Abstract
In the past decade, genome-sequencing projects have yielded a great amount of information on DNA sequences in several organisms. The release of the Drosophila melanogaster heterochromatin sequence by the Drosophila Heterochromatin Genome Project (DHGP) has greatly facilitated studies of mapping, molecular organization, and function of genes located in pericentromeric heterochromatin. Surprisingly, genome annotation has predicted at least 450 heterochromatic gene models, a figure 10-fold above that defined by genetic analysis. To gain further insight into the locations and functions of D. melanogaster heterochromatic genes and genome organization, we have FISH mapped 41 gene models relative to the stained bands of mitotic chromosomes and the proximal divisions of polytene chromosomes. These genes are contained in eight large scaffolds, which together account for ∼1.4 Mb of heterochromatic DNA sequence. Moreover, developmental Northern analysis showed that the expression of 15 heterochromatic gene models tested is similar to that of the vital heterochromatic gene Nipped-A, in that it is not limited to specific stages, but is present throughout all development, despite its location in a supposedly “silent” region of the genome. This result is consistent with the idea that genes resident in heterochromatin can encode essential functions.
HETEROCHROMATIN was originally defined cytologically as chromosomal regions that stained strongly at prophase and maintained a compact organization throughout all stages of the mitotic cell cycle (Heitz 1928). In a wide variety of eukaryotes, large chromosomal segments, or even entire chromosomes, can be composed of heterochromatin. Constitutive heterochromatin is a ubiquitous and abundant component of chromosomes of higher organisms, forming ∼5% of the genome in Arabidopsis thaliana, 30% in Drosophila, and 30% in humans, and up to 80% in certain nematodes and plants such as tomato (Moritz and Roth 1976; Peterson et al. 1998; Dimitri et al. 2005b). Despite these fluctuations in abundance, similar unusual structural properties characterize heterochromatin in virtually all animal and plant species, which together led to the view of heterochromatin as a “desert” of genetic functions (reviewed by John 1988). In the past 2 decades, however, genetic and molecular studies in Drosophila melanogaster have implicated constitutive heterochromatin in important cellular functions such as chromosome organization and inheritance and have shown it to contain genes required for viability and fertility (Gatti and Pimpinelli 1992; Williams and Robbins 1992; Weiler and Wakimoto 1995; Dernburg et al. 1996; Elgin 1996; Karpen et al. 1996; Eissenberg and Hilliker 2000; Henikoff et al. 2001; Coulthard et al. 2003; Dimitri et al. 2005b; Fitzpatrick et al. 2005). These genes were identified by recessive lethal mutations genetically linked to the heterochromatin of chromosomes 2 and 3 (Hilliker 1976; Marchant and Holm 1988) and were mapped to a specific region of mitotic heterochromatin (Dimitri 1991; Koryakov et al. 2002). Thus far, at least 32 essential genes have been mapped to the mitotic heterochromatin of chromosomes 2 and 3, but only a few of them—RpL5, light, concertina, rolled, RpL38, Nipped-B, Nipped-A, Parp, and RpL15—were satisfactorily characterized at the molecular level (Hilliker 1976; Devlin et al. 1990; Parks and Wieshous 1991; Biggs et al. 1994; Rollins et al. 1999; Tulin et al. 2002; Myster et al. 2004; Marygold et al. 2005; Schulze et al. et al. 2005). Notably, the presence of genes in heterochromatin, far from being a peculiarity of Drosophila, appears to be a conserved trait in the evolution of eukaryotic genomes. Heterochromatic genes have been recently identified in Saccharomyces cerevisiae, Schizosaccharomyces pombe, rice, A. thaliana, and humans (Kuln et al. 1991; Arabidopsis Genome Initiative 2000; Horvath et al. 2000; Brun et al. 2003; Nagaki et al. 2004).
The release of the sequence of D. melanogaster heterochromatin by the Berkeley Drosophila Genome Project has greatly facilitated the study of the molecular organization and function of heterochromatic genes. Initially, 3.8 Mb of ∼120 Mb of the D. melanogaster euchromatic genome sequence from release1 was found to correspond to heterochromatic sequences (Adams et al. 2000). Later, an improved whole-genome shotgun assembly (heterochomatic WGS3; Hoskins et al. 2002) was produced by the Drosophila Heterochromatin Genome Project (DHGP), which includes 20.7 Mb of draft-quality heterochromatic sequence. About 450 gene models (computational genes [CGs]) have been identified by the annotation of this sequence (Hoskins et al. 2002). In the recent release 5 of the D. melanogaster genome sequence, the number of genes predicted in heterochromatin was estimated to be even larger (DHGP; G. Karpen, personal communication). Several studies have concentrated on an effort to map gene models to the mitotic heterochromatin of D. melanogaster using BACs and P elements (Hoskins et al. 2002; Corradini et al. 2003; Yasuhara et al. 2003). However, the location of several heterochromatic gene models in the heterochromatin genome of D. melanogaster is still unclear and there is a need for further mapping studies.
To expand our current knowledge of genetic functions located in pericentromeric heterochromatin, we have performed detailed fluorescence in situ hybridization (FISH) mapping of 10 cDNA clones and two P-element insertions to mitotic and polytene chromosomes. The results of this analysis led to the cytological location in heterochromatin of eight large scaffolds, which together account for ∼1.4 Mb of sequence that contains 41 gene models. The results of this analysis also extend our understanding of the correspondence between mitotic and polytene chromosome heterochromatin. Finally, Northern analysis demonstrated that, similarly to known essential genes, gene models located in heterochromatin can be expressed throughout all development, despite their location in a supposedly “silent” region of the genome.
MATERIALS AND METHODS
Drosophila strains:
Genetic markers, mutations, and balancer chromosomes are described in Lindsley and Zimm (1992) and at FlyBase (http://www.flybase.org/). Cultures were maintained at 25° on standard cornmeal–sucrose–yeast–agar medium.
Cytology:
Mitotic and polytene chromosome preparations and FISH procedures were performed according to Dimitri (2004). The probes were labeled using a digoxigenin nick-translation kit (Boehringher Mannheim, Indianapolis); digoxigenin was revealed using an antidigoxigenin rhodamine-conjugated IgG as described in Dimitri (2004); plasmids carrying the cDNA inserts were used as probes.
cDNAs clones, gene models, and P-element insertions:
The cDNAs clones and heterochromatic gene models studied in this work are listed in Table 1. We chose 10 cDNAs from scaffolds containing at least two or more gene models whose cytological locations in heterochromatin were not yet determined or were still rough at the time that we started the experiments reported here. The complete sequences of the scaffolds are available at http://www.ncbi.nlm.nih.gov/. We mapped two P-element insertions: l(2)309 and EY09264. l(2)309 (Myster et al. 2004) are derived from KG01086 yellow+ and white+ SUPor-P element (Roseman et al. 1995). EY09264 is a EPgy2 P element carrying both the mini-white transgene, an intronless yellow transgene and the GAGA/Gal4-UAS enhancer. Both insertions are available at the Bloomington Stock Center.
TABLE 1.
Scaffolds, cDNAs, and gene models
| Scaffold (kb) | Location | cDNA clone | Gene model |
|---|---|---|---|
| AABU01002756 (199) | Unknown | RE24541 | CG12567 |
| CG40040 | |||
| CG40041 | |||
| SD19278 | CG40042 | ||
| AABU01002199 (160) | h39–h42 | CG40211 | |
| CG41264 | |||
| RE36623 | CG40218 | ||
| CG41265 | |||
| CG40220 | |||
| AABU01002549 (120) | h35–h46 | CG41063 | |
| h38–h41 | CG40190 | ||
| CG41066 | |||
| CG40192 | |||
| CG41065 | |||
| LD12604 | CG40191 | ||
| AABU01002750 (143) | Unknown | RE25729 | CG17691 |
| CG40068 | |||
| CG40388 | |||
| CG40390 | |||
| CG40067 | |||
| AABU01002711 (342) | 2R:41B | CG40130 | |
| CG40129 | |||
| CG17665 | |||
| CG40131 | |||
| GM16138 | CG40127 | ||
| CG40128 | |||
| CG40133 | |||
| RE37350 | CG17683 | ||
| AABU01002748 (134) | Unknown | CG40084 | |
| CG40081 | |||
| LD07633 | CG40080 (haspin) | ||
| CG40085 | |||
| CG41098 | |||
| AABU01002740 (252) | Unknown | CG17514 | |
| CG40103 | |||
| CG40163 | |||
| RE54950 | CG10837 (eIF-4B) | ||
| CG40105 | |||
| AABU01002752 (122) | Unknown | RE06111 | CG17419 |
| CG40065 | |||
| CG41101 |
Scaffold length is indicated in parentheses. Scaffold numbers and gene model annotations are according to release 5 sequence (http://www.dhgp.org; DHGP, unpublished results).
