Abstract
DNA methylation is a major determinant of epigenetic inheritance. DNA methyltransferase 1 (DNMT1) is the enzyme responsible for the maintenance of DNA methylation patterns during cell division, and deregulated expression of DNMT1 leads to cellular transformation. We show herein that AU-rich element/poly(U)-binding/degradation factor 1 (AUF1)/heterogenous nuclear ribonucleoprotein D interacts with an AU-rich conserved element in the 3′ untranslated region of the DNMT1 mRNA and targets it for destabilization by the exosome. AUF1 protein levels are regulated by the cell cycle by the proteasome, resulting in cell cycle-specific destabilization of DNMT1 mRNA. AUF1 knock down leads to increased DNMT1 expression and modifications of cell cycle kinetics, increased DNA methyltransferase activity, and genome hypermethylation. Concurrent AUF1 and DNMT1 knock down abolishes this effect, suggesting that the effects of AUF1 knock down on the cell cycle are mediated at least in part by DNMT1. In this study, we demonstrate a link between AUF1, the RNA degradation machinery, and maintenance of the epigenetic integrity of the cell.
DNA methylation patterns are a critical component of the epigenome, controlling gene expression in vertebrates (37, 38). The enzyme DNA methyltransferase 1 (DNMT1) is responsible for maintenance and propagation of DNA methylation patterns. These patterns are altered in tumorigenesis (2, 14). The overexpression of DNMT1 in NIH 3T3 mouse fibroblasts causes cell transformation (55), while DNMT1 overexpression in human fibroblasts results in aberrant methylation of endogenous CpG islands (51). In parallel, the down regulation of DNMT1 inhibits cancer growth in animal models (20, 28, 35). On the basis of these reports, DNMT1 was therefore proposed as a target for anticancer therapy (46, 47).
As was expected from its critical role in maintaining epigenomic integrity, DNMT1 expression was shown to be controlled by cell growth (39, 48, 49). Multiple mechanisms, such as transcriptional (3, 17, 27, 29, 40), posttranscriptional (11), and posttranslational (1, 12) mechanisms, ensure a tight regulation of its expression during the cell cycle. It was suggested that deregulated expression of DNMT1 during the cell cycle might be critical for cell growth control (39, 50) and DNA replication (18, 31). Deregulated cell cycle control of DNMT1 was observed in breast cancer and colorectal cancers (10, 33).
Our previous study showed that DNMT1 3′ untranslated region (3′-UTR) contains a highly conserved noncanonical AU-rich region, which is responsible for regulating its expression level during the cell cycle (11). Deletion of this conserved region resulted in cellular transformation. We also observed binding of a protein with an apparent size of ∼40 kDa on this region, which triggered the destabilization of DNMT1 transcript in G0/G1 phase.
Using affinity capture with the 3′UTR of DNMT1 mRNA and matrix-assisted laser desorption ionization-time of flight tandem mass spectrometry (MALDI-TOF-MS-MS) analysis, we identified AU-rich element (ARE)/poly(U)-binding/degradation factor (AUF1), which is also called heterogenous nuclear ribonucleoprotein D (hnRNP D) (5, 57) and determined its role in posttranscriptional regulation of DNMT1 mRNA through the exosome. AUF1 is expressed as four isoforms (p37, p40, p42, and p45) arising through alternative splicing of a common pre-mRNA (52, 54). While differences in the activities of the various AUF1 isoforms have been documented, all isoforms enhance target mRNA decay (22, 26). AUF1 was previously shown to influence the stability of many transcripts encoding proteins involved in mitogenic stimulation, immune response, such as interleukin 10 (6), stress response, and cell cycle, such as p16 (53) and p21 (21). In particular, cyclin D1 is present at low abundance in quiescent cells but rapidly accumulates after stimulation with serum or mitogens. It is suggested that its rapid cell cycle regulation requires AUF1 binding to the 3′-UTR of this mRNA (21, 25). We describe here a cell cycle-dependent regulation of AUF1 by the proteasome. We further show that cell cycle regulation of AUF1 can posttranscriptionally control DNMT1 mRNA and is critical for maintaining the integrity of genomic methylation levels.
MATERIALS AND METHODS
Materials and antibodies.
Serum-starved HeLa cell pellets were purchased from Cilbiotech. Recombinant AUF1 protein was obtained from Upstate Biotechnology. The following antibodies were used: anti-AUF1 (Upstate Biotechnology), anti-β-actin (Sigma), and anti-DNMT1 (New England Biolabs) antibodies. hRrp40p, hRrp41p, and hRrp46p, were a generous gift from G. Schilders and G. J. Pruijn; hRrp4 was provided by D. Tollervey, and PM-Scl 75 was provided by W. J. van Venrooij. Cycloheximide was purchased from Sigma, and MG-132 and streptavidin agarose suspension were purchased from Calbiochem.
Cell culture and transfections.
HEK-293, BALB/c, and HeLa cells were maintained in Dulbecco modified Eagle medium containing 10% fetal calf serum (FCS) and antibiotics (Life Technologies). MCF7 and T24 cells were maintained in minimal essential medium and McCoy medium, respectively. GM01887 human fibroblasts, provided by the Coriell Cell Repositories (Camden, NJ), were grown in minimal essential medium containing 10% FCS. HEK-293 cells were transfected using calcium phosphate, while Lipofectamine 2000 (Invitrogen) was used for transfection of HeLa cells and Lipofectin (Invitrogen) was used for transfection of MCF7 cells, GM01887 human fibroblasts, and T24 cells. For serum starvation studies, cells were grown for 2 to 14 days in their medium plus 0.5% FCS and then released from cell cycle arrest by the addition of serum (10%) for 6 to 48 h.
Plasmids and small interfering RNA (siRNA) oligonucleotides.
Construction of pBluescript SK 3′-UTR DNMT1-poly(A) vector: the pSK Δ5′259 vector, containing the human DNMT1 (hDNMT1) 3′-UTR conserved region from positions 5349 to 5405 (GenBank accession number NM001379.1) (11) was annealed with a poly(A) primer. PCR was performed using a 3′ poly(T) primer and 5′ primer containing an hDNMT1 3′-UTR complementary region flanked by a T3 promoter sequence. The PCR product was cloned into pcDNA3.1 V5-HIS-TOPO/A (Invitrogen). The pSP64 poly(A) vector was purchased from Promega.
pFLAG-CMV2-AUF1 isoform vectors were kindly provided by R. Schneider (43), and pSilencer 2.0-U6 and pSilencer AUF15 were kindly provided by M. Gorospe (53). pSuper AUF1 and pSuper CT were generated by inserting the following sequence into HindIII/BglII pSR-Neo sites (Oligoengine), respectively: 5′-AGCTTTTCCAAAAAGATCCTATCACAGGGCGATTCTCTTGAAATCGCCCTGTATAGGATC-3′and 5′-GATCCCCGACGACGACGACGACGATGTTTTCAAGAGAAACATCGTCGTCGTCGTCGTCTTTTTGGAAA-3′. 3′-UTR DNMT1 deletion constructs were generated by PCR and inserted in frame into pcDNA3.1 His B (Invitrogen). Oligonucleotide antisense for DNMT1 was previously described (18). Control (CT) siRNA, 5′-UGGAGAGCACCGUUCUCC-3′, hRrp40 P4, PM-Scl 75 siRNA (45), and AUF1 exon 3 siRNA (34) were purchased from Dharmacon.
3′-UTR DNMT1-poly(A) vector contained the hDNMT1 3′-UTR conserved region from positions 5349 to 5405 (GenBank accession number NM001379.1) (11). pcDNA3-hRrp4-TAP vector was provided by C. Y. Chen and used as described previously (8). pFLAG-CMV2-AUF1 isoforms vectors were kindly provided by R. Schneider (43), and pSilencer 2.0-U6 and pSilencer AUF15 were kindly provided by M. Gorospe (53). pSuper AUF1 and pSuper CT was generated by inserting the following sequence into HindIII/BglII pSR-Neo sites (Oligoengine), respectively: 5′-AGCTTTTCCAAAAAGATCCTATCACAGGGCGATTCTCTTGAAATCGCCCTGTATAGGATC-3′and 5′-GATCCCCGACGACGACGACGACGATGTTTTCAAGAGAAACATCGTCGTCGTCGTCGTCTTTTTGGAAA-3′. 3′-UTR DNMT1 deletion constructs were generated by PCR and inserted in frame into pcDNA3.1 His B (Invitrogen). Oligonucleotide antisense for DNMT1 was previously described (18). CT siRNA, 5′-UGGAGAGCACCGUUCUCC-3′, hRrp40 P4, PM-Scl 75 siRNA (45), AUF1 exon 3 siRNA (34) and p21 siRNA were purchased from Dharmacon.
In vitro RNA transcription.
pSP64 poly(A) and pBluescript SK 3′-UTR DNMT1-polyA vectors were in vitro transcribed with either T3 or SP6 polymerase using the in vitro transcription kit (Ambion). For affinity chromatography, larger quantities of RNA were synthesized using the MEGAscript kit (Ambion).
RNA affinity chromatography.
Five hundred micrograms of in vitro-transcribed RNA (DNMT1 3′-UTR or control) were hybridized in incubation buffer (10 mM HEPES [pH 8.0], 3 mM MgCl2, 40 mM KCl, 20% glycerol, 1 mM dithiothreitol, complete protease inhibitors [Roche Diagnostics]) with 500 mg of oligo(dT)-cellulose beads (Sigma). Whole-cell protein extracts (1 mg) from serum-starved HeLa cells were incubated with RNA probe or oligo(dT) beads in incubation buffer supplemented with tRNA. Proteins or RNA-bead complexes were pelleted, and unbound proteins were eliminated by two 50-ml washes in the incubation buffer and five washes with 50 ml washing buffer (10 mM HEPES [pH 8.0], 3 mM MgCl2, 40 mM KCl, 20% glycerol, 1 mM dithiothreitol, 500 mM NaCl, protease inhibitors). Bound proteins were eluted by a step-wise gradient (0.8 M to 4.3 M NaCl). The fractions from each NaCl concentration were desalted and concentrated using Amicon Ultra-4 centrifugal filters (Millipore). Five microliters of the concentrated fraction was loaded onto a 15% acrylamide gel and silver stained using the Bio-Rad silver staining kit (Bio-Rad). For mass spectrometry analysis, samples were stained by mass spectrometry-compatible Coomassie blue.
Mass spectrometry (MALDI-TOF-MS-MS).
