Abstract
The med-1 and med-2 genes encode small, highly similar proteins related to GATA-type transcription factors and have been proposed as necessary for specification of both the mesoderm and the endoderm of Caenorhabditis elegans. However, we have previously presented evidence that neither maternal nor zygotic expression of the med-1/2 genes is necessary to specify the C. elegans endoderm. Contradicting our conclusions, a recent report presented evidence, based on presumed transgene-induced cosuppression, that the med-1/2 genes do indeed show an endoderm-specifying maternal effect. In this article, we reinvestigate med-2(−); med-1(−) embryos using a med-2- specific null allele instead of the chromosomal deficiences used previously and confirm our previous results: the large majority (∼84%) of med-2(−); med-1(−) embryos express gut granules. We also reinvestigate the possibility of a maternal med-1/2 effect by direct injection of med dsRNA into sensitized (med-deficient) hermaphrodites using the standard protocol known to be effective in ablating maternal transcripts, but again find no evidence for any significant maternal med-1/2 effect. We do, however, show that expression of gut granules in med-1/2-deficient embryos is exquisitely sensitive to RNAi against the vacuolar ATPase-encoding unc-32 gene [present on the same multicopy med-1(+)-containing transgenic balancer used in support of the maternal med-1/2 effect]. We thus suggest that the experimental evidence for a maternal med-1/2 effect should be reexamined and may instead reflect cosuppression caused by multiple transgenic unc-32 sequences, not med sequences.
THE med-1 and med-2 genes encode a redundant pair of highly similar GATA-factor-related zinc-finger proteins that together are crucial for the early development of the Caenorhabditis elegans embryo (Maduro et al. 2001). The original model for med-1/2 action (Maduro et al. 2001) was that both genes were strictly zygotic, acting downstream of the maternally transcribed but zygotically translated transcription factor SKN-1 and essential for specifying E and MS fate (where the E and MS blastomeres give rise to all of the worm endoderm and much of the worm mesoderm, respectively). In this article, we are concerned only with the question of whether med-1/2 are indeed necessary to specify the C. elegans endoderm.
The evidence that med-1/2 are essential for endoderm specification was based on two different protocols for administering med-1/2 RNA interference (RNAi) (Maduro et al. 2001). In the first protocol, embryos were collected 7–9 hr following injection of concentrated double-stranded RNA (dsRNA) into the maternal gonad; the rationale for such an early and limited observation period was to more effectively target the transient expression of the med-1/2 genes in the early embryo (Maduro et al. 2001; Coroian et al. 2006). In the more standard protocol, the RNAi effect is found to be maximally effective from 1 to 3 days following injection (Fire et al. 1998; Zipperlen et al. 2001; Goszczynski and McGhee 2005; Ahringer 2006); however, these more usual methods of administering RNAi show no effect with the med-1/2 genes (Kamath et al. 2003; Goszczynski and McGhee 2005; Sonnichsen et al. 2005). A second method of administering med-1/2 RNAi (Maduro et al. 2001) was by means of a transgenic heat-inducible promoter driving expression of double-stranded med RNA (Tavernarakis et al. 2000). Together, these two protocols were reported to produce a small number of embryos that arrested with a distinctive phenotype (approximately twofold elongation with absence of posterior pharynx); ∼50% of these arrested embryos failed to show the standard marker for endoderm specification, the presence of birefringent gut granules (Maduro et al. 2001; Coroian et al. 2006).
