Abstract
Although multiple regulatory elements and protein factors are known to regulate the non-neuronal pathway of alternative processing of the calcitonin/calcitonin gene-related peptide (CGRP) pre-mRNA, the mechanisms controlling the neuron-specific pathway have remained elusive. Here we report the identification of Fox-1 and Fox-2 proteins as novel regulators that mediate the neuron-specific splicing pattern. Fox-1 and Fox-2 proteins function to repress exon 4 inclusion, and this effect depends on two UGCAUG elements surrounding the 3′ splice site of the calcitonin-specific exon 4. In neuron-like cells, mutation of a subset of UGCAUG elements promotes the non-neuronal pattern in which exon 4 is included. In HeLa cells, overexpression of Fox-1 or Fox-2 protein decreases exon 4 inclusion. Fox-1 and Fox-2 proteins interact with the UGCAUG elements specifically and regulate splicing by blocking U2AF65 binding to the 3′ splice site upstream of exon 4. We further investigated the inter-relationship between the UGCAUG silencer elements and the previously identified intronic and exonic splicing regulatory elements and found that exon 4 is regulated by an intricate balance of positive and negative regulation. These results define a critical role for Fox-1 and Fox-2 proteins in exon 4 inclusion of calcitonin/CGRP pre-mRNA and establish a regulatory network that controls the fate of exon 4.
Alternative RNA processing is a major contributor to proteomic complexity in eukaryotic organisms. Through this process, the majority of human pre-mRNA molecules generate more than one mRNA molecule, which are then translated into different protein isoforms that have distinct biological activities (6, 27). Tissue-specific alternative RNA processing plays an important role in regulating gene expression. It has been demonstrated that alternative splicing is controlled by a complex interplay between positive and negative splicing regulators that function through binding at their cognate splicing enhancer and silencer elements located in both the alternatively spliced exon and the adjacent introns (27). However, our knowledge of tissue-specific regulation of splicing is very limited, with only a handful of tissue-specific splicing regulators identified to date (6, 40).
The human calcitonin/calcitonin gene-related peptide (CGRP) gene is an excellent model to study tissue-specific regulation of alternative splicing. The calcitonin/CGRP gene contains six exons. In neurons, CGRP mRNA production results from joining of exons 1 to 3 to exons 5 to 6 accompanied by usage of a distal polyadenylation signal located at the 3′ end of the sixth exon (3, 35). In thyroid C cells, calcitonin mRNA is produced by joining exons 1 to 3 to exon 4, accompanied by usage of the proximal polyadenylation site located at the 3′ end of exon 4 (Fig. 1A) (38). Alternative processing of calcitonin/CGRP pre-mRNA is subject to complex control involving multiple cis-acting regulatory elements and trans-acting factors. Studies to date have identified several RNA sequence elements and protein factors that affect the ratio of calcitonin and CGRP mRNAs. All but one of these regulatory elements appear to function by directly affecting the calcitonin-specific exon 4 inclusion. Two enhancer elements that promote inclusion of exon 4 have been characterized. One is an exonic enhancer element (ESE) located on exon 4 and bound by Tra2β and SRp55 (42, 43, 46). The other is an intronic enhancer element located downstream of exon 4 that interacts with U1 snRNP, TIAR, SRp20, and PTB; at least one function of this intronic enhancer is to promote polyadenylation of exon 4 (21-25, 48). Our understanding of silencer elements that repress exon 4 inclusion remains limited. The noncanonical branchpoint at the 3′ splice site upstream of exon 4 (uridine in human and cytosine in rat and mouse) was demonstrated to function as a negative element. Mutation of this uridine residue into the canonical branchpoint adenosine residue resulted in a significant increase in calcitonin exon 4 inclusion in all of the cell lines tested (1, 2). In addition, a sequence motif residing in the intronic element discussed above appears to function as a suppressor sequence (21). Finally, a sequence at the 3′ splice site of exon 5 has been shown to promote exon 5 inclusion and, as a result, decreases the usage of exon 4 in the two cell lines tested (26, 32). To date, no trans-acting factor has been identified and documented as a cell-type-specific regulator that controls alternative processing of calcitonin/CGRP pre-mRNA. However, a brain-specific activity was identified that suppresses usage of exon 4, although the identity of the protein factor responsible for this activity remains unknown (8, 33).
FIG. 1.
