Abstract
Proteins encoded by the mdm2 gene, which has a pivotal role in the regulation of growth and differentiation, exist principally in human and murine cells as two isoforms that migrate in gels as 75-kDa and 90-kDa proteins. There is limited understanding of the respective biological roles of these isoforms, their molecular nature, and their mechanism of formation. We report here that human p75MDM2 is an N-terminally truncated mixture of protein isoforms produced by the initiation of translation at two distinct internal AUG codons. The p75MDM2 doublets and p90MDM2, which is the full-length MDM2 protein, are expressed in approximately equal amounts from transcripts initiated at the constitutive P1 promoter of mdm2. Unlike murine transcripts initiated at the p53-activated P2 promoter, human cell transcripts initiated at the P2 promoter preferentially produce p90MDM2. The ubiquitin enzyme variant protein TSG101, which interacts functionally with MDM2 in an autoregulatory loop that parallels the p53/MDM2 feedback control loop, interferes with degradation of both isoforms; however, only p90MDM2 promotes proteolysis of TSG101 and p53. Our results reveal the mechanism of formation of the principal MDM2 isoforms, the differential effects of p53 on the production of these isoforms, and the differential abilities of human MDM2 isoforms as regulators of the MDM2/TSG101 and p53/MDM2 feedback control loops.
mdm2 initially was identified in a screen for genes amplified on double minute chromosomes found in spontaneously transformed mouse 3T3 fibroblasts (14). Subsequent work, which has shown that elevated expression of the MDM2 protein can promote neoplastic transformation of cells in culture and tumor formation in animals (24) and that amplification of the human ortholog of mdm2 occurs in a wide variety of cancers (39, 41), suggests that mdm2 is an oncogene of major importance. Perhaps the most extensively studied biological function of MDM2 is regulation of the activity and abundance of p53, a tumor suppressor protein that modulates the expression of genes involved in DNA repair, cell cycle progression, and apoptosis (13). By binding to p53, MDM2 inhibits p53's transcription-promoting actions and additionally functions as an E3 ubiquitin ligase to accelerate degradation of the p53 protein (15, 21, 40); conversely, transcription of mdm2 is activated by p53 (3). Thus, MDM2 and p53 participate in an autoregulatory (“feedback control”) loop in which p53 modulates the production of a protein that inhibits its function (53).
The actions of other cellular genes implicated in tumorigenesis affect the workings of the MDM2/p53 feedback control loop (13). Among these is tumor susceptibility gene tsg101, which initially was identified in a genetic screen for mouse fibroblasts that undergo neoplastic transformation as a result of random chromosomal gene inactivation (28). Deficiency of TSG101 in NIH 3T3 cells reversibly results in colony formation in soft agar, focus formation in monolayer cultures, and the ability to form metastatic tumors in nude mice (28). The steady-state level of TSG101, which is an essential cellular protein (45, 52), is regulated posttranslationally within a narrow range (16), and overexpression of TSG101 from an adventitious promoter can also result in neoplastic transformation (28). Through its ubiquitin-conjugating E2 variant (UEV) domain, TSG101 interacts with MDM2, inhibits MDM2 ubiquitination, and prolongs the half-life of MDM2 protein; conversely, elevation of MDM2 promotes proteolysis of TSG101, as occurs for p53 (29). As the MDM2/TSG101 regulatory loop modulates the cellular levels of both proteins and consequently affects MDM2 control of p53 (29), TSG101 is both a regulator of and target of p53/MDM2 circuitry. TSG101 has also been shown to be a key component of complexes that mediate endocytic trafficking of cell surface receptors and the budding of medically important pathogenic viruses (18, 30, 32).
More than 40 splice variants of mdm2 mRNA and also multiple isoforms of MDM2 protein that interact differentially with p53 have been identified in tumors and normal tissues (4). However, two particular isoforms, which migrate in sodium dodecyl sulfate (SDS) polyacrylamide gels as 90-kDa and 75-kDa proteins, predominate in both mouse and human cells (6, 46). Earlier experiments from our laboratory have shown that ubiquitination and stability of at least one of these human isoforms, p90MDM2, is affected by TSG101 (29). There have been differing conclusions as to whether this isoform or p75MDM2 is the full-length protein (7, 37, 55). Here we report the results of investigations of the molecular nature and mechanism of production of these two principal human MDM2 isoforms and their respective roles in the MDM2/TSG101 and MDM2/p53 feedback control loops. Our findings confirm that p90MDM2 is an unconjugated full-length human MDM2 (HDM2) protein, demonstrate that this human MDM2 isoform is preferentially synthesized on transcripts initiated at the p53-regulated P2 promoter, and establish that p75MDM2 is a mixture of truncated proteins produced by the initiation of translation at two different internal AUG codons within transcripts encoded largely by P1. We further show that only p90MDM2 promotes proteolysis of either p53 or TSG101, but that TSG101 stabilizes both the p75MDM2 and p90MDM2 isoforms. Our results identify p90MDM2 as the human MDM2 isoform that controls the protein levels of p53, TSG101, and MDM2 itself through the actions of p53/MDM2 and MDM2/TSG101 feedback control loops.
MATERIALS AND METHODS
Plasmid and vector construction.
Exon1-MDM2-YFP and Exon2-MDM2-YFP are expressing constructs that produce transcripts similar to P1- and P2-derived mdm2 mRNAs, respectively. In order to isolate mdm2 transcripts derived from the P1 and P2 promoters, Saos-2 cells (p53 null) were transfected with the p53 expression construct pC53-SN3, and mRNA from transfected cells was isolated by FastTrack (Invitrogen). Following reverse transcription, cDNA was subjected to PCR amplification using specific primer sets that generate the mdm2 open reading frame (ORF) attached to either exon 1 or exon 2 as its 5′ untranslated region (UTR). The desired PCR products were cloned into p-EYFP-N1 (Clontech) between a cytomegalovirus (CMV) promoter and the ORF of yfp to create Exon1-MDM2-YFP and Exon2-MDM2-YFP. Exon1-MDM2-YFP-50AUC, Exon1-MDM2-YFP-62AUC, Exon1-MDM2-YFP-50, 62 AUC, Exon1-MDM2-YFP-62, 102 AUC, and Exon1-MDM2-YFP-50, 62, 102 AUC were modified from Exon1-MDM2-YFP by using the Quikchange site-directed mutagenesis kit (Stratagene) to generate an ATG-to-ATC point mutation at desired target sites. The absence of additional mutations in the ORF of fusion genes was confirmed by DNA sequencing. The full-length mdm2 ORF containing a hemagglutinin (HA) tag at the 5′ end was inserted into the pLLEXP1 vector 3′ to the CMV promoter and 5′ to a polyadenylation site to create HA-MDM2-FL. Other constructs that express truncated MDM2 tagged at the N-terminal end with HA (i.e., HA-MDM2-DN61 and HA-MDM2-DN101) were derived from HA-MDM2-FL by replacing the full-length mdm2 ORF with corresponding PCR products that lack the nucleotides for either the first 61 or 101 amino acids of MDM2. Vectors expressing human TSG101 (HA-TSG101), human wild-type p53 (pC53-SN3), and green fluorescent protein (pCMV-GFP) have been described previously (29).