DNMT1 3′-UTR-specific binding proteins were identified by comparing the DNMT1 3′-UTR and the control lanes. Gel slices were excised and digested with porcine trypsin on a MassPrep robotic workstation (Micromass). Tryptic peptides were analyzed on a QTrap 4000 ion trap mass spectrometer (Applied Biosystems). The tryptic peptides were applied Pico Frit column containing BioBasic C18 packing. Eluted peptides were electrosprayed as they exited the column, and doubly, triply, or quadruply charged ions were selected for passage into a collision cell. MS-MS data were analyzed by BioAnalyst 1.4 software (Applied Biosystems) and submitted to Mascot (Matrix Science) for identification by analysis against the NCBI nonredundant database. MS-MS analyses were performed by the Genome Quebec Proteomic Facility (Montreal, Quebec, Canada).
RNA-protein UV cross-linking.
Twenty micrograms of cell extract or 1 to 2 μg of purified AUF1 protein was subjected to RNA UV cross-linking using the indicated RNA probes as previously described (11).
Protein and RNA immunoprecipitation.
HEK-293 cells were transfected with Flag-tagged AUF1 or hRrp4p-TAP vectors. T24 cells or transfected HEK-293 cells were lysed, and protein precipitation of the cytoplasmic fraction was performed with hRrp4p, M2 anti-Flag antibody (Sigma), or pull down using TAP purification system. hRrp4p was immunoprecipitated from T24 cells. For AUF1 immunoprecipitation, antibodies were cross-linked to protein G agarose beads (Roche Diagnostics) using dimethyl pimelimidate. RNA was prepared from the supernatants and pellets following immunoprecipitation and subjected to reverse transcription-PCR (RT-PCR) as described below.
RT-PCR and quantitative real-time PCR (q-RT-PCR).
Total RNA was extracted using RNeasy kit (QIAGEN). cDNA was synthesized, and PCR were performed as previously described (11), using the following primers: DNMT1 and β-actin (11); DNMT3A forward (5′-ACCCTCCAAAGGTTTACCCACCTG-3′), DNMT3A reverse (5′-CATACCGGGAAGGTTACCCCAGAA-3′), DNMT3B forward (5′-GACTGGACCGTGCGCCTGCAGGCC-3′), DNMT3B reverse (5′-GAAGCGACGTACTTTCCTACCTTT-3′), p16 (19), AUF1 (56), and p21 (30) The numbers of cycles were selected so that the PCR amplification remained in the linear range after a series of amplification at different numbers of cycles. Each PCR was performed in triplicate; the intensity of signal obtained for each messenger was determined by densitometry (NIH Image and normalized to the intensity of the signal obtained for β-actin. For some PCR mixtures, nucleic acids were transferred by Southern blotting to a nylon membrane. An oligonucleotide specific for the DNMT1 mRNA sequence (5′-CCTCGAGGCCTAGAAACAAA GGGAAGGGCAAG-3′) was synthesized, radiolabeled, and then hybridized to the membranes, which were exposed to PhosphorImager screens. The screens were scanned by a PhosphorImager (Molecular Dynamics). Relative optical density readings were determined using a computer-assisted densitometry program (Molecular Dynamics). Real-time PCR analysis was performed using the Roche light cycler. PCR was performed in 25-μl reaction mixtures with SYBR green (SuperArray). All the primer sets used produced no signal in control reaction mixtures lacking template. Dissociation curve analysis showed that single products with the expected melting temperature values were generated by each primer set. Standard curves were determined for each primer set by dilution of the input DNA. The amount of each cDNA was calculated from the cycle threshold for each primer set using the standard curves. The relative units recovered for each primer set were determined by dividing the calculated amount of cDNA by the amount of β-actin cDNA. The following primer sequences were used: DNMT1 forward (5′-TTTGTATGTTGGCCAAAGCCCGAG-3′), DNMT1 reverse (5′-TTCATGTCAGCCA AGGCCACAAAC-3′), DNMT3A forward (5′-GACTCCATCACGGTGGGCATGG-3′), DNMT3A reverse (5′-TGTCCCTCTTGTCAC TAACGCC-3′), DNMT3B forward (5′-GAGTCCATTGCTGTTGGAACCG-3′), DNMT3B reverse (5′-ATGTCCCTCTTGTCGCCAACCT-3′) (16), β-actin forward (5′-AGATGTGGATCAGCAAGCAGGAGT-3′), and β-actin reverse (5′-GCAATCAAAGTCC TCGGCC ACATT-3′).
In vitro RNA degradation assay.
RNA decay rates were assessed as previously described (7). Following autoradiography, the relative signal strength of the 32P-labeled RNA was quantified by two-dimensional densitometric scanning with NIH imaging system. Quantitative decay of the RNA was calculated as the percentage of signal remaining compared to signal at time zero.
Adenoviral infection.
Adenoviral vectors encoding hDNMT1 and hDNMT1 lacking its 3′-UTR as well as the adenoviral particle production and infection were previously described (11).
Northern blot analysis and DNMT1 mRNA half-life measurement.
DNMT1, GFP, and neomycin mRNA levels were analyzed by Northern blot analysis (11). After transfection with a combination of pSilencer AUF15 and pSuperAUF1 or the corresponding CT siRNA vector, HEK-293 cells were treated with actinomycin D (5 μg/ml) for the indicated time. DNMT1 and neomycin mRNA levels were estimated by Northern blotting. Quantitative decay of the RNA was calculated as the percentage of signal remaining compared to signal at time zero. The following probes were used: hDNMT1 (11), green fluorescent protein (GFP) (pEGFP C2 [Clontech]), X-press (pcDNA3.1 His B [Invitrogen]), and neomycin (pcDNA3.1 His B). In T24 cells, detection of DNMT1 mRNA after actinomycin treatment was performed by RT-PCR, followed by oligonucleotide hybridization as described previously (11), and DNMT1 mRNA was quantified by q-RT-PCR.
Flow cytometry analysis.
Cells were stained with propidium iodide, and the DNA content was measured by flow cytometry. Data were analyzed using WinMDI v2.8 software.
CAT reporter activity assays.
T24 cells were transfected with the previously described pMet-P1ΔHX-CAT plasmid (3). As a control, a plasmid with the same fragment in the opposite orientation was used. Chloramphenicol acetyltransferase (CAT) assays were performed as described previously (41).
[3H]thymidine incorporation DNA synthesis assay.
Cells were incubated for 4 h with 1 μCi/ml of [3H]thymidine (Perkin Elmer). Cells were fixed in 10% trichloroacetic acid and then lysed with 1 N NaOH-1% sodium dodecyl sulfate (SDS). Lysates were collected and applied onto a liquid scintillation cocktail. [3H]thymidine incorporation was measured using a liquid scintillation counter (1211Rackbeta-LKB Wallac).
Assay of DNA methyltransferase activity.
DNA methyltransferase activity in nuclear extracts from human fibroblasts was assayed as described previously (48).
5-Methylcytosine quantification by nearest-neighbor analysis.
5-Methylcytosine level was quantified by nearest-neighbor analysis as described previously (15, 36). The intensities of 5-methylcytosine and cytosine mononucleotide spots were measured using a phosphorimager screen and ImageQuant quantification.
Statistical analysis.
Experiments were performed in triplicate. Averages and standard deviations were calculated. A Student's t test was performed, and critical values for statistical significance (P < 0.05 and P < 0.01) are indicated.
RESULTS
Isolation and identification of AUF1/hnRNP D as a 3′-UTR DNMT1 mRNA-binding protein.
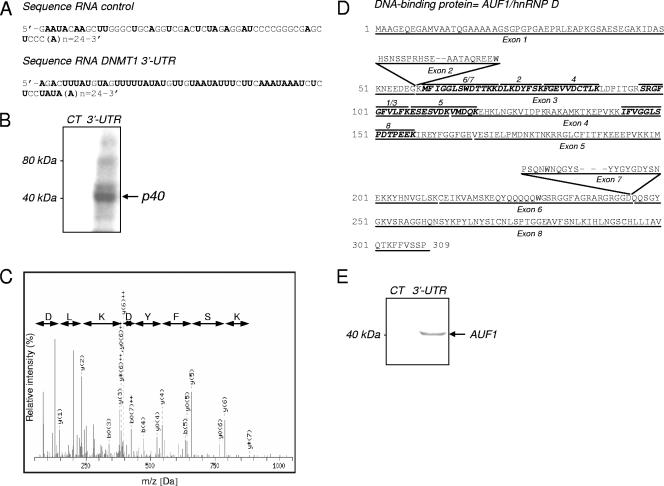
Our previous studies suggested that the depletion of DNMT1 mRNA during the G0/G1 phase of the cell cycle was mediated by a ∼40-kDa protein binding to a conserved DNMT1 3′-UTR (11). As expected, a binding protein at ∼40 kDa, which interacts specifically with the DNMT1 3′-UTR (Fig. 1A) was detected by UV cross-linking (Fig. 1B) in cytoplasmic extracts from serum-starved HeLa cells. Cell cycle regulation of DNMT1 was verified in this cell line (see Fig. S1 in the supplemental material). Using RNA affinity chromatography with either the 3′UTR sequence or the control RNA sequence as bait (Fig. 1A), we partially purified this protein from the extracts (see Fig. S2 in the supplemental material). In the 3.2 M NaCl fraction, a ∼40-kDa protein was found to interact specifically with the 3′-UTR DNMT1 RNA. This fraction was analyzed by UV cross-linking to confirm the presence of a ∼40-kDa DNMT1 3′-UTR specific-binding protein (see Fig. S3 in the supplemental material). Two bands from the 3.2 M fractions were excised together and analyzed by MALDI-TOF-MS-MS (Fig. 1C). Eight sequenced peptides corresponding to the AUF1/hnRNP D protein which were common to all of the four known AUF1 isoforms were identified (Fig. 1D and Table 1). The presence of AUF1 protein in the fractions eluted from DNMT1 3′-UTR-RNA matrix was confirmed by Western blot analysis (Fig. 1E).
FIG. 1.
Identification of AUF1/hnRNPD as a 3′-UTR DNMT1 mRNA-binding protein. (A) In vitro-transcribed RNA sequences encoding the AU-rich conserved element of DNMT1 3′-UTR (81 nt) and a control sequence (77 nt) extended by a 24-nucleotide poly(A) tail. Adenine and uracil nucleotides are shown in boldface type. (B) Cytoplasmic extracts of serum-starved HeLa cells were incubated and UV cross-linked with a 32P-labeled DNMT1 UTR (3′-UTR) or control RNA probe (CT). The position of an RNA-protein complex with an apparent size of ∼40 kDa is indicated by the arrow. (C) MS-MS spectrum of an identified peptide corresponding to the AUF1/hnRNPD sequence. (D) Peptide sequences of hnRNP D/AUF1 isoforms. Peptides sequence identified by MALDI-TOF-MS-MS are shown in boldface italic type. The positions of exons 2 and 7 are indicated above the peptide sequences. (E) Western blot analysis performed on the 3.2 M eluted fractions.