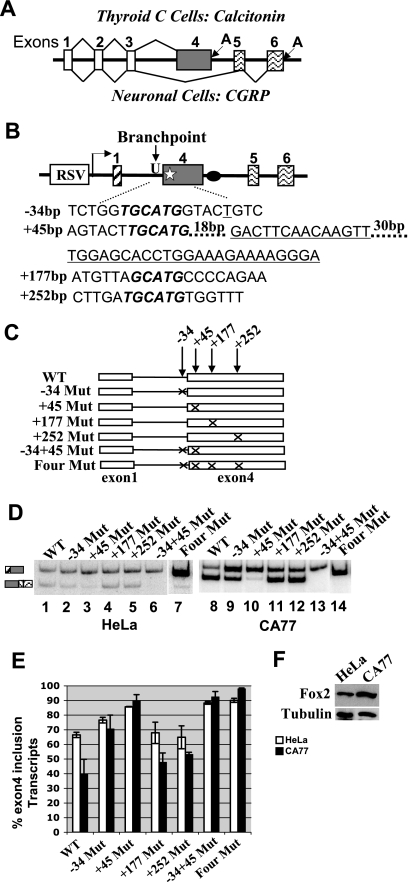
The UGCAUG elements at positions −34 and +45 surrounding the 3′ splice site of calcitonin-specific exon 4 are silencers for the calcitonin pathway. (A) Schematic diagram of the calcitonin/CGRP gene and its alternative RNA processing pathways. A, polyadenylation signal. (B) Diagram of the calcitonin/CGRP reporter minigene indicating the locations of the four (U)GCAUG elements. Numbers in front of nucleotides represent the base pair number upstream or downstream from the 3′ splice site of exon 4 (−, upstream; +, downstream). RSV, Rous sarcoma virus promoter. The black oval represents the intronic enhancer element. The star represents the exonic enhancer element that is shown as the underlined sequences. (C) Diagram illustrating various mutations generated to test the role of (U)GCAUG elements in exon 4 inclusion. An “X” indicates disruption of the wild-type (U)GCAUG to (U)GACUG. (D) Results of RT-PCR assay of total RNA isolated from HeLa and CA77 Cells transfected with the diagramed calcitonin/CGRP reporter gene with wild-type or mutated (U)GCAUG elements. (E) Quantification of the RT-PCR results shown in panel C. For each reporter, percentage of exon 4 inclusion [exon 4 inclusion/(exon 4 inclusion + exon 4 exclusion)] is calculated based on at least three independent transfections. (F) Western blot analysis of Fox-2 expression in HeLa and CA77 cells. The blot was probed for tubulin as a loading control and Fox-2 to detect the endogenous expression of Fox-2 protein in HeLa and CA77 cells.
The (U)GCAUG hexanucleotide repeat element has been characterized as an important sequence motif that regulates inclusion of a number of alternatively spliced exons. The element has been shown to play a important role in proper splicing regulation of fibronectin, c-src, non-muscle myosin II heavy chain B, fibroblast growth factor receptor (FGFR2) and 4.1R (9, 13, 17, 19, 29). A computational study indicates that the most common (U)GCAUG hexamers were found in the downstream introns of many neuron-specific exons, suggesting that this element may be a hallmark of neuronal splicing regulation (7, 28). Interestingly, it was noted that several (U)GCAUG repeats are present near the calcitonin-specific exon 4 (13). A study addressing the role of these repeats in the regulation of the rat calcitonin/CGRP pre-mRNA alternative processing yielded complicated results; although the authors suggested that these elements act to enhance the calcitonin-specific exon 4 inclusion, some deletion mutants nevertheless showed increased exon 4 inclusion in a CGRP-specific cell line (12).
Recently, a group of protein factors have been identified that interact with (U)GCAUG hexamers to regulate alternative splicing of a number of alternatively spliced exons (15). These proteins are Fox-1 and Fox-2, which share a common protein domain structure containing the four-stranded β-sheet of the RBD (4). Fox-1 and Fox-2 proteins are expressed in muscle, heart, and brain tissues (30). In the brain, these proteins are exclusively expressed in neurons (44). The alternatively spliced exons regulated by these proteins include neuronal N30 exon splicing in NMHC-B (15), the N1 exon in c-src (44), exon IIIb of FGFR2 (5), and exon 16 of protein 4.1R (31). In most cases, Fox-1 or Fox-2 proteins function to promote inclusion of an exon. However, the mechanism by which these proteins regulate splicing has not been addressed.