As reported in detail elsewhere (Goszczynski and McGhee 2005), we had only limited success producing med-type-arrested embryos by either of these two protocols. With the first protocol, we were able to produce fewer than one gut-granule-negative embryo for every two injected hermaphrodites, which is only marginally higher than that produced by injection of nonspecific control RNA (see also Coroian et al. 2006). In our hands, the second protocol produces either sterile mothers or embryos that arrest prior to the stage where they would ordinarily express gut granules. We thus proceeded with a more definitive test of the requirements of the med-1/2 genes for C. elegans endoderm specification, constructing worm strains segregating embryos that genetically lacked both med-1 and med-2 zygotic function. In these strains, the med-1 gene was removed by the gene-specific deletion ok804; the med-2 gene was removed by either of two chromosomal deficiencies, sDf127 or nDf16, both of which remove several hundred genes, including med-2. Our results were unexpected but clear: we found that only 16–17% or 0–3% of the doubly homozygous med-2(null); med-1(null) embryos did not express gut granules (depending on whether sDf127 or nDf16 was used to remove med-2). Both estimates were far lower than the >70% gut-granule-negative embryos predicted by the original model of Maduro et al. (2001). We thus concluded that the zygotic expression of the med-1 and med-2 genes could not play a major necessary role in specifying endoderm. Even though med transcripts cannot be detected in the maternal germline (Reinke et al. 2004), we nonetheless looked for (but did not find) any evidence for a possible maternal effect of the med-1/2 genes, using the standard protocol of dsRNA injection into sensitized med-1/2-deficient hermaphrodites. Using the same protocol even with nonsensitized strains, we routinely produce 100% arrested embryos when targeting the maternal skn-1 gene (Goszczynski and McGhee 2005) and 100% arrested larvae when targeting the relatively early zygotic gene elt-2 (Fukushige et al. 2005).
Two unresolved issues remained. First, what is the correct estimate for the percentage of gut-granule-negative med-2(−); med-1(−) embryos? If the 16–17% estimated with the sDf127-containing strain is correct (and hence the 0–3% estimated with the nDf16-containing strain is incorrect), can an “endoderm suppressor” be identified among the genes removed by nDf16 but not by sDf127? The second issue is that, in spite of our defense of chromosomal deficiencies (Goszczynski and McGhee 2005), use of gene-specific knockouts for both med-1 and med-2 would strengthen our conclusion that loss of the med-1/2 genes causes only a weakly impenetrant loss of endoderm.
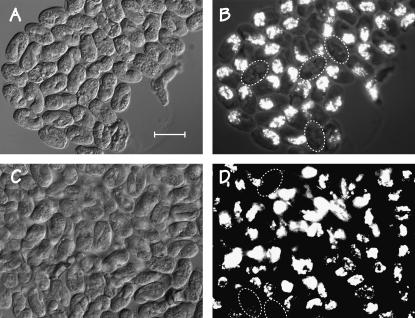
The recent availability of a Mos transposon insertion (Granger et al. 2004; Williams et al. 2005) into the med-2 gene (allele cxTi9744) provides a means to reassess the importance of the med-1/2 genes for specifying endoderm without relying on chromosomal deficiencies. Thus, we constructed the two balanced strains described in Table 1:JM142 [med-2(cxTi9744); + med-1(ok804)/lin-2 +)] lacks both copies of med-2(+) but has one copy of med-1(+), and JM143 [+ med-2(cxTi9744)/sma-3 +; med-1(ok804)] lacks both copies of med-1(+) but has one copy of med-2(+). Both med alleles are likely to be null (see footnote a to Table 1) and are hereafter referred to simply as med-1(−) or med-2(−). One-quarter of the embryos produced by either strain should be med-2(−); med-1(−) and should arrest. This expectation is accurately met with strain JM142 but 42.2% of the embryos produced by JM143 arrest prior to hatching, suggesting a degree of med-2 haplo-insufficiency (verified semiquantitatively by PCR on arrested embryos). In other words, an embryo with only a single copy of any med gene has a higher probability of surviving if that single copy is med-1 (∼100%) rather than med-2 (∼66%). The morphology of the arrested embryos is similar for both strains (Figure 1, A and C): many of the embryos are arrested at the twofold stage with a morphology like that originally reported by Maduro et al. (2001); others have arrested earlier and can be grossly vacuolated.
TABLE 1.