Inverse PCR, genomic DNA, and cDNA preparations:
Extraction of plasmids containing the cDNA inserts was performed as described in the protocol for midi and maxi plasmid kits (QIAGEN, Chatsworth, CA). Genomic DNA extraction and inverse PCR were performed according to the protocol of the Berkeley Drosophila Genome Project (http://www.fruitfly.org/about/methods/index.html). Primers were designed using the Primer3 program (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3.cgi). The following primers were used to amplify by inverse PCR the flanking sequences of the 19.74.3 PLacW insertion: FOR 5′-TCACTCGCACTT ATTGCA-3′ and REV 5′-TAACCCTTAGCATGTCCGTG-3′. PCR products were cleaned up using either the QIAquick PCR purification kit or the QIAquick Gel extraction kit (QIAGEN). Sequencing reactions were performed by CRIBI using an ABI 3730 DNA analyzer. Sequences were analyzed using Sequencher (Gene Codes, Ann Arbor, MI).
RNA preparations and Northern analysis:
Poly(A+) mRNA was partially purified from 200 mg material of different Drosophila developmental stages using the Quickprep mRNA purification kit (Amersham, Buckinghamshire, UK). The RNA was sized electrophoretically through a 1.2% agarose–formaldehyde gel, blotted to nylon membrane, and hybridized at 65° with 32P-labeled probes. The same filter was used with different probes after removing the hybridization signal by boiling in 0.1× SSC and checking for residual labeling. Autoradiographs were recorded with the Phosphorimager Storm 820 (Molecular Dynamics, Eugene, OR).
Microphotography:
Chromosome preparations were analyzed using a computer-controlled Zeiss Axioplan epifluorescence microscope equipped with a cooled CCD camera (Photometrics, Tucson, AZ). Fluorescence was visualized using the Pinkel no. 1 filter set combination (Chroma Technology, Brattleboro, VT). The fluorescent signals were recorded by IP Spectrum Lab Software and edited with Adobe PhotoShop 7.
RESULTS
FISH mapping to mitotic chromosomes:
FISH experiments with cDNA probes were performed to prometaphase chromosomes of third instar larvae neuroblasts from the y; cn bw sp strain used to generate the genome sequence (Adams et al. 2000). The results of this analysis are shown in Figure 1 and Figure 2A. We initially undertook a detailed mapping of 10 cDNAs and two P-element insertions located in eight scaffolds whose cytological locations were not yet determined or were still approximate (Table 1; see supplemental Table 1 in Hoskins et al. 2002; http://www.dhgp.org). Together, according to the recent release 5 (http://www.dhgp.org/; DHGP, unpublished results), the scaffolds defined by cDNAs carried 41 annotated gene models and covered ∼1.4 Mb of heterochromatic DNA (Table 1).
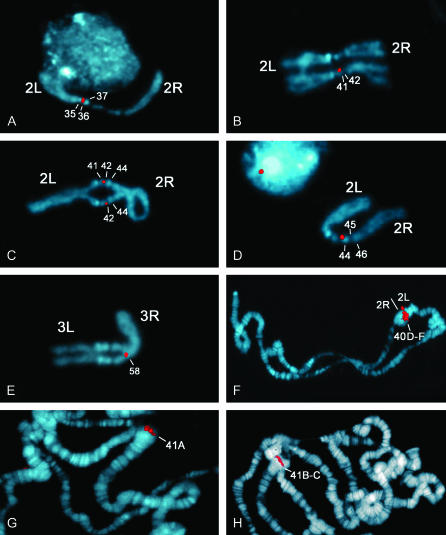
Figure 1.—
Examples of FISH mapping of cDNA probes to the mitotic and polytene heterochromatin of chromosome 2. Prometaphase chromosomes of larval brain cells and salivary gland polytene from the y; cn bw sp strain were stained with DAPI and pseudocolored in blue; cDNA signals were pseudocolored in red. (A) The RE24541 cDNA maps to the distal h35–proximal h36 of 2Lh. (B) The RE36623 cDNA signal maps to h41 of 2Rh; only one homolog is marked. (C) The LD12604 cDNA maps to h41 distal–h42 proximal. (D) The hybridization signal of RE25729 cDNA is located in the proximal edge of h44. (E) The hybridization signal of RE06111 maps to h58 in the distal heterochromatin of 3R; (F) The RE24541 cDNA signal maps to region 40D-F of the polytene chromosomes. (G) Multiple dots at 41A formed by the hybridization signal of RE36623 cDNA. (H) The probe containing a mixture of both GM16138 and RE37350 cDNAs maps to division 41B-C.
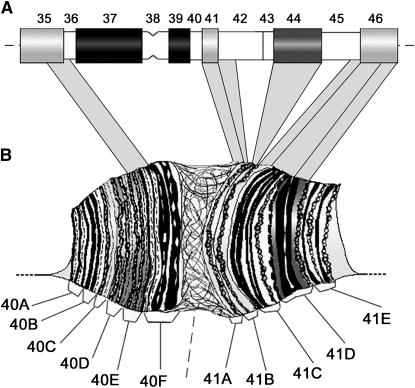
Figure 2.—
Diagram showing the mapping of cDNAs to the heterochromatin of mitotic and polytene chromosome 2. The mapping of X cDNA clones to mitotic (A) and polytene heterochromatin (B) is shown. Numbers from 35 to 46 indicate the cytological regions of the heterocromatin map of chromosme 2; numbers from 40 to 41 indicate different cytological divisions of the polytene chromosome map. Correspondences of cDNA localizations between mitotic and polytene chromosome regions are shown.
2Lh mitotic heterochromatin:
RE24541 and SD19278 cDNA clones from CG12567 and CG40042 gene models, respectively, were mapped to the mitotic heterochromatin of the left arm of chromosome 2 (2Lh). CG12567, CG40042, CG40040, and CG40041 are located in scaffold AABU01002756, which is ∼199 kb in length. The RE24541 cDNA produces a single FISH signal in heterochromatic bands h35 distal–h36 proximal (Figure 1A and Figure 2A). A similar result was obtained with SD19278. Thus, the FISH data indicate that AABU01002756 is located in region h35–h36.
2Rh mitotic heterochromatin:
Six cDNA clones were mapped to the mitotic heterochromatin of the right arm of chromosome 2 (2Rh). These cDNAs identify five different scaffolds: AABU01002199, AABU01002549, AABU01002750, AABU01002711, and AABU01002748, which together contain 29 gene models.
AABU01002199:
AABU01002199 (160 kb) carries five gene models: CG40211, CG41264, CG40218 (also called elica or Yeti), CG41265, and CG40220. This scaffold was previously localized to regions h40–h43 on the basis of cytological and molecular analysis of P-element insertion sites (Konev et al. 2003; see the list from the lab of G. Karpen at http://taputea.lbl.gov/research/het/HetPs/data/index.html and http://www.dhgp.org). To define the location of this scaffold more accurately, we performed FISH mapping using the RE36623 cDNA corresponding to CG4218. As shown in Figure 1B and Figure 2A, the RE36623 probe produced a weak, yet clear, fluorescence signal that maps to region h41 of 2Rh. This result is corroborated by our molecular analysis of the P-element insertion 19.74.3, which maps cytologically to h41 (Corradini et al. 2003). Inverse PCR and BLAST-n analysis of flanking sequences showed that the 19.74.3 insert is located in AABU01002199, ∼4 kb downstream of CG40218 (see materials and methods). Together, these data enabled us to restrict the localization of AABU01002199 with its five gene models to region h41.
