Abstract
Cellular information is encoded genetically in the DNA nucleotide sequence and epigenetically by the “histone code,” DNA methylation, and higher-order packaging of DNA into chromatin. Cells possess intricate mechanisms to sense and repair damage to DNA and the genetic code. However, nothing is known of the mechanisms, if any, that repair and/or compensate for damage to epigenetically encoded information, predicted to result from perturbation of DNA and histone modifications or other changes in chromatin structure. Here we show that primary human cells respond to a variety of small molecules that perturb DNA and histone modifications by recruiting HP1 proteins to sites of altered pericentromeric heterochromatin. This response is essential to maintain the HP1-binding kinetochore protein hMis12 at kinetochores and to suppress catastrophic mitotic defects. Recruitment of HP1 proteins to pericentromeres depends on histone H3.3 variant deposition, mediated by the HIRA histone chaperone. These data indicate that defects in pericentromeric epigenetic heterochromatin modifications initiate a dynamic HP1-dependent response that rescues pericentromeric heterochromatin function and is essential for viable progression through mitosis.
The eukaryotic genome is packaged into the cell nucleus in the form of chromatin, a composite of DNA, RNA, and protein. The basic repeating unit of chromatin structure is the core nucleosome, comprised of eight histone protein molecules and 146 bp of DNA. Nucleosomes are folded into multiple higher levels of organization, together with other proteins and RNA, to form chromatin (16). In chromatin, a complex array of histone modifications, histone variants, associated proteins, RNAs, higher-order folding conformations, and site-specific DNA methylation controls DNA accessibility and recruitment of specific regulatory molecules, thereby regulating most DNA functions, including transcription, replication, recombination, and repair. For example, histone acetylation acts to open up chromatin, thereby promoting transcription, and also recruits specific transcription activators, such as Brg1, through interactions between acetylated histones and bromodomains in the protein module (29).
Information that is not directly coded by the DNA sequence but instead is encoded by DNA methylation, histone modifications, or other aspects of chromatin structure, is epigenetically determined. The histone modifications that contribute to epigenetic information have been proposed to constitute a histone code, by analogy to the genetic code (68). Cells possess elaborate mechanisms to sense and repair damage to the genetic code (19). Extending the analogy between the genetic code and the histone code, if histone and DNA modifications encode information outside of the DNA sequence, the cell is likely to have mechanisms to sense and repair, or otherwise respond to, alterations in these modifications. How cells respond to alteration of chromatin and epigenetic modifications is poorly understood.
Kinetochores are large macromolecular protein assemblies on chromosomes (31) that serve as attachment sites for the spindle microtubules that pull apart the sister chromatids during mitosis. Foundation proteins—CenpA, CenpB, CenpC, CenpH, CenpI, and hMis12—set the location of the kinetochores. These proteins form the base of the kinetochore complex that ultimately includes proteins required for the initiation, maintenance, and monitoring of microtubule attachments, as well as suppression of chromosome segregation until all attachments are properly formed (31). Depending on the specific chromosome, human kinetochores are built on hundreds to thousands of kilobases of repetitive centromeric α-satellite DNA. Flanking these α-satellite repeats are additional extended repeat sequences, pericentromeric DNA. Pericentromeric repeats contain α-satellites, γ-satellites, satellite III sequences, and interspersed L1 LINE retrotransposons. Despite the sequence similarity underlying kinetochores, the existence of neocentromeres—fully functional kinetochores at noncanonical sites with no DNA sequence similarity to canonical centromeres—indicates that the assembly of kinetochores is primarily defined by the chromatin structure and not DNA (31).
Recent studies have defined a direct link between kinetochore assembly and function and the underlying chromatin structure. The pericentromeric DNA repeats that flank the centromeric α-satellite DNA are incorporated into compact constitutive heterochromatin. This pericentromeric heterochromatin contains specific histone modifications, such as a high level of methylated lysine 9 of histone H3 (H3K9Me) and low levels of acetylated lysine 9 of histone H3 (H3K9Ac) and methylated lysine 4 of histone H3 (H3K4Me) (31). In addition, pericentromeric heterochromatin is enriched in a family of low-molecular-weight proteins—HP1α, HP1β, and HP1γ—that bind to heterochromatin through an interaction with H3K9Me of heterochromatin and/or through the histone variant H2AZ (49) and/or an unknown RNA component (3, 18, 24, 32, 37). HP1 proteins are thought to contribute to the transcriptional silencing of heterochromatin (39, 41, 50, 53). In addition, HP1 proteins at pericentromeres physically interact with the kinetochore foundation protein, hMis12. Consequently, HP1 proteins are required for the recruitment of hMis12 to kinetochores and also to suppress formation of aberrant micronuclei which are thought to result from defective chromosome segregation in mitosis (40).
In recent years, much has been learnt of the mechanisms that protect a cell's genetic integrity, from studies with radiation and small molecules, such as hydroxyurea, aphidicolin, and methyl methanesulfonate, which induce genotoxic stress (19). In an analogous fashion, we investigated here the response of primary human cells to a panel of small molecules that disrupt DNA and histone modifications and, presumably, epigenetically encoded information. Specifically, we have utilized several histone deacetylase inhibitors and an inhibitor of DNA methylation, all of which antagonize formation of heterochromatin (25). In response to perturbation of heterochromatin by these small molecules, primary human cells mount a dynamic mobilization of HP1 proteins to further enrich these proteins at the pericentromeric regions. Relocation of these proteins depends upon deposition of the histone variant H3.3 in the pericentromeric chromatin, mediated by the histone chaperone HIRA. Although inactivation of HP1 proteins on their own had no detectable effect on the cells, simultaneous perturbation of heterochromatin with small molecules and inactivation of HP1 proteins caused gross defects in kinetochore structure. Specifically, kinetochores were depleted of the HP1-binding protein, hMis12. Ultimately, these cells died after aberrant progression through mitosis, with defects characteristic of defective kinetochore function. We conclude that perturbation of epigenetic heterochromatin modifications causes dynamic recruitment of HP1 proteins to pericentromeres, so as to maintain kinetochore structure and function through the recruitment of hMis12 to kinetochores. These results indicate that HP1 proteins are essential players in a dynamic “repair-like” response to perturbation of heterochromatin that preserves the functional integrity of heterochromatin.
MATERIALS AND METHODS
Cell culture, drug treatment, and plasmid construction.
Cells were cultured according to the American Type Culture Collection, in 2% (approximating physiological) or 21% (ambient) oxygen as indicated. All histone deacetylase inhibitors (HDIs) and 5-aza-2′-deoxycytidine (AzaC) were from Sigma and applied at the indicated doses and durations. All plasmids were generated by using standard molecular biology procedures; details are available on request.
FACS, TUNEL, immunofluorescence, and immunofluorescence-fluorescence in situ hybridization (FISH).
Fluorescence-activated cell sorting (FACS) and TUNEL (terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling) assays were performed as previously published (15). Anti-HP1α was a gift from William Earnshaw. Anti-HP1β (Chemicon), anti-HP1γ (Chemicon), anti-H3 (Upstate), and anti-acetyl-H3 (Upstate) were from the indicated suppliers. Anti-Bub1 and anti-macroH2A antibodies have been described previously (9). Anti-hMis12 antibody was raised in rabbit by using purified hMis12 protein as described previously (40). ACA antibody was a gift from J. B. Rattner, University of Calgary. Two- and three-color immunofluorescence was performed as described previously (74).
Immunofluorescence-FISH was performed as previously described (55). Briefly, after regular immunostaining, slides were fixed in 4% paraformaldehyde in phosphate-buffered saline for 10 min at room temperature and washed three times with double-distilled H2O and then further fixed in methanol-acetic acid (3:1) for 15 min at room temperature. The slides were dehydrated by sequential immersion in 70, 90, and 100% ethanol at room temperature for 2 min for each treatment, followed by complete drying of the slide at room temperature. The DNA was denatured by immersion of the slides in 83°C 70% formamide in 2× SCC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) for 4 min, followed by dehydrating the slides immediately in −20°C 70, 90, and 100% ethanol (2 min for each treatment). The slides were then dried completely at room temperature. The pericentromeric satellite III probe was PCR amplified from genomic DNA using 5′-TCCCTTTCGAGTCCATTCAATG-3′ (forward) and 5′-GATCATCATCGAATGGACCC--3′ (reverse) primers with Pfu polymerase and biotin labeled with Bioprimer DNA labeling kit (Invitrogen). The probe was dissolved in 50% formamide in 2× SSC solution and hybridized overnight at 37°C. After hybridization, the slides were washed once in 50% formamide in 2× SSC for 15 min at 43°C, followed by two washes in 2× SSC for 5 min each time at room temperature. The signal was detected by avidin-fluorescein isothiocyanate (Vector Laboratories) binding and amplified by biotinylated anti-avidin (Vector Laboratories), followed by another layer of avidin-FITC.