The majority (>80%) of med-2(−); med-1(−) embryos express the endoderm marker gut granules
| Strain | Genotypea | % arrested embryosb | Observed % gut-granule-negative arrested embryosc | % gut-granule- negative med-2(−); med-1(−) embryosd |
|---|---|---|---|---|
| JM142 | med-2(−); + med-1(−)/lin-2 + | 25.2 (111) | 14.1 (893) | 14.2 |
| JM143 | + med-2(−)/sma-3 +; med-1(−) | 42.2 (2338) | 10.2 (1652) | 17.3 |
| Average | 15.8 |
med-2(−) refers to Mos insertion allele med-2(cxTi9744), previously designated cxP9744, which was outcrossed five times; we confirmed the site and sequence of the insertion event. med-2(cxTi9744) is predicted to introduce a stop codon upstream of the MED-2 DNA-binding domain and is likely to be a null. The med-1(−) allele refers to deletion allele ok804, which removes the med-1-coding sequence and has now been outcrossed a total of five times. The markers used for balancing are lin-2(e1309) and sma-3(e491), located on cosmids adjacent to med-1 and med-2, respectively.
The Lin-2 phenotype can be slightly impenetrant so all the counts on percentage of arrest were performed on embryos produced by mothers whose genotype had been confirmed by PCR. The two strains med-2(−); lin-2 and sma-3; med-1(−) produce 1.3 and 3.4% arrested embryos, respectively (data not shown). Total number of scored embryos is shown in parentheses.
Birefringent gut granules were assayed as previously described (Goszczynski and McGhee 2005) by allowing balanced heterozygous mothers to lay eggs for several hours on a thin layer of seeded NGM agar poured on a microscope slide. After removal of mothers, embryos were incubated at 20° overnight; gut granules were scored by inspecting unhatched embryos with polarized light. In this manner, embryos are minimally manipulated and the possibility of selection of particular classes of embryos is avoided. The total number of arrested embryos scored is shown in parentheses.
Corrected by assuming that arrested embryos in excess of the expected 25% are all gut granule positive.
Figure 1.—
Images of arrested embryos produced by (A and B) strain JM142 [med-2(−); + med-1(−)/lin-2 +] and (C and D) strain JM143 [+ med-2(−)/sma-3 +; med-1(−)]. In A and C, differential interference contrast optics were used. In B and D, polarization optics reveal birefringent gut granules (Z-projection of four to five different focal planes). Bar, 50 μm. Dotted ellipses indicate arrested embryos that are gut granule negative. Embryos produced by JM143 appear more heterogenous (more than might be expected from the observed med-2 haplo-insufficiency) and more fragile than those produced by JM142.
Figure 1, B and D, show that only a minor fraction of the arrested embryos produced by either strain do not express gut granules, the standard assay for specified endoderm. After correcting for haplo-insufficiency (Table 1), the proportion of gut-granule-negative homozygous med-2(−); med-1(−) embryos is estimated at 14.2 and 17.3% for the two different strains, in excellent agreement with the 16–17.3% gut-granule-negative med-1/2-deficient embryos previously measured segregating from the sDf127-containing strain JM134 (Goszczynski and McGhee 2005). Thus, our combined results remain incompatible with the original model of Maduro et al. (2001, p. 481), which proposed that the med-1/2 genes “function downstream of SKN-1 in the EMS lineage and are essential to specify E (and MS) fates in any context.”
Our final estimate of ∼16% gut-granule-negative med-2(−); med-1(−) embryos differs significantly from the 0–3% gut-granule-negative med-2(−); med-1(−) embryos segregating from the nDf16-containing strain JM136 (Goszczynski and McGhee 2005). As noted above, this discrepancy raises the possibility that a potential “endoderm suppressor” resides among the ∼413 genes (excluding microRNAs) deleted by nDf16 but not by sDf127. We thus fed JM134 [dpy-17 sDf127 unc-32 III; sDp3 (III,f); med-1(−)] animals on individual Escherichia coli strains containing the 290 of these 413 genes that are present in the Ahringer RNAi library (Kamath et al. 2003). However, we were not able to identify any clone that could lower the proportion of gut-granule-negative embryos from the ∼16% seen with sDf127 to the <3% seen with nDf16.