TABLE 1.
Peptides sequenced by MS-MS from the excised gel (see Fig. S2 in the supplemental material)
| Peptide | m/z submitted | MH+ matched | δppm | Start position | End position | Peptide sequence |
|---|---|---|---|---|---|---|
| 1 | 913.44 | 913.51 | −0.006 | 118 | 125 | GFGFVLFK |
| 2 | 1,014.44 | 1,014.50 | −0.006 | 91 | 98 | DLKDYFSK |
| 3 | 1,156.57 | 1,156.64 | −0.007 | 116 | 125 | SRGFGFVLFK |
| 4 | 1,166.49 | 1,166.56 | −0.007 | 99 | 108 | FGEVVDCTLK |
| 5 | 1,393.56 | 1,393.64 | −0.008 | 126 | 137 | ESESVDKVMDQK |
| 6 | 1,354.59 | 1,354.66 | −0.007 | 79 | 89 | MFIGGLSWDTTK |
| 7 | 1,482.67 | 1,482.75 | −0.009 | 78 | 90 | MFIGGLSWDTTKK |
| 8 | 1,487.62 | 1,487.75 | −0.015 | 163 | 176 | IFVGGLSPDTPEEK |
Binding of AUF1 to DNMT1 3′-UTR results in a decrease of DNMT1 expression levels by destabilizing its mRNA.
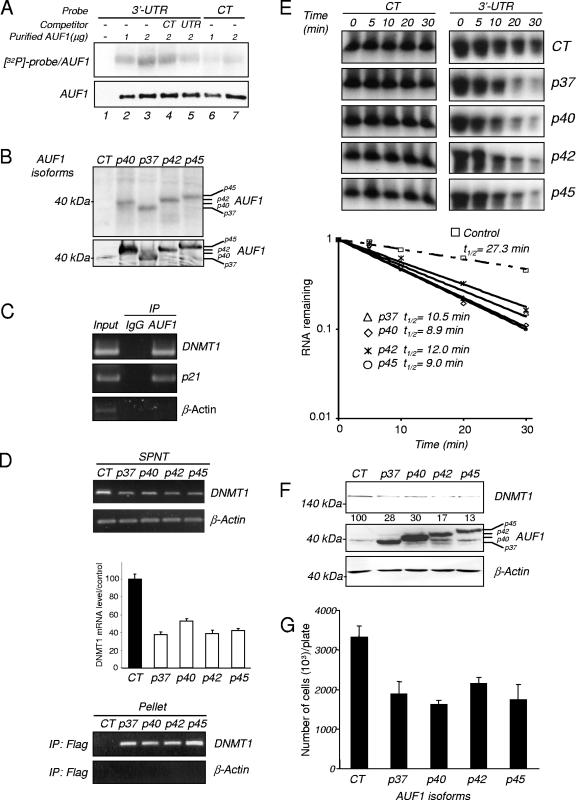
We confirmed that AUF1 binds to DNMT1 3′-UTR RNA by UV cross-linking purified AUF1 with a labeled DNMT1 3′-UTR RNA (Fig. 2A). Whereas no signal was observed with the probe alone (Fig. 2A, lane 1), AUF1 exhibited an enhanced interaction with the DNMT1 3′-UTR probe (3′-UTR) compared to the nonspecific probe (CT) (Fig. 2A, lanes 2 and 3 and lanes 6 and 7). In addition, the binding of AUF1 to the DNMT1 3′-UTR probe was weakly competed out by excess unlabeled control probe (Fig. 2A, lane 4), whereas the binding was much more effectively competed out by excess unlabeled DNMT1 3′-UTR probe (Fig. 2A, lane 5).
FIG. 2.
AUF1 binding to DNMT1 3′-UTR leads to a decrease in DNMT1 levels. (A) UV cross-linking assay. One microgram (lanes 2 and 6) or 2 μg (lanes 3 to 5 and 7) of purified AUF1 was incubated with a 32P-labeled 3′-UTR DNMT1 (3′-UTR) (lanes 2 to 5) or control RNA (CT) probe (lanes 6 and 7) and UV cross-linked. The DNMT1 3′UTR probe alone was used as a control (lane 1). RNA-protein complexes were separated on a 7.5% SDS-polyacrylamide gel and visualized by autoradiography (top gel). Purified AUF1 was incubated with the 32P-labeled 3′-UTR DNMT1 probe in the presence of 10-fold molar excess of the unlabeled control (lane 4) or 3′-UTR DNMT1 RNA probe (lane 5). The amounts of purified AUF1 per lane were controlled by Western blot analysis (bottom gel). (B) Cytoplasmic extracts of HEK-293 cells transfected with cDNAs of the different AUF1 isoforms or an empty vector (CT) were incubated in the presence of a 32P-labeled 3′-UTR DNMT1 (3′-UTR) and UV cross-linked. RNA-protein complexes were resolved by SDS-polyacrylamide gel electrophoresis and visualized by autoradiography (top gel). Overexpression of the AUF1 isoforms was verified by Western blotting (bottom gel). (C) RNA immunoprecipitation assay. AUF1 protein was immunoprecipitated from HEK-293 extracts using anti-AUF1 antibody. Immunoprecipitation (IP) using rabbit immunoglobulin G (IgG) was performed as a negative control. Immunoprecipitated RNAs were extracted and subjected to RT-PCR as well as RNA present in an aliquot of the initial extracts (Input). Detection of DNMT1 and p21 mRNA was performed by RT-PCR. β-Actin amplification was used as a negative control. (D) RNA immunoprecipitation (IP) assay. HEK-293 cells were transfected with cDNAs of the four different AUF1 isoforms or an empty vector (CT). Flag antibody-precipitated RNAs were extracted from the supernatants (SPNT) (top panel) and pellets (bottom panel). DNMT1 mRNAs were visualized by RT-PCR and quantified in SPNT samples by q-RT-PCR. β-Actin amplification was used as control. The graph represents the average percentage of DNMT1 mRNA expression in the SPNT samples relative to the control level. (E) In vitro degradation assay. Cytoplasmic extracts from transfected HEK-293 cells (CT or AUF1 cDNAs) were incubated with radiolabeled DNMT1 3′-UTR RNA transcript or control RNA probe for various lengths of time and electrophoresed. The signals were detected by autoradiography and quantified by densitometry. The half-lives were obtained by determining the time point at which 50% of the RNA had been degraded. The graph shows the half-lives of DNMT1 3′-UTR RNA probe in the presence of HEK-293 cell extracts. (F) HEK 293 cells were transfected with either the four different AUF1 isoforms or an empty vector (CT). DNMT1 (top gel) and AUF1 (middle gel) protein levels were estimated by Western blot analysis. β-Actin antibody was used as a loading control. The numbers below the DNMT1 gel indicate the percentages of DNMT1 protein expression relative to that of the control level (G) Transfected HEK-293 cells (CT or AUF1 cDNAs) were counted in the presence of trypan blue. The values presented are the averages plus standard deviations (error bars) of three measurements from three different plates.
To determine whether the different AUF1 isoforms could interact with DNMT1 mRNA in living cells, we resorted to transient transfection in human embryonic kidney HEK-293 cells which can be transfected by exogenous expression vectors at high efficiency. A UV cross-linking assay revealed that all four AUF1 isoforms bind the DNMT1 3′-UTR probe in HEK-293 cells (Fig. 2B). RNA immunoprecipitation followed by RT-PCR using AUF1 antibody revealed that AUF1 is endogenously associated with DNMT1 mRNA (Fig. 2C) as well as p21 mRNA, which is a known AUF1 target, but not with β-actin, which is not known to interact with AUF1. The same nonquantitative approach using anti-Flag antibody confirmed that endogenous DNMT1 mRNA can physically associate with all known AUF1 isoforms in living cells. Indeed, while both DNMT1 and β-actin mRNAs were detected in all the supernatant fractions (Fig. 2D, top panel), only DNMT1 mRNA was present in all tagged AUF1 immunoprecipitates (Fig. 2D, bottom panel). We further observed that AUF1 overexpression led to a reduction in the DNMT1 mRNA level (Fig. 2D, top panel).
We examined whether AUF1 altered the stability of DNMT1 mRNA. Using in vitro RNA degradation assays, we showed that extracts derived from HEK-293 cells ectopically expressing the AUF1 isoforms (p37, p40, p42, and p45) degraded DNMT1 3′-UTR RNA at an accelerated rate (half-life [t1/2]: 10.5, 8.9, 12.0, and 9.0 min, respectively) relative to control vector-transfected cell extracts that contained markedly lower levels of endogenous AUF1 (t1/2: 27.3 min) (Fig. 2E). Moreover, Western blot analysis revealed that the overexpression of AUF1 isoforms in HEK-293 cells led to a reduction of DNMT1 protein levels (Fig. 2F), which are in agreement with the mRNA levels (Fig. 2D, top panel).
Interestingly, overexpression of AUF1 isoforms, which decreased DNMT1 levels, also led to a decrease in the number of HEK-293 cells (Fig. 2G). This is consistent with previous data which showed that DNMT1 was part of the DNA replication complex (42) and that inhibition of DNMT1 inhibited DNA replication and cell growth (18).
Down regulation of AUF1 induces DNMT1 expression by stabilizing its mRNA.