Image collection was done by examining optical sections obtained with a confocal microscope or digital images obtained using an epifluorescence microscope and a cooled charge-coupled device camera. The details of these procedures are available on request.
ChIP analysis.
Chromatin immunoprecipitation (ChIP) analyses were performed as described previously (43). Cross-linking was performed with 1% formaldehyde. For each immunoprecipitation, 10 μg of antibody was used to immunoprecipitate sonicated chromatin from 5 × 105 cells. The anti-HIRA (D34 and WC15) antibodies were as described previously (15). Anti-HP1γ (Upstate), anti-acetyl-H3 (Upstate), anti-H3K9Me2 (Abcam), anti-H3K9Me3 (Abcam), anti-H3 (Abcam), and anti-HA (Y11; Santa Cruz) antibodies were from the indicated suppliers. The PCR primer sequences for pericentromeric repetitive satellite III were 5′-TCCCTTTCGAGTCCATTCAATG-3′ (forward) and 5′-GATCATCATCGAATGGACCC--3′ (reverse). PCR was performed with Pfu polymerase (Stratagene).
siRNAs, nucleofection, and retroviruses.
All small interfering RNAs (siRNAs) were purchased from Dharmacon. The sequences were as follows: siHIRA#2, sense strand, 5′-GGAGAUGACAAACUGAUUAUU-3′; siHIRA#4, sense strand, 5′-GAAAUUCUAGCUACUCUGAUU-3′; siH3.3A, sense strand, 5′-CUACAAAAGCCGCUCGCAAUU-3′; and siH3.3B, sense strand, 5′-GCUAAGAGAGUCACCAUCAUU-3′. Nucleofection of siRNAs, pBos-H2B-GFP, and pBCHGN-GFP-HP1γ was performed according to the manufacturer's instructions (Amaxa Biosystems). The following plasmids were used to generate retroviruses: pQCXIP-H2B-GFP, pQXCIP-HA-HP1β(103-185) (HP1βΔN), pQCXIP-HA-HP1β, pQCXIP-HA-H3.1, and pQCXIP-HA-H3.3. Retrovirus-mediated gene transfer was performed as described previously (74), using Phoenix cells to make the infectious viruses (Gary Nolan, Stanford University). Virus infections were performed at a multiplicity of infection of approximately 1.
Live cell imaging.
H2B-green fluorescent protein (GFP) was expressed in WI38 cells by nucleofection or retrovirus-mediated gene transfer. Cells were seeded the day before imaging. Two hours before imaging, the medium was changed from regular Dulbecco modified Eagle medium to phenol-free and HEPES-buffered Dulbecco modified Eagle medium. HDIs were added at the indicated time prior to imaging. Immediately prior to imaging, the surface of the medium was covered by mineral oil. Images were acquired automatically at multiple locations by using an inverted Nikon TE2000 microscope fitted with a ×10 Plan Fluor objective lens using a Cascade 650 monochrome camera (Photometrics). The microscope was housed in a 37°C chamber. Fluorescence and differential interference contrast images were obtained every 10 min for a period of 24 to 36 h. The images were acquired and processed by using MetaMorph software (Universal Imaging/Molecular Devices). Scale bars were set at 10 μm.
RESULTS
Chromatin perturbation recruits additional HP1 proteins to pericentromeres.
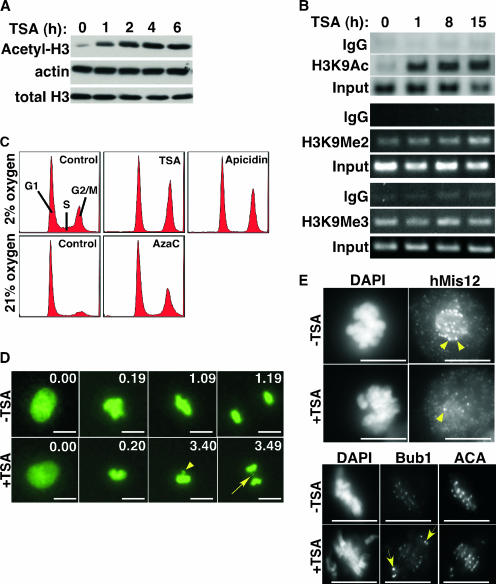
To investigate the cellular response to the perturbation of histone and DNA modifications, we made use of a panel of chromatin-disrupting agents (CDAs), namely, HDIs, such as trichostatin A (TSA), sodium butyrate, apicidin, and valproic acid (which increase histone hyperacetylation), as well as a DNA methylation inhibitor, AzaC, which decreases genomic DNA methylation (25). Just as small molecules such as hydroxyurea, aphidicolin, and methyl methanesulfonate are extensively used to induce genotoxic stress (19), all of these CDAs are predicted to perturb chromatin, specifically by antagonizing formation of heterochromatin. As expected, when applied to primary human WI38 fibroblasts, each of the HDIs caused a rapid increase in total genomic histone H3 acetylation (Fig. 1A and data not shown). Moreover, repetitive pericentromeric satellite III sequences (54), DNA sequences that are normally incorporated into constitutive heterochromatin containing hypoacetylated histones, showed a dramatic increase in acetylated histone H3 by ChIP assay (Fig. 1B). In contrast, HDIs induced no change in abundance of H3K9Me2 and H3K9Me3 at pericentromeres (Fig. 1B), a finding consistent with previous conclusions of Bickmore and coworkers (11). As reported previously (48, 73), after 24 h of treatment all of the HDIs induced a marked accumulation of cells in G2/M phase of the cell cycle (Fig. 1C and data not shown). Similarly, AzaC caused an increase in the G2/M phase of the cell cycle with a concomitant decrease in the level of genome wide DNA methylation, after 72 h of treatment (Fig. 1C and data not shown). To define the cause of the G2/M phase accumulation, we introduced a plasmid encoding H2B fused to GFP at its C terminus into WI38 cells by nucleofection, allowing us to visualize real-time chromosome dynamics in living cells. Time-lapse images showed that control cells took 1 h to progress from prometaphase to telophase, whereas TSA-treated cells took 4 h to make the same transition (Fig. 1D). Moreover, the TSA-treated cells showed “slow-to-align” or “unaligned” chromosomes prior to anaphase and “lagging” chromosomes during anaphase (Fig. 1D). These mitotic defects were accompanied by defects in kinetochore structure, as indicated by partial depletion of the inner kinetochore protein hMis12 (40), and activation of the mitotic spindle checkpoint, as indicated by enrichment of Bub1 at unattached kinetochores (31) (Fig. 1E). Together, these data indicate that perturbation of chromatin by CDAs disrupts kinetochore structure and microtubule-kinetochore interactions and consequently causes a checkpoint-dependent mitotic delay.
FIG. 1.
Chromatin-disrupting agents delay progression through M-phase, accompanied by defects in kinetochore structure and activation of the mitotic checkpoint. (A) WI38 cells were treated without and with TSA (1 μM) and Western blotted to detect total acetylated histone H3. (B) Same as in panel A, but sonicated chromatin was subjected to ChIP with anti-acetyl H3, anti-H3K9Me2, or anti-H3K9Me3 antibodies as indicated to detect associated pericentromeric satellite III DNA sequence. (C) FACS analysis of WI38 cells treated with 1 μM TSA and 8 μM apicidin for 24 h and 5 μM AzaC for 72 h in either 21% (ambient) or 2% (approximating physiological) oxygen. WI38 cells grow faster in 2% oxygen and so have a larger G2/M population in the absence of CDAs. (D) Time-lapse images of H2B-GFP-expressing WI38 cells treated with or without TSA (1 μM). The yellow arrowhead points to an unaligned chromosome in metaphase, and the yellow arrow points to a lagging chromosome in anaphase. Time is expressed as “hours.minutes”. (E) hMis12 and Bub1 staining of mitotic WI38 cells treated with or without TSA (1 μM) for 15 h. The yellow arrowheads point to hMis12 foci at kinetochores. The yellow arrows point to the enriched Bub1 foci at the kinetochores of unaligned chromosomes. Scale bars, 10 μm.
