Abstract
E2F-mediated control of gene expression is believed to have an essential role in the control of cellular proliferation. Using a conditional gene-targeting approach, we show that the targeted disruption of the entire E2F activator subclass composed of E2f1, E2f2, and E2f3 in mouse embryonic fibroblasts leads to the activation of p53 and the induction of p53 target genes, including p21CIP1. Consequently, cyclin-dependent kinase activity and retinoblastoma (Rb) phosphorylation are dramatically inhibited, leading to Rb/E2F-mediated repression of E2F target gene expression and a severe block in cellular proliferation. Inactivation of p53 in E2f1-, E2f2-, and E2f3-deficient cells, either by spontaneous mutation or by conditional gene ablation, prevented the induction of p21CIP1 and many other p53 target genes. As a result, cyclin-dependent kinase activity, Rb phosphorylation, and E2F target gene expression were restored to nearly normal levels, rendering cells responsive to normal growth signals. These findings suggest that a critical function of the E2F1, E2F2, and E2F3 activators is in the control of a p53-dependent axis that indirectly regulates E2F-mediated transcriptional repression and cellular proliferation.
A considerable body of in vitro and in vivo work carried out over the past decade has led to the view that the E2F family of transcription factors regulates cellular proliferation by controlling the transcription of a plethora of genes involved in DNA replication, DNA repair, mitosis, and cell cycle progression (4, 8, 31, 35). Mammalian E2F activity is composed of eight different family members encoded by distinct genes. Based on structure-function studies and amino acid sequence analysis, E2F family members can be divided into two main subclasses, repressor E2Fs and activator E2Fs. Members of the repressor subclass, E2F4, E2F5, and possibly E2F3b, are constitutively expressed and can associate with pocket proteins retinoblastoma (Rb), p107, and p130. E2F6, E2F7, and E2F8 also contribute to gene silencing, but in a manner that is independent of pocket proteins (5, 7, 10, 19-21, 24, 34). Recent experiments using chromatin immunoprecipitation (ChIP) assays carried out by Dynlacht and colleagues have shown that during G0, E2F4-5 repressor complexes associate with promoters containing E2F-binding elements (2, 4, 32). These complexes can recruit additional chromatin-modifying activities necessary for transcriptional repression. Upon reentry into the cell cycle and prior to peak E2F target gene expression, cyclin-dependent kinases mediate the phosphorylation of Rb-related proteins and facilitate the disruption of repressor complexes, displacing them from promoters of E2F target genes. Consistent with a key role for these complexes in the control of cellular proliferation, mouse embryonic fibroblasts (MEFs) deficient for the three pocket proteins have deregulated E2F target expression and fail to arrest under low-serum conditions (6, 28).
In contrast to E2F repressors, E2F1, E2F2, and E2F3a (E2F1-3a) are potent transactivators that can transiently bind and activate E2F target promoters (4, 8, 31, 35). These activator E2Fs are regulated by transcription and protein degradation in response to growth stimulation, and as a result their activities peak during G1/S (23). While these factors accumulate at G1/S during the first cell cycle following serum stimulation, it is interesting that E2F3a is the principal DNA-binding activity that reappears in subsequent G1/S transitions (17). During this period, E2F3a can be found transiently bound to many E2F target promoters and is presumably involved in controlling the cyclic nature of their expression (32). Not surprisingly, the combined disruption of E2f1, E2f2, and E2f3 (E2f1-3) in MEFs severely impedes E2F target expression and cell proliferation (38). Together, these results begin to solidify the long-standing belief that E2Fs regulate cell cycle-dependent gene expression through sequential interactions of repressor/activator E2Fs with their cognate E2F-binding elements on target promoters.
The complexity of E2F function has been further illustrated by recent studies suggesting that E2F-mediated transcriptional repression and activation are mechanistically linked. On one hand, disruption of E2F-mediated repression by the inactivation of Rb results in the accumulation of E2F1-3a proteins and the unscheduled entry of cells into S phase (8, 31, 35). On the other hand, disruption of E2F-mediated activation by the combined inactivation of E2F1-3 leads to an increase in p21CIP1 expression, an inhibition of CDK activity, and the hypophosphorylation of Rb-related pocket proteins (38), suggesting that activator E2Fs may indirectly control Rb/E2F-mediated repression through a p21CIP1-mediated negative regulatory feedback loop. These tight molecular interrelationships between the transcriptional repression and activation machineries have made dissecting these pathways particularly difficult.
In this study, conditional knockout strategies were used in studies with mice to elucidate the mechanism by which E2F1, E2F2, and E2F3s regulate gene expression and cellular proliferation via the p21CIP1-mediated feedback loop. We found that targeted disruption of E2f1-3 leads to the recruitment of p53 to p53-responsive promoters and the induction of p21CIP1 as well as many other p53 target genes, leading to a profound cell cycle arrest. Ablation of p53 in E2f1-3-deficient cells prevented the induction of p21CIP1 and, surprisingly, restored both the expression of E2F target genes and the capacity of these cells to proliferate. These results suggest an essential role for E2f1-3 in the regulation of p53.
MATERIALS AND METHODS
Cell culture and retroviral infection.
Primary MEFs were isolated from embryos at embryonic day 13.5 by use of standard methods. The MEF genotype E2f1−/− E2f2−/− E2f3f/f is herein designated 123f/f, and 123f/f cell lines 1 to 4 and the p53f/f 123f/f cell line were established using the NIH 3T9 protocol (33). All cells were grown in Dulbecco's minimal essential medium (DMEM) with 15% fetal bovine serum (FBS). For the production of retrovirus, the full-length cDNAs for human papillomavirus type 16 (HPV16) E6, HPV16 E6mut, HPV11 E6, and a myc-tagged E2F3a were subcloned into the pBabe-hygromycin retroviral vector. The cre recombinase cDNA was cloned into pBabe-puromycin. High-titered retroviruses were produced by transient transfection of retroviral constructs into the Phoenix-Eco packaging cell line as described previously (25). MEFs were infected by incubating the cells for 5 h with the supernatants containing 4 μg/ml of Sequabrene (Sigma) from the transfected cells. Subsequent to infection, cells were split and grown in selection media containing either 2.5 μg/ml puromycin (Sigma) or 400 μg/ml hygromycin (Roche) or both for 3 to 5 days. For 5-bromodeoxyuridine (BrdU) incorporation and real-time reverse transcriptase PCR (RT-PCR) experiments, subconfluent MEFs were synchronized by incubation in DMEM with 0.2% FBS for 72 h. Cells were then stimulated to proliferate by the addition of DMEM supplemented with 15% FBS and harvested at the indicated time points.
Proliferation assays.
For the growth curves of the colonies derived from 123f/f cell line 4, cells were plated at a density of 7 × 104 cells per 60-mm dish. Duplicate plates were counted daily and were replated every 72 h at the same density of the initial plating. Colony formation assays were performed by plating 500 and 2,500 cells per 100-mm dish. Once colonies formed, cells were fixed with 70% ethanol and stained with 5 mg/ml crystal violet in 20% methanol. Colonies from three separate plates at the appropriate density were counted, and the mean and standard deviation from one representative experiment are reported unless otherwise stated. For BrdU incorporation assays, proliferating or serum-stimulated cells were incubated with 50 μM BrdU for the indicated time and subsequently fixed with methanol and acetic acid in a 1:1 ratio. Cells were stained with α-BrdU antibody (Ab-3; Oncogene) as previously described (17) and counterstained with 4,6-diamidino-2-phenylindole (DAPI). A total of 500 DAPI-positive nuclei was scored for each time point.
Promoter reporter assay.
The pGL-mp21 reporter construct was generated by PCR amplifying a 3,043-bp promoter fragment with primers ST-30, GCGGTACCCCCCTTGGATTTCCTTTCTATCAGC, and ST-31, CCAGCTCGAGTTCCCCTAGACTCTGACACCGC, containing KpnI and XhoI sites, respectively, and subcloning it into the pGL3.1 luciferase reporter vector (Promega). The p53-binding elements were deleted in this construct by utilizing an internal EcoRV site located just downstream of the binding elements and a KpnI site within the vector. The construct was digested, the KpnI site was filled in, and the ends were religated. Cells from colonies of 123f/f cell line 4 were individually transfected with either pGL-mp21 or pGL-mp21Δp53 reporter vectors and the cytomegalovirus β-galactosidase plasmid as an internal control. Cells were harvested 48 h posttransfection, and luciferase and β-galactosidase assays were performed as described previously (29).