Cell culture and transfection.
Saos-2 and U2OS cells were obtained from the American Type Culture Collection. HCT116 cells were kindly provided by Jim Ford. Cells were cultured in Dulbecco's modified Eagle's medium (Saos-2 and U2OS) or McCoy's 5A medium (HCT116) supplemented with 10% fetal bovine serum. Transfections were carried out using LipofectAMINE Plus (Invitrogen), LipofectAMINE 2000 (Invitrogen), or FuGENE 6 (Roche) as described by the manufactures.
Antibodies.
Monoclonal antibodies against GFP (Clontech), β-tubulin (NeoMarkers), p53 (DO-1; Santa Cruz), TSG101 (Santa Cruz), and SUMO-1 (Zymed) were purchased. Monoclonal anti-MDM2 antibodies 2A9 (Oncogene) and 2A10 (Oncogene), used for immunoprecipitation, were purchased, and other MDM2 antibodies, such as 4B2, 3G5, and 4B11, were generously provided by A. Levine. These anti-MDM2 antibodies were generated against human MDM2 proteins, and the epitope recognized by each monoclonal antibody had been mapped individually (11). Purified rabbit polyclonal antibodies against RanGAP1 (a kind gift of Larry Gerace) were used for immunoprecipitation, and monoclonal RanGAP1 antibody (Zymed) was used in immunoblotting. Detection of HA-tagged proteins was carried out using anti-HA antibody (horseradish peroxidase labeled; Roche).
Immunoprecipitation and immunoblotting analyses.
Immunoprecipitation and immunoblotting analyses were performed as described previously (29), with minor modifications. Cultured cells were lysed with radio immunoprecipitation assay (RIPA) buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1% NP-40, 0.1% SDS, and 1% sodium deoxycholate) supplemented with 1 mM Na3VO4, 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, and protease inhibitor cocktail (Sigma). The collected lysates were precleaned with prewashed protein A agarose beads (Upstate) for 1 h at 4°C and then incubated with respective antibodies (1 μg) overnight at 4°C. Following incubation with prewashed protein A agarose beads for another 3 h, the beads were washed with RIPA buffer, TBS (10 mM Tris-HCl [pH 8.0] and 150 mM NaCl) containing 0.5 M LiCl, and washed two additional times with TBS. The immunocomplex was dissolved in SDS loading buffer and fractionated, followed by immunoblotting analysis with the indicated antibodies. To achieve better resolution for protein molecules having a mass greater than 100 kDa, SDS-5% polyacrylamide gels were used to fractionate precipitated proteins. For detection of proteins that coprecipitated with TSG101, transfected cell populations containing the indicated expression constructs were lysed in Nonidet P-40 lysis buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 0.5% NP-40, and 50 mM NaF), and the immunocomplex was washed with NP-40 lysis buffer instead of RIPA buffer. The signal for the immunoblotting was detected by ECL Plus Western Blotting Detection Reagents (Amersham) or by Western lighting (PerkinElmer Life Sciences). Protein size markers with known molecular masses ranging from 10 to 250 kDa were purchased from Bio-Rad (all blue; #161-0373) and used in estimating the molecular mass of proteins analyzed in gels.
Reverse transcription-PCR (RT-PCR).
After transfection with the indicated constructs, total RNA was isolated using TRIzol (Invitrogen). An equivalent amount of RNA from each sample was converted into cDNA by reverse transcriptase and subsequently amplified with PCR using Exon1 primer (5′-GAGTGGAATGATCCCCGAGG-3′) and Exon6 primer (5′-GACTACTACCAAGTTCCTGTAG-3′) for the Exon1-6 fragment and Exon2 (5′-CAGTGGCGATTGGAGGGTAG-3′) and Exon6 primers for the Exon2-6 fragment. The primer set of GAPDH-f (5′-AAGTATGACAACAGCCTCAAGA-3′) and GAPDH-r (5′-CACCACCTTCTTGATGTCATCA-3′) was used to detect the corresponding glyceraldehyde-3-phosphate dehydrogenase (GAPDH) PCR product with a size of 372 bp. The number of PCR amplification cycles for GAPDH, Exon1-6, and Exon2-6 were 21, 40, and 30, respectively.
RESULTS
Human p75MDM2 is an N-terminally truncated MDM2 protein.
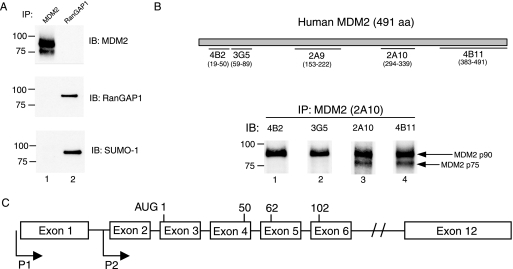
Notwithstanding a calculated molecular mass of 54 kDa for the full-length MDM2 protein, which contains 491 amino acid residues, the principal human MDM2 isoforms normally observed by SDS-polyacrylamide gel electrophoresis migrate at positions corresponding to 75-kDa and 90-kDa proteins. In murine cells, two MDM2 isoforms migrating at analogous positions have been discovered and extensively characterized (3, 46). As the amino acid sequences of the MDM2 proteins are highly conserved between human and murine cells, it has been generally assumed that the similarly migrating isoforms are formed by a similar mechanism in human and murine cells. Whereas recent reports have reached disparate conclusions about the nature of the human p75 and p90 MDM2 species and the role of sumoylation in their formation (7, 37, 55), our immunoblot studies of MDM2 proteins isolated from the human osterosarcoma cell line Saos-2 (Fig. 1A) support the view (37, 55) that p90MDM2 is an unconjugated full-length form of the protein. Additional immunoblot analyses using antibodies directed against SUMO-1 also did not detect this protein in the p75MDM2 band or the related SUMO-3 protein in either MDM2 isoform (data not shown).
FIG. 1.
N-terminally truncated forms of human and murine p75MDM2 originate differently. (A) Protein extracts of Saos-2 cells were subjected to immunoprecipitation (IP) using either RanGAP1 or MDM2 antibody (2A10), followed by immunoblotting (IB) and detection by MDM2 (4B11), RanGAP1, and SUMO-1 antibodies applied sequentially. The band positions for protein standards having known molecular masses of 75 kDa and 100 kDa (Bio-Rad) are indicated at the left of each panel. (B) Equal amounts of immunoprecipitated proteins by MDM2 antibodies (2A10) from whole-cell lysates of Saos-2 were applied to polyacrylamide gels, separated by SDS-polyacrylamide gel electrophoresis, and analyzed by immunoblotting using MDM2 antibodies 4B2, 3G5, 2A10, and 4B11 as shown. A schematic diagram of locations on the full-length MDM2 protein of epitopes recognized by the MDM2 monoclonal antibodies used in this study, including 4B2 (amino acids 19 to 50), 3G5 (amino acids 59 to 89), 2A9 (amino acids 153 to 222), 2A10 (amino acids 294 to 339), and 4B11 (amino acids 383 to 491) is shown on the top of figure. (C) Genomic organization of the human mdm2 gene with two distinct promoters designated P1 and P2. P1 is a constitutive promoter, and P2 is a p53-responsive promoter that is located between exon 1 and exon 2 of the gene. The first AUG codon for the initiation of translation of MDM2 protein resides in exon 3, and the corresponding positions for other potential translation start sites (AUG codons) are also indicated.