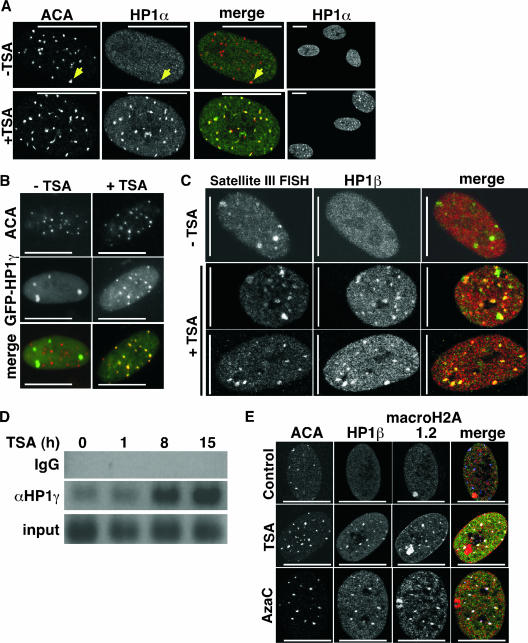
Recruitment of hMis12 to kinetochores depends on its interaction with HP1 proteins in pericentromeric heterochromatin (40). Moreover, inactivation of hMis12 causes mitotic defects, similar to those reported here after CDA treatment. Since CDAs alter the modification status of pericentromeric heterochromatin, as judged by increased histone acetylation in this region (Fig. 1B), we initially hypothesized that CDAs deplete HP1 proteins from pericentromeric heterochromatin and that this, in turn, depletes hMis12 from kinetochores, causing the mitotic defects. To test this, we investigated the effect of CDAs on pericentromeric heterochromatin in more detail. Growing WI38 cells were treated with HDIs, at doses that cause G2/M phase accumulation, and then stained with antibodies to HP1 proteins and anti-centromere antibodies (ACA; from patients with Raynaud's syndrome) (10). Three centromeric proteins are recognized by ACA: CENP-B, CENP-A, and CENP-C. In untreated cells, all three HP1 isoforms were predominantly evenly diffused throughout the nucleus in all cells, although weak colocalization could be detected between ACA and HP1 proteins, a finding consistent with known binding of HP1 proteins at the pericentromeres (Fig. 2A) (35). To our surprise, all HDIs caused a striking increase in pericentromeric staining of all three HP1 proteins (Fig. 2A and see Fig. S1 in the supplemental material). This was observed in several primary human cells, including WI38 fibroblasts, BJ fibroblasts, IMR90 fibroblasts, MRC5 fibroblasts, AG11132A primary mammary epithelial cells, and human ovarian surface epithelial cells (Fig. 2 and data not shown); it occurred 6 to 8 h after treatment with HDIs, and it occurred in close to 100% of the treated cells (Fig. 2A and data not shown). Importantly, the effect was observed with three different antibodies that specifically recognize the three different HP1 isoforms, strongly suggesting that it does not result from exposure of a single epitope due to a conformational change (Fig. 2A, Fig. S1 in the supplemental material, and data not shown). To more rigorously eliminate the epitope exposure possibility, we ectopically expressed HP1γ fused to GFP at its N terminus in WI38 cells. In the absence of HDIs, GFP-HP1γ was enriched in some large foci at unknown sites that did not colocalize with ACA. However, HDIs caused GFP-HP1γ to colocalize with ACA, similar to the endogenous proteins (Fig. 2B). FRAP analysis showed that the GFP-HP1γ recruited to foci by TSA was more stably bound to chromatin than GFP-HP1γ in the bulk nucleoplasm (data not shown), a characteristic of heterochromatic HP1 proteins (7). We also showed by double-label immunoFISH analysis that TSA treatment of WI38 cells caused the relocalization of HP1β to pericentromeric sequences, the latter defined by their hybridization to a pericentromeric satellite III probe (Fig. 2C). Moreover, ChIP analysis of endogenous, untagged HP1γ also showed that this protein is recruited to repetitive pericentromeric satellite III DNA sequences in response to HDIs (Fig. 2D and see Fig. S2 in the supplemental material).
FIG. 2.
Chromatin-disrupting agents cause relocalization of HP1 and macroH2A proteins to pericentromeres. (A) WI38 cells treated with or without TSA (1 μM) for 15 h and stained with the indicated antibodies. Yellow arrow marks weak colocalization between HP1α and ACA in control cells. (B) GFP-HP1γ-transfected WI38 cells were treated with or without TSA (1 μM) for 15 h, and GFP was directly visualized. (C) WI38 cells were treated with or without 1 μM TSA for 15 h and immunostained with anti-HP1β antibodies, followed by FISH to detect pericentromeric satellite III DNA sequences. (D) ChIP with control immunoglobulin G (IgG) or HP1γ antibody to detect the associated pericentromeric satellite III sequence in WI38 cells. (E) WI38 cells mock treated (control) or treated with TSA (5 μM) for 15 h or AzaC (5 μM) for 72 h and stained with the indicated antibodies. The large domain of macroH2A staining is the inactive X chromosome. Scale bars, 10 μm.
Another family of heterochromatic proteins, the histone H2A variant macroH2A (macroH2A1.1, -1.2, and -2) (9), also colocalized with ACA after CDA treatment (Fig. 2E and see Fig. S1B in the supplemental material). Importantly, the DNA methylation inhibitor, AzaC, caused a similar relocalization of both HP1 and macroH2A proteins to pericentromeres (Fig. 2E), confirming that the observed relocalization is a general response to the perturbation of heterochromatin and not a non-chromatin-related response to HDIs, such as the acetylation of p53 (17, 67). To further verify and characterize the response, we performed several additional controls. First, recruitment of HP1 proteins to pericentromeres was not a consequence of accumulation of CDA-treated cells in G2/M phase of the cell cycle, because vinblastine had no effect on HP1 localization, despite causing a similar cell cycle perturbation as the CDAs (data not shown). Also, some TSA-treated cells with HP1 foci were in S phase of the cell cycle, as judged by pulse-labeling with 5′-bromodeoxyuridine (data not shown). Second, the CDAs did not induce markers of DNA damage and repair (e.g., phosphorylation of p53, Chk1, Chk2, and histone H2AX), and DNA damage did not cause relocalization of HP1 proteins, excluding the possibility that the observed effect is a consequence of DNA damage (data not shown). Third, we confirmed by several criteria that CDA-treated cells are not senescent and that the observed HP1- and macroH2A-containing foci are distinct from the superficially similar senescence-associated heterochromatin foci (SAHF) that form in senescent cells (38, 74). Specifically, the CDA-induced HP1 and macroH2A foci are observed in cells lacking the dramatic chromatin condensation characteristic of SAHF (data not shown); the CDA-induced foci form in proliferating cells, in contrast to SAHF (data not shown); and the CDA-induced foci form at pericentromeres, whereas SAHF exclude pericentromeres (Fig. 2C) (38, 74). In sum, our data show that, in contrast to our initial expectation, acute treatment of primary human fibroblasts and epithelial cells with CDAs triggers rapid relocalization of HP1 and macroH2A proteins to pericentromeres, at a time when the cells exhibit partial defects in kinetochore structure and mitotic progression.
Recruitment of HP1 proteins depends on histone chaperone HIRA and histone variant H3.3.
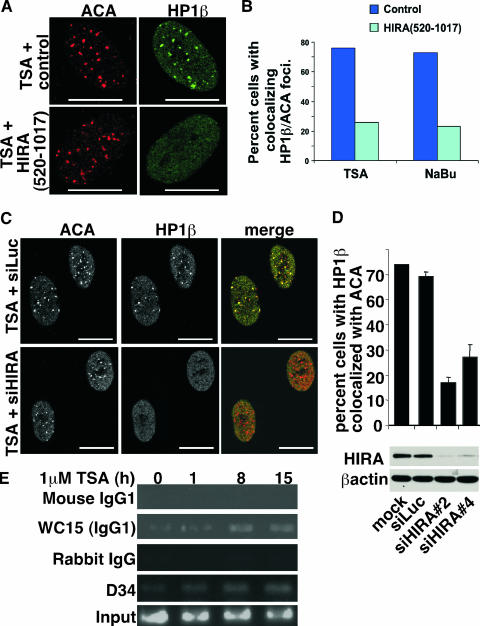
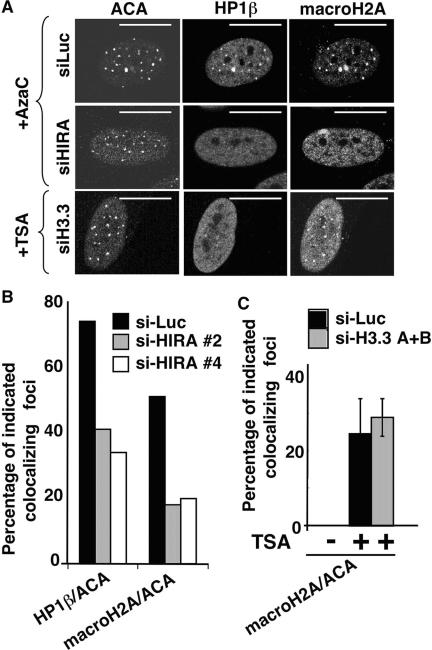
Next, we set out to determine the mechanism by which HP1 proteins are recruited to pericentromeres. Previously, we found that the histone chaperone, HIRA, drives the formation of domains of heterochromatin, SAHF, in senescent human cells (74). Significantly, HIRA's role in heterochromatin formation is conserved through evolution (4, 12, 20, 21, 46, 59, 74). Moreover, the orthologs of human HIRA in Saccharomyces cerevisiae and Schizosaccharomyces pombe are required for proper centromeric and pericentromeric chromatin structure, respectively (4, 60). In light of these observations, we sought to determine whether CDA-induced recruitment of HP1 proteins to pericentromeres in human cells depends on HIRA. First, we used a dominant-negative HIRA mutant to inhibit HIRA activity. This N-terminal truncation mutant, HIRA(520-1017), was fortuitously identified by virtue of its ability to block formation of SAHF in senescent human cells (X. Ye, B. Zerlanko, R. Zhang, N. Somaiah, M. Lipinski, and P. D. Adams, submitted for publication). When this mutant protein was expressed in WI38 cells, it blocked the recruitment of HP1 proteins to pericentromeres in response to CDAs (Fig. 3A and B). To more rigorously test whether HIRA is required for this response, we knocked down HIRA in WI38 cells using two different siRNAs targeted to HIRA. Knock down of HIRA did not affect the total abundance of endogenous HP1 proteins (data not shown) but did block their relocalization to pericentromeres (Fig. 3C and D). There was no effect of a control siRNA, confirming that HIRA is necessary for CDA-induced recruitment of HP1 proteins to pericentromeres.
