Abstract
L5Jcs1 is a perinatal lethal mutation uncovered in a screen for ENU-induced mutations on mouse chromosome 5. L5Jcs1 homozygotes exhibit posterior-to-anterior transformations of the vertebral column midsection, similar to mice deficient for Hoxc8 and Hoxc9. Positional cloning efforts identified a mutation in a novel, evolutionarily conserved, and ubiquitously expressed gene dubbed Tapt1 (Transmembrane anterior posterior transformation 1). TAPT1 is predicted to contain several transmembrane domains, and part of the gene is orthologous to an unusual alternatively spliced human transcript encoding the cytomegalovirus gH receptor. We speculate that TAPT1 is a downstream effector of HOXC8 that may act by transducing or transmitting extracellular information required for axial skeletal patterning during development.
THE axial skeleton consists of the skull, vertebral column, and thoracic rib cage. The vertebral column contains five types of vertebrae—cervical, thoracic, lumbar, sacral, and caudal—defined by anatomical features and position within the anterior–posterior axis. Variation within the vertebral column plays a significant role in morphological distinctions among species.
In vertebrate embryos, the somites are derived from the paraxial mesoderm and in turn give rise to the sclerotome and the dermomyotome. Chick/quail chimera experiments showed that the sclerotome gives rise to the axial skeleton (Huang et al. 2000). Grafting experiments in the chicken demonstrated that posterior portions of the paraxial mesoderm, when transplanted anteriorly, gave rise to posterior structures equivalent to those expected at the original site. Therefore, specification of the vertebral column occurs early in development, prior to the compaction of the paraxial mesoderm into somites (Kieny et al. 1972). Key factors known to play a role in this specification are the Hox genes. These encode transcription factors that can repress or activate downstream genes. A number of direct targets have been identified, particularly in Drosophila melanogaster, but it has been difficult to unequivocally identify mammalian target genes whose expression is directly controlled by Hox genes in vivo (Akin and Nazarali 2005; Pearson et al. 2005). Chromatin immunoprecipitation and microarray experiments are enabling progress in this area, although potential targets identified require functional validation. Similarly, the mechanisms of Hox gene regulation in mammals are quite complicated, prompting the use of elaborate chromosomal engineering strategies to understand the colinearity of expression within Hox clusters (Tarchini and Duboule 2006). Most of the individual regulators of Hox gene expression remain to be found.
Forward genetic screens are a powerful way to elucidate biological pathways, having the ability to uncover gene–phenotype correlations and developmental hierarchies. Recently, a region-specific ENU mutagenesis screen directed at proximal mouse chromosome 5 yielded 37 embryonic lethal mutations (Wilson et al. 2005). Here, we describe the characterization and positional cloning of one of these ENU-induced perinatal lethal mutations (L5Jcs1) that causes a homeotic transformation of the axial skeleton similar to that in Hoxc8 knockout mice (Le Mouellic et al. 1992). This novel, ubiquitously expressed gene, which otherwise would not be suspected to yield such a phenotype, uncovers a new aspect of vertebrate axial skeleton patterning.
MATERIALS AND METHODS
Phenotype analysis:
The L5Jcs1 mutation was maintained in trans to the Rump white (Rw) inversion (for details see Wilson et al. 2005). Homozygous embryos or neonates were obtained from intercrossing Rw/L5Jcs1 mice. Skeletal preparations were based on a published protocol (Kessel and Gruss 1991). Animals were skinned and eviscerated prior to being soaked in 100% ethanol for 4 days. They were then soaked in acetone for 3 days. Animals were rinsed in water and then stained (2 vol 0.14% alcian blue (Sigma, St. Louis) in 70% ethanol, 1 vol 0.12% alizarin red (Sigma) in 95% ethanol, 8 vol 100% glacial acetic acid, and 50 vol 70% ethanol) for 10 days. Stain was removed and replaced with 20% glycerol and 1% KOH and incubated at 37° for 16 hr and then at room temperature until the tissue cleared. Preparations were stored in 2:1:1 100% ethanol:glycerol:benzyl alcohol prior to photography.
Mapping L5Jcs1:
For recombination mapping, Rw/L5Jcs1 mice were crossed to CAST/Ei, and non-Rw F1 progeny (L5Jcs1/CAST) were intercrossed to produce F2's. These were genotyped with the following polymorphic markers: D5Mit353, D5Mit129, D5Mit78, D5Mit105, D5Mit231, D5Mit106, and D5Mit107. The sequences of primer pairs that we developed for mapping deletion breakpoints are as follows: D5Jcs608, TAAGTCTGGAGGTGGGGATG and GGCTGTCTTCCTCTCTGTGG; D5Jcs614, GCATGAACCTGCACACAGAT and ATGCTGGGAGCAGAACAGTC.
