Abstract
Chromosomal rearrangements can be triggered by recombination between distinct but related regions. Brassica napus (AACC; 2n = 38) is a recent allopolyploid species whose progenitor genomes are widely replicated. In this article, we analyze the extent to which chromosomal rearrangements originate from homeologous recombination during meiosis of haploid B. napus (n = 19) by genotyping progenies of haploid × euploid B. napus with molecular markers. Our study focuses on three pairs of homeologous regions selected for their differing levels of divergence (N1/N11, N3/N13, and N9/N18). We show that a high number of chromosomal rearrangements occur during meiosis of B. napus haploid and are transmitted by first division restitution (FDR)-like unreduced gametes to their progeny; half of the progeny of Darmor-bzh haploids display duplications and/or losses in the chromosomal regions being studied. We demonstrate that half of these rearrangements are due to recombination between regions of primary homeology, which represents a 10- to 100-fold increase compared to the frequency of homeologous recombination measured in euploid lines. Some of the other rearrangements certainly result from recombination between paralogous regions because we observed an average of one to two autosyndetic A–A and/or C–C bivalents at metaphase I of the B. napus haploid. These results are discussed in the context of genome evolution of B. napus.
ACCUMULATING evidence indicates that chromosomal rearrangements are commonplace within and among species and that they contribute to the evolution, adaptation, and speciation of plants and animals (Stebbins 1971; Rieseberg 2001; Coghlan et al. 2005). For instance, deletions and duplications modify gene copy number, which can induce change in gene expression (Song and Messing 2003) and phenotype (Guo et al. 1996; Veitia 2005). In plants, changes in chromosome and genome organization are prompted by interspecific hybridization and/or polyploidization (Wendel 2000). Studies of young natural and experimentally synthesized interspecific hybrids (Rieseberg et al. 1995; Baack et al. 2005) and polyploids (Song et al. 1995; Feldman et al. 1997; Lukens et al. 2006) have revealed that extensive and diverse genomic changes can arise immediately at the onset of genome merging or within a few generations. Subsequent rearrangements, which occur over longer periods of time, have been less extensively characterized even if they induce changes in karyotype and chromosome organization (Lysak et al. 2006) that blur the initial hybridization/polyploidization events (Wolfe 2001) and lead to genome downsizing (Leitch and Bennett 2004). Most rearrangements occur at meiosis and can be mediated by the homologous recombination pathway. Deletions, duplications, and translocations can then arise when information for the repair of double-strand breaks (DSBs) is taken from (i) the same chromosome (“intrachromosomal recombination,” Devos et al. 2002; Chantret et al. 2005), (ii) elsewhere in the genome (“ectopic recombination,” Petrov et al. 2003), or (iii) homeologous chromosomes in polyploids and interspecific hybrids (“homeologous recombination,” Udall et al. 2005). Homeologous exchanges have been shown to drive introgressive hybridization in Gossypium (Cronn et al. 2003), to be responsible for the adaptation of Helianthus homoploid species to new environments (Rieseberg et al. 2003), and to increase the range of genetic variation observed for important ecological and agronomic traits like flowering time (Pires et al. 2004) or seed yield (Osborn et al. 2003) in newly synthesized Brassica napus.
This study focuses on the oilseed crop (B. napus; AACC, 2n = 38), which is a young allopolyploid species resulting from multiple independent hybridization events between ancestors of the modern diploid B. oleracea (CC, 2n = 18) and B. rapa (AA, 2n = 20) (U 1935; Palmer et al. 1983; Song and Osborn 1992). It is now largely accepted that the diploid progenitors of B. napus are widely replicated although it is still not clear whether they have evolved from a hexaploid ancestor or via segmental duplication of one or two ancestral genomes (Truco et al. 1996; Parkin et al. 2003, 2005; Lukens et al. 2004; Lysak et al. 2005). Most genomic segments and genes of the B. napus genome are found in multiple copies, two of which are primary homeologues (regions from the A and C genomes that share the most recent common ancestry), while the remaining ones are paralogous to one or the other of these copies. Alignment of the A- (N1–N10) and C-genome (N11–N19) linkage groups allows identification of regions of primary homeology on the basis of the longest stretches of collinearity (Parkin et al. 2003). It is noteworthy that the degree of conserved synteny varies considerably among linkage groups, with some stretches of collinear marker loci extending to entire linkage groups (e.g., N1 and N11) while others extend only to half-linkage groups (e.g., N9 and N18). Identification of regions of primary homeology is confirmed by the localization of homeologous recombination events, resulting in de novo homeologous nonreciprocal translocations (HNRT) (Parkin et al. 1995; Sharpe et al. 1995), the latter called homeologous nonreciprocal transposition by Udall et al. (2005) with the same acronym. HNRT events have been reported by these authors at frequencies of 0.43–1.6% of total recombination events in several mapping populations of euploid B. napus (AACC, 2n = 38); the highest frequencies were observed when resynthesized B. napus was used as a parent. Preexisting reciprocal (Lombard and Delourme 2001; Osborn et al. 2003; Piquemal et al. 2005) and nonreciprocal homeologous translocations (Udall et al. 2005) have also been identified among different accessions of B. napus and shown to stimulate further rearrangements in their vicinities.
B. napus haploids (AC, n = 19) are attractive models with which to study the rate and pattern of homeologous recombination. First, these plants undergo a complete meiosis with variable and sometimes huge amounts of chromosome pairing at metaphase I (MI) (Olsson and Hagberg 1955; Renard and Dosba 1980; Attia and Röbbelen 1986; Jenczewski et al. 2003). Second, the extent of affinity between the homeologous chromosomes/regions of B. napus is revealed since there is no pairing between homologues. Third, these plants produce viable restituted gametes with somatic chromosome numbers (Morinaga and Fukushima 1933; Tai and Ikonen 1988), which is crucial for the occurrence and transmission of intergenomic exchanges. Finally, it should be noted that haploid plants arise spontaneously within natural populations at a frequency up to 6/1000 (Thompson 1969; Stringham and Downey 1973).
In this article, we analyze the extent and pattern of chromosomal rearrangements that occur during meiosis of B. napus haploids (AC, n = 19), with particular attention to the role of homeologous recombination. Our study focuses on three pairs of homeologous regions (N1/N11, N3/N13, and N9/N18) chosen to be representative of the different levels of synteny and homeologous recombination (Sharpe et al. 1995; Parkin et al. 2003, 2005; Udall et al. 2005). Our study shows that recombination preferentially occurs between regions of primary homeology and suggests that it may also occur between others regions showing intragenomic or intergenomic homology.
MATERIALS AND METHODS
Plant material:
The production of haploid plants from B. napus cv. Darmor-bzh and Yudal has been described by Jenczewski et al. (2003). Haploid plants were used as female parents with the male euploids donating haploid pollen. A first F1 population was produced by crossing 5 haploids of Darmor-bzh with 1 euploid plant of Yudal (DxHz F1 population, 172 plants). A second F1 population was obtained by crossing 9 haploids of Yudal with 1 euploid plant of Darmor-bzh (YxHz F1 population, 126 plants). Progeny were named according to their population with x designating the number of the maternal haploid plant and z designating the offspring number. One and 2 plants were chosen in the YxHz (Y9H11) and the DxHz (D1H29, D11H2) F1 populations, respectively, and backcrossed to their recurrent parent (i.e., Yudal when the parental haploid originated from Darmor-bzh and reciprocally). Each of these BC1 populations contained a minimum of 85 plants. Additionally, a doubled haploid line of B. oleracea, HDEM, and a B.napus cv. Darmor-bzh euploid line were used as controls for BAC–FISH analysis.
Cytology:
Flow cytometry was performed on 170 and 123 plants from the DxHz and the YxHz F1 populations, respectively, to assess chromosome number with an accuracy of plus or minus two chromosomes according to Eber et al. (1997).
For meiotic analyses, floral buds were fixed in Carnoy's solution (ethanol:chloroform:acetic acid, 6:3:1) for 24 hr and stored in 50% ethanol. Observations on pollen mother cells (PMCs) were performed on anthers squashed and stained in a drop of 1% acetocarmine solution; 37 PMCs were observed at MI to establish the average meiotic behavior of the Darmor-bzh haploid at this stage. A minimum of 10 PMCs per plant were observed at MI to verify the chromosome numbers of 59 F1 and 5 BC1 plants.
FISH with a genome-specific BAC clone:
FISH analyses were carried out on mitotic chromosomes of B. oleracea cv. HDEM and B. napus cv. Darmor-bzh and on anther meiocytes of the Darmor-bzh haploid at MI. Mitotic metaphase preparation from seedling root tips was as described by Snowdon et al. (1997). Meiotic chromosome preparation was the same as for mitotic observation. FISH was performed as extensively described in Leflon et al. (2006). Briefly, BAC clone BoB014O06 was labeled by random priming with biotin-14-dUTP (Invitrogen, San Diego; Life Technologies) and used as a probe at 100 ng/slide. Fluorescence images were captured using a CoolSnap HQ camera (Photometrics, Tucson, AZ) on an Axioplan 2 microscope (Zeiss, Oberkochen, Germany) and analyzed using MetaVue (Universal Imaging, Downington, PA).
Molecular markers:
DNA extraction was performed as described by Lombard and Delourme (2001) on a subset of 117 and 103 plants from the DxHz and the YxHz F1 populations, respectively; these all display 2n = 38 chromosomes (flow cytometry estimates confirmed by direct chromosome counting on a subset of plants). DNA extraction of backcross populations was performed on young leaves as described by Morales et al. (2005), using a Microlab Star Hamilton robot. All the markers used in this study have been previously placed on a reference map constructed using doubled haploid progeny of Darmor-bzh and Yudal euploid parents (Lombard and Delourme 2001; Delourme et al. 2006). Linkage groups N1, N3, and N9 represent chromosomes from the A genome of B. napus while groups N11, N13, and N18 represent chromosomes from the C genome of B. napus.
