Abstract
Cyclooxygenase 2 (COX-2) is a key enzyme in the conversion of arachidonic acid to prostaglandins, and COX-2 overexpression plays an important role in carcinogenesis. Exposure to UVB strongly increased COX-2 protein expression in mouse 308 keratinocytes, and this induction was inhibited by apigenin, a nonmutagenic bioflavonoid that has been shown to prevent mouse skin carcinogenesis induced by both chemical carcinogens and UV exposure. Our previous study suggested that one pathway by which apigenin inhibits UV-induced and basal COX-2 expression is through modulation of USF transcriptional activity in the 5′ upstream region of the COX-2 gene. Here, we found that apigenin treatment also increased COX-2 mRNA stability, and the inhibitory effect of apigenin on UVB-induced luciferase reporter gene activity was dependent on the AU-rich element of the COX-2 3′-untranslated region. Furthermore, we identified two RNA-binding proteins, HuR and the T-cell-restricted intracellular antigen 1-related protein (TIAR), which were associated with endogenous COX-2 mRNA in 308 keratinocytes, and apigenin treatment increased their localization to cell cytoplasm. More importantly, reduction of HuR levels by small interfering RNA inhibited apigenin-mediated stabilization of COX-2 mRNA. Cells expressing reduced TIAR showed marked resistance to apigenin's ability to inhibit UVB-induced COX-2 expression. Taken together, these results indicate that in addition to transcriptional regulation, another mechanism by which apigenin prevents COX-2 expression is through mediating TIAR suppression of translation.
Cyclooxygenases are key enzymes in the conversion of arachidonic acid to prostaglandins. The inducible isoform of cyclooxygenase, cyclooxygenase 2 (COX-2), is an early response gene and is regulated by growth factors, cytokines, and tumor promoters (64). Another cyclooxygenase isoform, COX-1, is constitutively expressed and mediates many physiological functions (54). COX-2 is overexpressed in many transformed cells and various malignancies (5, 31). COX-2-deficient mice exhibit a decreased intestinal tumor incidence (47). Moreover, inhibitors of cyclooxygenases, e.g., nonsteroidal antiinflammatory drugs, reduce the formation, development, and metastasis of tumors in many animal models as well as risk of colorectal carcinoma in humans (27, 59). Together, these studies clearly demonstrate a significant role of COX-2 in carcinogenesis.
Every year over 1 million cases of nonmelanoma skin cancer are diagnosed in the United States alone, and chronic UV light exposure is widely believed to be the cause (20). Solar UV radiation reaching the earth's surface is a mixture of UVB and UVA, and it is generally believed that UVB irradiation plays a causative role in skin cancer (20). Multiple lines of evidence demonstrate that induction of COX-2 expression and the subsequent increase in prostaglandins contribute to skin cancer development. For example, transgenic mice overexpressing COX-2 in basal epidermal cells develop skin tumors (46), selective COX-2 inhibitors reduce UV-induced skin tumors in mice (17, 50), and mice that are deficient in COX-2 show a dramatic decrease in skin tumorigenesis (60).
COX-2 expression can be regulated through both transcriptional and posttranscriptional mechanisms. Although transcriptional activation of COX-2 is an early event in the initiation of tumorigenesis (31), abnormal posttranscriptional regulation also has been shown to play a central role in overexpression of the COX-2 protein during tumorigenesis (12). Posttranscriptional regulation of COX-2 expression is linked to the AU-rich element (ARE) within the 3′-untranslated region (3′-UTR) of COX-2 mRNA that controls both mRNA stability and protein translation (9). The ARE of the COX-2 gene is intact in healthy individuals (37) as well as in tumor cells (55), and numerous reports in the literature have demonstrated that dysregulation of posttranscriptional regulation by altered ARE-binding proteins is primarily responsible for enhanced expression of the COX-2 protein in many instances (13, 14, 45, 53). A number of trans-acting ARE-binding proteins have been identified thus far. Several of these proteins promote mRNA stability, such as HuR (16, 49) and hnRNP A1 (21), while others destabilize mRNA, such as AU-binding factor 1 (AUF1) (38, 66) and tristetraprolin (4, 39). Other proteins have been characterized that suppress protein translation, including T-cell-restricted intracellular antigen 1 (TIA-1) (13, 51) and TIA-1-related protein (TIAR) (19, 22, 65). In addition, ARE-binding protein CUGBP2 has been reported to increase COX-2 mRNA stability while simultaneously inhibiting its translation (45).
Although many of these ARE-binding proteins are located predominantly in the nucleus, they can shuttle between the nucleus and cytoplasm, and it is believed that their translocation to cytoplasm plays an important role in regulating the expression of target genes. For instance, the cytoplasmic localization of both HuR and TIAR increased under conditions of stress, and studies suggested that HuR associates with target mRNAs and transports these mRNAs from nucleus to cytoplasm, thus stabilizing them; TIAR recruits ARE-containing mRNAs into stress granules (SGs) to block their translation (28, 62).
Previous studies in our laboratory and others have shown that expression of COX-2 protein in keratinocytes is induced by UV light (1, 61a). We have further shown that this induction is blocked by treatment of UV-irradiated cells with the chemopreventive bioflavonoid apigenin, possibly through modulation of USF transcriptional activity (61a), and there are also reports that apigenin down-regulates COX-2 expression in macrophages (36, 52). Apigenin is a nonmutagenic, naturally occurring flavonoid found in a variety of fruits and leafy vegetables (10). A large body of evidence suggests that apigenin shows promise as a chemopreventive agent by stimulation of gap junctional and intercellular communication (6), inhibition of transformation and angiogenesis (18, 30), stabilization of wild-type p53 (42), and induction of cell cycle arrest (35). Topical application of apigenin in mice has been shown to reduce the number and size of tumors in the skin induced by both chemical carcinogens (63) and UV exposure (3).
In the present study, we investigated the role of apigenin in modulating posttranscriptional control of COX-2 expression in keratinocytes, since COX-2 mRNA contains typical ARE in the 3′-UTR. We demonstrated that apigenin enhanced COX-2 mRNA stability although it blocked the accumulation of UVB-induced steady-state mRNA at the same time. The mechanisms underlying COX-2 mRNA stabilization and protein translation were further examined, and we found that UVB treatment resulted in increased reporter gene activity through the 3′-UTR of COX-2 mRNA, which was prevented by apigenin. RNA-binding proteins HuR and TIAR bound COX-2 mRNA, and apigenin increased their localization in the cytoplasm. Reduction of HuR with small interfering RNA (siRNA) suppressed apigenin's ability to enhance COX-2 mRNA stability, whereas reduction of TIAR by siRNA reversed the ability of apigenin to inhibit UVB-induced COX-2 protein expression. Our results provide the first evidence that, in addition to transcriptional regulation, one of the mechanisms by which apigenin inhibits COX-2 expression is through suppression of translation by TIAR.
MATERIALS AND METHODS
Cell culture, UV irradiation, and apigenin treatment.
The 308 mouse keratinocyte cell line (56) was maintained in suspension minimum essential medium (United States Biological, Swampscott, MA) supplemented with 8% chelexed (Bio-Rad Laboratories, Hercules, CA) fetal bovine serum and 0.05 mmol/liter Ca2+. Prior to UV exposure, cells were grown to 90% confluence, the culture medium was removed and saved, cells were rinsed with phosphate-buffered saline (PBS) and irradiated, and medium was replaced. UVB was provided by FS40T12-UVB lamps (National Biological Corporation, Twinsburg, OH) with peak emission at 313 nm, and a Kodacel K6808 filter (Eastman Kodak, Rochester, NY) was used to filter out wavelengths of UVC (wavelengths below 295 nm). Apigenin (Sigma, St. Louis, MO) stock solutions were prepared in dimethyl sulfoxide (DMSO) and added to the existing culture medium to achieve the desired final concentration immediately after irradiation. The concentration of DMSO in cell cultures was less than 0.1%.
Western blot analysis.
After treatment, cells were harvested at the indicated times in lysis buffer (20 mmol/liter Tris [pH 7.5], 150 mmol/liter NaCl, 2 mmol/liter EDTA, 10% glycerol, 1% Triton X-100, 1 mmol/liter phenylmethylsulfonyl fluoride, and protease inhibitor cocktail [Sigma]). Protein concentrations were determined by using the BCA reagent (Pierce, Rockford, IL), and proteins were resolved on 12% sodium dodecyl sulfate (SDS)-polyacrylamide gels followed by eletrophoretic transfer onto nitrocellulose membranes. The membranes were blocked with 5% nonfat dry milk in Tris-buffered saline-0.1% Tween 20 for 2 h and incubated at 4°C overnight with primary antibodies against COX-2 (C-20), HuR (3A2), TIAR (C-18), TIA-1 (C-20), actin (I-19), β-tubulin (N-20), and hnRNP C1/C2 (N-16) (Santa Cruz Biotechnology, Santa Cruz, CA) and AUF1 (Upstate, Lake Placid, NY). Following incubation with appropriate horseradish peroxidase-conjugated secondary antibodies, signals were detected with an enhanced chemiluminescence system (Amersham Biosciences, Piscataway, NJ).
PGE2 measurement.
Cells were grown in 12-well plates to near confluence. After treatment, cell-free culture media were collected and prostaglandin E2 (PGE2) levels were determined by competitive enzyme-linked immunosorbent assay (ELISA) as directed by the manufacturer (Cayman Chemical Co., Ann Arbor, MI). The production of PGE2 was normalized to protein concentrations.
Transient transfection.
