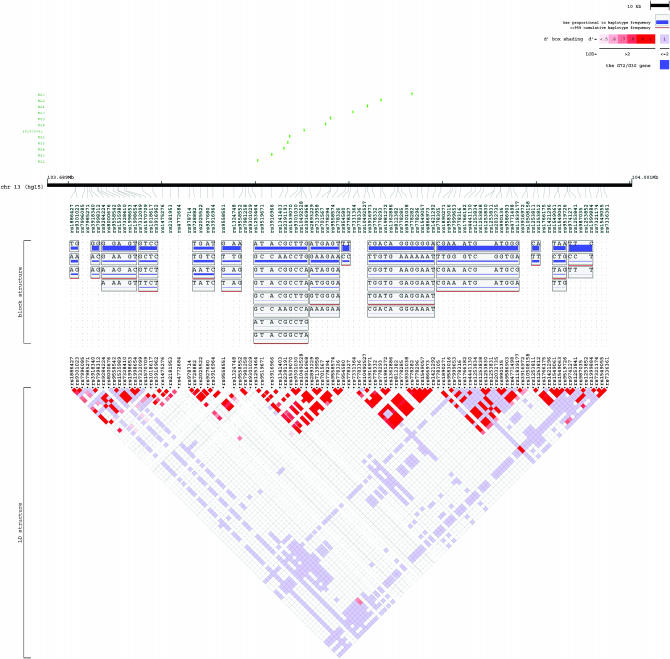
As for the LD and haplotype structure for the G72/G30 genes, the 13 polymorphisms, covering four blocks, were in two strong LD structures (Figure 3). Furthermore, the M12 and M15 polymorphisms were located in one block, while M23 and M24 were in another strong block, which was consistent with results of the retrospective analysis. For the DAAO gene, the three SNPs were in a strong LD block, as we have described elsewhere (Liu et al. 2004).
Figure 3.—
Representation of the LD structure. The LD structure for the G72/G30 genes, spanning 282 kb, was constructed using genotype data from 108 SNPs. The 12 polymorphisms are shown in green. Vertical tick marks above the name indicate the relative genomic position of each SNP. The LD structure represents the pairwise calculation of D′ for each possible combination of SNPs. D′ < 0.5 is shown in white, D′ = 1.0 in dark red, with increasing shades of red representing increasing D′ among the SNPs. It was produced by Locusview software (T. Petryshen, A. Kirby and M. Ainscow, unpublished results).
Although previous individual studies have reported strong associations in the allelic, genotypic, or haplotypic analyses using either family-based or case-control methods, the current meta-analysis confirmed only weak overall association with G72/G30, and there was considerable heterogeneity among the associated alleles. First, for most SNPs with heterogeneity, different alleles, genotypes, or haplotypes seemed to be associated in different studies. Heterogeneity is partly due to possible sampling bias, including population stratification due to ethnicities and diagnostic variations or considerable differences in allele frequencies. Second, the G72/G30 genes may be under a complex expression control. It is likely that different regulatory elements respond to different cell types and developmental stages and that different SNP combinations affect the regulation of expression. Third, environmental factors, such as the season of birth, may contribute to several psychiatric and neurological disorders, including schizophrenia.(Chotai et al. 2003) Schizophrenia may be characterized by genetic heterogeneity, and the genetic architecture in different populations can differ. Heterogeneity may also be a factor in explaining the results of the sensitivity analysis.
Only one meta-analysis on G72 has previously been performed (Detera-Wadleigh and McMahon 2006), and it combined data from different studies at the level of P-values. Compared with that study, this meta-analysis uses more extensive statistical methods, which can systematically incorporate information on specific alleles and genotypes. This study, therefore, is a valuable addition to the literature on G72. However, it has a number of limitations, principal among which is that no real genotypes were available, which prevented comparison of haplotype frequencies. Other limitations include the possible effects of variables such as age, ethnicity, and gender, which can be dealt with only by using a greater range of studies. For subsequent meta-analyses, the sample size needed may depend on the degree of association, linkage disequilibrium, accuracy of phenotypic data, and heterogeneity of allelic frequencies. More accurate phenotype definition, strict selection of samples, and uniformity of diagnosis and classification as well as use of standard demographic statistical methods would reduce the discrepancies and simplify collaboration and comparisons between studies.
Recent meta-analyses covering international populations have pointed to glutamate-related genes such as the NRG1 (Li et al. 2006) and DTNBP1 (our unpublished results) as promising candidates for schizophrenia. It may be that these genes confer susceptibility to schizophrenia in interaction with one another in either an epistatic or a polygenic manner. Further investigations are required to identify if other at-risk polymorphisms within G72/G30 and DAAO confer a risk of schizophrenia and to clarify the role of the G72/G30 genes.