How, then, are the two isoforms of human MDM2 produced? In murine cells, the presence or absence of the amino terminus of MDM2 protein has been proposed to account for differences in the apparent molecular mass of two MDM2-derived polypeptides migrating at approximately the same positions as the p90MDM2 and p75MDM2 human MDM2 isoforms. Using monoclonal antibodies that recognize different segments of the human MDM2 protein (11) to identify the domains present in p90MDM2 and p75MDM2, we found that human p90MDM2 includes amino acid residues from both ends of the protein, whereas p75MDM2 failed to interact with antibody specifically directed against the N-terminal portion antibodies (i.e., antibodies 4B2 and 3G5; Fig. 1B). However, this analysis also indicated that amino-terminally truncated human p75MDM2, which was not detected by antibody 3G5, is shorter than the corresponding murine protein, which was detected by this antibody (46). As antibody 3G5 requires amino acid residues 59 to 89 of MDM2 for binding, we infer that human p75MDM2 lacks at least the first 59 amino acids of the MDM2 protein.
Translation initiation at the internal start sites of P1-driven mdm2 transcript leads to protein synthesis of N-terminally truncated human MDM2 isoforms.
At least four mechanisms, individually or in combination, potentially could account for the generation of N-terminally truncated MDM2 isoform: initiation of mdm2 transcription at a downstream site, alternative splicing of mdm2 transcripts, internal initiation of translation, or proteolytic cleavage of full-length MDM2 protein (43, 46, 47). Earlier work has shown that transcription of both the human and murine mdm2 genes occurs at two distinct promoters, designated P1 and P2 (25, 58). P2 is a p53-responsive promoter located within the first intron of the gene, whereas the more upstream P1 promoter is constitutive and p53 independent. Human mdm2 transcripts initiated from P1 undergo removal of exon 2 by splicing and retain exon 1 as a 5′ UTR; conversely, transcripts initiated from P2 lack exon 1 but contain exon 2 (58). As the first AUG codon of the mdm2 translational open reading frame (ORF) is in exon 3 (Fig. 1C), mdm2 transcripts initiated at either P1 or P2 potentially can encode the synthesis of full-length MDM2 protein (2).
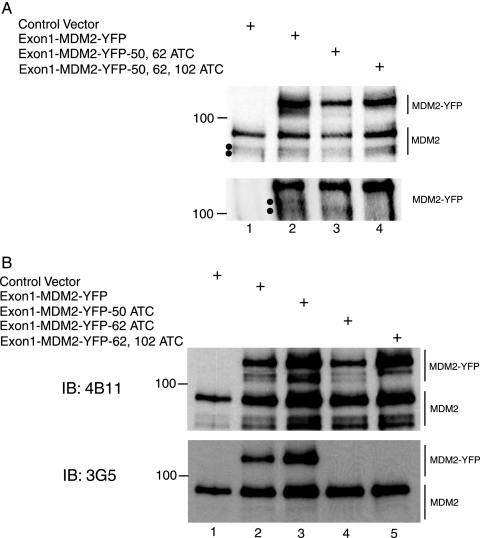
In murine cells, production of p75MDM2 requires the presence of exon 1 as a 5′ UTR and results from initiation of translation at an internal AUG codon at a site corresponding to codon position 50 of the full-length protein (35, 46). However, there is limited sequence conservation between exon 1 of the human and murine mdm2 genes (23), and while an AUG codon is present also at position 50 of full-length human MDM2 protein, our finding that the N-terminal end of the 75-kDa human MDM2 isoform is located distally to this codon suggested that human p75MDM2 is generated by a mechanism different from the one that generates its murine counterpart. To investigate the mechanism of formation of the N-terminally truncated isoform of human MDM2, we designed a construct that expresses, under control of a CMV promoter, MDM2 proteins fused in frame at the C-terminal end with yellow fluorescent protein (YFP) (Exon1-MDM2-YFP). In this construct, the exon 1 sequence of mdm2 transcripts serves as a 5′ UTR. As the C-terminal YFP tag increases the mass of MDM2 isoforms that are expressed from this construct and were detectable only in cells transfected with it, endogenous MDM2 isoforms and the adventitiously expressed YFP-tagged isoforms produced in the same cell can be probed concurrently with anti-MDM2 antibody 4B11 in immunoblotting experiments.
As shown in Fig. 2A, lanes 1 and 2, two analogous sets of bands having migration properties corresponding to YFP-tagged and native MDM2 isoforms were observed by Western blot analysis of lysates of Saos-2 cells transfected with the Exon1-MDM-YFP vector. Gels that were subjected to an extended period of electrophoresis in order to resolve protein species having only slightly different migration properties (Fig. 3A, lane 2) revealed that both the native and YFP-labeled proteins consist of doublet bands migrating at the positions expected for N-terminally truncated MDM2 isoforms. Fractionation of immunoprecipitated proteins in SDS-5% polyacrylamide gel followed by immunoblot analysis with anti-MDM2 antibody 4B11, which recognizes an epitope located between amino acid residues 383 and 491 of the full-length protein (11), confirmed that two distinct MDM2 isoforms having slightly different rates of migration were being produced by the Exon1-MDM2-YFP construct (Fig. 2A, lane 2 versus lane 1, bottom panel). In contrast, the 3G5 antibody, which interacts with a segment of MDM2 located between amino acids 59 and 89 (11), failed to detect either the adventitiously expressed or endogenous set of doublet bands while nevertheless detecting the 90-kDa native protein and an adventitiously expressed protein corresponding to YFP-tagged full-length MDM2 (Fig. 2B, lane 2, bottom panel), indicating absence of all or part of the 59- to 89-amino-acid region in human MDM2 protein isoforms produced from mdm2 transcripts containing exon 1 as a 5′ UTR.
FIG. 2.
Translation initiation at start sites internal to the mdm2 ORF is responsible for the production of N-terminally truncated MDM2 isoforms. (A) Saos-2 cells were transfected with the indicated YFP-tagged MDM2 constructs (5 μg). Following immunoprecipitation by a mixture of MDM2 antibodies 2A9 and 2A10, precipitated proteins from lysates of the transfected cells were analyzed by immunoblotting (IB) using monoclonal MDM2 antibody 4B11 as probe. The top panel shows gel locations of bands corresponding to endogenous as well as exogenous MDM2. Bands resulting from expression of YFP-tagged MDM2 constructs and which have been separated in a lower percentage gel to achieve better resolution are shown in the bottom panel. Dots in both panels indicate the positions of faster-migrating MDM2 isoforms in which the N-terminal domain of the protein is absent. (B) MDM2 proteins in Saos-2 cells transfected with indicated constructs were analyzed as described for panel A, except that MDM2 antibodies 4B11 as well as 3G5 were used for probing sequentially in immunoblotting. The 3G5 antibody, which requires a native MDM2 segment between amino acids 59 and 89, fails to detect mutated MDM2-YFP (lane 4 and lane 5, bottom panel), possibly because the target protein has a substituted amino acid at position 62.