RAPD markers (one-letter prefix) were described by Foisset et al. (1996). Microsatellite markers originated either from the BBSRC (prefixed with Na, Ol, or Ra; Lowe et al. 2004) or from the CELERA consortium (prefixed with CB, BRAS, or MR; Piquemal et al. 2005). PCR-specific markers derived from Arabidopsis are described by Sillito et al. (2000) or Fourmann et al. (2002). Physical functional markers (PFM) are prefixed CZ and have been developed from coding sequence of Arabidopsis and Brassica ESTs (H. Belcram and B. Chalhoub, unpublished data; primer sequences are available upon request to Genoplante). SINE markers (prefixed with JLP) have been described in Prieto et al. (2005).
PCR assays were conducted essentially as described in the quoted articles. Forward primers (5′) were tailed with M13 to be revealed by LICOR technology. PCR products were analyzed on a 5.5% acrylamide gel and dye detected with LICOR as described by Piquemal et al. (2005), on a 2.5% agarose gel as described by Foisset et al. (1996), or on a 16-capillary ABI Prism 3130xl as described by Esselink et al. (2004).
Determination of allele copy number:
Quantitative fluorescent-PCR analysis was used to determine the allele copy number of the haploid parents. PCR products were analyzed by capillary electrophoresis sequencer. Data were collected by Gene Scan v3.1 software and analyzed with GeneMapper v3.7 (Applied Biosystems, Foster City, CA). Peak height and area of alleles from multiallelic markers were estimated using GeneMapper. For haploid progeny alleles peak height and area ratios were shown to be highly correlated for some markers.
For each locus, there were three groups of plants, one with r = 0 copies of the haploid parent (HP) allele, one with r = 1 copy of the HP allele, and one with r = 2 copies of the HP allele. Plants with no peak associated with the HP allele were directly considered to satisfy r = 0. However, a more refined statistical analysis was needed to discriminate between plants where a peak was present (r = 1 or r = 2). For each locus, preliminary graphical representations of the data showed a clear linear relationship between the peak areas of the diploid and haploid parent alleles, with two distinct slopes corresponding to the cases r = 1 and r = 2. After these preliminary investigations, the following model was considered for each locus,
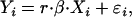 |
(1) |
where Yi is the peak area of the HP allele measured on plant i, Xi is the peak area of the diploid parent (DP) allele on plant i, β is the basic regression coefficient, and, for each plant, r = 1 with probability p or r = 2 with (lower) probability 1 − p. The error term ɛi was assumed to follow a normal distribution with standard error equal to 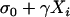 . The four unknown parameters β, p,
. The four unknown parameters β, p, 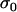 , and γ of the model were estimated by maximum likelihood, separately for each locus. In statistical terms the model (1) is a mixture of two regression models with probabilities p and 1 − p, where the components correspond to case r = 1 or r = 2. Once the parameters of such a model are estimated, it is possible to estimate the probability pi that each plant i satisfies r = 1, given the data and the Yi.-value for that plant (Everitt and Hand 1981; Jenczewski et al. 2003). The odds ratio
, and γ of the model were estimated by maximum likelihood, separately for each locus. In statistical terms the model (1) is a mixture of two regression models with probabilities p and 1 − p, where the components correspond to case r = 1 or r = 2. Once the parameters of such a model are estimated, it is possible to estimate the probability pi that each plant i satisfies r = 1, given the data and the Yi.-value for that plant (Everitt and Hand 1981; Jenczewski et al. 2003). The odds ratio 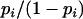 is the estimated ratio between the probabilities that plant i satisfies r = 1 or r = 2. For subsequent data analyses, a plant i was considered to have one copy of the HP allele (r = 1) when its odds ratio was >100. It was considered to have two copies of the HP allele when its odds ratio was <1/100. In the few intermediate cases (0–8% of plants depending on marker), information was considered insufficient to decide allele copy number and hence they were discarded from further analyses. This analysis was carried out for every plant in the DxHz population for loci on N1, N9, and N11 and in the progeny of Yudal haploids for loci on N3.
is the estimated ratio between the probabilities that plant i satisfies r = 1 or r = 2. For subsequent data analyses, a plant i was considered to have one copy of the HP allele (r = 1) when its odds ratio was >100. It was considered to have two copies of the HP allele when its odds ratio was <1/100. In the few intermediate cases (0–8% of plants depending on marker), information was considered insufficient to decide allele copy number and hence they were discarded from further analyses. This analysis was carried out for every plant in the DxHz population for loci on N1, N9, and N11 and in the progeny of Yudal haploids for loci on N3.
All statistical analyses were performed with R language on R version 2.2.1 software (R Development Core Team 2005) or on S-Plus 2000 software.
RESULTS
In situ hybridization survey of meiosis in haploid B. napus:
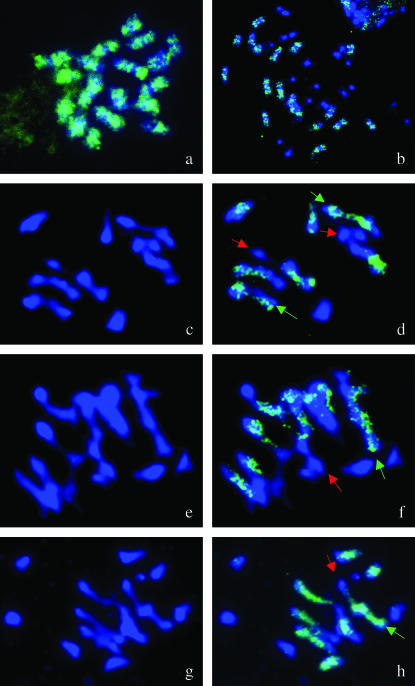
FISH analysis was first carried out using BoB014O06 to probe mitotic chromosomes of B. oleracea cv. HDEM and B. napus cv. Darmor-bzh. Only the nine C genome chromosomes in B. napus were labeled by this probe; however, the hybridization signal is not homogeneous along the chromosomes (Figure 1, a and b). At meiosis, MI chromosomes of haploid Darmor-bzh were labeled with the BoB014O06 probe to determine the occurrence of autosyndesis (A–A or C–C pairing) vs. allosyndesis (A–C pairing) (Figure 1, d, f, and h). Autosyndesis was shown to be commonplace at MI in Darmor-bzh haploids since 84% of the 49 observed PMCs contained at least one autosyndetic bivalent. Having classified chromosome configurations (univalent, I; bivalent, II; trivalent, III; and tetravalent, IV) and determined the genome origin of each chromosome, we calculate the average meiotic behavior of the Darmor-bzh haploid at MI to be: 3 IA + 2.2 IC + 5.2 IIC–A + 0.8 IIA–A + 0.7 IIC–C + 0.1 III + 0.04 IV. Autosyndetic bivalents therefore represented 22% of the bivalents observed. The multivalent configurations observed also included intragenomic pairing.
Figure 1.—
Detection of autosyndesis at metaphase I using FISH on pollen mother cells from haploid Darmor-bzh. Mitotic chromosomes of B. oleracea cv. HDEM (a) and B. napus cv. Darmor-bzh (b) are probed with BAC BoB014O06 (green) and counterstained with DAPI (blue), to show that this BAC stains only C genome chromosomes. DAPI staining is then used on anther meiocytes to establish the meiotic behavior of every Darmor-bzh PMC at MI (c, e, and g) and then combined with BoB014O06 FISH signals (d, f, and h) to distinguish autosyndetic and allosyndetic associations. Red and green arrows indicate autosyndetic bivalents between A and C chromosomes, respectively. (d) Seven bivalents including one ring bivalent (four autosyndetic and three allosyndetic) and five univalents. (e) Seven bivalents (two autosyndetic and five allosyndetic) and five univalents. (f) Six bivalents (two autosyndetic and four allosyndetic) and seven univalents.
Genomic structure of plants in the progeny of haploid B. napus:
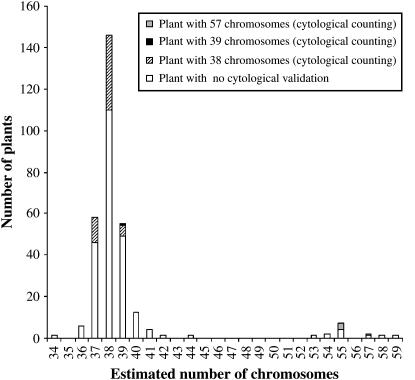
Flow cytometry showed that the vast majority of plants in the progeny of haploid B. napus had an estimated chromosome number ranging from 37 to 39 (Figure 2). Interestingly, 10 plants (3%) appear to be triploid (AAACCC) because they display an estimated chromosome number distributed around 57; these estimates were confirmed by direct cytological chromosome counting for 4 plants. To further corroborate the flow cytometry results, we directly counted the number of chromosomes of a subset of 55 plants with an estimated 2n = 38 chromosomes. Only 1 plant had 39 chromosomes while the remaining 54 had 38 as expected (Figure 2). Extending this estimate to the entire population, we predict that 77–93% of progeny plants of Darmor-bzh and Yudal haploids have 38 chromosomes. Ninety percent of these 54 plants displayed >75% of PMCs with 19 bivalents at MI, which demonstrates that the 2n = 38 plants originated from restituted female gametes that had one copy of each of the 19 chromosomes of the haploid mother plants. Most viable gametes produced by haploid plants were therefore analogous to first division restitution (FDR) gametes.