AABU01002549:
AABU01002549 (120 kb) contains six gene models: CG41063, CG40190, CG41066, CG40192, CG41065, and CG40191. This scaffold was associated with the heterochromatin of the second chromosome (Drosophila Heterochromatin Genome Project 2004), yet no precise localization was defined. The LD12604 (CG40191) cDNA produced a single FISH signal in h41 distal–h42 proximal, thus mapping AABU01002549 in this position in 2Rh (Figure 1C and Figure 2A).
AABU01002750:
AABU01002750 (143 kb) contains five gene models: CG17691, CG40068, CG40388, CG40390, and CG40067. As shown in Figure 1D and Figure 2A, the RE25729 (CG17691) cDNA clone produced a sharp hybridization signal located in the proximal portion of h44, placing AABU01002750 in this region.
AABU01002711:
AABU01002711 (342 kb) contains eight gene models: CG40130, CG40129, CG17665, CG40131, CG40127, CG40128, CG40133, and CG17683. AABU01002711 was localized to h45 distal–h46 proximal (Figure 2A) using a mixture of both RE37350 (CG17683) and GM16138 (CG40127) cDNA clones as the probe. This result was confirmed by FISH mapping of the EY09264 P insertion (assigned to AABU01002711 by the Gene Disruption Project) to h45 distal–h46 proximal (data not shown).
AABU01002748:
AABU01002748 (134 kb) contains five gene models: CG40084, CG40081, CG40080, CG40085, and CG41098. Using the cDNA clone (LD07633) from CG40080, a single FISH signal was mapped to h45 distal–h46 proximal (Figure 2A), indicating that AABU01002748, like AABU0100271, spans this segment. This observation is in agreement with the finding that AABU01002748 and AABU01002711 are physically linked by the BAC map (DHGP; R. Hoskins and G. Karpen, personal communication).
Chromosome 3 mitotic heterochromatin:
Two scaffolds, AABU01002740 and AABU01002752, were mapped to the heterochromatin of chromosome 3. The RE54950 cDNA probe (CG10837) localized AABU01002740 (252 kb) with its five gene models (CG17514, CG40103, CG40163, CG10837, and CG40105) to region h48 of 3L (data not shown). The RE06111 cDNA clone (CG17419) mapped to region h58 of 3R heterochromatin (Figure 1E) and allowed us to localize AABU01002752 (122 kb) and its three genes (CG17419, CG40065, and CG41101) into this region.
FISH mapping of cDNAs to polytene chromosomes:
FISH experiments using the cDNAs tested on mitotic chromosomes were also performed with salivary gland polytene chromosomes of the y; cn bw sp isogenic strain. The aim of these experiments was to confirm the chromosomal location of the clones that were mapped to mitotic heterochromatin and to learn more about the relationship between mitotic and polytene chromosome heterochromatin. The results are shown in Figure 1, F, G, and H; Figure 2B). The SD19278 (CG12567) cDNA (scaffold AABU01002756; h35 distal–h36 proximal) produced a diffuse hybridization signal that maps to division 40D-F of 2Lh (Figure 1F and Figure 2B). A similar result was obtained with RE24541 (CG12567).
Six cDNAs were assigned to the proximal regions of the right arm of chromosome 2. The hybridization signal of RE36623 (CG40218) cDNA (scaffold AABU01002199; h41 of 2Rh) is formed by multiple dots that map to division 41A and span the inner portions of the chromocenter (Figure 1G and Figure 2B). Similarly, the signal of LD12604 (AABU01002549; h42 proximal) appears as a cluster of dots and maps to 41A (Figure 2B). The RE25729 (CG40191) cDNA (scaffold AABU01002750; h44 proximal) gave a clear fluorescence signal mapping to the distal portion of 41A (Figure 2B). This result apparently conflicts with a previous finding suggesting that the mitotic regions h43–h44 do not undergo polytenization in salivary glands (Koryakov et al. 1996). The probe containing a mixture of both GM16138 (CG40127) and RE37350 (CG17683) cDNAs (scaffold AABU01002711; h45 distal–h46 proximal) maps to 41B-C (Figure 1H and Figure 2B), consistent with previous mapping (supplemental Table 1 in Hoskins et al. 2002). These results extend and confim our previous study (Corradini et al. 2003), indicating that division 41C of salivary gland chromosomes originates from polytenization of DNA sequences located in h46, the most distal region of 2R mitotic heterochromatin. Finally, on chromosome 3, the FISH signal of RE54950 cDNA mapped to the inner portions of the 3L chromocenter; FISH with RE06111 (CG17419) cDNA (scaffold AABU01002752; h58 of 3Rh) produced a hybridization signal mapping to 81F.
BLAST analysis:
Next, we wanted to identify human homologs of D. melanogaster heterochromatic gene models and study their chromosomal location in humans. To this aim, the putative protein sequence of 98 gene models assigned to heterochromatin (Hoskins et al. 2002; Corradini et al. 2003; Yasuhara et al. 2003) was used as query for a BLAST-p search against the human protein database. The results of this analysis are shown in Table 2. The 14 genes located at the hetero-euchromatic transition region were also included in this group. The results of this analysis indicate that ∼70% of the putative proteins encoded by heterochromatin-linked gene models studied in this work share significant similarity with human proteins. It is worth noting that all conserved hits identify genes that are located in euchromatic regions of the human genome. In particular, among the human genes that have been mapped in detail (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=OMIM), 35% were found to be associated with G bands and 65% with R bands. The observed distribution may reflect the average gene distribution found in human chromosomes, in which R bands are more gene enriched compared to G bands.
TABLE 2.