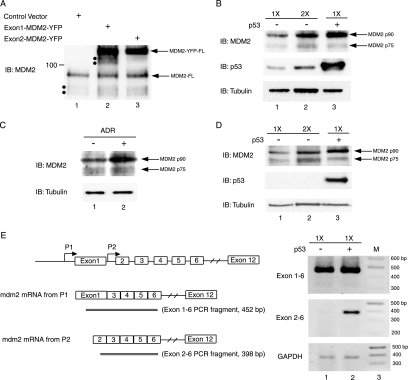
FIG. 3.
Full-length MDM2 protein is the principal translation product of mdm2 mRNA derived from the P2 promoter and is preferentially induced upon p53 activation. (A) Translation products of Exon1-MDM2-YFP and Exon2-MDM2-YFP that contain 5′ UTRs corresponding to native mdm2 mRNAs transcribed from the P1 and P2 promoter, respectively, were analyzed in Saos-2 cells as described in the legend to Fig. 2. (B) Expression of MDM2 protein isoforms in HCT116 cells transfected with an HA-tagged p53 construct was analyzed by immunoblotting using the indicated antibodies: anti-MDM2 (4B11), anti-p53 (DO-1), and anti-β-tubulin. Analysis of lysates of cells transfected with empty vector pcDNA3 was included as a control in lane 1 and lane 2. For the purpose of quantification, twice the amount of protein sample shown in lane 1 was loaded in lane 2 (designated 1× and 2×, respectively). (C) Expression of cellular MDM2 following treatment with the DNA damage agent adriamycin (ADR) at a concentration of 525 nM for 24 h was examined in HCT116 cells. (D) Saos-2, a p53-null cell line, was subjected to adventitious expression of p53 from a transfecting construct, and MDM2 abundance was analyzed as described in the legend to panel B. (E) The presence of alternatively spliced mdm2 mRNAs was examined by RT-PCR of mRNA isolated from Saos-2 cells transfected with an empty vector or a p53-expressing vector (lane 1 and 2). Using primer set Exon1 and Exon6 or Exon2 and Exon6, PCR products consisting of a 452-bp fragment or a 398-bp fragment are expected for intact mdm2 mRNA from P1 and P2, respectively. If exon 3 has been spliced out, it will give rise to PCR products 85 bp shorter than the intact ones. GAPDH was used as a control to assess the amounts of RNA applied in the RT-PCR analysis. A 100-bp DNA ladder (New England Biolabs) served as a molecular weight standard and was loaded in lane 3 of agarose gels for electrophoresis. IB designates the antibody used for immunoblotting.
Human mdm2 mRNA contains AUG codons at positions 62 and 102, in addition to one at position 50; using site-directed mutagenesis, we constructed plasmids that express Exon1-MDM2-YFP derivatives mutated to AUC at one or more of these positions. Immunoblot analysis of MDM2 isoforms produced from the resulting constructs in Saos-2 cells using 4B11 antibody as a probe (Fig. 2B, lanes 2 and 3, top panel) showed that mutation of AUG 50 did not affect either component of the MDM2-YFP fusion product doublet, indicating that the protein isoforms represented by the doublet are not generated by internal translation initiated at AUG 50. In contrast, mutation of AUG 62 to AUC resulted in replacement of the upper band of the doublet with a more slowly migrating band (Fig. 2B top panel, lane 4). This new protein species migrates at the same position as the truncated protein that lacks the first 49 amino acids of MDM2 fusion protein and is produced from the transcript in which AUG 62 and AUG 102 have both been mutated (Fig. 2B, lane 5, top panel). These results indicate that translation of promoter P1-initiated mdm2 transcripts can begin internally at AUG 50 when the normal translation initiation site at codon AUG 62 has been ablated. Furthermore, we observed that only one of the MDM2-YFP doublet bands was produced by a construct containing mutations at both AUG 50 and AUG 62 (Fig. 2A, lane 3, bottom panel) and that a construct having an additional change of AUG 102 to AUC failed to produce any truncated YFP-tagged species (Fig. 2A, lane 4, bottom panel). Collectively, these results argue strongly that the transcripts from promoter P1 have the ability to generate human MDM2 protein isoforms initiated at two distinct internal AUG codons located at positions 62 and 102.
mdm2 mRNA transcribed from the P2 promoter upon p53 activation preferentially produces full-length MDM2 proteins.
Whereas exon 1 is retained as a 5′ UTR in human mdm2 mRNA initiated at P1, human mdm2 mRNA initiated at P2 retains exon 2 as its 5′ UTR (58). The MDM2 proteins translated on transcripts initiated at P2 were analyzed using a construct, Exon2-MDM2-YPF, that was created by replacing the exon 1 5′ UTR of Exon1-MDM2-YFP with mdm2 exon 2 as the 5′ UTR. Comparison of the translation products of Exon2-MDM2-YFP and Exon1-MDM2-YFP in human Saos-2 cells (Fig. 3A) indicated that while Exon1-MDM2-YFP produced two shorter truncated fusion proteins as well as the full-length MDM2 isoform, the bulk of the fusion protein generated from Exon2-MDM2-YFP was full length. A similar observation for differential translation profiles of murine mdm2 transcripts has been reported during in vitro translation studies (2). Our finding suggests that the ratio of endogenous full-length versus N-terminally truncated human MDM2 protein should increase upon p53 activation of the P2 promoter, which has been reported to initiate transcripts containing a 5′ UTR similar or identical to the one present in transcripts produced by Exon2-MDM2-YFP. However, expression of p75MDM2 and p90MDM2 has been reported to increase in parallel upon p53 reactivation in a murine engineered cell line and upon p53 induction by UV irradiation (2, 46).
To investigate the basis for the disparate findings reported for murine versus human cells and to elucidate the role of p53 in the production of human MDM2 isoforms, we analyzed MDM2 proteins made in HCT116 human colon adenocarcinoma cells transfected with a p53 expression construct. Quantification of the p90MDM2 and p75MDM2 signals in lysates of these cells indicated that under the experimental conditions we used, p90MDM2 increased more than fivefold in response to p53 expression, while the increase in p75MDM2 was marginal (i.e., less than twofold) (Fig. 3B). Quantitatively similar differential production of p90MDM2 was also observed in HCT116 cells induced to produce p53 by treatment with the DNA-damaging agent adriamycin and in Saos-2 cells producing transcripts initiated at P2 after being transfected with a p53 expression construct (Fig. 3C and D).
A second reported mechanism for the production of murine p75MDM2 upon p53 activation is alternative splicing of mRNA initiated at P2, leading to the removal of exon 3 and the consequent positioning of AUG 50 as the most 5′ AUG codon in the message (46). However, the alternatively spliced form of P2-derived mdm2 mRNA observed in murine cells was not detected in human cells by RT-PCR analysis (Fig. 3E). Thus, in contrast to the concurrent induction of both the p75MDM2 and p90MDM2 MDM2 isoforms by p53 observed in the mouse, p90MDM2 is the principal MDM2 protein synthesized in response to p53 activation in human cells.
Differential participation of human MDM2 isoforms in the p53/MDM2 regulatory loop.
