Abstract
Emerging evidence supports the idea that the c-Jun N-terminal kinases (JNKs) possess overlapping but distinct functions. The potential roles of the ubiquitously expressed JNK1 and JNK2 in regulating expression of the central transcription initiation factor, TATA-binding protein (TBP), were examined. Relative to wild-type fibroblasts, TBP was decreased in Jnk1−/− cells and increased in Jnk2−/− cells. Similarly, reduction of JNK1 in human hepatoma cells decreased TBP expression, whereas reduction of JNK2 enhanced it. JNK-mediated regulation of TBP expression occurs at the transcriptional level through their ability to target Elk-1, which directly regulates the TBP promoter in response to epidermal growth factor stimulation. JNK1 increases, whereas JNK2 decreases, the phosphorylation state of Elk-1, which differentially affects Elk-1 occupancy at a defined site within the TBP promoter. These JNK-mediated alterations in TBP expression, alone, serve to regulate c-Jun expression and fibroblast proliferation rates. These studies uncovered several new molecular events that distinguish the functions of JNK1 and JNK2 that are critical for their regulation of cellular proliferation.
The TATA binding protein (TBP) is a central eukaryotic transcription component, as it is required by all three cellular RNA polymerases for transcription initiation. It associates with additional proteins to form at least three distinct complexes, which specifies its role in RNA polymerase I (pol I)-, II-, or III-dependent transcription (18). TBP can be a limiting factor for both RNA pol I- (40, 45) and RNA pol III-dependent promoters (34, 38, 39, 45). Thus, increased expression of TBP serves to increase production of rRNAs and tRNAs. TBP is differentially limiting for RNA pol II-dependent promoters, depending on the nature of the promoter and the composition and location of regulatory elements (5, 26, 29).
Substantial evidence supports the idea that the activation of certain oncogenic signaling pathways can induce TBP expression. Treatment of cells with 12-O-tetradecanoylphorbol-13-acetate (TPA), a potent activator of protein kinase C, enhances cellular TBP production(15, 16). Activation of epidermal growth factor receptor 1 (EGFR1) induces TBP expression through the activation of Ras (45). In addition to regulation of cellular concentrations of TBP, its function can also be regulated. For example, the tumor suppressor p53 binds to TBP and negatively regulates its ability to form functional TFIIIB complexes, producing selective changes in RNA pol III-dependent transcription (6).
The fact that tumor suppressor and oncogenic signaling pathways tightly regulate cellular concentrations of TBP or its function suggested the possibility that alterations in TBP levels may influence the transformation state of cells. Consistent with this idea, inhibiting Ras-mediated increases in TBP in NIH 3T3 cells abrogated Ras-induced transformation, while increasing TBP expression in rat 1A fibroblast cells induced anchorage-independent growth and tumor formation in mice (20, 21). Heterozygous disruption of the TBP gene in a chicken B-cell line caused abnormalities in cell growth and size (37). These studies demonstrate that small changes in the cellular concentrations of TBP produce substantial phenotypic effects on cells. Depending on the cell type, changes in TBP expression can alter cell growth and/or the transformation state, suggesting the intriguing idea that TBP plays a role in oncogenesis. In support of this idea, comparison of matched normal human colon epithelium and colon tumors revealed that TBP expression is increased in a subset of human colon cancers (20, 21).
To further identify the signaling events that regulate TBP expression, we examined the role of the c-Jun N-terminal kinases (JNKs). The JNKs are a class of mitogen-activated protein kinases (MAPKs) that are activated by stress, proinflammatory stimuli, and mitogenic factors (3, 9). The JNKs are encoded by three genes, Jnk1, Jnk2, and Jnk3, which give rise to multiple isoforms generated by alternative splicing (10, 23, 27). JNK1 and JNK2 are ubiquitously expressed, whereas JNK3 expression is restricted to brain, heart, and testis (27). As JNK1 and JNK2 possess structural and biochemical similarities, they have many overlapping functions. However, JNK1 and JNK2 have been shown to possess some differences in their substrate specificities and gene targets (2, 17, 25, 28). Functions unique to JNK1 include its selective interaction with ATF2 (28), its regulation of microtubule-associated proteins (2), and its ability to induce apoptosis in response to tumor necrosis factor alpha (25). The role of these JNKs in tumor development remains controversial (12, 24). Depending on the cell type and stimulus, the JNKs have been implicated in both stimulating oncogenic transformation as well as acting as potential tumor suppressors.
In this study, we identified a new target of the JNKs, the central transcription initiation factor, TBP. Our results elucidate novel functional differences between the ubiquitously expressed JNK1 and JNK2. Through their opposing effects on the phosphorylation of Elk-1, which directly regulates TBP promoter activity, JNK1 increases TBP expression, whereas JNK2 decreases TBP expression. The different effects these JNKs have on regulating cellular TBP concentrations serve to modulate proliferation rates of fibroblasts. Furthermore, we demonstrate that one mechanism by which TBP affects cell proliferation is through its regulation of c-Jun expression. Thus, we further identify a novel mechanism by which the JNKs regulate c-Jun.
MATERIALS AND METHODS
Plasmids and reagents.
The plasmids containing the human TBP promoter, the β3-integrin promoter, and a hemagglutinin (HA) human TBP (HA-hTBP) expression plasmid, were described previously (22). The antisense TBP expression construct contains a 991-bp fragment of the 3′ end of the mouse TBP cDNA as previously described (20, 21). The synthesized human small interfering RNA (siRNA) oligonucleotide sequences were described previously for JNK1 (8), JNK2 (7), and Elk-1 (41). The synthesized mouse c-Jun siRNA was obtained from Santa Cruz (catalog no. SC-29224).
RT-PCR and PCR analysis.
Analysis of TBP, JNK1, JNK2, and β-actin mRNA by reverse transcription-PCR (RT-PCR) was performed according to the protocol of SuperScript One-Step RT-PCR with Platinum tag (Invitrogen). TBP and β-actin primer sequences for RT-PCR were described previously (20, 21). The mouse JNK1 primer sequences were as follows: forward primer, 5′-GCCATTCTGGTAGAGGAAGTTTCTC-3′; and reverse primer, 5′-CGCCAGTCCAAAATCAAGAATC-3′. The mouse JNK2 primer sequences were as follows: forward primer, 5′-TTGTGCTGCTTTTGATACAGTTCTTGGG-3′; and reverse primer, 5′-CTGGAAAGAGCTCTTCAAATTTGAT-3′. The human JNK1 and JNK2 primer sequences used were described by Uciechowski et al. (36).
Real-time PCR was performed using SYBR green supermix (Bio-Rad) on an MX3000P system (Strategene). The following TBP primer sequences were used: −119 forward primer, 5′-GACCTATGCTCACACTTCTCATGG-3′; +5 reverse primer, 5′-GAACCTGCCCGACCTCACTGAA-3′; −3315 forward primer, 5′-CAGGAGTTGGAGGTTGCAGT-3′; and −3158 reverse primer, 5′-GGCAACTCAAGACAGCTAGCAA-3′. All CT values were normalized to the PCR efficiency using the calculation 1/(2 · PCR efficiency)ΔCT. Normalized CT values for antibody pull-downs were normalized to input using the calculation antibody immunoprecipitation (IP) · 10/input. Changes (fold) in promoter occupancy were calculated by setting the level of promoter occupancy in the cells transfected with mismatch (mm) siRNA and in the absence of EGF treatment at 1.
Transfection and reporter gene assays.