In the present study, we show that mutation of two of the four (U)GCAUG elements surrounding the 3′ splice site of the human calcitonin specific exon 4 dramatically increases exon 4 inclusion, indicating that these two UGCAUG repeats function as silencer elements. In HeLa cells, overexpression of Fox-1 or Fox-2 isoforms inhibits inclusion of exon 4, and this effect depends on the two UGCAUG silencer elements at −34 and +45 positions. Conversely, small interfering RNA (siRNA) knockdown of Fox-1 and Fox-2 proteins promotes calcitonin-specific splicing. To determine the mechanism of Fox-1/Fox-2-mediated regulation, we examined the binding of spliceosomal components to the 3′ splice site of exon 4. We show that the mutation of silencer elements at −34 and +45 increases U2AF65 binding to the 3′ splice site of the calcitonin-specific exon, and the addition of recombinant Fox-1 or Fox-2 protein blocks U2AF65 binding to the wild-type RNA. Furthermore, we demonstrate that exon 4 inclusion results from a balance between Fox-mediated silencing via the UGCAUG element and activation via U1 snRNP/TIAR/SRp20 binding to the intronic enhancer element. Mutation of the silencer element alleviates the need for the intronic enhancer element. In contrast, the deleterious effects of mutating the ESEs located on exon 4 cannot be rescued by mutation of the Fox-mediated silencer element, suggesting that the silencer element functions to specifically suppress ESE-dependent splicing. Finally, overexpression of Fox-1 or Fox-2 can still repress calcitonin-specific exon 4 splicing with a reporter in which the uridine branchpoint is changed to the canonical adenosine branchpoint.
These results reveal a critical role for Fox-1 and Fox-2 proteins in exon 4 definition of calcitonin/CGRP pre-mRNA and provide mechanistic insights into this regulation. Moreover, they establish a regulatory network that controls the fate of exon 4. Thus, Fox-1 and Fox-2 proteins represent one group of neuron-specific regulators in the calcitonin/CGRP system.
MATERIALS AND METHODS
Plasmids.
The human calcitonin/CGRP reporter constructs used in transfection experiments consist of calcitonin/CGRP gene exons 4 to 6 fused to a heterologous first exon from adenovirus (25). The mutated reporter constructs pCT-34, pCT+45, pCT+177 and pCT+252 were generated by PCR-directed mutagenesis and contain mutations (TGCATG to TGACTG) at the −34, +45, +177, and+252 positions surrounding the calcitonin-specific exon 4 3′ splice. The double-mutant construct pCT-34+45 and the quadruple mutant construct pCT-34+45+177+252 were generated by combining the single mutants. The pCT-5'ss and pCT-U-tract were previously described (25, 48). To create the pCT-34+5′ss, pCT45+5′ss, pCT-D+5′ss, pCT-34+U-tract, pCT45+U-tract, and pCT-D+U-tract plasmids, pCT-34, pCT+45, and pCT-34+45 were digested with XbaI and BsmI, and then fragments were cloned into pCT-5′ss and pCT-U-tract between the XbaI and BsmI sites, respectively. The reporter constructs pCT-ESEs and pCT-U-A were generated by PCR-directed mutagenesis and contain the mutations (GACTTCAACAAGTT—TGGAGCACCTGGAAAGAAAAGGGA to GAGTCGATGTACTT—TGAAGCTTATCCATTCTTATCCGA) and (TACTGTC to TACTGAC), respectively (the mutated nucleotides are underlined). The pCT-34+ESEs, pCT45+ESEs, and pCT-D+ESEs constructs were generated by combining the single mutants. To construct the plasmids pGEM-WT, pGEM-34, pGEM+45, and pGEM-34+45 used for in vitro transcription of RNA substrates in UV cross-linking assays, the pCT-WT and pCT-34+45 plasmids were digested with XbaI and NsiI and then subcloned into the pGEM-4 vector between the XbaI and PstI sites.
The pGEX-F011 and pGEX-F402 plasmids used to make recombinant Fox-2 proteins were generated by PCR-mediated cloning using pF011 and pF402 as templates. The plasmid for expression of GST-Fox-1 was generated through PCR mediated cloning into the pGEX-2TK vector using a Fox-1 cDNA (18).
Cell transfection and RNA and protein analysis.
HeLa cells transfection was carried out as previously described (22, 23). For each transfection, 2 μg of the calcitonin/CGRP reporter plasmid and 50, 200, or 400 ng of plasmids expressing different Fox-1 or Fox-2 isoforms were used. The procedures for total RNA and protein isolation and reverse transcription-PCR (RT-PCR) analysis were described previously (22). Use of low-cycle (18 to 20 cycles) PCR permitted determination of the relative abundance of individual RNA species. Quantification of exon inclusion was determined by PhosphorImager. The results shown are representative of at least three independent transfections for each experiment. The CA77 cell transfection was carried out similar to HeLa cells, except that Lipofectamine 2000 (Invitrogen) was used instead of regular Lipofectamine and that RNA was isolated at 72 h instead of at 48 h posttransfection. To analyze RNA isolated from CA77 cells, 22 to 24 cycles of RT-PCR were carried out.
siRNA-mediated knockdown of Fox-1 or Fox-2.
The Fox-1 and Fox-2 siRNA duplexes were synthesized (Dharmacon) based on the method of Underwood et al. (44) and were targeted the 3′ untranslated region of Fox-1 or Fox-2. The target sequences are CCGUUUUGCUCCAUAUUAA (Fox-1) and CCUGGCUAUUGCAAUAUUU (Fox-2), respectively. siRNA transfections were performed with same cell transfection method. Amounts of 20 and 100 pmol of siRNA were used in these transfections.