Identification of the Tapt1 mutation and RT–PCR:
RNA was prepared from whole E12.5 embryos using a QIAGEN (Chatsworth, CA) RNeasy kit. Primer pairs used for RT–PCR of the 5′-end of Tapt1 (Transmembrane anterior posterior transformation 1) were Tapt1.1F (CAATGTAGCTTTTAACTCTCACAACA) and Tapt1.1R (TGGCTTCCTTCACGTATTGA). These same primers were used to evaluate tissue-specific expression, using a normalized cDNA panel (BD Biosciences Clontech). Genomic DNA was amplified using the primers Tapt1.G2F (TGACAAAGATGGTATTTCTGTTGC) and Tapt1.G2R (TCAACATTTGCTTATGGTAATGAG) to sequence the mutation.
In silico analyses:
Multiple sequence alignments were carried out using ClustalW, and the output was formatted using Multiple Align Show (http://bioinformatics.org/sms/multi_align.html). Programs used to detect potential nuclear localization were PredictNLS (http://cubic.bioc.columbia.edu/predictNLS/) and NucPred (http://www.sbc.su.se/∼maccallr/nucpred/). Predicted transmembrane domains were identified by the programs TMPRED (http://www.ch.embnet.org/software/TMPRED_form.html) and TMHMM (http://www.cbs.dtu.dk/services/TMHMM/).
RNA in situ hybridization:
Embryos were rinsed in 1× phosphate buffered saline (PBS) plus Tween 20 before fixation overnight in 4% paraformaldehyde (PFA). They were then dehydrated by 25, 50, and 75% methanol in PBS before being stored in 100% methanol at −20°. A Tapt1 riboprobe vector was generated by amplifying a 608-bp RT–PCR product corresponding to nucleotides 1760–2368 of the cDNA, using the following primers that contained SP6 and T7 primer sites, respectively: H3RS7F, GCGATTTAGGTGACACTATAGAATACTGGATGCTGACACTTGAACA; H3RS7R GCGTAATACGACTCACTATAGGGAGACCTTTAACTTGGAATAACCAATG. The amplimer was in vitro transcribed, incorporating digoxygenin-dUTP.
LacZ staining in Hoxc8 knockin mice:
E12.5 embryos were harvested, rinsed in 1× PBS, and fixed in 4% PFA in PBS at 4° for 5 min. Embryos were stained in X-Gal overnight at 37° and then postfixed in 4% PFA in PBS at 4° overnight. Embryos were placed in 100% glycerol for photography and storage.
RESULTS
The L5Jcs1 mutation causes homeotic-like transformations of the axial skeleton and perinatal lethality:
L5Jcs1 is one of 37 lethal mutations recovered in a region-specific ENU mutagenesis screen directed at the Rw inversion region of proximal mouse chromosome 5 (Wilson et al. 2005). The majority of animals homozygous for L5Jcs1 survived to approximately embryonic day 18.5 (E18.5), but either were born dead or survived until ∼1 day postpartum. Newborns or C-section-derived E18.5 homozygous mutants were observed to breath, turn pink, and move actively. Rare homozygotes survived up to several days postpartum, but they were smaller than littermates and never survived to weaning age. The cause of lethality remains unknown (but see discussion).
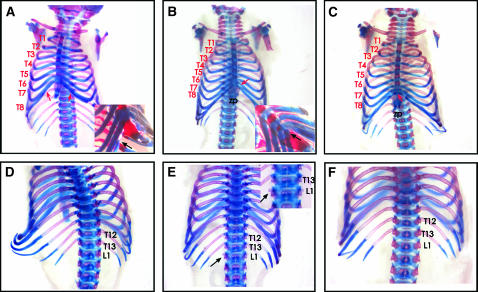
As first reported anecdotally (Wilson et al. 2005), skeletons from L5Jcs1/L5Jcs1 mice exhibited certain posterior-to-anterior transformations (Figure 1; Table 1). Specifically, the axial skeleton showed posterior-to-anterior transformations of the thoracic and lumbar vertebrae. Unlike normal mice in which the eighth rib (thoracic segment 8–T8) terminus is free floating, it typically becomes attached to the sixth sternebra in L5Jcs1 homozygotes. In nearly half the animals, the first lumbar vertebra (L1) showed the development of a 14th pair of ribs. Judging from rib length, it appears that the 9th–13th thoracic vertebrae (T9–13) are transformed into more anterior structures. In addition, the xiphoid process was overgrown and splayed in most mutants (Figure 1, B and C; supplemental Figure 1, B and C, at http://www.genetics.org/supplemental/; Table 1).
Figure 1.—
Homeotic-like transformations of L5Jcs1 skeletons. Shown are E18.5 skeletons stained with alcian blue and alizarin red. The top and bottom rows are ventral and dorsal views, respectively. The lumbar (“L”) and thoracic (“T”) vertebrae are numbered. (A and D) Wild type. The inset in A is a ventrilateral closeup of T6 and T7 attachment to the sternum; the black arrow points to the terminus of T8, which is unattached to the sternum. (B and E) Homozygous mutant. The inset in B is a ventrilateral closeup of T8 attachment to the sternum (black arrow). The inset in E shows a close-up of the dorsal vertebral column exhibiting partial transformation of L1 into a thoracic-like identity (arrows). (C and F) Homozygous mutant showing the fusion of T8 to the sternum (red arrow in C), but not the L1 partial transformation. Note the splayed xiphoid processes (zp) in B and C.
