Abstract
The basement membrane is important for proper tissue development, stability, and physiology. Major components of the basement membrane include laminins and type IV collagens. The type IV procollagens Col4a1 and Col4a2 form the heterotrimer [α1(IV)]2[α2(IV)], which is ubiquitously expressed in basement membranes during early developmental stages. We present the genetic, molecular, and phenotypic characterization of nine Col4a1 and three Col4a2 missense mutations recovered in random mutagenesis experiments in the mouse. Heterozygous carriers express defects in the eye, the brain, kidney function, vascular stability, and viability. Homozygotes do not survive beyond the second trimester. Ten mutations result in amino acid substitutions at nine conserved Gly sites within the collagenous domain, one mutation is in the carboxy-terminal noncollagenous domain, and one mutation is in the signal peptide sequence and is predicted to disrupt the signal peptide cleavage site. Patients with COL4A2 mutations have still not been identified. We suggest that the spontaneous intraorbital hemorrhages observed in the mouse are a clinically relevant phenotype with a relatively high predictive value to identify carriers of COL4A1 or COL4A2 mutations.
THE basement membrane is an extracellular matrix associated with overlying cells and is important for proper tissue development, stability, and physiology. Basement membrane biosynthesis is initiated by cell surface assembly and anchorage of laminins followed by the incorporation of type IV collagens, nidogens, and perlecans (Li et al. 2005). There are six type IV procollagen genes, Col4a1–Col4a6, arranged as three head-to-head gene pairs in the mammalian genome. The type IV procollagens form specific heterotrimer molecules, and their proper incorporation into the extracellular matrix is important for basement membrane stability. The participation of type IV procollagen genes in basement membrane biosynthesis is developmentally stage and tissue specific (Miner and Sanes 1994; Kühn 1995; Sado et al. 1998). In contrast to Col4a3–Col4a6, which are expressed in specific tissues at later developmental stages, the major isoform from the Col4a1 and Col4a2 genes is the heterotrimer [α1(IV)]2[α2(IV)] and it is ubiquitously expressed in basement membranes during early stages of development.
There is an extensive mutant database of human type IV procollagen genes. Mutations in the COL4A3, COL4A4, and COL4A5 genes are causative in patients with Alport syndrome (renal disease with or without deafness and/or eye abnormalities), whereby COL4A5 mutations predominate (Lemmink et al. 1997; Jais et al. 2000; Badenas et al. 2002; Pescucci et al. 2004). Familial porencephaly due to COL4A1 mutations has been recently identified (Gould et al. 2005; Breedveld et al. 2006). Involvement of COL4A6 in hereditary disease has been shown only in intergenic rearrangements simultaneously affecting the closely linked COL4A5 and COL4A6 genes in patients expressing Alport syndrome with leiomyomatosis (Zhou et al. 1993; Renieri et al. 1994; Garcia-Torres et al. 2000; Mothes et al. 2002; Anker et al. 2003). In the mouse, mutations of Col4a1 (Gould et al. 2005; Van Agtmael et al. 2005), Col4a3 (Cosgrove et al. 1996; Miner and Sanes 1996), and Col4a5 (Rheault et al. 2004) have been recovered or constructed. In addition, a targeted mutation that ablates the closely linked Col4a1 and Col4a2 genes (Pöschl et al. 2004) and an insertional mutation that simultaneously disrupts the closely linked Col4a3 and Col4a4 genes (Lu et al. 1999) have been generated. The phenotypic abnormalities associated with the mouse mutations mirror the human genetic defects. To date intragenic mutations affecting the Col4a2/COL4A2 or Col4a6/COL4A6 genes have not been identified. We present the genetic, molecular, and phenotypic characterization of nine Col4a1 and the first three Col4a2 missense mutations. Heterozygous carriers express a variable phenotype affecting the eye, vascular stability, the brain, kidney function, and survival in embryonic or postnatal stages.
MATERIALS AND METHODS
Mutations, animals, and mapping:
All original mutants were recovered in random mutagenesis screens in the offspring of (102xC3H)F1, C3H/HeJ, or DBA/2 males exposed to ethylnitrosourea, procarbazine, or γ-irradiation. In all experiments the progeny screened were derived from exposure of stem cell spermatogonia. The treated parental generation males were mated to untreated Oak Ridge test-stock females that are homozygous for recessive mutant alleles at seven loci affecting coat pigmentation or the size of the external ear (Russell 1951). Ophthalmological examinations were conducted as previously described (Favor 1983). Before initiating the present studies congenic C3H/HeJ mutant lines were established. To map the mutations congenic mutant C3H mice were outcrossed to wild-type strain C57BL/6El. Outcross generation mutant heterozygotes were backcrossed to wild-type strain C57BL/6El, the resultant offspring were phenotyped by slit lamp biomicroscopy, and liver tissue was collected for genomic DNA extraction. Segregation analyses relative to 42 autosomal MIT-microsatellite markers were carried out according to our standard laboratory protocol (Favor et al. 1997). After identification of linkage to proximal chromosome (Chr) 8, mice were genotyped for additional MIT-microsatellite markers within the region. Segregation data were analyzed with Map Manager version 2.6.5 (Manly 1993), and the gene order was determined by minimizing the number of multiple crossovers. Animals were bred and maintained in our animal facility according to the German law for the protection of animals. All inbred strain C3H/HeJ and C57BL/6El animals were obtained from breeding colonies maintained by the GSF–National Research Center for Environment and Health, Department of Animal Resources at Neuherberg.
RT–PCR and sequence analysis:
Heterozygous and wild-type embryos were prepared at E15 and heads and livers snap frozen on dry ice for RNA or genomic DNA extraction. Total RNA was isolated from heads with the RNeasy kit (QIAGEN, Hilden, Germany). RNA was reverse transcribed with a Titan one tube RT–PCR kit (Roche Diagnostics, Mannheim, Germany) or the Access RT–PCR System (Promega, Madison, WI). Primer sets were used to amplify 9–11 overlapping regions across the Col4a1 or Col4a2 cDNAs (supplemental Table 1 at http://www.genetics.org/supplemental/). The PCR products were electrophoretically separated on 1% agarose gels, extracted with a QIAquick gel extraction kit (QIAGEN), and used as templates for sequencing with a Taq Dye-Deoxy terminator cycle sequencing kit on an ABI 3100 DNA sequencer (Applied Biosystems, Foster City, CA). The mutations were confirmed by sequencing from genomic DNA of the originally analyzed heterozygous embryos and additional heterozygous carriers. All sites harboring mutations in the Col4a1 or the Col4a2 genes were sequenced in the strains C3H/HeJ, 102/El, DBA/2, and test stock, which represent all potential Chr 8 parental haplotypes in the matings that produced the original mutants. All mutant allele symbols were submitted to the Mouse International Nomenclature Committee.
Histology, gross embryo and lens morphology, and slit lamp photography:
Pregnant females were killed by cervical dislocation. Embryos were carefully freed from placentas and embryonic membranes in room temperature PBS, phenotyped under a dissecting microscope (MZ APO; Leica, Bensheim, Germany), and photographed. Embryos were fixed in 10% buffered formalin or Carnoy's solution, and the heads were embedded in paraffin and serial sectioned (coronal) at 5 μm. Sections were stained with hematoxylin and eosin. P21 mice were killed by cervical dislocation. The eyes were removed, fixed for 24 hr in Carnoy's solution, transferred to 70% EtOH, dehydrated, and embedded in JB4 (Polysciences, Eppelheim, Germany). Serial transverse 3-μm sections were stained with methylene blue and basic fuchsin and evaluated by light microscopy (Axioplan; Carl Zeiss, Hallbergmoos, Germany). Digital photos were acquired (Axiocam and Axiovision; Carl Zeiss, Hallbergmoos, Germany) and imported into Adobe Photoshop CS (Adobe Systems, Unterschleissheim, Germany).
P35 mice were killed by cervical dislocation, and the eyes were dissected and placed in room temperature PBS. Lenses were immediately dissected from the eyes under a dissecting microscope (MZ APO; Leica), carefully freed from remnants of the ciliary body, and photographed. Embryo and lens photos were converted to digital images (Nikon LS-2000 slide scanner) and imported into Adobe Photoshop CS.