For transient-transfection assays, mouse embryonic fibroblasts (MEFs) (2 × 105 cells per ml) or Huh-7 cells (1 × 106 cells per ml) were seeded. Fifteen hours later, cells were transfected using 1 μl of Lipofectin/μg of DNA (Invitrogen). Serum-free medium was added to each dish with Lipofectin-DNA complexes, and cells were further incubated at 37°C for 4 h. The wild-type and Jnk2−/− MEF cell lines expressing hTBP or antisense mTBP were constructed by cotransfecting pLTRe-hTBP gene or pcDNA-antisense mTBP plasmids containing a G418 resistance gene as previously described (20, 21). Selective medium containing 200 μg of G418/ml was added to propagate the cells. Huh-7 cells were transfected with 100 nM annealed double-stranded RNA interference, using 2 μl of Lipofectamine 2000 (Invitrogen)/μg of siRNA, and DNA mix. Two days after transfection, RNA and protein were isolated from the transfected cells. Total cell lysates were prepared from transfected cells to determine TBP or c-Jun promoter activity as previously described (45). The resultant luciferase activities were normalized to the amount of protein in each lysate. The values shown are the mean ± the standard error of the mean (SEM). The change (fold) in promoter activity was calculated by setting the level of luciferase activity in the absence of EGF or anisomycin at 1. For experiments comparing promoter activities in the presence or absence of TBP expression plasmid, the level of luciferase activity in the presence of empty vector was set at 1. Differences in promoter activity are expressed as the mean ± SEM.
Immunoblot analysis.
Mouse embryo fibroblasts and Huh-7 cells were grown to subconfluence (approximately 80%) in 5% fetal bovine serum in Dulbecco's modified Eagle's medium and then in 0.1% fetal bovine serum in Dulbecco's modified Eagle's medium for 12 h. Cells were incubated with either 12.5 ng/ml EGF or 25 ng/mg anisomycin for 30 min at 37°C unless otherwise indicated. Lysates (100 μg of protein) were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis and immunoblot analysis. Membranes were probed with specific polyclonal antibodies directed against TBP (Upstate Biotechnology), Elk-1 (Santa Cruz), c-Jun, and phosphorylated JNKs (catalog no. 9251S; Cell Signaling). Mouse monoclonal antibodies directed against β-actin or JNK1 (Santa Cruz) were used as indicated. Bound primary antibody was visualized using horseradish peroxidase-conjugated secondary antibody (Vector Laboratories) and enhanced chemiluminescence reagents (Pierce).
ChIP assay.
Chromatin immunoprecipitation (ChIP) assays were performed as described previously (44). Cells were treated with 50 ng/ml EGF for 5 min prior to cross-linking with formaldehyde. Briefly, chromatin was precleared with protein A/G plus agarose beads for 30 min at 4°C prior to addition of Elk-1 or control antibody overnight. Protein A/G plus agarose beads were added for 3 h, and the immunocomplex was isolated. The cross-links were reverse by incubating the mixture with 0.3 M NaCl at 65°C overnight. Eluents were desalted and concentrated following the Qiaex II (QIAGEN) protocol. The DNA was then subjected to PCR.
Cell proliferation and apoptosis assays.
Approximately 2 × 103 cells per ml of the MEFs were seeded in six-well plates in triplicate. The degree of confluence of the cells over a 6-day period was between 2 and 70%. Cells were assayed for viability and counted each day for 6 days using a Coulter counter. Bromodeoxyuridine (BrdU) labeling was performed using 100 μM of BrdU (Sigma) for 3 h. The percentage of cells in S phase was determined using anti-BrdU-tetramethyl rhodamine isocyanate (TRITC) conjugate according to the manufacturer's instructions (Guava Technologies Inc). The percentage of BrdU-labeled cells in S phase was measured by DNA profiling determined by flow cytometry. Cells in early apoptosis were analyzed using an annexin V kit following the manufacturer's instructions (Guava Technologies, Inc.), with flow cytometry to quantify the percentage of annexin V-labeled cells.
RESULTS
Activation of JNK1 increases TBP expression, whereas JNK2 reduces TBP expression in mouse embryo fibroblasts.
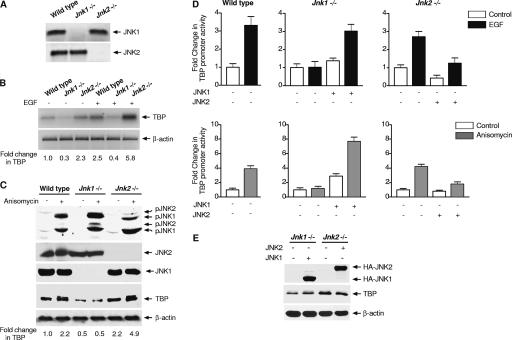
The potential roles of JNK1 and JNK2 in regulating TBP expression were first examined using MEFs with homozygous deletions in Jnk1 or Jnk2. RT-PCR analysis confirmed that JNK1 was expressed only in wild-type and Jnk2−/− cells, whereas JNK2 was expressed only in wild-type and Jnk1−/− cells (Fig. 1A). The relative levels of TBP mRNA were determined by RT-PCR. Compared to nonstimulated wild-type cells, TBP mRNA expression was decreased in Jnk1−/− cells, whereas TBP mRNA levels were increased in Jnk2−/− cells (Fig. 1B). We next determined how TBP expression would be affected upon EGF treatment, previously shown to induce TBP expression (45). EGF enhanced TBP mRNA levels in wild-type and Jnk2−/− cells, while no significant increase in TBP mRNA was observed upon EGF treatment of Jnk1−/− cells (Fig. 1B).
FIG. 1.
TBP expression is differentially regulated in mouse embryo fibroblasts deficient for JNK1 or JNK2 (A) JNK1 and JNK2 expression in MEFs. RT-PCR was performed using RNA isolated from each of the MEFs and specific primers for JNK1 and JNK2. (B) TBP mRNA expression in MEFs. Serum-starved cells were treated with or without EGF, and RT-PCR was performed using total RNA and specific primers for TBP or β-actin as indicated. (C) TBP levels in MEFs. Cells were treated with or without anisomycin. Protein lysates from each cell line were isolated, and immunoblot analysis was performed to detect the two phosphorylated isoforms each of JNK1 (p-JNK1) and JNK2 (p-JNK2), total JNK1 and JNK2, or β-actin antibodies as designated. A representative result of at least three independent determinations is shown. (D) Induction of TBP promoter activity requires JNK1. Wild-type, Jnk1−/−, or Jnk2−/− MEFs were transfected with a human TBP promoter-luciferase construct (p−4500/+66hTBP-Luc) alone or together with either JNK1 or JNK2 expression plasmids as indicated. Cells were treated with or without EGF (top panel) or anisomycin (bottom panel). Protein lysates were prepared, and luciferase activity was measured. The change in TBP promoter activity was calculated relative to that in nontreated cells. (E) Expression of HA-JNK1, HA-JNK2, TBP, and β-actin in cells deficient for JNK1 or JNK2. Jnk1−/− or Jnk2−/− MEFs were transfected with expression plasmids for HA-JNK1 or HA-JNK2. Protein lysates were isolated, and immunoblot analysis was used to detect HA-JNKs using antibodies against HA. The membranes were stripped and reprobed with antibodies against TBP or β-actin.
To further examine the role of activated JNKs in TBP expression, the MEFs were treated with anisomycin, a potent activator of JNK1 and JNK2, and immunoblot analysis was used to determine the amount of TBP (Fig. 1C). In nonstimulated cells, TBP levels were reduced in Jnk1−/− cells but increased in Jnk2−/− cells compared to wild-type cells. Anisomycin treatment produced an increase in TBP in both wild-type and Jnk2−/− cells, but not in Jnk1−/− cells. However, no changes in the amounts of JNK1 or JNK2 were observed. Together, these results support the idea that TBP expression is differentially regulated by JNK1 and JNK2. JNK1 is required for EGF- or anisomycin-induced TBP expression. In contrast, JNK2 represses TBP expression, even in nonstimulated cells.
JNKs regulate TBP expression at the transcriptional level.