TABLE 1.
Penetrance of skeletal abnormalities
| Genotype | T8 > T7 | L1 > T14 | Splayed XP |
|---|---|---|---|
| L1/L1 | 10/13 | 6a/13 | 11/13 |
| Rw/+ or L1/+ | 1/30b | 1/30b | 5/30 |
| Fisher's exact test | P < 0.0001 | P = 0.00165 | P < 0.0001 |
XP, xiphoid process. L1, L5Jcs1. All animals were E18.5–1 day postpartum.
Five of the six had bilateral extra ribs.
The affected animal was exencephalic, and the L1 > T14 transformation was unilateral. This animal also had a splayed XP.
Positional cloning of L5Jcs1:
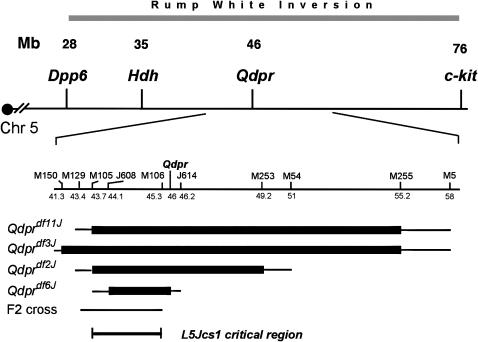
As previously reported, L5Jcs1 was never observed to recombine in trans to the Rw inversion, which spans the ∼26- to 74-Mb interval on chromosome 5. The chromosomal deletion Qdprdf3J failed to complement L5Jcs1 (Wilson et al. 2005). Specifically, no L5Jcs1/Qdprdf3J weanlings were observed of 23 produced by Rw/L5Jcs1 × Qdprdf3J/+ intercrosses, thus localizing the mutation to this ∼15- to 17-Mb interval (Figure 2). To refine the critical region, complementation tests were performed with three other deletions in the Qdpr complex: Qdprdf2J, Qdprdf6J, and Qdprdf11J (Figure 2). None complemented L5Jcs1; of the 22, 11, and 31 non-Rw progeny, respectively, examined from crosses of each deletion to L5Jcs1/Rw, none contained the deletion. This localized the mutation to the 2.5-Mb region between D5Mit105 and D5Jcs614.
Figure 2.—
Map of the L5Jcs1 region. (Top) Map of the Rw inversion region of mouse chromosome 5. Some gene loci are indicated. (Bottom) The region surrounding the Qdpr deletion complex is expanded. The extents of deletions are indicated as thick horizontal bars. The thinner bars at the ends of the thicker bars indicate the intervals in which the breakpoints lie. The region to which L5Jcs1 was mapped in the F2 cross is indicated. Microsatellite loci are abbreviated with the prefix “M” instead of D5Mit. Numbers under these markers are megabase coordinates according to the February 2006 mouse genome assembly Build 36. The L5Jcs1 critical region is indicated.
To confirm that the failure to obtain L5Jcs1/deletion animals was due to noncomplementation rather than to synthetic lethality, timed matings were conducted between Qdprdf2J/+ and Rw/L5Jcs1 mice, and the skeletal phenotypes of three Qdprdf2J/L5Jcs1 E18.5 pups were examined. All three showed the same thoracic rib transformation observed in L5Jcs1 homozygotes, not only demonstrating that L5Jcs1 lies within the Qdprdf2J deletion, but also suggesting that it is a null mutation (supplemental Figure 1 at http://www.genetics.org/supplemental/).
To further narrow the interval, recombination mapping was conducted by intercrossing L5Jcs1/Cast F1 heterozygotes and genotyping unaffected F2 progeny. Regions that were homozygous for the B6 markers could not contain L5Jcs1. These data, combined with data from deletion mapping, localized the mutation to a 1.6-Mb region (between D5Mit105 and D5Mit106) containing 10 Refseq genes (Table 2). Of these, we eliminated several as L5Jcs1 candidates on the basis of their known mutant phenotypes. Two (Cd38 and Ldb2) have been knocked out in mice, and homozygotes for both are viable. Mutations in Prom1 cause retinal degeneration in humans, while RNAi inhibition of Ppih and 5730509K17Rik in Caenorhabditis elegans gives wild-type viable phenotypes.
TABLE 2.