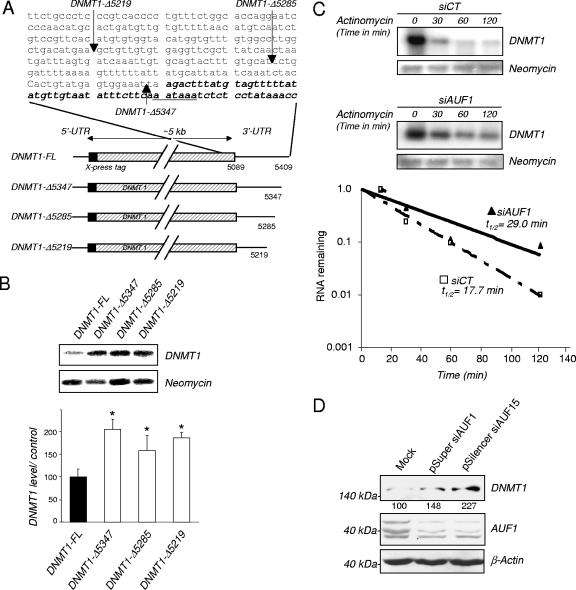
To ascertain the cellular role of the 3′-UTR in mediating destabilization of DNMT1 mRNA by AUF1, different X-press-tagged DNMT1 3′-UTR deletion constructs were transiently transfected into HEK-293 cells (Fig. 3A). Northern blot analysis revealed that the deletion of the 54-nucleotide (nt) AUF1 binding sequence flanked by an additional sequence 64 nt upstream (construct DNMT1-Δ5285) increased the steady-state mRNA levels (Fig. 3B). The strict deletion of the AUF1 binding site (construct DNMT1-Δ5347) also resulted in a similar increase, which strongly suggests that the AUF1 binding region in the DNMT1 3′-UTR confers instability to the DNMT1 mRNA. We also determined the effect of AUF1 depletion mediated by AUF1 siRNA on the half-life of DNMT1 mRNA. As visualized by Western blotting (data not shown), down regulation of the level of AUF1 protein extended the half-life of DNMT1-FL mRNA (t1/2: 29.0 min) compared to the half-life in control cells (t1/2: 17.7 min) (Fig. 3C). AUF1 knock down also increased the levels of endogenous DNMT1 protein levels (Fig. 3D).
FIG. 3.
AUF1 depletion stabilizes DNMT1 mRNA. (A) Schematic representations of DNMT1 3′-UTR deletion constructs and DNMT1 3′-UTR sequence (GenBank accession number NM001379.1). The AU-rich highly conserved region of DNMT1 3′-UTR is indicated by boldface italic type. (B) The levels of expression of the different DNMT1 3′-UTR deletion construct mRNAs were estimated by Northern blotting using an X-press tag probe normalized by hybridization with neomycin as the loading control probe. The graph indicates the percentage of normalized DNMT1 mRNA expression of the deletion constructs relative to the normalized expression of the full-length construct DNMT1 mRNA (100%). Values that were statistically significantly different from the DNMT1-FL value as tested by the Student t test at a P of <0.05 are indicated by an asterisk. (C) DNMT1 mRNA half-life measurements. The DNMT1-FL construct was cotransfected into HEK-293 cells with plasmids encoding AUF1 siRNA (siAUF1) or control siRNA (siCT). Cells were treated with actinomycin D for the indicated period of time. X-press-tagged DNMT1 and neomycin mRNA levels were determined by Northern blot analysis, and the normalized levels of DNMT1-FL were calculated. The autoradiogram is representative of three different assays. (D) HEK-293 cells were transfected with pSuper siAUF1, pSilencer siAUF15, or mock vector. Proteins were extracted, and the levels of DNMT1 and AUF1 were determined by Western blot analysis. Measurement of β-actin levels was used as a loading control. The values below the DNMT1 gel are the percentages of normalized DNMT1 (to β-actin) relative to the control levels.
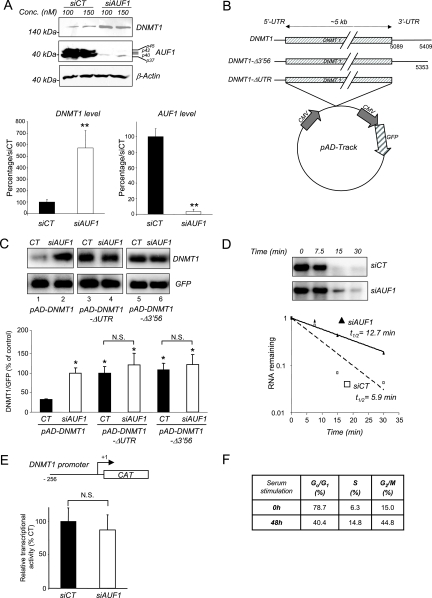
We further confirmed that AUF1 down regulates DNMT1 mRNA using a different human cell line, bladder carcinoma T24 cell line, in which DNMT1 was shown to be regulated by the cell cycle (39). We verified by RNA immunoprecipitation that AUF1 is also associated with endogenous DNMT1 mRNA in this cell line (data not shown). A siRNA knock down of AUF1 (Fig. 4A, middle panel) resulted in an increase in endogenous DNMT1 protein levels (Fig. 4A, top panel). We then examined whether the effect of AUF1 required the 3′-UTR. T24 cells were infected with adenoviral vectors expressing DNMT1 mRNA that either contain the entire 3′-UTR (pAD-DNMT1) or lack this region (pAD-DNMT1-ΔUTR). An adenoviral vector expressing DNMT1 mRNA with a UTR lacking the AUF1 binding site (pAD-DNMT1-Δ3′56) was also used (11) (Fig. 4B). Northern blot analysis indicated that DNMT1 transcripts that lacked the 3′-UTR or the AUF1 binding site were more abundant than those containing the entire 3′-UTR (Fig. 4C, lanes 1, 3, and 5). Moreover, AUF1 depletion increased the level of DNMT1 mRNA containing the 3′-UTR (Fig. 4C, lanes 1 and 2) but had no significant effect on the level of DNMT1 mRNA that does not contain the 3′-UTR (Fig. 4C, lanes 3 and 4) or the AUF1 binding site (Fig. 4C, lanes 5 and 6). These data show that AUF1 modulation of DNMT1 mRNA expression in T24 cells requires the 3′-UTR. In addition, an in vitro degradation assay revealed an increased half-life of the DNMT1 3′-UTR RNA probe incubated with a cytosolic extract from AUF1-depleted T24 cells from 5.9 to 12.7 min (Fig. 4D), confirming that, in T24 cells, AUF1 is involved in destabilization of the DNMT1 transcripts.
FIG.4.
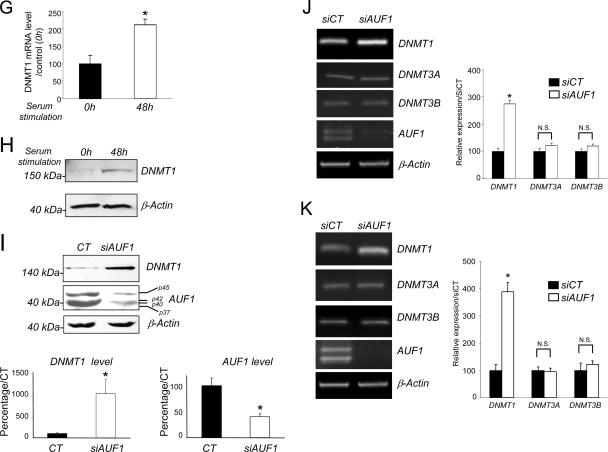
AUF1 depletion leads to an increased expression and increased stability of DNMT1 mRNA but not other DNMT mRNAs. (A) T24 cells were transfected with AUF1 siRNA (siAUF1) or control siRNA (siCT). Levels of DNMT1 and AUF1 proteins were determined by Western blot analysis. The graph shows averages plus standard errors of the means (error bars) for three independent determinations. Values that were statistically significantly different from the value for siCT by the Student t test at a P of <0.05 are indicated (**). (B) Schematic representations of the adenoviral constructs pAd-DNMT1, pAd-DNMT1-Δ3′56, and pAD-DNMT1-ΔUTR. CMV, cytomegalovirus. (C) AUF1-depleted or wild-type T24 cells were infected with the indicated adenoviral constructs. DNMT1 and GFP mRNA levels were measured by Northern blot analysis. DNMT1 mRNA levels were normalized to GFP mRNA as a control for infection efficiency. The graph shows the averages plus standard errors of the means (error bars) for three different adenoviral infections relative to the control pAD-DNMT1 siAUF1 (100%). Statistical significance (indicated by an asterisk) was tested by the Student t test at a P of <0.05. Values that were not statistically significantly different (N.S.) are indicated by the brackets. (D) In vitro degradation assay. Radiolabeled DNMT1 3′-UTR RNA transcript was incubated with cytoplasmic extracts from AUF1 siRNA-transfected T24 cells (siAUF1) or control siRNA-transfected cells (siCT) for the indicated lengths of time. The signals were detected by autoradiography and quantified by phosphorimaging densitometry. (E) Analysis of DNMT1 promoter activity. pMet-P1ΔHX-CAT construct was transfected into AUF1-depleted T24 cells (siAUF1) or siRNA control cells (siCT). CAT activity was measured as described in Materials and Methods. Each value represents the mean plus standard deviation (error bar) for three independent transfections. The Student t test confirmed no statistical difference (N.S.) between the values for the treatment groups. (F) Percentages of human fibroblasts in the different stages of the cell cycle after serum starvation (14 days) and serum supplementation (serum release) (48 h) were determined by flow cytometry. (G) Human fibroblasts were serum starved (14 days) and released to enter into the cell cycle for the indicated period of time. The level of DNMT1 mRNA was determined by q-RT-PCR. β-Actin was used as control. Statistical significance was tested by a Student t test. The difference for DNMT1 was significant (P < 0.05). (H) DNMT1 protein level in serum-starved or serum-released fibroblasts was determined by Western blotting. β-Actin was used as loading control. (I) Human fibroblasts were transfected with AUF1 siRNA (siAUF1) or control siRNA (siCT). Levels of DNMT1 and AUF1 were determined by Western blot analysis. (J and K) Measurement of DNMT1, DNMT3A, and DNMT3B mRNA levels in AUF1-depleted T24 cells (J) and human fibroblasts (K) following AUF1 depletion by reverse transcription and q-RT-PCR analyses. Graphs represent the averages plus standard deviations (error bars) from three different amplifications. Statistical significance was tested by a Student t test. The difference for DNMT1 was significant (P < 0.05 [*]). No statistical significance (N.S.) was observed for the other DNMT mRNAs (P > 0.5).
In order to exclude the possibility that AUF1 could also act at a transcription level by indirectly modulating DNMT1 promoter activity, we tested whether AUF1 depletion would increase hDNMT1 promoter-driven CAT activity (Fig. 4E) (3). There was no observed change in DNMT1 promoter activity.
To test whether the regulation of DNMT1 by AUF1 is specific to cancer cells or whether it also applies to nontransformed cells, we investigated the effects of knocking down AUF1 protein on DNMT1 expression in nontransformed human skin GM01887 fibroblasts. We first established that DNMT1 expression was also regulated by the cell cycle in human fibroblasts. Indeed, culture in serum-starved conditions for 14 days led to an arrest of a large population of cells in G0/G1 phase measured by fluorescence-activated cell sorting (FACS) analysis, whereas a subsequent serum supplementation for 48 h triggered an entry into S/G2M phase (Fig. 4F). DNMT1 mRNA and protein expression was measured in these two populations and demonstrated that DNMT1 was also regulated in a cell cycle-dependent manner in this human fibroblast cell line (Fig. 4G and H). AUF1 down regulation triggered a marked increase in DNMT1 protein as determined by Western blotting (Fig. 4I, top panel), indicating that AUF1 also controls DNMT1 level in nontransformed cells.