Western blot analysis.
Total cell lysate used in Western blot analysis was prepared in lysis buffer (50 mM Tris [pH 7.6], 150 mM NaCl, 0.1% sodium dodecyl sulfate [SDS], 1% NP-40) (22). Western blot analysis was carried out with anti-Fox-2 (1:250), antitubulin (1:5,000), and anti-Myc (1:5,000) antibodies.
In vitro assays.
UV cross-linking reaction was carried out in a volume of 50 μl containing 44% (vol/vol) HeLa cell nuclear extract, 20 mM creatine phosphate, 2 mM ATP, 0.6 mM MgCl2, 1.5% polyethylene glycol, 0.15 mM dithiothreitol, and 5 × 105 cpm of 32P-labeled RNA in the absence or presence of 20 or 60 ng of Fox-1 or Fox-2 protein/μl for the binding assay or for the cross-linking reactions, respectively. Reaction mixtures were incubated at 30°C for 10 min, and heparin was added to a final concentration of 2 μg/μl, followed by UV irradiation (254 nm) at 4°C for 15 min. Reaction mixtures were subsequently treated with 30 μg of RNase A at 37°C for 40 min. Cross-linked polypeptides were immunoprecipitated using monoclonal antibodies against Fox-1, Fox-2, or U2AF65. Immunoprecipitated proteins were separated on SDS-10% polyacrylamide gel electrophoresis (PAGE) gels.
Expression of GST-Fox-1 and GST-Fox-2 proteins.
Glutathione S-transferase (GST)-Fox-1 and GST-Fox-2 proteins were purified as previously described (5, 41).
RESULTS
The UGCAUG elements at positions −34 and+45 surrounding the 3′ splice site of the calcitonin-specific exon 4 suppress exon 4 inclusion.
Four (U)GCAUG elements are present on or surrounding the calcitonin-specific exon 4 in the human calcitonin/CGRP pre-mRNA. They are localized at positions −34, +45, +177, and +252 relative to the 3′ splice site of the calcitonin-specific exon 4. Three elements match the consensus hexamer UGCAUG, whereas the element at +177 site has the sequence of AGCAUG (Fig. 1B). To determine whether these elements regulate calcitonin/CGRP alternative splicing, the (U)GCAUG sequence was mutated individually or in combination by site-directed mutagenesis, changing (U)GCAUG to (U)GACUG. Mutated minigene constructs were tested by transient transfection of HeLa and CA77 cells that process the wild-type reporter pre-mRNA via the calcitonin- or CGRP-specific pathway, respectively. CA77 is a cell line derived from rat medullary thyroid carcinomas and exhibits a number of neuronal features (37). Importantly, calcitonin/CGRP is expressed endogenously in these cells, and its pre-mRNA is processed to produce predominantly CGRP mRNA.
A complete panel of mutants is illustrated in Fig. 1C. These reporter plasmids were transfected into both HeLa and CA77 cells, and RNA processing of their pre-mRNAs was assayed by RT-PCR using total RNA isolated from transfected cells (Fig. 1D). For all of the reporters, the results of at least three independent transfections are illustrated in the bar graph in Fig. 1E. As shown in Fig. 1D and E, the hexanucleotides at positions −34 and +45 are important for regulated exon 4 inclusion. In HeLa cells, exon 4 inclusion increased from 66.5% of the wild-type transcript to 76.5% for the −34 Mut and 85.7% for the +45 Mut transcript, respectively (compare lanes 2 or 3 to lane 1). Interestingly, a dramatic increase of exon 4 inclusion was observed in CA77 cells, where exon 4 is normally included at 20 to 30% for the wild-type transcript (compare lane 9 or 10 to lane 8). In contrast, mutation of the hexanucleotides at +177 or +252 position has little effect on alternative processing of the reporter pre-mRNA (compare lanes 4 and 5 to lane 1 for HeLa cell transfection and lanes 11 and 12 to lane 8 for CA77 cell transfection). When the mutations at the −34 and +45 hexanucleotide are combined (−34+45 Mut), exon 4 inclusion showed a slight increase over the +45 single mutants to 88% in HeLa cells and 93% in CA77 cells. The combination of mutations at all four positions gave results similar to the double mutant (Fig. 1D and E). These data demonstrate that the UGCAUG elements at −34 and +45 positions function as silencer elements for calcitonin-specific exon 4 inclusion.
Because the UGCAUG elements are potential binding targets of the Fox-1 and Fox-2 proteins, we next examined the expression levels of these proteins in HeLa and CA77 cells. As shown in Fig. 1F, although both cell lines produce Fox-2 protein, there is significantly more Fox-2 expression in CA77 cells than in HeLa cells, a result consistent with the report by Underwood et al., which showed no expression of Fox-1 and very low expression of Fox-2 protein in HeLa cells and high levels of both proteins in cell lines of neuronal origin (44). These results suggest that Fox-1 and Fox-2 may negatively regulate calcitonin/CGRP exon 4 inclusion and thus promote the neuron-specific pattern of alternative processing.