Firefly luciferase reporter gene constructs were generated as described previously by Morrison and coworkers (9). 308 cells were transiently transfected using Lipofectamine PLUS reagent (Invitrogen, Carlsbad, CA). Cells were plated in 24-well plates and grown to 80 to 90% confluence, and 0.2 μg reporter gene plasmid DNA and 1 ng control Renilla luciferase plasmid DNA (pRL-TK; Promega, Madison, WI) were prepared in 25 μl of serum-free medium and incubated with 4 μl of PLUS reagent at room temperature for 15 min, followed by 1 μl of Lipofectamine in an additional 25 μl serum-free medium. The mixture was incubated for another 15 min and layered onto the cells. After 3 h of incubation, normal medium containing 2× fetal bovine serum was added, and cells were incubated overnight for gene expression.
Luciferase assay.
Luciferase activity was determined using the Dual-Luciferase reporter assay system (Promega) according to the manufacturer's protocol. Briefly, cells were rinsed with PBS and removed by scraping into 100 μl of passive lysis buffer. The lysate was then transferred into a tube and subjected to one or two freeze-thaw cycles to accomplish complete lysis of cells. Assays were performed by using a Monolight 3010 luminometer (Analytical Luminescence Laboratory, Ann Arbor, MI). Firefly luciferase activity is expressed as relative light units and was normalized to Renilla luciferase activity.
Real-time PCR assays.
Total RNA was isolated at various times after treatment using TRIzol reagent (Invitrogen) and treated with a DNA-free kit (Ambion, Austin, TX) to eliminate genomic DNA contamination. RNA was reverse transcribed using the SuperScript III first-strand synthesis system with random hexamer primers (Invitrogen). After first-strand synthesis, for quantification of COX-2 cDNA, real-time PCR was performed using the TaqMan Gene Express assay (assay ID Mm00478374_m1; Applied Biosystems, Foster City, CA) specific for the COX-2 gene. Fluorescence was detected with an ABI Prism 7900HT real-time PCR system and normalized to rRNA as measured using a TaqMan eukaryotic 18S rRNA endogenous control (Applied Biosystems). Relative amounts of cDNA were calculated by the relative quantification (ΔΔCt) study method. For quantification of luciferase cDNA, real-time PCR was performed using luciferase gene-specific primers and the double-stranded DNA-binding dye SYBR Green I as described previously (9) and normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) content.
Cell fractionation.
Cytoplasmic and nuclear cell fractions were prepared as described elsewhere (8) with modification. Cells were washed in ice-cold PBS, scraped into 200 μl of hypotonic buffer A (10 mmol/liter HEPES [pH 7.5], 10 mmol/liter KCl, 1 mmol/liter EDTA, 1 mmol/liter dithiothreitol [DTT], 1 mmol/liter MgCl2, 5% glycerol, 0.1 mmol/liter Na3VO4) supplemented with protease inhibitors as described above, incubated on ice for 10 min, and lysed by adding of 5 μl of buffer A containing 10% Nonidet P-40 (buffer B). Nuclei were isolated by centrifugation (1,500 × g for 10 min at 4°C). The supernatant was further centrifuged (16,000 × g for 5 min at 4°C) and saved as the cytoplasmic fraction. Nuclei were washed twice by centrifugation (1,500 × g for 10 min at 4°C) in buffer C (10 mmol/liter HEPES [pH 7.5], 10 mmol/liter KCl, 1 mmol/liter DTT, 1 mmol/liter MgCl2) and resuspended in nuclear lysis buffer D (20 mmol/liter HEPES [pH 7.5], 0.4 mol/liter NaCl, 1 mmol/liter EDTA, 1 mmol/liter DTT, 1 mmol/liter MgCl2, 25% glycerol, and protease inhibitors) on ice for 15 min with frequent vortexing. Lysed nuclei were centrifuged at 15,000 × g for 5 min at 4°C, and supernatant was saved as the nuclear fraction. For polysome separation, cell cytoplasmic fraction lysates were layered onto an ice-cold buffer (20 mmol/liter HEPES, pH 7.5, 50 mmol/liter KO-acetate, 5 mmol/liter MgO-acetate, 1 mmol/liter DTT, 100 U/ml RNase Out, and protease inhibitors) containing 30% sucrose. After ultracentrifugation at 100,000 × g for 2 h at 4°C in a Beckman SW55 Ti rotor, the supernatant was saved as the cytosolic fraction. The pellet was resuspended in buffer A containing 0.3 mol/liter NaCl, incubated on ice for 1 h, and centrifuged at 10,000 × g for 15 min at 4°C, and the resulting supernatant was saved as the polysomal fraction (40).
Immunofluorescence.
Cells were cultured, treated, washed with PBS, and fixed for 30 min in PBS containing 4% paraformaldehyde at room temperature. Cell preparations were washed with PBS again and incubated in PBS containing 0.1% Triton X-100 for 10 min. After incubation in blocking buffer (PBS containing 0.1% Tween 20, 3% bovine serum albumin, and 3% donkey serum) for 1 h, the slides were incubated with either goat anti-TIAR or mouse anti-HuR antibody (Santa Cruz Biotechnology) in PBS containing 3% donkey serum for 48 h at 4°C, washed with PBS containing 0.1% Tween 20, and further incubated with Alexa Fluor 594 donkey anti-goat immunoglobulin G (IgG) or Alexa Fluor 594 donkey anti-mouse IgG (Invitrogen) for 2 h in the dark. After a final wash with PBS, the slides were mounted in Vectashield with diamidinophenylindole (Vector Laboratories) and visualized with a Zeiss Axiovert 200 (Carl Zeiss MicroImaging, Inc.) fluorescence microscope.
Immunoprecipitation of mRNP complex and reverse transcription-PCR (RT-PCR).
Cytoplasmic extracts (400 μg, harvested as described above except adding 100 U/ml RNase Out [Invitrogen] in buffer A) were mixed with 15 μg of HuR, TIAR, or normal mouse IgG antibody. After incubation on ice for 2 h, 30 μl protein A/G Plus-agarose beads (Santa Cruz Biotechnology) was added to each sample, followed by mixture for a further 1.5 h at 4°C. Samples were centrifuged at 2,000 × g for 2 min at 4°C, the pelleted beads were washed four times with cold buffer A supplemented with protease and RNase inhibitors, and RNA was isolated using the TRIzol reagent. DNase I treatment and reverse transcription were performed as described above. The cDNA was used as template for PCR (30 s at 94°C, 30 s at 56°C, and 1 min at 72°C, for 35 cycles) with COX-2-specific primers (forward, 5′-GCTGTACAAGCAGTGGCAAA-3′; reverse, 5′-CCCCAAAGATAGCATCTGGA-3′) and β-actin-specific primers (forward, 5′-TATGGAATCCTGTGGCATCC-3′; reverse, 5′-GTACTTGCGCTCAGGAGGAG-3′). These primers produce amplicons that span different introns, allowing the discrimination of cDNA- and genomic DNA-related amplification. PCR products were visualized by using an ethidium bromide-stained 1.2% agarose gel.
RNA interference experiment.
For TIAR knockdown, 308 cells were transfected by using Lipofectamine PLUS (Invitrogen) with pSM2-TIAR-shRNA plasmid (RMM1766-96883937; Open Biosystems, Huntsville, AL), which contains a short hairpin RNA (shRNA) sequence targeting mouse TIAR mRNA. After 48 h of incubation, cells were treated with puromycin (2 μg/ml; Sigma), single clones of puromycin-resistant cells were selected, and TIAR expression was examined by immunoblotting. Clones with the highest knockdown were used for subsequent experiments. An shRNA nonsilencing control vector (Open Biosystems) was used as a control. The control vector contains nonsilencing shRNA sequence that has no homology to any known mammalian genes. For HuR knockdown, siRNA duplex targeting mouse HuR mRNA and nontargeting control siRNA (Santa Cruz Biotechnology) were used. The transfection of siRNA into 308 cells was performed by using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions, and 48 h after transfection, cell density was adjusted so that cell confluence would be 90% after the transfected cells were kept in culture for another 48 h.
Analysis of nascent COX-2 protein.
For metabolic labeling, 308 cells were incubated for 1 h in Dulbecco's modified Eagle's medium without methionine and cysteine (Sigma) before treatment (UVB irradiation or UVB plus apigenin). At 8 h posttreatment, each sample was labeled with 500 μCi l-[35S]methionine and l-[35S]cysteine (GE Healthcare, Piscataway, NJ) for 20 min, cells were harvested in lysis buffer, and immunoprecipitations were carried out overnight at 4°C using COX-2 antibody or normal goat IgG. Following extensive washes in washing buffer (50 mmol/liter Tris-HCl [pH 7.5], 250 mmol/liter NaCl, 5 mmol/liter EDTA, and 0.5% NP-40), the immunoprecipitated materials were resolved by SDS-polyacrylamide gel electrophoresis and assessed by autoradiography.
RESULTS
Apigenin inhibits UVB-induced COX-2 protein expression.
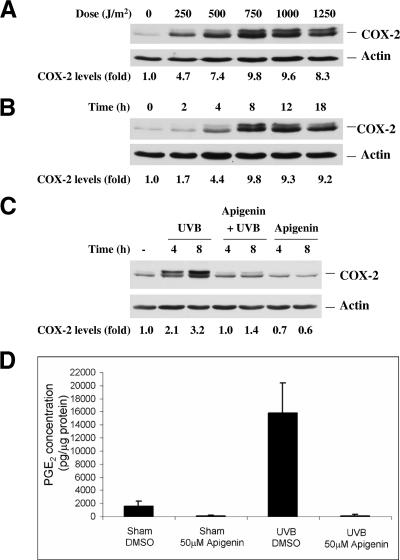
The mouse 308 keratinocyte cell line, which was derived from BALB/c mouse skin initiated by dimethylbenz[α]anthracene, contains a point-mutated Ha-ras gene and a wild-type p53 gene (56). COX-2 protein expression was examined in cells irradiated with UVB at various doses, and Western blot analysis demonstrated that UVB induced COX-2 protein expression dramatically, with a peak at doses from 750 to 1,000 J/m2 (Fig. 1A). To further determine the time of maximal COX-2 protein induction, cells were irradiated with 1,000 J/m2 UVB and harvested at different time points. The increase of COX-2 protein expression occurred rapidly after irradiation, with an approximately 10-fold maximal increase at 8 h after treatment. This level of COX-2 protein expression was sustained for more than 18 h postirradiation (Fig. 1B). Two COX-2 bands were detected in the Western blot analysis. The lower band (72 kDa) corresponds to COX-2 that is N-glycosylated at three sites, while the upper band (74 kDa) was identified as corresponding to the COX-2 form that is N-glycosylated at four sites, as reported previously (48) and confirmed by disappearance of the upper band after treatment with the glycosylation inhibitor tunicamycin (data not shown).