Eleven-month-old mice were anesthetized with 137 mg ketamine and 6.6 mg xylazine/kg body weight and quickly photographed with a slit lamp microscope (Zeiss SL 120) equipped with a compact video camera. Images were captured in Axiovision (Zeiss) and imported into Adobe Photoshop CS. After photography ophthalmic salve (Regepithel, Alcon) was applied to the eyes of the anaesthetized mice to prevent eye injury due to dehydration and the animals were caged individually until fully recuperated.
Hematology, clinical chemistry, and urine analysis:
Urine and blood were sampled from a total of 301 mice and analyzed for hematology and clinical chemical parameters (Rathkolb et al. 2000; Gailus-Durner et al. 2005). Approximately 15 heterozygous carriers (range 14–22) per mutant line as well as wild-type littermates were assayed. The sex ratio within the genotype groups was ∼1:1. All mutant lines with the exception of Col4a1Acso and Col4a1D456 were included in the analysis.
Urine was collected from 11-week-old, nonfasted mice at 9:00 am in sterile petri dishes. Total protein and albumin in the undiluted urine was determined with an Olympus (Hamburg, Germany) AU 400 autoanalyzer.
Blood samples were taken from ether-anesthetized 12-week-old, nonfasted mice by puncturing the retroorbital sinus with a nonheparinized capillary tube (0.8 mm diameter) at 9:00 am. Fifty microliters of blood were collected in EDTA-coated tubes (KABE, Nümbrecht, Germany) and analyzed with an ABC-Blood analyzer (Scil Animal Care Company, Viernheim, Germany).
Six hundred microliters of blood were collected in a heparinized tube (Li-heparin; KABE, Nümbrecht, Germany) for clinical–chemical analyses. The tubes were held at room temperature for 2 hr and centrifuged (10 min, 4656 × g; Biofuge, Heraeus, Hanau, Germany). One hundred thirty microliters of plasma were diluted with an equal volume of aqua dist and centrifuged as above, and creatinine and urea concentrations were measured (Olympus AU400 autoanalyzer, Hamburg, Germany).
The data were analyzed for mean differences among the mutant lines and between genotypes (heterozygote vs. wild type) by two-way ANOVA (SigmaStat 3.1; Systat Software, Richmond, CA).
RESULTS
Recovery, mapping, and molecular characterization of mutations:
Presumed mutants were identified with eye abnormalities ranging from microphthalmia, to anterior polar opacity, to lens vacuoles, to capsule opacity (Table 1). The frequencies of mutants in the irradiation and procarbazine experiments were very low. One mutant (Col4a1F247) was recovered in 23,182 progeny of (102xC3H)F1 males exposed to 3 Gy (our unpublished results), one mutant (Col4a1D456) was recovered in 15,931 progeny of DBA/2 males exposed to 3 + 3 Gy (Favor et al. 1987), and one mutant (Col4a1Acso) was recovered in 37,146 progeny of (102xC3H)F1 males exposed to 600 mg procarbazine/kg body weight (Kratochvilova et al. 1988). By comparison, the frequencies of mutants following ethylnitrosourea treatment were higher. One mutant (Col4a1ENU911) was recovered in 5090 offspring of (102xC3H)F1 males exposed to 80 mg ENU/kg body weight (Favor 1986) and three (Col4a1ENU4004, Col4a2ENU4003, and Col4a2ENU4020) mutants were recovered in 10,697 offspring of DBA/2 males exposed to 80 mg ENU/kg body weight (Favor and Neuhäuser-Klaus 2000). One mutant (Col4a2ENU415) was recovered in 9352 progeny of (102xC3H)F1 males exposed to 250 mg ENU/kg body weight (Favor 1983). The highest yield of mutants was observed in the progeny of C3H/HeJ males exposed to 3 × 100 mg ENU/kg body weight, four mutants (Col4a1ENU6005, Col4a1ENU6009, Col4a1ENU6019, and Col4a1ENU6024) in 2303 progeny screened (our unpublished results).
TABLE 1.
Origin of the mouse Col4a1 and Col4a2 mutations
| Allele symbola | Phenotype in original mutant | Reference |
|---|---|---|
| Col4a1Acso | Ant. capsule and suture opacity | Kratochvilova et al. (1988) |
| Col4a1ENU911 | Ant. pole opacity, lens vacuoles | Favor (1986) |
| Col4a1ENU4004 | Ant. pole opacity, lens vacuoles | Favor and Neuhäuser-Klaus (2000) |
| Col4a1ENU6005 | Ant. pole opacity, lens vacuoles | Our unpublished data |
| Col4a1ENU6009 | Ant. pole opacity | Our unpublished data |
| Col4a1ENU6019 | Ant. pole opacity | Our unpublished data |
| Col4a1ENU6024 | Ant. pole opacity | Our unpublished data |
| Col4a1D456 | Lens vacuoles | Favor et al. (1987) |
| Col4a1F247 | Capsule opacity | Our unpublished data |
| Col4a2ENU415 | Microphthalmia | Favor (1983) |
| Col4a2ENU4003 | Ant. pole opacity | Favor and Neuhäuser-Klaus (2000) |
| Col4a2ENU4020 | Ant. pole opacity, lens vacuoles | Favor and Neuhäuser-Klaus (2000) |
Ant., anterior.
Superscripts are the original mutant symbols. Superscripts with “ENU” designate mutants from ethylnitrosourea, with D456 and F247 those from γ-irradiation, and with Acso those from procarbazine mutagenesis experiments.
The loci responsible for the dominantly expressed eye phenotype in all 12 mutant lines mapped between 2 and 8 cM proximal to the marker D8Mit124 located on Chr 8, at 14.7 Mb (Table 2). Our mapping results indicate the mutant locus/loci to be in the extreme proximal region of Chr 8 at approximately cM 5. On the basis of chromosomal location and expression in the eye, three candidate genes were considered for mutation analysis, Sox1, Col4a1, and Col4a2. Sequencing of the Sox1 gene indicated no differences of all 12 mutant lines with wild type (data not shown). In 9 mutant lines mutations in the Col4a1 procollagen gene were identified and 3 mutant lines were associated with mutations in the Col4a2 procollagen gene (Table 3). We observed some differences between the sequences from our wild-type C3H/HeJ and the sequences given in the NCBI Gene Bank (Col4a1: cG2568C, Glu813Gln; cT2651C, Gly840Gly; cC3794T, Pro1221Pro; cC4007G, Pro1292Pro. Col4a2: cCG2012/2013GA, Arg626Glu; cC2646G, Ala837Gly; cG4126C, Gly1330Gly; cT5003C, Tyr1623His). The sequences at these eight sites in all mutant lines were identical to our wild-type C3H/HeJ.
TABLE 2.
Segregation analysis with proximal Chr 8 markers in offspring from the cross (C3H–Mut/B6 − +) × (B6 − +/B6 − +)
| Linea | Locusb | Haplotype inherited from the (C3H–Mut/B6 − +) parentc | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Col4a1/a2 | ▪ | □ | ▪ | □ | ▪ | □ | ▪ | □ | |
| D8Mit124 | ▪ | □ | □ | ▪ | ▪ | □ | □ | ▪ | |
| D8Mit335 | ▪ | □ | □ | ▪ | □ | ▪ | ▪ | □ | |
| Col4a1Acso | 44 | 45 | 0 | 1 | 2 | 5 | 0 | 0 | |
| Col4a1ENU911 | 43 | 59 | 2 | 4 | 1 | 0 | 0 | 0 | |
| Col4a1ENU4004 | 46 | 70 | 2 | 3 | 1 | 2 | 0 | 0 | |
| Col4a1ENU6005 | 50 | 87 | 1 | 1 | 1 | 4 | 0 | 0 | |
| Col4a1ENU6009 | 45 | 59 | 4 | 19 | 3 | 3 | 0 | 1 | |
| Col4a1ENU6019 | 48 | 85 | 3 | 3 | 2 | 1 | 0 | 0 | |
| Col4a1ENU6024 | 51 | 70 | 0 | 0 | 3 | 2 | 0 | 0 | |
| Col4a1D456 | 42 | 103 | 1 | 1 | 0 | 6 | 0 | 0 | |
| Col4a1F247 | 45 | 61 | 4 | 4 | 1 | 2 | 0 | 0 | |
| Totald | 369 | 580 | 13 | 17 | 11 | 22 | 0 | 0 | |
| Col4a2ENU415 | 47 | 61 | 0 | 1 | 2 | 3 | 0 | 0 | |
| Col4a2ENU4003 | 56 | 51 | 2 | 0 | 2 | 3 | 0 | 0 | |
| Col4a2ENU4020 | 53 | 49 | 2 | 1 | 0 | 2 | 0 | 0 | |
| Total | 156 | 161 | 4 | 2 | 4 | 8 | 0 | 0 | |
Due to the misclassification of mutant Col4a1ENU6009 offspring as wild type resulting from incomplete penetrance of the mutant phenotype (see results), mapping analysis of this mutant line was only with offspring classified as mutants.