FIG. 3.
The recruitment of HP1 proteins to pericentromeres is mediated by the histone chaperone HIRA. (A) WI38 cells were infected with a retrovirus encoding HIRA(520-1017), treated for 8 h with TSA (1 μM), and stained with antibodies to ACA or HP1β as indicated. (B) A total of 100 cells from panel A, after treatment with TSA or sodium butyrate (NaBu), were scored for colocalizing ACA and HP1β foci. (C) WI38 cells were transfected with siRNAs to HIRA or luciferase by nucleofection, treated with TSA (1 μM) for 8 h, and then stained with antibodies to ACA or HP1β. (D) A total of 100 cells from panel C were scored for colocalizing HP1β and ACA foci. The mean of three independent experiments is given. Cell extracts were Western blotted with antibodies to HIRA and actin. siHIRA#2 and siHIRA#4 are two distinct siRNAs to HIRA. (E) WI38 cells were treated with TSA as indicated and subjected to ChIP analysis with antibodies to HIRA (WC15 and D34) or appropriate controls to detect association with satellite III pericentromeric DNA. Scale bars, 10 μm.
Since HIRA is a histone chaperone, its requirement for HP1 recruitment is likely to reflect a direct role at pericentromeric heterochromatin. If so, we ought to be able to detect CDA-induced recruitment of HIRA to pericentromeric DNA sequences. To test this, we performed ChIP analyses with two separate antibodies to HIRA, a mouse monoclonal antibody and a rabbit polyclonal antibody, and then sought to determine whether HIRA coimmunoprecipitated with satellite III pericentromeric sequences. Indeed, CDA treatment increased the specific association of HIRA with these DNA sequences; importantly, the increase was observed with both anti-HIRA antibodies but neither control antibody (Fig. 3E). Taken together, these results show that recruitment of HP1 proteins to pericentromeric DNA sequences depends on a known heterochromatin-forming protein (4, 12, 20, 21, 46, 59, 74), the histone chaperone HIRA.
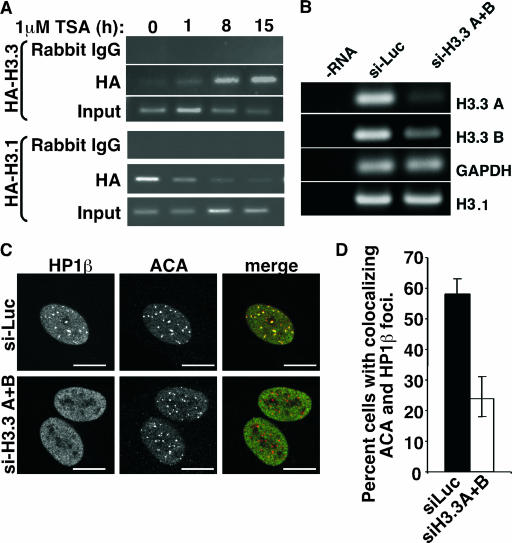
The histone chaperone HIRA is known to favor the histone variant, histone H3.3, as a deposition substrate (63). Therefore, we sought to determine whether CDAs induce deposition of this histone variant into pericentromeres and whether this is required for recruitment of HP1 proteins. Since histone H3.3 and the canonical histone H3.1 are not immunologically distinguishable, to determine whether CDAs induce deposition of histone H3.3 and/or histone H3.1 at pericentromeres we ectopically expressed HA-tagged histone H3.3 or histone H3.1 in WI38 cells, treated the cells with TSA, and then performed ChIP analysis with an anti-HA antibody and scored for the coprecipitation of satellite III sequences. TSA induced a striking recruitment of HA-tagged histone H3.3 to pericentromeres and a reciprocal depletion of canonical histone H3.1 (Fig. 4A). Significantly, histone H3.3 was recruited to pericentromeres before HP1γ, a finding consistent with the proposal that the deposition of histone H3.3 is required for the recruitment of HP1γ (compare Fig. 2D and 4A and see also Fig. S2 in the supplemental material). To directly ask whether recruitment of histone H3.3 is required for recruitment of HP1 proteins to pericentromeres, we used two siRNAs to simultaneously knock down the endogenous products of the two genes coding for histone H3.3, H3.3A, and H3.3B, without affecting canonical histone H3.1 (Fig. 4B). Strikingly, an ∼50% knock down of histone H3.3 mRNA resulted in a 50% block to the recruitment of HP1 proteins to pericentromeres after TSA treatment (Fig. 4C and D). We conclude that the preferred substrate of the histone chaperone HIRA, histone H3.3, is also required for the recruitment of HP1 proteins to pericentromeres and the deposition of histone H3.3 occurs, directly or indirectly, via its exchange with histone H3.1.
FIG. 4.
Recruitment of HP1 proteins to pericentromeres is mediated by the histone variant, histone H3.3. (A) WI38 cells were infected with retroviruses encoding HA-histone H3.1 and HA-histone H3.3, treated with or without TSA (1 μM), and then subjected to ChIP analysis with anti-HA or control antibodies to detect association with satellite III pericentromeric DNA. (B) WI38 cells were transfected, by nucleofection, with a pool of two siRNAs targeted to the histone H3.3A and H3.3B mRNAs or a control siRNA to luciferase. Expression of the indicated genes was analyzed by reverse transcription-PCR. (C) WI38 cells from panel B were treated with 1 μM TSA for 8 h and stained with antibodies to HP1β and ACA. (D) A total of 100 cells from panel C were scored for colocalizing HP1β and ACA foci. The mean of three independent experiments is given. Scale bars, 10 μm.
Recruitment of macroH2A depends on histone chaperone HIRA but not on histone variant H3.3.
Next, we sought to determine whether the recruitment of macroH2A to pericentromeres also requires HIRA and histone H3.3. siRNAs targeted to HIRA, histone H3·3A, H3·3B, or luciferase as a negative control were introduced into cells by nucleofection, and the cells were treated with TSA or AzaC. Similar to HP1 proteins, siRNA-mediated knockdown of HIRA blocked the recruitment of macroH2A to the pericentromeres (Fig. 5A and B). However, knockdown of the histone H3.3A and H3.3B variants had no effect on the relocalization of macroH2A (Fig. 5A and C). This suggests that HIRA activates at least two effector pathways. Specifically, HIRA-dependent deposition of histone H3.3 is required for loading of HP1 proteins, whereas HIRA either directly recruits macroH2A or does so via another HIRA-activated effector pathway that does not require histone 3.3.
FIG. 5.
Relocalization of macroH2A caused by chromatin-disrupting agents is mediated by the histone chaperone HIRA but is independent of histone H3 variant, H3.3. (A) WI38 cells were transfected with siRNAs to HIRA, H3.3, or a control luciferase siRNA by using nucleofection and then treated with 1 μM TSA for 8 h (siH3.3) or 5 μM AzaC for 72 h (siHIRA and siLuc) and subjected to immunostaining with the indicated antibodies. (B and C) A total of 100 cells from panel A were scored for colocalizing macroH2A1.2 and ACA foci. Comparable results were obtained, regardless of whether the cells were treated with TSA or AzaC. Scale bars, 10 μm.
HP1 proteins protect cells from the lethal consequences of CDAs.