FIG. 1.
Apigenin inhibits UVB-induced COX-2 expression and subsequent PGE2 production. 308 mouse keratinocytes were grown to 90% confluence and irradiated with UVB; control cells were sham irradiated. (A) Western blot analysis showing dose-dependent induction of COX-2 protein at 8 h postirradiation. (B) Time-dependent induction of COX-2 protein expression by UVB. Cells were irradiated with 1,000 J/m2 UVB, harvested at the indicated time points, and analyzed by Western blotting. (C) Apigenin inhibits basal COX-2 protein expression as well as UVB-induced COX-2 expression. Immediately after UVB (1,000 J/m2) or sham irradiation, apigenin was added to culture medium to a final concentration of 50 μmol/liter, and cells were harvested at the indicated times. “-” indicates untreated control. (D) UVB-induced PGE2 synthesis is inhibited by apigenin. 308 cells were treated with UVB, apigenin, or UVB plus apigenin as indicated above, and PGE2 levels in the culture medium were measured by competitive ELISA.
To confirm our previous results that apigenin could block UVB-induced COX-2 protein expression, apigenin was added to the culture medium immediately after UVB or sham irradiation and incubated for various times. As shown in Fig. 1C, apigenin inhibited basal COX-2 protein expression as well as UVB-induced COX-2 expression.
In view of the fact that apigenin treatment inhibited UVB-induced COX-2 protein levels, we next determined the effect of apigenin treatment on PGE2, a downstream product of COX-2. PGE2 levels were measured by ELISA in cell culture medium after cells were treated with UVB, apigenin, or UVB plus apigenin. Consistent with the observed increase of COX-2 expression induced by UVB and inhibition of COX-2 expression by apigenin, we found that DMSO (as vehicle) or apigenin-treated cells produced little PGE2 (Fig. 1D). In contrast, cells exposed to UVB were shown to have a 10-fold increase in PGE2 that was reduced to even less than the basal level by adding apigenin (Fig. 1D). Overall, these results demonstrated that apigenin not only suppresses UVB-induced COX-2 protein expression but also inhibits the level of its downstream product, PGE2.
UVB irradiation and apigenin treatment stabilize COX-2 mRNA.
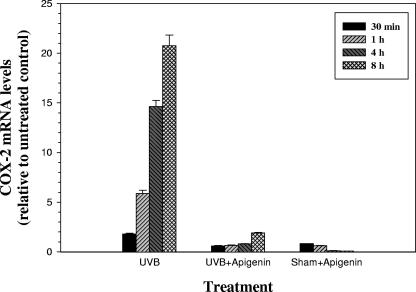
To further investigate the mechanism responsible for the inhibitory effect of apigenin on COX-2 expression, we examined the steady-state levels of COX-2 mRNA by real-time RT-PCR assay. Total RNAs were purified from cells that had been treated with UVB, apigenin, or UVB plus apigenin and harvested at different times posttreatment. As shown in Fig. 2, UVB induced a dramatic increase in COX-2 mRNA levels and apigenin significantly decreased the levels of COX-2 mRNA in both UVB- and sham-irradiated cells. The concentration of mRNA is determined by rates of both synthesis and degradation, and previous studies in our laboratory have demonstrated that UVB exposure induces COX-2 expression by transcriptional up-regulation of the COX-2 promoter and apigenin blocks this induction through modulation of USF transcriptional activity (61a). In the present report, we further investigated whether message stabilization/destabilization and modulation of protein translation could play a role in apigenin-mediated inhibition of COX-2 expression, because COX-2 mRNA contains typical ARE in the 3′-UTR and there is growing evidence that the ARE within the 3′-UTR of COX-2 mRNA can affect both mRNA stability and protein translation (9).
FIG. 2.
Real-time PCR analysis of steady-state COX-2 mRNA levels. 308 cells were treated with 1,000 J/m2 UVB, 50 μmol/liter apigenin, or UVB plus apigenin, and total RNA was harvested at the indicated times. TaqMan real-time quantitative PCR was carried out as described in Materials and Methods. The graph represents the results of three independent experiments.
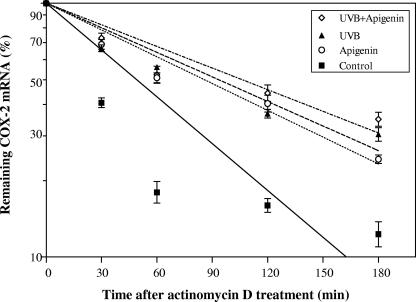
To this end, cells were treated with UVB, apigenin, or UVB plus apigenin, and subsequent transcription was stopped by adding actinomycin D immediately after the treatment. RNA was isolated at different time points after adding actinomycin D and subjected to TaqMan real-time PCR analysis. As shown in Fig. 3, treatment of cells with UVB caused a significant increase in the half-life of COX-2 mRNA (from less than 50 min to approximately 100 min). Surprisingly, apigenin treatment also increased COX-2 mRNA stability significantly, suggesting that the observed inhibition of COX-2 protein expression by apigenin treatment is not only the result of transcriptional regulation but also may reflect effects of apigenin on posttranscriptional events, including suppression of translation of COX-2 mRNA.
FIG. 3.
UVB and apigenin enhance the stability of COX-2 mRNA. Cells were treated with 750 J/m2 UVB, 50 μmol/liter apigenin, or UVB plus apigenin. Actinomycin D (10 μg/ml) was added immediately, and cells were incubated for 0, 30, 60, 120, or 180 min. Total RNA was isolated, reverse transcribed, and analyzed for COX-2 mRNA using TaqMan real-time PCR. The amount of COX-2 mRNA is expressed as a percentage of the level measured at the 0-min time point. Data represent the means ± standard error for three to six independent experiments.
The ARE in the COX-2 3′-UTR is important for the stability of COX-2 mRNA and translation efficiency.
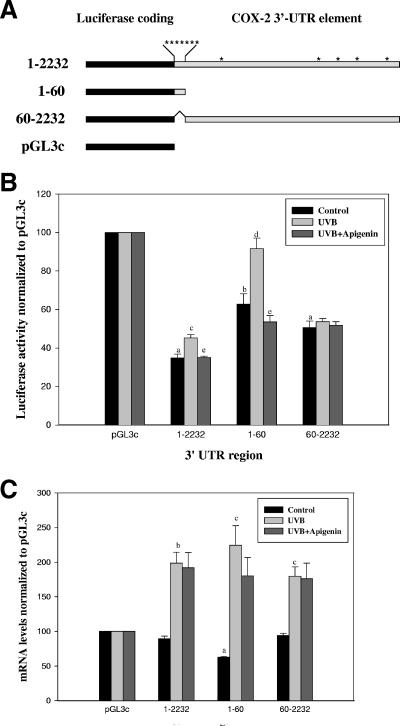
In view of the fact that the ARE is important for modulating COX-2 mRNA stability and protein translation (9), we next determined whether the presence of the ARE mediated the effects of UVB exposure or apigenin treatment. Transient transfections were performed with reporter gene constructs containing luciferase fused to the full-length COX-2 3′-UTR (nucleotides [nt] 1 to 2232), the ARE region (nt 1 to 60), or the ARE deleted from the full-length 3′-UTR (nt 60 to 2232) (Fig. 4A). As illustrated in Fig. 4B, the full-length 3′-UTR, nt 1 to 2232, reduced basal luciferase activity by ∼60%. The ARE region nt 1 to 60 or the ARE deleted from the full-length 3′-UTR (nt 60 to 2232) also reduced reporter activity ∼40% and ∼50%, respectively. These results suggested that the 60-bp ARE region was responsible for altering reporter gene expression and that additional cis-elements (not exclusively those in the nt 1 to 60 ARE region) are present that alter message stability and translation efficiency as reported previously (9). However, when we examined the effects of UVB irradiation on the luciferase activity of each of these constructs in 308 cells, we found that UVB increased luciferase activity by ∼30% and ∼40% in the case of the full-length 3′-UTR (nt 1 to 2232) and the ARE region (nt 1 to 60), respectively, whereas the ARE deleted from the full-length 3′-UTR (nt 60 to 2232) appeared to be unresponsive to UVB (Fig. 4B). When apigenin treatment was combined with UVB exposure, it suppressed the effect of UVB on luciferase reporter gene expression (Fig. 4B). Taken together, these results suggested that the nt 1 to 60 ARE region of COX-2 mRNA is important for both UVB response and apigenin inhibition.
FIG. 4.