Figure 2.—
Analysis of chromosome number in the progenies of B. napus haploids. Flow cytometry was used to estimate the chromosome number for 293 haploid-derived offspring. Cytological observations at MI of meiosis on a mean number of 17 PMCs per plant were performed on 55 plants selected to have chromosome estimates ranging from 37 to 39 (open and solid bars).
Detection of homeologous exchanges with PCR molecular markers:
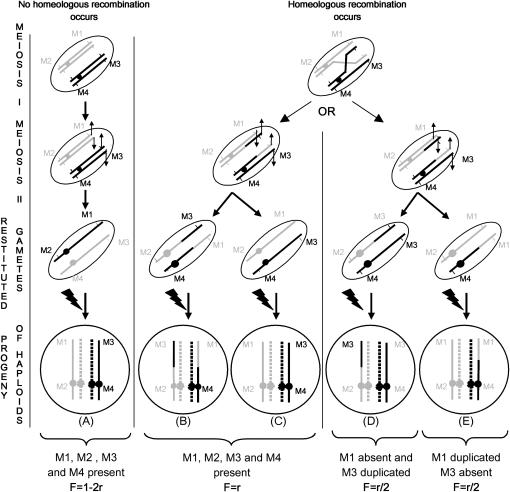
In F1 plants (2n = 38) derived from these FDR-like gametes all the markers from the HP are expected to be present when no recombination occurs during HP meiosis (Figure 3, offspring A). Reciprocally, HNRTs, which originate from disjunction of reciprocal homeologous exchanges, can be detected in the progeny of haploid × euploid B. napus by looking for plants that lack the HP alleles for a given set of loci and had duplicate HP alleles at corresponding homeologous loci (Figure 3, offspring D and E). In this study, we analyzed transmission and/or duplication of molecular markers spanning three pairs of homeologous regions that represent a range of synteny levels (described in Parkin et al. 2003, 2005) and homeologous recombination rates in euploid lines (Sharpe et al. 1995; Udall et al. 2005): (i) the bottom sections of linkage groups N3 and N13 are structurally very different and do not recombine with one another, (ii) group N18 is collinear with one end of group N9 and they display moderate levels of homeologous recombination with each other, and (iii) groups N1 and N11 are syntenous along their entire length and display the highest frequencies of HNRTs.
Figure 3.—
Segregation of molecular markers in the progenies of B. napus haploids. This chromatid model of homeologous exchanges assumes that B. napus haploids produce restituted gametes with 19 chromosomes. When no homeologous recombination occurs during meiosis of the haploid, then all markers from the haploid parent (HP) are transmitted to its progeny (offspring A). When homeologous recombination occurs during meiosis of the haploid parent, recombinant chromatids may either (i) move to the same pole of anaphase, meaning all the markers from the HP are transmitted to its progeny (offspring B and C) despite the presence of a homeologous reciprocal translocation (HRT), or (ii) move to opposite anaphase poles, meaning some of the markers from the HP are not transmitted to its progeny while others, carried by the (partially homeologous) substituting chromosomal segment are duplicated (offspring D and E). These latter exchanges are commonly termed homeologous nonreciprocal translocations (HNTRs). r, frequency of homeologous recombination; jagged arrowhead, fertilization by male gametes (n = 19) produced by the euploid male parent.
HP allele loss in the progeny of B. napus haploids:
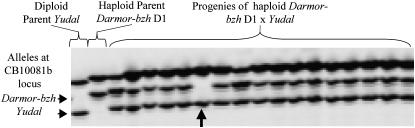
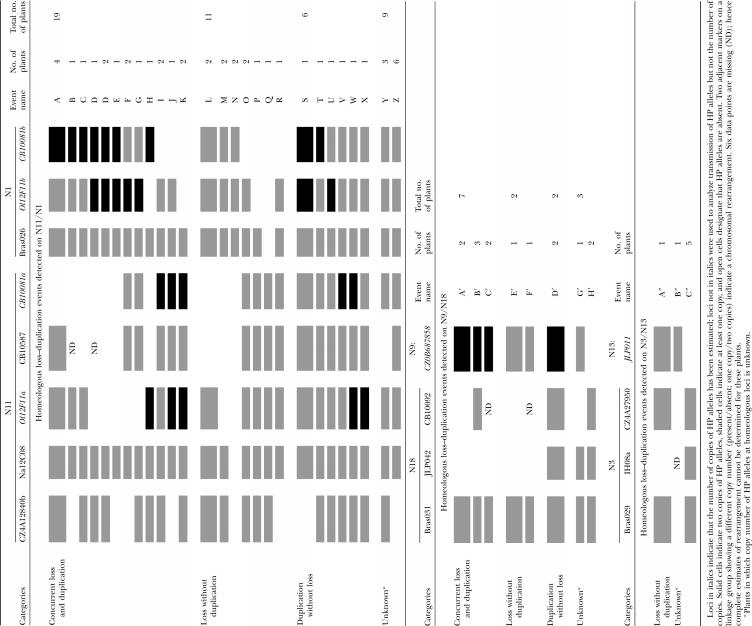
We analyzed transmission of 20 molecular markers spanning the six linkage groups described above. We genotyped the haploid parents and their progeny with codominant markers to be sure that absence of HP alleles was due to nontransmission of markers. The proportion of plants in the progeny of Darmor-bzh haploids that lacked HP alleles (e.g., Figure 4) varied from 0 to 14%, depending upon the locus (Table 1). The highest frequencies were obtained for markers located at opposing ends of N11. We confirmed that markers located on the bottom section of N3 rarely displayed loss of HP alleles by genotyping an additional 103 progeny of Yudal haploids with the same markers; it was observed that only one plant (Y9H11) lacked the HP alleles for marker IH08a (Table 1). Looking at all progeny plants in all but three cases offspring that lacked the HP allele at one or more loci on a linkage group displayed HP allele(s) present at other loci on the same linkage group (Table 2); this demonstrates that nontransmission of the HP allele was not due to the whole-chromosome loss.
Figure 4.—
Inheritance of the HP allele at CB10081b in the F1 progeny of the Darmor-bzh haploid, D1. One plant lacks the Darmor-bzh HP allele (arrow).
TABLE 1.
Loss and duplication of haploid parent (HP) alleles at corresponding homeologous loci in progeny plants
| Loss of haploid parent (HP) alleles
|
Duplication of HP alleles at homeologous loci
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Haploid parent | LG | PI | Loci | No. of progeny lacking HP allele | % of progeny lacking HP allele | LG | PI | Loci | No. of progeny with duplicated HP allele | % of progeny with duplicated HP allele |
| Darmor-bzh | N1 | 0 | CB10081b | 10 | 9 | N11 | 0.02 | CB10081a | 7 | 6 |
| 0.41 | Ol12F11b | 5 | 4 | 0.43 | Ol12F11a | 5 | 4 | |||
| 0.66 | Bras026 | 1 | 1 | — | — | — | — | |||
| N11 | 0.02 | CB10081a | 16 | 14 | N1 | 0 | CB10081b | 13 | 11 | |
| 0.24 | CB10587 | 14 | 13 | — | — | — | — | |||
| 0.43 | Ol12F11a | 11 | 10 | 0.41 | Ol12F11b | 9 | 8 | |||
| 0.56 | Na12C08 | 2 | 2 | — | — | — | ||||
| 0.83 | CZ4A12840b | 14 | 12 | — | — | — | ||||
| N9 | 0.09 | CZ0B687858 | 2 | 2 | N18 | — | — | — | — | |
| N18 | 0.03 | CB10092 | 4 | 6 | N9 | — | — | — | — | |
| 0.39 | JLP042 | 9 | 8 | 0.09 | CZ0B687858 | 9 | 10 | |||
| 1 | Bras031 | 0 | 0 | — | — | — | — | |||
| N3 | 0.28 | Bras029 | 1 | 1 | N13 | |||||
| 0.65 | IH08a | 0 | 0 | 0.85 | JLP011 | 0 | 0 | |||
| 0.97 | CZ4A27950 | 1 | 1 | — | — | — | — | |||
| N13 | 0.85 | JLP011 | 5 | 4 | N3 | — | — | — | — | |
| Yudal | N3 | 0.28 | Bras029 | 0 | 0 | N13 | — | — | — | — |
| 0.65 | IH08a | 1 | 1 | 0.85 | JLP011 | 0 | 0 | |||
| 0.97 | CZ4A27950 | 0 | 0 | — | — | — | — | |||
| N13 | 0.85 | JLP011 | 0 | 0 | N3 | — | — | — | — | |
LG, linkage group; PI, position index, relative position of loci on linkage group calculated by dividing the genetic position of loci by the size of the LG on the reference map: 0 designates the top of the linkage group and 1 the bottom. Markers appearing on the same line are exact homeologues or located in homeologous regions.
TABLE 2.
Chromosomal rearrangements detected by simultaneous analysis of loss and duplication of haploid parent (HP) alleles at different homeologous loci
Duplicated HP alleles in the progeny of B. napus haploids:
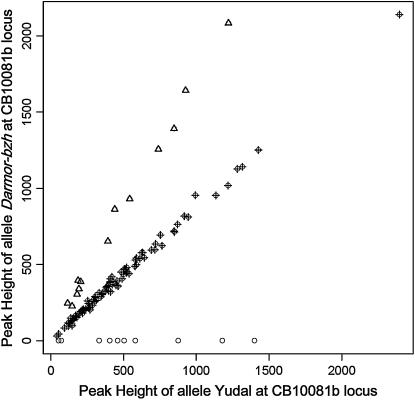
Detection of HP allele duplication (e.g., Figure 5) was achieved for four markers that revealed six loci in regions homeologous to segments where missing HP alleles had been identified (Table 1). Exact correspondence between homeologous loci was ascertained when marker assays detected multiple loci mapping to collinear blocks (Ol12F11a/b, CB10081a/b). When these markers were not available, we used markers anchored in Arabidopsis to find the most likely homeologues within collinear blocks (data not shown). The proportion of plants in the progeny of B. napus haploids having two copies of HP alleles varied from 0 to 11.5% depending on both the linkage group and the position of the markers (Table 1). The highest frequencies were obtained for markers located at the top of N1 and N9 while the lowest was at JLP011, located at the distal end of N13 (Table 1).