Heterochromatic genes of D. melanogaster and evolutionary conservation of their protein products in humans
| Scaffold | Location | CG (alleles) | D. melanogaster molecular function (GO) | Closest BLASTp human hit (P(n)) |
|---|---|---|---|---|
| AABU1002768 | h35 | CG17540 (1) | Pre-mRNA splicing factor | NP_116294.1, RNA-binding motif protein 17 (2e-53) |
| CG41117 (1) | Unknown | No significant similarity found | ||
| CG18140 (1) | Chitin binding, chitinase | AAG60019.1, acidic mammalian chitinase precursor (8e-75) chitotriosidase 1e-17 | ||
| concertina (14) | GTPase; GTP binding; signal transducer | NP_006563.2, guanine nucleotide- binding protein (6e-108) | ||
| CG40005 (1) | GTPase | AAA74235.1, guanine nucleotide regulatory protein α 13 (3e-26) | ||
| light (47) | Ubiquitin-protein ligase; zinc ion binding | P49754, vacuolar assembly protein VPS41 homolog (S53) (3e-147) | ||
| CG41118 (1) | Unknown | No significant similarity found | ||
| CG17715 (1) | Unknown | AAH32396.1, LOC157378 hypothetical protein (8e-30) | ||
| CG41121 (1) | Unknown | No significant similarity found | ||
| CG40439 (1) | Unknown | No significant similarity found | ||
| CG41120 (1) | Unknown | No significant similarity found | ||
| CG40006 (1) | Serine-type endopeptidase inhibitor; receptor | AAH80647.1, scavenger receptor class B member 1 (8e-49) | ||
| CG17494 (1) | Unknown | NP_009090.2, sarcolemma-associated protein (3e-66) | ||
| CG17493 (1) | Calmodulin binding; calcium ion binding | AAP35920.1, centrin EF hand protein 2 (7e-62) caltractin 1e-62 | ||
| CG17490 (1) | Unknown | No significant similarity found | ||
| RpL5 (5) | Ribosomal protein L5 | NP_000960.2, ribosomal protein L5 (1e-119) | ||
| AABU01002756 | h35–h36 | CG12567 (1) | Thiamin diphosphokinase | No significant similarity found |
| CG40040 (1) | Unknown | No significant similarity found | ||
| CG40041 (1) | Hormone | AA033390.1, glycoprotein hormone β-subunit (7e-10) | ||
| CG40042 (1) | Carrier; protein transporter | AAR14723.1, mitochondrial inner membrane translocase 23 (5e-36) | ||
| AABU01002199 | h41 | CG40211 (1) | Unknown | No significant similarity found |
| CG41264 (1) | Unknown | No significant similarity found | ||
| CG40218 (2) | Kinesin binding | AAP88821.1, craniofacial development protein 1 (4e-13) | ||
| CG41265 (1) | Nucleic acid binding; damaged DNA binding | XP_371082.1, predicted: hypothetical protein FLJ20753 (6e-25) | ||
| CG40220 (1) | Unknown | No significant similarity found | ||
| AABU01001947 | h41 | CG40241 (1) | Unknown | No significant similarity found |
| CG12552 (1) | Unknown | No significant similarity found | ||
| CG40498 (1) | Unknown | No significant similarity found | ||
| CG40239 (1) | Unknown | No significant similarity found | ||
| rolled (90) | MAP kinase | AAH17832.1, mitogen activated protein kinase 1 (3e-171) | ||
| CG40244 (1) | Unknown | No significant similarity found | ||
| AABU01002549 | h41–h42 | CG41063 (1) | Unknown | No significant similarity found |
| CG40190 (1) | Protein kinase | CAD97888.1, hypothetical protein (2e-25) | ||
| CG41066 (1) | Unknown | No significant similarity found | ||
| CG40192 (1) | Unknown | No significant similarity found | ||
| CG41065 (1) | Unknown | No significant similarity found | ||
| CG40191 (1) | Unknown | AAD34052.1, CGI-57 protein (6e-22) | ||
| AABU01002750 | h44 | CG17691 (1) | 3 methyl-2-oxobutanoate dehydrogenase | CAI15049.1, branched chain keto acid dehydrogenase E1 (2e-140) |
| CG40068 (1) | Nucleic acid binding; translation factor | AAM70196.1, translation initiation factor 2 (5e-33) | ||
| CG40388 (1) | Unknown | No significant similarity found | ||
| CG40390 (1) | Unknown | No significant similarity found | ||
| CG40067 (1) | Unknown | No significant similarity found | ||
| AABU01002748 | h45–h46 | CG40084 (1) | Unknown | NP_951058.1, cyclin M2 isoform 2 (3e-124) |
| CG40081 (1) | Unknown | No significant similarity found | ||
| CG40080 (1) | ATP binding; protein serine/threonine kinase | NP_114171.2, serine/threonine- protein kinase Haspin (4e-69) | ||
| CG40085 (1) | Unknown | No significant similarity found | ||
| CG41098 (1) | Unknown | No significant similarity found | ||
| AABU01002711 | h45–h46 | CG40130 (1) | Unknown | No significant similarity found |
| CG40129 (1) | ATP binding; G-protein- coupled receptor kinase | NP_005151.1, β-adrenergic receptor kinase 2 (0.0) | ||
| CG17665 (2) | Unknown | NP_075391.3, TPA_exp: integrator complex subunit 3 (2e-148) | ||
| CG40131 (1) | Unknown | No significant similarity found | ||
| CG40127 (1) | Unknown | NP_001004333.1, hypothetical protein LOC440400 (2e-13) | ||
| CG40128 (1) | Unknown | No significant similarity found | ||
| CG40133 (1) | Unknown | No significant similarity found | ||
| CG17683 (1) | Oxidoreductase; ferredoxin hydrogenase | NP_071938.1, nuclear prelamin A recognition factor-like (1e-117) | ||
| AABU01002769 | h46 | RpL38 (6) | Ribosomal protein 38 | NP_000990.1, ribosomal protein L38 (7e-15) |
| CG40293 (1) | ATP binding; protein serine/threonine kinase | AAG48269.1, serologically defined breast cancer antigen NY-BR-96 8e-43 | ||
| CG40290 (1) | Unknown | No significant similarity found | ||
| p120ctn (12) | Adherens junction protein | AAB97957.1, arm-repeat protein NPRAP/neurojungin (2e-131) 1e-126 | ||
| CG17486 (1) | Ligase; asparagine synthase | AAX88843.1, unknown (7e-76) | ||
| CG17478 (1) | Unknown | No significant similarity found | ||
| Nipped-B (8) | Transcriptional activator; sister-chromatid cohesion | DAA05331.1, TPA: TPA_exp: transcriptional regulator (0.0) | ||
| CG17706 (2) | Unknown | No significant similarity found | ||
| CG17082 (1) | Unknown | AAI07417.1, Rho GTPase activating protein 18 (1e-31) | ||
| CG12547 (1) | Unknown | CAH73013.1, novel NHL repeat domain containing protein (1e-134) | ||
| CG17528 (1) | Microtubule binding; ATP binding | NP_004725.1, doublecortin and CaM kinase-like 1 (5e-132) | ||
| CG40285 (1) | Unknown | AAH47775.1, chromosome 11 open reading frame 46 (2e-13) | ||
| AE003788 | h46 | CR30260 | tRNA | |
| CR30505 | tRNA | |||
| CG33492 (1) | Ionotropic glutamate receptor | CAH73378.1, glutamate receptor, ionotropic, delta 1 (2e-07) | ||
| TpnC41C (1) | Calcium ion binding | AAH08437.1, calmodulin 2 (phosphorylase kinase, delta) (4e-25) | ||
| AE003787 | h46 | CG3107(1) | Metalloendopeptidase | AAC67244.1, metalloprotease 1 (0.0) |
| CG2944 (8) | Oocyte anterior/posterior axis determination | BAD92796.1, SPRY domain-containing SOCS box protein SSB-1 variant (7e-117) | ||
| CG3136 (2) | DNA binding; protein homodimerization | AAB47487.1, cAMP response-element- binding protein-related (3e-13) | ||
| Nipped-A (47) | Transcription regulator; cytokinesis | BAD92726.1, Transformation/transcription domain-associated protein variant (0.0) | ||
| CG2682(1) | Transcription factor; ubiquitin-protein ligase | NP_006259.1, D4, zinc and double PHD fingers family 2 (5e-49) | ||
| AE003787 | Hetero- euchromatin transition region | CG10392 (1) | Transferase activity, transferring glycosyl groups | NP_858059.1, O-linked GlcNAc transferase isoform 2 (0.0) |
| CG10465 (2) | Voltage-gated potassium channel; protein binding | BAB55188.1, unnamed protein product (9e-100) | ||
| CG10395 (1) | HIT zinc-finger protein domain | NP_112578.2, high-mobility group AT-hook 1-like 4 (5e-14) | ||
| CG30441 (1) | Unknown | AAP50265.1, intraflagellar transport protein 20-like protein (9e-12) | ||
| CG10396 (1) | Cytochrome-c oxidase | AAH62437.1, cytochrome c oxidase subunit IV isoform 1 (1e-23) | ||
| CG10417 (1) | Protein serine/threonine phosphatase | BAD92434.1, protein phosphatase 1G variant (1e-61) | ||
| AE003786 | Hetero- euchromatin transition region | CG30437 (7) | Laccase; copper ion binding | No significant similarity found |
| CG32838 (1) | Nother laccase; 75% identical to CG30437 | AAQ24260.1, MY 01 protein (2e-04) | ||
| CG30440 (1) | Guanyl-nucleotide exchange factor | CAI39715.1, MCF.2 cell-line-derived transforming sequence (3e-77) | ||
| CG30438 (1) | Transferase activity, transferring glycosyl groups | AAC51187.1, ceramide UDP galactosyltransferase (5e-68) | ||
| TpnC4 (1) | Calcium ion binding | AAH08437.1, calmodulin 2 (7e-25) | ||
| CG17510 (1) | Unknown | NP_057152.1, tetratricopeptide repeat domain 11 (5e-12) | ||
| CG17508 (1) | Unknown | CAC12716.1, C20orf108 (7e-24) | ||
| CG11665 (1) | Monocarboxylic acid transporter | CAI12383.1, solute carrier family 16 (3e-33) | ||
| AABU01002740 | h48 | CG17514 (1) | Translation activator; kinase egulator | NP_006827.1, GCN1 general control of amino-acid synthesis 1-like 1 (0.0) |
| CG40103 (1) | Unknown | No significant similarity found | ||
| CG40163 (1) | Unknown | No significant similarity found | ||
| CG10837 (eIF-4B) (8) | RNA binding; translation initiation factor | CAA39265.1, initation factor 4B (2e-15) | ||
| CG40105 (1) | Specific RNA pol II transcription factor | CAI14825.1, ventral anterior homeobox 1 (1e-10) | ||
| AABU01002752 | h58 | CG17419 (1) | Receptor signaling protein; zinc ion binding | BAA86569.2, KIAA1255 protein (0.0) |
| CG40065 (1) | Unknown | No significant similarity found | ||
| CG41101 (1) | Unknown | No significant similarity found |
In the scaffold column, length is indicated in parentheses; in the gene column, the number of alleles is indicated in parentheses. Only BLASTp hits with e < 10−3 were selected. The locations refer to the mitotic heterochromatin map. The cytological border of the hetero-euchromatin transition region was established by FISH mapping of BACs (Corradini et al. 2003) and is approximate; some of the genes assigned to this region may be actually located in heterochromatin. Scaffolds numbers and gene model annotations were according to release 5 sequence (http://www.dhgp.org; DHGP, unpublished results).