Changes in TBP mRNA levels in the MEFs suggested that JNK1 and JNK2 could regulate TBP expression at the transcriptional level. To test this possibility, a 4,500-bp genomic fragment containing the human TBP promoter linked to a luciferase reporter was transiently expressed in the MEFs. Cells were treated with either EGF or anisomycin. Consistent with the relative amounts of TBP mRNA and protein expressed in these cells, TBP promoter activity was induced by EGF or anisomycin in wild-type and Jnk2−/− cells, but not in Jnk1−/− cells (Fig. 1D). To confirm that these results were specifically due to the JNK status of these cells, JNK1 and JNK2 expression was restored in Jnk1−/− and Jnk2−/− cells, respectively. Expression of JNK1 in Jnk1−/− cells reestablished TBP promoter induction by EGF and anisomycin, whereas expression of JNK2 in Jnk2−/− cells resulted in an overall decrease in TBP promoter activity (Fig. 1D). Immunoblot analysis of the lysates prepared from the transfected cells showed that the ectopically expressed HA-tagged JNK1 and HA-JNK2 were expressed at comparable levels (Fig. 1E). Given the approximate 30% transfection efficiency, reintroduction of JNK1 into JNK1-deficient cells still resulted in a small increase in cellular TBP concentrations, whereas expression of JNK2 in JNK2-deficient cells reduced TBP levels. Together, these results demonstrate that at least one mechanism by which the JNKs regulate cellular TBP concentrations is through their ability to regulate TBP promoter activity. JNK1, but not JNK2, is required for EGF- and anisomycin-mediated TBP promoter induction, while JNK2 produces an overall reduction in TBP promoter activity.
JNK1 and JNK2 differentially regulate TBP expression in human hepatoma cells.
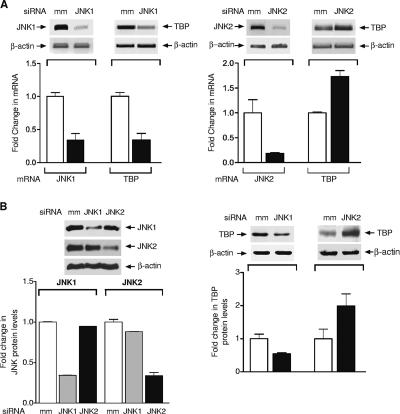
To further determine whether JNK-mediated regulation of TBP was cell type or species specific, we examined the functions of JNK1 and JNK2 in regulating TBP expression in the human Huh-7 cell line, derived from a hepatocellular carcinoma. Synthesized siRNAs selective for either JNK1 or JNK2 were transiently expressed in these cells. Compared to a 3-bp-mismatch (mm) siRNA, transfection of either the JNK1-specific or the JNK2-specific siRNAs into Huh-7 cells produced an approximately four- to fivefold decrease in JNK mRNA expression (Fig. 2A) as well as a selective decrease in the levels of these proteins (Fig. 2B, left panel). Changes in TBP expression upon the reduction of amounts of cellular JNK1 or JNK2 were further examined. Reduction in JNK1 produced a decrease in TBP mRNA, whereas reduction in JNK2 produced an increase in TBP mRNA (Fig. 2A), corresponding with changes in the amounts of TBP (Fig. 2B, right panel). Together, these results indicate that JNK1 positively regulates TBP expression in Huh-7 cells, whereas JNK2 negatively regulates TBP expression. Given that these results are observed in two distinct cell types, this indicates that the functions of the JNKs in regulating TBP expression are not likely to be cell type specific.
FIG. 2.
TBP expression is differentially regulated by reducing JNK1 or JNK2 expression in Huh-7 cells (A) Analysis of changes in TBP mRNA expression upon reduction of JNK1 or JNK2. Huh-7 cells were transfected with an siRNA selective for JNK1, JNK2, or mismatch siRNAs. RT-PCR was performed using RNA isolated from the Huh-7 cells and specific primers for JNK1, JNK2, TBP, or β-actin. (B) Reduction of JNK1 or JNK2 has opposing effects on TBP levels. Protein lysates derived from the siRNA-transfected cells were subjected to immunoblot analysis using antibodies against JNKs and β-actin (left) or TBP and β-actin (right) as indicated.
JNK1- and JNK2-mediated regulation of the TBP promoter requires a putative Ets binding site.
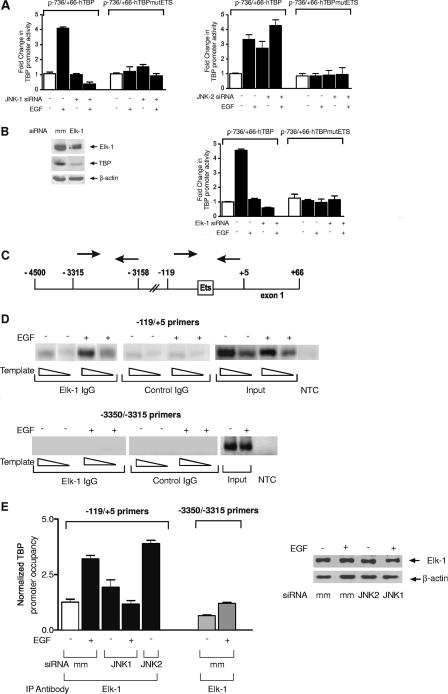
To determine the mechanism by which the JNKs differentially regulate TBP promoter activity, sequences within the TBP promoter required for JNK-mediated effects on TBP transcription were identified. When 5′ flanking sequences were deleted, 736 bp upstream from the transcription start site was found to be sufficient to confer both JNK1 and JNK2 responsiveness in MEFs (data not shown) and Huh-7 cells (Fig. 3A). As our previous studies revealed that a putative Ets transcription factor binding site between −50 and −41 bp, relative to the transcription start site, was required for EGF-mediated TBP promoter induction (45), we tested whether this site was also required for JNK1- and JNK2-mediated effects on TBP promoter activity. Huh-7 cells were cotransfected with −736/+66-hTBP or −736/+66mutEts-hTBP promoter constructs together with either mismatch siRNA or siRNAs specific for either JNK1 or JNK2 (Fig. 3A). Reduction in JNK1 expression or mutation of the putative Ets site abolished EGF-induced TBP promoter activity (Fig. 3A, left panel). Furthermore, in the absence of EGF treatment, decreased expression of JNK1 had no effect on TBP promoter activity. In contrast, decreased expression of JNK2 induced TBP promoter activity even in the absence of EGF stimulation, and this induction required the putative Ets binding site (Fig. 3A, right panel). Together, these results indicate that both JNK1 and JNK2 target the same site within the TBP promoter to regulate its activity.
FIG. 3.
Elk-1 directly modulates EGF-induced TBP promoter activity and is differentially regulated by JNK1 and JNK2. (A) JNK1 and JNK2 regulate TBP promoter activity through a common Ets transcription factor binding site. Huh-7 cells were transfected with either a p−736/+66hTBP-Luc or p−736/+66hEtsmutTBP-Luc construct. In addition, cells were transfected with mismatch siRNA (−) or JNK1 siRNA (+) (left panel) or mismatch siRNA (−) or JNK2 siRNA (+) (right panel). Where designated, cells were additionally treated with EGF. Protein lysates were prepared, and luciferase activity was measured. Changes (fold) were calculated based on the control (TBP promoter plus mm siRNA) and normalized to total protein. (B) Elk-1 regulates TBP expression. Huh7 cells were transfected with either mismatch siRNA or Elk-1 siRNA. Immunoblot analysis was performed using lysates prepared from transfected cells using antibodies against Elk-1, TBP, and β-actin (left panel). Cells were additionally transfected with either p−736/+66hTBP-Luc or p−736/+66hEtsmutTBP-Luc (right panel), and TBP promoter activity was measured as described for panel A. (C) Schematic diagram of a genomic fragment containing the human TBP promoter. The boxed Ets motif designates the Ets binding site that is mutated in the p−736/+66hEtsmutTBP-Luc construct used. The arrows depict the relative location of the PCR primers used for the ChIP analysis. (D) Elk-1 binds to the TBP promoter upon EGF stimulation. ChIP assays were performed as described in Materials and Methods. The resultant chromatin was immunoprecipitated with Elk-1 antibody or a control antibody. Specific DNA fragments were quantified by PCR using primers targeting the regions within the TBP promoter specified in panel C. “Input” represents primer-specific amplification of 10% of total chromatin isolated for each sample. “NTC” designates reactions performed without added template. IgG, immunoglobulin G. (E) JNK1 increases, whereas JNK2 decreases, Elk-1 occupancy on the TBP promoter. Huh-7 cells were transfected with mismatch, JNK1, or JNK2 siRNAs. Where designated, cells were additionally treated with EGF. Immunoblot analysis was performed using lysates derived from the transfected cells (right panel). ChIP assays were performed using the transfected cells, and real-time PCR was used to quantify the amplified DNA. The change (fold) in TBP occupancy was calculated based on the control (TBP promoter plus mm siRNA). The results shown were derived from three independent chromatin preparations.