Figure 5.—
Determination of copy number of the Darmor-bzh HP allele at CB10081b. A quantitative value for the Yudal allele peak area (x-axis) was used as a baseline to which the Darmor-bzh allele peak area (y-axis) has been compared by maximum-likelihood analyses: O, plants with no Darmor-bzh allele at the CB10081b locus (loss); +, plants with a single Darmor-bzh allele at the CB10081b locus (no duplication, no loss); Δ, plants with two Darmor-bzh alleles at the CB10081b locus (duplication).
Concurrent loss and duplication of homeologous HP alleles by HNRTs:
Simultaneous analysis of homeologous loci showed that nontransmission of HP alleles was frequently associated with the duplication of HP alleles in homeologous regions (Table 2). For example, 14 of 19 plants lacked Darmor-bzh alleles at CB10081a and/or Ol12F11a on N11 and had two copies of Darmor-bzh alleles at homeologous loci CB10081b and/or Ol12F11b on N1 (Table 2, events A–H). Taking into account that H has two independent rearrangements, 20 and 7 instances of concurrent loss and duplication of linked loci were observed for N1/N11 and N18/N9, respectively. Subsequent analysis of other loci on the same linkage groups revealed that concurrent loss and duplication were rarely limited to interstitial loci (Table 2, events F–H) and usually extended to the end of linkage groups (Table 2, events A–E, H–K, and A′). Six plants had a pattern of duplication and loss that was compatible with the occurrence of two distinct simultaneous rearrangements on a single chromosome (Table 2, events B, E, F, H, and J). Finally, we observed over twice as many plants with simultaneous loss of HP alleles on N11 and duplication on N1 than the number of plants with concurrent loss of HP alleles on N1 and duplication on N11 (14 vs. 6; Table 2, events A–H vs. H–K).
In summary, concurrent loss and duplication of homeologous alleles represented 51% of events for N1/N11 and 64% for N18/N9 (Table 2), indicating that approximately half the chromosomal rearrangements we detected are HNRTs. These HNRTs can be verified by checking the inheritance of HP alleles in controlled backcross progeny.
Independent loss and duplication of homeologous HP alleles:
Both loss without duplication and duplication without loss was observed on N1/N11 and N18/N9. Likewise, the only plant (Y9H11) that lacked a HP allele at a N3 marker did not display HP allele duplication on the homeologous region of N13. We observed twice as many instances of loss without duplication than duplication without loss on N1/N11, but similar levels of these two kinds of rearrangements on N18/N9 (Table 2). The number and position of independently duplicated or lost loci were variable and nonrandom since independent loss and duplication of HP alleles occurred more frequently at the distal ends of linkage groups than interstitially (Table 2, events U, X, and A″).
Characterizing chromosomal rearrangements by backcrossing:
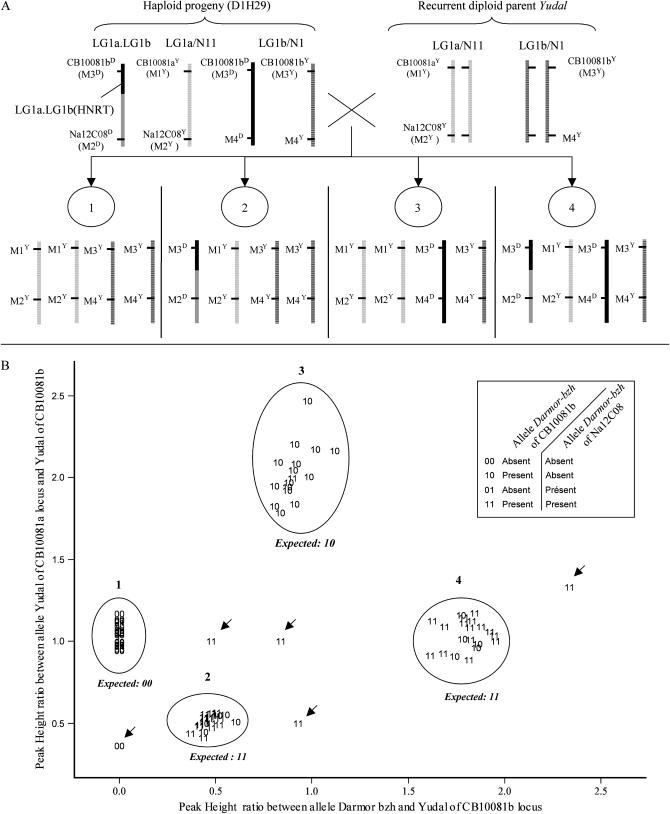
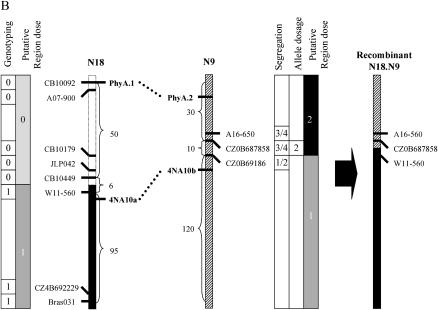
Two plants D1H29 (Table 2, event D) and D11H2 (Table 2, event A′) that displayed simultaneous loss and duplication of Darmor-bzh alleles at homeologous loci on N1/N11 and N9–N18, respectively, were backcrossed to Yudal to analyze marker segregation on implicated chromosomes (Figure 6).
Figure 6.—
Expected and observed segregation of the N11.N1 homeologous nonreciprocal translocation in the backcross progeny of D1H29. (A) Expected segregation of the N11.N1 HNRT assuming no further recombination event in the genetic intervals carrying the breakpoints of this HNRT. Three markers are considered: M1, CB10081a; M2, Na12C08; and M3, CB10081b. D1H29 lacks the Darmor-bzh allele at CB10081a (M1D) and had two copies of the Darmor-bzh allele at CB10081b (M3D). Chromatid sorting results in four groups of BC1 offspring that can be unambiguously recognized by comparing allelic composition of Darmor-bzh and Yudal alleles at CB10081a and CB10081b (M1Y/M3D/M3Y). If the cosegregation pattern of the Darmor-bzh alleles at CB10081b (M3D) and Na12C08 (M2D) is used, groups 1 and 3 can be identified by absence–absence (00) and presence–absence (10), respectively; however, groups 2 and 4 cannot be distinguished using these two marker alleles. (B) Observed segregation. Physical location on this plot reveals the four expected groups of BC1 offspring (circled) from analysis of copy number ratios at CB10081a and CB10081b loci. Five outlying individuals (arrows) display abnormal allelic composition at CB10081a and/or CB10081b. Each plant is symbolized by its cosegregation pattern for two Darmor-bzh alleles at CB10081b and Na12C08 loci, respectively. The expected cosegregation pattern in the absence of recombination at the Na12C08–CB10081b interval is indicated above each of the four circles. Plants that display discrepancies between the expected cosegregation pattern (00 in group 1, 01 in group 3, and 11 in groups 2 and 4) and that observed are considered recombinant at the Na12C08–CB10081b interval. Plants in agreement with expected copy number ratios are considered nonrecombinant.
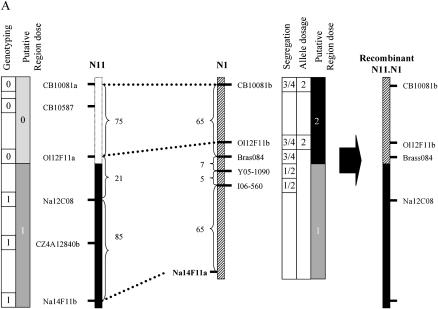
The first plant, D1H29 had a pattern of duplication and loss that was compatible with the presence of a translocation of N1 (duplicated) on N11 (substituted) [N11.N1 (T)]. Using molecular markers distributed along these groups, we observed that markers located at the distal end of N11 did not display the Darmor-bzh allele while three markers from the corresponding part of N1 were duplicated (Figure 7A). The latter markers segregated at 3:1 in the (D1H29 × Yudal) backcross population but at 1:1 in another BC1 progeny that was free from any [N11.N1 (T)]. We then analyzed the extent of linkage disequilibrium between markers flanking the breakpoints of this putative HNRT (Brass084, Ol12F11b, and CB10081b on N1 and Na12C08 on N11) and observed that these markers cosegregated more often than expected by chance (χ2 < 0.01%; data not shown). These results confirmed that D1H29 carried a distal [N11.N1 (T)] HNRT (Figure 7A). Estimating the size of genetic intervals carrying the breakpoints of this HNRT was not straightforward. Figure 6A assumes that no further recombination event took place in this interval during the backcrossing process. In reality recombination is likely to have occurred and considerable thought must go into detecting these events when analyzing marker segregation data. For example, if one were to look for backcross recombination events purely by looking at presence of M3D CB10081b and absence of M2D Na12C08 without considering the segregation of other markers, one-quarter of backcross progeny (group 3) could be misclassified as having recombined during backcrossing. Fortunately all recombination between [N11.N1 (T)] and N1 and half of recombination between [N11.N1 (T)] and N11 can be detected by combining allelic composition and cosegregation analysis (Figure 6B: plants with unexpected cosegregation pattern 01 in group 1, 10 in groups 2 and 4, and 11 in group 3). Overall, 13 of 81 plants were detected as recombinant in the [N11.N1 (T)] interval of D1H29, which represents a map distance of 15 cM. Allele composition information also identifies 5 additional plants (highlighted by arrows in Figure 6) that displayed abnormal allelic composition: 2 plants lacked one locus CB10081a and 3 had an extra copy of the CB10081b locus. Among these 5 outliers, 2 plants were aneuploid (2n = 37 and 2n = 39) while the remaining 3 had a very irregular meiotic behavior with a high number of either univalents (1 plant, 60% of PMCs) or multivalents (2 plants, 75 and 50% of PMCs).