PCR genotyping.
For the colony PCR genotyping analysis, DNA was extracted from single colonies isolated from 96-well culture plates set up in parallel with a colony formation assay. DNA was extracted from cells and tumor samples by use of standard techniques. E2F3 PCR genotyping was performed by combining three primers, E2F3C, AGCAAAAGGCAATAGTCACTCCAG, E2F3K, GTCCACAACTCCAAACACACACAG, and E2F3W/F, AGGAGAGGCATCACGCTGC, at a final concentration of 0.125 mM with 1× Perkin-Elmer Buffer II, 2 mM MgCl2, 0.25 mM deoxynucleoside triphosphates, and 0.5 U Amplitaq Gold (Perkin-Elmer). Reactions were performed in a Perkin-Elmer 9600 instrument with the following cycles: 94°C for 8 min and 40 cycles of 94°C for 30 s, 54°C for 30 s, and 72°C for 45 s. The floxed allele (E2f3f/f) produces a 184-bp PCR fragment, and the knockout E2f3 allele produces a 416-bp fragment. p53 PCR genotyping was performed as previously described (22). The wild-type and knockout reactions were performed separately due to the similarity in sizes. The p53 wild-type and knockout alleles produce 460-bp and 612-bp fragments, respectively. All PCR products were separated on 2% agarose gels.
Western blot and kinase assays.
Protein lysates were separated on sodium dodecyl sulfate-polyacrylamide gels and blotted on polyvinylidene fluoride membranes. Blots were incubated overnight at 4°C with 1 to 2% skim milk in TBS-T buffer (Tris-buffered saline with 0.2% Tween 20) with the following antibodies: anti-E2F3 (SC-878; Santa Cruz), anti-p21CIP1 (M-19 and C-19; Santa Cruz), anti-tubulin (T-9026; Sigma), anti-cdk4 (C-22; Santa Cruz), anti-p19ARF (NB200-106; Novus Biologicals), anti-p53-ser15 (9286; Cell Signaling), and anti-p53 (NCL-p53-CM5p; Novocastra). The primary antibodies were then detected using horseradish peroxidase-conjugated secondary antibodies and ECL reagent (Amersham) as described by the manufacturer. Kinase assays using histone 1 were performed as described previously (17).
Real-time RT-PCR.
Approximately 1 × 106 cells were harvested at the indicated time point and total RNA was isolated using a QIAGEN RNA miniprep column as described by the manufacturer, including a DNase treatment before elution from the column. Reverse transcription of 2 μg of total RNA was performed by combining 1 μl of Superscript III reverse transcriptase (Invitrogen), 4 μl of 10× buffer, 0.5 μl of 100 mM oligo(dT) primer, 0.5 μl of 25 mM deoxynucleoside triphosphates, 1.0 μl of 0.1 M dithiothreitol, 1.0 μl of RNase inhibitor (Roche), and water up to a volume of 20 μl. Reaction mixtures were incubated at 50°C for 60 min and then diluted fivefold with 80 μl of water. Real-time RT-PCR was performed using a Bio-Rad iCycler PCR machine. Each PCR mixture contained 0.5 μl of cDNA template and primers at a concentration of 100 nM in a final volume of 25 μl of SYBR green reaction mix (Bio-Rad). Each PCR generated only the expected amplicon as shown by the melting-temperature profiles of the final products and by gel electrophoresis. Standard curves were calculated using cDNA to determine the linear range and PCR efficiency of each primer pair. Reactions were done in triplicate, and relative amounts of cDNA were normalized to GAPDH. Primer sequences are available upon request.
ChIP assays.
For ChIP assays, cells were harvested for antibody and the no-antibody control after selection. Formaldehyde was added directly to the culture medium at a final concentration of 1%. Cross-linking was allowed to proceed for 10 min at room temperature and was then stopped by the addition of glycine to a final concentration of 0.125 M. Cross-linked cells were washed twice with phosphate-buffered saline, scraped off the plate, and lysed in sodium dodecyl sulfate buffer. Lysates were sonicated in order to shear the genomic DNA into fragments of between 200 and 1,000 bp. Samples were immunoprecipitated overnight at 4°C with polyclonal antibodies specific for either acetyl-histone H4 (1 μg; 06-866; Upstate), E2F4 (2 μg; sc-1084x), or p130 (2 μg; sc-317x). Antibody-protein-DNA complexes were recovered by adding 60 μl of salmon sperm DNA-protein A agarose slurry and incubated for 1 h at 4°C. Following extensive washing, the complexes were eluted and cross-links were reversed by heating the samples to 65°C for 4 h. The eluted material was phenol-chloroform extracted, ethanol precipitated, and resuspended in 30 μl of water. PCR was performed with 1 μl of DNA, and 100 nM primers were diluted to a final volume of 25 μl in SYBR green reaction mix (Bio-Rad). Accumulation of fluorescent products was monitored by real-time PCR using a Bio-Rad iCycler PCR machine. Reactions were done in triplicate and normalized using the cycle threshold number for the total input sample. No PCR product was observed for the mock and no-antibody control reactions.
RESULTS
Loss of E2f1-3 in primary MEFs leads to the activation of p53 target genes.
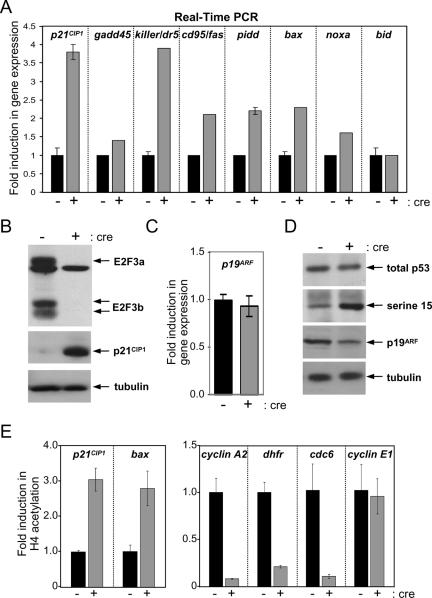
Using cre-LoxP technology and homologous recombination in mice, we have generated a conditional allele of E2f3 (E2f3f/f) that can be subsequently deleted in tissue culture by the infection of cells with a retroviral vector that expresses cre recombinase (38). By intercrossing E2f1−/−, E2f2−/−, and E2f3f/f mice, we generated primary E2f1−/− E2f2−/− E2f3f/f MEFs (this genotype is herein designated 123f/f). This conditional gene knockout strategy enabled us to delete the entire E2f1-3 activator subclass in fibroblasts and study its role in proliferation. Expression of cre inthese primary 123f/f MEFs led to the ablation of E2f3 and prevented the activation of E2F target gene expression and the entry of quiescent cells into S phase (Fig. 1A to C). As we have previously shown, the inactivation of E2f1-3 also resulted in a marked elevation of p21CIP1 protein levels that could be accounted for by a corresponding increase in mRNA levels as determined by real-time PCR analysis (Fig. 1D and data not shown) (38). Since p21CIP1 is a known transcriptional target of the p53 tumor suppressor (18, 36), we explored whether other p53-regulated genes were similarly induced in E2f1-3-deficient cells. Real-time PCR analyses demonstrated that numerous p53 targets, including killer (dr5), pidd, cd95 (fas), bax, and noxa, were also significantly elevated in these primary cells (Fig. 1D). These results suggest that in addition to a decrease in E2F target gene expression, loss of E2f1-3 leads to the activation of p53 transcriptional activity.
FIG. 1.