JNK1 and JNK2 differentially regulate the phosphorylation state of Elk-1 and its binding to the TBP promoter.
To identify the specific Ets protein that regulates TBP promoter activity, we considered Elk-1. Elk-1 is ubiquitously expressed and it is a direct target of all three classes of MAPKs (reviewed in reference 33). Transient expression of an siRNA selective for Elk-1 reduced both Elk-1 and TBP levels in Huh-7 cells (Fig. 3B, left). In addition, reduction of Elk-1 expression inhibited EGF-mediated TBP promoter induction (Fig. 3B, right). However, no effect on promoter activity was produced when Elk-1 expression was repressed if the Ets binding site was mutated. To determine whether Elk-1 directly targeted the TBP promoter, ChIP assays were used. Two sets of primers were used to amplify different regions flanking the 5′ region of the human TBP gene (Fig. 3C). Elk-1 was not significantly bound to the TBP promoter in the absence of EGF. However, as early as 5 min after EGF treatment, the occupancy of Elk-1 was strongly enhanced at sequences amplified between −119 and +5 containing the Ets binding site, but not at the region between −3315 and −3158 (Fig. 3D). Together, these results identify Elk-1 as a transcription factor responsible for directly inducing TBP promoter activity through EGF.
To determine if JNK1 and JNK2 regulate the binding of Elk-1 to the TBP promoter, Huh-7 cells were transfected with either mismatched siRNA or siRNAs specific for JNK1 or JNK2 to inhibit their expression. Reduction in either JNK1 or JNK2 expression did not change Elk-1 protein levels in these cells (Fig. 3E, right). ChIP analysis and real-time RT-PCR were used to quantify Elk-1 interactions with the TBP promoter. EGF-mediated increases in Elk-1 occupancy were abrogated when JNK1 expression was reduced (Fig. 3E, left). However, decreases in JNK1 expression did not significantly alter Elk-1 occupancy in nonstimulated cells. In contrast, reduction of JNK2 expression increased the occupancy of Elk-1 on the TBP promoter in the absence of EGF stimulation.
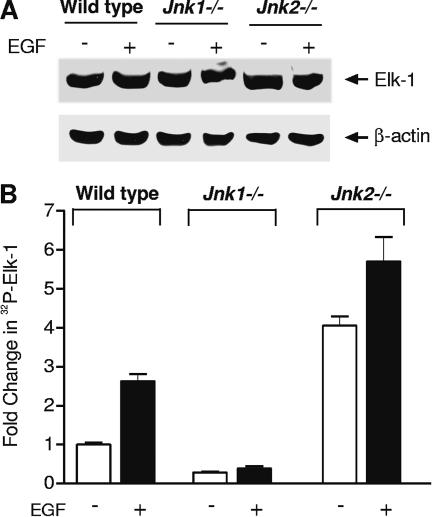
To further determine how the JNKs might regulate Elk-1 binding to the promoter, we examined the levels of Elk-1 and its phosphorylation state in the JNK-deficient MEFs. Elk-1 levels were comparable in wild-type, Jnk1−/−, and Jnk2−/− cells (Fig. 4A). To determine the relative phosphorylation state of Elk-1, cells were labeled with 32P and Elk-1 was immunoprecipitated from the resultant lysates. The total phosphorylation state of Elk-1 was significantly decreased in JNK1-deficient MEFs but increased in JNK2-deficient MEFs compared to wild-type cells (Fig. 4B). Upon EGF treatment, the phosphorylation state of Elk-1 was increased in only wild-type and JNK2-deficient cells. Together, these results support the idea that JNK1 and JNK2 have antagonizing functions in regulating the overall phosphorylation state of Elk-1, thereby mediating its interaction with the TBP promoter.
FIG. 4.
JNK1 and JNK2 differentially regulate Elk-1 phosphorylation (A) JNK1 and JNK2 do not change Elk-1 protein levels. Wild-type, Jnk1−/−, and Jnk2−/− MEFs were serum starved overnight and then incubated with or without EGF. Protein lysates were isolated, and immunoblot analysis was performed using Elk-1 or β-actin antibodies. (B) JNK1 increases, whereas JNK2 decreases, the total phosphorylation state of Elk-1. MEFs were cultured as described for panel A. In addition, cells were incubated with 32Pi (0.5 mCi/ml) for 3 h prior to EGF treatment. The resultant cell lysates were immunoprecipitated with Elk-1 antibody and subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Autoradiography was used to examine the amount of 32P-incorporated Elk-1. The values shown are the means ± SEM of three independent experiments where the values were normalized to the total amount of Elk-1 protein.
Cellular TBP concentrations regulate cell proliferation rates of mouse embryo fibroblasts.
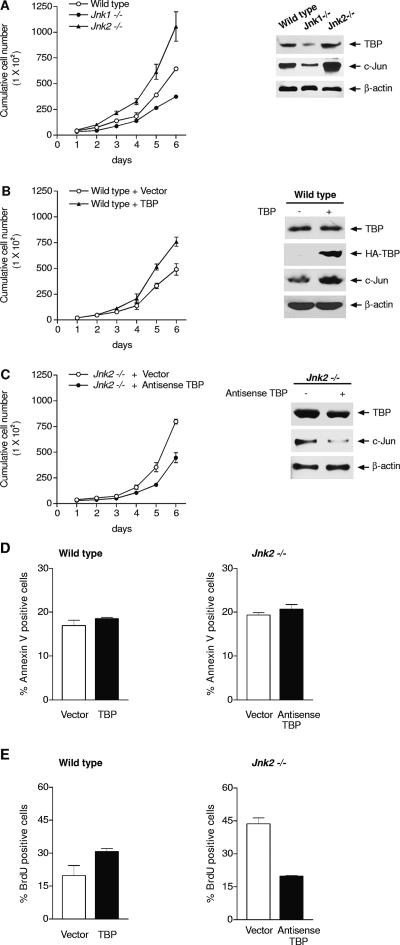
We next determined whether JNK-mediated regulation of TBP levels, alone, could alter the proliferation rates of the MEFs. Comparison of these cells revealed that the absence of JNK1 resulted in decreased accumulation of the cells, whereas JNK2-deficient cells exhibited an increase in the number of cells as compared to wild-type cells (Fig. 5A, left panel), consistent with previous studies (28, 32). As the accumulation rate of these cells correlated with the levels of TBP (Fig. 5A, right panel), we directly altered TBP expression in these cells and assessed potential changes. As we were unsuccessful at generating stable Jnk1−/− cell lines, and to complement our TBP loss of function effects in the Jnk2−/− cells, we constructed wild-type MEFs that were stably transfected with a TBP expression plasmid and pooled populations of selected cells were analyzed. Immunoblot analysis of lysates derived from these cells showed that the ectopically introduced HA-tagged TBP was expressed in these cells without altering the levels of endogenous TBP (Fig. 5B, right panel). The stable cell lines engineered to express increased amounts of TBP accumulated at a faster rate relative to wild-type cells (Fig. 5B, left panel). We next determined whether reducing expression of TBP would decrease the accumulation rate of Jnk2−/− cells, which contain higher levels of TBP relative to wild-type cells. The Jnk2−/− cells were stably transfected with an antisense TBP expression vector, and pooled populations of cells were analyzed. An approximately twofold decrease in TBP was observed in the antisense TBP-expressing Jnk2−/− cells (Fig. 5C, right panel). This reduction in TBP resulted in a decrease in the accumulation rate of these cells (Fig. 5C, left panel). These results support the idea that alterations in the cellular concentrations of TBP produce corresponding changes in the rate at which MEFs accumulate in culture.
FIG. 5.