As noted above, we had previously performed med RNAi in the sensitized strain JM134 [one copy of med-2(+) and no copies of med-1(+)] using the standard protocol that efficiently ablates maternal transcripts (Goszczynski and McGhee 2005). We found no significant increase in the proportion of gut-granule-negative embryos following injection of dsRNA targeting both med-1 and med-2, compared to injection of GFP dsRNA, and concluded that the med-1/2 genes do not show a maternal effect. However, Maduro et al. (2007) have recently made the opposite claim, namely that the med-1/2 genes show a previously unreported maternal rescue of endoderm specification (although apparently not maternal rescue of mesoderm specification). Maduro et al. (2007) did not address our RNAi results (Goszczynski and McGhee 2005) that contradict their proposal. However, because our previous results could possibly have been influenced by the use of chromosomal deficiencies and could always be criticized as being “negative” (i.e., no effect was observed, even though both positive and negative controls behaved as expected), we repeated our experiments using a strain carrying the two gene-specific med-1/2 nulls. med-1 dsRNA (which should target both med-1 and med-2 transcripts) was injected into JM143 [med-1(−); + med-2(−)/sma-3 +] mothers, GFP dsRNA was injected as a control, and arrested embryos were scored (blind) for expression of gut granules. The results are collected in Table 2 and our conclusions remain unchanged: we could find no significant evidence that the med-1/2 genes show a maternal effect.
TABLE 2.
RNAi against med-1 and med-2 has no significant effect but RNAi against unc-32 greatly increases the percentage of gut-granule-negative med-2(−); med-1(−) embryos
| Recipient strain | dsRNA injecteda | nb | % gut granule negative (±SD)c |
|---|---|---|---|
| JM143 [+ med-2(−)/sma-3 +; med-1(−)] | med-1 | 620 | 21.9 ± 5.8 |
| JM143 [+ med-2(−)/sma-3 +; med-1(−)] | GFP | 450 | 18.6 ± 6.6 |
| JM143 [+ med-2(−)/sma-3 +; med-1(−)] | unc-32 | 1144 | 83.6 ± 7.1 |
| N2 wild-type control | unc-32 | 1008 | 26.0 ± 3.4 |
med-1 dsRNA was injected at a concentration of ∼1 mg/ml by the protocol described previously (Goszczynski and McGhee 2005), which is highly effective against either maternal or zygotic transcripts; i.e., it produces 100% arrested embryos/larvae when skn-1/elt-2 dsRNA is injected (Fukushige et al. 2005; Goszczynski and McGhee 2005). Because of their high sequence similarity (Maduro et al. 2001), injection of med-1 dsRNA should also target med-2 transcripts. Integrity and concentration of dsRNA was verified by gel electrophoresis following the injection.
Total number of embryos scored.
Presence or absence of birefringent gut granules was scored blind in unhatched embryos. Results are presented as the average (± standard deviation) of the percentage of gut-granule-negative embryos scored in two independent experiments; a total of 19–20 injected hermaphrodites and five to six separate broods were collected between 1 and 3 days following injection. The average values have not been corrected for possible haplo-insufficiency of med-2. The 83.6 ± 7.1% gut-granule-negative embryos produced by injection of unc-32 dsRNA into strain JM143 is close to the 81.5% expected if unc-32 RNAi completely inhibits gut granule formation in med-2(−); med-1(−) homozygous embryos and in med-2(−)/med-2(+); med-1(−) heterozygous embryos and in only 26% of med-2(+); med-1(−) embryos (as in the wild-type controls).
We thus want to consider possible explanations for the differences between our results and those of Maduro et al. (2007). In spite of the large difference in interpretations and implications, the numerical differences between our observations are rather modest. We routinely observe that <20% of med-2(−); med-1(−) embryos do not express gut granules (Goszczynski and McGhee 2005; Table 1) and for all but two of their strains, Maduro et al. (2007) record a similar number. (Their numbers are usually slightly higher than ours, in the range of 20–25% gut granule negative; we ascribe no significance to this slight difference and suggest that it may be due to our gentler observation technique; see footnote c to Table 1.) We also note that when Maduro et al. (2007) assay embryos segregated from our strain JM134, they reproduce our results. Only two strains of Maduro et al. (2007), MS162 and MS247, produce med-2(−); med-1(−) embryos that are in the range of 40–50% gut granule negative. This ∼20–30% difference in the proportion of gut-granule-negative embryos is the basis for their conclusion that the med-1/2 genes show a maternal effect. We will propose an alternative explanation that is both simpler and experimentally supported.