The positions of the Chr 8 markers D8Mit124 and D8Mit335 employed for mapping were 14.7 and 19.2 Mb, respectively.
Solid boxes represent the marker alleles carried by strain C3H and the Col4a1 or Col4a2 mutant phenotype. Open boxes represent the marker alleles carried by strain C57BL/6 and the Col4a1 or Col4a2 wild-type phenotype.
The total does not include results for the mutant Col4a1ENU6009.
TABLE 3.
Twelve independent mutations of the Col4a1 or Col4a2 procollagen genes in the mouse
| Line | Nucleotide | Codon | Amino acid |
|---|---|---|---|
| Col4a1Acso | cG2104A | GGC → GAC | Gly658Asp |
| Col4a1ENU911 | cG2866T | GGC → GTC | Gly912Val |
| Col4a1ENU4004 | cG1312T | GGT → GTT | Gly394Val |
| Col4a1ENU6005 | cG3670A | GGC → GAC | Gly1180Asp |
| Col4a1ENU6009 | cT4875C | TCC → CCC | Ser1582Pro |
| Col4a1ENU6019 | cG4162A | GGT → GAT | Gly1344Asp |
| Col4a1ENU6024 | cG3243A | GGC → AGC | Gly1038Ser |
| Col4a1D456 | cG2991A | GGA → AGA | Gly954Arg |
| Col4a1F247 | cG4152C | GGT → CGT | Gly1341Arg |
| Col4a2ENU415 | cG227T | GTC → TTC | Val31Phe |
| Col4a2ENU4003 | cG2073A | GGT → GAT | Gly646Asp |
| Col4a2ENU4020 | cG2073A | GGT → GAT | Gly646Asp |
The 12 mutations associated with mutant phenotypes were distributed among 11 mutant sites, with identical (cG2073A) mutations in Col4a2ENU4003 and Col4a2ENU4020. This unexpected observation may be due to a breeding error or a preexisting mutation in the parental generation. In these cases, the two mutant lines with the cG2073A base pair substitution represent a single mutational event. Alternatively, the mutant lines may have been recovered from two independent repeat mutations. A breeding error is not likely since the first mutant, Col4a2ENU4003, was born 27.7.1989 and the mutant line was established and bred in an animal room separate from the room housing the parental generation mutagenesis matings. The second mutant, Col4a2ENU4020, was born 1.6.1990 in the animal room housing the parental generation mutagenesis matings, 10 months after the first mutant had been removed. A preexisting mutation in the parental generation mutagenesis matings is unlikely since the Col4a2ENU4003 and Col4a2ENU4020 mutations arose in different parental matings in the mutagenesis screen. A total of 50 and 71 offspring were screened in the parental matings giving rise to the Col4a2ENU4003 and Col4a2ENU4020 mutants, respectively, and in both cases a single mutant was recovered from the mating. This observation indicates that the mutants arose de novo and did not segregate from a heterozygous or a homozygous carrier parent. Further, before initiating the mutagenesis screen, potential parental generation animals were ophthalmologically screened and any individuals with an eye phenotype deviating from wild type were excluded from the experiment. Thus, we conclude the mutations to be repeat mutational events.
All 11 mutant sites were sequenced from genomic DNA of mutant carriers in all mutant lines as well as from the strains C3H/HeJ, 102/El, DBA/2J, and test stock. With the exception of the two repeat (cG2073A) mutations, each mutation was observed only in the designated mutant line. None of the observed mutations were observed in the wild-type strains examined, which represent all potential parental chromosomes in the mutagenesis studies in which the original mutants were recovered. All mutations were base pair substitutions and at the codon level 10 mutations result in amino acid substitutions at nine glycine sites and one each at a serine and a valine site.
Eye morphology and histology:
Characterization of heterozygous mutants from congenic C3H/HeJ mutant lines indicated that the phenotypes of carriers ranged from microphthalmia, to buphthalmos (especially in older animals), to anterior polar opacity with or without corneal-lens adhesions, to corneal opacities with or without hyperplasia and neovascularization, to lens vacuoles, to total lens opacity, to red floaters. Often the phenotypes observed in the two eyes of an individual were different. Thus all mutations were concluded to have variable expressivity. Examples of eye phenotypes are given in Figures 1 and 2.
Figure 1.—
Slit lamp microscopy documenting variable eye phenotypes associated with Col4a1 and Col4a2 mutations. Eyes from 11-month-old mice are shown. (A) C3H wild type. (B) Col4a2ENU4003 +/− expressing microphthalmia and total lens opacity. (C) Col4a2ENU4003 +/− expressing buphthalmos. (D) Col4a2ENU4020 +/− with total lens opacity, eye size normal. (E) Col4a1ENU911 +/− with microphthalmia. (F) Contralateral eye to E expressing corneal opacity, hyperplasia, and neovascularization (arrowhead). (G) Col4a1ENU4004 +/− with total lens opacity, eye size normal. (H) Col4a1ENU4004 +/− with total lens opacity and intraorbital hemorrhage (arrowhead). All eyes are photographed at 20× magnification. Bar in B, 1 mm.
Figure 2.—
Histology and lens morphology documenting the variable eye phenotypes associated with Col4a1 and Col4a2 mutations. (A–C) Eyes of E14 embryos. (A) Wild type with well-developed cornea (cor), lens (le), and retina (ret); (B) Col4a2ENU415 −/+ eye of normal size, thickened cornea, and reduced distance separating the cornea and the anterior surface of the lens (arrowhead); (C) Col4a2ENU415 −/− eye expressing microphthalmia and the lumen of the lens has failed to fill with primary lens fibers (arrowhead). (D–F) Eyes of P21 Col4a2ENU4020 −/+ mutant mice. (D) Area from the anterior surface of the lens showing disorganization of the lens epithelial layer (ep) and vacuoles in the underlying secondary fiber cell region (arrowhead). (E) Corneal-lens adhesion (arrowhead). (F) Preretinal blood vessel (arrowhead). (G–I) Eyes of P21 Col4a2ENU4003 −/+ mutant mice. (G) Preretinal blood vessel (arrowhead) in an eye diagnosed to have intraorbital hemorrhage and red floaters. (H) Anterior section of the same eye as in G demonstrating a hernia in the corneal basement (arrowhead) and the anterior chamber is filled with red blood cells. (I) Higher magnification of the same eye as in G demonstrating the disrupted preretinal blood vessel (arrowhead) and a plume of red blood cells in the posterior chamber. (J–L) Lenses of P35 Col4a1 or Col4a2 −/+ mutant mice. (J) Total lens opacity in a Col4a2ENU415 −/+ heterozygote. Eye size is normal and the lens is intact. (K) Lens of a Col4a1F247 −/+ carrier is totally opaque and disrupted. Eye size is normal. (L) Lenses from both eyes of a Col4a1D456 −/+ carrier. To the left is an intact and totally opaque lens from a normal-size eye. To the right is the lens from the contralateral eye of the same carrier expressing total opacity and microphthalmia. Bars in A–C, 100 μm. Bar in D, 20 μm. Bars in E–I, 50 μm.