JNK-mediated changes in TBP levels alter the accumulation rates of MEFs (A) The accumulation rates of wild-type, Jnk1−/−, and Jnk−/− MEFs correlate with c-Jun and TBP levels. Wild-type, Jnk1−/−, and Jnk2−/− MEFs were plated in triplicate, and cell viability and total cell numbers were measured daily for 6 days (left panel). The results represent three independent experiments. Protein lysates derived from each of the cell lines were subjected to immunoblot analysis using antibodies against TBP, c-Jun, or β-actin as indicated (right panel). (B) Increased TBP expression enhances c-Jun expression and the accumulation of wild-type MEFs. Wild-type MEFs were stably transfected with an HA-tagged human TBP expression vector (Wild type + TBP) or empty vector (Wild type + vector). The accumulation rates of these cells were measured as described for panel A (left panel). The results represent two independent experiments resulting from two nonclonally selected populations of each stable cell line. Protein lysates derived from these cells were subjected to immunoblot analysis using antibodies against TBP, c-Jun, hemagglutinin, or β-actin as designated (right panel). (C) Reduced expression of TBP decreases the c-Jun expression and accumulation rate of Jnk2−/− MEFs. Stable cell lines were established by transfecting Jnk2−/− MEFs with an antisense TBP expression construct (Jnk2−/− + Antisense TBP) or vector alone (Jnk2−/− + Vector). The accumulation rates of these cells were measured as described for panel A (left panel). TBP, c-Jun, and β-actin protein levels were determined by immunoblot analysis (right panel). (D) Alterations in TBP levels do not affect apoptotic rates of MEFs. Stable cells of the wild-type-plus-TBP (left panel) and Jnk2−/−-plus-antisense TBP cell lines (right panel) were stained with annexin V, and rates of apoptosis were assessed as described in Materials and Methods. (E) Alterations in TBP levels change cellular proliferation rates of MEFs. Stable cells of the wild-type-plus-TBP (left panel) and Jnk2−/−-plus-antisense TBP cell lines (right panel) were labeled with BrdU, and S-phase-labeled cells were quantified by flow cytometry. The values shown are the means ± SEM of three independent experiments.
To distinguish whether the TBP-mediated changes in the accumulation rates of these cells were due to alterations in proliferation rates or apoptotic rates, the MEFs were labeled with BrdU and annexin V, respectively. Flow cytometry was used to quantify the number of labeled cells. No significant differences in the spontaneous cell death rates were observed between the various cells (Fig. 5D). However, wild-type cells expressing increased amounts of TBP incorporated 11% more BrdU compared to the parental wild-type cells, whereas Jnk2−/− cells expressing the antisense TBP construct incorporated 24% less BrdU than the parental Jnk2−/− cells (Fig. 5E). These results indicate that the cellular levels of TBP determine the rate at which MEFs proliferate.
TBP regulates c-Jun expression and AP-1-dependent promoter activity.
To determine how TBP regulates the proliferation rates of these cells, we considered the possibility that TBP might regulate c-Jun expression, as previous studies showed that the levels of c-Jun dictate MEF proliferation rates (28). Immunoblot analysis was used to compare the levels of c-Jun in the MEFs. In accordance with the amounts of TBP in these cells, Jnk1−/− cells express reduced amounts of c-Jun, whereas Jnk2−/− cells express increased amounts of c-Jun, compared to wild-type cells (Fig. 5A, right panel). In addition, wild-type stably transfected MEFs expressing increased amounts of TBP also contained increased amounts of c-Jun (Fig. 5B, right panel). In contrast, Jnk2−/− cells expressing the antisense TBP construct and reduced TBP levels displayed a corresponding decrease in c-Jun amounts (Fig. 5C, right panel). These results indicate that alterations in TBP expression produce corresponding changes in c-Jun expression.
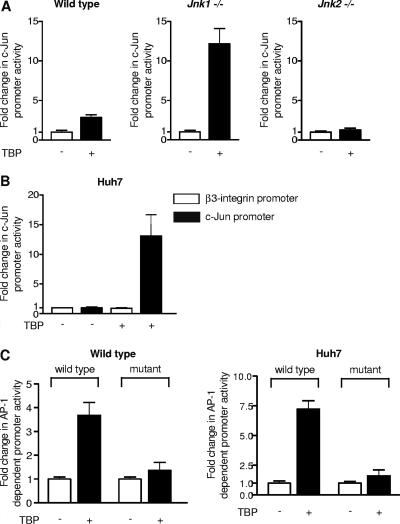
To further examine whether TBP regulates c-Jun at the transcription level, a c-Jun promoter-reporter construct was transiently transfected in the MEFs with a TBP expression plasmid or empty vector (Fig. 6A). Increased expression of TBP into wild-type MEFs resulted in a modest stimulation of the c-Jun promoter. However, in Jnk1−/− cells, increased expression of TBP strongly induced the c-Jun promoter. In contrast, increased expression of TBP did not induce the c-Jun promoter in Jnk2−/− cells. These results indicate that the c-Jun promoter is differentially sensitive to increases in TBP in the MEFs based on their endogenous levels of TBP. In Huh-7 cells, the c-Jun promoter is strongly stimulated by increased expression of TBP in the Huh-7 cells (Fig. 6B). This can be compared to the activity of the β3-integrin promoter, which is not affected by increased expression of TBP. To further determine whether the TBP-mediated increase in c-Jun could enhance c-Jun function, AP-1-dependent promoter activity was measured in cells expressing increased amounts of TBP. Transcription of an AP-1-responsive promoter was induced upon increased TBP expression in MEFs and Huh-7 cells, whereas mutation of the AP-1 binding site eliminated TBP-mediated induction (Fig. 6C). These results indicate that alterations in the cellular concentrations of TBP serve to regulate c-Jun expression and AP-1-dependent promoter activity.
FIG. 6.
Cellular TBP amounts regulate c-Jun and AP-1 promoter activities in Huh-7 cells and MEFs (A) c-Jun promoter activity is differentially responsive to increases in TBP in wild-type and Jnk1- and Jnk2-deficient cells. MEFs were transiently cotransfected with human c-jun promoter-luciferase and human TBP expression plasmids, and luciferase activity was measured. The values shown are the means ± SEM of at least four independent experiments. (B) Increasing TBP expression enhances c-Jun promoter activity in Huh-7 cells. Human hepatoma Huh-7 cells were transiently cotransfected with the c-jun promoter-luciferase construct or a β3-integrin-luciferase construct together with the human TBP expression plasmid or empty vector. The change in c-jun promoter activity was calculated relative to that with vector alone. (C) Increasing TBP expression enhances AP-1-dependent promoter activity in wild-type MEFs and Huh-7 cells. Wild-type MEFs or Huh-7 cells were transiently cotransfected with a human AP-1-dependent promoter-luciferase construct, or the same promoter containing a mutated AP-1 site and the human TBP expression plasmid. The change in AP-1-dependent promoter activity was calculated relative to that with vector alone. The values shown are the means ± SEM of three independent experiments.
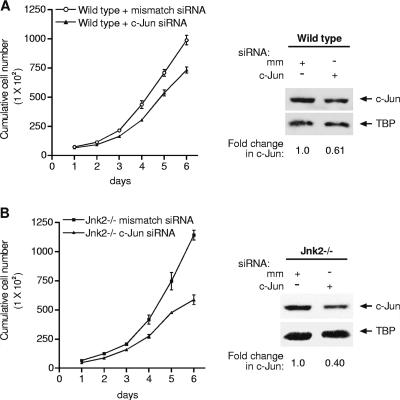
Since these results revealed that TBP concentrations regulate c-Jun levels, we further assessed whether c-Jun might also regulate TBP. We tested whether reducing expression of c-Jun would decrease expression of TBP. Wild-type and Jnk2−/− MEFs were transfected with either c-Jun siRNAs or mismatched siRNAs, and immunoblot analysis was performed using the derived cell lysates (Fig. 7A and B, right). An approximately twofold reduction in c-Jun in either the wild-type or Jnk2−/− cells did not affect the amount of TBP expressed. These results are consistent with results demonstrating that increased expression of c-Jun in either MEFs or Huh-7 cells does not result in TBP promoter stimulation. However, under these conditions, AP-1-dependent promoter activity was strongly induced (data not shown). We further determined how this reduction in c-Jun might affect the accumulation rate of these cells. Decreased expression of c-Jun in both the wild-type and Jnk2−/− cells diminished the accumulation rates of these cells. Together, these results demonstrate that JNK-mediated regulation of TBP expression serves to regulate c-Jun expression, but that c-Jun expression does not regulate TBP expression. Furthermore, these results provide evidence that TBP-induced changes in cellular proliferation occur through its ability to regulate c-Jun levels.