AUF1 specifically destabilizes DNMT1 mRNA in T24 cells and human fibroblasts and not other DNMT mRNAs.
It was previously shown that DNMT3A and DNMT3B expression and DNMT1 expression are regulated by the cell cycle in T24 cells (39). We found no significant changes in DNMT3A and DNMT3B levels in either T24 (Fig. 4J) or human fibroblast cells (Fig. 4K) upon AUF1 knock down. This indicates that AUF1 selectively targets DNMT1 mRNA. The cell cycle regulation of DNMT3A and DNMT3B must entail other cell-regulating mechanisms.
Biological consequences of AUF1 depletion on DNA replication are partially mediated by DNMT1 in T24 cells.
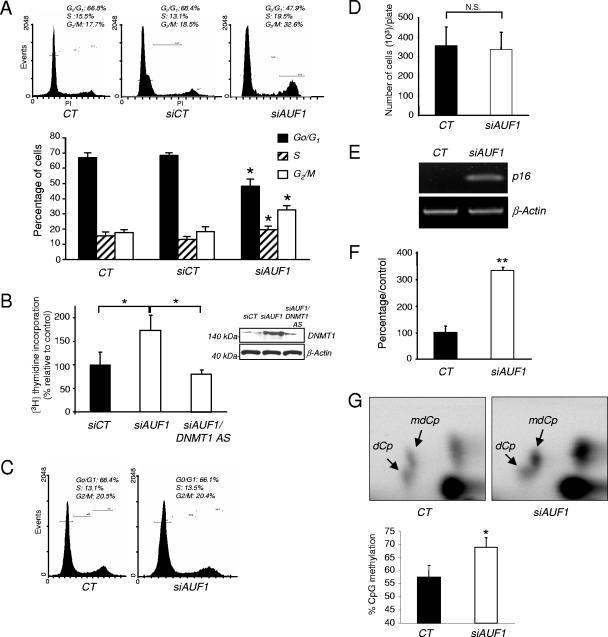
We previously showed that an acute knock down of DNMT1 leads to arrest of firing of the origin of replication (18) and results in an intra-S-phase arrest of DNA replication (31). We therefore tested whether AUF1 knock down would affect cell cycle kinetics (Fig. 5A). AUF1 depletion in T24 cells resulted in an increase in the fraction of cells in the S and G2/M phases compared to control siRNA-transfected cells (siCT) (19.5% in S and 32.6% in G2/M and 13.1% in S and 18.5% in G2/M, respectively) and a decrease in the number of cells in G0/G1 (47.9% versus 68.4%). AUF1 regulates a number of transcripts important for cell cycle regulation, such as p16 (53) and p21 (21). We determined whether DNMT1 was a downstream effector of AUF1 action on DNA replication by concurrent knock down of AUF1 and DNMT1. AUF1 knock down stimulated DNA synthesis as predicted from the FACS analysis, and this increase was diminished by concurrent DNMT1 knock down with a DNMT1 antisense oligonucleotide (18) (Fig. 5B). The fact that double knock down of AUF1 and DNMT1 did not result in an increase in DNA replication suggests that AUF1 knock down effects on DNA replication require the presence of DNMT1. In the absence of DNMT1, AUF1 does not induce DNA replication. However, this does not rule out the possibility that the effect of the double knock down could be a combination of two independent effects as well. On the other hand, simultaneous knock down of p21 and AUF1 led to stimulation of DNA replication and increased the fraction of cells in the S phase of the cell cycle (see Fig. S4 in the supplemental material). Thus, the effects of AUF1 knockdown on the cell cycle are dependent on different effectors. Some of the AUF1 targets, such as DNMT1, enhance entry into the S phase, while others, such as p16 and p21, repress cell cycle progression.
FIG. 5.
Biological consequences of AUF1 depletion in T24 cells and human fibroblasts. (A) The fraction of untransfected (CT) cells or cells transfected with a control siRNA (siCT) or AUF1 siRNA (siAUF1) at different stages of the cell cycle was determined by flow cytometry. The graph represents the averages from three different experiments. Values that were statistically significantly different from the control values as tested by the Student t test at a P of <0.05 are indicated (*). (B) T24 cells were transfected with CT siRNA (siCT), AUF1 siRNA (siAUF1), or a combination of AUF1 siRNA and DNMT1 antisense (DNMT1 AS) oligonucleotide. [3H] thymidine uptake was measured as described in Materials and Methods. The results are averages plus standard deviations (error bars) from three independent experiments. Values that are statistically significantly different (*) as tested by a Student t test at a P of <0.05 are indicated by the brackets. The levels of DNMT1 after the different treatments were measured by Western blot analysis. (C) The percentage of AUF1 siRNA (siAUF1)- or CT siRNA (siCT)-transfected human fibroblasts at different stages of the cell cycle was determined by flow cytometry. (D) AUF1 siRNA (siAUF1)- or CT siRNA (siCT)-transfected human fibroblasts were counted 3 days posttransfection. The number of cells represents the average plus standard deviation (error bar) for three measurements from three different plates. A Student t test showed no statistically significant difference (N.S.) between the values for the treatment groups at a P of >0.5. (E) p16 mRNA expression in AUF1 siRNA (siAUF1)- or CT siRNA (siCT)-transfected human fibroblasts was evaluated by semiquantitative RT-PCR. This amplification is representative of three different experiments. (F) DNA methyltransferase assay. DNA methyltransferase activity in AUF1 siRNA (siAUF1)- or CT siRNA (siCT)-transfected human fibroblasts was measured as described in Materials and Methods. The graph represents the averages plus standard deviations (error bars) from three different estimations. An asterisk indicates statistical significance as tested by a Student t test at a P of <0.05. (G) The level of CpG methylation in AUF1 siRNA (siAUF1)- or CT siRNA (siCT)-transfected human fibroblasts was measured by nearest-neighbor analysis. The graph represents the average plus standard deviations (error bars) from three different determinations. An asterisk indicates statistical significance as tested by a Student t test at a P of <0.05.
AUF1 depletion induces DNMT1 expression in human fibroblasts and increases global DNA methylation.
Since AUF1 depletion in T24 cells modified the cell cycle, we next asked whether a similar effect would be observed in nontransformed cells. Interestingly, up regulation of DNMT1 generated by an AUF1 knock down observed in Fig. 4I did not result in a distinct change in cell cycle kinetics (Fig. 5C) or cell proliferation (Fig. 5D) in nontransformed human fibroblasts.
However, ectopic expression of DNMT1 was previously shown to transform immortalized mouse fibroblast cells (55). Since an AUF1 knock down did not result in cellular transformation of human fibroblasts (data not shown), we hypothesized that knocking down AUF1 would unleash other control mechanisms that would override DNMT1 action on cell cycle kinetics. One possibility is that AUF1 knock down activates tumor suppressor genes that counteract the impact of increased DNMT1. It was previously shown that AUF1 depletion triggers p16 gene expression in human fibroblasts (53). We confirmed that AUF1 knock down induced tumor suppressor gene p16 mRNA expression in these particular human fibroblasts (Fig. 5E).
Next, we tested whether knocking down AUF1 would result in an overall increase in DNA methylation of genomic DNA, since DNMT1 expression is elevated in the absence of concomitant increase in DNA synthesis. We first showed that DNA methyltransferase activity on hemimethylated DNA substrate in nuclear extract prepared from AUF1 siRNA-treated cells was elevated in comparison with extracts from control siRNA-treated cells (Fig. 5F). This increase in DNA methyltransferase activity was associated with a significant global increase in CpG methylation level as measured by nearest-neighbor analysis (Fig. 5G). These data suggest that AUF1 depletion leads to an increased methylation capacity of human fibroblasts and increased genomic methylation by up regulating DNMT1 mRNA. Our earlier findings that DNMT3A and DNMT3B are not regulated by AUF1 (Fig. 4J and K) are consistent with the hypothesis that the increased DNA methyltransferase activity and global methylation observed are primarily caused by increased DNMT1 mRNA and protein.
AUF1 expression is inversely correlated with DNMT1 expression during the cell cycle.
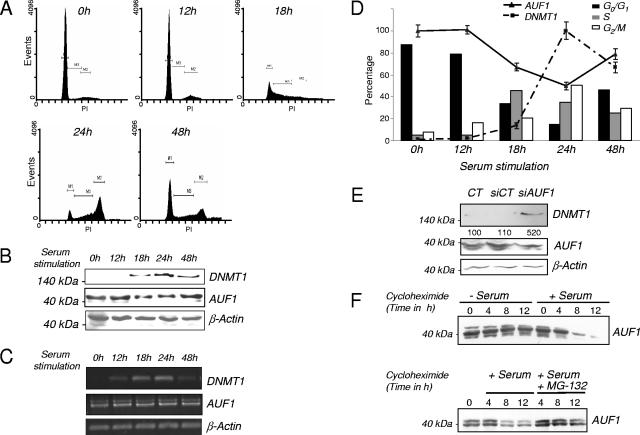
Our in vitro and cell culture experiments showed that AUF1 interacted with DNMT1 mRNA to negatively regulate its expression. We tested the hypothesis that AUF1 was regulated in a cell cycle-dependent manner which regulated, in turn, DNMT1 mRNA expression in T24 cells. T24 cells were arrested by serum starvation, and entry into the cell cycle was induced by serum supplementation. The fractions of cells at the different stages of the cell cycle were determined by FACS analysis (Fig. 6A). AUF1 and DNMT1 protein levels during the progression of the cell cycle were measured by Western blot analysis (Fig. 6B) and RT-PCR (Fig. 6C). As seen in Fig. 6A, serum deprivation arrested a large fraction of the cells at G0/G1. Serum supplementation induced entry into the S phase of the cell cycle. As expected, an increase in the level of DNMT1 protein was observed 18 h after serum stimulation (Fig. 6B) and reached a maximum level after 24 h. In contrast, a simultaneous decrease in AUF1 protein level was observed upon entry into the cell cycle. After 48 h, the levels of DNMT1 and AUF1 proteins returned to basal levels. In contrast to AUF1 protein levels, no changes were observed in AUF1 mRNA concentration during the cell cycle (Fig. 6C), indicating that AUF1 regulation in the course of the cell cycle is most likely posttranslational. The graph in Fig. 6D shows the levels of DNMT1 mRNA and AUF1 in cell populations at different stages of the cell cycle. This cell cycle regulation of AUF1 was also observed in other healthy and cancer cell lines from human and mouse origin (see Fig. S5 in the supplemental material), suggesting that this mechanism of regulation of AUF1 is not an idiosyncrasy of T24 cells. siRNA knock down of AUF1 in serum-starved cells increased DNMT1 levels, suggesting that the higher levels of AUF1 observed in G0/G1 cells are, at least in part, responsible for the down regulation of DNMT1 during G0/G1 (Fig. 6E). Our data are therefore consistent with the hypothesis that the changes in DNMT1 during the cell cycle could be partially caused by inverse cell cycle regulation of AUF1 levels.