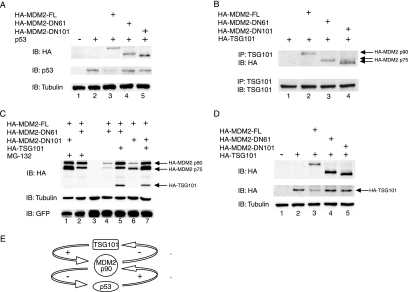
Interaction of p53 with sequences that are located near the N-terminal end of the MDM2 protein (i.e., amino acid residues 19 to 102), and consequently are partially absent in p75MDM2, is necessary for both MDM2-mediated inhibition of the transcription-promoting actions of p53 and MDM2-mediated degradation of p53 (11, 19, 26). As in vitro-translated human MDM2 lacking the first 61 amino acid residues cannot bind p53 (51), we anticipated that p75MDM2, which as shown by the data presented above consists a mixture of isoforms lacking the first 61 or first 101 amino acids of MDM2 protein, would be unable to modulate the decay of p53. Consistent with this prediction, we observed that in Saos-2 cells that have been cotransfected with separate constructs expressing p53 and specific MDM2 isoforms (Fig. 4A), the steady-state level of p53 protein was reduced by adventitious overproduction of full-length MDM2, consistent with previous reports (19, 29), but was unaffected by analogous overproduction of either component of the p75MDM2 doublet (Fig. 4A).
FIG. 4.
Modulation of abundance of the p90MDM2 and p75MDM2 isoforms and of p53 and TSG101 by the p53/MDM2 and MDM2/TSG101 control loops. (A) Effect of p90MDM2 and p75MDM2 on p53 protein abundance. Saos-2 cells were cotransfected with a construct expressing p53 (25 ng) and with vectors expressing HA-tagged MDM2 proteins, including full-length (HA-MDM2-FL) and N-terminal truncation proteins as indicated by the position of the last amino acid in the deleted portion. HA-tagged MDM2, p53, and β-tubulin were detected by immunoblotting (IB) using anti-HA, anti-p53, and anti-β-tubulin antibodies as indicated. (B) Physical interaction of p90MDM2 and p75MDM2 with TSG101. The indicated constructs were introduced into Saos-2 cells, and protein extracts from these cells were subjected to immunoprecipitation with anti-TSG101 antibody, as indicated by the designation IP. Coprecipitated proteins were examined by immunoblotting with antibodies against TSG101 or HA epitope. (C) Effect of TSG101 expression on the 26S proteasome-mediated degradation of MDM2. U2OS cells were cotransfected with MDM2 constructs expressing full-length as well as N-terminally truncated proteins. Forty-eight hours posttransfection, cells were left untreated or were treated with proteasome inhibitor MG132 (40 uM) for another 8 h, and cellular protein extracts were analyzed by immunoblotting (lanes 1, 2, 4, and 6). The indicated MDM2 constructs and a construct expressing HA-tagged TSG101 were cointroduced into U2OS cells by transfection (lanes 5 and 7). HA-tagged MDM2 as well as TSG101, β-tubulin, and GFP were detected by immunoblotting with anti-HA, anti-β-tubulin, and anti-GFP antibodies, respectively. An empty vector was introduced into cells as a control (lane 3). To ensure comparable transfection efficiency among these samples, a GFP expression construct, pCMV-GFP (0.1 μg), was included, and its protein level is shown in the bottom panel. (D) Effect of p90MDM2 and p75MDM2 on the protein stability of TSG101. A constant amount of HA-tagged TSG101 construct (0.5 μg) was cotransfected with constructs expressing HA-tagged MDM2 isoforms in Saos-2 cells. Cellular protein levels of HA-MDM2 and HA-TSG101 were detected by immunoblotting with anti-HA antibody. (E) Model showing the proposed role of p90MDM2 in the p53/MDM2 and MDM2/TSG101 feedback control loops.
TSG101 modulates the protein stability of both full-length and truncated MDM2 isoforms, but only full-length MDM2 promotes the decay of TSG101 in the TSG101/MDM2 feedback loop.
As noted earlier, the ability of full-length MDM2, but not p75MDM2, to function as an E3 ligase in promoting the degradation of p53 is associated with the presence of a p53 binding site within the full-length MDM2 protein (11). In contrast to the inability of p53 to interact with p75MDM2, our immunoprecipitation experiments indicated that TSG101 can bind to p90MDM2 and both of the p75MDM2 isoforms in Saos-2 cells (Fig. 4B), indicating that a site of interaction between MDM2 and TSG101 is located distally to amino acid residue 101.
Decay of p90MDM2 is mediated through the ubiquitin-proteasome pathway, yielding a half-life of less than 20 min (10, 15, 21). Previously reported results indicate that elevation of TSG101 expression can double the half-life of p90MDM2 by interfering with its ubiquitination and consequent decay by the 26S proteasome (29). To learn whether the stability of p75MDM2 is similarly modulated by TSG101, we determined the steady-state level of p75MDM2 during ectopic expression of TSG101 or in the presence of the proteasome inhibitor MG132. As shown in Fig. 4C (lane 1 versus 6 and lane 2 versus 4), p75MDM2 and p90MDM2 accumulated dramatically in the presence of MG132, suggesting that decay of both proteins is mediated by the ubiquitin-proteasome pathway and indicating that the N-terminal region of MDM2 is not required for its ubiquitination and consequent degradation by 26S proteasome. Additionally, both p75MDM2 and p90MDM2 were stabilized by overproduction of TSG101 (Fig. 4C, lanes 4 to 7), indicating that at least the first 61 amino acids of MDM2 are also dispensable for TSG101 modulation of MDM2 protein decay.
Just as TSG101 modulates the cellular level of MDM2 by negatively regulating its ubiquitination and decay, MDM2 has a parallel role in the proteolytic degradation of TSG101 by the 26S proteasome (29). Whereas TSG101 stabilized both full-length and truncated isoforms of MDM2, we found that only full-length MDM2 promoted degradation of TSG101. Consistent with previous observations (29), examination of the steady-state level of adventitiously produced TSG101 in cells cotransfected with various MDM2 constructs indicated that TSG101 abundance was decreased by more than 50% when p90MDM2 was overexpressed (Fig. 4D, lane 2 versus 3). Conversely, no change in the level of TSG101 was observed in cells cotransfected with constructs overexpressing p75MDM2 (Fig. 4D, lane 2 versus 4 and 5), suggesting that the biological actions of the previously identified MDM2/TSG101 autoregulatory loop can be modulated by the differential production of the p75MDM2 and p90MDM2 isoforms.
DISCUSSION
The mdm2 gene and the protein(s) it encodes are well recognized as having an important role in the regulation of multiple cellular processes and signaling pathways related to cancer. Not only does MDM2 protein control cell cycle progression through regulation of p53 and the pRb/E2F complex (22, 31, 50), but it also functions as an activator of certain steroid hormone receptors (36, 44), an inhibitor of the transforming growth factor beta pathway (49, 56), a modulator of phosphatidylinositol 3-kinase/AKT signaling (33, 59), and a regulator of degradation of the ubiquitin enzyme variant (UEV) protein, TSG101, which mediates cell proliferation (45, 52), endosomal trafficking (30, 38), and other diverse cellular activities (18, 20, 32, 54). Just as the cellular abundance of the p53 and TSG101 proteins can be controlled by MDM2, the steady-state level of MDM2 in turn is modulated by both p53 and TSG101 in separate and parallel autoregulatory feedback control loops (5, 19, 26, 27, 29, 53).