Loss of E2f1-3 leads to the activation of p53 target genes in primary cells. (A) Primary E2F1−/− E2F2−/− E2F3f/f (123f/f) cells were infected with control (−) or cre (+) retroviruses. Genomic DNA was then extracted from control- and cre-treated cells and used for PCR to determine the extent of E2f3 deletion. (B) BrdU incorporation of the primary 123f/f cell line. Primary 123f/f cells were treated as described for panel A and used for a BrdU incorporation assay. Cells were brought to quiescence by serum starvation and stimulated to proliferate by the addition of serum. Cells were harvested for BrdU incorporation at 0 and 18 h after serum stimulation. At least 500 cells were counted at each time point. (C) Real-time PCR analysis of E2F target genes. Total RNA was harvested from control-treated (−) or cre-treated (+) 123f/f cells and used to produce cDNA. Real-time PCR analysis was done to determine the relative levels of the indicated E2F target genes. Results are shown as the induction (n-fold) of gene expression, where levels for control-treated samples were standardized to equal 1. (D) Real-time PCR analysis of p53 target genes in the 123f/f cell line. Primary 123f/f cells were treated as described for panel A, and cells were harvested under proliferating conditions. Total RNA was extracted and used to produce cDNA that was then used to look at the relative levels of the indicated p53 target genes. Results are shown as the induction (n-fold) of gene expression in the E2f1−/− E2f2−/− E2f3−/− cells (+) compared to that in the control-treated E2f1−/− E2f2−/− E2f3f/f cells (−). (E) p19ARF mRNA levels are elevated in TKO cells. RNA from cells treated as described for panel C was used for real-time analysis. Shown are levels of p19ARF in control-treated or cre-treated 123f/f cells. (F) Increased levels of p19ARF protein in TKO cells. Cellular lysates from 123f/f cells infected with either control-expressing or cre-expressing retroviruses were used for a Western blot probed with the p19ARF, p53, and tubulin antibodies.
Previous work showing that E2F1 overexpression could induce p19ARF (9, 39) and that E2F3b, one of the two isoforms encoded by the E2f3 locus, could directly bind to E2F-binding elements in the p19ARF promoter (1) provided a possible connection between E2F activity and the control of the p53 tumor suppressor axis. Therefore, we initially considered the possibility that an increase in p19ARF protein, which is known to promote p53 protein stability, could represent the underlying basis for the dramatic changes in p53 and E2F target gene expression observed in E2f1-3-deficient cells. Whereas real-time PCR analysis revealed a small increase in p19ARF mRNA levels in E2f1-3-deficient cells, Western blot assays indicated a much more robust accumulation of p19ARF protein (Fig. 1E and F). Consistent with a role for p19ARF in the regulation of p53 protein stability (37), we also found a concomitant accumulation of p53 protein in these cells (Fig. 1F). These findings raised the possibility that loss of E2f1-3 may lead to a derepression of p19ARF expression, an accumulation of p53 protein, and the activation of p53-regulated targets. Because an increase in p19ARF protein levels is associated with the entry of primary MEFs into senescence (30), it is possible that the accumulation of p19ARF protein in E2f1-3-deleted primary MEFs may be an indirect consequence of their compromised proliferation and not due to a direct role of E2F3a/b in controlling p19ARF expression.
Loss of E2f1-3 leads to the activation of p53 target genes independent of p19ARF induction.
To obviate the potential confounding effects that premature entry into senescence may have on the interpretation of the results described above, we established four independent E2f1−/− E2f2−/− E2f3f/f MEF lines (designated 123f/f cell lines 1 through 4) by use of the 3T9 protocol (33). The established 123f/f cell lines 1 through 4 retained an intact p53 pathway, since these cells continued to express both p19ARF and p53 and retained the ability to arrest in response to γ-irradiation (data not shown). In addition, sequencing of p53 cDNA derived from these cells failed to identify any mutations in its coding sequence (data not shown). Consistent with the analysis of primary MEFs, cre expression in each of the four established 123f/f cell lines led to the efficient ablation of both E2F3a and E2F3b proteins, yielding triple knockout (TKO) cells that failed to proliferate (Fig. 2B and reference 38). Real-time PCR analysis of RNA isolated from control- or cre-treated 123f/f MEFs revealed a failure of TKO cells to induce, in response to mitogenic stimulation, the timely expression of most E2F targets that normally peak during the G1/S transition (see Fig. 6B and reference 36). As in the analysis of primary E2f1-3-deficient MEFs, cre-mediated ablation of E2f3 in the established 123f/f MEF lines 1 through 4 led to a marked induction in p53 target gene expression (Fig. 2A and B). In contrast what was seen for primary MEFs, however, loss of E2f1-3 in established cell lines did not result in an increase of p19ARF mRNA or protein levels or in an increase of total p53 protein (Fig. 2C and D), even though p53 transcriptional activity was markedly induced (Fig. 2A). In fact, p19ARF protein levels in cre-treated 123f/f established cells were found to be slightly lower than those in control-treated cells (Fig. 2D), consistent with previous reports describing a negative autoregulatory loop for the regulation of p19ARF expression by p53 (26). These findings suggest that p19ARF is unlikely to play a causal role in the severe block of proliferation we observe for established E2f1-3-deficient cells.
FIG. 2.
Loss of E2f1-3 leads to p53 phosphorylation on serine 15. (A) Real-time PCR analysis of p53 target genes in the 123f/f cell line. Established 123f/f MEFs were infected with either control-expressing or cre-expressing (+) retrovirus and selected with puromycin. The cells were harvested under proliferating conditions. Total RNA was extracted and used to produce cDNA that was then used to look at relative levels of the indicated p53 target genes. Results are shown as induction (n-fold) of gene expression in the E2f1−/− E2f2−/− E2f3−/− cells compared to that in the control-treated E2f1−/− E2f2−/− E2f3f/f cells. (B) p21 levels are increased in TKO cells. The 123f/fcell line was infected with either control-expressing or cre-expressing retroviruses and selected with puromycin. Protein was then extracted from these cells, and equal amounts of lysates from control- and cre-treated 123f/f cells were used for Western blot analysis with antibodies against E2F3 (top panel), p21CIP1 (middle panel), and tubulin as a loading control (bottom panel). (C) p19ARF mRNA levels in TKO cells. RNA from cells treated as described for panels A and B was used for real-time PCR analysis. Shown here are levels of p19ARF in control-treated or cre-treated 123f/f cells. (D) Phosphorylated p53 protein levels are increased in TKO cells. Lysates from cells treated as described for panels A and B were used for Western blot analysis. For the Western blot, antibodies against total p53, p53 (ser15) and p19ARF were used along with tubulin as a loading control. (E) Acetylation of histone H4. Cell lysates from experiments similar to those described above were harvested and used for chromatin immunoprecipitation assays with antibodies against acetylated histone H4. Real-time PCR was then performed using primers around either the p53-binding sites of the p21 and bax promoters or the E2F sites in the cyclin A2, dhfr, cdc6, and cyclin E1 promoters. Results are shown as inductions (n-fold) of histone H4 acetylation in control versus TKO cells from one representative experiment.
FIG. 6.
E2F target genes are expressed in QKO cells. (A) cDNA prepared for the experiment shown in Fig. 5 was also used to determine the expression levels of E2F target genes in proliferating TKO versus QKO cells. In contrast to the results shown below, this graph demonstrates repression (n-fold) of gene expression in the cre-treated cells relative to that in the cells infected with control virus. (B) Induction of S phase and real-time PCR analysis of E2F target genes in TKO and QKO cells. Cells were synchronized by serum starvation, stimulated by the addition of serum, and harvested at the indicated time points. Total RNA was extracted from the cells and subsequently used to produce cDNA and determine the levels of the indicated E2F target genes. In order to determine the timing of S-phase entry in these cells relative to that of gene expression, the incorporation of 5-bromodeoxyuridine was also analyzed. BrdU was added to the plates at the same time as the serum, and harvesting took place at the zero time point as well as at the 8-hour time point. A total of 500 DAPI-stained nuclei from each cell line was counted, and the percent positive for BrdU incorporation is shown. Percent BrdU incorporation is indicated by the dashed lines, and solid lines represent induction (n-fold) of target genes. Symbols: □, cells treated with control virus; •, cre-treated cells.