Overexpression of Fox-1 or Fox-2 protein inhibits exon 4 inclusion.
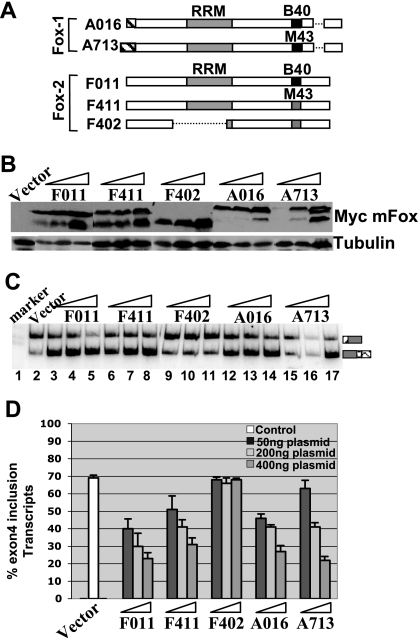
As a more direct test of our hypothesis, we overexpressed Fox proteins to determine whether this would modulate calcitonin/CGRP exon 4 inclusion. Fox-1 and Fox-2 proteins exist in many different isoforms. Nakahata and Kawamoto (30) found that the mouse A2BP (Fox-1) and Fxh (Fox-2) genes undergo brain- and muscle-specific alternative splicing, thereby generating at least five isoforms of Fox-1 and three isoforms of Fox-2. To evaluate the role of different Fox isoforms in calcitonin/CGRP alternative splicing, we overexpressed two Fox-1 isoforms (A016 and A713) and three Fox-2 isoforms (F011, F411, and F402) in HeLa cells. Of these proteins, A016 and F011 are brain specific and A713, F411, and F402 are muscle specific. However, F402 lacks the RRM domain and therefore serves as a negative control (Fig. 2A). We cotransfected a constant amount of the wild-type calcitonin/CGRP reporter construct with increasing amounts of different Fox protein expression plasmids. Western blot analysis using the anti-tag (myc) antibody shows the dose-dependent expression of different Fox protein isoforms (Fig. 2B). Figure 2C and D demonstrate that as the amount of A016, A713, F011, and F411 isoform expression was increased, the level of the exon 4 inclusion decreased in a dose-dependent manner. For example, with 50 ng of transfected plasmid, exon 4 inclusion decreased to 40% (lanes 3, 6, 12, and 15) and when 400 ng of expression plasmid was transfected, exon 4 inclusion decreased to 20 to 30% (lanes 5, 8, 14, and 17). As expected, overexpression of F402 did not affect exon 4 inclusion. These results demonstrate that both brain-specific and muscle-specific isoforms of Fox-1 or Fox-2 have the ability to inhibit the calcitonin-specific exon 4 inclusion. At 400 ng, no difference was detected between Fox-1 and Fox-2 in their ability to repress exon 4 inclusion.
FIG. 2.
Overexpression of Fox-1 and Fox-2 represses the calcitonin-specific exon 4 inclusion. (A) Schematic diagram of Fox-1 and Fox-2 isoforms used in the present study. The broken lines indicate that the sequence is not included in a specific isoform. This diagram is recreated based on the findings of Nakahata and Kawamoto (30). (B) Western blot analysis showing overexpression of different Myc-tagged Fox-1 and Fox-2 isoforms in HeLa cells. The blot was probed with antitubulin (control) and anti-Myc antibodies. For each isoform, increasing amounts (50, 200, or 400 ng) of the expression plasmid was used in the transfections. (C) RT-PCR assay as in Fig. 1D. (D) Quantification the RT-PCR results shown in panel C.
The effect of Fox-1 and Fox-2 proteins on exon 4 inclusion depends on the UGCAUG elements at −34 and+45 positions.