Northern blotting analysis of heterochromatic gene models in the wild-type Oregon-R strain of D. melanogaster:
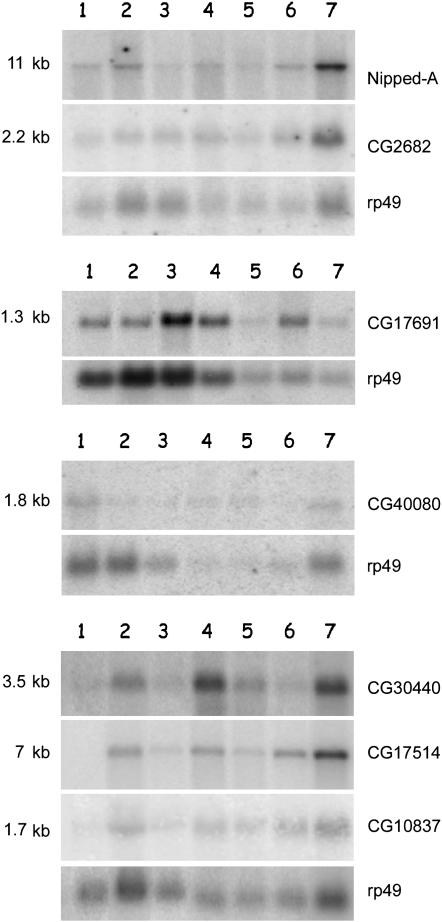
To learn more about the function of the genes resident in heterochromatin, we investigated transcription of the vital gene Nipped-A and of the following 15 gene models: CG12567 and CG40042 of 2Lh, CG40218, CG41063, CG17691, CG40080, CG40130, CG40129, CG17665, CG2944, CG2682, and CG30440 of 2Rh, CG17514, CG10837 of 3Lh, and CG17419 of 3Rh (Tables 1 and 2). Notably, the protein products encoded by 13 of these gene models are conserved in humans (see Table 2). Transcription was studied throughout developmental stages in the wild-type Oregon-R strain by Northern blotting (see examples in Figure 3). Using the Nipped-A cDNA probe, a transcript of ∼11 kb was detected in all stages, suggesting that the expression of this essential gene is required throughout all development. A similar picture emerges from the analysis of the gene models tested: for example, homologous transcripts to cDNAs of CG17514, CG10837, CG30440, CG17691, CG40080, and CG2682 are present at different stages of development (Figure 3); the transcript of CG17514 was not seen in embryos. In each case, the molecular weight of bands is fully consistent with the predicted mRNA size.
Figure 3.—
Developmental Northern analysis of heterochromatic gene models Autoradiographic exposure of Northern blots containing poly(A+) RNA from different developmental stages. Different filters were probed with several cDNA probes. As a loading control, the rp49 ribosomal protein gene probe was used (O'Connel and Rosbash 1984). Cytosolic and plastid rRNA of Triticum durum were used as markers of molecular weight. Lane 1, embryos; lane 2, first instar larvae; lane 3, third instar larvae; lane 4, early pupae; lane 5, late pupae; lane 6, adult males; lane 7, adult females.
DISCUSSION
In previous work we mapped 22 D. melanogaster gene models, defined in release 3 of the D. melanogaster genome, to the distal heterochromatin of chromosome 2 (Corradini et al. 2003). The results of this analysis expand our knowledge of the genetic functions located in the heterochromatin of this chromosome. Here, we have performed a detailed FISH mapping to the mitotic heterochromatin of 41 additional gene models of D. melanogaster, whose location was unclear at the time that we started our experiments. Moreover, transcription of the vital gene Nipped-A and 15 heterochromatic gene models was also investigated by Northern blotting analysis.
Known genes and gene models in the heterochromatin of chromosome 2:
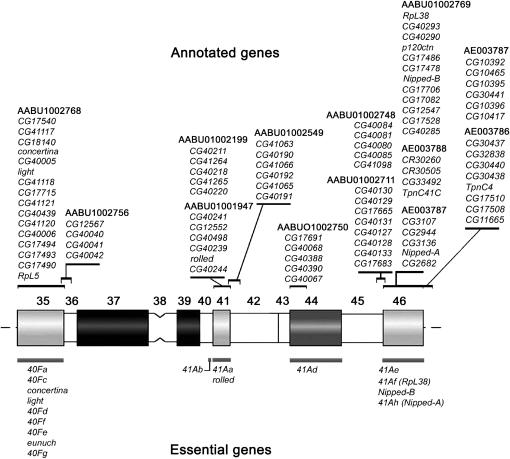
The mapping results provided by this work complement those obtained in previous studies (Dimitri 1991; Hoskins et al. 2002; Corradini et al. 2003; Yasuhara et al. 2003), enabling us to construct a map of the heterochromatic genes located in chromosome 2 (Figure 4 and Table 2). To date, 11 scaffolds, which together account for ∼6 Mb of genomic DNA, have been mapped to the pericentric heterochromatin of chromosome 2. Thus, it emerges that this region contains at least 76 annotated genes, including 17 genes defined as essential by genetic analysis (Hilliker 1976; Dimitri 1991; reviewed by Coulthard et al. 2003 and by Dimitri et al. 2005b). Recently, Myster et al. (2004) have found additional lethals on 2Rh, but it is still unclear whether or not these lethals actually correspond to new vital genes.
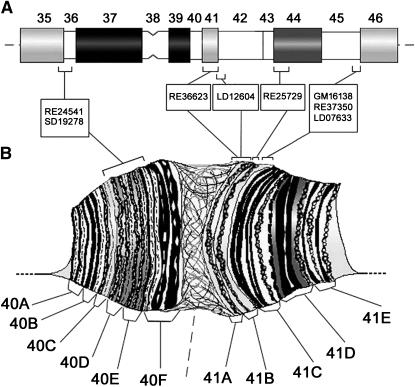
Figure 4.—
Mapping of essential genes and gene models to the heterochromatin of chromosome 2. Cytogenetic map showing the location of both essential genes (below) and gene models (above) within the heterochromatin of chromosome 2; hues of shading correspond to the intensity of DAPI staining, solid blocks represent regions with strong fluorescence intensity, and open blocks are nonfluorescent regions. Scaffold designation is shown at the top of each gene model list. Essential genes and gene models are nonrandomly distributed in the constitutive heterochromatin of chromosome 2; most of them are located in weakly DAPI-fluorescent chromosomal regions, which harbor clusters of transposable-element homologous sequences and lack highly repetitive satellite DNAs. In addition, most of the genes are grouped within regions h35 and h46, which represent the most distal portions of mitotic heterochromatin. Additional 2Rh lethals defined by Myster et al. (2004) have not been included in the map, because it is still unclear whether or not these lethals actually correspond to new vital genes.