The observations described above then raise the question of how the inactivation of E2f1-3 results in the induction of p53 target genes without a concomitant increase in p53 protein levels. We explored whether p53 transcriptional activity in E2f1-3-deficient cells might be modulated by posttranslational modifications. Indeed, Western blot assays employing p53-ser15-specific antibodies revealed higher levels of serine 15-phosphorylated p53 in E2f1-3-deficient cells than in control-treated samples (Fig. 2D). While additional mechanisms may be at play here, these data raise the possibility that the activation of p53 and its target genes in E2f1-3-deficient established MEFs may be due to the phosphorylation of p53.
To investigate further the possible mechanism underlying the dramatic changes in the regulation of p53 and E2F target gene expression observed for E2f1-3-deficient cells, we utilized ChIP assays to determine the status of histone acetylation in nucleosomes positioned on p53- and E2F-regulated promoters. Acetylation of histone H4 (K5, K8, K12, K16) corresponds with an open chromatin structure that is permissive for E2F-dependent transcription activation, whereas deacetylation corresponds with transcriptional silencing (12, 16). Analysis of nucleosomes positioned near the E2F-binding elements of the cyclin A and dhfr promoters, two E2F-regulated genes severely impacted by the loss of E2f1-3, revealed significant decreases (10- and 5-fold, respectively) in the levels of acetyl-histone H4 associated with these promoters (Fig. 2E, right panel). In contrast, histone H4 acetylation on nucleosomes near the −1800 and −1500 p53-binding elements of the p21CIP1 and bax promoters, respectively, increased significantly (Fig. 2E, left panel). While this increase was not dramatic, it was consistently observed in multiple experiments. From these results, we conclude that the E2F1-3 factors are necessary for both the cell cycle-dependent activation of E2F target genes and the silencing of numerous p53-regulated genes. The fact that the p53-regulated promoters analyzed as described above, except for p21CIP1, lack E2F-binding elements and do not respond to E2F overexpression suggests that E2F is unlikely to directly regulate their expression. The fact that the acetylation of histones positioned near the p53-binding elements of p53-responsive promoters is enhanced in TKO cells suggests that E2F1-3 might regulate the expression of p21CIP1 and other p53 target genes through the modulation of p53 activity.
Loss of E2f1-3 results in the recruitment of E2F4-p130 complexes to target promoters.
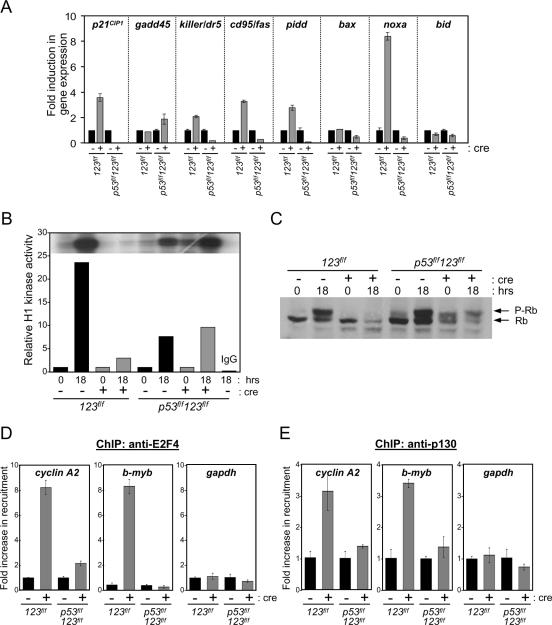
Inactivation of E2f1-3 results in a marked elevation of p21CIP1 protein that can be accounted for by a corresponding increase in its mRNA levels (Fig. 2A and B). Along with the induction of p21CIP1 expression, we could also measure a corresponding decrease in mitogen-activated cyclin-dependent kinase activity and a concomitant increase in hypophosphorylated Rb and the related p130 pocket protein family member (see Fig. 5B and C and reference 38). Because hypophosphorylated pocket proteins can associate with the E2F4 family member to form a transcriptional repressor complex, we investigated whether p130 and E2F4 in TKO cells might be recruited to E2F-binding sites on known E2F target promoters. ChIP assays revealed that loss of E2f1-3 led to the loading of E2F4 and p130 onto E2F-binding elements of two known E2F target promoters, cyclin A2 and b-myb (see Fig. 5D and E). Thus, the decrease in E2F target expression observed in TKO cells could stem from the absence of E2F activators, the accumulation of E2F repressor complexes on E2F target promoters, or both.
FIG. 5.
Loss of p53 in TKO cells prevents the activation of p53-regulated genes and restores kinase activity. (A) Real-time PCR analysis of p53 target genes in TKO and QKO cells. The 123f/f and p53f/f 123f/f cell lines were infected with either control-expressing (−) or cre-expressing (+) retroviruses and selected with puromycin. Total RNA was extracted from proliferating cells and then used to produce cDNA to determine relative levels of the indicated p53 target genes. Results are shown as induction (n-fold) of gene expression in the cre-treated versus the control-treated cells from one representative experiment. (B) Quadruple knockout cells have normal kinase activity. Relative activities of cyclin E-dependent kinase in protein lysates derived from TKO and QKO cells were determined by harvesting cells after 72 h of serum starvation (0 hrs) or 18 h of serum stimulation (18 hrs) and using histone 1 as a substrate. The blot shown in the inlay was scanned and quantified, and relative activity is shown. (C) Rb phosphorylation status of cells used in the experiment described for panel B as determined by Western blot analysis. (D and E) Cell lysates from TKO and QKO cells treated with either control (−) or cre (+) retrovirus were harvested and then used for chromatin immunoprecipitation assays with antibodies against E2F4 and p130. Real-time PCR was then performed using primers around the E2F sites in the cyclin A2, bmyb, and gapdh promoters. Results are shown as recruitment (n-fold) in cre-treated cells relative to that in the cells infected with control virus from one representative experiment.
TKO cells harboring loss of heterozygosity and mutations in p53 can proliferate.
In three out of the four cell lines (123f/f cell lines 1 through 3) used in the studies described above, ablation of E2f3 rendered 100% of TKO cells unresponsive to normal mitogenic signals (Fig. 3A, left panel; also data not shown). Surprisingly, the disruption of E2f3 in the fourth cell line (123f/f cell line 4) consistently yielded TKO cells that could proliferate and form colonies, albeit at a very low frequency (Fig. 3A and B). The appearance of cells that acquired the capacity to proliferate in the absence of E2F1, E2F2, and E2F3 suggested that the requirement for E2Fs in proliferation is not absolute but rather can be overcome by alteration(s) of parallel or downstream pathways. We reasoned that the identification of the putative genetic or epigenetic changes that must have occurred in this particular set of TKO clones could provide invaluable insight towards the understanding of E2F function, and thus, we analyzed these TKO colonies further.
FIG. 3.
Analysis of proliferating TKO colonies. (A) Colony formation assay of TKO cells. Cells treated as described in the legend for Fig. 2A were counted and plated on 100-mm plates. After 2 weeks, the colonies were fixed, stained, and counted. The mean and standard deviation of colony counts from triplicate plates are shown. Colonies from the cre-infected cells have been corrected for deletion of E2f3 using the method described in panel B. (B) To determine the efficiency of E2f3 deletion, cells treated with the cre retrovirus were also plated into 96-well plates. DNA was extracted from wells containing single colonies and analyzed for deletion of E2f3 by PCR genotyping. A total of 32 colonies for each cell line was analyzed. Shown are results for seven representative colonies. (C) Colonies obtained from 123f/f cell line 4 from an experiment similar to the one described above were expanded and subjected to various assays. E2f3 PCR genotyping of genomic DNA extracted from the 123f/f cell line 4 colonies is shown. PCR on genomic DNA from the original population of cells is shown in lanes 2 and 3. Lanes 4 to 15 represent 12 different colonies. Asterisks identify TKO colonies. Colonies without asterisks represent cells that were infected with the cre retrovirus and survived selection but failed to delete E2f3. (D) Western blot analysis of colonies for several cell cycle regulators. Protein lysates were made from the original cell populations and the 12 colonies. After electrophoresis of equal amounts of protein, the ensuing Western blots were probed with antibodies against p21CIP1 (top panel), p53 (middle panel), and cdk4 (bottom panel) proteins. (E) p53 promoter activity in colonies of 123f/f cell line 4. Cells were transfected with either a wild-type p21CIP1 promoter construct (pGL3.1-mp21) or a mutant construct deleted for the p53-binding sites (pGL3.1-mp21Δp53). The graph demonstrates the relative activity obtained from the wild-type construct versus that from the mutated construct for each colony. 123f/f1, 123f/f cell line 1; 123f/f4, 123f/f cell line 4.