Mutant viability and segregation analysis in embryonic stages:
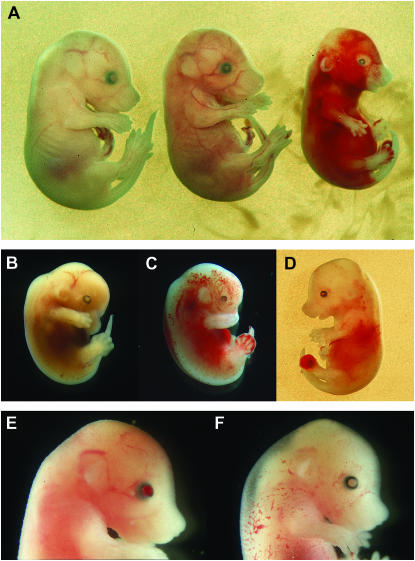
We attempted to produce homozygous mutant mice by inter se matings of heterozygotes (Table 4, Figure 3). Analysis of embryos at E14–E16 indicated a reduction in the number of embryos, an increase in the number of resorption sites (indicative of early embryo death), an increase in the number of dead embryos, and an approximate 1:2 ratio of wild-type to heterozygous mutant embryos (χ2 = 0.35 for the six Col4a1 mutant lines examined; χ2 = 0.00 for the three Col4a2 mutant lines). The number of dead embryos corresponds approximately to the expected number of homozygous mutant embryos. The increase in the number of resorbtion sites is not accompanied by a distortion in the ratio of homozygous wild-type to heterozygous mutant embryos and suggests that there is an increased probability of early embryonic death regardless of embryonic genotype in maternal mutant heterozygotes. Mutant embryos were often observed with hemorrhages in the eye as well as in other parts of the body (Figure 3). In eight of the nine mutant lines analyzed no homozygotes were observed. An extreme homozygous phenotype was observed only for Col4a2ENU415 (Table 4, Figure 3). However, the number of homozygous mutants observed was reduced and no homozygous mutants were observed at weaning (data not shown). Thus we concluded that all mutations are homozygous lethal.
TABLE 4.
Classification of embryos at E14–E16 in inter se crosses of heterozygous mutant or wild-type strain C3H/HeJ mice
| No. litters | Live embryos | Resorption sites | Dead embryos | Phenotype of live embryosa | |||
|---|---|---|---|---|---|---|---|
| Line | +/+ | M/+ | M/M | ||||
| C3H (wt) | 6 | 48 | 5 | 3 | 48 | 0 | 0 |
| Col4a1Acso | 7 | 43 | 25 | 4 | 3 | 9 | 0 |
| Col4a1ENU911 | 4 | 18 | 19 | 2 | 7 | 9 | 0 |
| Col4a1ENU4004 | 1 | 8 | 5 | 0 | 4 | 4 | 0 |
| Col4a1ENU6005 | 2 | 11 | 5 | 5 | 4 | 7 | 0 |
| Col4a1ENU6009 | 3 | 21 | 7 | 0 | 7 | 14 | 0 |
| Col4a1ENU6024 | 4 | 15 | 14 | 12 | — | — | — |
| Col4a1D456 | 1 | 3 | 8 | 0 | 1 | 2 | 0 |
| Col4a1F247 | 4 | 10 | 13 | 2 | — | — | — |
| Total | 26 | 129 | 96 | 25 | 26 | 45 | 0 |
| Col4a2ENU415 | 12 | 77 | 26 | 8 | 13 | 27 | 5 |
| Col4a2ENU4003 | 3 | 20 | 9 | 1 | 7 | 13 | 0 |
| Col4a2ENU4020 | 6 | 33 | 30 | 2 | 5 | 10 | 0 |
| Total | 21 | 130 | 65 | 11 | 25 | 50 | 5 |
Embyos from all litters were not classified.
Figure 3.—
Phenotypes in E14 or E15 embryos associated with the Col4a2ENU415 mutation. (A) +/+ (left), −/+ (middle), and −/− (right) E15 embryos. The heterozygous mutant expresses microphthalmia and a small hemorrhage on the right hind paw. The homozygous mutant expresses microphthalmia and extensive hemorrhaging over the entire body. (B) E14 −/+ embryo expressing total cataract. Eye size is normal. (C) E14 −/+ embryo expressing microphthalmia and extensive hemorrhages over the entire body. (D) E15 −/+ embryo expressing microphthalmia and hemorrhaging in the eye and body. (E) E15 −/+ embryo with hemorrhaging in a normal-sized eye. (F) E15 −/+ embryo with a normal eye and hemorrhaging in the skin.
Segregation analysis at weaning:
Segregation analyses of wild-type and heterozygous mutant offspring assessed at weaning (3 weeks) in the mapping backcrosses are given in Table 2. There was an overall deviation from the expected 1:1 ratio in the Col4a1 mutant lines (619:393, χ2 = 50.47). In contrast, the observed segregation of wild-type and heterozygous Col4a2 mutant offspring was as expected (171:164, χ2 = 0.15). Thus we concluded that most Col4a1 mutations are subvital and that the loss of heterozygous carriers occurs in late gestation or postnatal stages prior to weaning.
One mutation had reduced penetrance. In the mapping results from line Col4a1ENU6009 (Table 2), there was an abnormally high frequency of the crossover haplotype, wild type–D8Mit124C3H–D8Mit335C3H. Among the 52 backcross offspring classified as mutants there were four crossovers between the mutant locus and the marker D8Mit124, whereas in the 82 backcross offspring classified as wild type there were 20 presumed crossovers in the same region (P = 0.02, Fisher's exact test). We hypothesized a high rate of misclassification of mutant carriers as wild type and sequenced 18 presumed crossovers between the mutant locus and the marker D8Mit124, which were classified as having the haplotype wildtype–D8Mit124C3H for the cT4875C Col4a1ENU6009 mutant site. Seventeen were shown to be heterozygous carriers of the Col4a1ENU6009 mutation and represent misclassifications. One animal was homozygous wild-type Col4a1 and represents a crossover between Col4a1 and the marker D8Mit124.
Brain pathology:
Brains from a total of 3 wild-type, 12 Col4a1 heterozygous mutants and 11 Col4a2 heterozygous mutants were histologically evaluated (Figure 4). Twelve mutant heterozygotes (2 Col4a1F247, 2 Col4a1D456, 2 Col4a1ENU6005, 2 Col4a2ENU415, 3 Col4a2ENU4003, and 1 Col4a2ENU4020) were associated with abnormalities ranging from irregularities of lamina I with protrusions into the subarachnoid space and adhesion to the arachnoid, to pseudocysts in the upper cortical plate, to hemorrhages surrounding small blood vessels, to focal hemorrhagic necroses. In 14 embryos (3 +/+, 2 Col4a1Acso, 2 Col4a1ENU6005, 2 Col4a1ENU911, 3 Col4a2ENU415, 1 Col4a2ENU4003, and 1 Col4a2ENU4020) no histological defects were observed.
Figure 4.—
Brain histology in wild-type and heterozygous mutant Col4a1 or Col4a2 mouse embryos. (A) Coronal section of the dorsal telencephalon of an E15 +/+ embryo showing a well-developed arachnoid (ar), an intact and continuous lamina I (arrow) below the subarachnoid space, normal organization of the underlying laminae II–VI, and the lateral ventricular lumen (vl). (B) Dorsal telencephalon in an E17 Col4a2ENU415 heterozygote. There are multiple porencephalic lesions (*), a floating blood vessel (arrowhead) within a porencephalic lesion, disruptions of lamina I, and focal adhesions of lamina I to the arachnoid (arrow) with attachments of neuroblasts. (C) Thalamus of an E17 Col4a2ENU4003 heterozygote with extensive diapedesis of erythrocytes through the vascular wall (arrowhead). (D) Coronal section of the dorso-lateral telencephalon in an E17 Col4a2ENU4003 heterozygote. There is a large pseudocyst (*) situated between lamina I and the underlying cerebral cortex, clustering of migrating neurons at the margins of the pseudocyst (arrowhead), and disruptions in lamina I with a focal adhesion to the arachnoid (arrow). (E) Coronal section of the thalamic region in an E16 Col4a1D456 +/− embryo. There is a large hemorrhagic necrosis (*) at the lateral surface of the epithalamus. Nonpathological blood-filled meningeal vessels are shown (arrow). tha, thalamus; tel, telencephalon; vl III, third ventricular lumen. (F) Coronal section of the thalamic region in an E15 Col4a1F247 +/− embryo. There is disruption in the ventricular layer extending laterally through the neural parenchyma (arrowhead). A–E, 200× magnification. F, 400× magnification. Bars in B and F, 50 μm.