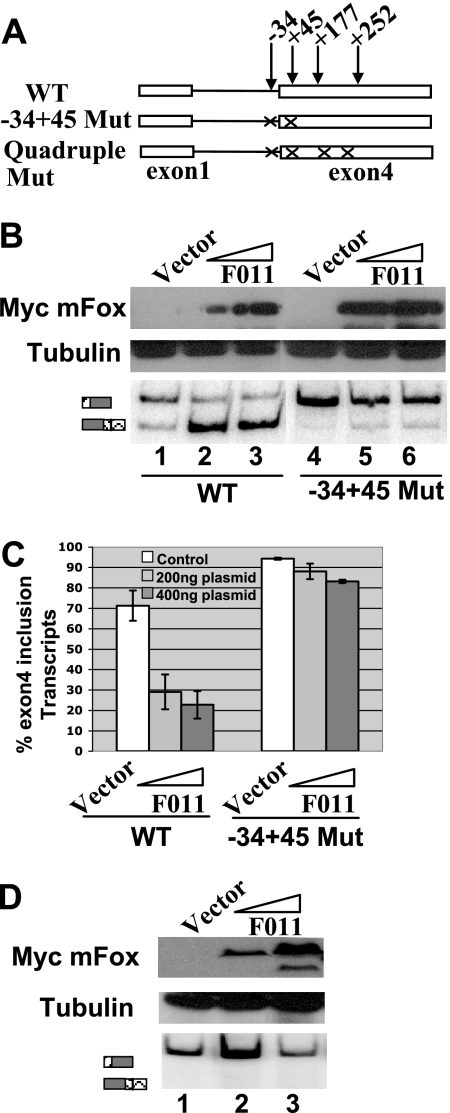
To test whether the repression of exon 4 inclusion by Fox-1 and Fox-2 depends on the presence of UGCAUG at −34 and +45 positions, we cotransfected F011 with the double mutant reporter minigene in which both of the UGCAUG elements at −34 and +45 positions were disrupted. While F011 (Fox-2) overexpression resulted in a dramatic decrease in exon 4 inclusion with the wild-type calcitonin/CGRP reporter, the −34+45 Mut reporter showed a very slight change (from 94 to 83%, Fig. 3B and C). A similar result was observed with A016 (Fox-1) protein (data not shown). These data demonstrate that the UGCAUG elements at the −34 and +45 positions are necessary for repression of exon 4 mediated by Fox-1 and Fox-2. The slight decrease of exon 4 inclusion when Fox-1 or Fox-2 protein was cotransfected with the double mutant suggests that Fox-1 and Fox-2 may still have an effect by interacting with other UGCAUG elements when the −34 and +45 UGCAUG elements are mutated. Consistent with this hypothesis, when the quadruple mutant was cotransfected with increasing amounts of F011 (Fox-2), no change in inclusion of exon 4 was observed (Fig. 3D). Exon 4 inclusion of the quadruple mutant transcript is 93% when cotransfected with a control vector and 93 to 94% when cotransfected with F011 (Fox-2) expression plasmids. Nevertheless, these results in aggregate show that the UGCAUG elements at the −34 and +45 positions are the key elements for the repression of calcitonin-specific exon 4 splicing mediated by Fox-1 and/or Fox-2 protein.
FIG. 3.
Repression of exon 4 inclusion mediated by Fox-1 and Fox-2 protein depends on the UGCAUG elements at −34 and +45 positions. (A) Diagram of reporter minigenes used in these experiments. (B) RT-PCR assay and Western blot analysis. (C) Quantification of the RT-PCR analysis of the cotransfection of wild-type or −34+45 Mut calcitonin/CGRP reporter minigene with increasing amounts of F011 (Fox-2) isoform expression plasmid (200 or 400 ng). (D) RT-PCR and Western blot analysis of the cotransfection of the quadruple mutant of the calcitonin/CGRP reporter minigene with increasing amounts of F011 (Fox-2) expression plasmid (200 or 400 ng).
RNAi-mediated Fox-2 knockdown promotes exon 4 inclusion in HeLa cells.
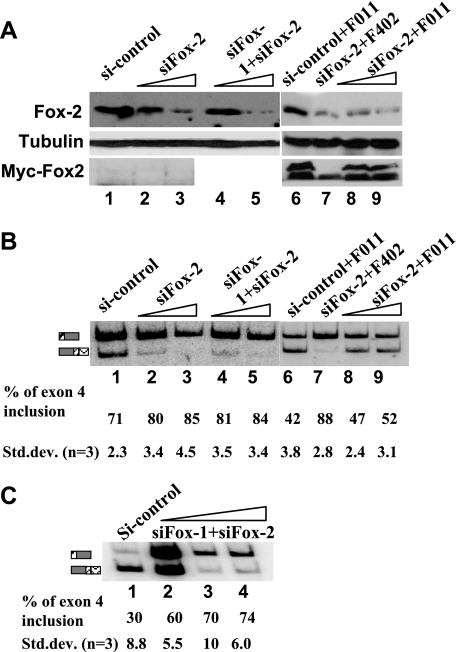
We next tested whether inclusion of exon 4 is altered when the levels of endogenous Fox proteins are decreased by siRNA-mediated knockdown. We used both HeLa and CA77 cells in this experiment. It was shown previously that HeLa cells have only a low level of Fox-2 and no detectable Fox-1 protein (44). When HeLa cells were treated with a specific siRNA duplex that targets Fox-2 mRNA, the level of Fox-2 protein was decreased significantly, and exon 4 inclusion of the wild-type calcitonin/CGRP reporter increased from 70 to 85% in a dose-dependent manner (Fig. 4A and B, lanes 1 to 3). A control siRNA did not alter Fox-2 expression, nor did it change exon 4 inclusion. When HeLa cells were treated with both Fox-1 and Fox-2 siRNA duplex, results were observed similar to those of the Fox-2-only siRNA duplex treatment (Fig. 4A and B, lanes 1, 4, and 5). Since the siRNA specific for Fox-2 targets a region in 3′-untranslated region of Fox-2, the expressed protein from the F011 (Fox-2) expression plasmid is resistant to this siRNA. When this RNAi-resistant F011 (Fox-2) expression plasmid was cotransfected with the siRNA duplex and the calcitonin/CGRP reporter, the