An initial characterization of 21 of these TKO colonies revealed that they fell into two groups with distinct growth rates. One group consisting of 17 colonies exhibited rapid proliferation (colonies 2, 4, and 7 in Fig. 3C and D; also data not shown), and the other group of 4 colonies maintained a low growth rate even after many cell doublings (>30 doublings) (colonies 9 to 12 in Fig. 3C and D). The isolation of TKO colonies with two different growth properties suggested that at least two compensatory pathways capable of overcoming an E2f1-3 deficiency might exist. In an attempt to identify the genetic alterations that might be contributing to the ability of TKO clones of 123f/f cell line 4 to proliferate, we analyzed various cell cycle-regulatory components in these cells. Given our previous results, we analyzed p21CIP1 and p53 protein in control- and cre-treated 123f/f cell line 4 populations prior to clonal selection, in colonies that escaped E2f3 deletion (nondeleted), and in the rare TKO colonies. Western blot analysis demonstrated that while no significant change in the protein expression of cyclin E, cdk2, or cdk4 was observed, p21CIP1 protein was significantly reduced in nondeleted and fast-growing TKO colonies relative to what was seen either for slow-growing TKO colonies (colonies 9 to 12) or for TKO cell populations prior to clonal selection (Fig. 3D). Moreover, cdk2-dependent kinase activity was markedly elevated in the fast-growing TKO colonies (data not shown), consistent with a decrease in p21CIP1 protein in these cells. Relatively low levels of p53 were present in control- and cre-treated E2f1−/− E2f2−/− E2f3f/f cell populations and in nondeleted or slow-growing TKO colonies. In contrast, fast-growing TKO colonies accumulated high levels of p53 protein. Because stabilization of p53 protein in human cancers is often associated with missense mutations in its coding sequence, we cloned and sequenced p53 cDNA from the various TKO colonies. As suspected, this analysis revealed a single missense p53 mutation (569G→C) at codon 193 (R193P) in colonies with high levels of p53 protein. In each case, close inspection of the sequencing histograms revealed a single cytosine peak at the first position of codon 193, suggesting that the remaining wild-type p53 allele underwent loss of heterozygosity. Interestingly, this mutation was found only in the TKO colonies with high levels of p53 and not in the remaining clones or in cells that had escaped cre-mediated deletion of E2f3 (data not shown).
Amino acid 193 is located within the DNA-binding domain of p53, and mutations in this region are predicted to impair the ability of the corresponding protein to bind and activate target genes, including p21CIP1 (13). To test this possibility, a p21CIP1-luciferase reporter construct containing the two previously characterized p53-binding sites within a 3.2-kb fragment of the mouse p21CIP1 promoter was used to directly measure p53 activity in fast- and slow-growing TKO colonies. A p21CIP1 reporter lacking the p53-binding elements was used as a negative internal control. As predicted, these p21CIP1 reporter assays confirmed the absence of functional p53 in the fast-growing TKO colonies (Fig. 3E). Similar results were obtained when an artificial p53-responsive reporter construct containing tandem repeats of wild-type or mutant p53-binding elements was used to assess p53 function in fast- and slow-growing TKO colonies (data not shown). These data show that the presence of the inactivating R193P mutation in the p53 gene correlates with the loss of p21CIP1 expression and the ability of TKO cells to proliferate rapidly. While this analysis suggests that mutation of p53 may be a key genetic alteration allowing cells to grow in the absence of E2f1-3, our data do not rule out the possibility that additional genetic changes in these cells could be contributing to their ability to proliferate. Moreover, the fact that p53 activity remained intact in slow-growing TKO colonies suggests that additional genetic changes in downstream or parallel pathways may also overcome the requirement for E2F1-3 in cellular proliferation.
Targeted disruption of p53 suppresses the severe growth defect of TKO MEFs.
The results presented above raise the possibility that E2F1-3 activators may promote cell cycle-dependent gene expression and proliferation through the inhibition of the p53 axis. To test this hypothesis, we sought to genetically disrupt p53 in TKO cells in order to examine whether p53 inactivation is sufficient to bypass a requirement for E2F1-3 in proliferation. To this end, mice containing a conditional p53 allele (p53f/f) were interbred with E2f1−/− E2f2−/− E2f3f/f mice in order to generate p53f/f E2f1−/− E2f2−/− E2f3f/f (p53f/f 123f/f) MEFs. The introduction of cre recombinase into p53f/f 123f/f cells would therefore yield quadruple knockout (QKO) MEFs. We initially analyzed the proliferation capacities of control- and cre-treated primary p53f/f E2f1−/− E2f2−/− E2f3f/f MEFs as well as those of two p53f/f E2f1−/− E2f2−/− E2f3f/f MEF lines established using the 3T9 protocol as before. In each case, PCR and Western blot analysis confirmed the efficient cre-mediated deletion of p53 and E2f3 (Fig. 4A and data not shown). Established QKO cells incorporated BrdU into genomic DNA equally well as control-treated p53f/f 123f/f or 123f/f cells or cre-treated 123f/f cells reconstituted with a myc-tagged version of E2F3a, indicating that cells deficient for E2f1-3 and p53 were able to replicate their DNA efficiently (Fig. 4B). Moreover, cre-treated p53f/f 123f/f established cells could form colonies that were indeed deleted for both E2f3 and p53 (Fig. 4C and D). While a few of the colonies analyzed were deleted for p53 only, none of the colonies were deleted for E2f3 alone (data not shown). Parallel experiments using primary MEFs demonstrated that quiescent control- and cre-treated p53f/f 123f/f MEFs could be stimulated to enter the cell cycle equally well (Fig. 4E). We also compared the expression levels of a panel of p53-regulated genes in primary and established TKO and QKO cells by real-time PCR techniques. This analysis revealed that the induction of p21CIP1, killer (dr5), cd95 (fas), pidd, and noxa expression typically observed in E2f1-3-deficient cells is inhibited by the deletion of p53 (Fig. 5A and data not shown). Together, these results suggest that the activation of p53 in E2f1-3-deficient cells is the critical event leading to their block in proliferation.
FIG. 4.

Genetic deletion of p53 rescues the cellular proliferation defect of E2f1−/− E2f2−/− E2f3−/− cells. The 123f/f and p53f/f 123f/f established cell lines (passage number, >20) were infected first with control- or E2F3a-expressing retroviruses and then with either control- or cre-expressing retroviruses, selected for hygromycin and puromycin resistance, and then used in the following assays. (A) E2f3 (top panel) and p53 (bottom two panels) PCR genotyping of the population of cells from the experiment described above showing efficient deletion of both E2f3 and p53. The floxed allele (E2f3f/f) produces a 184-bp PCR fragment, and the knockout E2f3 allele produces a 416-bp fragment. The p53 conditional and knockout alleles produce 584-bp and 612-bp fragments, respectively. (B) BrdU incorporation of established 123f/f and p53f/f 123f/f cell lines. Proliferating cells were incubated with BrdU for 12 h and then fixed and stained. A total of 500 DAPI-stained nuclei from each cell line was counted, and the percent positive for BrdU incorporation is shown. (C) Cells from the experiment described above were also tested for their long-term growth potentials by colony formation assay. Values shown have been corrected for deletion of E2f3 and p53 by colony PCR. (D) E2f3 (top panel) and p53 (bottom two panels) PCR genotyping on genomic DNA from the cre-infected 123f/f and p53f/f 123f/f colonies. E2f3 was not deleted in colonies arising from the 123f/f cell line but was deleted in all the colonies arising from the cre-infected p53f/f 123f/f population of cells. (E) BrdU incorporation of the primary p53f/f 123f/f cell line. Primary p53f/f 123f/f cells were infected with control-expressing (−) or cre-expressing (+) retroviruses. Cells were synchronized by serum starvation, stimulated by the addition of serum, and harvested at the indicated time points. At least 500 cells were counted at each time point, and the percent positive for BrdU incorporation is shown.