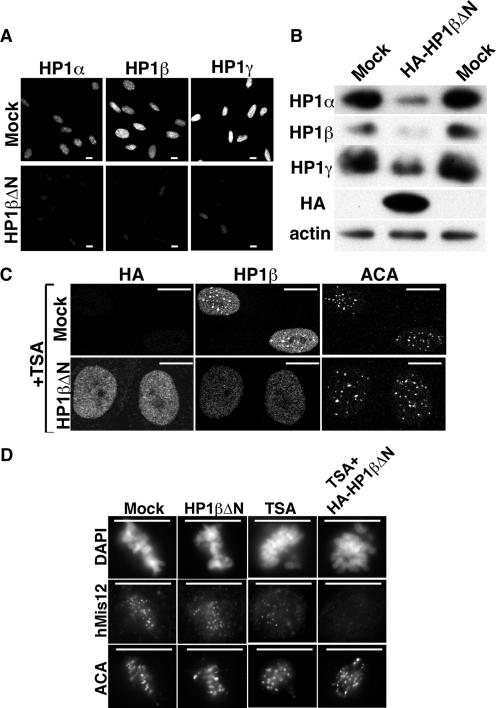
Next, we wanted to know the functional significance of HP1 and/or macroH2A's recruitment to pericentromeres. Since pericentromeric HP1 proteins are thought to promote kinetochore assembly and function (40), our results suggested that the dynamic recruitment and enrichment of HP1 proteins at pericentromeres might reflect a protective response to rescue defects in kinetochore structure and function caused by CDAs, thereby preventing an otherwise much more severe defect in kinetochore function and mitotic progression than that observed in these normal cells (Fig. 1). To test this idea, we set out to determine whether functional disruption of HP1 proteins exacerbates the mitotic defects caused by HDIs. To do this, we generated a putative dominant-negative HP1 mutant. HP1 proteins have been reported and/or suggested to bind to heterochromatin by binding to H3K9Me (3, 18, 24), to RNA (32, 37), and/or to the histone variant, H2AZ (49). HP1 proteins have an N-terminal chromodomain, which binds to H3K9Me, and a C-terminal shadowdomain, which can hetero- or homodimerize with other HP1 proteins. Thus, HP1 proteins are suggested to cross-link and compact H3K9Me-containing heterochromatin. Rauscher and coworkers hypothesized that an N-terminally truncated mutant of HP1 would hetero- and homodimerize with endogenous HP1 proteins but not bind to H3K9Me, thereby sequestering the endogenous proteins away from chromatin and inhibiting their function (26). To test this, an N-terminally truncated HP1β lacking the chromodomain (HP1βΔN) was generated and stably, ectopically expressed in WI38 cells by retrovirus infection and drug selection. As predicted, the amount of all three chromatin-bound endogenous HP1 isoforms was reduced by ca. 80% based on immunofluorescence and Western blot assays, indicating that this mutant functions efficiently as a dominant-negative mutant (Fig. 6A and B). Remarkably, stable ectopic expression of HP1βΔN had no effect on the cell growth, viability, or life span of primary human fibroblasts (Fig. 7A, B, and D and data not shown). The apparent lack of any phenotype in these cells might be because the residual 20% of chromatin-bound HP1 proteins is sufficient to mediate all essential HP1 functions under unstressed conditions. Regardless, if CDA-induced relocalization of HP1 proteins reflects an essential dynamic protective function of these proteins in the presence of altered heterochromatin modifications, HP1 proteins ought to be essential in the presence of CDAs.
FIG. 6.
A dominant-negative HP1 mutant exacerbates the effect of chromatin-disrupting agents on kinetochore structure. (A and B) Detergent preextracted mock or HA-tagged HP1βΔN stably expressing WI38 cells were immunostained or Western blotted with the indicated antibodies. None of the anti-HP1 antibodies recognized truncated HP1βΔN (see Fig. S3A in the supplemental material). (C) Mock or HA-tagged HP1βΔN stably expressing WI38 cells treated with 1 μM TSA for 15 h and stained with antibodies to HA, ACA, and HP1β. (D) Mock or HP1βΔN stably expressing WI38 cells treated with or without 1 μM TSA for 15 h and stained with antibodies to ACA and hMis12. Scale bars, 10 μm.
FIG. 7.
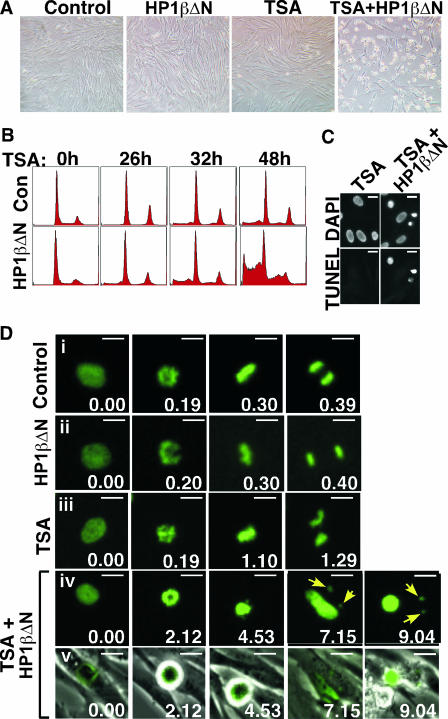
Depletion of chromatin-bound HP1 proteins makes primary cells exquisitely sensitive to apoptotic killing by HDIs. (A) Phase-contrast images of control or HP1βΔN stably expressing WI38 cells treated with or without 0.5 μM TSA for 48 h. (B) FACS analysis of control or HP1βΔN stably expressing WI38 cells treated with or without 0.5 μM TSA for the indicated times. (C) TUNEL analysis of control or HP1βΔN stably expressing WI38 cells treated with 0.5 μM TSA for 30 h. (D) Time-lapse images of H2B-GFP-infected control or HP1βΔN stably expressing WI38 cells treated with or without 1 μM TSA. Yellow arrows point to the lost chromosomes in TSA-treated HP1βΔN stably expressing cells. Scale bars, 10 μm.
We used the HP1βΔN mutant to determine whether functional HP1 proteins protect cells from the debilitating effects of HDIs. Control primary human fibroblasts or the same cells stably expressing HP1βΔN were treated with or without TSA. After TSA treatment, HP1βΔN failed to localize to pericentromeres itself and blocked the relocalization of endogenous HP1 proteins (Fig. 6C). Strikingly, the hMis12 signal at kinetochores was almost completely abolished in the TSA-treated HP1βΔN-expressing cells, below the level observed with TSA alone (Fig. 6D and 1E). Moreover, compared to the control cells, cells expressing HP1βΔN and unable to recruit HP1 to pericentromeres were exquisitely sensitive to killing by HDIs (Fig. 7A). HDI-treated HP1βΔN-expressing cells died by apoptosis, as judged by the presence of sub-G1-phase and TUNEL-positive cells (Fig. 7B and C). To test whether cell killing was a consequence of extreme defects in mitotic progression, in other words a worsening of the defects seen with TSA alone (Fig. 1), the cells stably expressing HP1βΔN were infected with a retrovirus encoding H2B fused to GFP at its C terminus and then treated with or without TSA. Consistent with the apparently normal growth of the HP1βΔN-expressing cells in culture (Fig. 7A, B, and D and data not shown), mitosis in these cells in the absence of HDIs was indistinguishable from the control cells; in both cases cells completed mitosis in about 40 min (Fig. 7Di and ii). As shown previously, the TSA-treated control cells also completed mitosis, even though they took more than 1 h to do so and exhibited some unaligned and lagging chromosomes in the process (Fig. 1D and 7Diii). In stark contrast, the TSA-treated HP1βΔN-expressing cells typically failed to progress from prometaphase to metaphase, and then, after a long delay of several hours, returned to interphase, often losing chromosomes in the process and eventually dying from apoptosis (Fig. 7Div and v). Together, these data show that disruption of HP1 protein function worsens the HDI-induced defects in kinetochore structure, as indicated by a complete loss of hMis12, and prevents alignment of chromosomes at the metaphase plate, ultimately causing an aborted mitosis and cell death. The fact that WI38 cells stably expressing HP1βΔN exhibited no detectable defects when repeatedly analyzed over 20 to 30 population doublings shows that the profound effect of TSA on these cells is not due to worsening of a phenotype that already exists at low level in the HP1βΔN cells. We conclude that, in the presence of HDIs, recruitment of HP1 proteins to pericentromeres maintains the levels of hMis12 at kinetochores and protects cells from the lethal damaging effects of HDIs.
DISCUSSION
We have shown that the perturbation of DNA and histone modifications by a variety of chromatin disrupting agents triggers a dynamic relocalization of HP1 and macroH2A proteins to sites of heterochromatin alteration at the pericentromeres. Consequently, the amount of HP1 proteins bound to these sites increases over the normal level in untreated cells. Recruitment of HP1 proteins to these sites of altered heterochromatin is preceded by and depends on deposition of the histone variant H3.3 and its apparent replacement of canonical H3.1, mediated by the histone chaperone HIRA. In contrast, recruitment of macroH2A depends on HIRA, but not histone H3.3. Recruitment of HP1 proteins to the pericentromeres is essential to maintain kinetochore integrity, as judged by the loading of the HP1-binding protein hMis12 onto kinetochores, and ultimately for viable progression through mitosis. We conclude that HP1 proteins relocalize to sites of perturbed pericentromeric heterochromatin to maintain hMis12 at kinetochores, thereby protecting the cells from a catastrophic and lethal mitosis. This indicates that, at least in response to the small molecule CDAs used here, HP1 proteins can serve as dynamic responders to the perturbation of heterochromatic modifications, thereby protecting the cells from the disruptive effects of such molecules. Significantly, some of these small molecule CDAs occur naturally in the environment (see below).