Genes in the L5Jcs1 critical region
| Gene | Exons | KO | Sequenced |
|---|---|---|---|
| C1qtnf7 (C1q and TNF-related protein 7) | 6 | Yes | |
| 5730509K17Rik | 40 | ||
| Fbxl5 (F-box and leucine-rich-repeat protein 5) | 9 | Yes | |
| Bst1 (bone marrow stromal antigen 1) | 9 | Yesc | |
| Cd38 | 8 | Yesa | |
| Ppih (peptidyl prolyl isomerase H) | 1 | ||
| Fgfbp1 (fibroblast growth factor binding protein 1) | 2 | Yes | |
| Prom1 (prominin 1) | 27 | ||
| 4932414K18Rik (Tapt1) | 14 | Yes | |
| Ldb2 (LIM homeodomain binding 2) | 9 | Yesb |
C1qtnf7 is subject to alternative splicing; all isoforms were sequenced. Primers used for sequencing are in supplemental Table 1 at http://www.genetics.org/supplemental/. KO, knockout allele available.
Mutants are viable.
A Lexicon viable mutant.
All coding regions but exon 5 were sequenced.
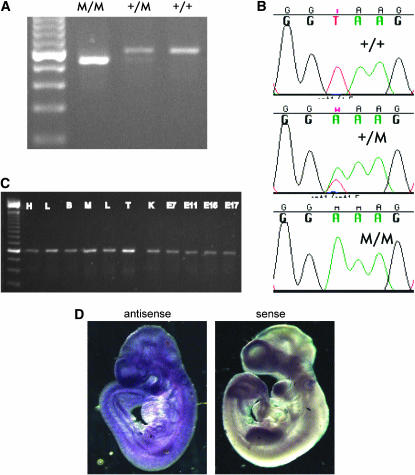
To identify possible mutations in the remaining six genes, we began generating RT–PCR products from L5Jcs1/L5Jcs1, L5Jcs1/+, and +/+ E12.5 embryos. No mutations were found in C1qtnf7, Fbxl5, and Fgfbp1 (Table 2). In the case of the novel 14-exon mouse gene 4932414K18Rik, a size difference was observed between L5Jcs1/L5Jcs1 and wild-type-derived products when amplifying between exons 6 and 9. Two bands were observed in heterozygotes (Figure 3A). Sequencing the products revealed that the mutant amplimers lacked exon 7. Analysis of exon 7 and flanking genomic sequence in the mutant revealed a T-to-A transition in the 5′ splice site of intron 7 (3′ of exon 7); this is likely responsible for the skipping of exon 7 during splicing (Figure 3B and Figure 4). This causes an altered reading frame (exon 7 is 70 nucleotides long), resulting in the introduction of 12 anomalous amino acids after N279, followed by a premature termination that probably causes a severe disruption of function. However, the mutant message may also be subject to nonsense-mediated decay; the mutant transcript appears to be present at much lower levels compared to the wild-type transcript in heterozygotes (Figure 3A).
Figure 3.—
Identification of mutation in Tapt1 and RT–PCR analysis. (A) RT–PCR analysis of the Tapt1 gene using primers flanking exon 7. M, mutant (L5Jcs1 allele). Note the fainter mutant (lower) band in the heterozygote. The leftmost lane is a 100-bp ladder. (B) DNA sequence traces of indicated genotypes of the distal exon 7 splice junction. (C) PCR analysis of cDNAs from various tissues. (D) In situ hubridization of antisense or sense (control) RNAs to E9.5 wild-type embyros. H, heart; L, lung; B, brain; M, skeletal muscle; L, liver; T, testis; K, kidney; E, embryonic day.
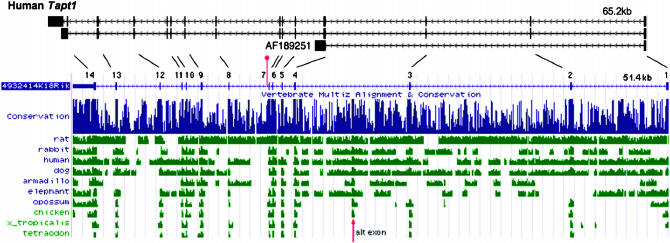
Figure 4.—
Structure of the mouse and human Tapt1 genes. The genomic structures of mouse Tapt1 (4932414K18Rik) and human Tapt1 (FLJ90013), as represented in the University of California at Santa Cruz (UCSC) Genome Browser as of April 2006 are shown, as are three human mRNAs from GenBank. The AF189251 mRNA was reported to encode the HCMV gH receptor (Baldwin et al. 2000). Exons are represented by vertical bars along the horizontal lines and are numbered. Transcription is to the left, toward the centromere in both species. The location of the L5Jcs1 mutation, distal to exon 7, is indicated by a vertical red bar/ball. The lines drawn between the human and mouse genes connect orthologous exons. (Bottom) A depiction of conservation between the mouse sequence and other vertebrates, as displayed by the indicated UCSC browser track. The red arrow indicates the location of a highly conserved region that corresponds to an alternative exon included in some ESTs (not shown).
The presence of this mutation in the small, genetically defined mutant interval strongly implicates 4932414K18Rik as L5Jcs1. It is unlikely that another locus in the critical region is the causative gene; Keays et al. (2006) demonstrated that the probability of a cosegregating ENU-induced mutation in a 5-Mb interval (the Tapt1 interval is only 1.6 Mb) is exceedingly small (P < 0.002).