FIG. 7.
Decreased expression of c-Jun in MEFs reduces accumulation rates but does not affect TBP expression. Wild-type (A) or Jnk2−/− (B) MEFs were transfected with either c-Jun siRNA or an mm siRNA. MEFs were plated in triplicate, and cell viability and total cell numbers were measured daily for 6 days (left panel). The results represent three independent experiments. Protein lysates derived from each of the cell lines 72 h after transfection with the siRNAs were subjected to immunoblot analysis using antibodies against TBP and c-Jun. The changes in c-Jun levels were normalized to the total amount of TBP.
DISCUSSION
The JNKs play an important role in regulating the stability and activity of certain transcription activators. Once activated, these proteins phosphorylate a variety of cellular targets, including transcription factors such as c-Jun, JunD, ATF2, and Elk-1, resulting in their activation and subsequent alterations in cellular gene activity (30). In the absence of external stimuli, however, the JNKs may enhance the degradation of their substrates (13). Our results have uncovered new functions and roles for the JNKs in regulating the expression of the transcription initiation factor, TBP. TBP is a central transcription initiation factor and changes in its levels produce pleiotropic but selective effects on transcription by all three nuclear RNA polymerases. Increases in TBP induce the transcription of genes encoding tRNAs and rRNAs (34, 38-40, 45), while protein-encoding genes are differentially regulated (5, 26, 29). Since TBP levels influence the activities of such a diverse array of genes, the JNK-mediated regulation of cellular TBP expression is likely to be a key mechanism by which the JNKs significantly impact cellular gene expression. Consistent with this idea, we find that the JNKs regulate RNA polymerase III-dependent gene activity (S. Zhong and D. L. Johnson, unpublished results).
Our findings further reveal that JNK1 and JNK2 have opposing roles in regulating TBP expression. The selective activation of JNK1, but not JNK2, is responsible for inducing TBP mRNA and protein expression in both mouse and human cell lines. Furthermore, JNK2 negatively regulates TBP expression in nonstimulated cells. This is consistent with previous studies demonstrating that JNK2, but not JNK1, undergoes autophosphorylation in the absence of upstream-activated kinases (35). While previous work has demonstrated that TBP expression can be induced by the activation of select signaling molecules, our results are the first to demonstrate that TBP expression can be negatively regulated. Together, these results support the idea that the balance between JNK1 and JNK2 is an important mechanism responsible for exquisitely regulating cellular TBP amounts.
Our results demonstrate that JNK1 and JNK2 mediate their effects on TBP expression by transcriptional regulation of the TBP promoter through a common Elk-1 binding site. Elk-1 does not appreciably bind to this site in nonstimulated cells, but upon EGF stimulation, Elk-1 is recruited to the promoter, correlating with its induction. Thus, we have identified a new Elk-1-targeted promoter. Interestingly, our results reveal that the JNKs have antagonizing functions in regulating Elk-1 phosphorylation and Elk-1 binding to the TBP promoter. Consistent with these results, JNK1 can directly phosphorylate Elk-1 (1, 40a), and increased phosphorylation of Elk-1 enhances its DNA binding function (42) and its binding to the coactivator p300 (24a). Surprisingly, in contrast to previous studies demonstrating that JNK2 directly phosphorylates Elk-1 in vitro (43), cells deficient for JNK2 display an increase in the phosphorylation state of Elk-1, which can be further augmented upon EGF stimulation. Together, these results demonstrate that these JNK-mediated effects on Elk-1 are critical determinants in dictating TBP promoter activity.
Together, our results support the idea that JNK2 does not phosphorylate Elk-1 in vivo and that it acts to negatively regulate the phosphorylation state of Elk-1. Elk-1 is a direct substrate for all three classes of MAPKs (33). It is conceivable that JNK2 specifically inhibits the ability of JNK1, p38, and/or ERK1/2 to phosphorylate Elk-1. Alternatively, JNK2 may prevent a potential priming kinase from phosphorylating Elk-1, thereby decreasing subsequent phosphorylation of Elk-1 by the other MAPKs. However, the fact that JNK2 acts to decrease the phosphorylation state of Elk-1 in nonstressed cells, suggests the idea that, even in the absence of activation of the MAPKs, JNK2 is still capable of reducing Elk-1 phosphorylation. Protein phosphatase 2B has been shown to negatively regulate Elk-1 by antagonizing the phosphorylation induced by MAPK activation (31). Therefore, it is also plausible that JNK2 could function to facilitate dephosphorylation of Elk-1 through its ability to enhance the function of an Elk-1-targeted phosphatase. In either case, our studies support a novel role for JNK2 in negatively regulating the phosphorylation state and function of Elk-1.
The recent use of a chemical genetic approach to selectively inhibit the function of JNK2 in MEFs suggests that the use of knockout cells is insufficient to derive conclusions regarding the function of JNK2 (19). This study proposes an alternative explanation for the observed opposing functions of JNKs in MEFs and reveals that, contrary to previous studies (28, 32) and our current studies, JNK2 positively, rather than negatively, regulates c-Jun expression and cellular proliferation. The explanation proposed for the basis of these observed differences was that long-term loss of gene function can lead to an adaptive response and changes in the functions of other genes and that there is a compensatory increase in JNK1 function in Jnk2−/− cells. However, our study has importantly used an additional approach and another cell line to examine the functions of both JNK1 and JNK2. Transient repression of JNK2 expression by approximately fourfold by an siRNA approach in the Huh-7 cell line, for less than 2 days, had functional consequences on TBP expression similar to those observed in the Jnk2−/− MEFs. Interestingly, treatment of the Jnk2−/− MEFs with the chemical inhibitor of JNK2 was conducted within a similar time frame (19). This argues against the notion that the functional differences observed for JNK2 in MEFs are due to the long-term consequences of gene disruption. Nevertheless, further work will be needed to examine how long-term loss of gene function might affect the expression or function of other genes and the length of time required for these potential compensatory changes in genes to occur.
The present study identifies a new mechanism by which the JNKs regulate cellular concentrations of c-Jun, through their ability to regulate TBP expression. These results demonstrate that small changes in TBP levels, alone, serve to regulate c-Jun expression, supporting the idea that this is an important mechanism by which the JNKs affect c-Jun concentrations. c-Jun has been extensively shown to be regulated by the JNKs (11). While both JNK1 and JNK2 bind to and phosphorylate c-Jun, leading to its activation, the JNKs have also been shown to target c-Jun for ubiquitination and degradation (14). Further analysis of the individual roles of the JNKs revealed that JNK1 increases c-Jun activity and stability, whereas JNK2 preferentially binds to c-Jun in nonstimulated cells and targets it for degradation (25, 28). The tumor suppressor p16INK4a, which selectively interacts with JNK1 and JNK3, prevents c-Jun phosphorylation (4). Thus, these studies, and our current results demonstrate that the JNKs regulate c-Jun levels and activity through multiple distinct mechanisms.
The JNKs play essential roles in organogenesis during development and regulate a wide range of cellular functions in mammalian cells, including cell proliferation, survival, and apoptosis. They also are important mediators in the immune responses by regulating cytokine gene expression. While we have so far shown the EGFR1- and anisomycin-mediated activation of JNK1 induces TBP expression, it remains to be tested whether other cellular stress conditions that activate JNK1, such as hypoxia, also regulate TBP expression. The ability of the JNKs to regulate TBP expression prompted us to consider potential phenotypic consequences. The JNKs have been shown to differentially regulate fibroblast proliferation (28, 32). As the levels of TBP correlated with the rate of accumulation of these cells, we examined whether directly altering TBP levels could modulate cellular proliferation. These results indicate that in MEFs, cellular TBP concentrations dictate the proliferation rates of these cells. Examining potential TBP-targeted genes that might contribute to the altered growth rate, we find that changes in TBP levels regulate c-Jun promoter activity and expression and that decreases in c-Jun result in diminished proliferation of MEFs. Thus, the ability of the JNKs to regulate TBP expression and thereby modulate cellular gene expression, including that of c-Jun, is a key mechanism for controlling cellular proliferation.