Figure 7.—
Schematic of three rearrangements detected in B. napus haploid-derived progeny. Rearrangements A and B are HNRTs that originated from homeologous exchanges between DY1a/N11 and DY1b/N1 (A, D1H29) and between DY5/N9 and DY8/N18 (B, D11H2). The origin of the third rearrangement (C, Y9H11) is unknown. The positions of breakpoints and the sizes of the lost and duplicated fragments (in centimorgans) are indicated on the parental linkage groups. Genotyping data (present, 1; absent, 0), the number of copies of the HP allele (solid bar, two copies; shaded bar, one copy; dotted bar, no HP allele), and the segregation pattern (e.g., 1:1) are indicated. Dotted lines between multilocus PCR markers indicate a syntenic relationship between homeologous regions; solid markers to the left of DY linkage groups indicate homeologous relationship between DY linkage groups but were not used in our molecular analysis.
The same analyses were performed on D11H2 and confirmed that this plant carried a distal HNRT of N9 on N18 [N18.N9 (T)] (Figure 7B). As allele composition information could not be determined for our markers on N9 and N18, the size of this HNRT was determined using cosegregation patterns 01 and 00 that can be unambiguously recognized as recombinant or not recombinant, respectively. We thus estimated that 5 cM lay between W11.560 and CZ0B687858 in the interval carrying the N18.N9 (T) breakpoint of D11H2 (Figure 7B).
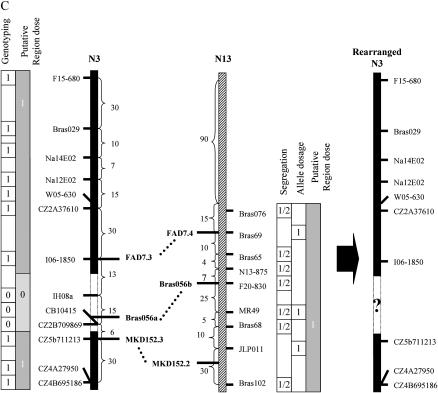
Finally we tried to characterize the rearrangement carried by plant Y9H11 that lacked Yudal alleles at 3 loci on N3 and did not display two copies of the HP allele at 9 loci in its homeologous region on N13 (Figure 7C). Using 13 markers from N3, we demonstrated that the missing segment was interstitial and encompassed a region between 28 cM (12% of linkage group length) and 34 cM (24%) on N3, depending on breakpoint positioning (Figure 7C). We then tested for marker duplication on other regions displaying intragenomic (N5, N1) or intergenomic (N11) homology to the missing part of N3. Part of N15 is also paralogous to the missing part of N3; however, polymorphic markers could not be found and hence this possibility remains unexamined. Allelic segregation ratios for markers on N1 and N11 did not deviate from the 1:1 ratio, indicating absence of duplication. In contrast, a marker on N5 segregated 3:1 (excess of Yudal allele) but no evidence that this locus cosegregated with CZ5b711213 on N3 (Figure 7C) was found, indicating that this region of N5 is duplicated but has not been translocated onto N3.
Comparison of recombination rates between homeologous and homologous:
Considering that a minimum of 50% of the detected losses of HP alleles resulted from crossovers between homeologous regions and that half the products of homeologous recombination can be detected in the progenies of B. napus haploids (Figure 3), we compared the frequencies of homeologous recombination measured in this study to the frequencies of homologous recombination estimated for the same intervals from the genetic mapping data. The ratio between the proportions of homeologous and homologous recombinants was lower for two intervals on N1(CB10081b–Ol12F11b) and N18 (JLP042–Bras031) because the corresponding markers were genetically independent on the linkage groups (Table 3). Notwithstanding this point, we observed that this ratio averaged a constant 10-fold decrease of homeologous vs. homologous recombination for most intervals and linkage groups, with the notable exception of N11, which showed a higher relative proportion of homeologous recombinants (Table 3).
TABLE 3.
Comparison of the rate of homologous vs. homeologous recombination measured on the same intervals
| Linkage groups | Locus interval | % homeologous recombination | % homologous recombinationa | Ratiobc |
|---|---|---|---|---|
| N11 | CB10081a–CB10587 | 7 | 22 | 0.3** |
| CB10587–Ol12F11a | 11 | 15 | 0.7 | |
| Ol12F11a–Na12C08 | 8 | 15 | 0.5* | |
| Na12C08–CZ4A12840b | 12 | 15 | 0.8 | |
| N1 | CB10081b–Ol12F11b | 6 | 46 | 0.1** |
| Ol12F11b–Bras026 | 3 | 11 | 0.2** | |
| N18 | CB10092–JLP042 | 5 | 20 | 0.2** |
| JLP042–Bras031 | 8 | 42 | 0.2** | |
| N3 | CZ4A27950–IH08a | 1 | 20 | 0.1** |
| IH08a–Bras029 | 1 | 50 | 0** |
The proportion of homologous recombination at each location was obtained by analyzing the DH mapping population used to build the reference map.
This ratio, which reflects the rate of decrease of homeologous vs. homologous recombination, is estimated by dividing the percentage of homeologous recombination by the percentage of homologous recombination for the same interval.
χ2-tests were performed to compare the relative proportion of homologous and homeologous recombinants at each location: **P < 1%; *P < 5%.
We also took advantage of the fact that our approach was similar to that of Udall et al. (2005), by comparing the frequency of HNRTs generated per chromosome arm during meiosis of haploid vs. euploid lines. We observed that the frequency of homeologous recombination was increased 10- to 100-fold during meiosis of B. napus haploids when compared to that of the natural or synthetic euploid B. napus (Table 4). This ratio varied considerably among the linkage groups, the highest increase in the rate of homeologous recombination in haploid vs. euploid lines being observed at the top of N18–N9.
TABLE 4.
Comparison of de novo viable HNRT frequencies generated during meiosis of haploid vs. euploid lines
| Linkage group carrying the HNRT | Origin of duplicated segment | Progenies (female × male) | Interval observed | No. of de novo HNRTs detected | Mean no. of de novo HNRTs per plant | Ratioab |
|---|---|---|---|---|---|---|
| Top N1 | Top N11 | Haploid × euploid | CB10081b–Brass026 | 6 | 0.052 | 12** |
| Euploid × euploid (Udall et al. 2005) | pW169d–pX128e | 6 | 0.009 | |||
| Top N11 | Top N1 | Haploid × euploid | CB10081a–Na12C08 | 14 | 0.121 | 32** |
| Euploid × euploid (Udall et al. 2005) | pW125c–pC128g | 5 | 0.008 | |||
| Top N18 | Top N9 | Haploid × euploid | CB10092–JLP042 | 7 | 0.080 | 106** |
| Euploid × euploid (Udall et al. 2005) | pW123d–pW227 | 1 | 0.002 | |||
| Bottom N3 | Bottom N13 | Haploid × euploid | Brass29–Cz4A27950 | 0 | 0.000 | — |
| Euploid × euploid (Udall et al. 2005) | pW147b–pX136a | 0 | 0.000 | — |
This ratio, which reflects the rate of increase of haploid vs. euploid homeologous recombination, is estimated by dividing twice the mean number of de novo HNRTs per plant detected in the haploid × euploid progenies by the mean number of de novo HNRTs per plant detected in the euploid × euploid progenies (data from Udall et al. 2005). Magnification of the numerator by a factor of 2 aims at compensating for the presence of two homologous chromosomes in the euploids while there is only one in the haploids.
χ2-tests were performed to compare the relative proportion of homologous and homeologous recombinants at each location: **P < 1%; *P < 5%.
DISCUSSION
In this study, we have analyzed in progeny of haploid plants (2n = 19) transmission and/or duplication of molecular markers spanning three pairs of homeologous regions (N1/N11, N3/N13, and N9/N18) that represented a range of levels of synteny and homeologous recombination. We have shown that duplication and/or loss of chromosomal regions frequently occurs during meiosis of B. napus haploids and that half of the changes are due to recombination between regions of primary homeology. We also observed an unexpected number of autosyndetic bivalents, which strongly suggests that recombination can also occur between paralogous regions, leading to part of chromosomal rearrangements. These results raise the question of the role of haploids in the long-term evolution of the B. napus genome.
High occurrence and transmission of homeologous recombination products during meiosis of B. napus haploids:
Meiosis and gamete production in haploid forms of B. napus has been well documented (Morinaga and Fukushima 1933; Olsson and Hagberg 1955; Renard and Dosba 1980; Attia and Röbbelen 1986; Tai and Ikonen 1988). Our study moves two steps further in demonstrating that (i) homeologous exchanges occur during meiosis of B. napus haploids and that (ii) recombined chromosomes can be transmitted to the progeny of B. napus haploids by unreduced FDR-like gametes.