Multiple isoforms of the murine and human MDM2 proteins have been observed and have been reported to result from transcript initiation at different promoters (2, 58), the production of splice variant transcripts (46, 47, 51), and/or different types of posttranslation modifications of MDM2 proteins (1, 34, 37, 43, 55). In human cells, species migrating as 75 kDa and 90 kDa are the most abundant MDM2 protein products. In mouse cells, a 75-kDa MDM2 isoform has been identified as an N-terminally truncated protein that lacks the first 49 amino acid residues; this protein is produced both by the initiation of translation at AUG 50 of the same transcript that directs the synthesis of full-length MDM2 protein and by the translation of alternatively spliced variant transcripts in which exon 3 is removed and AUG 50 becomes the most 5′ translation initiation site (46). As the genomic organization and amino acid sequence of murine and human mdm2 genes are highly conserved (14, 25, 41, 58), the production of a human MDM2 isoform similar in size to murine p75MDM2 has been assumed to occur by the same mechanism reported for murine p75MDM2 (37). However, another study has concluded that the molecular weight difference between human p90MDM2 and p75MDM2 is due to posttranslational conjugation of full-length MDM2 with the small ubiquitin-like protein SUMO-1 (7), although the earlier notion that sumoylation of MDM2 interferes with ubiquitination (6) appears to be incorrect (17).
We found that human p75MDM2 is in fact a mixture of two separate N-terminally truncated proteins that, respectively, lack either the first 61 or 101 amino acids and are produced by initiation of protein synthesis at internal AUG codons at position 62 or 102; one of these sites (i.e., AUG 62) was observed previously to function as an internal initiation site within mdm2 transcripts that contain a novel exon and are expressed specifically in certain human tissues (51). We additionally observed that the production of full-length p90MDM2 occurs preferentially during p53 activation of mdm2 gene transcription at the p53-responsive P2 promoter. Importantly, we observed that the two major protein isoforms of MDM2 behaved differently in the MDM2/TSG101 feedback control loop: whereas the steady-state level of both p90MDM2 and p75MDM2 is altered by adventitious overexpression of the TSG101 protein, only the full-length MDM2 species (i.e., p90MDM2), which contains the MDM2 segment required for its interaction with and degradation of p53, promoted degradation of TSG101. Collectively, these findings suggest that p90MDM2 is the form of MDM2 that mediates destabilization of both p53 and TSG101 in their feedback control loops with MDM2 (Fig. 4E). The production of protein isoforms having distinct biological roles in the control of cell growth or differentiation has been reported previously for c-Myc, p53, SCL, and C/EBP (8, 9, 12, 48, 57). In the case of c-Myc and p53, truncated proteins can modulate the functions of the full-length protein by acting as dominant-negative inhibitors (12, 48, 57). Whereas our data indicate that the mechanism underlying the production of the N-terminally truncated MDM2 isoform differs for human and murine cells, internal initiation of translation in the absence of a typical internal ribosome entry site sequence (23) occurs in both instances. As exon 1 is required for the production of murine p75MDM2 from transcripts initiated at P1, it has been suggested that this exon may contain a cis-acting element or unique structural feature that facilitates the internal initiation of translation (35, 46). Consistent with this notion, two small upstream open reading frames (uORFs) located within exon 1 of the mdm2 gene have been identified in both mouse and human cells, and it has been shown that these uORFs have a regulatory role in translation (23). As uORFs in mRNAs of the SCL and C/EBP genes can function as cis-regulatory elements for translation initiation from internal start sites (8, 9), such uORFs may have a similar role in the production of the truncated MDM2 isoforms we have identified.
In murine cells, both the p75 and p90 MDM2 isoforms are induced equally by p53 (46), and overexpression of p75MDM2, which cannot bind p53, has been found to interfere with the ability of p90MDM2 to promote p53 decay (42). In contrast to what has been found for murine MDM2, our data indicate that in human cells the p90MDM2 isoform is produced preferentially during p53 activation of the mdm2 P2 promoter. As p90 is the MDM2 isoform that participates in the p53/MDM2 and MDM2/TSG101 feedback control loops, the differential activation of p90MDM2 production by p53 in human versus murine cells potentially may lead to differences in the effects of MDM2 in these two biological species.
Acknowledgments
We thank A. Levine and L. Gerace for MDM2 and RanGAP1 antibodies, J. Ford for HCT116 cells, and C. Contag for the pEYFP-N1 plasmid. We acknowledge the helpful comments and advice of L. Li as well as members of the Cohen laboratory.
These studies were supported by grants from the National Foundation for Cancer Research and the California Breast Cancer Research Program to S.N.C. and in part by a grant from the National Science Council of Taiwan (NSC-93-2320-B-010-063) to T.H.C.
Footnotes
Published ahead of print on 23 October 2006.