FIG. 6.
AUF1 expression is inversely correlated with DNMT1 level during the cell cycle and is controlled by the proteasome. (A) The percentages of T24 cells in the different stages of the cell cycle were determined by flow cytometry. (B) T24 cells were serum starved and released to enter the cell cycle for the indicated lengths of time. DNMT1 and AUF1 protein levels were measured by Western blot analysis. β-Actin was used as a loading control. (C) T24 cells were serum starved and released to enter the cell cycle for the indicated period of time. DNMT1 and AUF1 mRNA levels were visualized by semiquantitative RT-PCR. (D) Graphical representation of DNMT1 and AUF1 protein levels and the percentage of T24 cells at the different stages of the cell cycle at different time points after serum release (serum supplementation). DNMT1 mRNA level was measured by q-RT-PCR. (E) Serum-starved T24 cells (CT) were transfected with CT siRNA (siCT) or AUF1 siRNA (siAUF1). DNMT1 and AUF1 protein levels were analyzed by Western blotting. The numbers below the DNMT1 gel indicate the levels of DNMT1 protein as a percentage of the control value. (F) In vivo degradation assay of AUF1 protein at G0/G1 and S phases of the cell cycle. Serum-starved (− Serum) or 18-h serum-released (+ Serum) T24 cells were treated with cycloheximide for the indicated period of time. Levels of AUF1 were then determined by Western blotting (top gel). Eighteen-hour serum-released cells treated with cycloheximide were also treated with MG-132, a proteasome inhibitor (bottom gel). AUF1 levels were determined by Western blotting.
The proteasome machinery is responsible for an increased AUF1 degradation rate during S phase.
Since we observed a decrease in AUF1 protein levels during S phase, which was not explained by reduced transcription of AUF1 mRNA, we determined whether differences in protein stability were responsible. We therefore compared AUF1 protein stability after a cycloheximide block of de novo protein synthesis in serum-starved T24 cells (mostly in Go/G1 phase) and in 18-h serum-released T24 cells (mostly in S phase). This analysis revealed a shorter AUF1 half-life of AUF1 in the S-phase cell population (Fig. 6F, top panel) than in the serum-starved cells. Several studies have previously demonstrated that AUF1 isoforms were subjected to ubiquitination-mediated degradation by the proteasome machinery (22-24). We observed that the AUF1 degradation rate in the S-phase cell population was clearly diminished by the addition of the proteosome inhibitor MG-132 (Fig. 6F, bottom panel), suggesting the participation of the proteasome in this cell cycle-dependent regulation. Furthermore, we wanted to evaluate the consequences of MG-132 treatment on DNMT1 mRNA in T24 cells. Unfortunately, upon further investigation, it was determined that MG-132 treatment itself significantly modified the cell cycle and would bias the measurement of cell cycle-regulated mRNAs, such as DNMT1 mRNA (see Fig. S6 in the supplemental material). Nevertheless, these MG-132 effects were not observed in the T24 cell populations in which protein translation was chemically blocked by cycloheximide (see Fig. S7 in the supplemental material).
The exosome participates in the AUF1-triggered degradation of DNMT1 mRNA.
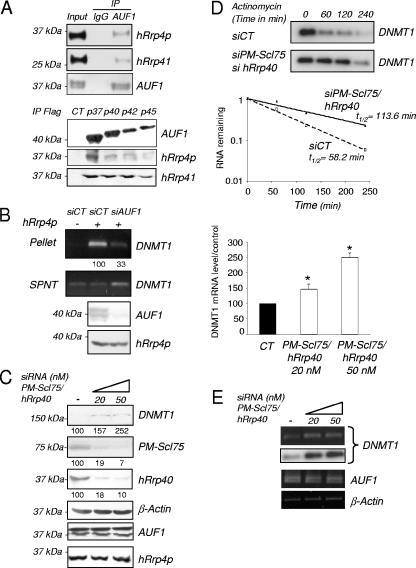
AUF1 was shown to interact with exosome and to be responsible for the targeting of ARE-containing mRNA to this RNA degradation machinery (8). Immunoprecipitation experiments confirmed a physical interaction between the four AUF1 isoforms and some component of the exosome complex, such as hRrp4p or hRrp41 (Fig. 7A). In parallel, immunoprecipitation of hRrp4p in T24 cells showed the presence of DNMT1 mRNA in the precipitated complex (Fig. 7B). As anticipated if AUF1 were to mediate the interaction between the 3′-UTR and the exosome, an AUF1 knockdown led to a reduced amount of DNMT1 mRNA detected in the hRrp4p immunoprecipitate (Fig. 7B). These data demonstrate for the first time that AUF1 links DNMT1 mRNA and the exosome complex and provide a mechanism for how AUF1 affects DNMT1 mRNA stability.
FIG.7.
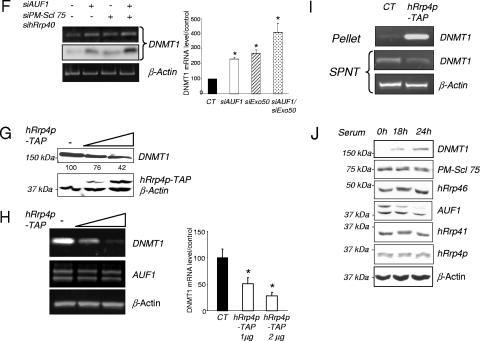
The exosome participates in the AUF1-triggered degradation of DNMT1 mRNA. (A) AUF1 proteins were immunoprecipitated from HEK-293 whole-cell extracts. The presence of hRrp4p and hRrp41 in the immunoprecipitates was determined by Western blot analysis (top panel). A preimmune antibody (immunoglobulin G [IgG]) was used as control for immunoprecipitation (IP). HEK-293 cells were transfected with the different tagged AUF1 cDNA-encoding vectors. AUF1 isoforms were immunoprecipitated using a Flag antibody (bottom panel). The presence of hRrp4p and hRrp41 in the immunocomplexes was determined by Western blotting. (B) hRrp4p was immunoprecipitated from T24 cells treated (+) with either control (SiCT) or AUF1 siRNA (SiAUF1). Precipitated RNAs were extracted from the supernatants (SPNT) and pellets. DNMT1 mRNA was detected by RT-PCR and quantified by q-RT-PCR. The numbers below the pellet DNMT1 gel indicate the levels of DNMT1 mRNA expression relative to the level measured in siCT (as a percentage). The levels of AUF1 and hRrp4p following AUF1 siRNA treatment were determined by Western blotting. (C) T24 cells were simultaneously transfected with PM-Scl 75/hRrp40 siRNAs at the indicated concentrations (0 [−], 20, or 50 nM). Levels of the indicated proteins were determined by Western blotting. Numbers indicate the levels as a percentage of DNMT1, PM-Scl75, and hRrp40 protein expression relative to the control value. (D) RNA degradation assay. Control or PM-Scl 75/hRrp40 siRNA-transfected T24 cells were treated with actinomycin D for the indicated period of time. DNMT1 mRNA levels were visualized by RT-PCR followed by specific oligonucleotide hybridization and quantified using q-RT-PCR. The graph shows the results of quantification for three different experiments. (E) DNMT1 and AUF1 mRNA levels corresponding to the experiment shown in panel C were visualized by RT-PCR analysis and quantified by real-time PCR. The graph to the right of the gels shows the level as a percentage of DNMT1 mRNA relative to the control level. An asterisk indicates statistical significance as tested by a Student t test at a P of <0.05. (F) T24 cells were separately and simultaneously transfected with PM-Scl 75/hRrp40 or AUF1 siRNAs. The level of DNMT1 mRNA was visualized by RT-PCR (top gel) followed by specific oligonucleotide hybridization (middle gel) and quantified using q-RT-PCR. The graph shows the results of quantification by real-time PCR of DNMT1 mRNA levels relative to the control value. An asterisk indicates statistical significance as tested by a Student t test at a P of <0.05. (G) HEK-293 cells were transiently transfected with hRr4p-TAP vector at different concentrations. DNMT1 and hRrp4p protein levels were determined by Western blotting. (H) DNMT1 and AUF1 mRNA levels corresponding to the experiment shown in panel G were visualized by RT-PCR analysis. The graph shows the results of quantification of DNMT1 mRNA levels by q-RT-PCR relative to the control value (100%). An asterisk indicates statistical significance as tested by a Student t test at a P of <0.05. (I) Pull down of hRrp4p-TAP protein from hRrp4p-transfected HEK-293 cells. Precipitated RNAs were extracted from the supernatants (SPNT) and pellets. Detection of DNMT1 mRNA was performed by RT-PCR. β-Actin amplification was used as a control. (J) T24 cells were serum starved for 6 days (0 h) and supplemented with serum for 18 h and 24 h. Levels of the indicated protein were determined by Western blotting.
To confirm the role of the exosome in DNMT1 mRNA degradation, we decided to disrupt exosome function using a siRNA strategy. In brief, we targeted PM-Scl 75 protein, a core exonuclease of the exosome (45) and hRrp40, another component of the exosome (Fig. 7C). The consequence of this exosome knock down in T24 cells was a slow down in the DNMT1 mRNA degradation rate (Fig. 7D), subsequently resulting in an increase in the level of protein (Fig. 7C) and mRNA expression (Fig. 7E). In addition, combined knock down of the two DNMT1 mRNA-destabilizing agents, AUF1 and the exosome, potentiated DNMT1 mRNA expression (Fig. 7F). Inversely, hRrp4 overexpression of the exosome protein hRrp4p led to a decrease of DNMT1 protein (Fig. 7G) and mRNA expression (Fig. 7H) in HEK-293 cells. Pull down of the hRrp4p protein revealed the presence of DNMT1 mRNA, indicating that DNMT1 mRNA is physically associated with this component of the exosome in HEK-293 cells (Fig. 7I).