The AU-rich element of the COX-2 3′-UTR is important for the stability of COX-2 mRNA and translation efficiency. (A) Structure of luciferase reporter gene constructs. Various regions of the 3′-UTR of COX-2 were fused to reporter gene luciferase to generate the constructs containing the full-length 3′-UTR (nt 1 to 2232), the AU-rich element (nt 1 to 60), the AU-rich element deleted from the full-length 3′-UTR (nt 60 to 2232), or luciferase control (pGL3c) without the 3′-UTR. The locations of the AUUUA sequence are indicated by asterisks. (B) Various regions from the 3′-UTR of COX-2 cause a decrease in luciferase activity, and the AU-rich element confers UVB induction and apigenin inhibition. 308 cells were cotransfected with the different reporter genes and pRL-TK plasmid, and 24 h after transfection, cells were treated with 1,000 J/m2 UVB or UVB plus 50 μmol/liter apigenin for another 8 h. Luciferase activity was expressed relative to the pGL3c control. All values were normalized to Renilla luciferase activity, and results are the means ± standard errors for at least three independent experiments, each performed in duplicate. a, significantly different from pGL3c, P < 0.001; b, significantly different from pGL3c, P < 0.003; c, significantly different from unirradiated self, P < 0.02; d, significantly different from unirradiated self, P < 0.03; e, significantly different from irradiated self, P < 0.02. (C) Effects of various regions from the 3′-UTR of COX-2 on luciferase reporter gene message levels. Cells were transfected and treated as described for panel B. Total RNA was extracted and assayed for luciferase and GAPDH mRNAs by using real-time PCR as described in Materials and Methods. Luciferase message levels were normalized to GAPDH mRNA and expressed relative to the pGL3c control. a, significantly different from pGL3c, P < 0.001; b, significantly different from unirradiated self, P < 0.003; c, significantly different from unirradiated self, P < 0.005.
Changes in luciferase activity could be due to either alterations in message stability or rates of protein translation. To distinguish between these two possibilities, we further measured the steady-state levels of luciferase mRNA using real-time RT-PCR assay. As shown in Fig. 4C, UVB enhanced luciferase reporter gene mRNA stability significantly, but apigenin treatment had no effect on the UVB-induced increase in luciferase mRNA stability. These findings are consistent with COX-2 mRNA decay analysis (Fig. 3). Comparison of the results from luciferase activity experiments (Fig. 4B) and measurements of luciferase mRNA stability (Fig. 4C) further indicated that apigenin inhibition of COX-2 expression could be through suppression of protein translation.
Apigenin increases levels of HuR and TIAR in the cytoplasm.
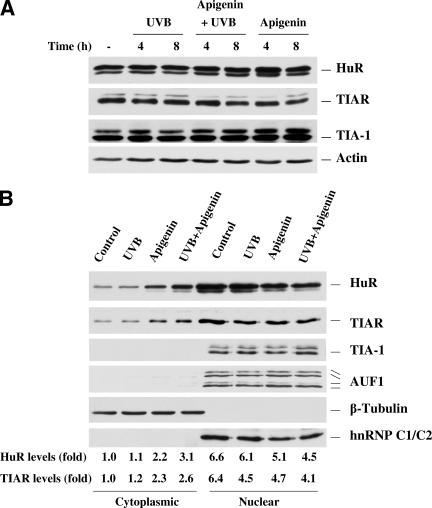
Posttranscription regulation mediated by AREs is primarily governed by RNA-binding proteins (RBPs) that bind to mRNAs containing ARE and ensure their proper processing and export, as well as subcytoplasmic transit, stability, and translation rate (11, 15). Among these RBPs, three ARE-RBPs (HuR, TIA-1, and TIAR) have been confirmed to influence translation, and numerous reports have demonstrated that cytoplasmic localization is associated with their functional activity (22, 40, 51). Therefore, it was our primary interest to examine the subcellular localization of HuR, TIA-1, and TIAR under conditions in which COX-2 expression is induced by UVB and inhibited by apigenin. Western blot analysis using whole-cell lysates demonstrated that there was no change in total HuR, TIA-1, and TIAR expression during UVB and apigenin treatment of 308 cells (Fig. 5A). When subcellular fractions were further analyzed, we observed a higher level of cytoplasmic HuR and TIAR in apigenin-treated and UVB plus apigenin-treated cells, but not in control or UVB-irradiated cells (Fig. 5B). Verification that nuclear proteins did not leak into the cytoplasmic fractions during the fractionation process was obtained by immunoblotting the same membranes to detect the nuclear marker hnRNP C1/C2 protein. hnRNP C1/C2 protein was found only in the nuclear fraction, while the cytoplasmic marker β-tubulin was detected only in the cytoplasmic fraction (Fig. 5B). TIA-1 and another RNA-binding protein AUF1 were also examined, and they were exclusively located in the nucleus irrespective of culture treatment (Fig. 5B).
FIG. 5.
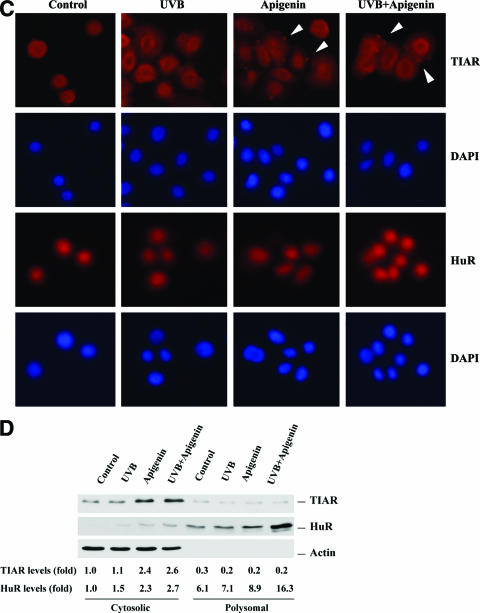
Effect of UVB and apigenin treatment on the subcellular location of HuR and TIAR. (A) Whole-cell lysates were prepared and subjected to Western blot analysis for HuR, TIAR, and TIA-1. Immediately after UVB (1,000 J/m2) or sham irradiation, apigenin was added to culture medium to a final concentration 50 μmol/liter, and cells were harvested at the indicated times. The “-” indicates untreated control. (B) Four hours after UVB or apigenin treatment, cytoplasmic and nuclear lysates were prepared and Western blot analysis was used to monitor the expression of HuR, TIAR, TIA-1, and AUF1. Expression levels of the cytoplasmic marker β-tubulin and the nuclear marker hnRNP C were also monitored. (C) The subcellular localizations of TIAR and HuR were also monitored by immunofluorescence, and stress granules (arrowheads) were observed in apigenin- and apigenin plus UVB-treated TIAR-stained cells. The slides were also stained with diamidinophenylindole for visualization of nuclei. (D)Western blot showing subcellular localization of TIAR and HuR in cytosol and polysomal fractions.
TIAR acts as a translation silencer by binding to AREs of the target mRNAs and diverting them from polysomes to SGs, where TIAR accumulates along with untranslated mRNAs, resulting in translation inhibition of SG-associated mRNA (28). To assess whether UVB or apigenin treatment induces the formation of SGs, immunofluorescence studies were carried out with anti-HuR or anti-TIAR antibodies. As illustrated in Fig. 5C, under apigenin or apigenin plus UVB treatment, TIAR was found to accumulate at several cytoplasmic foci which represented SGs, and no SGs were found when anti-HuR antibody was used. To further monitor the cytoplasmic distribution of TIAR and HuR, cytosolic and polysomal fractions were separated by ultra-centrifugation of the cytoplasm, and Western blotting was performed to determine the location of TIAR and HuR. As shown in Fig. 5D, TIAR was almost exclusively found associated with the cytosolic fraction, and apigenin or apigenin plus UVB treatment increased the levels of TIAR protein. In contrast, HuR was found mainly located in the polysomal fraction, and apigenin or apigenin plus UVB treatment increased the HuR levels in both the polysomal and cytosolic fractions.
HuR and TIAR bind endogenous COX-2 mRNA in 308 cells.
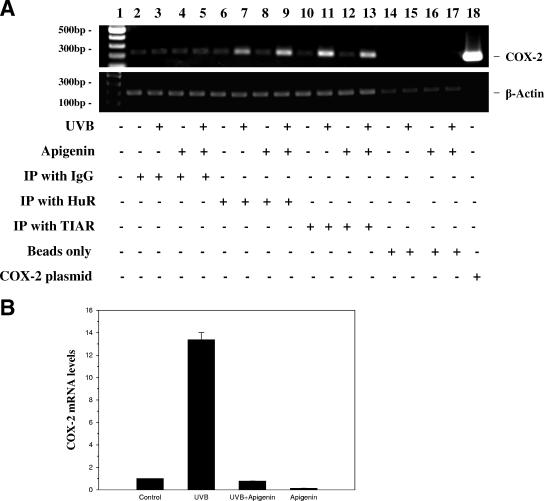
The fact that both HuR and TIAR increased their localization to the cytoplasm under apigenin treatment suggested the possibility that they may be responsible for suppression of COX-2 expression by apigenin. To investigate the endogenous association between these RNA-binding proteins and COX-2 mRNA in 308 cells, we used an immunoprecipitation assay followed by measurement of COX-2 mRNA by RT-PCR. As shown in Fig. 6A, HuR and TIAR coimmunoprecipitated with COX-2 mRNA in a UVB-dependent manner when using anti-HuR and anti-TIAR antibodies (lanes 7 and 11, respectively, for UVB-treated cells, and lanes 9 and 13, respectively, for UVB plus apigenin treatment). Control samples treated with normal IgG showed only very weak amplification, and no amplification was observed in the absence of antibody (beads only). Lane 18 is a positive control to confirm the amplification from COX-2 cDNA plasmid. Based on the size of all amplicons, there is no genomic contamination, because the PCR product would be much larger if the intron sequence were amplified as well. The levels of the housekeeping β-actin mRNA were also examined as a sample input control. Since treatment of cells with UVB, UVB plus apigenin, or apigenin alone resulted in a dramatic change of steady-state COX-2 mRNA levels (Fig. 2), the levels of COX-2 mRNA were monitored in parallel samples by real-time PCR (Fig. 6B); UVB treatment increased the steady-state level of COX-2 mRNA dramatically, and apigenin treatment down-regulated the steady-state level of COX-2 mRNA to the same level as that in untreated cells. Taken together, we propose that the increase of HuR- and TIAR-bound COX-2 mRNA after UVB treatment (lanes 7 and 11, respectively) is due to the increased expression of COX-2 mRNA induced by UVB, since we did not observe an increase in cytoplasmic HuR and TIAR following UVB treatment.(Fig. 5B). In contrast we propose that the increase of HuR- and TIAR-bound COX-2 mRNA after UVB plus apigenin treatment (Fig. 6A, lanes 9 and 13, respectively) is due to markedly increased levels of cytoplasmic HuR and TIAR in UVB plus apigenin-treated cells (Fig. 5B), since the level of COX-2 mRNA was nearly the same in UVB plus apigenin-treated cells as in untreated cells (Fig. 6B). Overall, these date provide definitive evidence that HuR and TIAR bind COX-2 mRNA in 308 keratinocyte cells.