REFERENCES
- 1.Ashcroft, M., R. L. Ludwig, D. B. Woods, T. D. Copeland, H. O. Weber, E. J. MacRae, and K. H. Vousden. 2002. Phosphorylation of HDM2 by Akt. Oncogene 21:1955-1962. [DOI] [PubMed] [Google Scholar]
- 2.Barak, Y., E. Gottlieb, T. Juven-Gershon, and M. Oren. 1994. Regulation of mdm2 expression by p53: alternative promoters produce transcripts with nonidentical translation potential. Genes Dev. 8:1739-1749. [DOI] [PubMed] [Google Scholar]
- 3.Barak, Y., T. Juven, R. Haffner, and M. Oren. 1993. mdm2 expression is induced by wild type p53 activity. EMBO J. 12:461-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartel, F., H. Taubert, and L. C. Harris. 2002. Alternative and aberrant splicing of MDM2 mRNA in human cancer. Cancer Cell 2:9-15. [DOI] [PubMed] [Google Scholar]
- 5.Bond, G. L., W. Hu, and A. J. Levine. 2005. MDM2 is a central node in the p53 pathway: 12 years and counting. Curr. Cancer Drug Targets 5:3-8. [DOI] [PubMed] [Google Scholar]
- 6.Buschmann, T., S. Y. Fuchs, C. G. Lee, Z. Q. Pan, and Z. Ronai. 2000. SUMO-1 modification of Mdm2 prevents its self-ubiquitination and increases Mdm2 ability to ubiquitinate p53. Cell 101:753-762. [DOI] [PubMed] [Google Scholar]
- 7.Buschmann, T., D. Lerner, C. G. Lee, and Z. Ronai. 2001. The Mdm-2 amino terminus is required for Mdm2 binding and SUMO-1 conjugation by the E2 SUMO-1 conjugating enzyme Ubc9. J. Biol. Chem. 276:40389-40395. [DOI] [PubMed] [Google Scholar]
- 8.Calkhoven, C. F., C. Muller, and A. Leutz. 2000. Translational control of C/EBPalpha and C/EBPbeta isoform expression. Genes Dev. 14:1920-1932. [PMC free article] [PubMed] [Google Scholar]
- 9.Calkhoven, C. F., C. Muller, R. Martin, G. Krosl, H. Pietsch, T. Hoang, and A. Leutz. 2003. Translational control of SCL-isoform expression in hematopoietic lineage choice. Genes Dev. 17:959-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang, Y. C., Y. S. Lee, T. Tejima, K. Tanaka, S. Omura, N. H. Heintz, Y. Mitsui, and J. Magae. 1998. mdm2 and bax, downstream mediators of the p53 response, are degraded by the ubiquitin-proteasome pathway. Cell Growth Differ. 9:79-84. [PubMed] [Google Scholar]
- 11.Chen, J., V. Marechal, and A. J. Levine. 1993. Mapping of the p53 and mdm-2 interaction domains. Mol. Cell. Biol. 13:4107-4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Courtois, S., G. Verhaegh, S. North, M. G. Luciani, P. Lassus, U. Hibner, M. Oren, and P. Hainaut. 2002. DeltaN-p53, a natural isoform of p53 lacking the first transactivation domain, counteracts growth suppression by wild-type p53. Oncogene 21:6722-6728. [DOI] [PubMed] [Google Scholar]
- 13.Daujat, S., H. Neel, and J. Piette. 2001. MDM2: life without p53. Trends Genet. 17:459-464. [DOI] [PubMed] [Google Scholar]
- 14.Fakharzadeh, S. S., S. P. Trusko, and D. L. George. 1991. Tumorigenic potential associated with enhanced expression of a gene that is amplified in a mouse tumor cell line. EMBO J. 10:1565-1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fang, S., J. P. Jensen, R. L. Ludwig, K. H. Vousden, and A. M. Weissman. 2000. Mdm2 is a RING finger-dependent ubiquitin protein ligase for itself and p53. J. Biol. Chem. 275:8945-8951. [DOI] [PubMed] [Google Scholar]
- 16.Feng, G. H., C. J. Lih, and S. N. Cohen. 2000. TSG101 protein steady-state level is regulated posttranslationally by an evolutionarily conserved COOH-terminal sequence. Cancer Res. 60:1736-1741. [PubMed] [Google Scholar]
- 17.Fuchs, S. Y., C. G. Lee, Z. Q. Pan, and Z. Ronai. 2002. SUMO-1 modification of Mdm2 prevents its self-ubiquitination and increases Mdm2 ability to ubiquitinate p53. Cell 110:531. [DOI] [PubMed] [Google Scholar]
- 18.Garrus, J. E., U. K. von Schwedler, O. W. Pornillos, S. G. Morham, K. H. Zavitz, H. E. Wang, D. A. Wettstein, K. M. Stray, M. Cote, R. L. Rich, D. G. Myszka, and W. I. Sundquist. 2001. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell 107:55-65. [DOI] [PubMed] [Google Scholar]
- 19.Haupt, Y., R. Maya, A. Kazaz, and M. Oren. 1997. Mdm2 promotes the rapid degradation of p53. Nature 387:296-299. [DOI] [PubMed] [Google Scholar]
- 20.Hittelman, A. B., D. Burakov, J. A. Iniguez-Lluhi, L. P. Freedman, and M. J. Garabedian. 1999. Differential regulation of glucocorticoid receptor transcriptional activation via AF-1-associated proteins. EMBO J. 18:5380-5388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Honda, R., H. Tanaka, and H. Yasuda. 1997. Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett. 420:25-27. [DOI] [PubMed] [Google Scholar]
- 22.Hsieh, J. K., F. S. Chan, D. J. O'Connor, S. Mittnacht, S. Zhong, and X. Lu. 1999. RB regulates the stability and the apoptotic function of p53 via MDM2. Mol. Cell 3:181-193. [DOI] [PubMed] [Google Scholar]
- 23.Jin, X., E. Turcott, S. Englehardt, G. J. Mize, and D. R. Morris. 2003. The two upstream open reading frames of oncogene mdm2 have different translational regulatory properties. J. Biol. Chem. 278:25716-25721. [DOI] [PubMed] [Google Scholar]
- 24.Jones, S. N., A. R. Hancock, H. Vogel, L. A. Donehower, and A. Bradley. 1998. Overexpression of Mdm2 in mice reveals a p53-independent role for Mdm2 in tumorigenesis. Proc. Natl. Acad. Sci. USA 95:15608-15612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Juven, T., Y. Barak, A. Zauberman, D. L. George, and M. Oren. 1993. Wild type p53 can mediate sequence-specific transactivation of an internal promoter within the mdm2 gene. Oncogene 8:3411-3416. [PubMed] [Google Scholar]
- 26.Kubbutat, M. H., S. N. Jones, and K. H. Vousden. 1997. Regulation of p53 stability by Mdm2. Nature 387:299-303. [DOI] [PubMed] [Google Scholar]
- 27.Lahav, G., N. Rosenfeld, A. Sigal, N. Geva-Zatorsky, A. J. Levine, M. B. Elowitz, and U. Alon. 2004. Dynamics of the p53-Mdm2 feedback loop in individual cells. Nat. Genet. 36:147-150. [DOI] [PubMed] [Google Scholar]
- 28.Li, L., and S. N. Cohen. 1996. Tsg101: a novel tumor susceptibility gene isolated by controlled homozygous functional knockout of allelic loci in mammalian cells. Cell 85:319-329. [DOI] [PubMed] [Google Scholar]
- 29.Li, L., J. Liao, J. Ruland, T. W. Mak, and S. N. Cohen. 2001. A TSG101/MDM2 regulatory loop modulates MDM2 degradation and MDM2/p53 feedback control. Proc. Natl. Acad. Sci. USA 98:1619-1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lu, Q., L. W. Hope, M. Brasch, C. Reinhard, and S. N. Cohen. 2003. TSG101 interaction with HRS mediates endosomal trafficking and receptor down-regulation. Proc. Natl. Acad. Sci. USA 100:7626-7631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martin, K., D. Trouche, C. Hagemeier, T. S. Sorensen, N. B. La Thangue, and T. Kouzarides. 1995. Stimulation of E2F1/DP1 transcriptional activity by MDM2 oncoprotein. Nature 375:691-694. [DOI] [PubMed] [Google Scholar]