Ablation of p53 leads to derepression of E2F-regulated genes in TKO cells.
To determine whether the p53-dependent induction of p21CIP1 and the consequent inhibition of cdk activity and loading of E2F-Rb repressor complexes on E2F-responsive promoters might be responsible for the inhibition of E2F target gene expression in TKO cells, we also examined these events in QKO cells. To this end, 123f/f and p53f/f 123f/f cells were treated with control- or cre-expressing retroviruses, brought to quiescence by serum deprivation, and then restimulated by the addition of serum. Whereas cells deficient for E2f1-3 failed to induce cdk2-dependent kinase activity in response to serum stimulation, cells deficient for p53 and E2f1-3 responded normally (Fig. 5B). As a result, the relative levels of phosphorylated to hypophosphorylated Rb in cre-treated p53f/f 123f/f cells increased upon serum stimulation (Fig. 5C). Importantly, ChIP assays using E2F4- and p130-specific antibodies showed that in contrast to the loss of E2f1-3, the ablation of p53 and E2f1-3 prevented the loading of E2F4 and p130 onto E2F target promoters (Fig. 5D and E). We then asked whether E2F target expression might also be restored in these QKO cells. Indeed, cre-mediated ablation of p53 and E2f3 from p53f/f 123f/f cells restored E2F target gene expression to levels normally found in wild-type proliferating cells (Fig. 6A). Together, these results demonstrate that inactivation of p53 in E2f1-3-deficient cells is sufficient to prevent the accumulation of p21CIP1 protein and restore the normal mitogen-mediated regulation of cdk2 activity, culminating in pocket protein phosphorylation, derepression of E2F target genes, and cell cycle progression.
Ablation of p53 leads to a derepression of E2F-regulated genes in TKO cells.
E2F target gene expression oscillates during the cell cycle in a growth factor-dependent manner (17). While overall levels of E2F targets in cells lacking E2f1-3 could be restored by the loss of p53, it remained possible that the exact timing of their expression during the cell cycle could be altered. Thus, we also evaluated E2F target expression in quiescent TKO and QKO cells that were serum induced and harvested over a period of 24 h (Fig. 6B). Cell cycle progression and target gene expression were evaluated by BrdU incorporation and real-time PCR, respectively. Two types of E2F targets were evaluated: targets normally induced late in G1, such as cdc6, mcm3, and dhfr, and targets induced later in the cell cycle, such as cyclin A2. Quiescent control-treated 123f/f and p53f/f 123f/f cells responded equally well to serum, leading to the timely G1/S activation of cdc6, mcm3, tk, and dhfr expression followed by the induction of cyclin A2 expression (Fig. 6B). Multiple cell lines for each genotype yielded similar results. Quiescent cre-treated 123f/f cells did not enter the cell cycle in response to serum stimulation to any appreciable extent, and E2F target expression remained low throughout the time course of the experiment (Fig. 6B, left panel). In contrast, quiescent cre-treated p53f/f 123f/f cells stimulated by the addition of serum entered S phase either at the same time as or slightly earlier than control-treated p53f/f 123f/f cells (Fig. 6B, right panel). The pattern of E2F target gene expression in QKO cells, however, was dramatically different from that seen for TKO cells. Surprisingly, this expression pattern was also significantly compromised relative to that for control-treated p53f/f 123f/f cells. First, the peak levels of expression for most E2F targets, except for tk and cyclin E1 (not shown), were attenuated in cre-treated relative to control-treated p53f/f 123f/f cells (Fig. 6B, right panels). Second, in cre-treated p53f/f 123f/f cells the late-G1-phase-specific targets (cdc6, mcm3, dhfr) were maximally induced subsequent to S-phase entry, and their peak induction was markedly reduced relative to that seen for control-treated p53f/f 123f/f cells. Finally, the timings of activation of S-phase-specific targets (cyclin A2) were similar in cre- and control-treated p53f/f 123f/f cells, but this activation in the cre-treated cells could be considered delayed compared to the entry of cells into S phase. From these results, we can make several conclusions. First, in cells containing wild-type p53, E2F activators are essential for both the amplitude and timing of expression of E2F target genes and consequently are required for cellular proliferation. Second, in p53-deficient cells, E2F activators are required for the timely induction of late-G1-phase-regulated E2F targets that normally peak prior to the G1/S transition but are not required for the overall accumulation of targets in asynchronous proliferating populations. In general, the above data suggest that the basal level of E2F targets is largely determined by E2F-mediated repression but that the precise oscillatory nature of E2F target gene expression during the cell cycle is facilitated by E2F-mediated activation.
Activation of p53 precedes E2F-mediated transcriptional repression in TKO cells.
From the results presented above, we propose that the activation of p53 is a primary consequence of E2f1-3 ablation. The subsequent accumulation of p21CIP1 and other p53 target genes leads to a cascade of events that begins with a decrease in cdk activity, followed by hypophosphorylation of pocket proteins, pocket protein-mediated repression of E2F target genes, and eventual cell cycle arrest. This hypothesis predicts that p53 activation in TKO cells occurs prior to the observed repression of E2F target genes. To test this model, we directly analyzed the chromatin state around p53-binding sites of p53-responsive promoters and around E2F-binding sites of E2F-responsive promoters at early and late time points (2 and 4 days) after cre-mediated deletion of E2f3 from 123f/f cells. We chose to analyze TKO cells after 2 days of cre treatment, which we considered as the early time point since this represents the earliest time point when we could confidently confirm the complete deletion of E2f3 from cre-treated 123f/f cells (data not shown). As shown by the ChIP assay results presented in Fig. 7A (2 days, right panel), there was an acute increase in histone H4 acetylation around p53-binding sites of the p21CIP1 and bax promoters soon after the ablation of E2f3. This was consistent with the activation of these genes at the early time point (Fig. 7C, right panel). In contrast, there was no significant decrease in H4 acetylation at E2F-binding sites of the cyclin A2, dhfr, and cdc6 promoters (Fig. 7A, left panel), consistent with the relatively unaffected expression of these genes at this early time point (compare left panels of Fig. 7C and D). By 4 days following cre treatment, however, there was a pronounced decrease in H4 acetylation on E2F-responsive promoters that was followed by a robust decrease in their expression (Fig. 7B and D, left panels). The acute increase of H4 acetylation on p53-responsive promoters was attenuated by 4 days after cre treatment, even though their expression remained elevated (Fig. 7B and D, right panels). Both the early induction of H4 acetylation on p53-responsive promoters and the late reduction in H4 acetylation on E2F-responsive promoters were dependent on p53, since these effects were completely abolished in E2f1-3-deficient cells that also lacked functional p53 protein (Fig. 7 E and F; compare cre-treated 123f/f and p53f/f 123f/f samples). The fact that the activation of p53 in cre-treated 123f/f MEFs was followed by the repression of E2F target genes is consistent with the notion that the control of p53 activity is the primary event regulated by E2F1-3.
FIG. 7.
Histone H4 acetylation status of E2F and p53 target promoters. (A) Cell lysates from TKO cells treated with either control (−) or cre (+) retrovirus were harvested after 2 days (left panel) and 4 days (right panel) of selection and then used for chromatin immunoprecipitation assays with antibodies against acetylated histone H4. Real-time PCR was then performed using primers around either the E2F sites in the cyclin A2, dhfr, and cdc6 promoters or the p53-binding sites of the p21 and bax promoters. Results are shown as induction (n-fold) of histone H4 acetylation in control versus TKO cells from one representative experiment. (B) In addition to cell lysates from cells in the experiment described above, total RNA was extracted and used to produce cDNA. Real-time PCR analysis was then used to look at the relative levels of the indicated E2F and p53 target genes. Results are shown as induction (n-fold) of gene expression. (C) Repeat of the experiment described above with the addition of QKO cells treated with control or cre retrovirus.