FIG. 6.
HuR and TIAR associate with COX-2 mRNA in 308 cells. (A) Four hours after treatment, cytoplasmic extracts were prepared from control cells (lanes 2, 6, 10, and 14), UVB (1,000 J/m2)-treated cells (lanes 3, 7, 11, and 15), apigenin (50 μmol/liter)-treated cells (lanes 4, 8, 12, and 16), and UVB plus apigenin-treated cells (lanes 5, 9, 13, and 17). Protein-RNA complexes were immunoprecipitated using anti-HuR antibody, anti-TIAR antibody, normal IgG, or beads only, followed by RT-PCR analysis to detect endogenous COX-2 mRNA and β-actin mRNA. Lane 1, DNA marker; lane 18, COX-2 cDNA plasmid as a positive control. (B) Cells were treated as described above and subjected to real-time PCR for analysis of COX-2 mRNA levels at 4 h posttreatment.
Knockdown of HuR expression inhibits apigenin-mediated COX-2 mRNA stabilization.
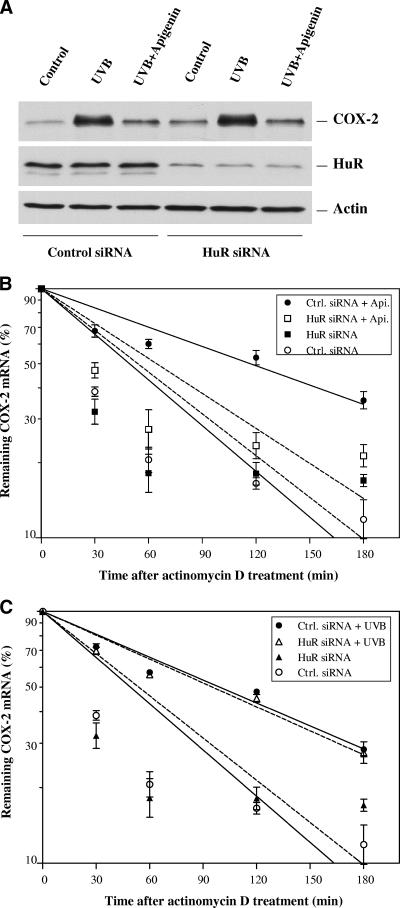
Having demonstrated that apigenin treatment increased the HuR protein level in cytoplasm (Fig. 5B), we next determined whether HuR was required for the stabilization of COX-2 mRNA induced by apigenin. We employed specific reduction of HuR expression in 308 keratinocytes by using siRNA technology. As shown in Fig. 7A, depletion of >75% of HuR protein was achieved by transfection of HuR siRNA duplex into 308 cells, with no effect on actin level. Transfection using a control nontargeting siRNA duplex did not change either HuR or actin expression. Interestingly, reduced HuR had no effect on COX-2 protein expression, whether it was induced by UVB or inhibited by apigenin (Fig. 7A). However, when control siRNA- and HuR siRNA-transfected cells were subjected to apigenin or UVB treatment and COX-2 mRNA decay was analyzed by quantitative real-time PCR following addition of actinomycin D, we found that depletion of HuR markedly reduced the apigenin-mediated stabilization of COX-2 mRNA (Fig. 7B) but had no effect on UVB-induced COX-2 mRNA stabilization (Fig. 7C). These findings demonstrated that HuR is essential for apigenin-mediated COX-2 mRNA stabilization.
FIG. 7.
Effect of reduced HuR on apigenin- and UVB-mediated COX-2 mRNA stabilization. (A) 308 cells were transfected with HuR siRNA duplex or control nontargeting siRNA duplex, and 96 h after transfection, cells were exposed to UVB (1,000 J/m2) and apigenin (50 μmol/liter) treatment. COX-2, HuR, and actin expression levels were analyzed by Western blotting after another 8 h of culture. (B and C) Cells transfected as described above were subjected to apigenin (B) or UVB (C) treatment, and COX-2 mRNA decay was analyzed by quantitative real-time PCR as described in Materials and Methods.
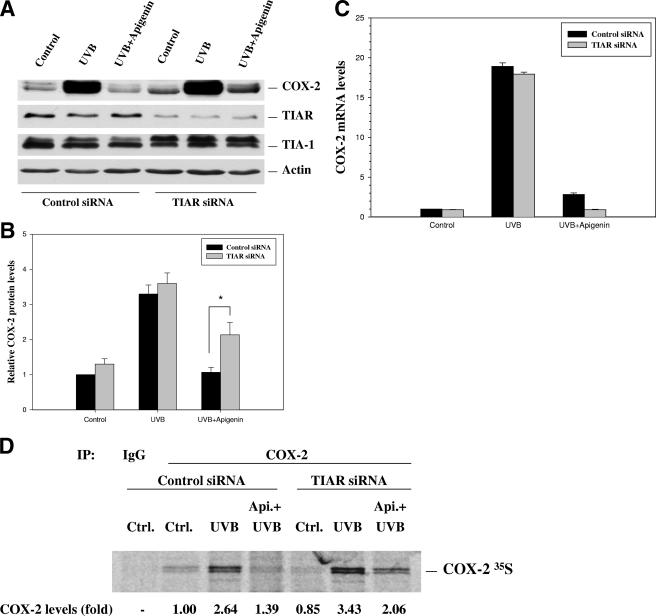
TIAR knockdown reverses the ability of apigenin to inhibit COX-2 protein expression.
Based on the results indicated above and the recent report that in macrophages HuR synergizes with TIA-1 to increase tumor necrosis factor (TNF) mRNA stability but suppresses protein translation (23), we hypothesized that while cytoplasm-located HuR promoted COX-2 mRNA stabilization, the TIAR protein was functioning to suppress protein translation in 308 cells under apipenin treatment. To test this hypothesis, we knocked down TIAR expression by transfecting cells with a plasmid expressing siRNA that specifically targeted TIAR. As shown in Fig. 8A, TIAR expression was reduced substantially by approximately 70%, to ∼30% of the amount of the nonsilencing control. Our results further demonstrated that reduction of TIAR protein levels resulted in a blockade of apigenin's inhibitory effect on UVB-induced COX-2 protein expression, with approximately twofold-greater COX-2 protein expression in apigenin-treated cells under TIAR knockdown than in cells transfected with control siRNA (Fig. 8A). Quantitative analysis showed the change was significant, with a P value of <0.05 (Fig. 8B). Given the extensive sequence homology between TIAR and TIA-1, we also examined the levels of TIA-1 in the TIAR knockdown cells and demonstrated that TIA-1 levels remained unchanged (Fig. 8A), verifying the specificity of TIAR knockdown. As determined by real-time RT-PCR analysis, TIAR knockdown did not significantly alter COX-2 mRNA levels in untreated or UVB-treated cells, but cells treated with UVB plus apigenin showed significant reduction in COX-2 mRNA level when TIAR was knocked down (Fig. 8C). We next performed biosynthetic labeling studies with [35S]methionine-cysteine to measure nascent translation of COX-2 protein. In keeping with TIAR's role as a translation silencer, our results showed that TIAR knockdown partially rescued the apigenin-mediated translational suppression of COX-2 protein in UVB-treated cells (Fig. 8D). Together, these results confirmed our hypothesis that one of the mechanisms by which apigenin treatment results in inhibition of COX-2 expression is through mediating TIAR suppression of protein translation.
FIG. 8.
TIAR knockdown blocks apigenin's inhibitory effect on COX-2 protein expression. (A) 308 cells were transfected with plasmids containing either control shRNA or shRNA targeted to TIAR, and stable cell lines were selected. Eight hours after UVB (1,000 J/m2) exposure and apigenin (50 μmol/liter) plus UVB treatment, COX-2, TIAR, TIA-1, and actin expression levels were analyzed by Western blot analysis using whole-cell lysates. (B) Quantitative analysis of the TIAR knockdown effect on apigenin's inhibition of COX-2 expression. COX-2 bands were scanned by densitometry and normalized to their corresponding actin bands. The graph represents the results of three independent experiments. *, P < 0.05. (C) Levels of COX-2 mRNA were determined by real-time PCR analysis at 8 h posttreatment. (D) Cells were treated with UVB or apigenin plus UVB for 8 h, followed by a brief incubation (20 min) with l-[35S]methionine and l-[35S]cysteine. Newly translated COX-2 protein was assessed by immunoprecipitation using anti-COX-2 antibody, SDS-polyacrylamide gel electrophoresis, and autoradiography. Normal IgG was used to determine the specificity of the immunoprecipitation reaction.