Acknowledgments
We acknowledge the anonymous reviewers for their insightful comments and suggestions for our manuscript. This work was supported by grants from the Ministry of Education, People's Republic of China, the national 973 and 863 programs, the National Natural Science Foundation of China, the Shanghai Municipal Commission for Science and Technology, and research grant MH44292 from the National Institutes of Health, Bethesda, Maryland.
References
- Addington, A. M., M. Gornick, A. L. Sporn, N. Gogtay, D. Greenstein et al., 2004. Polymorphisms in the 13q33.2 gene G72/G30 are associated with childhood-onset schizophrenia and psychosis not otherwise specified. Biol. Psychiatry 55: 976–980. [DOI] [PubMed] [Google Scholar]
- Blouin, J. L., B. A. Dombroski, S. K. Nath, V. K. Lasseter, P. S. Wolyniec et al., 1998. Schizophrenia susceptibility loci on chromosomes 13q32 and 8p21. Nat. Genet. 20: 70–73. [DOI] [PubMed] [Google Scholar]
- Brzustowicz, L. M., W. G. Honer, E. W. Chow, D. Little, J. Hogan et al., 1999. Linkage of familial schizophrenia to chromosome 13q32. Am. J. Hum. Genet. 65: 1096–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon, T. D., J. Kaprio, J. Lonnqvist, M. Huttunen and M. Koskenvuo, 1998. The genetic epidemiology of schizophrenia in a Finnish twin cohort. A population-based modeling study. Arch. Gen. Psychiatry 55: 67–74. [DOI] [PubMed] [Google Scholar]
- Chen, Y. S., N. Akula, S. D. Detera-Wadleigh, T. G. Schulze, J. Thomas et al., 2004. Findings in an independent sample support an association between bipolar affective disorder and the G72/G30 locus on chromosome 13q33. Mol. Psychiatry 9: 87–92; image 85. [DOI] [PubMed] [Google Scholar]
- Cho, H. J., I. Meira-Lima, Q. Cordeiro, L. Michelon, P. Sham et al., 2005. Population-based and family-based studies on the serotonin transporter gene polymorphisms and bipolar disorder: a systematic review and meta-analysis. Mol. Psychiatry 10: 771–781. [DOI] [PubMed] [Google Scholar]
- Chotai, J., A. Serretti, E. Lattuada, C. Lorenzi and R. Lilli, 2003. Gene-environment interaction in psychiatric disorders as indicated by season of birth variations in tryptophan hydroxylase (TPH), serotonin transporter (5-HTTLPR) and dopamine receptor (DRD4) gene polymorphisms. Psychiatry Res. 119: 99–111. [DOI] [PubMed] [Google Scholar]
- Chumakov, I., M. Blumenfeld, O. Guerassimenko, L. Cavarec, M. Palicio et al., 2002. Genetic and physiological data implicating the new human gene G72 and the gene for d-amino acid oxidase in schizophrenia. Proc. Natl. Acad. Sci. USA 99: 13675–13680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Detera-Wadleigh, S. D., and F. J. McMahon, 2006. G72/G30 in schizophrenia and bipolar disorder: review and meta-analysis. Biol. Psychiatry 60: 106–114. [DOI] [PubMed] [Google Scholar]
- Egger, M., G. Davey Smith, M. Schneider and C. Minder, 1997. Bias in meta-analysis detected by a simple, graphical test. BMJ 315: 629–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall, D., J. A. Gogos and M. Karayiorgou, 2004. The contribution of three strong candidate schizophrenia susceptibility genes in demographically distinct populations. Genes Brain Behav. 3: 240–248. [DOI] [PubMed] [Google Scholar]
- Harrison, P. J., and M. J. Owen, 2003. Genes for schizophrenia? Recent findings and their pathophysiological implications. Lancet 361: 417–419. [DOI] [PubMed] [Google Scholar]
- Hattori, E., C. Liu, J. A. Badner, T. I. Bonner, S. L. Christian et al., 2003. Polymorphisms at the G72/G30 gene locus, on 13q33, are associated with bipolar disorder in two independent pedigree series. Am. J. Hum. Genet. 72: 1131–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jablensky, A., 2000. Epidemiology of schizophrenia: the global burden of disease and disability. Eur. Arch. Psychiatry Clin. Neurosci. 250: 274–285. [DOI] [PubMed] [Google Scholar]