Hematology and clinical chemistry:
Since mutant embryos were often observed with hemorrhages in the skin and eye, we hypothesized that heterozygous carriers suffer from chronic bleeding and we conducted hematological analyses of 12-week-old mutants. In comparison to wild-type littermates, heterozygotes of all mutant lines showed a reduction in erythrocyte numbers, hematocrit, and hemoglobin content (supplemental Table 2 at http://www.genetics.org/supplemental/; Figure 5). We also hypothesized that the collagen defects might result in kidney dysfunction and measured total protein and albumin concentrations in the urine as well as creatinine and urea concentrations in the plasma of wild-type and heterozygous mutants. Results indicated an increase in the concentration of total protein in the urine and urea in the plasma of heterozygous Col4a1 or Col4a2 mutants as compared to wild-type littermates (supplemental Table 2; Figure 5). The kidneys of two heterozygous carriers each from the Col4a2ENU415, Col4a2ENU4003, and Col4a2ENU4020 lines (age range 6–11 months) were examined histologically and by electron microscopy. No pathological alterations in kidney histology or basal membrane thickness were observed (data not shown).
Figure 5.—
Hematology and clinical chemistry analysis of plasma and urine of wild-type and heterozygous mutant Col4a1 or Col4a2 mice. Each bar represents the mean with ±SEM from an approximately equal number of male and female mice. WT represents the pooled wild-type littermates from all Col4a1 and Col4a2 mutant lines (129 animals). Col4a1 consists of the pooled heterozygous carriers of the procollagen Col4a1 mutant lines (115 animals). Col4a2 represents the pooled heterozygous carriers from the procollagen Col4a2 mutant lines (57 animals). (A) Red blood cell count, showing a slight but consistent reduction in heterozygous mutants as compared to wild type. (B) Hematocrit, showing a reduction in the mutants as compared to wild type. (C) Hemoglobin, showing a reduction in the heterozygous mutants as compared to wild type. (D) Total protein in urine, showing an increase in the mutant lines as compared to wild type. (E) Albumin in urine, with no differences among the genotypes. (F) Plasma urea, showing a slight but significant increase in the mutant lines as compared to wild type.
Predicted disruption of the signal peptide cleavage site in Col4a2ENU415:
The Col4a2ENU415 mutation, Val31Phe, is in the extreme amino-terminal region of the Col4a2 procollagen protein. We submitted the sequence of the first 50 amino acids of the Col4a2 wild-type and Col4a2ENU415 mutant to SignalP 3.0 (Nielsen and Krogh 1998; Bendtsen et al. 2004) to determine if the signal peptide cleavage site is affected. In wild type (Val31) the signal peptide cleavage site is predicted to be between Gly33 and Gly34, while in the Col4a2ENU415 mutant sequence (Phe31) the signal peptide cleavage site is predicted to be disrupted (Figure 6).
Figure 6.—
Predicted location of signal peptide cleavage sites in the mouse Col4a2 wild-type and Col4a2ENU415 (Val31Phe) amino-terminal sequences. The sequence of the first 50 amino acids was submitted to SignalP 3.0 for evaluation. Red bars indicate the cleavage site scores for each amino acid position. (A) The wild-type sequence was predicted to have a signal peptide cleavage site between amino acid positions 33 and 34, with a cleavage site probability of 0.786. (B) Col4a2ENU415 sequence. The Val31Phe amino acid substitution disrupts the predicted cleavage site between positions 33 and 34. Low cleavage-site scores were obtained between positions 28 and 29, 30 and 31, and 32 and 33, with the maximum cleavage site probability of 0.252 between positions 28 and 29.
DISCUSSION
We report 12 missense mutations distributed among 11 sites in the mouse, which represent nine mutant alleles of the Col4a1 gene and the first documented mutant alleles at two sites of the Col4a2 gene. The mutations were associated with abnormalities of the lens, the cornea, vascular stability, the brain, kidney function, and survival in the embryonic or the postnatal period. These observations are consistent with the ubiquitous expression of the Col4a1 and Col4a2 genes, with specific expression documented in the cornea, the lens, the lens capsule, and the retina (Kleppel and Michael 1990; Sarthy 1993; Cheong et al. 1994; Qin et al. 1997; Kelley et al. 2002); capillaries (Bell et al. 2001; Tilling et al. 2002; Urabe et al. 2002); the brain (Kleppel et al. 1989; Pöschl et al. 2004); the kidney (Desjardins et al. 1990; Gunwar et al. 1998; Boutaud et al. 2000); and the embryonic membrane, Reichert's membrane, and the placenta (Kleppel et al. 1989; Miner et al. 2004; Pöschl et al. 2004). The nine Col4a1 mutations extend the mouse allelic series as well as the five COL4A1 missense mutations in humans. The mutant phenotypes were variable, possibly due to an interaction between genetic predisposition and random environmental trauma. Gould et al. (2005) reported that heterozygous carriers of a splice-site mutation leading to exon 40 skipping in the mouse express porencephaly and semilethality due to cerebral hemorrhage and also stated that carriers often expressed ocular and renal abnormalities, smaller body size, and reduced fertility. Homozygous mutants did not survive beyond midgestation. Van Agtmael et al. (2005) observed heterozygous Col4a1 missense mutants to express bruising at birth, buphthalmos, corneal opacity, malformed pupils, lens vacuoles, iris/corneal adhesions, and renal basement membrane defects especially in Bowman's capsule. The five human COL4A1 missense mutations were all identified in heterozygous patients from families with hereditary porencephaly caused by perinatal hemorrhages in the brain (Gould et al. 2005; Breedveld et al. 2006). Additional examination in one family also identified the risk of COL4A1 carriers to recurrent stroke and cataract in adults (van der Knaap et al. 2006). All of the Col4a1/COL4A1 missense mutations as well as the in-frame deletion of exon 40 likely encode for dominant negative gene products. In contrast, the targeted mutation that ablates Col4a1 and Col4a2 is not associated with phenotypic abnormalities in heterozygotes and suggests that a single copy of the Col4a1 and Col4a2 genes is sufficient for normal development (Pöschl et al. 2004). Homozygotes die at midgestation and express disrupted basement membranes, bleeding in the pericardium, reduced vascular development, and impaired placental development.
We present the first Col4a2 mutations and demonstrate defects in heterozygotes in the eye, the brain, vessel stability, and kidney function, similar to that documented for carriers of Col4a1 mutations. However, in contrast to Col4a1 mutants there was no reduction in survival to weaning in Col4a2 mutant heterozygotes. The major isoform of type IV collagen produced by the procollagens Col4a1 and Col4a2 is the heterotrimer [α1(IV)]2[α2(IV)]. If the participation in trimer formation of the mutant procollagens is not affected, 25% of the trimers will be normal in a heterozygous Col4a1 missense mutant. In a heterozygous Col4a2 missense mutant 50% of the trimers will be normal. The larger portion of normal collagen trimers in Col4a2 heterozygous mutants can explain the milder phenotypes as compared to Col4a1 heterozygous mutants. A milder phenotypic abnormality in Col4a2 mutant heterozygotes could also explain the predominance of Col4a1 mutations as compared to Col4a2 mutations recovered in the extensive mutagenesis screens for eye abnormalities (Favor 1983, 1986; Favor et al. 1987; Kratochvilova et al. 1988; Favor and Neuhäuser-Klaus 2000; Thaung et al. 2002; this study), although the target size of the two genes is similar.