Since we showed that AUF1 protein level varied during the cell cycle, we also measured the levels of expression of different components of the exosome. Western blot analysis did not show dramatic changes in terms of protein levels during the cell cycle (Fig. 7J). We therefore hypothesized that the AUF1 protein variations during the cell cycle may control, at least in part, the relative amount of DNMT1 mRNA or protein during the cell cycle.
DISCUSSION
AUF1 was first characterized on the basis of its property of binding AU-rich elements and was shown to affect mRNA stability in a number of genes involved in growth control. The observation that DNMT1 is regulated by AUF1, a factor that also regulates a number of other growth control genes, such as p16 (53) and p21 (21), provides a mechanism of coordination of DNMT1 levels with other cell cycle events. Since DNMT1 has a critical function in copying the epigenetic information during cell division, it is crucial that its expression is tightly coordinated with the state of cell division.
The manner by which AUF1 maintains a dynamic balance of growth control by coordinating the expression of genes with opposite functions is illustrated by the AUF1 siRNA knock down experiments. AUF1 knockdown in nontransformed human fibroblasts results in an induction of DNMT1 mRNA and protein levels (Fig. 4K and I), which has been shown to have a growth-promoting effect in other human cell lines. However, the AUF1 knock down also results in an induction of the tumor suppressor p16 gene, which may counter the growth-promoting effects of DNMT1 (Fig. 5A and E). Therefore, by coordinately destabilizing the expression of both tumor suppressor genes (p16 [53] and p21 [21]) and growth-promoting genes, such as DNMT1, AUF1 may maintain balanced growth. In other words, AUF1 effects on the cell cycle may result from a simultaneous destabilization of target mRNA encoding proteins with important function in the cell cycle regulation. It is therefore tempting to speculate that the ability of DNMT1 to transform cells requires the elimination of the counteracting action of tumor suppressor genes by regional hypermethylation, which was shown to be a late consequence of DNMT1 overexpression (51), or by mutation.
In this study, we demonstrated for the first time that AUF1 expression itself is regulated by the cell cycle in T24 cells (Fig. 6B) and other cell lines (see Fig. S5 in the supplemental material) at a posttranslational level. An in vivo degradation assay revealed a higher degradation rate of AUF1 in S-phase cells, which is dependent on proteasome activity. This observation complements previous findings for the role of the proteasome machinery in AUF1 degradation (22-24). However, although all four AUF1 isoforms are regulated by the cell cycle, an interesting question raised by our data is why all AUF1-controlled (ARE-containing) mRNAs are not inversely regulated by the cell cycle. An explanation might be the dual role of AUF1, in either stabilizing or destabilizing mRNAs depending on the (ARE-containing) mRNA sequence context (9). Another possible explanation is that other proteins are involved in regulating AUF1 to certain AU-containing mRNAs. One recent example is the Pin1 protein, a cis-trans isomerase which was recently shown to regulate the association of AUF1 isoforms with the granulocyte-macrophage colony-stimulating factor mRNA, accelerating or slowing mRNA decay (44). Moreover, we do not exclude the possibility that other ARE-binding proteins, such as HuR, could participate in DNMT1 mRNA decay.
The exosome functions in several processes involving the 3′-to-5′ processing or degradation of RNA. Among them are the maturation of 5.8 S rRNA, the processing of many small nuclear and nucleolar RNAs, and the turnover of different type of mRNAs, especially the ARE-containing mRNA (8, 32). We show that a similar mechanism is involved in DNMT1 regulation by AUF1. Since the levels of different exosome elements do not vary during the cell cycle, we suggest that modulation of AUF1 expression is mostly responsible for the cell cycle-specific targeting of DNMT1 mRNA to the exosome and its degradation. The regulation of DNMT1 mRNA stability described here is to our knowledge, the first example of a cell cycle-dependent regulation of an mRNA implicating the exosome machinery.
Finally, we demonstrate that depletion of AUF1 protein in nontransformed human fibroblasts leads to increased levels of DNMT1 protein and global genomic methylation (Fig. 4I and 5G). DNMT1 protein overexpression and tumor suppressor gene hypermethylation characterize a number of different tumors (2, 14), and these elevated levels are believed to contribute directly to tumorigenicity. Changes in AUF1 protein expression have been observed in tumor progression in neoplastic lung tissue (4), which could have potential implications for DNMT1 mRNA regulation. Moreover, Morello's group stated that the “AUF1 p37 transgene induces tumors in mice” (13). These interesting findings suggest that in certain cell types, the AUF1 p37 isoform negatively controls more tumor suppressor gene mRNAs than growth-promoting gene mRNAs. The overproduction of the AUF1 p37 in these mice might create an increased destabilization of these cell cycle repressor gene mRNAs, resulting in aberrant cell growth.
Although many different mechanisms are believed to contribute to enhanced DNMT1 expression, we show for the first time that alteration of posttranscriptional regulation and the exosome function could play an important role in maintenance of the genome DNA methylation level and therefore tumorigenesis.
Supplementary Material
Acknowledgments
This work was supported by a grant from National Cancer Institute of Canada. Jerome Torrisani was supported by a fellowship of the Fondation pour la Recherche Médicale (France).
We thank R. Schneider (Department of Microbiology, New York University School of Medicine) and M. Gorospe (Laboratory of Cellular and Molecular Biology, NIH,Baltimore, MD) for kindly providing pFLAG-CMV2-AUF1 (p37, p40, p42, and p45) expression plasmids and pSilencer 2.0-U6/AUF15 constructs, respectively. We also thank G. Schilders andG. J. Pruijn (Department of Biochemistry, Nijmegen, The Netherlands), D. Tollervey (Wellcome Trust Centre for Cell Biology, University of Edinburgh), and W. J. van Venrooij (Department of Biochemistry, Nijmegen, The Netherlands) for providing the hRrp4p-TAP vector and the hRrp4p, hRrp40, hRrp41, hRrp46, and PM-Scl 75 antibodies. We thank Jing Ni Ou and Nadine Provençal for their technical assistance and Costandina Arvanitis for critical reading of the manuscript.
We declare no conflicts of interests.
Footnotes
Published ahead of print on 9 October 2006.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Agoston, A. T., P. Argani, S. Yegnasubramanian, A. M. De Marzo, M. A. Ansari-Lari, J. L. Hicks, N. E. Davidson, and W. G. Nelson. 2005. Increased protein stability causes DNA methyltransferase 1 dysregulation in breast cancer. J. Biol. Chem. 280:18302-18310. [DOI] [PubMed] [Google Scholar]
- 2.Baylin, S. B., and J. G. Herman. 2000. DNA hypermethylation in tumorigenesis: epigenetics joins genetics. Trends Genet. 16:168-174. [DOI] [PubMed] [Google Scholar]
- 3.Bigey, P., S. Ramchandani, J. Theberge, F. D. Araujo, and M. Szyf. 2000. Transcriptional regulation of the human DNA methyltransferase (dnmt1) gene. Gene 242:407-418. [DOI] [PubMed] [Google Scholar]
- 4.Blaxall, B. C., L. D. Dwyer-Nield, A. K. Bauer, T. J. Bohlmeyer, A. M. Malkinson, and J. D. Port. 2000. Differential expression and localization of the mRNA binding proteins, AU-rich element mRNA binding protein (AUF1) and Hu antigen R (HuR), in neoplastic lung tissue. Mol. Carcinog. 28:76-83. [PubMed] [Google Scholar]
- 5.Brewer, G. 1991. An A+U-rich element RNA-binding factor regulates c-myc mRNA stability in vitro. Mol. Cell. Biol. 11:2460-2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brewer, G., S. Saccani, S. Sarkar, A. Lewis, and S. Pestka. 2003. Increased interleukin-10 mRNA stability in melanoma cells is associated with decreased levels of A + U-rich element binding factor AUF1. J. Interferon Cytokine Res. 23:553-564. [DOI] [PubMed] [Google Scholar]
- 7.Buzby, J. S., G. Brewer, and D. J. Nugent. 1999. Developmental regulation of RNA transcript destabilization by A + U-rich elements is AUF1-dependent. J. Biol. Chem. 274:33973-33978. [DOI] [PubMed] [Google Scholar]
- 8.Chen, C. Y., R. Gherzi, S. E. Ong, E. L. Chan, R. Raijmakers, G. J. Pruijn, G. Stoecklin, C. Moroni, M. Mann, and M. Karin. 2001. AU binding proteins recruit the exosome to degrade ARE-containing mRNAs. Cell 107:451-464. [DOI] [PubMed] [Google Scholar]
- 9.Dean, J. L., G. Sully, A. R. Clark, and J. Saklatvala. 2004. The involvement of AU-rich element-binding proteins in p38 mitogen-activated protein kinase pathway-mediated mRNA stabilisation. Cell Signal. 16:1113-1121. [DOI] [PubMed] [Google Scholar]
- 10.De Marzo, A. M., V. L. Marchi, E. S. Yang, R. Veeraswamy, X. Lin, and W. G. Nelson. 1999. Abnormal regulation of DNA methyltransferase expression during colorectal carcinogenesis. Cancer Res. 59:3855-3860. [PubMed] [Google Scholar]
- 11.Detich, N., S. Ramchandani, and M. Szyf. 2001. A conserved 3′-untranslated element mediates growth regulation of DNA methyltransferase 1 and inhibits its transforming activity. J. Biol. Chem. 276:24881-24890. [DOI] [PubMed] [Google Scholar]
- 12.Ding, F., and J. R. Chaillet. 2002. In vivo stabilization of the Dnmt1 (cytosine-5)-methyltransferase protein. Proc. Natl. Acad. Sci. USA 99:14861-14866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gouble, A., S. Grazide, F. Meggetto, P. Mercier, G. Delsol, and D. Morello. 2002. A new player in oncogenesis: AUF1/hnRNPD overexpression leads to tumorigenesis in transgenic mice. Cancer Res. 62:1489-1495. [PubMed] [Google Scholar]