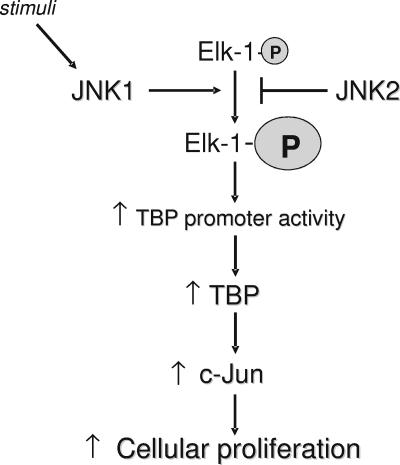
Our results corroborate recent findings that, despite their structural and biochemical similarities, JNK1 and JNK2 possess distinct functions. Our results define several new and opposing functions of the JNKs that link regulation of Elk-1 phosphorylation and TBP expression with their capacity to control c-Jun expression (Fig. 8). In MEFs, these events ultimately affect cellular proliferation rates. The JNKs have been shown to induce apoptosis or enhance cellular proliferation and/or transformation, depending on the cell type (12, 24). As small increases in cellular TBP levels have been shown to promote transformation of rat 1A cells, without changing cellular proliferation rates (20, 21), the cell-type-dependent phenotypic effects of the JNKs may be a consequence of their ability to regulate TBP expression. Together, these results support the idea that the balance between JNK1 and JNK2 is an important mechanism that is responsible for tightly regulating cellular TBP amounts. The opposing functions of JNK1 and JNK2 in regulating TBP levels suggest that JNK1-mediated increases in TBP could promote oncogenesis, whereas JNK2-mediated decreases in TBP could act to suppress oncogenesis. While the regulation of the JNKs is complex, our study has discovered a critical function of JNK1 and JNK2 in regulating the cellular concentrations of TBP, which ultimately contribute to the growth potential and transformation state of cells.
FIG. 8.
Model for opposing functions of JNK1 and JNK2. JNK1 promotes Elk-1 phosphorylation in response to EGF stimulation, whereas JNK2 negatively regulates the phosphorylation state of Elk-1 in the absence of stimuli. Phosphorylation of Elk-1 enhances it ability to be recruited to the TBP promoter, thereby inducing TBP promoter activity and its expression. Increased TBP levels then modulate c-Jun promoter activity and expression, ultimately dictating cellular proliferation rates.
Acknowledgments
We thank Annette Woiwode, Sandra Johnson, Michael Stallcup, and Ebrahim Zandi for critical reading of the manuscript; Zigang Dong for the JNK-deficient MEFs; Anning Lin for JNK1 and JNK2 expression plasmids; Wayne Vedeckis for the c-Jun promoter; and the USC Norris Comprehensive Cancer Center Gene Regulation Group for helpful discussions. We greatly appreciate Mary Perry for her support and helpful discussions.
This work was supported by Public Health Service grant CA074138 from the National Cancer Institute to D.L.J.
Footnotes
Published ahead of print on 30 October 2006.
REFERENCES
- 1.Cavigelli, M., F. Dolfi, F. X. Claret, and M. Karin. 1995. Induction of c-fos expression through JNK-mediated TCF/Elk-1 phosphorylation. EMBO J. 14:5957-5964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chang, L., Y. Jones, M. H. Ellisman, L. S. Goldstein, and M. Karin. 2003. JNK1 is required for maintenance of neuronal microtubules and controls phosphorylation of microtubule-associated proteins. Dev. Cell 4:521-533. [DOI] [PubMed] [Google Scholar]
- 3.Chang, L., and M. Karin. 2001. Mammalian MAP kinase signalling cascades. Nature 410:37-40. [DOI] [PubMed] [Google Scholar]
- 4.Choi, B. Y., H. S. Choi, K. Ko, Y. Cho, F. Zhu, B. S. Kang, S. P. Ermakova, W. Y. Ma, A. M. Bode, and Z. Dong. 2005. The tumor suppressor p16(INK4a) prevents cell transformation through inhibition of c-Jun phosphorylation and AP-1 activity. Nat. Struct. Mol. Biol. 12:699-707. [DOI] [PubMed] [Google Scholar]
- 5.Colgan, J., and J. L. Manley. 1992. TFIID can be rate limiting in vivo for TATA-containing but not TATA-lacking, RNA pol II promoters. Genes Dev. 6:304-315. [DOI] [PubMed] [Google Scholar]
- 6.Crighton, D., A. Woiwode, C. Zhang, N. Mandavia, J. P. Morton, L. J. Warnock, J. Milner, R. J. White, and D. L. Johnson. 2003. p53 represses RNA polymerase III transcription by targeting TBP and inhibiting promoter occupancy by TFIIIB. EMBO J. 22:2810-2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Curtin, J. F., and T. G. Cotter. 2004. JNK regulates HIPK3 expression and promotes resistance to fas-mediated apoptosis in DU 145 prostate carcinoma cells. J. Biol. Chem. 279:17090-17100. [DOI] [PubMed] [Google Scholar]
- 8.Dai, Y., M. Rahmani, and S. Grant. 2003. Proteasome inhibitors potentiate leukemic cell apoptosis induced by the cyclin-dependent kinase inhibitor flavopiridol through a SAPK/JNK- and NF-kB-dependent process. Oncogene 22:7108-7122. [DOI] [PubMed] [Google Scholar]
- 9.Davis, R. J. 2000. Signal transduction by the JNK group of MAP kinases. Cell 103:239-252. [DOI] [PubMed] [Google Scholar]
- 10.Derijard, B., M. Hibi, I. H. Wu, T. Barrett, B. Su, T. Deng, M. Karin, and R. J. Davis. 1994. JNK1, a protein kinase stimulated by UV light and Ha-Ras that binds and phosphorylates the c-Jun activation domain. Cell 76:1025-1037. [DOI] [PubMed] [Google Scholar]
- 11.Dunn, C., C. Wiltshire, A. MacLaren, and D. A. Gillespie. 2002. Molecular mechanism and biological functions of c-Jun N-terminal kinase signalling via the c-Jun transcription factor. Cell Signal. 14:585-593. [DOI] [PubMed] [Google Scholar]
- 12.Engelberg, D. 2004. Stress-activated protein kinases—tumor suppressors or tumor initiators? Semin. Cancer Biol. 14:271-282. [DOI] [PubMed] [Google Scholar]
- 13.Fuchs, S.Y., V. Adler, T. Buschmann, Z. Yin, X. Wu, S. N. Jones, and Z. Ronai. 1998. JNK targets p53 ubiquitination and degradation in nonstressed cells. Genes Dev. 12:2658-2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fuchs, S.Y., L. Dolan, R. J. Davis, and Z. Ronai. 1996. Phosphorylation-dependent targeting of c-Jun ubiquitination by Jun N-kinase. Oncogene 13:1531-1535. [PubMed] [Google Scholar]
- 15.Garber, M., S. Panchanathan, R. S. Fan, and D. L. Johnson. 1991. The phorbol ester, TPA induces specific transcription by RNA polymerase III in Drosophila Schneider cells. J. Biol. Chem. 266:20598-20601. [PubMed] [Google Scholar]
- 16.Garber, M. E., A. Vilalta, and D. L. Johnson. 1994. Induction of Drosophila RNA polymerase III gene expression by the phorbol ester 12-O-tetradecanoylphorbol-13-acetate (TPA) is mediated by transcription factor IIIB. Mol. Cell. Biol. 14:339-347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gupta, S., T. Barrett, A. J. Whitmarsh, J. Cavanagh, H. K. Sluss, B. Derijard, and R. J. Davis. 1996. Selective interaction of JNK protein kinase isoforms with transcription factors. EMBO J. 15:2760-2770. [PMC free article] [PubMed] [Google Scholar]
- 18.Hernandez, N. 1993. TBP, a universal eukaryotic transcription factor? Genes Dev. 7:1291-1308. [DOI] [PubMed] [Google Scholar]
- 19.Jaeschke, A., M. Karasarides, J. J. Ventura, A. Ehrhardt, C. Zhang, R. A. Flavell, K. M. Shokat, and R. J. Davis. 2006. JNK2 is a positive regulator of the cJun transcription factor. Mol. Cell 23:899-911. [DOI] [PubMed] [Google Scholar]