Our finding that HDIs induce localization of additional HP1 proteins to pericentromeres is in apparent contrast to previous studies, which showed that HDIs cause a depletion of HP1 proteins from pericentromeres (8, 51, 62). Confirming our findings, we obtained the same result by immunofluorescence with three different specific antibodies to the three HP1 family members HP1α, HP1β, and HP1γ, with ectopically expressed GFP-tagged HP1γ, by ChIP analysis, and by immunoFISH. In addition, we showed that inhibition of DNA methylation by 5′-AzaC triggered the same relocalization, showing that the effect of the HDIs is due to the disruption of chromatin rather than another nonchromatin target of the HDIs, e,g., p53 (17, 67). Significantly, the previous studies examining the effects of HDIs on HP1 proteins at the pericentromeres were typically carried out after a much longer duration of HDI treatment and, most notably, typically in marsupial, murine, or transformed human cells. In fact, none of the previous reports showed raw data from primary human cells (8, 51, 62). Moreover, we, too, have found that the acute treatment of murine cells with TSA or chronic treatment of transformed human cells with TSA, replicating the conditions of a previous study (62), results in the loss of HP1 proteins from pericentromeres (see Fig. S3B and C in the supplemental material). In sum, the behavior of HP1 proteins after acute HDI treatment of primary human cells is profoundly different from results reported previously in other cell types with different treatment protocols.
Our data indicate that the recruitment of HP1 proteins to the pericentromeres depends on HIRA-mediated deposition of the histone variant, histone H3.3, into chromatin, seemingly by exchange with the canonical histone H3.1 subtype, since the abundance of the canonical histone in pericentromeric DNA decreases at the same time that the variant increases. Interestingly, newly synthesized histone H3 in human cells has been reported to be unacetylated (23, 61). Moreover, a recent study by Almouzni and coworkers has shown that a proportion of “free” non-chromatin-bound histone H3.3 is methylated to create H3K9Me2 (30). Thus, HP1 proteins might be recruited to pericentromeres by their binding to newly deposited histone H3.3 that is methylated on lysine 9 prior to its deposition or becomes methylated after its incorporation into chromatin. Since the abundance of H3K9Me2 and H3K9Me3 at the pericentromeres does not increase in HDI-treated cells, deposition of newly synthesized histone H3.3 may serve to maintain the preexisting level of H3K9Me in the face of rising histone acetylation, implying that more HP1 proteins bind to the same number of H3K9Me binding sites in HDI-treated cells. Conceivably, H3K9Me moieties are more accessible to HP1 in the context of hyperacetylated “open” chromatin (64). The idea that histone H3.3 recruits HP1 proteins appears to contradict several recent reports that link histone H3.3 to transcription activation (1, 34, 36, 57, 70). However, it should be noted that histone H3.3 per se has not been shown to cause or contribute to transcription activation. Moreover, a proportion of histone H3.3 does carry transcription modifications characteristic of silent chromatin, such as H3K9Me (14, 30, 34). Also, histone H3.3 accumulates in nondividing, differentiated cells and fibroblasts approaching senescence, in some cases to ca. 90% of the total histone H3, with presumably the majority being in inactive chromatin (5, 6, 13, 22, 44, 47, 52, 65, 71). Moreover, upon egg fertilization in flies and worms, the sperm chromatin is remodeled prior to DNA replication using histone variant H3.3, seemingly in a transcription-independent manner (28, 42). Therefore, an alternative view of histone H3.3 is that it is a replacement variant histone, associated with any “change in chromatin state,” that is incorporated in a replication-independent manner during transcription, sperm chromatin remodeling or, as proposed here, as part of a dynamic response to rectify defects in chromatin modifications.
Many of the proteins now known to contribute to the cellular response to DNA damage, such as ATM, BRCA1, and the Fanconi's anemia gene products, were initially implicated in this process based on their dynamic relocalization in response to genotoxic stress (27, 33, 58) and/or the sensitivity of cells lacking these proteins to DNA-damaging agents (2, 69). In some respects, the response of HP1 proteins to CDAs is analogous to the response of these DNA repair proteins to DNA damage: HP1 proteins relocalize to sites of altered modifications in response to CDAs, and cells deficient in HP1 proteins are extremely sensitive to killing by CDAs. Thus, it is tempting to speculate that one function of HP1 proteins is to serve as dynamic responders to the perturbation of heterochromatin modifications, thereby rescuing heterochromatin function and protecting the cells from those perturbations. We note that HDIs occur naturally in the environment. For example, the concentration of butyrate in the colon can reach millimolar concentrations as a consequence of the bacterial fermentation of carbohydrate (56). Thus, one function of HP1 proteins might be to protect cells from the detrimental effects of such toxins.
Extending this view, HP1 proteins might also respond to alterations in chromatin structure caused by physiological nuclear processes. This might be why HP1 proteins are dynamically bound to chromatin, exhibiting surprisingly high off rates and on rates (7). This dynamic binding behavior would be expected to facilitate their recruitment to sites of altered chromatin. Also consistent with this idea, it was recently shown that HP1γ is recruited to transcriptionally active genes during the elongation phase (66). HP1γ might play a dynamic role in reestablishing transcriptionally silent chromatin after disruption of the chromatin structure by the passing RNA polymerase. Interestingly, previous reports have shown that the activation of gene expression by HDIs is a transient response, suggesting that cells are somehow able to suppress the transcription-activating effects of HDIs (45). Consistent with the idea that HP1 proteins rescue defects in chromatin structure and function outside of pericentromeres, a proportion of TSA-treated HP1βΔN-expressing cells appeared to die during interphase (data not shown). Together, these results indicate that HP1 proteins might play a more general role in the dynamic rescue of chromatin defects than that described here.
In summary, we have shown that, in response to a variety of small molecules that perturb heterochromatin, HP1 proteins are dynamically relocalized to sites of altered heterochromatin at the pericentromeres. This relocalization is essential to maintain the HP1-binding protein hMis12 at the kinetochores and suppress lethal defects in mitosis. We propose that these results point to a new function of HP1 proteins as dynamic responders to the perturbation of chromatin modifications and as essential players in a chromatin “repair-like” process.
Supplementary Material
Acknowledgments
We thank Sandy Jablonski at the Fox Chase Cancer Center Cell Imaging Facility for technical help, William Earnshaw for the HP1α antibody, J. B. Rattner for the ACA antibody, Richard Katz for providing apicidin and valproic acid, Inma Ibanez and Paul Cairns for AzaC, Andy Godwin for human ovarian surface epithelial cells, and Kenneth Zaret for helpful comments.
This study was supported by NIH grant GM062281 and LLS grant 1520-04 to P.D.A.; an AFAR grant to R.Z.; NIH grant GM49351 to J.P.; and NIH grants GM44762, CA99423, and CA75138 and core grant CA06927 to T.J.Y. and S.-T.L.