We have demonstrated pathological defects in the brains of mice heterozygous for missense mutations of the Col4a1 or Col4a2 genes. The porencephalic lesions that we observed may be the consequence of brain hemorrhages and subsequent cell death due to a reduced regional blood supply or improper neuronal migration. In the brain, the covering membrane limitans functions as an anchoring point for the radial glial cells that extend from the ventricular zone to the pia and form a scaffold to traffic the migration of the differentiating neurons from the ventricular zone to the cortex. Proper attachment of the glial endfeet to the membrane limitans requires an intact basement membrane. We observed defects in mutant heterozygotes that reflect disturbances in the proper migration of neurons: in the areas of defective migration pseudocysts occur in the cortical plate and the neurons that normally would populate this area either migrate abnormally around this area with pathological attachments to the arachnoid or remain trapped basal to the cyst. Similar defects including neuronal ectopias often extending into the subarachnoid space, neuronal heteropias in the cortex, and hemorrhages were associated with mutations of basement membrane components in which the radial glia scaffold was disrupted such as in the Col4a1/a2 knockout (Pöschl et al. 2004), a mutation in the nidogen binding site of gamma 1 laminin (Halfter et al. 2002; Haubst et al. 2006), mutations in reelin and its adaptor molecule disabled homolog 1 (Förster et al. 2002; Frotscher et al. 2003; Hartfuss et al. 2003) and in perlecan (Costell et al. 1999; Haubst et al. 2006), and mutations of alpha 6 or beta 1 integrin (Georges-Labouesse et al. 1998; Graus-Porta et al. 2001; Haubst et al. 2006).
The Col4a1 and Col4a2 procollagens contain three domains, the amino-terminal 7s domain, a long collagenous domain, and a carboxy-terminal noncollagenous (NCl) domain (Muthukumaran et al. 1989; Saus et al. 1989). The collagenous domain is characterized by Gly-Xaa-Yaa amino acid repeats, with the highly conserved Gly sites critical for the proper biosynthesis of the collagen trimers (Prockop and Kivirikko 1995). Missense mutations at Gly sites within the collagenous domain of Col4a1/COL4A1 or Col4a2 predominate. Ten of 12 (this study), 2 of 3 (Van Agtmael et al. 2005), and 4 of 5 (Gould et al. 2005; Breedveld et al. 2006) mutations were identified as amino acid substitutions at Gly sites, which is consistent with previous observations in the extensive COL4A5 mutation database (Lemmink et al. 1997). Three missense mutations have been identified that are not at Gly sites of the collagenous domain. Col4a1Raw is within the collagenous domain, results in a Lys950Glu amino acid substitution, and was associated with a mild mutant phenotype (Van Agtmael et al. 2005). The Col4a1ENU6009 Ser1582Pro mutation is within the NC1 domain and also results in a milder mutant phenotype (this study). The mutated Ser site is completely conserved. A similar observation has been reported for a Lys1649Arg missense mutation at a highly conserved site within the NC1 domain of the COL4A5 procollagen, but results in a mild phenotype (Barker et al. 1996). The Col4a2 Ser1582 site has been designated to reside either in the IIβ2 or in the adjacent β3′ β-strand (authors' designations) of the NC1 domain (Sundaramoorthy et al. 2002; Than et al. 2002). A substitution of serine by a proline with a cyclic side chain within a β-strand would interfere with hydrogen bonding in the formation of a β-sheet. Two scenarios may be considered in reconciling the fact that an amino acid substitution at a highly conserved site with predicted effects on a β-strand motif is associated with a mild phenotype. The amino acid substitution may result in an extremely modified procollagen and reduce its participation in collagen trimer formation. Under this scenario the mutation behaves more like a null mutation as is seen in the targeted ablation of the Col4a1 and Col4a2 genes (Pöschl et al. 2004) and frameshift mutations in the carboxy terminus of the Col1a1 or Col1a2 genes (Pihlajaniemi et al. 1984; Willing et al. 1990). Assignment of the Col4a2ENU6009 mutation to either motif is consistent with this hypothesis. The Col4a2 β3′ strand is contained in a 6-strand β-sheet essential for trimer structure. The Col4a2 IIβ2 strand is contained in a 3-strand β-sheet important for the structure and stability of the β-barrel-like core (Sundaramoorthy et al. 2002; Than et al. 2002). It has been recently shown that the Col4a2 NCl domain “plays a regulatory role in directing chain composition in the assembly of (α1)2α2 triple-helical molecule” (Khoshnoodi et al. 2006, p. 6058). Alternatively, the globular NC1 domain contains such a large number of motifs (20 β-strands and three 310 helices) that a mutation in one motif may only slightly alter the overall globular structure with reduced effects on collagen network formation in the basement membrane.
The Col4a2ENU415 Val31Phe mutation is in the signal peptide sequence and is associated with a strong mutant phenotype. The Val31Phe mutant site is at position −3 relative to the signal peptidase I cleavage site. A bulky and aromatic Phe at this position violates the “−3, −1 rule” whereby small, neutral residues strongly prevail at these sites (von Heijne 1983) and signal peptide cleavage was predicted to be disrupted. Our characterization of the Col4a2ENU415 Val31Phe mutation indicates a strong abnormal phenotype likely due to impaired secretion of the Col4 heterotrimers similar to that demonstrated for two COL10A1 mutations that disrupt the signal peptide cleavage site. The procollagen peptides are translocated to the ER lumen, heterotrimer formation is not affected but cleavage of the signal peptide is inhibited, and the heterotrimers remain anchored to the ER membrane (Chan et al. 2001).
Our present results extend and complement previously published studies and associate phenotype defects with mutations of Col4a1 or Col4a2. We have still not identified human patients carrying COL4A2 mutations. On the basis of our results in the mouse we suggest that the spontaneous intraorbital hemorrhages in the eye are a clinically significant phenotype with a relatively high predictive value to identify COL4A1 or COL4A2 mutations. In our extensive eye screen in the mouse, this phenotype has been observed only in association with Col4a1 or Col4a2 mutants.
Acknowledgments
We thank Laure Bally-Cuif for critically reading the manuscript. The authors thank Sybille Frischolz, Bianca Hildebrand, Brigitta May, Elenore Samson, and Irmgard Zaus for their expert technical assistance and Utz Linzner, GSF–National Research Center for Environment and Health, Institute of Pathology, for the synthesis of the oligonucleotide primers. This research was supported in part by National Institutes of Health grant no. R01EY10321.