The two strains in this study and Maduro et al. (2007) that can be most closely compared are our strain JM134 [dpy-17(e164) sDf127(s2428) unc-32(e189) III; sDp3(III,f); med-1(ok804) X], producing <20% gut-granule-negative embryos, and their strain MS162 [dpy-17(e164) sDf127(s2428) unc-32(e189) III; irDp1(III,f); med-1(ok804) X], producing 40–50% gut-granule-negative embryos. Doubly homozygous arrested embryos produced by either strain should have exactly the same genotype: dpy-17(e164) sDf127(s2428) unc-32(e189) III; med-1(ok804) X. The difference lies in the manner in which the two strains are balanced. Our strain JM134 is balanced by the well-characterized free duplication sDp3 (III,f) (Rosenbluth et al. 1985; Hedgecock and Herman 1995), which contains a single copy of the wild-type med-2 gene in its normal chromosomal context. Strain MS162 used by Maduro et al. (2007) is balanced by irDp1, a derivative of sDp3 containing a spontaneously integrated transgenic array composed of multiple copies of unc-119∷YFP, med-1(+) and a plasmid containing the wild-type unc-32 gene (Pujol et al. 2001). Thus, the key observation is that med-2(−); med-1(−) embryos segregating from irDp1-containing MS162 mothers are 40–50% gut granule negative whereas genotypically identical arrested embryos segregating from sDp3-containing JM134 mothers are <20% gut granule negative. Maduro et al. (2007) interpret this difference to mean that putative maternal med transcripts are being removed by the RNAi-related phenomenon called cosuppression (Dernburg et al. 2000; Robert et al. 2005) caused by the multiple transgenic copies of med-1(+) integrated into irDp1. However, Maduro et al. (2007) did not test their model by standard methods of med RNAi. In contrast, as we have noted above, we performed med RNAi in sensitized strains carrying only a single copy of a med gene under conditions that should ablate maternal transcripts, but the proportion of gut-granule-negative embryos is not significantly increased (Goszczynski and McGhee 2005; Table 2). Thus, the interpretation of Maduro et al. (2007) requires that the putative maternal med transcripts are somehow susceptible to cosuppression and susceptible to RNAi for only a narrow window of several hours following injection (Maduro et al. 2001), but are not susceptible to conventional (and usually much more powerful) RNAi effects that routinely persist for days.
We will now propose a simple alternative explanation for the difference between our results and those of Maduro et al. (2007). We suggest that cosuppression caused by the multiple transgenic copies of the unc-32 gene (not the med-1 gene) present on the irDp1 balancer is the reason that strain MS162 produces the higher levels (40–50%) of gut-granule-negative med-2(−); med-1(−) embryos. unc-32 is a complex locus with multiple transcripts expressed widely in the worm, including in the maternal germline and throughout the early embryo (Pujol et al. 2001); the null allele of unc-32 is associated with a strict maternal-effect lethality and the arrested embryos often show vacuolated intestines (Pujol et al. 2001) [unc-32(e189), present in both JM134 and MS162, is a weak allele primarily affecting the nervous system]. unc-32 encodes a subunit of a vacuolar ATPase involved in acidifying intracellular organelles (Pujol et al. 2001), and since gut granules are lysosome derivatives (Clokey and Jacobson 1986; Hermann et al. 2005), it seemed possible that unc-32-mediated cosuppression in the maternal germline could weaken the subsequent formation of embryonic gut granules. To test this possibility, we synthesized dsRNA corresponding to a portion of the unc-32 gene (completely included in the transgenic unc-32 sequences present on irDp1), injected it into strain JM143 [+ med-2(−)/sma-3 +; med-1(−)] hermaphrodites (with wild-type hermaphrodites as controls), and then assayed gut granule formation in the subsequently produced embryos. The results are collected in Table 2 and strongly support our hypothesis. The three key observations are: (i) unc-32 RNAi is effective and causes essentially complete embryonic arrest in both strains, as expected from the genetic experiments of Pujol et al. (2001); (ii) gut granule birefringence in arrested wild-type control embryos is markedly weakened such that ∼25% of the embryos are scored as gut granule negative; and (iii) gut granules in embryos produced by the med-deficient strain JM143 [+ med-2(−)/sma-3 +; med-1(−)] are far more sensitive to unc-32 