We have first proved that concurrent loss and duplication of homeologous loci can be used to detect de novo homeologous nonreciprocal translocations (HNRTs), also called homeologous nonreciprocal transposition by Udall et al. (2005), by demonstrating in two plants that the haploid parent alleles located in the vicinity of the missing part of chromosomes no longer segregated independently from those that are duplicated on the corresponding homeologous region. This is due to the fact that translocations create “recombined linkage groups” that usually display reduced recombination in the vicinity of exchange breakpoints (Sybenga 1975; Parker et al. 1982), therefore promoting linkage disequilibrium between markers initially carried by different linkage groups. Assuming that simultaneous loss and duplication of linked loci in one plant originated from the same homeologous exchange, we detected a total of 18 distal and 7 interstitial HNRTs resulting from single and double homeologous crossovers, respectively (Table 2). These HNRTs represent approximately half the rearrangements we detected. We also observed patterns of loss and duplication that are compatible with occurrence of three crossovers spaning over two homeologous regions (Table 2, events B, E, F, H, and J). All these results indicate that crossovers frequently and preferentially occur between homeologous chromosomes during meiosis of B. napus haploids. Therefore a large proportion, if not all, of the bivalents observed at MI, which appear alike, are in fact chiasmatic.
Our results also confirm what Morinaga and Fukushima (1933), Olsson and Hagberg (1955), and Tai and Ikonen (1988) had previously observed, i.e., that most of the functional and viable gametes produced by B. napus haploid contain one copy of each of the 19 chromosomes of B. napus and are therefore genetically similar to first division restitution gametes. Our study is the first to demonstrate that recombined chromosomes can be transmitted by these FDR-like restituted gametes, allowing the immediate restoration of euploid chromosome number (2n = 38) and fertility (data not shown) in the progenies of haploids. However, our results indicate that HNRTs can promote abnormal segregation of chromosomes in subsequent generations, probably by inducing multivalent formation (Sybenga 1975; Ramsey and Schemske 2002); indeed, 6–28% of PMCs observed at MI in the progeny of haploids carrying HNRTs displayed quadrivalents (D11H2 and D1H29 revealed approximately one-quarter of PMCs with quadrivalents). Disjunction of these multivalents can lead to further rearrangements or to aneuploidy, as probably exemplified by the occurrence of five plants in the D1H29 backcross progeny with abnormal allelic composition (Figure 6).
Frequency of homeologous recombination is greatly enhanced in B. napus haploids:
Quantifying the number of concurrent losses and duplications of HP alleles can be used to estimate the proportion of chromosomal rearrangements transmitted by FDR-like gametes that are due to homeologous recombination. However, this approach leads to underestimating the rate of homeologous recombination. Meiotic homeologous exchanges lead both to homeologous reciprocal translocations (HRTs) and to HNRTs (Figure 3), but only the latter can be detected in the progeny of B. napus haploids. Likewise the patterns of segregation described in Figure 3 do not take into account the occurrence of trivalents and quadrivalents that represent 5–10% of associated chromosomes per MI (Jenczewski et al. 2003). Disjunction in these multivalents can generate gametes carrying uncoupled losses and duplications despite the occurrence of recombination between regions of primary homeology. Finally, homeologous recombination may generate lethal or deleterious chromosomal assortments that cannot be detected in the progenies of haploids because the individuals derived from these gametes, or the gametes themselves, do not survive or are counterselected. In that respect, HRTs and HNRTs are not expected to have the same consequences because HRTs do not change gene content or copy number while HNRTs lead to duplication and loss of genes that have been retained differently among homeologous segments or whose expression can be subfunctionalized (e.g., Adams et al. 2004).
With that proviso, we estimate that the frequency of homeologous recombination is 10- to 100-fold increased during meiosis of B. napus haploids as compared to that of the natural or synthetic euploid B. napus (Table 4). Homeologous recombination also shows <10-fold reduction when compared to homologous recombination measured on the same intervals (Table 3). It must be emphasized here that the values of these different ratios have been computed using the frequency of homeologous recombination measured in Darmor-bzh haploids, which display high numbers of bivalents at MI and have a permissive allele at the PrBn locus (Jenczewski et al. 2003). These comparisons can then be readily interpreted to reflect the suppression of pairing competition between homologues and homeologues during meiosis of haploids and the overall reduction of recombination between divergent genomes. Differences in the magnitude of change in recombination frequency among linkage groups we examined provide considerably more information.
We observe only a slight reduction of homeologous vs. homologous recombination for most intervals and linkage groups (Table 3). This confirms that the A and C homeologous regions of B. napus, which diverged ∼4 MYA (Inaba and Nishio 2002), remain very closely related. This assertion is in agreement with the fact that very few gene rearrangements differentiate the homeologous regions of B. napus (Rana et al. 2004). The slightest reduction is observed at opposing ends of N11, suggesting that these regions are the least divergent against their homeologues on N1. This assertion is not confirmed when looking at N1, which shows a lower relative proportion of homeologous recombinants (Table 3) and a lower number of HNRTs (Table 2). This asymmetry indicates either that a double-strand break is more frequently resolved by crossing over rather than by conversion on N11 than on N1 (e.g., Jeffreys and May 2004) or that plants carrying N11.N1 (T) are more viable than those carrying N1.N11 (T). Surprisingly, such an asymmetry is not observed for the other chromosomal rearrangements we have detected.
The increase of haploid vs. euploid homeologous recombination shows considerable variation among the three pairs of homeologous regions analyzed in this study (Table 4). Part of this variation is certainly due to the fact that Udall et al.'s data originate from a variable number of segregating populations, which can bias comparison and lead to underestimating the frequency of HNRTs in some regions/linkage groups as compared to others. Notably this could be the case for N9 for which Udall et al. (2005) estimated the frequency of HNRTs using only two of four populations. Correcting for this bias, more than a twofold difference persists between linkage groups. This variation occurs even if the numbers of HNRTs detected for the three pairs of homeologous regions analyzed in this study are ranked in the same order as that in Udall et al. (2005) (N1/N11 > N9/N18 ≫ bottom N3/N13). Specifically we observe no increase of haploid vs. euploid homeologous recombination at the bottom of N3/N13 that shows a constant very low number of rearrangements. We have confirmed that these regions are not prone to rearrange during haploid meiosis by detecting a similarly low amount of loss and duplication in two progenies derived from very different haploid genotypes (Darmor-bzh and Yudal haploids; Table 1). Characterization of the Y9H11 rearrangement indicates that the bottoms of N3 and N13 do not recombine with one another. Overall, these results show that a local disruption of synteny between otherwise extensively collinear chromosomes (Parkin et al. 2003, 2005) is sufficient to preclude homeologous recombination in that rearranged region. Likewise, recombination data confirm that the homeologous regions between N9 and N18, which consist of juxtaposed genomic blocks from different Arabidopsis chromosomes (Parkin et al. 2005) are slightly more divergent than the two arms of N1 and N11, which are collinear to a single region of Arabidopsis chromosomes, respectively. All these results suggest that the level of (macro)synteny can be used as a rough indicator of the likelihood of homeologous recombination occurring and give hints toward the recombination frequency (see N1/N11 asymmetry).
Does recombination occur between paralogous regions?
Half the losses and duplications of homeologous loci detected in this study occurred independently. Although some of these rearrangements may have resulted from the disjunction of multivalents including homeologous chromosomes (see above), most of them do not result from recombination between regions of primary homeology. Several mechanisms have been described that generate losses and duplications: centric misdivision of univalents (e.g., Friebe et al. 2005), unequal breakage of the anaphase bridge (e.g., Siroky et al. 2003), ectopic recombination between interspersed repeated sequences (e.g., Montgomery et al. 1991), and transposition (McClintock 1953; Bennetzen 2000). Our FISH results point to a very appealing alternative to these mechanisms, which echoes the highly replicated structure of the B. napus genome, i.e., that recombination may occur between regions that show intragenomic (paralogues) or intergenomic homology (intergenomic homologues do not equal homeologues). Our FISH data show that autosyndesis is quasi-systematically observed at MI and represents 20% of the bivalents. These autosyndetic bivalents are likely to be chiasmatic (see above), indicating that recombination may occur between paralogous regions. However, paralogous regions seem to recombine less than homeologous regions; this is in agreement with the fact that paralogous regions within B. oleracea and B. rapa genomes diverged four to five times earlier than homeologous regions. Then, more gene rearrangements differentiated paralogous from homeologous regions (Inaba and Nishio 2002; Rana et al. 2004; Town et al. 2006; Yang et al. 2006).
We attempted to test for the occurrence of exchanges between regions showing intragenomic (and intergenomic) homology. Although we used a very dense genetic map with a mean and a maximum distance between adjacent markers of 5.5 and 28 cM, respectively (R. Delourme, unpublished data), complete examination could not be achieved at all locations, due to the lack of markers with suitable polymorphisms (e.g., N18 remained unexamined in the case of Y9H11) and/or the lack of accurate identification of all the paralogous regions. The most saturated marker survey was achieved for Y9H11 that showed concurrent loss and duplication of paralogous regions but revealed independent segregation of these regions; Y9H11 displayed a few multivalents at MI (10.5% of PMCs), suggesting that the missing part of N3 was not lost from a simple deletion but from a more complex chromosome reshuffling.
A surprising picture emerges from our data. Our study has shown that recombination occurs preferentially between homeologous regions and it is striking to observe that autosyndesis occurs more frequently within B. napus haploids than in monoploid B. rapa and B. oleracea in which autosyndesis was observed in <20 and 45% of PMCs, respectively (Armstrong and Keller 1981, 1982). Preferential homeologous pairing should effectively compete with autosyndesis and reduce, not increase, its level in B. napus haploids. This point deserves further consideration, notably by taking into account the variability of meiotic behaviors in the haploid forms of B. napus (Jenczewski et al. 2003).