DISCUSSION
In this report, we have demonstrated that UVB increases COX-2 protein expression in a dose- and time-dependent manner, and apigenin inhibits UVB-induced COX-2 expression and subsequent PGE2 production. Our conclusion that HuR increases COX-2 mRNA stability but TIAR suppresses protein translation in 308 cells under apigenin treatment is based on the following observations: first, although apigenin could inhibit COX-2 expression through modulating transcription as we reported recently (61a), on the other hand, after stopping transcription by actinomycin D treatment, apigenin increased the half-life of COX-2 mRNA dramatically (Fig. 3). Second, the ARE within the 3′-UTR of COX-2 mRNA increased luciferase reporter gene activity after UVB irradiation, and apigenin inhibited this activity (Fig. 4B) but had no significant effect on message stability (Fig. 4C). Interestingly, previous studies by Cok and coworkers also demonstrated that the same ARE region of the 3′-UTR of COX-2 was a major translational control element that reduced reporter gene activity under normal culture conditions due to decreased translational efficiency (9). Third, it is widely reported that ARE-binding proteins need transport from the nucleus to the cytoplasm to regulate their targeted mRNAs (12), and in agreement with this observation, we detected that cytoplasmic HuR and TIAR increased only after apigenin and apigenin plus UVB treatment (Fig. 5B). Moreover, we found that TIAR accumulated in SGs that were induced by apigenin or apigenin plus UVB treatment (Fig. 5C), and TIAR protein was almost exclusively located in the cytosol when cytosolic and polysomal fractions were further separated from the cytoplasm. Apigenin treatment increased TIAR's levels in cytosol (Fig. 5D), which also agrees with the findings that TIAR inhibits protein translation by diverting the target mRNAs from the polysome to SGs and accumulates in SGs (28). Fourth, experiments of immunoprecipitation of HuR and TIAR followed by RT-PCR to detect COX-2 mRNA demonstrated that endogenous HuR and TIAR were associated with COX-2 mRNA in cultured 308 cells (Fig. 6A). These results are consistent with the previous report by Cok and coworkers demonstrating in an antibody supershift-electrophoretic mobility shift assay that HuR and TIAR bind the ARE region of the 3′-UTR of murine COX-2 in vitro (7). Although HuR and TIAR are reported to be primarily located in the nucleus, in 308 cells we found a low level of HuR and TIAR also located in the cytoplasm under normal culture conditions (Fig. 5B). However, after apigenin plus UVB treatment, we observed increased COX-2 mRNA in pull-down experiments with anti-HuR and anti-TIAR antibodies, although steady-state COX-2 mRNA was almost the same level as in the control, indicating that the increased cytoplasmic HuR and TIAR are responsible for the increased COX-2 mRNA pull-down. Fifth, depletion of HuR expression by using siRNA showed substantial reduction on apigenin-mediated COX-2 mRNA stabilization but had no effect on UVB-mediated message stability (Fig. 7), demonstrating HuR is a required component of apigenin-induced COX-2 mRNA stabilization. Finally, knockdown of TIAR expression reversed apigenin's ability to inhibit COX-2 protein expression and increased nascent COX-2 translation (Fig. 8), which is clear evidence suggesting that TIAR works downstream of HuR to prevent COX-2 expression. Similarly, a recent study demonstrated that in the absence of TIA-1, the translational silencing imposed by HuR overexpression on TNF mRNA is also abolished (23). Interestingly, Kawai et al. reported that global mRNA stabilization is preferentially linked to translational repression during the endoplasmic reticulum stress response (24). Together, our current study and these reports strongly support the long-standing hypothesis that mRNA stability and translation might be functionally linked.
TIAR has been reported to participate in the regulation of alternative pre-mRNA splicing (34) and promote apoptosis (58). However, TIAR has been best characterized as a silencer of translation (19, 32, 41, 65). It was cloned based on similarity to TIA-1 (another suppressor of translation through binding target ARE-bearing mRNAs), and the two proteins share 80% amino acid identity (26). Like TIA-1, TIAR is normally localized in the nucleus and translocated to the cytoplasm under stress conditions, and it has also been shown to bind the ARE of COX-2 mRNA in vitro (7). Unlike TIA-1−/− mice, however, the TIAR−/− mouse is highly embryonic lethal (2), and so the exact role of TIAR in COX-2 expression has not been firmly established. In this study, we demonstrated that TIAR associates with endogenous COX-2 mRNA by using an immunoprecipitation-RT-PCR assay, and we further showed that knockdown of TIAR reversed apigenin's ability to inhibit UVB-induced COX-2 protein expression and knockdown of TIAR enhanced nascent COX-2 translation, thus providing the first evidence that TIAR functionally inhibits COX-2 protein expression by modulating translation.
It should be pointed out that RNA-binding protein CUGBP2 itself can increase COX-2 mRNA stability while simultaneously inhibiting its translation, and this function is dependent on CUGBP32's binding to the COX-2 ARE and translocation from the nucleus to cytoplasm (45). However, unlike HuR or TIAR, we did not detect any change in expression of CUGB2 in whole-cell extracts or cytoplasm under UVB or apigenin treatment (data not shown), indicating this protein may not be responsible for the inhibition of COX-2 expression by apigenin in 308 cells. Meanwhile, recent reports have shown that RBPs can influence their targets without changes in abundance themselves (25, 41); thus additional studies may be needed to address the role of CUGBP32 in regulation of COX-2 expression by UVB/apigenin.
Previous reports demonstrated that UVC increases p21 mRNA stability and promotes p53 expression through increasing localization of HuR in cytoplasm (40, 62). In the present study, UVB also enhanced COX-2 mRNA stability. However, we did not observe an increase in the HuR level in the cytoplasm after UVB irradiation alone (Fig. 5), and reduced HuR had no effect on UVB-induced COX-2 protein expression and message stability (Fig. 7). These findings indicate that there may be another RNA-binding protein(s) that is required for UVB-induced COX-2 expression. Alternatively, it is possible that the dramatic UVB-induced COX-2 protein expression seen in Fig. 1 is mainly the result of increased transcription, rather than substantial modulation at the 3′ end, as reflected by the relatively modest effects of the 3′-UTR on luciferase reporter activity in cells treated with UVB alone.
Although studies have predominantly demonstrated that cytoplasmic HuR can stabilize mRNA by binding to AREs within the 3′-UTR of specific mRNAs and thus enhance protein expression (53, 57), growing evidence indicates that HuR's functions are not definite. For example, HuR binds the 5′-UTR of insulin-like growth factor I receptor or p27 to inhibit translation (29, 43), binds the 3′-UTR of p53 to enhance translation (40), synergizes with TIA-1 to stabilize TNF mRNA but inhibits protein translation (23), and coordinates with TIA-1 to regulate cytochrome c translation (25). Here, we show that HuR stabilizes COX-2 mRNA but TIAR works downstream to suppress protein translation, indicating that the final effect of HuR's function not only is dependent on the cis-elements of target mRNA that it binds but also is regulated by other molecules that work together with HuR.
The fact that both HuR and TIAR bound COX-2 mRNA and increased their cytoplasmic localization after apigenin treatment suggested that they may be functionally linked through association each other or association with common target mRNA. To test these possibilities, we employed protein coimmunoprecipitation techniques, but we found that HuR and TIAR did not bind each other through protein-protein interactions (data not shown), and so it is possible that they are just functionally linked through association with COX-2 mRNA in the current system. Although antibody supershift-electrophoretic mobility shift assay results show that HuR and TIAR associate with the same ARE of COX-2 (7), at present, we do not know whether they bind the COX-2 mRNA simultaneously or sequentially. Polysomal/cytosolic distribution (Fig. 5D) showed that TIAR was almost exclusively found in the cytosol, which is consistent with its function as a translation repressor. In contrast, HuR was found mainly located in the polysomal fraction as reported previously (33). Apigenin treatment or apigenin plus UVB treatment increased the HuR levels at both the polysome and cytosol, which also reflects the multifunctions of HuR's influence on target mRNA stability or translation.
As mentioned above, COX-2 overexpression plays an important role in carcinogenesis. While chemotherapy is a main approach to treat cancer, many chemotherapeutic agents, such as taxanes, induce COX-2 expression (44), raising the possibility that inhibiting COX-2 will greatly augment the anticancer effect of chemotherapy if the agents are coadministered with a selective COX-2 inhibitor. However, the gastrointestinal toxicity of nonsteroidal antiinflammatory drugs and the recent findings that selective COX-2 inhibitors can produce potentially adverse side effects on the cardiovascular system (61) may reduce the enthusiasm for using them as cancer chemopreventive agents. Here, we demonstrate that apigenin, a diet-derived agent, can inhibit inducible COX-2 expression by either modulating transcription or suppressing protein translation. Meanwhile, apigenin at up to 100 μmol/liter does not affect COX-2 enzyme activity and has no effect on constitutive COX-1 expression (J. Pelling, A. Abu-Yousif, et al., unpublished data), thus providing an attractive alternative for chemoprevention and chemotherapy.
Acknowledgments
This work was supported by NIH grant CA104768 (to J.C.P.).
Footnotes
Published ahead of print on 30 October 2006.