- Korostishevsky, M., M. Kaganovich, A. Cholostoy, M. Ashkenazi, Y. Ratner et al., 2004. Is the G72/G30 locus associated with schizophrenia? Single nucleotide polymorphisms, haplotypes, and gene expression analysis. Biol. Psychiatry 56: 169–176. [DOI] [PubMed] [Google Scholar]
- Levinson, D. F., P. Holmans, R. E. Straub, M. J. Owen, D. B. Wildenauer et al., 2000. Multicenter linkage study of schizophrenia candidate regions on chromosomes 5q, 6q, 10p, and 13q: schizophrenia linkage collaborative group III. Am. J. Hum. Genet. 67: 652–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, D., and L. He, 2006. Further clarification of the contribution of the tryptophan hydroxylase (TPH) gene to suicidal behavior using systematic allelic and genotypic meta-analyses. Hum. Genet. 119: 233–240. [DOI] [PubMed] [Google Scholar]
- Li, D., D. A. Collier and L. He, 2006. Meta-analysis shows strong positive association of the neuregulin 1 (NRG1) gene with schizophrenia. Hum. Mol. Genet. 15: 1995–2002. [DOI] [PubMed] [Google Scholar]
- Lin, M. W., P. Sham, H. G. Hwu, D. Collier, R. Murray et al., 1997. Suggestive evidence for linkage of schizophrenia to markers on chromosome 13 in Caucasian but not Oriental populations. Hum. Genet. 99: 417–420. [DOI] [PubMed] [Google Scholar]
- Liu, X., G. He, X. Wang, Q. Chen, X. Qian et al., 2004. Association of DAAO with schizophrenia in the Chinese population. Neurosci. Lett. 369: 228–233. [DOI] [PubMed] [Google Scholar]
- Mothet, J. P., A. T. Parent, H. Wolosker, R. O. Brady, Jr., D. J. Linden et al., 2000. D-serine is an endogenous ligand for the glycine site of the N-methyl-d-aspartate receptor. Proc. Natl. Acad. Sci. USA 97: 4926–4931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulle, J. G., K. V. Chowdari, V. Nimgaonkar and A. Chakravarti, 2005. No evidence for association to the G72/G30 locus in an independent sample of schizophrenia families. Mol. Psychiatry 10: 431–433. [DOI] [PubMed] [Google Scholar]
- O'Donovan, M. C., N. M. Williams and M. J. Owen, 2003. Recent advances in the genetics of schizophrenia. Hum. Mol. Genet. 12 (Spec. no. 2): R125–R133. [DOI] [PubMed] [Google Scholar]
- Owen, M. J., N. M. Williams and M. C. O'Donovan, 2004. The molecular genetics of schizophrenia: new findings promise new insights. Mol. Psychiatry 9: 14–27. [DOI] [PubMed] [Google Scholar]
- Rapoport, J. L., A. M. Addington, S. Frangou and M. R. Psych, 2005. The neurodevelopmental model of schizophrenia: update 2005. Mol. Psychiatry 10: 434–449. [DOI] [PubMed] [Google Scholar]
- Schumacher, J., R. A. Jamra, J. Freudenberg, T. Becker, S. Ohlraun et al., 2004. Examination of G72 and d-amino-acid oxidase as genetic risk factors for schizophrenia and bipolar affective disorder. Mol. Psychiatry 9: 203–207. [DOI] [PubMed] [Google Scholar]
- Shaw, S. H., M. Kelly, A. B. Smith, G. Shields, P. J. Hopkins et al., 1998. A genome-wide search for schizophrenia susceptibility genes. Am. J. Med. Genet. 81: 364–376. [DOI] [PubMed] [Google Scholar]
- Shirts, B. H., and V. Nimgaonkar, 2004. The genes for schizophrenia: Finally a breakthrough? Curr. Psychiatry Rep. 6: 303–312. [DOI] [PubMed] [Google Scholar]
- Tsai, G., and J. T. Coyle, 2002. Glutamatergic mechanisms in schizophrenia. Annu. Rev. Pharmacol. Toxicol. 42: 165–179. [DOI] [PubMed] [Google Scholar]
- Wang, X., G. He, N. Gu, J. Yang, J. Tang et al., 2004. Association of G72/G30 with schizophrenia in the Chinese population. Biochem. Biophys. Res. Commun. 319: 1281–1286. [DOI] [PubMed] [Google Scholar]
- Zou, F., C. Li, S. Duan, Y. Zheng, N. Gu et al., 2005. A family-based study of the association between the G72/G30 genes and schizophrenia in the Chinese population. Schizophr. Res. 73: 257–261. [DOI] [PubMed] [Google Scholar]
- DerSimonian, R., and N. Laird, 1986. Meta-analysis in clinical trials. Control Clin. Trials. 7: 177–188. [DOI] [PubMed] [Google Scholar]