- 32.Martin-Serrano, J., T. Zang, and P. D. Bieniasz. 2001. HIV-1 and Ebola virus encode small peptide motifs that recruit Tsg101 to sites of particle assembly to facilitate egress. Nat. Med. 7:1313-1319. [DOI] [PubMed] [Google Scholar]
- 33.Mayo, L. D., and D. B. Donner. 2001. A phosphatidylinositol 3-kinase/Akt pathway promotes translocation of Mdm2 from the cytoplasm to the nucleus. Proc. Natl. Acad. Sci. USA 98:11598-11603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meek, D. W., and U. Knippschild. 2003. Posttranslational modification of MDM2. Mol. Cancer Res. 1:1017-1026. [PubMed] [Google Scholar]
- 35.Mendrysa, S. M., M. K. McElwee, and M. E. Perry. 2001. Characterization of the 5′ and 3′ untranslated regions in murine mdm2 mRNAs. Gene 264:139-146. [DOI] [PubMed] [Google Scholar]
- 36.Metivier, R., G. Penot, R. P. Carmouche, M. R. Hubner, G. Reid, S. Denger, D. Manu, H. Brand, M. Kos, V. Benes, and F. Gannon. 2004. Transcriptional complexes engaged by apo-estrogen receptor-alpha isoforms have divergent outcomes. EMBO J. 23:3653-3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miyauchi, Y., S. Yogosawa, R. Honda, T. Nishida, and H. Yasuda. 2002. Sumoylation of Mdm2 by protein inhibitor of activated STAT (PIAS) and RanBP2 enzymes. J. Biol. Chem. 277:50131-50136. [DOI] [PubMed] [Google Scholar]
- 38.Moberg, K. H., S. Schelble, S. K. Burdick, and I. K. Hariharan. 2005. Mutations in erupted, the Drosophila ortholog of mammalian tumor susceptibility gene 101, elicit non-cell-autonomous overgrowth. Dev. Cell. 9:699-710. [DOI] [PubMed] [Google Scholar]
- 39.Momand, J., D. Jung, S. Wilczynski, and J. Niland. 1998. The MDM2 gene amplification database. Nucleic Acids Res. 26:3453-3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Momand, J., G. P. Zambetti, D. C. Olson, D. George, and A. J. Levine. 1992. The mdm-2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell 69:1237-1245. [DOI] [PubMed] [Google Scholar]
- 41.Oliner, J. D., K. W. Kinzler, P. S. Meltzer, D. L. George, and B. Vogelstein. 1992. Amplification of a gene encoding a p53-associated protein in human sarcomas. Nature 358:80-83. [DOI] [PubMed] [Google Scholar]
- 42.Perry, M. E., S. M. Mendrysa, L. J. Saucedo, P. Tannous, and M. Holubar. 2000. p76(MDM2) inhibits the ability of p90(MDM2) to destabilize p53. J. Biol. Chem. 275:5733-5738. [DOI] [PubMed] [Google Scholar]
- 43.Pochampally, R., B. Fodera, L. Chen, W. Shao, E. A. Levine, and J. Chen. 1998. A 60 kd MDM2 isoform is produced by caspase cleavage in non-apoptotic tumor cells. Oncogene 17:2629-2636. [DOI] [PubMed] [Google Scholar]
- 44.Reid, G., M. R. Hubner, R. Metivier, H. Brand, S. Denger, D. Manu, J. Beaudouin, J. Ellenberg, and F. Gannon. 2003. Cyclic, proteasome-mediated turnover of unliganded and liganded ERalpha on responsive promoters is an integral feature of estrogen signaling. Mol. Cell 11:695-707. [DOI] [PubMed] [Google Scholar]
- 45.Ruland, J., C. Sirard, A. Elia, D. MacPherson, A. Wakeham, L. Li, J. L. de la Pompa, S. N. Cohen, and T. W. Mak. 2001. p53 accumulation, defective cell proliferation, and early embryonic lethality in mice lacking tsg101. Proc. Natl. Acad. Sci. USA 98:1859-1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Saucedo, L. J., C. D. Myers, and M. E. Perry. 1999. Multiple murine double minute gene 2 (MDM2) proteins are induced by ultraviolet light. J. Biol. Chem. 274:8161-8168. [DOI] [PubMed] [Google Scholar]
- 47.Sigalas, I., A. H. Calvert, J. J. Anderson, D. E. Neal, and J. Lunec. 1996. Alternatively spliced mdm2 transcripts with loss of p53 binding domain sequences: transforming ability and frequent detection in human cancer. Nat. Med. 2:912-917. [DOI] [PubMed] [Google Scholar]
- 48.Spotts, G. D., S. V. Patel, Q. Xiao, and S. R. Hann. 1997. Identification of downstream-initiated c-Myc proteins which are dominant-negative inhibitors of transactivation by full-length c-Myc proteins. Mol. Cell. Biol. 17:1459-1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sun, P., P. Dong, K. Dai, G. J. Hannon, and D. Beach. 1998. p53-independent role of MDM2 in TGF-beta1 resistance. Science 282:2270-2272. [DOI] [PubMed] [Google Scholar]
- 50.Uchida, C., S. Miwa, K. Kitagawa, T. Hattori, T. Isobe, S. Otani, T. Oda, H. Sugimura, T. Kamijo, K. Ookawa, H. Yasuda, and M. Kitagawa. 2005. Enhanced Mdm2 activity inhibits pRB function via ubiquitin-dependent degradation. EMBO J. 24:160-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Veldhoen, N., S. Metcalfe, and J. Milner. 1999. A novel exon within the mdm2 gene modulates translation initiation in vitro and disrupts the p53-binding domain of mdm2 protein. Oncogene 18:7026-7033. [DOI] [PubMed] [Google Scholar]
- 52.Wagner, K. U., A. Krempler, Y. Qi, K. Park, M. D. Henry, A. A. Triplett, G. Riedlinger, I. E. Rucker, and L. Hennighausen. 2003. Tsg101 is essential for cell growth, proliferation, and cell survival of embryonic and adult tissues. Mol. Cell. Biol. 23:150-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu, X., J. H. Bayle, D. Olson, and A. J. Levine. 1993. The p53-mdm-2 autoregulatory feedback loop. Genes Dev. 7:1126-1132. [DOI] [PubMed] [Google Scholar]
- 54.Xie, W., L. Li, and S. N. Cohen. 1998. Cell cycle-dependent subcellular localization of the TSG101 protein and mitotic and nuclear abnormalities associated with TSG101 deficiency. Proc. Natl. Acad. Sci. USA 95:1595-1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xirodimas, D. P., J. Chisholm, J. M. Desterro, D. P. Lane, and R. T. Hay. 2002. P14ARF promotes accumulation of SUMO-1 conjugated (H)Mdm2. FEBS Lett. 528:207-211. [DOI] [PubMed] [Google Scholar]
- 56.Yam, C. H., W. Y. Siu, T. Arooz, C. H. Chiu, A. Lau, X. Q. Wang, and R. Y. Poon. 1999. MDM2 and MDMX inhibit the transcriptional activity of ectopically expressed SMAD proteins. Cancer Res. 59:5075-5078. [PubMed] [Google Scholar]
- 57.Yin, Y., C. W. Stephen, M. G. Luciani, and R. Fahraeus. 2002. p53 Stability and activity is regulated by Mdm2-mediated induction of alternative p53 translation products. Nat. Cell. Biol. 4:462-467. [DOI] [PubMed] [Google Scholar]
- 58.Zauberman, A., D. Flusberg, Y. Haupt, Y. Barak, and M. Oren. 1995. A functional p53-responsive intronic promoter is contained within the human mdm2 gene. Nucleic Acids Res. 23:2584-2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhou, B. P., Y. Liao, W. Xia, Y. Zou, B. Spohn, and M. C. Hung. 2001. HER-2/neu induces p53 ubiquitination via Akt-mediated MDM2 phosphorylation. Nat. Cell. Biol. 3:973-982. [DOI] [PubMed] [Google Scholar]