Tapt1, the gene mutated in L5Jcs1, encodes a highly conserved putative transmembrane protein that overlaps the human cytomegalovirus gH receptor:
4932414K18Rik encodes a 565-amino-acid protein annotated as LOC231225 in GenBank. Due to the likelihood that mutation of this gene underlies the L5Jcs1 phenotype, we have renamed it Tapt1 (Transmembrane anterior posterior transformation 1), thus making the allele designation Tapt1L5Jcs1.
The predicted TAPT1 protein is evolutionarily conserved. Highly similar orthologs exist from vertebrates to yeast (Figure 4; Figure 5). The overall genomic structure and sequence of Tapt1 and its vertebrate orthologs are conserved (Figure 4). A variety of ESTs led to the derivation of a consensus RefSeq structure for mouse Tapt1, with the exception of one alternatively spliced 66-bp exon that is present in some ESTs and that corresponds to conserved vertebrate sequences in homology plots (Figure 4).
Figure 5.—
Alignment of TAPT1 orthologs. Identical amino acids are on a black background only in those cases where four or more residues have identity or similarity. Similar residues are shaded. The amino and carboxy terminal ends of the proteins, where homology decays markedly, are not aligned here. Some landmark amino acid positions in mice are indicated. Dashes are introduced to maximize alignment by the ClustalW algorithm. A bracket above the sequence corresponds to those amino acids encoded by mouse exon 7, which is skipped in the L5Jcs1 allele. Mm, mouse; Hs, human; Gg, chicken; Dr, zebrafish; Dm, D. melanogaster; Sc, Saccharomyces cerevisiae.
The intron/exon structure of the human ortholog aligns well with mouse, with the exception of AF189251, a cDNA corresponding to exon 1, part of exon 4, and a portion of intron 4, in which it terminates. This cDNA was reported to encode the human cytomegalovirus (HCMV) gH receptor (Baldwin et al. 2000). A BLAST search of the mouse TAPT1 amino acid sequence against human proteins revealed a high similarity to that part of the AF189251 protein encoded by exons 1 and 4. However, our analysis with the now-available human genome sequence shows that much of the predicted AF189251 protein corresponds to that encoded by the intron 4 sequence. Interestingly, this intron-4-encoded amino acid sequence includes the receptor for the HCMV gH glycoprotein ligand and is essential for virus infectivity (Baldwin et al. 2000). The ability of anti-AF189251 monoclonal antibodies to block viral infection provides experimental support for the prediction that the intron-4-encoded epitopes of the protein are present at the cell surface (Baldwin et al. 2000), but it is not clear which amino acids are responsible for placing the protein at the cell surface. Interestingly, the AF189251 cDNA is unique; no mouse or human ESTs exhibit a similarly spliced form. Furthermore, VISTA analysis (Mayor et al. 2000) indicates that the intron-4-coding region is not evolutionarily conserved (not shown).
Other than this unusual AF189251 transcript, no other information on the cellular function of mammalian TAPT1 exists. Analysis with the TMPRED and TMHMM protein analysis algorithms indicates that TAPT1 contains as many as six transmembrane domains (supplemental Figure 2 at http://www.genetics.org/supplemental/). Alignment of the predicted amino acid sequence reveals high levels of conservation from mammals to yeast, especially near its central region (Figure 5). The L5Jcs1 mutation lies within this highly conserved region, near the end of a 133-amino-acid stretch that (with one residue exception) is perfectly identical among humans, mice, chickens, and zebrafish.
Yeast deleted for their Tapt1 ortholog (an uncharacterized ORF called YER140W) are viable, although C. elegans RNA interference (RNAi) screens (summarized in WormBase) report that one of five RNAi molecules directed against the worm ortholog cause 24–100% embryonic lethality. The YER140W gene product was found to interact, in two-hybrid screens, with Mch1p, Dhh1p, and Tma19p (Ito et al. 2001). Mch1p has similarity to mammalian monocarboxylate permeases, which are involved in transport of monocarboxylic acids across plasma membranes. However, mch1 mutants are not deficient in monocarboxylic acid transport (Makuc et al. 2001). Dhh1p is a putative DexD/H-box helicase, involved in mRNA export and translation. TMA19 is a nonessential gene encoding a putative ribosomal protein (according to the Saccharomyces Genome Database). In sum, while there have been no directed studies of Tapt1 or its homologs in experimental organisms, evidence suggests that it is a transmembrane protein involved in transporting molecules across membranes and may be required for normal embryonic development in both worms and mice.
Tapt1 expression:
Despite the specific skeletal defect exhibited by mutant embryos, Tapt1 is widely expressed. Public databases contain 223 Tapt1 ESTs stemming from 30 different cDNA libraries, according to Unigene and the Stanford S.O.U.R.C.E. (Diehn et al. 2003). We conducted RT–PCR analysis to confirm the apparently ubiquitous pattern of Tapt1 expression in both adult tissues and whole E7–E17 embryos (Figure 3C). All samples yielded similar amounts of Tapt1 product from normalized cDNA sources (see materials and methods). To determine if Tapt1 is expressed in distinct embryonic structures that might correspond to the observed skeletal defects, whole-mount RNA in situ hybridization was performed on E9.5 embryos. Tapt1 message, as detected by two nonoverlapping probes (Figure 3D; data not shown), was present throughout the embryo.