REFERENCES
- 1.Bachelor, M. A., A. L. Silvers, and G. T. Bowden. 2002. The role of p38 in UVA-induced cyclooxygenase-2 expression in the human keratinocyte cell line, HaCaT. Oncogene 21:7092-7099. [DOI] [PubMed] [Google Scholar]
- 2.Beck, A. R., I. J. Miller, P. Anderson, and M. Streuli. 1998. RNA-binding protein TIAR is essential for primordial germ cell development. Proc. Natl. Acad. Sci. USA 95:2331-2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Birt, D. F., D. Mitchell, B. Gold, P. Pour, and H. C. Pinch. 1997. Inhibition of ultraviolet light induced skin carcinogenesis in SKH-1 mice by apigenin, a plant flavonoid. Anticancer Res. 17:85-91. [PubMed] [Google Scholar]
- 4.Carballo, E., W. S. Lai, and P. J. Blackshear. 1998. Feedback inhibition of macrophage tumor necrosis factor-alpha production by tristetraprolin. Science 281:1001-1005. [DOI] [PubMed] [Google Scholar]
- 5.Chan, G., J. O. Boyle, E. K. Yang, F. Zhang, P. G. Sacks, J. P. Shah, D. Edelstein, R. A. Soslow, A. T. Koki, B. M. Woerner, J. L. Masferrer, and A. J. Dannenberg. 1999. Cyclooxygenase-2 expression is up-regulated in squamous cell carcinoma of the head and neck. Cancer Res. 59:991-994. [PubMed] [Google Scholar]
- 6.Chaumontet, C., C. Droumaguet, V. Bex, C. Heberden, I. Gaillard-Sanchez, and P. Martel. 1997. Flavonoids (apigenin, tangeretin) counteract tumor promoter-induced inhibition of intercellular communication of rat liver epithelial cells. Cancer Lett. 114:207-210. [DOI] [PubMed] [Google Scholar]
- 7.Cok, S. J., S. J. Acton, and A. R. Morrison. 2003. The proximal region of the 3′-untranslated region of cyclooxygenase-2 is recognized by a multimeric protein complex containing HuR, TIA-1, TIAR, and the heterogeneous nuclear ribonucleoprotein U. J. Biol. Chem. 278:36157-36162. [DOI] [PubMed] [Google Scholar]
- 8.Cok, S. J., S. J. Acton, A. E. Sexton, and A. R. Morrison. 2004. Identification of RNA-binding proteins in RAW 264.7 cells that recognize a lipopolysaccharide-responsive element in the 3′-untranslated region of the murine cyclooxygenase-2 mRNA. J. Biol. Chem. 279:8196-8205. [DOI] [PubMed] [Google Scholar]
- 9.Cok, S. J., and A. R. Morrison. 2001. The 3′-untranslated region of murine cyclooxygenase-2 contains multiple regulatory elements that alter message stability and translational efficiency. J. Biol. Chem. 276:23179-23185. [DOI] [PubMed] [Google Scholar]
- 10.Czeczot, H., B. Tudek, J. Kusztelak, T. Szymczyk, B. Dobrowolska, G. Glinkowska, J. Malinowski, and H. Strzelecka. 1990. Isolation and studies of the mutagenic activity in the Ames test of flavonoids naturally occurring in medical herbs. Mutat. Res. 240:209-216. [DOI] [PubMed] [Google Scholar]
- 11.Derrigo, M., A. Cestelli, G. Savettieri, and I. Di Liegro. 2000. RNA-protein interactions in the control of stability and localization of messenger RNA. Int. J. Mol. Med. 5:111-123. [PubMed] [Google Scholar]
- 12.Dixon, D. A. 2004. Dysregulated post-transcriptional control of COX-2 gene expression in cancer. Curr. Pharm. Des. 10:635-646. [DOI] [PubMed] [Google Scholar]
- 13.Dixon, D. A., G. C. Balch, N. Kedersha, P. Anderson, G. A. Zimmerman, R. D. Beauchamp, and S. M. Prescott. 2003. Regulation of cyclooxygenase-2 expression by the translational silencer TIA-1. J. Exp. Med. 198:475-481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dixon, D. A., N. D. Tolley, P. H. King, L. B. Nabors, T. M. McIntyre, G. A. Zimmerman, and S. M. Prescott. 2001. Altered expression of the mRNA stability factor HuR promotes cyclooxygenase-2 expression in colon cancer cells. J. Clin. Investig. 108:1657-1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dreyfuss, G., V. N. Kim, and N. Kataoka. 2002. Messenger-RNA-binding proteins and the messages they carry. Nat. Rev. Mol. Cell Biol. 3:195-205. [DOI] [PubMed] [Google Scholar]
- 16.Fan, X. C., and J. A. Steitz. 1998. Overexpression of HuR, a nuclear-cytoplasmic shuttling protein, increases the in vivo stability of ARE-containing mRNAs. EMBO J. 17:3448-3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fischer, S. M., C. J. Conti, J. Viner, C. M. Aldaz, and R. A. Lubet. 2003. Celecoxib and difluoromethylornithine in combination have strong therapeutic activity against UV-induced skin tumors in mice. Carcinogenesis 24:945-952. [DOI] [PubMed] [Google Scholar]
- 18.Fotsis, T., M. S. Pepper, E. Aktas, S. Breit, S. Rasku, H. Adlercreutz, K. Wahala, R. Montesano, and L. Schweigerer. 1997. Flavonoids, dietary-derived inhibitors of cell proliferation and in vitro angiogenesis. Cancer Res. 57:2916-2921. [PubMed] [Google Scholar]
- 19.Gueydan, C., L. Droogmans, P. Chalon, G. Huez, D. Caput, and V. Kruys. 1999. Identification of TIAR as a protein binding to the translational regulatory AU-rich element of tumor necrosis factor alpha mRNA. J. Biol. Chem. 274:2322-2326. [DOI] [PubMed] [Google Scholar]
- 20.Hall, E. J., M. Astor, J. Bedford, C. Borek, S. B. Curtis, M. Fry, C. Geard, T. Hei, J. Mitchell, N. Oleinick, et al. 1988. Basic radiobiology. Am. J. Clin. Oncol. 11:220-252. [DOI] [PubMed] [Google Scholar]
- 21.Hamilton, B. J., C. M. Burns, R. C. Nichols, and W. F. Rigby. 1997. Modulation of AUUUA response element binding by heterogeneous nuclear ribonucleoprotein A1 in human T lymphocytes. The roles of cytoplasmic location, transcription, and phosphorylation. J. Biol. Chem. 272:28732-28741. [DOI] [PubMed] [Google Scholar]
- 22.Kandasamy, K., K. Joseph, K. Subramaniam, J. R. Raymond, and B. G. Tholanikunnel. 2005. Translational control of β2-adrenergic receptor mRNA by T-cell-restricted intracellular antigen-related protein. J. Biol. Chem. 280:1931-1943. [DOI] [PubMed] [Google Scholar]
- 23.Katsanou, V., O. Papadaki, S. Milatos, P. J. Blackshear, P. Anderson, G. Kollias, and D. L. Kontoyiannis. 2005. HuR as a negative posttranscriptional modulator in inflammation. Mol. Cell 19:777-789. [DOI] [PubMed] [Google Scholar]
- 24.Kawai, T., J. Fan, K. Mazan-Mamczarz, and M. Gorospe. 2004. Global mRNA stabilization preferentially linked to translational repression during the endoplasmic reticulum stress response. Mol. Cell. Biol. 24:6773-6787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kawai, T., A. Lal, X. Yang, S. Galban, K. Mazan-Mamczarz, and M. Gorospe. 2006. Translational control of cytochrome c by RNA-binding proteins TIA-1 and HuR. Mol. Cell. Biol. 26:3295-3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kawakami, A., Q. Tian, X. Duan, M. Streuli, S. F. Schlossman, and P. Anderson. 1992. Identification and functional characterization of a TIA-1-related nucleolysin. Proc. Natl. Acad. Sci. USA 89:8681-8685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kawamori, T., C. V. Rao, K. Seibert, and B. S. Reddy. 1998. Chemopreventive activity of celecoxib, a specific cyclooxygenase-2 inhibitor, against colon carcinogenesis. Cancer Res. 58:409-412. [PubMed] [Google Scholar]
- 28.Kedersha, N., and P. Anderson. 2002. Stress granules: sites of mRNA triage that regulate mRNA stability and translatability. Biochem. Soc. Trans. 30:963-969. [DOI] [PubMed] [Google Scholar]
- 29.Kullmann, M., U. Gopfert, B. Siewe, and L. Hengst. 2002. ELAV/Hu proteins inhibit p27 translation via an IRES element in the p27 5′UTR. Genes Dev. 16:3087-3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kuo, M. L., and N. C. Yang. 1995. Reversion of v-H-ras-transformed NIH 3T3 cells by apigenin through inhibiting mitogen activated protein kinase and its downstream oncogenes. Biochem. Biophys. Res. Commun. 212:767-775. [DOI] [PubMed] [Google Scholar]
- 31.Kutchera, W., D. A. Jones, N. Matsunami, J. Groden, T. M. McIntyre, G. A. Zimmerman, R. L. White, and S. M. Prescott. 1996. Prostaglandin H synthase 2 is expressed abnormally in human colon cancer: evidence for a transcriptional effect. Proc. Natl. Acad. Sci. USA 93:4816-4820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lal, A., K. Abdelmohsen, R. Pullmann, T. Kawai, S. Galban, X. Yang, G. Brewer, and M. Gorospe. 2006. Posttranscriptional derepression of GADD45α by genotoxic stress. Mol. Cell 22:117-128. [DOI] [PubMed] [Google Scholar]
- 33.Lal, A., K. Mazan-Mamczarz, T. Kawai, X. Yang, J. L. Martindale, and M. Gorospe. 2004. Concurrent versus individual binding of HuR and AUF1 to common labile target mRNAs. EMBO J. 23:3092-3102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Le Guiner, C., F. Lejeune, D. Galiana, L. Kister, R. Breathnach, J. Stevenin, and F. Del Gatto-Konczak. 2001. TIA-1 and TIAR activate splicing of alternative exons with weak 5′ splice sites followed by a U-rich stretch on their own pre-mRNAs. J. Biol. Chem. 276:40638-40646. [DOI] [PubMed] [Google Scholar]