DISCUSSION
In an attempt to rigorously assess the role and mechanism of E2F family members in the control of gene expression and cellular proliferation, we have conditionally targeted the disruption of the entire E2F subclass of activators comprised of E2f1, E2f2, and E2f3 in mice. This genetic approach led us to suggest that E2f1-3 impact two separate mechanisms that operate in synergy to stimulate E2F target gene expression. First, E2F1-3 factors could function directly as bona fide activators of transcription. Second, they could prevent repression of target genes by promoting the dissociation of pocket protein-E2F4-5 repressor complexes through a p21CIP1-dependent mechanism. We now demonstrate that the persistent p130-E2F4-mediated repression of E2F target genes in E2f1-3-deficient cells temporally follows the activation of p53. Importantly, the targeted inactivation of p53 was sufficient to lift E2F-dependent repression, leading to the reactivation of E2F target genes and the proliferation of E2f1-3-deficient cells. These results suggest a novel role for E2F1-3 in the negative regulation of p53 activity that is necessary for cell proliferation.
E2F1-3 controls E2F-dependent transcriptional repression and S-phase entry via the p53 axis.
The functions of E2F family members include mechanisms that involve both E2F-dependent repression and activation (4, 8, 11, 14, 15, 23, 31, 35). Because of the complex interrelationships between E2F-dependent activation and repression, and vice versa, it has been difficult to rigorously test how these two processes are coordinated during the cell cycle. The importance of the E2F1-3 activators in the control of cellular proliferation was previously demonstrated by our group via analysis of fibroblasts derived from mice lacking E2f1-3 (38). The experiments presented here now suggest that the first detectable event occurring immediately after the ablation of E2f1-3 is the recruitment of p53 to its target genes, including p21CIP1, followed by their activation. As p21CIP1 protein accumulates and cdk activity decreases, Rb and other pocket proteins become progressively hypophosphorylated, promoting the formation of pocket protein-E2F4 complexes and the repression of E2F target genes (38).
Several lines of evidence support a role for E2f1-3 in the control of a p53-dependent axis that is critical for G1/S-regulated gene expression and cellular proliferation. First, characterization of E2f1−/− E2f2−/− E2f3f/f cell lines led to the isolation of rare TKO cells harboring p53-inactivating mutations that were capable of proliferating. Second, we could show that the papillomavirus E6 oncogenic product (from HPV16 and HPV18) can promote the proliferation of TKO cells and that this rescue was dependent on its p53-inactivating function (data not shown). Finally, the cre-mediated deletion of a conditional p53 allele suppressed the growth arrest in TKO MEFs. In each case, the ablation of p53 mitigated the activation of p53 target genes, restored E2F target gene expression, and promoted the proliferation of TKO cells. These results solidify the connection between the functions of E2F1, E2F2, and E2F3, the control of p53 activity, and the p21CIP1-dependent negative feedback loop controlling E2F-dependent repression. Considering the large and diverse set of target genes that the E2F1-3 factors has been proposed to regulate, it is striking that the inactivation of a single negative regulator of proliferation—p53—appears to be sufficient to reverse the severe growth defect of E2f1-3-deficient cells. Together, these observations argue that at least in fibroblasts, E2F-mediated repression is the main mode of regulating most E2F target genes that are critical for cellular proliferation.
The mechanism by which E2F1, E2F2, and E2F3 regulate p53 activity remains unclear. Recent work by Aslanian et al. suggested that E2F3b might be directly involved in repressing p19ARF expression, which is known to mediate p53 protein stabilization (1). These authors report that E2F3b binds to the E2F-binding elements present in the p19ARF promoter at a time when p19ARF is normally repressed (G0). Furthermore, they show that loss of E2f3 results in a twofold increase in p19ARF mRNA levels and a dramatic increase in p19ARF protein (>10-fold). Consistent with these findings, we find that the acute loss (via cre-mediated recombination) of E2f1-3 from primary MEFs leads to a very mild induction of p19ARF mRNA levels and a disproportionate accumulation of p19ARF protein (Fig. 1). In both studies there is a clear disconnect between p19ARF mRNA and protein levels in MEFs lacking E2f3. It is not clear how a 1.5- or 2-fold increase in p19ARF mRNA levels could account for the >10-fold increase in p19ARF protein. Given these observations, we would suggest that the marked accumulation of p19ARF protein is associated with the premature entry of these primary E2f1-3 MEFs into a senescent state and not due to a direct role of E2F3a or E2F3b in controlling p19ARF expression.
To obviate the inadvertent consequences of early entry into a senescent state, the studies presented here also measured the acute inactivation of E2f3 in established 3T9 cells that retain an intact p19ARF-p53 pathway. We feel that this latter setting circumvents the confounding effects of senescence and thus better reflects the immediate effects resulting from the inactivation of E2f3. In this setting, acute loss of E2f3 does not significantly affect p19ARF mRNA and protein levels, even though E2f1-3deficiency results in a robust induction of p53 activity and p53-responsive genes. While the exact mechanism by which E2F1-3 controls p53 activity remains to be determined (1, 3, 27), our data suggest that the increase in p53 activity in E2f1-3-deficient cells is associated with posttranslational mechanisms, which could include the phosphorylation of p53 at serine 15.
E2F1-3 facilitates the cell cycle-dependent expression of E2F target genes.
The results presented in this study also indicate a role for E2F1-3 in determining the extent and precise timing of E2F target gene expression during cell cycle entry. We suggest that E2F target expression during the cell cycle is regulated by E2F1-3 via two synergistic mechanisms involving E2F-mediated repression and activation. Whereas repression and derepression appear to be the principal mechanism of setting the overall pattern of expression, transcriptional activation would serve to accentuate and define the precise timing of E2F target expression at the G1-S boundary. Our data support a role for E2F1-3 in controlling the E2F-mediated repression arm via a p53-p21CIP1 feedback loop that regulates the formation and accumulation of p130-E2F4 repressor complexes. Hence, E2F1-3 could be viewed as an activity necessary for coordinating repression and activation, first by promoting the cdk-dependent dissociation of Rb/E2F repressor complexes and then by directly activating the “timely and acute” induction of G1-S gene expression—a time in the cell cycle when E2F target gene products are needed the most.
Acknowledgments
This work was funded by NIH grants to G.L. (R01CA85619, R01CA82259, R01HD047470, P01CA097189), C.T. (ACS Fellowship), L.W. (K01CA102328), and H.I.S. (K01CA104079). G.L. is the recipient of The Pew Charitable Trusts Scholar Award and the Leukemia & Lymphoma Society Scholar Award.
We declare that we have no competing financial interests.
Footnotes
Published ahead of print on 30 October 2006.