Hox gene expression in L5Jcs1 mutants:
This skeletal phenotype is remarkably similar to mice with a knockout of Hoxc8, and to a somewhat lesser degree, Hoxc9 (Le Mouellic et al. 1992; Suemori et al. 1995). Mutations in genes that cause axial skeletal patterning defects typically disrupt the expression pattern of a Hox gene(s) that, when mutated, causes a similar phenotype (see discussion). We therefore explored whether mutation of Tapt1 disrupts Hoxc8 expression, exploiting an allele of Hoxc8 containing a knocked-in LacZ reporter that is expressed in a pattern that reflects endogenous Hoxc8 transcription (Le Mouellic et al. 1992). Double heterozygotes (L5Jcs1/+; Hoxc8−/+) were crossed to L5Jcs1 heterozygotes, and the embryos were stained for LacZ and genotyped. There was no alteration in LacZ expression in embryos homozygous for the L5Jcs1 mutation and that contained the Hoxc8 allele (Figure 6).
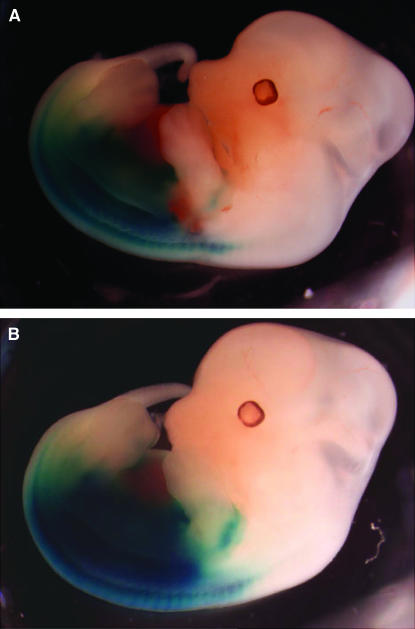
Figure 6.—
Hoxc8 expression in L5Jcs1 mutant and control embryos. Shown are whole-mount E12.5 embryos stained for LacZ activity. The LacZ gene is under the control of the Hoxc8 promoter. (A) L5Jcs1/+; Hoxc8+/−. (B) L5Jcs1/L5Jcs1; Hoxc8+/−.
DISCUSSION
Function of TAPT1 in development:
All or most known mouse mutations that cause homeotic skeletal transformations either are in the Hox genes themselves or affect Hox gene expression. The latter include polycomb and trithorax genes (Hanson et al. 1999), the E2f6 transcription factor (Storre et al. 2002), the homeobox gene Cdx1 (Subramanian et al. 1995), the TGF-β family member Bmp11 (McPherron et al. 1999), Plzf (Barna et al. 2000), and others (van der Lugt et al. 1996; Oh and Li 1997; Collins et al. 2002). The TAPT1 protein sequence itself does not contain any apparent nuclear localization signals or DNA-binding motifs, suggesting that it does not directly modulate gene expression. Indeed, we found that Hoxc8 expression appeared unaffected in L5Jcs1 mutants, although it remains possible that TAPT1 affects the expression of other Hox genes. Thus, it appears that TAPT1 functions at a unique level in the regulatory hierarchy of axial skeletal patterning among molecules known to be involved in the process.
The most enigmatic aspect of Tapt1 is that its mutation causes a specific developmental defect yet its expression is widespread. There are at least two models that we can envision to reconcile this paradox. One is that TAPT1 is a ubiquitously expressed receptor that enables developmentally specific functions by transducing extracellular signals that are acted upon by effectors downstream of Hox genes. Specificity of action at particular times and places during development would be conferred by the ligands. The idea that TAPT1 is a cell surface receptor is consistent with the presence of several transmembrane domains and with the fact that an alternatively spliced human ortholog encodes the HCMV gH cell surface receptor (with many caveats related to the “normal” TAPT1 as discussed below).
The second model is that Tapt1 is not truly ubiquitously expressed during development, but rather its expression is modulated in certain cell types at particular times that are important in patterning the skeleton. The Hox family of transcription factors is the most well-studied group of genes affecting segmental identity of the axial skeleton (reviewed by Favier and Dolle 1997). In mouse, the Hox genes are expressed in partially overlapping regions including mesoderm, neuroectoderm, and endoderm from the base of the head to the tail. As indicated earlier, disruption of both Hoxc8 and Hoxc9 leads to posterior-to-anterior transformations of the thoracic and lumbar vertebrae similarly to L5Jcs1, that is, attachment of T8 to the sternum, formation of a partial 14th pair of ribs on the first lumbar segment, and a malformed xiphoid process (Le Mouellic et al. 1992; Suemori et al. 1995). In addition to other axial skeleton abnormalities, Hoxc4 mutant mice often display the T8 sternum attachment and xiphoid process malformations similarly to L5Jcs1 homozygotes (Saegusa et al. 1996). The L5Jcs1 phenocopy raises the possibility that one or more Hox genes regulate the pattern of Tapt1 in development and that Tapt1, possibly acting at the cell surface, affects specification of developing axial skeletal components.