RNAi than in control embryos. Over 80% of arrested JM143 unc-32 RNAi embryos were scored as gut granule negative. We do not know whether the unc-32 RNAi effect is directly on the formation of the gut granule marker (e.g., a block in lysosome maturation because of aberrant acidification) or is indirect because it causes early embryonic arrest; in either case, the end result would be the same, namely that the affected embryos would be scored as gut granule negative. Thus, because of the strong effect caused by unc-32 RNAi and because of the absence of any effect caused by med RNAi (Goszczynski and McGhee 2005; Table 2), we suggest that even mild cosuppression caused by the multiple transgenic unc-32 sequences present on irDp1 is a more likely explanation for the results of Maduro et al. (2007) than is cosuppression caused by med sequences.
One obvious experiment to distinguish between the two cosuppression-based explanations is to balance med-2(−); med-1(−) with a multicopy transgenic array containing med-1(+) sequences but not containing unc-32 sequences. Maduro et al. (2007) have already performed this experiment. Their strain MS290 (med-2(cxTi9744); med-1(ok804); Ex[med-1(+); unc-119∷CFP]) contains transgenic med-1(+) sequences but does not contain transgenic unc-32 sequences. Strain MS290 clearly segregates low levels (∼17%) of gut-granule-negative arrested embryos, agreeing precisely with our prediction. Maduro et al. (2007) find this result “unexpected” and suggest that there must be intrinsic differences in the cosuppression abilities of individual transgenic arrays. However, their explanation contradicts the findings of Dernburg et al. (2000), who showed that cosuppression by individual transgenic arrays, if it does occur, is highly reliable and reproducible.
The results of a second experiment performed by Maduro et al. (2007) also agree with our hypothesis but not with theirs. Maduro et al. (2007) identified eight MS290 hermaphrodites that had lost the Ex[med-1(+); unc-119∷CFP] balancing array from the maternal germline, as well as a single hermaphrodite from a different strain that had lost the balancing sDp3 duplication; in other words, none of these nine mothers should have any maternal wild-type med-1/2 genes in their germline. Again, contrary to the expectation of Maduro et al. (2007), these germline mosaic mothers produce arrested embryos that are only 27% gut granule negative, accepted by the authors as clearly in the low category. According to our model, this result is exactly what would be predicted, simply because the med genes show no maternal effect. In contrast, Maduro et al. (2007) introduce an unusual ad hoc hypothesis, namely that the putative med transcripts in the maternal germline are not actually produced in the germline but rather are imported from the anterior intestine [although these cells were not previously reported to express a med-1∷GFP transgene (Maduro et al. 2001)]. In defense of their hypothesis, Maduro et al. (2007) point out that such a somatic transport model could resolve a major discrepancy in their results, namely why an in situ hybridization signal ascribed to med transcripts in the maternal germline is ablated by SKN-1 RNAi when, as the authors point out, there is no evidence that SKN-1 functions in the maternal germline.
In summary, the results of this study completely confirm our previous conclusions (Goszczynski and McGhee 2005): the large majority (>80%) of embryos that lack both copies of med-1 and med-2 nonetheless still express markers of endoderm specification. Thus, the original model of Maduro et al. (2001) in which med-1/2 were the sole (or even the major) downstream effectors of SKN-1 in specifying endoderm must be ruled out. Our present results also contradict the revised model of Maduro et al. (2007) in which it is claimed that the med genes show a maternal effect. However, their claim is based on the assumption that a particular balancing array causes cosuppression involving multiple transgenic copies of the med-1 gene. We suggest that their results are more likely to be caused by cosuppression associated with multiple transgenic copies of the unc-32 gene present on the same balancing array; indeed, we show that expression of gut granules by med-deficient embryos appears exquisitely sensitive to unc-32 RNAi. This suggestion provides a much simpler explanation for several other experiments performed by Maduro et al. (2007) that otherwise require the introduction of ad hoc hypotheses.