- 35.Lepley, D. M., and J. C. Pelling. 1997. Induction of p21/WAF1 and G1 cell-cycle arrest by the chemopreventive agent apigenin. Mol. Carcinog. 19:74-82. [DOI] [PubMed] [Google Scholar]
- 36.Liang, Y. C., Y. T. Huang, S. H. Tsai, S. Y. Lin-Shiau, C. F. Chen, and J. K. Lin. 1999. Suppression of inducible cyclooxygenase and inducible nitric oxide synthase by apigenin and related flavonoids in mouse macrophages. Carcinogenesis 20:1945-1952. [DOI] [PubMed] [Google Scholar]
- 37.Lin, H. J., K. M. Lakkides, T. O. Keku, S. T. Reddy, A. D. Louie, I. H. Kau, H. Zhou, J. S. Gim, H. L. Ma, C. F. Matthies, A. Dai, H. F. Huang, A. M. Materi, J. H. Lin, H. D. Frankl, E. R. Lee, S. I. Hardy, H. R. Herschman, B. E. Henderson, L. N. Kolonel, L. Le Marchand, R. M. Garavito, R. S. Sandler, R. W. Haile, and W. L. Smith. 2002. Prostaglandin H synthase 2 variant (Val511Ala) in African Americans may reduce the risk for colorectal neoplasia. Cancer Epidemiol. Biomarkers Prev. 11:1305-1315. [PubMed] [Google Scholar]
- 38.Loflin, P., C. Y. Chen, and A. B. Shyu. 1999. Unraveling a cytoplasmic role for hnRNP D in the in vivo mRNA destabilization directed by the AU-rich element. Genes Dev. 13:1884-1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mahtani, K. R., M. Brook, J. L. Dean, G. Sully, J. Saklatvala, and A. R. Clark. 2001. Mitogen-activated protein kinase p38 controls the expression and posttranslational modification of tristetraprolin, a regulator of tumor necrosis factor alpha mRNA stability. Mol. Cell. Biol. 21:6461-6469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mazan-Mamczarz, K., S. Galban, I. Lopez de Silanes, J. L. Martindale, U. Atasoy, J. D. Keene, and M. Gorospe. 2003. RNA-binding protein HuR enhances p53 translation in response to ultraviolet light irradiation. Proc. Natl. Acad. Sci. USA 100:8354-8359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mazan-Mamczarz, K., A. Lal, J. L. Martindale, T. Kawai, and M. Gorospe. 2006. Translational repression by RNA-binding protein TIAR. Mol. Cell. Biol. 26:2716-2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McVean, M., H. Xiao, K. Isobe, and J. C. Pelling. 2000. Increase in wild-type p53 stability and transactivational activity by the chemopreventive agent apigenin in keratinocytes. Carcinogenesis. 21:633-639. [DOI] [PubMed] [Google Scholar]
- 43.Meng, Z., P. H. King, L. B. Nabors, N. L. Jackson, C. Y. Chen, P. D. Emanuel, and S. W. Blume. 2005. The ELAV RNA-stability factor HuR binds the 5′-untranslated region of the human IGF-IR transcript and differentially represses cap-dependent and IRES-mediated translation. Nucleic Acids Res. 33:2962-2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moos, P. J., D. T. Muskardin, and F. A. Fitzpatrick. 1999. Effect of taxol and taxotere on gene expression in macrophages: induction of the prostaglandin H synthase-2 isoenzyme. J. Immunol. 162:467-473. [PubMed] [Google Scholar]
- 45.Mukhopadhyay, D., C. W. Houchen, S. Kennedy, B. K. Dieckgraefe, and S. Anant. 2003. Coupled mRNA stabilization and translational silencing of cyclooxygenase-2 by a novel RNA binding protein, CUGBP2. Mol. Cell 11:113-126. [DOI] [PubMed] [Google Scholar]
- 46.Muller-Decker, K., G. Neufang, I. Berger, M. Neumann, F. Marks, and G. Furstenberger. 2002. Transgenic cyclooxygenase-2 overexpression sensitizes mouse skin for carcinogenesis. Proc. Natl. Acad. Sci. USA 99:12483-12488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Oshima, M., J. E. Dinchuk, S. L. Kargman, H. Oshima, B. Hancock, E. Kwong, J. M. Trzaskos, J. F. Evans, and M. M. Taketo. 1996. Suppression of intestinal polyposis in Apc Δ716 knockout mice by inhibition of cyclooxygenase 2 (COX-2). Cell 87:803-809. [DOI] [PubMed] [Google Scholar]
- 48.Otto, J. C., D. L. DeWitt, and W. L. Smith. 1993. N-glycosylation of prostaglandin endoperoxide synthases-1 and -2 and their orientations in the endoplasmic reticulum. J. Biol. Chem. 268:18234-18242. [PubMed] [Google Scholar]
- 49.Peng, S. S., C. Y. Chen, N. Xu, and A. B. Shyu. 1998. RNA stabilization by the AU-rich element binding protein, HuR, an ELAV protein. EMBO J. 17:3461-3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pentland, A. P., J. W. Schoggins, G. A. Scott, K. N. Khan, and R. Han. 1999. Reduction of UV-induced skin tumors in hairless mice by selective COX-2 inhibition. Carcinogenesis 20:1939-1944. [DOI] [PubMed] [Google Scholar]
- 51.Piecyk, M., S. Wax, A. R. Beck, N. Kedersha, M. Gupta, B. Maritim, S. Chen, C. Gueydan, V. Kruys, M. Streuli, and P. Anderson. 2000. TIA-1 is a translational silencer that selectively regulates the expression of TNF-alpha. EMBO J. 19:4154-4163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Raso, G. M., R. Meli, G. Di Carlo, M. Pacilio, and R. Di Carlo. 2001. Inhibition of inducible nitric oxide synthase and cyclooxygenase-2 expression by flavonoids in macrophage J774A.1. Life Sci. 68:921-931. [DOI] [PubMed] [Google Scholar]
- 53.Sengupta, S., B. C. Jang, M. T. Wu, J. H. Paik, H. Furneaux, and T. Hla. 2003. The RNA-binding protein HuR regulates the expression of cyclooxygenase-2. J. Biol. Chem. 278:25227-25233. [DOI] [PubMed] [Google Scholar]
- 54.Smith, W. L., R. M. Garavito, and D. L. DeWitt. 1996. Prostaglandin endoperoxide H synthases (cyclooxygenases)-1 and -2. J. Biol. Chem. 271:33157-33160. [DOI] [PubMed] [Google Scholar]
- 55.Spirio, L. N., D. A. Dixon, J. Robertson, M. Robertson, J. Barrows, E. Traer, R. W. Burt, M. F. Leppert, R. White, and S. M. Prescott. 1998. The inducible prostaglandin biosynthetic enzyme, cyclooxygenase 2, is not mutated in patients with attenuated adenomatous polyposis coli. Cancer Res. 58:4909-4912. [PubMed] [Google Scholar]
- 56.Strickland, J. E., D. A. Greenhalgh, A. Koceva-Chyla, H. Hennings, C. Restrepo, M. Balaschak, and S. H. Yuspa. 1988. Development of murine epidermal cell lines which contain an activated ras Ha oncogene and form papillomas in skin grafts on athymic nude mouse hosts. Cancer Res. 48:165-169. [PubMed] [Google Scholar]
- 57.Subbaramaiah, K., T. P. Marmo, D. A. Dixon, and A. J. Dannenberg. 2003. Regulation of cyclooxgenase-2 mRNA stability by taxanes: evidence for involvement of p38, MAPKAPK-2, and HuR. J. Biol. Chem. 278:37637-37647. [DOI] [PubMed] [Google Scholar]
- 58.Taupin, J. L., Q. Tian, N. Kedersha, M. Robertson, and P. Anderson. 1995. The RNA-binding protein TIAR is translocated from the nucleus to the cytoplasm during Fas-mediated apoptotic cell death. Proc. Natl. Acad. Sci. USA 92:1629-1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Thun, M. J., M. M. Namboodiri, and C. W. Heath, Jr. 1991. Aspirin use and reduced risk of fatal colon cancer. N. Engl. J. Med. 325:1593-1596. [DOI] [PubMed] [Google Scholar]
- 60.Tiano, H. F., C. D. Loftin, J. Akunda, C. A. Lee, J. Spalding, A. Sessoms, D. B. Dunson, E. G. Rogan, S. G. Morham, R. C. Smart, and R. Langenbach. 2002. Deficiency of either cyclooxygenase (COX)-1 or COX-2 alters epidermal differentiation and reduces mouse skin tumorigenesis. Cancer Res. 62:3395-3401. [PubMed] [Google Scholar]
- 61.Ulrich, C. M., J. Bigler, and J. D. Potter. 2006. Non-steroidal anti-inflammatory drugs for cancer prevention: promise, perils and pharmacogenetics. Nat. Rev. Cancer 6:130-140. [DOI] [PubMed] [Google Scholar]
- 61a.Van Dross, R., X. Hong, S. Essengue, S. M. Fischer, and J. C. Pelling. Modulation of UVB-induced and basal cyclooxygenase-2 (COX-2) expression by apigenin in mouse keratinocytes: role of USF transcription factors. Mol. Carcinog., in press. [DOI] [PubMed]
- 62.Wang, W., H. Furneaux, H. Cheng, M. C. Caldwell, D. Hutter, Y. Liu, N. Holbrook, and M. Gorospe. 2000. HuR regulates p21 mRNA stabilization by UV light. Mol. Cell. Biol. 20:760-769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wei, H., L. Tye, E. Bresnick, and D. F. Birt. 1990. Inhibitory effect of apigenin, a plant flavonoid, on epidermal ornithine decarboxylase and skin tumor promotion in mice. Cancer Res. 50:499-502. [PubMed] [Google Scholar]
- 64.Williams, C. S., M. Mann, and R. N. DuBois. 1999. The role of cyclooxygenases in inflammation, cancer, and development. Oncogene 18:7908-7916. [DOI] [PubMed] [Google Scholar]
- 65.Yu, Q., S. J. Cok, C. Zeng, and A. R. Morrison. 2003. Translational repression of human matrix metalloproteinases-13 by an alternatively spliced form of T-cell-restricted intracellular antigen-related protein (TIAR). J. Biol. Chem. 278:1579-1584. [DOI] [PubMed] [Google Scholar]
- 66.Zhang, W., B. J. Wagner, K. Ehrenman, A. W. Schaefer, C. T. DeMaria, D. Crater, K. DeHaven, L. Long, and G. Brewer. 1993. Purification, characterization, and cDNA cloning of an AU-rich element RNA-binding protein, AUF1. Mol. Cell. Biol. 13:7652-7665. [DOI] [PMC free article] [PubMed] [Google Scholar]