REFERENCES
- 1.Aslanian, A., P. J. Iaquinta, R. Verona, and J. A. Lees.2004. . Repression of the Arf tumor suppressor by E2F3 is required for normal cell cycle kinetics. Genes Dev. 18:1413-1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balciunaite, E., A. Spektor, N. H. Lents, H. Cam, H. Te Riele, A. Scime, M. A. Rudnicki, R. Young, and B. D. Dynlacht.2005. . Pocket protein complexes are recruited to distinct targets in quiescent and proliferating cells. Mol. Cell. Biol. 25:8166-8178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bates, S., A. C. Phillips, P. A. Clark, F. Stott, G. Peters, R. L. Ludwig, and K. H. Vousden.1998. . p14ARF links the tumour suppressors RB and p53.Nature 395:124-125. (Letter.) [DOI] [PubMed] [Google Scholar]
- 4.Cam, H., and B. D. Dynlacht. 2003. Emerging roles for E2F: beyond the G1/S transition and DNA replication. Cancer Cell 3:311-316. [DOI] [PubMed] [Google Scholar]
- 5.Christensen, J., P. Cloos, U. Toftegaard, D. Klinkenberg, A. P. Bracken, E. Trinh, M. Heeran, L. Di Stefano, and K. Helin.2005. . Characterization of E2F8, a novel E2F-like cell-cycle regulated repressor of E2F-activated transcription.Nucleic Acids Res. 33:5458-5470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dannenberg, J. H., A. van Rossum, L. Schuijff, and H. te Riele.2000. . Ablation of the retinoblastoma gene family deregulates G(1) control causing immortalization and increased cell turnover under growth-restricting conditions. Genes Dev. 14:3051-3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Bruin, A., B. Maiti, L. Jakoi, C. Timmers, R. Buerki, and G. Leone.2003. . Identification and characterization of E2F7, a novel mammalian E2F family member capable of blocking cellular proliferation.J. Biol. Chem. 278:42041-42049. [DOI] [PubMed] [Google Scholar]
- 8.DeGregori, J. 2002. The genetics of the E2F family of transcription factors: shared functions and unique roles.Biochim. Biophys. Acta 1602:131-150. [DOI] [PubMed] [Google Scholar]
- 9.DeGregori, J., G. Leone, A. Miron, L. Jakoi, and J. R. Nevins.1997. . Distinct roles for E2F proteins in cell growth control and apoptosis. Proc. Natl. Acad. Sci. USA 94:7245-7250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Di Stefano, L., M. R. Jensen, and K. Helin.2003. . E2F7, a novel E2F featuring DP-independent repression of a subset of E2F-regulated genes. EMBO J. 22:6289-6298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dyson, N. 1998. The regulation of E2F by pRB-family proteins.Genes Dev. 12:2245-2262. [DOI] [PubMed] [Google Scholar]
- 12.Eberharter, A., and P. B. Becker. 2002. Histone acetylation: a switch between repressive and permissive chromatin. Second in review series on chromatin dynamics. EMBO Rep. 3:224-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.El-Deiry, W. S. 1998. Regulation of p53 downstream genes. Semin. Cancer Biol. 8:345-357. [DOI] [PubMed] [Google Scholar]
- 14.Frolov, M. V., D. S. Huen, O. Stevaux, D. Dimova, K. Balczarek-Strang, M. Elsdon, and N. J. Dyson.2001. . Functional antagonism between E2F family members.Genes Dev. 15:2146-2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Helin, K. 1998. Regulation of cell proliferation by the E2F transcription factors. Curr. Opin. Genet. Dev. 8:28-35. [DOI] [PubMed] [Google Scholar]
- 16.Jenuwein, T., and C. D. Allis. 2001. Translating the histone code. Science 293:1074-1080. [DOI] [PubMed] [Google Scholar]
- 17.Leone, G., J. DeGregori, Z. Yan, L. Jakoi, S. Ishida, R. S. Williams, and J. R. Nevins. 1998. E2F3 activity is regulated during the cell cycle and is required for the induction of S phase. Genes Dev. 12:2120-2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Levine, A. J. 1997. p53, the cellular gatekeeper for growth and division. Cell 88:323-331. [DOI] [PubMed] [Google Scholar]
- 19.Logan, N., L. Delavaine, A. Graham, C. Reilly, J. Wilson, T. R. Brummelkamp, E. M. Hijmans, R. Bernards, and N. B. La Thangue. 2004. E2F-7: a distinctive E2F family member with an unusual organization of DNA-binding domains.Oncogene 23:5138-5150. [DOI] [PubMed] [Google Scholar]
- 20.Logan, N., A. Graham, X. Zhao, R. Fisher, B. Maiti, G. Leone, and N. B. La Thangue. 2005. E2F-8: an E2F family member with a similar organization of DNA-binding domains to E2F-7.Oncogene 24:5000-5004. [DOI] [PubMed] [Google Scholar]
- 21.Maiti, B., J. Li, A. de Bruin, F. Gordon, C. Timmers, R. Opavsky, K. Patil, J. Tuttle, W. Cleghorn, and G. Leone. 2005. Cloning and characterization of mouse E2F8, a novel mammalian E2F family member capable of blocking cellular proliferation. J. Biol. Chem. 280:18211-18220. [DOI] [PubMed] [Google Scholar]
- 22.Marino, S. 2000. Induction of medulloblastomas in p53-null mutant mice by somatic inactivation of Rb in the external granular layer cells of the cerebellum. Genes Dev. 14:994-1004. [PMC free article] [PubMed] [Google Scholar]
- 23.Nevins, J. R. 1998. Toward an understanding of the functional complexity of the E2F and retinoblastoma families.Cell Growth Differ. 9:585-593. [PubMed] [Google Scholar]
- 24.Ogawa, H., K. Ishiguro, S. Gaubatz, D. M. Livingston, and Y. Nakatani. 2002. A complex with chromatin modifiers that occupies E2F- and Myc-responsive genes in G0 cells.Science 296:1132-1136. [DOI] [PubMed] [Google Scholar]
- 25.Pear, W. S., G. P. Nolan, M. L. Scott, and D. Baltimore. 1993. Production of high-titer helper-free retroviruses by transient transfection. Proc. Natl. Acad. Sci. USA 90:8392-8396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Robertson, K. D., and P. A. Jones. 1998. The human ARF cell cycle regulatory gene promoter is a CpG island which can be silenced by DNA methylation and down-regulated by wild-type p53.Mol. Cell. Biol. 18:6457-6473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rowland, B. D., S. G. Denissov, S. Douma, H. G. Stunnenberg, R. Bernards, and D. S. Peeper.2002. . E2F transcriptional repressor complexes are critical downstream targets of p19(ARF)/p53-induced proliferative arrest.Cancer Cell 2:55-65. [DOI] [PubMed] [Google Scholar]
- 28.Sage, J., G. J. Mulligan, L. D. Attardi, A. Miller, S. Chen, B. Williams, E. Theodorou, and T. Jacks. 2000. Targeted disruption of the three Rb-related genes leads to loss of G(1) control and immortalization. Genes Dev. 14:3037-3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sears, R., K. Ohtani, and J. R. Nevins. 1997. Identification of positively and negatively acting elements regulating expression of the E2F2 gene in response to cell growth signals.Mol. Cell. Biol. 17:5227-5235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Serrano, M., A. W. Lin, M. E. McCurrach, D. Beach, and S. W. Lowe. 1997. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell 88:593-602. [DOI] [PubMed] [Google Scholar]
- 31.Stevaux, O., and Dyson, N. J. 2002. A revised picture of the E2F transcriptional network and RB function. Curr. Opin. Cell Biol. 14:684-691. [DOI] [PubMed] [Google Scholar]
- 32.Takahashi, Y., J. B. Rayman, and B. D. Dynlacht.2000. . Analysis of promoter binding by the E2F and pRB families in vivo: distinct E2F proteins mediate activation and repression. Genes Dev. 14:804-816. [PMC free article] [PubMed] [Google Scholar]
- 33.Todaro, G. J., and H. Green. 1963. Quantitative studies of the growth of mouse embryo cells in culture and their development into established lines. J. Cell Biol. 17:299-313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Trimarchi, J. M., B. Fairchild, J. Wen, and J. A. Lees.2001. . The E2F6 transcription factor is a component of the mammalian Bmi1-containing polycomb complex. Proc. Natl. Acad. Sci. USA 98:1519-1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Trimarchi, J. M., and J. A. Lees. 2002. Sibling rivalry in the E2F family. Nat. Rev. Mol. Cell Biol. 3:11-20. [DOI] [PubMed] [Google Scholar]
- 36.Vogelstein, B., D. Lane, and A. J. Levine. 2000. Surfing the p53 network. Nature 408:307-310. [DOI] [PubMed] [Google Scholar]
- 37.Weber, J. D., L. J. Taylor, M. F. Roussel, C. J. Sherr, and D. Bar-Sagi. 1999. Nuclear Arf sequesters Mdm2 and activates p53. Nat. Cell Biol. 1:20-26. [DOI] [PubMed] [Google Scholar]
- 38.Wu, L., C. Timmers, B. Maiti, H. I. Saavedra, L. Sang, G. T. Chong, F. Nuckolls, P. Giangrande, F. A. Wright, S. J. Field, M. E. Greenberg, S. Orkin, J. R. Nevins, M. L. Robinson, and G. Leone.2001. . The E2F1-3 transcription factors are essential for cellular proliferation. Nature 414:457-462. [DOI] [PubMed] [Google Scholar]
- 39.Wu, X., and A. J. Levine. 1994. p53 and E2F-1 cooperate to mediate apoptosis. Proc. Natl. Acad. Sci. USA 91:3602-3606. [DOI] [PMC free article] [PubMed] [Google Scholar]