References
- Anker, M. C., J. Arnemann, K. Neumann, P. Ahrens, H. Schmidt et al., 2003. Alport syndrome with diffuse leiomyomatosis. Am. J. Med. Genet. A 119: 381–385. [DOI] [PubMed] [Google Scholar]
- Badenas, C., M. Praga, B. Tazón, L. Heidet, C. Arrondel et al., 2002. Mutations in the COL4A4 and COL4A3 genes cause familial benign hematuria. J. Am. Soc. Nephrol. 13: 1248–1254. [DOI] [PubMed] [Google Scholar]
- Barker, D. F., C. J. Pruchno, X. Jiang, C. L. Atkin, E. M. Stone et al., 1996. A mutation causing Alport syndrome with tardive hearing loss is common in the western United States. Am. J. Hum. Genet. 58: 1157–1165. [PMC free article] [PubMed] [Google Scholar]
- Bell, S. E., A. Mavila, R. Salazar, K. J. Bayless, S. Kanagala et al., 2001. Differential gene expression during capillary morphogenesis in 3D collagen matrices: regulated expression of genes involved in basement membrane matrix assembly, cell cycle progression, cellular differentiation and G-protein signaling. J. Cell Sci. 114: 2755–2773. [DOI] [PubMed] [Google Scholar]
- Bendtsen, J. D., H. Nielsen, G. von Heijne and S. Brunak, 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340: 783–795. [DOI] [PubMed] [Google Scholar]
- Boutaud, A., D. B. Borza, O. Bondar, S. Gunwar, K. O. Netzer et al., 2000. Type IV collagen of the glomerular basement membrane. Evidence that the chain specificity of network assembly is encoded by the noncollagenous NC1 domains. J. Biol. Chem. 275: 30716–30724. [DOI] [PubMed] [Google Scholar]
- Breedveld, G., I. F. de Coo, M. H. Lequin, W. F. Arts, P. Heutink et al., 2006. Novel mutations in three families confirm a major role of COL4A1 in hereditary porencephaly. J. Med. Genet. 43: 490–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, D., M. S. Ho and K. S. Cheah, 2001. Aberrant signal peptide cleavage of collagen X in Schmid metaphyseal chondrodysplasia. Implications for the molecular basis of the disease. J. Biol. Chem. 276: 7992–7997. [DOI] [PubMed] [Google Scholar]
- Cheong, H. I., C. E. Kashtan, Y. Kim, M. M. Kleppel and A. F. Michael, 1994. Immunohistologic studies of type IV collagen in anterior lens capsules of patients with Alport syndrome. Lab. Invest. 70: 553–557. [PubMed] [Google Scholar]
- Cosgrove, D., D. T. Meehan, J. A. Grunkemeyer, J. M. Kornak, R. Sayers et al., 1996. Collagen COL4A3 knockout: a mouse model for autosomal Alport syndrome. Genes Dev. 10: 2981–2992. [DOI] [PubMed] [Google Scholar]
- Costell, M., E. Gustafsson, A. Aszódi, M. Mörgelin, W. Bloch et al., 1999. Perlecan maintains the integrity of cartilage and some basement membranes. J. Cell Biol. 147: 1109–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desjardins, M., F. Gros, J. Wieslander, M. C. Gubler and M. Bendayan, 1990. Heterogeneous distribution of monomeric elements from the globular domain (NC1) of type IV collagen in renal basement membranes as revealed by high resolution quantitative immunocytochemistry. Lab. Invest. 63: 637–646. [PubMed] [Google Scholar]
- Favor, J., 1983. A comparison of the dominant cataract and recessive specific-locus mutation rates induced by treatment of male mice with ethylnitrosourea. Mutat. Res. 110: 367–382. [DOI] [PubMed] [Google Scholar]
- Favor, J., 1986. The frequency of dominant cataract and recessive specific-locus mutations in mice derived from 80 or 160 mg ethylnitrosourea per kg body weight treated spermatogonia. Mutat. Res. 162: 69–80. [DOI] [PubMed] [Google Scholar]
- Favor, J., and A. Neuhäuser-Klaus, 2000. Saturation mutagenesis for dominant eye morphological defects in the mouse Mus musculus. Mamm. Genome 11: 520–525. [DOI] [PubMed] [Google Scholar]
- Favor, J., A. Neuhäuser-Klaus and U. H. Ehling, 1987. Radiation-induced forward and reverse specific locus mutations and dominant cataract mutations in treated strain BALB/c and DBA/2 male mice. Mutat. Res. 177: 161–169. [DOI] [PubMed] [Google Scholar]
- Favor, J., P. Grimes, A. Neuhäuser-Klaus, W. Pretsch and D. Stambolian, 1997. The mouse Cat4 locus maps to chromosome 8 and mutants express lens-corneal adhesion. Mamm. Genome 8: 403–406. [DOI] [PubMed] [Google Scholar]
- Förster, E., A. Tielsch, B. Saum, K. H. Weiss, C. Johanssen et al., 2002. Reelin, Disabled 1, and beta 1 integrins are required for the formation of the radial glial scaffold in the hippocampus. Proc. Natl. Acad. Sci. USA 99: 13178–13183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frotscher, M., C. A. Haas and E. Förster, 2003. Reelin controls granule cell migration in the dentate gyrus by acting on the radial glial scaffold. Cereb. Cortex 13: 634–640. [DOI] [PubMed] [Google Scholar]
- Gailus-Durner, V., H. Fuchs, L. Becker, I. Bolle, M. Brielmeier et al., 2005. Introducing the German Mouse Clinic: open access platform for standardized phenotyping. Nat. Methods 2: 403–404. [DOI] [PubMed] [Google Scholar]
- Garcia-Torres, R., D. Cruz, L. Orozco, L. Heidet and M. C. Gubler, 2000. Alport syndrome and diffuse leiomyomatosis. Clinical aspects, pathology, molecular biology and extracellular matrix studies. A synthesis. Nephrologie 21: 9–12. [PubMed] [Google Scholar]
- Georges-Labouesse, E., M. Mark, N. Messaddeq and A. Gansmüller, 1998. Essential role of alpha 6 integrins in cortical and retinal lamination. Curr. Biol. 8: 983–986. [DOI] [PubMed] [Google Scholar]
- Gould, D. B., F. C. Phalan, G. J. Breedveld, S. E. van Mil, R. S. Smith et al., 2005. Mutations in Col4a1 cause perinatal cerebral hemorrhage and porencephaly. Science 308: 1167–1171. [DOI] [PubMed] [Google Scholar]
- Graus-Porta, D., S. Blaess, M. Senften, A. Littlewood-Evans, C. Damsky et al., 2001. Beta1-class integrins regulate the development of laminae and folia in the cerebral and cerebellar cortex. Neuron 31: 367–379. [DOI] [PubMed] [Google Scholar]
- Gunwar, S., F. Ballester, M. E. Noelken, Y. Sado, Y. Ninomiya et al., 1998. Glomerular basement membrane. Identification of a novel disulfide-cross-linked network of alpha3, alpha4, and alpha5 chains of type IV collagen and its implications for the pathogenesis of Alport syndrome. J. Biol. Chem. 273: 8767–8775. [DOI] [PubMed] [Google Scholar]
- Halfter, W., S. Dong, Y. P. Yip, M. Willem and U. Mayer, 2002. A critical function of the pial basement membrane in cortical histogenesis. J. Neurosci. 22: 6029–6040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartfuss, E., E. Förster, H. H. Bock, M. A. Hack, P. Leprince et al., 2003. Reelin signaling directly affects radial glia morphology and biochemical maturation. Development 130: 4597–4609. [DOI] [PubMed] [Google Scholar]
- Haubst, N., E. Georges-Labouesse, A. De Arcangelis, U. Mayer and M. Götz, 2006. Basement membrane attachment is dispensable for radial glial cell fate and for proliferation, but affects positioning of neuronal subtypes. Development 133: 3245–3254. [DOI] [PubMed] [Google Scholar]
- Jais, J. P., B. Knebelmann, I. Giatras, M. De Marchi, G. Rizzoni et al., 2000. X-linked Alport syndrome: natural history in 195 families and genotype- phenotype correlations in males. J. Am. Soc. Nephrol. 11: 649–657. [DOI] [PubMed] [Google Scholar]
- Kelley, P. B., Y. Sado and M. K. Duncan, 2002. Collagen IV in the developing lens capsule. Matrix Biol. 21: 415–423. [DOI] [PubMed] [Google Scholar]
- Khoshnoodi, J., K. Sigmundsson, J. P. Cartailler, O. Bondar, M. Sundaramoorthy et al., 2006. Mechanism of chain selection in the assembly of collagen IV: a prominent role for the {alpha}2 chain. J. Biol. Chem. 281: 6058–6069. [DOI] [PubMed] [Google Scholar]
- Kleppel, M. M., and A. F. Michael, 1990. Expression of novel basement membrane components in the developing human kidney and eye. Am. J. Anat. 187: 165–174. [DOI] [PubMed] [Google Scholar]
- Kleppel, M. M., P. A. Santi, J. D. Cameron, J. Wieslander and A. F. Michael, 1989. Human tissue distribution of novel basement membrane collagen. Am. J. Pathol. 134: 813–825. [PMC free article] [PubMed] [Google Scholar]