- 20.Johnson, S. A., L. Dubeau, M. Kawalek, A. Dervan, A. Schonthal, C. V. Dang, and D. L. Johnson. 2003. Overexpression of the TATA-binding protein contributes to oncogenesis. Mol. Cell. Biol. 23:3043-3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson, S. A., L. Dubeau, R. J. White, and D. L. Johnson. 2003. The TATA-binding protein as a regulator of cellular transformation. Cell Cycle 2:442-444. [PubMed] [Google Scholar]
- 22.Johnson, S. A., N. Mandavia, H. D. Wang, and D. L. Johnson. 2000. Transcriptional regulation of the human TATA-binding protein by Ras cellular signaling. Mol. Cell. Biol. 20:5000-5009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kallunki, T., B. Su, I. Tsigelny, H. K. Sluss, B. Derijard, G. Moore, R. J. Davis, and M. Karin. 1994. JNK2 contains a specificity-determining region responsible for efficient c-Jun binding and phosphorylation. Genes Dev. 8:2996-3007. [DOI] [PubMed] [Google Scholar]
- 24.Kennedy, N. J., and R. J. Davis. 2003. Role of JNK in tumor development. Cell Cycle 3:199-201. [PubMed] [Google Scholar]
- 24a.Li, Q. J., S. H. Yang, Y. Maeda, F. M. Sladek, A. D. Sharrocks, and M. Martins-Green. 2003. MAP kinase phosphorylation-dependent activation of Elk-1 leads to activation of the co-activator p300. EMBO J. 22:281-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu, J., Y. Minemoto, and A. Lin. 2004. c-Jun N-terminal protein kinase 1 (JNK1), but not JNK2, is essential for tumor necrosis factor alpha-induced c-Jun kinase activation and apoptosis. Mol. Cell. Biol. 24:10844-10856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Majello, B., G. Napolitano, P. De Luca, and L. Lania. 1998. Recruitment of human TBP selectively activates RNA polymerase II TATA-dependent promoters. J. Biol. Chem. 273:16509-16516. [DOI] [PubMed] [Google Scholar]
- 27.Mohit, A. A., J. H. Martin, and C. A. Miller. 1995. p493F12 kinase: a novel MAP kinase expressed in a subset of neurons in the human nervous system. Neuron 14:67-78. [DOI] [PubMed] [Google Scholar]
- 28.Sabapathy, K., K. Hochedlinger, S. Y. Nam, A. Bauer, M. Karin, and E. F. Wagner. 2004. Distinct roles for JNK1 and JNK2 in regulating JNK activity and c-Jun-dependent cell proliferation. Mol. Cell 15:713-725. [DOI] [PubMed] [Google Scholar]
- 29.Sadovsky, Y., P. Webb, G. Lope, J. D. Baxter, P. M. Fitzpatrick, E. Gizang-Ginsberg, V. Cavailles, M. G. Parker, and P. J. Kushner. 1995. Transcriptional activators differ in their responses to overexpression of TATA-box-binding protein. Mol. Cell. Biol. 15:1554-1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shaulian, E., and M. Karin. 2002. AP-1 as a regulator of cell life and death. Nat. Cell Biol. 4:E131-E136. [DOI] [PubMed] [Google Scholar]
- 31.Tian, J., and M. Karin. 1999. Stimulation of Elk1 transcriptional activity by mitogen-activated protein kinases is negatively regulated by protein phosphatase 2B (calcineurin). J. Biol. Chem. 274:15173-15180. [DOI] [PubMed] [Google Scholar]
- 32.Tournier, C., P. Hess, D. D. Yang, J. Xu, T. K. Turner, A. Nimnual, D. Bar-Sagi, S. N. Jones, R. A. Flavell, and R. J. Davis. 2000. Requirement of JNK for stress-induced activation of the cytochrome c-mediated death pathway. Science 288:870-874. [DOI] [PubMed] [Google Scholar]
- 33.Treisman, R. 1996. Regulation of transcription by MAP kinase cascades. Curr. Opin. Cell Biol. 8:205-215. [DOI] [PubMed] [Google Scholar]
- 34.Trivedi, A., A. Vilalta, S. Gopalan, and D. L. Johnson. 1996. TATA-binding protein is limiting for both TATA-containing and TATA-lacking RNA polymerase III promoters in Drosophila cells. Mol. Cell. Biol. 16:6909-6916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tsuiki, H., M. Tnani, I. Okamoto, L. C. Kenyon, D. R. Emlet, M. Holgado Madruga, I. S. Lanham, C. J. Joynes, K. T. Vo, and A. J. Wong. 2003. Constitutively active forms of c-Jun NH2-terminal kinase are expressed in primary glial tumors. Cancer Res. 63:250-255. [PubMed] [Google Scholar]
- 36.Uciechowski, P., J. Saklatvala, J. von der Ohe, K. Resch, M. Szamel, and M. Kracht. 1996. Interleukin 1 activates jun N-terminal kinases JNK1 and JNK2 but not extracellular regulated MAP kinase (ERK) in human glomerular mesangial cells. FEBS Lett. 394:273-278. [DOI] [PubMed] [Google Scholar]
- 37.Um, M., J. Yamauchi, S. Kato, and J. L. Manley. 2001. Heterozygous disruption of the TATA-binding protein gene in DT40 cells reduced cdc25B phosphatase expression and delayed mitosis. Mol. Cell. Biol. 21:2435-2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang, H.-D., A. Trivedi, and D. L. Johnson. 1997. Hepatitis B virus X protein induces RNA polymerase III-dependent gene transcription and increases cellular TATA-binding protein by activating the Ras signaling pathway. Mol. Cell. Biol. 17:6838-6846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang, H.-D., A. Trivedi, and D. L. Johnson. 1998. Regulation of RNA polymerase I-dependent promoters by the hepatitis B virus X protein, activated Ras, and the TATA-binding protein. Mol. Cell. Biol. 18:7086-7094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang, H.-D., C.-H. Yuh, C. V. Dang, and D. L. Johnson. 1995. The hepatitis B virus X protein increases the cellular level of TATA-binding protein, which mediates transactivation of RNA polymerase III genes. Mol. Cell. Biol. 15:6720-6728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40a.Whitmarsh, A. J., P. Shore, A. D. Sharrocks, and R. J. Davis. 1995. Integration of MAP kinase signal transduction pathways at the serum response element. Science 269:403-407. [DOI] [PubMed] [Google Scholar]
- 41.Yang, S. H., and A. D. Sharrocks. 2004. SUMO promotes HDAC-mediated transcriptional repression. Mol. Cell 13:611-617. [DOI] [PubMed] [Google Scholar]
- 42.Yang, S. H., P. Shore, N. Willingham, J. H. Lakey, and A. D. Sharrocks. 1999. The mechanism of phosphorylation-inducible activation of the ETS-domain transcription factor Elk-1. EMBO J. 18:5666-5674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang, S. H., A. J. Whitmarsh, R. J. Davis, and A. D. Sharrocks. 1998. Differential targeting of MAP kinases to the ETS-domain transcription factor Elk-1. EMBO J. 17:1740-1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zheng, C, L. Comai, and D. L. Johnson. 2005. PTEN represses RNA polymerase I-dependent transcription through S6 kinase by disrupting the SL1 complex. Mol. Cell. Biol. 25:6899-6911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhong, S., C. Zheng, and D. L. Johnson. 2004. Epidermal growth factor enhances cellular TATA binding protein levels and induces RNA polymerase I- and III-dependent gene activity. Mol. Cell. Biol. 24:5119-5129. [DOI] [PMC free article] [PubMed] [Google Scholar]