Concluding remarks:
Numerous studies have now demonstrated that homeologous reciprocal and nonreciprocal translocations exist in the genome of B. napus (Parkin et al. 1995; Lombard and Delourme 2001; Piquemal et al. 2005; Udall et al. 2005). Yet, it is not known whether these rearrangements have occurred at the onset of B. napus polyploid formation or whether they have progressively accumulated during the subsequent generations (Udall et al. 2005). Our study provides strong evidence that duplication or loss of chromosomal regions occurs at high frequencies during meiosis of B. napus haploids. One may therefore envisage that B. napus haploids, which arise spontaneously in wild populations at a frequency up to 6/1000 (Thompson 1969; Stringham and Downey 1973), could serve as a recurrent source of chromosomal rearrangements. The B. napus haploid × euploid cross could therefore be instrumental for both evolutionary and agricultural purposes by changing genomic structure (e.g., duplication of regions of interest), and so this would generate new intraspecific variability on which natural or human selection can act (Ramanna and Jacobsen 2003; Pires et al. 2004). More generally, our study points to the role that restituted gametes and sexual polyploidy can play in the genome evolution of nascent polyploids (Ramsey and Schemske 2002). Lukens et al. (2006) recently indicated that sequence loss occurred at very low frequency in the S0 generation of colchicine-doubled and spontaneously doubled resynthesized B. napus polyploids. It would be worthwhile to test if more numerous rearrangements are triggered when the S0-resynthesized B. napus polyploids originate from restituted gametes produced by the A × C hybrids. Finally, the fact that homeologous recombination is sharply increased in haploids make these plants even more attractive models to gain insights into the genetic and genomic factors that influence the rate and pattern of intergenomic recombination. A key step will be to compare homeologous recombination frequencies between genotypes that have been shown to display different meiotic behaviors at MI (Jenczewski et al. 2003).
Acknowledgments
We thank Jean-Claude Letanneur for his great contribution to the production of plant material, Tomasz Ksiazczyk and Jolanta Maluszynska (University of Silesia, Poland) for providing a stab of BoB014O06, and Maria Manzanares-Dauleux [Unité Mixte de Recherche (UMR), Institut National de la Recherche Agronomique (INRA)-Agrocampus, France] for providing HDEM seeds. We greatly thank Harry Belcram and Karine Budin for developing and screening of physical functional markers and Genoplante for funding their development. Karine Alix (UMR de Génétique Végétale du Moulon, France) and Mathilde Grelon and Christine Mézard (INRA Versailles, France), as well as two anonymous referees, are gratefully acknowledged for their critical reading and valuable comments on the manuscript. Stéphane Nicolas is supported by a Centre Technique Interprofessionnel des Oléagineux Métropolitain and INRA-Génétique et Amélioration des Plantes fellowship and this work is supported by a grant from INRA and Agence Nationale de la Recherche (Biodiversité: ANR-05-BDIV-015).
References
- Adams, K. L., R. Percifield and J. F. Wendel, 2004. Organ-specific silencing of duplicated genes in a newly synthesized cotton allotetraploid. Genetics 168: 2217–2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong, K. C., and W. A. Keller, 1981. Chromosome pairing in haploids of Brassica campestris. Theor. Appl. Genet. 59: 49–52. [DOI] [PubMed] [Google Scholar]
- Armstrong, K. C., and W. A. Keller, 1982. Chromosome pairing in haploids of Brassica oleracea. Can. J. Genet. Cytol. 24: 735–739. [DOI] [PubMed] [Google Scholar]
- Attia, T., and G. Röbbelen, 1986. Meiotic pairing in haploids and amphihaploids of spontaneous versus synthetic origin in rape, Brassica napus L. Can. J. Genet. Cytol. 28: 330–334. [Google Scholar]
- Baack, E. J., K. D. Whitney and L. H. Rieseberg, 2005. Hybridization and genome size evolution: timing and magnitude of nuclear DNA content increases in Helianthus homoploid hybrid species. New Phytol. 167: 623–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennetzen, J. L., 2000. Transposable element contributions to plant gene and genome evolution. Plant Mol. Biol. 42: 251–269. [PubMed] [Google Scholar]
- Chantret, N., J. Salse, F. Sabot, S. Rahman, A. Bellec et al., 2005. Molecular basis of evolutionary events that shaped the Hardness locus in diploid and polyploid wheat species (Triticum and Aegilops). Plant Cell 17: 1033–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coghlan, A., E. E. Eichler, S. G. Oliver, A. H. Paterson and L. Stein, 2005. Chromosome evolution in eukaryotes: A multi-kingdom perspective? Trends Genet. 21: 673–682. [DOI] [PubMed] [Google Scholar]
- Cronn, R., R. L. Small, T. Haselkorn and J. F. Wendel, 2003. Cryptic repeated genomic recombination during speciation in Gossypium gossypioides. Evolution 57: 2475–2489. [DOI] [PubMed] [Google Scholar]
- Delourme, R., C. Falentin, V. Huteau, V. Clouet, R. Horvais et al., 2006. Genetic control of oilcontent in oilseed rape (Brassica napus L.). Theor. Appl. Genet. 113: 1331–1345. [DOI] [PubMed] [Google Scholar]
- Devos, K. M., J. K. M. Brown and J. L. Bennetzen, 2002. Genome size reduction through illegitimate recombination counteracts genome expansion in Arabidopsis. Genome Res. 12: 1075–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eber, F., J. C. Letanneur and A. M. Chevre, 1997. Chromosome number of oilseed rape (B. napus L.)-wild radish (R. Raphanistrum) spontaneous hybrids and their progeny estimated by flow cytometry. Cruciferae Newsl. 19: 17–18. [Google Scholar]
- Esselink, G. D., H. Nybom and B. Vosman, 2004. Assignment of allelic configuration in polyploids using the MAC-PR (microsatellite DNA allele counting-peak ratios) method. Theor. Appl. Genet. 109: 402–408. [DOI] [PubMed] [Google Scholar]
- Everitt, B. S., and D. J. Hand, 1981. Finite Mixture Distributions. Chapman & Hall, London.
- Feldman, M., B. Liu, G. Segal, S. Abbo, A. A. Levy et al., 1997. Rapid elimination of low-copy DNA sequences in polyploid wheat: a possible mechanism for differentiation of homoeologous chromosomes. Genetics 147: 1381–1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foisset, N., R. Delourme, P. Barret, N. Hubert, B. S. Landry et al., 1996. Molecular-mapping analysis in Brassica napus using isozyme, RAPD and RFLP markers on a doubled haploid progeny. Theor. Appl. Genet. 93: 1017–1025. [DOI] [PubMed] [Google Scholar]
- Fourmann, M., P. Barret, N. Froger, C. Baron, F. Charlot et al., 2002. From Arabidopsis thaliana to Brassica napus: development of amplified consensus genetic markers (ACGM) for construction of a gene map. Theor. Appl. Genet. 105: 1196–1206. [DOI] [PubMed] [Google Scholar]
- Friebe, B., P. Zhang, G. Linc and B. S. Gill, 2005. Robertsonian translocations in wheat arise by centric misdivision of univalents at anaphase I and rejoining of broken centromeres during interkinesis of meiosis II. Cytogenet. Genet. Res. 109: 293–297. [DOI] [PubMed] [Google Scholar]
- Guo, M., D. Davis and J. A. Birchler, 1996. Dosage effects on gene expression in a maize ploidy series. Genetics 142: 1349–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inaba, R., and T. Nishio, 2002. Phylogenetic analysis of Brassiceae based on the nucleotide sequences of the S-locus related gene, SLR1. Theor. Appl. Genet. 105: 1159–1165. [DOI] [PubMed] [Google Scholar]
- Jenczewski, E., F. Eber, A. Grimaud, S. Huet, M. O. Lucas et al., 2003. PrBn, a major gene controlling homeologous pairing in oilseed rape (Brassica napus) haploids. Genetics 164: 645–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffreys, A. J., and C. A. May, 2004. Intense and highly localized gene conversion activity in human meiotic crossover hot spots. Nat. Genet. 36: 151–156. [DOI] [PubMed] [Google Scholar]
- Leflon, M., F. Eber, J. C. Letanneur, L. Chelysheva, O. Coriton et al., 2006. Pairing and recombination at the meiosis of Brassica rapa (AA) x Brassica napus (AACC) hybrids. Theor. Appl. Genet. 113: 1467–1480. [DOI] [PubMed] [Google Scholar]
- Leitch, I. J., and M. D. Bennett, 2004. Genome downsizing in polyploid plants. Biol. J. Linn. Soc. 82: 651–663. [Google Scholar]
- Lombard, V., and R. Delourme, 2001. A consensus linkage map for rapeseed (Brassica napus L.): construction and integration of three individual maps from DH populations. Theor. Appl. Genet. 103: 491–507. [Google Scholar]
- Lowe, A. J., C. Moule, M. Trick and K. J. Edwards, 2004. Efficient large-scale development of microsatellites for marker and mapping applications in Brassica crop species. Theor. Appl. Genet. 108: 1103–1112. [DOI] [PubMed] [Google Scholar]
- Lukens, L. N., P. A. Quijada, J. Udall, J. C. Pires, M. E. Schranz et al., 2004. Genome redundancy and plasticity within ancient and recent Brassica crop species. Biol. J. Linn. Soc. 82: 665–674. [Google Scholar]
- Lukens, L. N., J. C. Pires, E. Leon, R. Vogelzang, L. Oslach et al., 2006. Patterns of sequence loss and cytosine methylation within a population of newly resynthesized Brassica napus allopolyploids. Plant Physiol. 140: 336–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lysak, M. A., M. A. Koch, A. Pecinka and I. Schubert, 2005. Chromosome triplication found across the tribe Brassiceae. Genome Res. 15: 516–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lysak, M. A., A. Berr, A. Pecinka, R. Schmidt, K. Mcbreen et al., 2006. Mechanisms of chromosome number reduction in Arabidopsis thaliana and related Brassicaceae species. Proc. Natl. Acad. Sci. USA 103: 5224–5229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mcclintock, B., 1953. Induction of instability at selected loci in maize. Genetics 38: 579–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery, E. A., S. M. Huang, C. H. Langley and B. H. Judd, 1991. Chromosome rearrangement by ectopic recombination in Drosophila melanogaster: genome structure and evolution. Genetics 129: 1085–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales, M., G. Orjeda, C. Nieto, H. Van Leeuwen, A. Monfort et al., 2005. A physical map covering the nsv locus that confers resistance to melon necrotic spot virus in melon (Cucumis melo L). Theor. Appl. Genet. 111: 914–922. [DOI] [PubMed] [Google Scholar]
- Morinaga, T., and E. Fukushima, 1933. Karyological studies on a spontaneous haploid mutant of Brassica napella. Cytologia 4: 457–460. [Google Scholar]
- Olsson, G., and A. Hagberg, 1955. Investigations on haploid rape. Hereditas 41: 227–237.