The estimated DNA content in heterochromatin of chromosome 2 is ∼20 Mb (Hoskins et al. 2002), 70% of which (14 Mb) is made up by satellite DNA (Lohe and Hilliker 1996). Thus, ∼6 Mb of the remaining heterochromatin should correspond to islands of nonsatellite, complex DNAs enriched in transposons and genes. The observation that the 11 heterochromatic scaffolds mapped to chromosome 2 account for ∼6 Mb of genomic DNA suggests that almost the entire amount of nonsatellite heterochromatic DNA of chromosome 2 has been assembled. Notably, four scaffolds molecularly linked by the BAC map (Hoskins et al. 2002) map to region h46 (Figure 4 and Table 2) and together account for 3.5 Mb, which is reasonably close to the inferred size of this region (∼2.5 Mb).
From Figure 4 it emerges that most gene models are located in distal heterochromatin; together, 54 gene models, carried by seven scaffolds, map to regions h35–h36 on 2Lh and h45 (distal)–h46 on 2Rh, which together represent ∼30% of chromosome 2 heterochromatin. By contrast, only 22 gene models, carried by four scaffolds, are present in the large h37–h45 (proximal) segment that accounts for ∼70% of chromosome 2 heterochromatin. Finally, 14 more genes (scaffolds AE003787 and AE003786) have been included in the map, which are likely to be scattered somewhere at the transition region between heterochromatin and euchromatin.
Among the 76 heterochromatic gene models annotated on chromosome 2, 7 corresponded to the known essential genes defined by genetic analyses: RpL5, light, concertina, rolled, RpL38, Nipped-B, and Nipped-A (Hilliker 1976; Devlin et al. 1990; Parks and Wieshous 1991; Biggs et al. 1994; Rollins et al. 1999; Tulin et al. 2002; Myster et al. 2004; Marygold et al. 2005). Thus, a significant portion of essential genes that mapped to chromosome 2 heterochromatin (Figure 4) still need to be identified molecularly. Notably, among those genes, l(2)41Ad is the only known vital gene mapping to region h44 (Dimitri 1991) and may be implicated in proper morphogenesis of the legs and wings (Hilliker 1976; Dimitri et al. 2005b). Region h44 contains at least five gene models (CG17691, CG40067, CG40068, CG40388, and CG40390) carried by AABU01002750, but, at present, there are no molecular data linking any of them to l(2)41Ad. Notably, CG17691, CG40068, and CG40390 encode polypeptides that share similarity with known human proteins, which are apparently not related to morphogenetic functions (Table 2). Interestingly, l(2)41Ad is highly mutable in I-R hybrid dysgenesis (Dimitri et al. 1997) and most 41Ad lethal alleles are associated with cytologically visible deletions of h43–h44 spanning up to ∼1 Mb of DNA (Dimitri et al. 2005a). These genetic and cytological features suggest that l(2)41Ad is a large locus. The largest gene model found in AABU01002750 is CG40388, which is ∼12 kb, a size similar to that of the light gene, but smaller compared to that of other vital heterochromatic genes of chromosome 2 such as Nipped-B, rolled, and Nipped-A, which are ∼39, 50, and 73 kb long, respectively (http://www.flybase.org). Together, these observations do not support any obvious correspondence between l(2)41Ad and any given gene model mapped to h44. More studies are required to establish the molecular identity and function of l(2)41Ad.
Previous works identified vital genes and gene models in the heterochromatin of chromosome 3 (Marchant and Holm 1988; Hoskins et al. 2002; Koryakov et al. 2002; Corradini et al. 2003; reviewed by Fitzpatrick et al. 2005). Here we mapped eight additional gene models: CG17514, CG40103, CG40163, CG10837, CG40105 to region h48 of 3Lh and CG17419, CG40065, and CG411013 to region h58 of 3Rh. Interestingly, five vital genes were mapped to these regions by Koryakov et al. (2002): l(3)80Fa, l(3)80Fb, and l(3)80Fc, which lie in h48 and l(3)81Fa and l(3)81Fb located in h58. On the basis of the complementation behavior of mutant alleles, l(3)81Fb was suggested as corresponding to the zeppelin (zep) locus (Fitzpatrick et al. 2005). At present it is unclear whether or not the eight gene models that mapped to h48 and h58 are candidates for essential genes in these regions; a possible exception may be CG17419. This gene model encodes a protein carrying ankyryn repeats (Table 2) and may correspond to zep. In fact, the zep product was suggested to be involved in cell–cell adhesion (Fitzpatrick et al. 2005), which is consistent with the finding that ankyrin repeats can be found in cell–cell interaction proteins (Mosavi et al. 2004).
As shown in Table 2, some of the products encoded by gene models share significant identity with known proteins from humans and other organisms. However, gene models and essential genes in D. melanogaster heterochromatin apparently do not have molecular functions that would distinguish them from genes located in euchromatin (Hoskins et al. 2002). Essential genes are on average significantly larger than euchromatic genes, due to the occurrence of long introns enriched in transposable element (TE)-related DNA sequences (Devlin et al. 1990; Hoskins et al. 2002; Tulin et al. 2002; Dimitri et al. 2003, 2005a). Surprisingly, although some of the gene models mapped to heterochromatin are also large, such as CG40130 (54 kb), CG40129 (55 kb), CG17514 (45 kb), and CG40080 (34 kb), this does not seem to be a general rule. For example, with the exception of rolled (CG12559), which is large (∼50 kb), most genes carried by scaffolds AABU01002199 and AABU01001947 (region h41) are <6 kb, and six of them are even very short (<600 bp). A similar situation is found in scaffolds AABU01002549 and AABU01002750.
Several hypotheses could explain these observations. First, during evolution, older genes in heterochromatin may increase their size due to TE insertions in the introns. If that is the case, then it would follow that short gene models in heterochromatin are younger, or, in other words, have recently “moved” to heterochromatin. Second, genes in heterochromatin may be differentially targeted by TEs, with some genes being somehow more refractory than others. Third, there might be a selective pressure to maintain some genes of short size in heterochromatin due to particular functional properties. Highly expressed genes indeed tend to have substantially shorter introns than those expressed at low levels (Castillo-Davis et al. 2002). In this regard, it is worth noting that three highly expressed ribosomal protein genes, i.e., RpL38, RpL5, and RpL15, all of short size, are located in heterochromatin (Marygold et al. 2005; Schulze et al. 2005). However, we did not detect significant differences in the expression of the short and large gene models tested by Northern blotting. Finally, some predictions may represent different portions of the same gene, or may be spurious, leading to an underestimate of the actual size of genes in heterochromatin and to an overestimate of the actual gene number.
Are D. melanogaster heterochromatic genes also found in heterochromatin in other organisms? It recently has been found that genes located in D. melanogaster heterochromatin have euchromatic orthologs in D. pseudoobscura and D. virilis (Yasuhara et al. 2005). Here, we report that 70% of heterochromatic gene models of chromosome 2 encode putative proteins sharing significant similarity with human proteins (Table 2). However, the corresponding human orthologs are invariably located in euchromatin. Although the function and regulation of known heterochromatic genes and their homologs in other eukaryotes is far from being elucidated, these data suggest that a heterochromatic location may not be crucial for the proper function of a given gene.