- Kratochvilova, J., J. Favor and A. Neuhäuser-Klaus, 1988. Dominant cataract and recessive specific-locus mutations detected in offspring of procarbazine-treated male mice. Mutat. Res. 198: 295–301. [DOI] [PubMed] [Google Scholar]
- Kühn, K., 1995. Basement membrane (type IV) collagen. Matrix Biol. 14: 439–445. [DOI] [PubMed] [Google Scholar]
- Lemmink, H. H., C. H. Schröder, L. A. Monnens and H. J. Smeets, 1997. The clinical spectrum of type IV collagen mutations. Hum. Mutat. 9: 477–499. [DOI] [PubMed] [Google Scholar]
- Li, S., P. Liquari, K. K. McKee, D. Harrison, R. Patel et al., 2005. Laminin-sulfatide binding initiates basement membrane assembly and enables receptor signaling in Schwann cells and fibroblasts. J. Cell Biol. 169: 179–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, W., C. L. Phillips, P. D. Killen, T. Hlaing, W. R. Harrison et al., 1999. Insertional mutation of the collagen genes Col4a3 and Col4a4 in a mouse model of Alport syndrome. Genomics 61: 113–124. [DOI] [PubMed] [Google Scholar]
- Manly, K. F., 1993. A Macintosh program for storage and analysis of experimental genetic mapping data. Mamm. Genome 4: 303–313. [DOI] [PubMed] [Google Scholar]
- Miner, J. H., and J. R. Sanes, 1994. Collagen IV alpha 3, alpha 4, and alpha 5 chains in rodent basal laminae: sequence, distribution, association with laminins, and developmental switches. J. Cell Biol. 127: 879–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miner, J. H., and J. R. Sanes, 1996. Molecular and functional defects in kidneys of mice lacking collagen alpha 3(IV): implications for Alport syndrome. J. Cell Biol. 135: 1403–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miner, J. H., C. Li, J. L. Mudd, G. Go and A. E. Sutherland, 2004. Compositional and structural requirements for laminin and basement membranes during mouse embryo implantation and gastrulation. Development 131: 2247–2256. [DOI] [PubMed] [Google Scholar]
- Mothes, H., L. Heidet, C. Arrondel, K. K. Richter, M. Thiele et al., 2002. Alport syndrome associated with diffuse leiomyomatosis: COL4A5–COL4A6 deletion associated with a mild form of Alport nephropathy. Nephrol. Dial. Transplant. 17: 70–74. [DOI] [PubMed] [Google Scholar]
- Muthukumaran, G., B. Blumberg and M. Kurkinen, 1989. The complete primary structure for the alpha 1-chain of mouse collagen IV. Differential evolution of collagen IV domains. J. Biol. Chem. 264: 6310–6317. [PubMed] [Google Scholar]
- Nielsen, H., and A. Krogh, 1998. Prediction of signal peptides and signal anchors by a hidden Markov model. Proc. Int. Conf. Intell. Syst. Mol. Biol. 6: 122–130. [PubMed] [Google Scholar]
- Pescucci, C., F. Mari, I. Longo, P. Vogiatzi, R. Caselli et al., 2004. Autosomal-dominant Alport syndrome: natural history of a disease due to COL4A3 or COL4A4 gene. Kidney Int. 65: 1598–1603. [DOI] [PubMed] [Google Scholar]
- Pihlajaniemi, T., L. A. Dickson, F. M. Pope, V. R. Korhonen, A. Nicholls et al., 1984. Osteogenesis imperfecta: cloning of a pro-alpha 2(I) collagen gene with a frameshift mutation. J. Biol. Chem. 259: 12941–12944. [PubMed] [Google Scholar]
- Pöschl, E., U. Schlötzer-Schrehardt, B. Brachvogel, K. Saito, Y. Ninomiya et al., 2004. Collagen IV is essential for basement membrane stability but dispensable for initiation of its assembly during early development. Development 131: 1619–1628. [DOI] [PubMed] [Google Scholar]
- Prockop, D. J., and K. I. Kivirikko, 1995. Collagens: molecular biology, diseases, and potentials for therapy. Annu. Rev. Biochem. 64: 403–434. [DOI] [PubMed] [Google Scholar]
- Qin, P., M. Piechocki, S. Lu and M. A. Kurpakus, 1997. Localization of basement membrane-associated protein isoforms during development of the ocular surface of mouse eye. Dev. Dyn. 209: 367–376. [DOI] [PubMed] [Google Scholar]
- Rathkolb, B., T. Decker, E. Fuchs, D. Soewarto, C. Fella et al., 2000. The clinical-chemical screen in the Munich ENU Mouse Mutagenesis Project: screening for clinically relevant phenotypes. Mamm. Genome 11: 543–546. [DOI] [PubMed] [Google Scholar]
- Renieri, A., M. T. Bassi, L. Galli, J. Zhou, M. Giani et al., 1994. Deletion spanning the 5′ ends of both the COL4A5 and COL4A6 genes in a patient with Alport's syndrome and leiomyomatosis. Hum. Mutat. 4: 195–198. [DOI] [PubMed] [Google Scholar]
- Rheault, M. N., S. M. Kren, B. K. Thielen, H. A. Mesa, J. T. Crosson et al., 2004. Mouse model of X-linked Alport syndrome. J. Am. Soc. Nephrol. 15: 1466–1474. [DOI] [PubMed] [Google Scholar]
- Russell, W. L., 1951. X-ray-induced mutations in mice. Cold Spring Harbor Symp. Quant. Biol. 16: 327–336. [DOI] [PubMed] [Google Scholar]
- Sado, Y., M. Kagawa, I. Naito, Y. Ueki, T. Seki et al., 1998. Organization and expression of basement membrane collagen IV genes and their roles in human disorders. J. Biochem. 123: 767–776. [DOI] [PubMed] [Google Scholar]
- Sarthy, V., 1993. Collagen IV mRNA expression during development of the mouse retina: an in situ hybridization study. Invest. Ophthalmol. Vis. Sci. 34: 145–152. [PubMed] [Google Scholar]
- Saus, J., S. Quinones, A. MacKrell, B. Blumberg, G. Muthukumaran et al., 1989. The complete primary structure of mouse alpha 2(IV) collagen. Alignment with mouse alpha 1(IV) collagen. J. Biol. Chem. 264: 6318–6324. [PubMed] [Google Scholar]
- Sundaramoorthy, M., M. Meiyappan, P. Todd and B. G. Hudson, 2002. Crystal structure of NC1 domains. Structural basis for type IV collagen assembly in basement membranes. J. Biol. Chem. 277: 31142–31153. [DOI] [PubMed] [Google Scholar]
- Than, M. E., S. Henrich, R. Huber, A. Ries, K. Mann et al., 2002. The 1.9-A crystal structure of the noncollagenous (NC1) domain of human placenta collagen IV shows stabilization via a novel type of covalent Met-Lys cross-link. Proc. Natl. Acad. Sci. USA 99: 6607–6612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thaung, C., K. West, B. J. Clark, L. McKie, J. E. Morgan et al., 2002. Novel ENU-induced eye mutations in the mouse: models for human eye disease. Hum. Mol. Genet. 11: 755–767. [DOI] [PubMed] [Google Scholar]
- Tilling, T., C. Engelbertz, S. Decker, D. Korte, S. Hüwel et al., 2002. Expression and adhesive properties of basement membrane proteins in cerebral capillary endothelial cell cultures. Cell Tissue Res. 310: 19–29. [DOI] [PubMed] [Google Scholar]
- Urabe, N., I. Naito, K. Saito, T. Yonezawa, Y. Sado et al., 2002. Basement membrane type IV collagen molecules in the choroid plexus, pia mater and capillaries in the mouse brain. Arch. Histol. Cytol. 65: 133–143. [DOI] [PubMed] [Google Scholar]
- Van Agtmael, T., U. Schlötzer-Schrehardt, L. McKie, D. G. Brownstein, A. W. Lee et al., 2005. Dominant mutations of Col4a1 result in basement membrane defects which lead to anterior segment dysgenesis and glomerulopathy. Hum. Mol. Genet. 14: 3161–3168. [DOI] [PubMed] [Google Scholar]
- van der Knaap, M. S., L. M. Smit, F. Barkhof, Y. A. Pijnenburg, S. Zweegman et al., 2006. Neonatal porencephaly and adult stroke related to mutations in collagen IV A1. Ann. Neurol. 59: 504–511. [DOI] [PubMed] [Google Scholar]
- von Heijne, G., 1983. Patterns of amino acids near signal-sequence cleavage sites. Eur. J. Biochem. 133: 17–21. [DOI] [PubMed] [Google Scholar]
- Willing, M. C., D. H. Cohn and P. H. Byers, 1990. Frameshift mutation near the 3′ end of the COL1A1 gene of type I collagen predicts an elongated Pro alpha 1(I) chain and results in osteogenesis imperfecta type I. J. Clin. Invest. 85: 282–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, J., T. Mochizuki, H. Smeets, C. Antignac, P. Laurila et al., 1993. Deletion of the paired alpha 5(IV) and alpha 6(IV) collagen genes in inherited smooth muscle tumors. Science 261: 1167–1169. [DOI] [PubMed] [Google Scholar]