Footnotes
Published ahead of print on 13 November 2006.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Ahmad, K., and S. Henikoff. 2002. The histone variant h3.3 marks active chromatin by replication-independent nucleosome assembly. Mol. Cell 9:1191-1200. [DOI] [PubMed] [Google Scholar]
- 2.Arlett, C. F., and S. A. Harcourt. 1980. Survey of radiosensitivity in a variety of human cell strains. Cancer Res. 40:926-932. [PubMed] [Google Scholar]
- 3.Bannister, A. J., P. Zegerman, J. F. Partridge, E. A. Miska, J. O. Thomas, R. C. Allshire, and T. Kouzarides. 2001. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410:120-124. [DOI] [PubMed] [Google Scholar]
- 4.Blackwell, C., K. A. Martin, A. Greenall, A. Pidoux, R. C. Allshire, and S. K. Whitehall. 2004. The Schizosaccharomyces pombe HIRA-like protein Hip1 is required for the periodic expression of histone genes and contributes to the function of complex centromeres. Mol. Cell. Biol. 24:4309-4320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bosch, A., and P. Suau. 1995. Changes in core histone variant composition in differentiating neurons: the roles of differential turnover and synthesis rates. Eur. J. Cell Biol. 68:220-225. [PubMed] [Google Scholar]
- 6.Brown, D. T., S. E. Wellman, and D. B. Sittman. 1985. Changes in the levels of three different classes of histone mRNA during murine erythroleukemia cell differentiation. Mol. Cell. Biol. 5:2879-2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheutin, T., A. J. McNairn, T. Jenuwein, D. M. Gilbert, P. B. Singh, and T. Misteli. 2003. Maintenance of stable heterochromatin domains by dynamic HP1 binding. Science 299:721-725. [DOI] [PubMed] [Google Scholar]
- 8.Cimini, D., M. Mattiuzzo, L. Torosantucci, and F. Degrassi. 2003. Histone hyperacetylation in mitosis prevents sister chromatid separation and produces chromosome segregation defects. Mol. Biol. Cell 14:3821-3833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Costanzi, C., and J. R. Pehrson. 2001. MACROH2A2, a new member of the MARCOH2A core histone family. J. Biol. Chem. 276:21776-21784. [DOI] [PubMed] [Google Scholar]
- 10.Earnshaw, W., B. Bordwell, C. Marino, and N. Rothfield. 1986. Three human chromosomal autoantigens are recognized by sera from patients with anti-centromere antibodies. J. Clin. Investig. 77:426-430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gilchrist, S., N. Gilbert, P. Perry, and W. A. Bickmore. 2004. Nuclear organization of centromeric domains is not perturbed by inhibition of histone deacetylases. Chromosome Res. 12:505-516. [DOI] [PubMed] [Google Scholar]
- 12.Greenall, A., E. S. Williams, K. A. Martin, J. M. Palmer, J. Gray, C. Liu, and S. K. Whitehall. 2006. Hip3 interacts with the HIRA proteins Hip1 and Slm9 and is required for transcriptional silencing and accurate chromosome segregation. J. Biol. Chem. 281:8732-8739. [DOI] [PubMed] [Google Scholar]
- 13.Grove, G. W., and A. Zweidler. 1984. Regulation of nucleosomal core histone variant levels in differentiating murine erythroleukemia cells. Biochemistry 23:4436-4443. [DOI] [PubMed] [Google Scholar]
- 14.Hake, S. B., B. A. Garcia, E. M. Duncan, M. Kauer, G. Dellaire, J. Shabanowitz, D. P. Bazett-Jones, C. D. Allis, and D. F. Hunt. 2006. Expression patterns and posttranslational modifications associated with mammalian histone H3 variants. J. Biol. Chem. 281:559-568. [DOI] [PubMed] [Google Scholar]
- 15.Hall, C., D. M. Nelson, X. Ye, K. Baker, J. A. DeCaprio, S. Seeholzer, M. Lipinski, and P. D. Adams. 2001. HIRA, the human homologue of yeast Hir1p and Hir2p, is a novel cyclin-cdk2 substrate whose expression blocks S-phase progression. Mol. Cell. Biol. 21:1854-1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Horowitz-Scherer, R. A., and C. L. Woodcock. 2006. Organization of interphase chromatin. Chromosoma 115:1-14. [DOI] [PubMed] [Google Scholar]
- 17.Ito, A., C. H. Lai, X. Zhao, S. Saito, M. H. Hamilton, E. Appella, and T. P. Yao. 2001. p300/CBP-mediated p53 acetylation is commonly induced by p53-activating agents and inhibited by MDM2. EMBO J. 20:1331-1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jacobs, S. A., S. D. Taverna, Y. Zhang, S. D. Briggs, J. Li, J. C. Eissenberg, C. D. Allis, and S. Khorasanizadeh. 2001. Specificity of the HP1 chromo domain for the methylated N terminus of histone H3. EMBO J. 20:5232-5241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kastan, M. B., and J. Bartek. 2004. Cell-cycle checkpoints and cancer. Nature 432:316-323. [DOI] [PubMed] [Google Scholar]
- 20.Kaufman, P. D., J. L. Cohen, and M. A. Osley. 1998. Hir proteins are required for position-dependent gene silencing in Saccharomyces cerevisiae in the absence of chromatin assembly factor I. Mol. Cell. Biol. 18:4793-4806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krawitz, D. C., T. Kama, and P. D. Kaufman. 2002. Chromatin assembly factor I mutants defective for PCNA binding require Asf1/Hir proteins for silencing. Mol. Cell. Biol. 22:614-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krimer, D. B., G. Cheng, and A. I. Skoultchi. 1993. Induction of H3.3 replacement histone mRNAs during the precommitment period of murine erythroleukemia cell differentiation. Nucleic Acids Res. 21:2873-2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuo, M. H., J. E. Brownell, R. E. Sobel, T. A. Ranalli, R. G. Cook, D. G. Edmondson, S. Y. Roth, and C. D. Allis. 1996. Transcription-linked acetylation by Gcn5p of histones H3 and H4 at specific lysines. Nature 383:269-272. [DOI] [PubMed] [Google Scholar]
- 24.Lachner, M., D. O'Carroll, S. Rea, K. Mechtler, and T. Jenuwein. 2001. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 410:116-120. [DOI] [PubMed] [Google Scholar]
- 25.Laird, P. W. 2005. Cancerepigenetics. Hum. Mol. Genet. 14:(Spec. No. 1)R65-R76. [DOI] [PubMed] [Google Scholar]
- 26.Lechner, M. S., G. E. Begg, D. W. Speicher, and F. J. Rauscher III. 2000. Molecular determinants for targeting heterochromatin protein 1-mediated gene silencing: direct chromoshadow domain-KAP-1 corepressor interaction is essential. Mol. Cell. Biol. 20:6449-6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li, M. J., M. C. Peakman, E. I. Golub, G. Reddy, D. C. Ward, C. M. Radding, and N. Maizels. 1996. Rad51 expression and localization in B cells carrying out class switch recombination. Proc. Natl. Acad. Sci. USA 93:10222-10227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Loppin, B., E. Bonnefoy, C. Anselme, A. Laurencon, T. L. Karr, and P. Couble. 2005. The histone H3.3 chaperone HIRA is essential for chromatin assembly in the male pronucleus. Nature 437:1386-1390. [DOI] [PubMed] [Google Scholar]
- 29.Loyola, A., and G. Almouzni. 2004. Bromodomains in living cells participate in deciphering the histone code. Trends Cell Biol. 14:279-281. [DOI] [PubMed] [Google Scholar]
- 30.Loyola, A., T. Bonaldi, D. Roche, A. Imhof, and G. Almouzni. 2006. PTMs on H3 variants before chromatin assembly potentiate their final epigenetic state. Mol. Cell 24:309-316. [DOI] [PubMed] [Google Scholar]
- 31.Maiato, H., J. DeLuca, E. D. Salmon, and W. C. Earnshaw. 2004. The dynamic kinetochore-microtubule interface. J. Cell Sci. 117:5461-5477. [DOI] [PubMed] [Google Scholar]
- 32.Maison, C., D. Bailly, A. H. Peters, J. P. Quivy, D. Roche, A. Taddei, M. Lachner, T. Jenuwein, and G. Almouzni. 2002. Higher-order structure in pericentric heterochromatin involves a distinct pattern of histone modification and an RNA component. Nat. Genet. 30:329-334. [DOI] [PubMed] [Google Scholar]
- 33.Maser, R. S., K. J. Monsen, B. E. Nelms, and J. H. Petrini. 1997. hMre11 and hRad50 nuclear foci are induced during the normal cellular response to DNA double-strand breaks. Mol. Cell. Biol. 17:6087-6096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McKittrick, E., P. R. Gafken, K. Ahmad, and S. Henikoff. 2004. Histone H3.3 is enriched in covalent modifications associated with active chromatin. Proc. Natl. Acad. Sci. USA 101:1525-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Minc, E., Y. Allory, H. J. Worman, J. C. Courvalin, and B. Buendia. 1999. Localization and phosphorylation of HP1 proteins during the cell cycle in mammalian cells. Chromosoma 108:220-234. [DOI] [PubMed] [Google Scholar]
- 36.Mito, Y., J. G. Henikoff, and S. Henikoff. 2005. Genome-scale profiling of histone H3.3 replacement patterns. Nat. Genet. 37:1090-1097. [DOI] [PubMed] [Google Scholar]