Progress in identifying downstream targets of HOX proteins in mice has been limited. Notably, Lei et al. (2005) identified potential targets of Hoxc8 regulation by overexpressing Hoxc8 in mouse embryonic fibroblasts and then performing microarray analysis. They identified 34 genes that were up-or downregulated, but none were Tapt1.
Other roles in homeostasis and development:
A major unanswered question about the biological role(s) of Tapt1 concerns the cause of lethality. It is unlikely to be due to the skeletal defects per se, since mice with similar transformations (such as Hoxc8−/−) can be viable and fertile. Because the gene is expressed ubiquitously, defects in other tissues are probably responsible. In an attempt to gain insight into the cause of death, we completely sectioned the head and thoracic region of a single mutant sickly animal that survived 3 days postpartum. Abnormalities noted were an infarction in brain, nasal congestion, pneumonia, and a potential swelling of heart vessels that might underlie the other phenotypes. Additional work will reveal if these defects are direct manifestations of the L5Jcs1 mutation.
On the role of TAPT1 as a HCMV receptor:
A very interesting finding is that an unusual isoform of the human TAPT1 gene encodes the HCMV gH receptor. The identity of this receptor was uncovered by screening of cDNA expression libraries with monoclonal antibodies against the gH receptor (Baldwin et al. 1996, 2000). The DNA sequences of these clones were determined before the human genome sequence and EST collections were available. As we describe in the results, the clones identified by Baldwin and colleagues represent an alternatively spliced form of TAPT1 that is not found in any EST sequenced to date. Given that only three clones containing this gH receptor isoform were identified from two human embryonic lung cell cDNA libraries, it is likely that this is an extremely rare isoform and/or is restricted to a small number of tissues.
Our observation that much of the HCMV coding region lies in what is an intron with respect to the “normal” transcription unit, and that this region is apparently not conserved, raises interesting questions about the mechanisms by which receptors for pathogens can arise. One possibility is that this alternative coding region predated the mammalian radiation, serving some function that became important exclusively for humans, only to be exploited by CMV after the divergence of species. Alternatively, it may have arisen in the primate lineage and fortuitously (for CMV) encoded a protein variant (essential or not) that CMV exploited.
During the course of scrutinizing the amino acid sequence of TAPT1 for similarity to known conserved motifs using Pfam, we noted that unfortunately this database lists the central half of the sequence as being a member of the “DUF747, eukaryotic membrane protein (cytomegalovirus gH-receptor) family.” However, most of this DUF747 region consists of amino acids that are not encoded by the gH receptor cDNA isoform, but rather correspond to the highly conserved region of mouse TAPT1 stretching from residues 148 to 459. This annotation is therefore potentially very misleading to those studying any of the TAPT1 orthologs. There are no apparent paralogs in the mouse genome.
Acknowledgments
The authors thank Tim O'Brien for experimental advice and Rod Bronson for evaluation of pathology. This work was funded by grant HD35984 to J.C.S. from the National Institute of Child Health and Human Development.
References
- Akin, Z. N., and A. J. Nazarali, 2005. Hox genes and their candidate downstream targets in the developing central nervous system. Cell. Mol. Neurobiol. 25: 697–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin, B., M. Kleinberg and S. Keay, 1996. Molecular cloning and expression of receptor peptides that block human cytomegalovirus/cell fusion. Biochem. Biophys. Res. Commun. 219: 668–673. [DOI] [PubMed] [Google Scholar]
- Baldwin, B., C. Zhang and S. Keay, 2000. Cloning and epitope mapping of a functional partial fusion receptor for human cytomegalovirus gH. J. Gen. Virol. 81: 27–35. [DOI] [PubMed] [Google Scholar]
- Barna, M., N. Hawe, L. Niswander and P. P. Pandolfi, 2000. Plzf regulates limb and axial skeletal patterning. Nat. Genet. 25: 166–172. [DOI] [PubMed] [Google Scholar]