Developmental expression of heterochromatic gene models:
We also analyzed heterochromatic gene expression throughout the Drosophila life cycle in Northern experiments (Figure 3). RNAs for the vital gene Nipped-A and 15 gene models were found to be transcribed in embryos, larvae, pupae, and adults. Although much remains to be learned about the number, structure, regulation, and evolution of heterochromatic genes, some interesting features are beginning to emerge from the developmental expression of gene models resident in pericentric heterochromatin. In particular, our observations indicate that, similarly to Nipped-A, the expression of the heterochromatic gene models studied here is not limited to specific stages, but is present throughout development. This is also true for vital heterochromatic genes such as light, rolled, RpL5, RpL38, RpL15, Nipped-B, and Parp (Biggs et al. 1994; Rollins et al. 1999; Tulin et al. 2002; Marygold et al. 2005; Schulze et al. 2005). This finding is consistent with the idea that genes resident in a heterochromatic environment can code for essential functions.
Correspondence between mitotic and polytene chromosome heterochromatin:
In D. melanogaster polytene tissues, the heterochromatic regions from all chromosomes aggregate to form a cytological structure called the chromocenter. Two cytological domains of heterochromatin can be distinguished in the chromocenter (Heitz 1934): α-heterochromatin, which corresponds to a small compact region located in the middle of the chromocenter and undergoes little if any replication during polytenization (Gall et al. 1971), and β-heterochromatin, a diffusely banded mesh-like material that lies between euchromatin and α-heterochromatin and is extensively polytenized (Zhang and Spradling 1994; Berghella and Dimitri 1996). The assembly and organization of the chromocenter represents one of the most intriguing aspects in the mitotic-to-polytene chromosome transition. Although the structural relationships between mitotic and polytene chromosomes have been analyzed in several studies, only a few have focused on the correspondence between mitotic and polytene chromosome heterochromatin (Koryakov et al. 1996).
Here, we report the mapping of 10 cDNAs to mitotic and polytene chromosome heterochromatin. All tested probes on polytene chromosomes produced hybridization signals of significant fluorescence intensity, indicating that the corresponding gene models undergo polytenization, at least to some extent. This observation is consistent with previous findings showing that single-copy genes located in deep heterochromatin are extensively polytenized (Devlin et al. 1990; Berghella and Dimitri 1996), in contrast with satellite DNA blocks that fail to undergo polytenization and exemplify the well-established paradigm of heterochromatin underreplication. In addition, several cDNA probes produced heterochromatic FISH signals split into two. A similar pattern was produced by some PZ heterochromatic insertions (Zhang and Spradling 1994) and by the rolled cDNA (Berghella and Dimitri 1996) and suggests that sister chromatids retain their identity within the chromocenter (Zhang and Spradling 1994).
Our cDNA mapping may have an intersting outcome for studying the correspondence between mitotic and polytene chromosome heterochromatin of chromosome 2 (Figure 2 and Figure 5). On 2Lh, polytenization of sequences in region h35 distal and h36 proximal contributes to division 40D-F. This correspondence is consistent with the previous finding by Koryakov et al. (1996) showing that region h35 corresponds to division 40D-F. In the same study (Koryakov et al. 1996), the authors also suggested that h46, the most distal region of 2Rh, corresponds to division 41C. More recently, we have extended this observation and shown that polytenization of DNA sequences located in h46 contribute to form division 41C-D (Corradini et al. 2003). Moreover, we found that, in the very proximal heterochromatin of 2R, sequences from the large h41–44 portion contribute to form polytene division 41A, while sequences located at the h45–h46 transition can contribute to polytene division 41B.
Figure 5.—
Mitotic (A) and polytene chromosome heterochromatin (B) correspondences: summary map, based on results from this and previous work (Devlin et al. 1990; Koryakov et al. 1996; Corradini et al. 2003; Yasuhara et al. 2003), depicting the physical correspondence between the mitotic and polytene chromosome heterochromatin of chromosome 2. Shaded areas indicate correspondences; open areas identify regions where correspondences are not clearly defined.
Acknowledgments
We are grateful to Gary Karpen and Patrizia Lavia for critical reading of the manuscript and suggestions; to the Drosophila Heterochromatin Genome Project (Roger Hoskins, Chris Smith, Joe Carlson, and Gary Karpen) for sharing unpublished data; and to Rachele Alicandro, Katia Gazzetti, Cinzia Rizzo, and Laura Somma for helpful assistence with BLAST analysis and polytene chromosome preparations. This work was supported by grants from Ministero dell'Università e della Ricerca “Progetti di ricerca di interesse nazionale 2004–2006” and from Istituto Pasteur-Fondazione Cenci-Bolognetti.
References
- Adams, M. D., S. E. Celniker, R. A. Holt, C. A. Evans, J. D. Gocayne et al., 2000. The genome sequence of Drosophila melanogaster. Science 287: 2185–2195. [DOI] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative, 2000. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815. [DOI] [PubMed] [Google Scholar]
- Berghella, L., and P. Dimitri, 1996. The heterochromatic rolled gene of Drosophila melanogaster is extensively polytenized and transcriptionally active in the salivary gland chromocenter. Genetics 144: 117–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggs, W. H., III, K. H. Zavitz, B. Dickson, A. van der Straten, D. Brunner et al., 1994. The Drosophila rolled locus encodes a MAP kinase required in the sevenless signal transduction pathway. EMBO J. 13: 1628–1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brun, M. E., M. Ruault, M. Ventura, G. Roizes and A. De Sario, 2003. Juxtacentromeric region of human chromosome 21: a boundary between centromeric heterochromatin and euchromatic chromosome arms. Gene 312: 41–50. [DOI] [PubMed] [Google Scholar]
- Castillo-Davis, C. I., S. L. Mekhedov, D. L. Hartl, E. V. Koonin and F. A. Kondrashov, 2002. Selection for short introns in highly expressed genes. Nat. Genet. 31: 415–418. [DOI] [PubMed] [Google Scholar]
- Corradini, N, F. Rossi, F. VernÌ and P. Dimitri, 2003. FISH analysis of Drosophila melanogaster heterochromatin using BACs and P elements. Chromosoma 112: 26–37. [DOI] [PubMed] [Google Scholar]
- Coulthard, A. B., D. F. Eberl, C. B. Sharp and A. J. Hilliker, 2003. Genetic analysis of the second chromosome centromeric heterochromatin of Drosophila melanogaster. Genome 46: 343–352. [DOI] [PubMed] [Google Scholar]
- Dernburg, A. F., J. W. Sedat and R. S. Hawley, 1996. Direct evidence of a role for heterochromatin in meiotic chromosome segregation. Cell 86: 135–146. [DOI] [PubMed] [Google Scholar]
- Devlin, R. H., B. Bingham and B. T. Wakimoto, 1990. The organization and expression of the light gene, a heterochromatic gene of Drosophila melanogaster. Genetics 125: 129–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitri, P., 1991. Cytogenetic analysis of the second chromosome heterochromatin of Drosophila melanogaster. Genetics 127: 553–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitri, P., 2004. Fluorescent in situ hybridization with transposable element probes to mitotic chromosomal heterochromatin of Drosophila. Methods Mol. Biol. 260: 29–39. [DOI] [PubMed] [Google Scholar]
- Dimitri, P., B. Arcà, L. Berghella and E. Mei, 1997. High genetic instability of heterochromatin after transposition of the LINE-like I factor in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 94: 8052–8057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitri, P., N. Junakovic and B. Arcà, 2003. Colonization of heterochromatic genes by transposable elements in Drosophila. Mol. Biol. Evol. 20: 503–512. [DOI] [PubMed] [Google Scholar]
- Dimitri, P., N. Corradini, F. Rossi, I. F. Zhimulev and F. VernÌ, 2005. a Transposable elements as artisans of the heterochromatin genome in Drosophila. Cytogenet. Genome Res. 110: 165–172. [DOI] [PubMed] [Google Scholar]
- Dimitri, P., N. Corradini, F. Rossi and F. VernÌ, 2005. b The paradox of functional heterochromatin. BioEssays 27: 29–41. [DOI] [PubMed] [Google Scholar]
- Drosophila Heterochromatin Genome Project, 2004. http://www.dhgp.org.