- 14.Herman, J. G. 1999. Hypermethylation of tumor suppressor genes in cancer. Semin. Cancer Biol. 9:359-367. [DOI] [PubMed] [Google Scholar]
- 15.Jackson, M., A. Krassowska, N. Gilbert, T. Chevassut, L. Forrester, J. Ansell, and B. Ramsahoye. 2004. Severe global DNA hypomethylation blocks differentiation and induces histone hyperacetylation in embryonic stem cells. Mol. Cell. Biol. 24:8862-8871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jinawath, A., S. Miyake, Y. Yanagisawa, Y. Akiyama, and Y. Yuasa. 2005. Transcriptional regulation of the human DNA methyltransferase 3A and 3B genes by Sp3 and Sp1 zinc finger proteins. Biochem. J. 385:557-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kimura, H., T. Nakamura, T. Ogawa, S. Tanaka, and K. Shiota. 2003. Transcription of mouse DNA methyltransferase 1 (Dnmt1) is regulated by both E2F-Rb-HDAC-dependent and -independent pathways. Nucleic Acids Res. 31:3101-3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Knox, J. D., F. D. Araujo, P. Bigey, A. D. Slack, G. B. Price, M. Zannis-Hadjopoulos, and M. Szyf. 2000. Inhibition of DNA methyltransferase inhibits DNA replication. J. Biol. Chem. 275:17986-17990. [DOI] [PubMed] [Google Scholar]
- 19.Kondo, Y., L. Shen, and J. P. Issa. 2003. Critical role of histone methylation in tumor suppressor gene silencing in colorectal cancer. Mol. Cell. Biol. 23:206-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laird, P. W., L. Jackson-Grusby, A. Fazeli, S. L. Dickinson, W. E. Jung, E. Li, R. A. Weinberg, and R. Jaenisch. 1995. Suppression of intestinal neoplasia by DNA hypomethylation. Cell 81:197-205. [DOI] [PubMed] [Google Scholar]
- 21.Lal, A., K. Mazan-Mamczarz, T. Kawai, X. Yang, J. L. Martindale, and M. Gorospe. 2004. Concurrent versus individual binding of HuR and AUF1 to common labile target mRNAs. EMBO J. 23:3092-3102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Laroia, G., R. Cuesta, G. Brewer, and R. J. Schneider. 1999. Control of mRNA decay by heat shock-ubiquitin-proteasome pathway. Science 284:499-502. [DOI] [PubMed] [Google Scholar]
- 23.Laroia, G., B. Sarkar, and R. J. Schneider. 2002. Ubiquitin-dependent mechanism regulates rapid turnover of AU-rich cytokine mRNAs. Proc. Natl. Acad. Sci. USA 99:1842-1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laroia, G., and R. J. Schneider. 2002. Alternate exon insertion controls selective ubiquitination and degradation of different AUF1 protein isoforms. Nucleic Acids Res. 30:3052-3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin, S., W. Wang, G. M. Wilson, X. Yang, G. Brewer, N. J. Holbrook, and M. Gorospe. 2000. Down-regulation of cyclin D1 expression by prostaglandin A2 is mediated by enhanced cyclin D1 mRNA turnover. Mol. Cell. Biol. 20:7903-7913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Loflin, P., C. Y. Chen, and A. B. Shyu. 1999. Unraveling a cytoplasmic role for hnRNP D in the in vivo mRNA destabilization directed by the AU-rich element. Genes Dev. 13:1884-1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.MacLeod, A. R., J. Rouleau, and M. Szyf. 1995. Regulation of DNA methylation by the Ras signaling pathway. J. Biol. Chem. 270:11327-11337. [DOI] [PubMed] [Google Scholar]
- 28.MacLeod, A. R., and M. Szyf. 1995. Expression of antisense to DNA methyltransferase mRNA induces DNA demethylation and inhibits tumorigenesis. J. Biol. Chem. 270:8037-8043. [DOI] [PubMed] [Google Scholar]
- 29.McCabe, M. T., J. N. Davis, and M. L. Day. 2005. Regulation of DNA methyltransferase 1 by the pRb/E2F1 pathway. Cancer Res. 65:3624-3632. [DOI] [PubMed] [Google Scholar]
- 30.Milutinovic, S., J. D. Knox, and M. Szyf. 2000. DNA methyltransferase inhibition induces the transcription of the tumor suppressor p21(WAF1/CIP1/sdi1). J. Biol. Chem. 275:6353-6359. [DOI] [PubMed] [Google Scholar]
- 31.Milutinovic, S., Q. Zhuang, A. Niveleau, and M. Szyf. 2003. Epigenomic stress response. Knockdown of DNA methyltransferase 1 triggers an intra-S-phase arrest of DNA replication and induction of stress response genes. J. Biol. Chem. 278:14985-14995. [DOI] [PubMed] [Google Scholar]
- 32.Mukherjee, D., M. Gao, J. P. O'Connor, R. Raijmakers, G. Pruijn, C. S. Lutz, and J. Wilusz. 2002. The mammalian exosome mediates the efficient degradation of mRNAs that contain AU-rich elements. EMBO J. 21:165-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nass, S. J., A. T. Ferguson, D. El-Ashry, W. G. Nelson, and N. E. Davidson. 1999. Expression of DNA methyl-transferase (DMT) and the cell cycle in human breast cancer cells. Oncogene 18:7453-7461. [DOI] [PubMed] [Google Scholar]
- 34.Raineri, I., D. Wegmueller, B. Gross, U. Certa, and C. Moroni. 2004. Roles of AUF1 isoforms, HuR and BRF1 in ARE-dependent mRNA turnover studied by RNA interference. Nucleic Acids Res. 32:1279-1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ramchandani, S., A. R. MacLeod, M. Pinard, E. von Hofe, and M. Szyf. 1997. Inhibition of tumorigenesis by a cytosine-DNA, methyltransferase, antisense oligodeoxynucleotide. Proc. Natl. Acad. Sci. USA 94:684-689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ramsahoye, B. H. 2002. Nearest-neighbor analysis. Methods Mol. Biol. 200:9-15. [DOI] [PubMed] [Google Scholar]
- 37.Razin, A. 1998. CpG methylation, chromatin structure and gene silencing—a three-way connection. EMBO J. 17:4905-4908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Razin, A., and M. Szyf. 1984. DNA methylation patterns. Formation and function. Biochim. Biophys. Acta 782:331-342. [DOI] [PubMed] [Google Scholar]
- 39.Robertson, K. D., K. Keyomarsi, F. A. Gonzales, M. Velicescu, and P. A. Jones. 2000. Differential mRNA expression of the human DNA methyltransferases (DNMTs) 1, 3a and 3b during the G0/G1 to S phase transition in normal and tumor cells. Nucleic Acids Res. 28:2108-2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rouleau, J., A. R. MacLeod, and M. Szyf. 1995. Regulation of the DNA methyltransferase by the Ras-AP-1 signaling pathway. J. Biol. Chem. 270:1595-1601. [DOI] [PubMed] [Google Scholar]
- 41.Rouleau, J., G. Tanigawa, and M. Szyf. 1992. The mouse DNA methyltransferase 5′-region. A unique housekeeping gene promoter. J. Biol. Chem. 267:7368-7377. [PubMed] [Google Scholar]
- 42.Rountree, M. R., K. E. Bachman, and S. B. Baylin. 2000. DNMT1 binds HDAC2 and a new co-repressor, DMAP1, to form a complex at replication foci. Nat. Genet. 25:269-277. [DOI] [PubMed] [Google Scholar]
- 43.Sarkar, B., Q. Xi, C. He, and R. J. Schneider. 2003. Selective degradation of AU-rich mRNAs promoted by the p37 AUF1 protein isoform. Mol. Cell. Biol. 23:6685-6693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shen, Z. J., S. Esnault, and J. S. Malter. 2005. The peptidyl-prolyl isomerase Pin1 regulates the stability of granulocyte-macrophage colony-stimulating factor mRNA in activated eosinophils. Nat. Immunol. 6:1280-1287. [DOI] [PubMed] [Google Scholar]
- 45.Stoecklin, G., T. Mayo, and P. Anderson. 2006. ARE-mRNA degradation requires the 5′-3′ decay pathway. EMBO Rep. 7:72-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Szyf, M. 1994. DNA methylation properties: consequences for pharmacology. Trends Pharmacol. Sci. 15:233-238. [DOI] [PubMed] [Google Scholar]
- 47.Szyf, M. 2003. Targeting DNA methylation in cancer. Ageing Res. Rev. 2:299-328. [DOI] [PubMed] [Google Scholar]
- 48.Szyf, M., V. Bozovic, and G. Tanigawa. 1991. Growth regulation of mouse DNA methyltransferase gene expression. J. Biol. Chem. 266:10027-10030. [PubMed] [Google Scholar]
- 49.Szyf, M., F. Kaplan, V. Mann, H. Giloh, E. Kedar, and A. Razin. 1985. Cell cycle-dependent regulation of eukaryotic DNA methylase level. J. Biol. Chem. 260:8653-8656. [PubMed] [Google Scholar]
- 50.Szyf, M., D. J. Knox, S. Milutinovic, A. D. Slack, and F. D. Araujo. 2000. How does DNA methyltransferase cause oncogenic transformation? Ann. N. Y. Acad. Sci. 910:156-177. [DOI] [PubMed] [Google Scholar]
- 51.Vertino, P. M., R. W. Yen, J. Gao, and S. B. Baylin. 1996. De novo methylation of CpG island sequences in human fibroblasts overexpressing DNA (cytosine-5-)-methyltransferase. Mol. Cell. Biol. 16:4555-4565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wagner, B. J., C. T. DeMaria, Y. Sun, G. M. Wilson, and G. Brewer. 1998. Structure and genomic organization of the human AUF1 gene: alternative pre-mRNA splicing generates four protein isoforms. Genomics 48:195-202. [DOI] [PubMed] [Google Scholar]
- 53.Wang, W., J. L. Martindale, X. Yang, F. J. Chrest, and M. Gorospe. 2005. Increased stability of the p16 mRNA with replicative senescence. EMBO Rep. 6:158-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wilson, G. M., and G. Brewer. 1999. The search for trans-acting factors controlling messenger RNA decay. Prog. Nucleic Acid Res. Mol. Biol. 62:257-291. [DOI] [PubMed] [Google Scholar]
- 55.Wu, J., J. P. Issa, J. Herman, D. E. Bassett, Jr., B. D. Nelkin, and S. B. Baylin. 1993. Expression of an exogenous eukaryotic DNA methyltransferase gene induces transformation of NIH 3T3 cells. Proc. Natl. Acad. Sci. USA 90:8891-8895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yasuda, S., S. Wada, Y. Arao, M. Kogawa, F. Kayama, and S. Katayama. 2004. Interaction between 3′ untranslated region of calcitonin receptor messenger ribonucleic acid (RNA) and adenylate/uridylate (AU)-rich element binding proteins (AU-rich RNA-binding factor 1 and Hu antigen R). Endocrinology 145:1730-1738. [DOI] [PubMed] [Google Scholar]
- 57.Zhang, W., B. J. Wagner, K. Ehrenman, A. W. Schaefer, C. T. DeMaria, D. Crater, K. DeHaven, L. Long, and G. Brewer. 1993. Purification, characterization, and cDNA cloning of an AU-rich element RNA-binding protein, AUF1. Mol. Cell. Biol. 13:7652-7665. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.