- Osborn, T. C., D. V. Butrulle, A. G. Sharpe, K. J. Pickering, I. A. P. Parkin et al., 2003. Detection and effects of a homeologous reciprocal transposition in Brassica napus. Genetics 165: 1569–1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer, J. D., C. R. Shields, D. B. Cohen and T. J. Orton, 1983. Chloroplast DNA evolution and the origin of amphidiploid Brassica species. Theor. Appl. Genet. 65: 181–189. [DOI] [PubMed] [Google Scholar]
- Parker, J. S., R. W. Palmer, M. A. F. Whitehorn and L. A. Edgar, 1982. Chiasma frequency effects of structural chromosome change. Chromosoma 85: 673–686. [Google Scholar]
- Parkin, I. A. P., A. G. Sharpe, D. J. Keith and D. J. Lydiate, 1995. Identification of the A and C genomes of amphidiploid Brassica napus (oilseed rape). Genome 38: 1122–1131. [DOI] [PubMed] [Google Scholar]
- Parkin, I. A. P., A. G. Sharpe and D. J. Lydiate, 2003. Patterns of genome duplication within the Brassica napus genome. Genome 46: 291–303. [DOI] [PubMed] [Google Scholar]
- Parkin, I. A. P., S. M. Gulden, A. G. Sharpe, L. Lukens, M. Trick et al., 2005. Segmental structure of the Brassica napus genome based on comparative analysis with Arabidopsis thaliana. Genetics 171: 765–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrov, D. A., Y. T. Aminetzach, J. C. Davis, D. Bensasson and A. E. Hirsh, 2003. Size matters: non-LTR retrotransposable elements and ectopic recombination in Drosophila. Mol. Biol. Evol. 20: 880–892. [DOI] [PubMed] [Google Scholar]
- Piquemal, J., E. Cinquin, F. Couton, C. Rondeau, E. Seignoret et al., 2005. Construction of an oilseed rape (Brassica napus L.) genetic map with SSR markers. Theor. Appl. Genet. 111: 1514–1523. [DOI] [PubMed] [Google Scholar]
- Pires, J. C., J. W. Zhao, M. E. Schranz, E. J. Leon, P. A. Quijada et al., 2004. Flowering time divergence and genomic rearrangements in resynthesized Brassica polyploids (Brassicaceae). Biol. J. Linn. Soc. 82: 675–688. [Google Scholar]
- Prieto, J. L., N. Pouilly, E. Jenczewski, J. M. Deragon and A. M. Chevre, 2005. Development of crop-specific transposable element (SINE) markers for studying gene flow from oilseed rape to wild radish. Theor. Appl. Genet. 111: 446–455. [DOI] [PubMed] [Google Scholar]
- R Development Core Team, 2005. R: A Language and Environment for Statistical Computing, Version 2.2.1. R Foundation for Statistical Computing, Vienna (http://www.R-project.org).
- Ramanna, M. S., and E. Jacobsen, 2003. Relevance of sexual polyploidization for crop improvement: a review. Euphytica 133: 3–8. [Google Scholar]
- Ramsey, J., and D. W. Schemske, 2002. Neopolyploidy in flowering plants. Annu. Rev. Ecol. Syst. 33: 589–639. [Google Scholar]
- Rana, D., T. Boogaart, C. M. O'Neill, L. Hynes, E. Bent et al., 2004. Conservation of the microstructure of genome segments in Brassica napus and its diploid relatives. Plant J. 40: 725–733. [DOI] [PubMed] [Google Scholar]
- Renard, M., and F. Dosba, 1980. Etude de l'haploidie chez le colza (Brassica napus L. var oleifera Metzger). Ann. Amélior. Plant. 30: 191–209. [Google Scholar]
- Rieseberg, L. H., 2001. Chromosomal rearrangements and speciation. Trends Ecol. Evol. 16: 351–358. [DOI] [PubMed] [Google Scholar]
- Rieseberg, L. H., C. Vanfossen and A. M. Desrochers, 1995. Hybrid speciation accompanied by genomic reorganization in wild sunflowers. Nature 375: 313–316. [Google Scholar]
- Rieseberg, L. H., O. Raymond, D. M. Rosenthal, Z. Lai, K. Livingstone et al., 2003. Major ecological transitions in wild sunflowers facilitated by hybridization. Science 301: 1211–1216. [DOI] [PubMed] [Google Scholar]
- Sharpe, A. G., I. A. P. Parkin, D. J. Keith and D. J. Lydiate, 1995. Frequent nonreciprocal translocations in the amphidiploid genome of oilseed rape (Brassica napus). Genome 38: 1112–1121. [DOI] [PubMed] [Google Scholar]
- Sillito, D., I. A. P. Parkin, R. Mayerhofer, D. J. Lydiate and A. G. Good, 2000. Arabidopsis thaliana: a source of candidate disease-resistance genes for Brassica napus. Genome 43: 452–460. [DOI] [PubMed] [Google Scholar]
- Siroky, J., J. Zluvova, K. Riha, D. Shippen and B. Vyskot, 2003. Rearrangements of ribosomal DNA clusters in late generation telomerase-deficient Arabidopsis. Chromosoma 112: 116–123. [DOI] [PubMed] [Google Scholar]
- Snowdon, R. J., W. Köhler, W. Friedt and A. Kölher, 1997. Genomic in situ hybridisation in Brassica amphidiploids and hybrids. Theor. Appl. Genet. 95: 1320–1324. [Google Scholar]
- Song, K., and T. C. Osborn, 1992. Polyphyletic origins of Brassica napus: new evidence based on organelle and nuclear RFLP analyses. Genome 35: 992–1001. [Google Scholar]
- Song, K. M., P. Lu, K. L. Tang and T. C. Osborn, 1995. Rapid genome change in synthetic polyploids of brassica and its implications for polyploid evolution. Proc. Natl. Acad. Sci. USA 92: 7719–7723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, R., and J. Messing, 2003. Gene expression of a gene family in maize based on noncollinear haplotypes. Proc. Natl. Acad. Sci. USA 100: 9055–9060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stebbins, G. L., 1971. Chromosomal Evolution in Higher Plants. Addison-Wesley, Reading, MA.
- Stringham, G. R., and R. K. Downey, 1973. Haploid frequencies in Brassica napus. Can. J. Plant Sci. 53: 229–231. [Google Scholar]
- Sybenga, J., 1975. Meiotic Configurations. Springer-Verlag, Berlin.
- Tai, W., and H. Ikonen, 1988. Incomplete bivalent pairing in dihaploids of Brassica napus L. Genome 30: 450–457. [Google Scholar]
- Thompson, K. F., 1969. Frequencies of haploids in spring oilseed rape (Brassica napus). Heredity 24: 318–319. [Google Scholar]
- Town, C. D., F. Cheung, R. Maiti, J. Crabtree, B. J. Haas et al., 2006. Comparative genomics of Brassica oleracea and Arabidopsis thaliana reveal gene loss, fragmentation, and dispersal after polyploidy. Plant Cell 18: 1348–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truco, M. J., J. Hu, J. Sadowski and C. F. Quiros, 1996. Inter- and intra-genomic homology of the Brassica genomes: implications for their origin and evolution. Theor. Appl. Genet. 93: 1225–1233. [DOI] [PubMed] [Google Scholar]
- U, N., 1935. Genomic analysis in Brassica with special reference to the experimental formation of B. napus and peculiar mode of fertilization. Jpn. J. Bot. 7: 389–452. [Google Scholar]
- Udall, J. A., P. A. Quijada and T. C. Osborn, 2005. Detection of chromosomal rearrangements derived from homeologous recombination in four mapping populations of Brassica napus L. Genetics 169: 967–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veitia, R. A., 2005. Paralogs in polyploids: One for all and all for one? Plant Cell 17: 4–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendel, J. F., 2000. Genome evolution in polyploids. Plant Mol. Biol. 42: 225–249. [PubMed] [Google Scholar]
- Wolfe, K. H., 2001. Yesterday's polyploids and the mystery of diploidization. Nat. Rev. Genet. 2: 333–341. [DOI] [PubMed] [Google Scholar]
- Yang, T.-J., J. S. Kim, S.-J. Kwon, K.-B. Lim, B.-S. Choi et al., 2006. Sequence-level analysis of the diploidization process in the triplicated FLOWERING LOCUS C region of Brassica rapa. Plant Cell 18: 1339–1347. [DOI] [PMC free article] [PubMed] [Google Scholar]