- 37.Muchardt, C., M. Guilleme, J. S. Seeler, D. Trouche, A. Dejean, and M. Yaniv. 2002. Coordinated methyl and RNA binding is required for heterochromatin localization of mammalian HP1α. EMBO Rep. 3:975-981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Narita, M., S. Nunez, E. Heard, A. W. Lin, S. A. Hearn, D. L. Spector, G. J. Hannon, and S. W. Lowe. 2003. Rb-mediated heterochromatin formation and silencing of E2F target genes during cellular senescence. Cell 113:703-716. [DOI] [PubMed] [Google Scholar]
- 39.Nielsen, S. J., R. Schneider, U. M. Bauer, A. J. Bannister, A. Morrison, D. O'Carroll, R. Firestein, M. Cleary, T. Jenuwein, R. E. Herrera, and T. Kouzarides. 2001. Rb targets histone H3 methylation and HP1 to promoters. Nature 412:561-565. [DOI] [PubMed] [Google Scholar]
- 40.Obuse, C., O. Iwasaki, T. Kiyomitsu, G. Goshima, Y. Toyoda, and M. Yanagida. 2004. A conserved Mis12 centromere complex is linked to heterochromatic HP1 and outer kinetochore protein Zwint-1. Nat. Cell Biol. 6:1135-1141. [DOI] [PubMed] [Google Scholar]
- 41.Ogawa, H., K. Ishiguro, S. Gaubatz, D. M. Livingston, and Y. Nakatani. 2002. A complex with chromatin modifiers that occupies E2F- and Myc-responsive genes in G0 cells. Science 296:1132-1136. [DOI] [PubMed] [Google Scholar]
- 42.Ooi, S. L., J. R. Priess, and S. Henikoff. 2006. Histone H3.3 variant dynamics in the germline of Caenorhabditis elegans. PLoS Genet. 2:e97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Orlando, V., H. Strutt, and R. Paro. 1997. Analysis of chromatin structure by in vivo formaldehyde cross-linking. Methods 11:205-214. [DOI] [PubMed] [Google Scholar]
- 44.Pantazis, P., and W. M. Bonner. 1984. Specific alterations in the pattern of histone-3 synthesis during conversion of human leukemic cells to terminally differentiated cells in culture. Differentiation 28:186-190. [DOI] [PubMed] [Google Scholar]
- 45.Peart, M. J., G. K. Smyth, R. K. van Laar, D. D. Bowtell, V. M. Richon, P. A. Marks, A. J. Holloway, and R. W. Johnstone. 2005. Identification and functional significance of genes regulated by structurally different histone deacetylase inhibitors. Proc. Natl. Acad. Sci. USA 102:3697-3702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Phelps-Durr, T. L., J. Thomas, P. Vahab, and M. C. Timmermans. 2005. Maize rough sheath2 and its Arabidopsis orthologue ASYMMETRIC LEAVES1 interact with HIRA, a predicted histone chaperone, to maintain knox gene silencing and determinacy during organogenesis. Plant Cell 17:2886-2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pina, B., and P. Suau. 1987. Changes in histones H2A and H3 variant composition in differentiating and mature rat brain cortical neurons. Dev. Biol. 123:51-58. [DOI] [PubMed] [Google Scholar]
- 48.Qiu, L., A. Burgess, D. P. Fairlie, H. Leonard, P. G. Parsons, and B. G. Gabrielli. 2000. Histone deacetylase inhibitors trigger a G2 checkpoint in normal cells that is defective in tumor cells. Mol. Biol. Cell 11:2069-2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rangasamy, D., I. Greaves, and D. J. Tremethick. 2004. RNA interference demonstrates a novel role for H2A.Z in chromosome segregation. Nat. Struct. Mol. Biol. 11:650-655. [DOI] [PubMed] [Google Scholar]
- 50.Richards, E. J., and S. C. Elgin. 2002. Epigenetic codes for heterochromatin formation and silencing: rounding up the usual suspects. Cell 108:489-500. [DOI] [PubMed] [Google Scholar]
- 51.Robbins, A. R., S. A. Jablonski, T. J. Yen, K. Yoda, R. Robey, S. E. Bates, and D. L. Sackett. 2005. Inhibitors of histone deacetylases alter kinetochore assembly by disrupting pericentromeric heterochromatin. Cell Cycle 4:717-726. [DOI] [PubMed] [Google Scholar]
- 52.Rogakou, E. P., and K. E. Sekeri-Pataryas. 1999. Histone variants of H2A and H3 families are regulated during in vitro aging in the same manner as during differentiation. Exp. Gerontol. 34:741-754. [DOI] [PubMed] [Google Scholar]
- 53.Roopra, A., R. Qazi, B. Schoenike, T. J. Daley, and J. F. Morrison. 2004. Localized domains of G9a-mediated histone methylation are required for silencing of neuronal genes. Mol. Cell 14:727-738. [DOI] [PubMed] [Google Scholar]
- 54.Rudd, M. K., and H. F. Willard. 2004. Analysis of the centromeric regions of the human genome assembly. Trends Genet. 20:529-533. [DOI] [PubMed] [Google Scholar]
- 55.Saffery, R., D. V. Irvine, B. Griffiths, P. Kalitsis, L. Wordeman, and K. H. Choo. 2000. Human centromeres and neocentromeres show identical distribution patterns of >20 functionally important kinetochore-associated proteins. Hum. Mol. Genet. 9:175-185. [DOI] [PubMed] [Google Scholar]
- 56.Scheppach, W., and F. Weiler. 2004. The butyrate story: old wine in new bottles? Curr. Opin. Clin. Nutr. Metab. Care 7:563-567. [DOI] [PubMed] [Google Scholar]
- 57.Schwartz, B. E., and K. Ahmad. 2005. Transcriptional activation triggers deposition and removal of the histone variant H3.3. Genes Dev. 19:804-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Scully, R., J. Chen, R. L. Ochs, K. Keegan, M. Hoekstra, J. Feunteun, and D. M. Livingston. 1997. Dynamic changes of BRCA1 subnuclear location and phosphorylation state are initiated by DNA damage. Cell 90:425-435. [DOI] [PubMed] [Google Scholar]
- 59.Sharp, J. A., E. T. Fouts, D. C. Krawitz, and P. D. Kaufman. 2001. Yeast histone deposition protein Asf1p requires Hir proteins and PCNA for heterochromatic silencing. Curr. Biol. 11:463-473. [DOI] [PubMed] [Google Scholar]
- 60.Sharp, J. A., A. A. Franco, M. A. Osley, P. D. Kaufman, D. C. Krawitz, T. Kama, E. T. Fouts, and J. L. Cohen. 2002. Chromatin assembly factor I and Hir proteins contribute to building functional kinetochores in Saccharomyces cerevisiae. Genes Dev. 16:85-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sobel, R. E., R. G. Cook, C. A. Perry, A. T. Annunziato, and C. D. Allis. 1995. Conservation of deposition-related acetylation sites in newly synthesized histones H3 and H4. Proc. Natl. Acad. Sci. USA 92:1237-1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Taddei, A., C. Maison, D. Roche, and G. Almouzni. 2001. Reversible disruption of pericentric heterochromatin and centromere function by inhibiting deacetylases. Nat. Cell Biol. 3:114-120. [DOI] [PubMed] [Google Scholar]
- 63.Tagami, H., D. Ray-Gallet, G. Almouzni, and Y. Nakatani. 2004. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell 116:51-61. [DOI] [PubMed] [Google Scholar]
- 64.Tse, C., T. Sera, A. P. Wolffe, and J. C. Hansen. 1998. Disruption of higher-order folding by core histone acetylation dramatically enhances transcription of nucleosomal arrays by RNA polymerase III. Mol. Cell. Biol. 18:4629-4638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Urban, M. K., and A. Zweidler. 1983. Changes in nucleosomal core histone variants during chicken development and maturation. Dev. Biol. 95:421-428. [DOI] [PubMed] [Google Scholar]
- 66.Vakoc, C. R., S. A. Mandat, B. A. Olenchock, and G. A. Blobel. 2005. Histone H3 lysine 9 methylation and HP1γ are associated with transcription elongation through mammalian chromatin. Mol. Cell 19:381-391. [DOI] [PubMed] [Google Scholar]
- 67.Vaziri, H., S. K. Dessain, E. Ng Eaton, S. I. Imai, R. A. Frye, T. K. Pandita, L. Guarente, and R. A. Weinberg. 2001. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell 107:149-159. [DOI] [PubMed] [Google Scholar]
- 68.Wang, Y., W. Fischle, W. Cheung, S. Jacobs, S. Khorasanizadeh, and C. D. Allis. 2004. Beyond the double helix: writing and reading the histone code. Novartis Found. Symp. 259:3-21, 163-169. [PubMed] [Google Scholar]
- 69.Weichselbaum, R. R., J. Nove, and J. B. Little. 1980. X-ray sensitivity of fifty-three human diploid fibroblast cell strains from patients with characterized genetic disorders. Cancer Res. 40:920-925. [PubMed] [Google Scholar]
- 70.Wirbelauer, C., O. Bell, and D. Schubeler. 2005. Variant histone H3.3 is deposited at sites of nucleosomal displacement throughout transcribed genes while active histone modifications show a promoter-proximal bias. Genes Dev. 19:1761-1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wunsch, A. M., and J. Lough. 1987. Modulation of histone H3 variant synthesis during the myoblast-myotube transition of chicken myogenesis. Dev. Biol. 119:94-99. [DOI] [PubMed] [Google Scholar]
- 72.Reference deleted.
- 73.Yoshida, M., and T. Beppu. 1988. Reversible arrest of proliferation of rat 3Y1 fibroblasts in both the G1 and G2 phases by trichostatin A. Exp. Cell Res. 177:122-131. [DOI] [PubMed] [Google Scholar]
- 74.Zhang, R., M. V. Poustovoitov, X. Ye, H. A. Santos, W. Chen, S. M. Daganzo, J. P. Erzberger, I. G. Serebriiskii, A. A. Canutescu, R. L. Dunbrack, J. R. Pehrson, J. M. Berger, P. D. Kaufman, and P. D. Adams. 2005. Formation of MacroH2A-containing senescence-associated heterochromatin foci and senescence driven by ASF1a and HIRA. Dev. Cell 8:19-30. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.