- Eissenberg, J. C., and A. J. Hilliker, 2000. Versatility of convinction: heterochromatin as both repressor and an activator of transcription. Genetica 109:19–24. [DOI] [PubMed] [Google Scholar]
- Elgin, S. C. R., 1996. Heterochromatin and gene regulation in Drosophila. Curr. Opin. Genet. Dev. 6: 193–202. [DOI] [PubMed] [Google Scholar]
- Fitzpatrick, K. A., D. A. Sinclair, S. R. Shulze, M. Syrzycka and B. M. Honda, 2005. A genetic and molecular profile of third chromosome centric heterochromatin in Drosophila melanogaster. Genome 48: 571–584. [DOI] [PubMed] [Google Scholar]
- Gall, J. G., E. H. Cohen and M. L. Polan, 1971. Repetitive DNA sequences in Drosophila. Chromosoma 33: 319–344. [DOI] [PubMed] [Google Scholar]
- Gatti, M., and S. Pimpinelli, 1992. Functional elements in Drosophila melanogaster heterochromatin. Annu. Rev. Genet. 26: 239–275. [DOI] [PubMed] [Google Scholar]
- Heitz, E., 1928. Das Heterochromatin der Moose. Jb. Wiss. Bot. 69: 762–818. [Google Scholar]
- Heitz, E., 1934. Uber alpha-heterochromatin sowie konstanz und bau der chromomeren bei Drosophila. Biol. Zentralbl. 45: 588–609. [Google Scholar]
- Henikoff, S., K. Ahmad and H. S. Malik, 2001. The centromere paradox: stable inheritance with rapidly evolving DNA. Science 293: 1098–1102. [DOI] [PubMed] [Google Scholar]
- Hilliker, A. J., 1976. Genetic analysis of the centromeric heterochromatin of chromosome 2 of Drosophila melanogaster: deficiency mapping of EMS-induced lethal complementation groups. Genetics 83: 765–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath, J.E., S. Schwartz and E. E. Eichlr, 2000. The mosaic structure of human pericentromeric DNA: a strategy for characterizing complex regions of the human genome. Genome Res. 10: 839–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoskins, R. A., C. D. Smith, J. W. Carlson, A. B. Carvalho, A. Halpern et al., 2002. Heterochromatic sequences in a Drosophila whole-genome shotgun assembly. Genome Biol. 3: RESEARCH0085. [DOI] [PMC free article] [PubMed]
- John, B., 1988. The biology of heterochromatin, pp. 1–128 in Heterochromatin: Molecular and Structural Aspects, edited by R. S. Verma. Cambridge University Press, Cambridge, UK/London/New York.
- Karpen, G. H., M. G. Le and H. Le, 1996. Centric heterochromatin and the efficiency of achiasmate disjunction in Drosophila female meiosis. Science 273: 118–122. [DOI] [PubMed] [Google Scholar]
- Konev, A. Y., C. M. Yan, D. Acevedo, C. Kennedy, E. Ward et al., 2003. Genetics of P-element transposition into Drosophila melanogaster centric heterochromatin. Genetics 165: 2039–2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koryakov, D. E., E. S. Belyaeva, A. A. Alekseyenko and I. F. Zhimulev, 1996. Alpha and beta heterochromatin in polytene chromosome 2 of Drosophila melanogaster. Chromosoma 105: 310–319. [DOI] [PubMed] [Google Scholar]
- Koryakov, D. E., I. F. Zhimulev and P. Dimitri, 2002. Cytogenetic analysis of the third chromosome heterochromatin of Drosophila melanogaster. Genetics 160: 509–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuln, R. M., L. Clarke and J. Carbon, 1991. Clustered tRNA genes in Schizosaccharomyces pombe centromeric DNA sequence repeats. Proc. Natl. Acad. Sci. USA 88: 1306–1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsley, D. L., and G. G. Zimm, 1992. The Genome of Drosophila melanogaster. Academic Press, San Diego.
- Lohe, A. R., and A. J. Hilliker, 1996. Return of the H-word (heterochromatin). Curr. Opin. Genet. Dev. 5: 746–755. [DOI] [PubMed] [Google Scholar]
- Marchant, G. E., and D. G. Holm, 1988. Genetic analysis of the heterochromatin of chromosome 3 in Drosophila melanogaster. II. Vital loci identified through EMS mutagenesis. Genetics 120: 519–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marygold, S. J., C. M. Coelho and S. J. Leevers, 2005. Genetic analysis of RpL38 and RpL5, two minute genes located in the centric heterochromatin of chromosome 2 of Drosophila melanogaster. Genetics 169: 683–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moritz, K. B., and G. E. Roth, 1976. Complexity of germline and somatic DNA in Ascaris. Nature 259: 55–57. [DOI] [PubMed] [Google Scholar]
- Mosavi, L. K., T. J. Cammet, D. C. Desrosiers and Z-Y Peng, 2004. The ankyrin repeat as molecular architecture for protein recognition. Protein Sci. 13: 1435–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myster, S. H., F. Wang, R. Cavallo, W. Christian, S. Bhotika et al., 2004. Genetic and bioinformatic analysis of 41C and the 2R heterochromatin of Drosophila melanogaster: a window on the heterochromatin-euchromatin junction. Genetics 166: 807–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaki, K., Z. Cheng, S. Ouyang, P. B. Talbert, M. Kim et al., 2004. Sequencing of a rice centromere uncovers active genes. Nat.. Genet. 36:138–45. [DOI] [PubMed] [Google Scholar]
- O'Connel, P., and M. Rosbash, 1984. Sequence, structure, and codon preference of the Drosophila ribosomal protein 49 gene. Nucleic Acids Res. 12: 5495–5513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks, S., and E. Wieschaus, 1991. The Drosophila gastrulation gene concertina encodes a Ga-like protein. Cell 64: 447–458. [DOI] [PubMed] [Google Scholar]
- Peterson, D. G., W. R. Pearson and S. M. Stack, 1998. Characterization of the tomato (Lycopersicon esculentum) genome using in vitro and in situ DNA reassociation. Genome 41: 346–356. [Google Scholar]
- Rollins, R. A., P. Morcillo and D. Dorsett, 1999. Nipped-B, a Drosophila homologue of chromosomal adherins, participates in activation by remote enhancers in the cut and Ultrabithorax genes. Genetics 152: 577–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roseman, R. R., E. A. Johnson, C. K. Rodesh, M. Bjerke, R. N. Nagoshi et al., 1995. A P element containing suppressor of hairy-wing binding regions has novel properties for mutagenesis in Drosophila melanogaster. Genetics 141: 1061–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze, S. R., D. A. Sinclair, K. A. Fitzpatrick and B. M. Honda, 2005. A genetic and molecular characterization of two proximal heterochromatic genes on chromosome 3 of Drosophila melanogaster. Genetics 169: 2165–2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tulin, A., D. Stewart and A. C. Spradling, 2002. The Drosophila heterochromatic gene encoding poly(ADP-ribose) polymerase (PARP) is required to modulate chromatin structure during development. Genes Dev. 16: 2108–2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiler, K. S., and B. T. Wakimoto, 1995. Heterochromatin and gene expression in Drosophila. Annu. Rev. Genet. 29: 577–605. [DOI] [PubMed] [Google Scholar]
- Williams, S. M., and L. G. Robbins, 1992. Molecular genetic analysis of Drosophila rDNA arrays. Trends Genet. 8: 335–340. [DOI] [PubMed] [Google Scholar]
- Yasuhara, J. C., M. Marchetti, L. Fanti, S. Pimpinelli and B. T. Wakimoto, 2003. A strategy for mapping the heterochromatin of chromosome 2 of Drosophila melanogaster. Genetica 117: 217–226. [DOI] [PubMed] [Google Scholar]
- Yasuhara, J. C., C. H. De Crease and B. T. Wakimoto, 2005. Evolution of heterochromatic genes of Drosophila. Proc. Natl. Acad. Sci. USA 102: 10958–10963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, P., and A. C. Spradling, 1994. Insertional mutagenesis of Drosophila heterochromatin with single P elements. Proc. Natl. Acad. Sci. USA 91: 3539–3543. [DOI] [PMC free article] [PubMed] [Google Scholar]