If loss of both med-1/2 genes causes only a weakly penetrant loss of endoderm and if no maternal med-1/2 effect exists, what is the major regulatory pathway specifying endoderm? As originally suggested by Zhu et al. (1997) and as we had pointed out previously (Goszczynski and McGhee 2005), all evidence points to the SKN-1 transcription factor having the major direct role in specifying endoderm. In particular, Maduro et al. (2005) have shown that the activity of the end-1 promoter (thought to be critically involved in endoderm specification) is severely decreased (“++++” to “+”) when an upstream region containing multiple SKN-1 sites is deleted. In contrast, the short proximal end-1 promoter region containing MED-1/2 sites drives the low (“+”) residual level of end-1 activity.
We certainly do not question the major role that the med-1/2 genes must play in C. elegans development; the severely deranged morphology of med-2(−); med-1(−) embryos seen in Figure 1 is convincing evidence of their importance. Nonetheless, the fact remains that >80% of these arrested med-2(−); med-1(−) embryos still express endoderm markers and there is no convincing evidence for any med-1/2 maternal effect.
Acknowledgments
We thank Laurent Segalat (Université Lyon Claude Bernard) for providing the med-2 (cxTi9744) Mos insertion allele, the C. elegans Genetics Center (funded by the National Center for Research Resources) for providing several strains, and Paul Mains (University of Calgary) for critical reading of the manuscript. This work was supported by an operating grant from the Canadian Institutes of Health Research (CIHR) to J.D.M. V.V.C. received partial support from a CIHR Institute of Genetics training grant. J.D.M. is a Medical Scientist of the Alberta Heritage Foundation for Medical Research and a Canada Research Chair.
References
- Ahringer, J., 2006. Reverse Genetics, edited by The C. elegans Research Community. WormBook (doi/10.1895/wormbook.1.47.1, http://www.wormbook.org).
- Clokey, G. V., and L. A. Jacobson, 1986. The autofluorescent “lipofuscin granules” in the intestinal cells of Caenorhabditis elegans are secondary lysosomes. Mech. Ageing Dev. 35: 79–94. [DOI] [PubMed] [Google Scholar]
- Coroian, C., G. Broitman-Maduro and M. F. Maduro, 2006. Med-type GATA factors and the evolution of mesendoderm specification in nematodes. Dev. Biol. 289: 444–455. [DOI] [PubMed] [Google Scholar]
- Dernburg, A. F., J. Zalevsky, M. P. Colaiacovo and A. M. Villeneuve, 2000. Transgene-mediated cosuppression in the C. elegans germ line. Genes Dev. 14: 1578–1583. [PMC free article] [PubMed] [Google Scholar]
- Fire, A., S. Xu, M. K. Montgomery, S. A. Kostas, S. E. Driver et al., 1998. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391: 806–811. [DOI] [PubMed] [Google Scholar]
- Fukushige, T., B. Goszczynski, J. Yan and J. D. McGhee, 2005. Transcriptional control and patterning of the pho-1 gene, an essential acid phosphatase expressed in the C. elegans intestine. Dev. Biol. 279: 446–461. [DOI] [PubMed] [Google Scholar]