levels of exon 4 inclusion were reduced to 47 and 52% with 20 and 100 nM Fox-2 siRNA duplex, respectively (Fig. 4B, lanes 6 to 9). This level is lower than that of the reporter in the absence of any siRNA (lane 1, 71%) because the protein level of overexpressed F011 is higher than that of the endogenous Fox-2 (not shown). However, the treatment of a control siRNA in the presence of F011 resulted in even less exon 4 inclusion (compare lanes 6 to 9, 42% versus 52%), indicating that the siRNA effect is an on-target, specific effect. As expected, knockdown of Fox-1 and Fox-2 in the CA77 cell line resulted in a dramatically dose-dependent increase of the exon 4 inclusion (from 30 to 74%) (Fig. 4C). These results suggest that the Fox-2 protein is the negative trans-acting factor driving the basal activity of the UGCAUG silencer in human calcitonin/CGRP processing in both HeLa and CA77 cells.
FIG. 4.
RNAi-mediated Fox-2 knockdown increases exon 4 inclusion and RNAi-resistant Fox-2 expression restores exon 4 repression. (A) Western blot analysis of the whole-cell lysate isolated after siRNA transfection of HeLa cells. The blot was probed with anti-tubulin as loading control, anti-Fox-2 antibodies to show knockdown of endogenous Fox-2 protein, and anti-myc to demonstrate expression of RNAi-resistant Fox-2 expression plasmid. Transfection of siRNA duplex was carried out in increasing amounts. Lane 1, control siRNA, 100 nM; lanes 2 and 3 and lanes 8 and 9, siRNA targeting Fox-2 (20 pmol or 100 pmol); lanes 4 and 5, siRNA targeting Fox-1 and Fox-2 (20 pmol + 20 pmol or 100 pmol + 100 pmol). (B) RT-PCR assay and quantification analysis. (C) RNAi-mediated Fox protein knockdown in CA77 cells. Transfection of siRNA duplex was carried out as in panel B.
Fox-1 and Fox-2 interact specifically with the UGCAUG elements at the −34 and +45 positions and inhibit U2AF65 binding to the 3′ splice site.
Previously, it has been shown that Fox-1 and Fox-2 proteins bind to the element (U)GCAUG with high specificity (15, 44). To confirm that the Fox-1 or Fox-2 protein does bind to the (U)GCAUG element in the context of the human calcitonin/CGRP pre-mRNA, we performed a UV cross-linking-immunoprecipitation (cross-linking/IP) assay using 32P-labeled in vitro-transcribed RNA substrate that contains all of the sequences up to the middle of exon 4 from the calcitonin/CGRP reporter minigene. HeLa nuclear extract contains low levels of Fox-2 and no Fox-1 protein, so we added 20 ng of A016 (Fox-1) or F011 (Fox-2)/μl into HeLa nuclear extract. In addition, F402 (Fox-2) was used as a negative control. The RNA substrates containing wild type, −34 mutant, +45 mutant, or the −34+45 double mutant were compared in these assays.
When the wild-type RNA was incubated in HeLa cell nuclear extract supplemented with the GST-F011 protein, one protein of 70 kDa, the size expected for the recombinant protein, was observed on SDS-PAGE when anti-Fox-2 antibodies were used (Fig. 5A, lane 2). However, when the wild-type RNA was incubated in HeLa extract supplemented with the GST-F402 protein that lacks the RRM domain, no protein was brought down by the anti-Fox-2 antibodies (Fig. 5A, lane 1). Note that the antibody does recognize the F402 (not shown). To determine the binding specificity of Fox-2 to the UGCAUG elements, we incubated the double-mutant RNA in HeLa nuclear extract supplemented with the GST-F011 protein. In this case, binding was dramatically reduced (Fig. 5A, lane 3).