- Collins, E. C., A. Appert, L. Ariza-McNaughton, R. Pannell, Y. Yamada et al., 2002. Mouse Af9 is a controller of embryo patterning, like Mll, whose human homologue fuses with Af9 after chromosomal translocation in leukemia. Mol. Cell. Biol. 22: 7313–7324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diehn, M., G. Sherlock, G. Binkley, H. Jin, J. C. Matese et al., 2003. SOURCE: a unified genomic resource of functional annotations, ontologies, and gene expression data. Nucleic Acids Res. 31: 219–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favier, B., and P. Dolle, 1997. Developmental functions of mammalian Hox genes. Mol. Hum. Reprod. 3: 115–131. [DOI] [PubMed] [Google Scholar]
- Hanson, R. D., J. L. Hess, B. D. Yu, P. Ernst, M. van Lohuizen et al., 1999. Mammalian Trithorax and polycomb-group homologues are antagonistic regulators of homeotic development. Proc. Natl. Acad. Sci. USA 96: 14372–14377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, R., Q. Zhi, C. Schmidt, J. Wilting, B. Brand-Saberi et al., 2000. Sclerotomal origin of the ribs. Development 127: 527–532. [DOI] [PubMed] [Google Scholar]
- Ito, T., T. Chiba, R. Ozawa, M. Yoshida, M. Hattori et al., 2001. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl. Acad. Sci. USA 98: 4569–4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keays, D. A., T. G. Clark and J. Flint, 2006. Estimating the number of coding mutations in genotypic- and phenotypic-driven N-ethyl-N-nitrosourea (ENU) screens. Mamm. Genome 17: 230–238. [DOI] [PubMed] [Google Scholar]
- Kessel, M., and P. Gruss, 1991. Homeotic transformations of murine vertebrae and concomitant alteration of Hox codes induced by retinoic acid. Cell 67: 89–104. [DOI] [PubMed] [Google Scholar]
- Kieny, M., A. Mauger and P. Sengel, 1972. Early regionalization of somitic mesoderm as studied by the development of axial skeleton of the chick embryo. Dev. Biol. 28: 142–161. [DOI] [PubMed] [Google Scholar]
- Lei, H., H. Wang, A. H. Juan and F. H. Ruddle, 2005. The identification of Hoxc8 target genes. Proc. Natl. Acad. Sci. USA 102: 2420–2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Mouellic, H., Y. Lallemand and P. Brulet, 1992. Homeosis in the mouse induced by a null mutation in the Hox-3.1 gene. Cell 69: 251–264. [DOI] [PubMed] [Google Scholar]
- Makuc, J., S. Paiva, M. Schauen, R. Kramer, B. Andre et al., 2001. The putative monocarboxylate permeases of the yeast Saccharomyces cerevisiae do not transport monocarboxylic acids across the plasma membrane. Yeast 18: 1131–1143. [DOI] [PubMed] [Google Scholar]
- Mayor, C., M. Brudno, J. R. Schwartz, A. Poliakov, E. M. Rubin et al., 2000. VISTA: visualizing global DNA sequence alignments of arbitrary length. Bioinformatics 16: 1046–1047. [DOI] [PubMed] [Google Scholar]
- McPherron, A. C., A. M. Lawler and S. J. Lee, 1999. Regulation of anterior/posterior patterning of the axial skeleton by growth/differentiation factor 11. Nat. Genet. 22: 260–264. [DOI] [PubMed] [Google Scholar]
- Oh, S. P., and E. Li, 1997. The signaling pathway mediated by the type IIB activin receptor controls axial patterning and lateral asymmetry in the mouse. Genes Dev. 11: 1812–1826. [DOI] [PubMed] [Google Scholar]
- Pearson, J. C., D. Lemons and W. McGinnis, 2005. Modulating Hox gene functions during animal body patterning. Nat. Rev. Genet. 6: 893–904. [DOI] [PubMed] [Google Scholar]
- Saegusa, H., N. Takahashi, S. Noguchi and H. Suemori, 1996. Targeted disruption in the mouse Hoxc-4 locus results in axial skeleton homeosis and malformation of the xiphoid process. Dev. Biol. 174: 55–64. [DOI] [PubMed] [Google Scholar]
- Storre, J., H. P. Elsasser, M. Fuchs, D. Ullmann, D. M. Livingston et al., 2002. Homeotic transformations of the axial skeleton that accompany a targeted deletion of E2f6. EMBO Rep. 3: 695–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian, V., B. I. Meyer and P. Gruss, 1995. Disruption of the murine homeobox gene Cdx1 affects axial skeletal identities by altering the mesodermal expression domains of Hox genes. Cell 83: 641–653. [DOI] [PubMed] [Google Scholar]
- Suemori, H., N. Takahashi and S. Noguchi, 1995. Hoxc-9 mutant mice show anterior transformation of the vertebrae and malformation of the sternum and ribs. Mech. Dev. 51: 265–273. [DOI] [PubMed] [Google Scholar]
- Tarchini, B., and D. Duboule, 2006. Control of Hoxd genes' collinearity during early limb development. Dev. Cell 10: 93–103. [DOI] [PubMed] [Google Scholar]
- van der Lugt, N. M., M. Alkema, A. Berns and J. Deschamps, 1996. The Polycomb-group homolog Bmi-1 is a regulator of murine Hox gene expression. Mech. Dev. 58: 153–164. [DOI] [PubMed] [Google Scholar]
- Wilson, L., Y.-H. Ching, M. Farias, S. A. Hartford, G. Howell et al., 2005. Random mutagenesis of proximal mouse chromosome 5 uncovers predominantly embryonic lethal mutations. Genome Res. 15: 1095–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]