- Goszczynski, B., and J. D. McGhee, 2005. Reevaluation of the role of the med-1 and med-2 genes in specifying the Caenorhabditis elegans endoderm. Genetics 171: 545–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granger, L., E. Martin and L. Segalat, 2004. Mos as a tool for genome-wide insertional mutagenesis in Caenorhabditis elegans: results of a pilot study. Nucleic Acids Res. 32: e117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedgecock, E. M., and R. K. Herman, 1995. The ncl-1 gene and genetic mosaics of Caenorhabditis elegans. Genetics 141: 989–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermann, G. J., L. K. Schroeder, C. A. Hieb, A. M. Kershner, B. M. Rabbitts et al., 2005. Genetic analysis of lysosomal trafficking in Caenorhabditis elegans. Mol. Biol. Cell 16: 3273–3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath, R. S., A. G. Fraser, Y. Dong, G. Poulin, R. Durbin et al., 2003. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421: 231–237. [DOI] [PubMed] [Google Scholar]
- Maduro, M. F., M. D. Meneghini, B. Bowerman, G. Broitman-Maduro and J. H. Rothman, 2001. Restriction of mesendoderm to a single blastomere by the combined action of SKN-1 and a GSK-3beta homolog is mediated by MED-1 and -2 in C. elegans. Mol. Cell 7: 475–485. [DOI] [PubMed] [Google Scholar]
- Maduro, M. F., J. J. Kasmir, J. Zhu and J. H. Rothman, 2005. The Wnt effector POP-1 and the PAL-1/caudal homeoprotein collaborate with SKN-1 to activate C. elegans endoderm development. Dev. Biol. 285: 510–523. [DOI] [PubMed] [Google Scholar]
- Maduro, M. F., G. Broitman-Maduro, I. Mengarelli and J. H. Rothman, 2007. Maternal deployment of the embryonic SKN-1→MED-1,2 cell specification pathway in C. elegans. Dev. Biol. 301: 590–601. [DOI] [PubMed] [Google Scholar]
- Pujol, N., C. Bonnerot, J. J. Ewbank, Y. Kohara and D. Thierry-Mieg, 2001. The Caenorhabditis elegans unc-32 gene encodes alternative forms of a vacuolar ATPase a subunit. J. Biol. Chem. 276: 11913–11921. [DOI] [PubMed] [Google Scholar]
- Reinke, V., I. S. Gil, S. Ward and K. Kazmer, 2004. Genome-wide germline-enriched and sex-biased expression profiles in Caenorhabditis elegans. Development 131: 311–323. [DOI] [PubMed] [Google Scholar]
- Robert, V. J., T. Sijen, J. van Wolfswinkel and R. H. Plasterk, 2005. Chromatin and RNAi factors protect the C. elegans germline against repetitive sequences. Genes Dev. 19: 782–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbluth, R. E., C. Cuddeford and D. L. Baillie, 1985. Mutagenesis in Caenorhabditis elegans. II. A spectrum of mutational events induced with 1500 r of gamma-radiation. Genetics 109: 493–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnichsen, B., L. B. Koski, A. Walsh, P. Marschall, B. Neumann et al., 2005. Full-genome RNAi profiling of early embryogenesis in Caenorhabditis elegans. Nature 434: 462–469. [DOI] [PubMed] [Google Scholar]
- Tavernarakis, N., S. L. Wang, M. Dorovkov, A. Ryazanov and M. Driscoll, 2000. Heritable and inducible genetic interference by double-stranded RNA encoded by transgenes. Nat. Genet. 24: 180–183. [DOI] [PubMed] [Google Scholar]
- Williams, D. C., T. Boulin, A. F. Ruaud, E. M. Jorgensen and J. L. Bessereau, 2005. Characterization of Mos1-mediated mutagenesis in Caenorhabditis elegans: a method for the rapid identification of mutated genes. Genetics 169: 1779–1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, J., R. J. Hill, P. J. Heid, M. Fukuyama, A. Sugimoto et al., 1997. end-1 encodes an apparent GATA factor that specifies the endoderm precursor in Caenorhabditis elegans embryos. Genes Dev. 11: 2883–2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipperlen, P., A. G. Fraser, R. S. Kamath, M. Martinez-Campos and J. Ahringer, 2001. Roles for 147 embryonic lethal genes on C.elegans chromosome I identified by RNA interference and video microscopy. EMBO J. 20: 3984–3992. [DOI] [PMC free article] [PubMed] [Google Scholar]