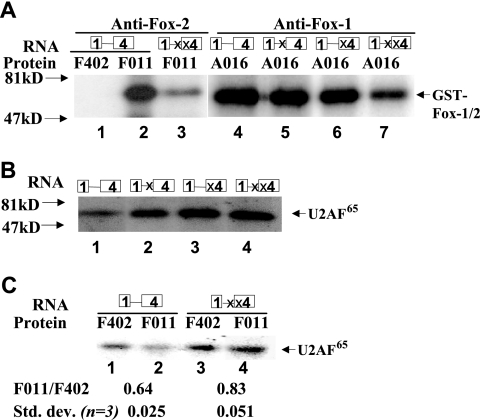
FIG. 5.
Fox-1 and Fox-2 proteins bind specifically to the UGCAUG elements at −34 and +45 positions and inhibit U2AF65 binding. (A) Interaction of GST-Fox-1 or GST-Fox-2 with the silencer elements in HeLa nuclear extract. The 32P-labeled in vitro-transcribed RNA substrate containing wild-type or mutated UGCAUG sequences at the −34, +45, or −34+45 position shown above the gel was UV cross-linked in HeLa cell nuclear extract supplemented with 20 ng of F402 (Fox-2)/μl (lanes 1), 20 ng of F011 (Fox-2)/μl (lanes 2 to 3), or 20 ng of A016 (Fox-1)/μl (lanes 4 to 7), followed by IP with antibodies specific to Fox-2 (lanes 1 to 3) or Fox-1 (lanes 4 to 7). Molecular mass markers are indicated in kilodaltons. (B) U2AF65 cross-linking with RNA substrate containing wild-type or mutated UGCAUG sequences at the −34, +45, or −34+45 position. The 32P-labeled, in vitro-transcribed RNAs were UV cross-linked in HeLa cell nuclear extract and immunoprecipitated with the anti-U2AF65 antibody. The positions of molecular mass markers and U2AF65 are indicated. (C) Addition of GST-Fox-2 decreases U2AF65 binding in a UGCAUG-dependent manner. Cross-linking/IP assays were carried out as in panel B. Lanes 1 and 3, 60 ng of GST-F402/μl; lanes 2 and 4, 60 ng of GST-F011 (Fox-2)/μl. The F011/F402 ratio shown below the gel represents the U2AF65 binding signal in lane 2 divided by that in lane 1 for the wild-type RNA and the signal in lane 4 divided by that in lane 3 for the double-mutant RNA.
To determine the relative contribution of individual Fox protein binding sites to the overall binding of RNA to Fox protein, we compared the binding of wild-type RNA to single- or double-mutant RNA substrates. Interestingly, cross-linking of the A016 (Fox-1) protein to −34 mutant and +45 mutant is similar to and only slightly less than the wild-type sequence, whereas the −34+45 double mutant resulted in strong reduction of cross-linking (Fig. 5A, lanes 4 to 7). These results suggest that Fox proteins are capable of binding to all of four (U)GCAUG elements at −34, +45, +177, and +252 positions, which are all included in the RNA substrate. There are still three (U)GCAUG elements in the two single-mutant RNA substrates, which resulted in strong cross-linking of the Fox-1 protein. However, the cell transfection experiments discussed in Fig. 1 indicate that binding of the Fox proteins to −34 or +45 UGCAUG elements leads to functional consequences. Nevertheless, these results demonstrate that both Fox-1 and Fox-2 proteins can bind specifically to the UGACUG elements at the −34 and +45 positions.
Next, we were interested in determining how Fox-1 or Fox-2 regulates the calcitonin/CGRP exon 4 inclusion. U2 auxiliary factor (U2AF) binds to the polypyrimidine tract at an early step in spliceosome assembly and promotes the ATP-dependent binding of U2 snRNP to the pre-mRNA branchpoint, thus playing a central role in maturation of the spliceosome (36). It has been shown that some splicing factors such as SR proteins regulate splicing by affecting U2AF large-subunit binding to the polypyrimidine tract upstream from the 3′ AG dinucleotide (16, 50). We performed a UV cross-linking/IP assay to determine whether Fox-1 and Fox-2 affect U2AF binding in the 3′ splice site of exon 4 of the calcitonin/CGRP pre-mRNA. The same RNA substrates described above were used in this assay. When the wild-type RNA was incubated in HeLa extract, binding of the 65-kDa U2AF65 protein was observed upon SDS-PAGE (Fig. 5B, lane 1). Interestingly, U2AF65 cross-linking was significantly increased in all of three mutant RNA substrates (Fig. 5B, lanes 2 to 4), suggesting that the UGCAUG silencer elements at the −34 and +45 positions are the important determinants of U2AF65 binding. However, the increase in U2AF65 cross-linking is significantly stronger for the +45 than for the −34 mutant (compare Fig. 5B, lane 2, to lane 3), a finding consistent with the RT-PCR result that +45 Mut results in more exon 4 inclusion than −34 Mut (Fig. 1E). To analyze the effect of Fox-1 and Fox-2 on U2AF65 binding, UV cross-linking was performed under splicing conditions in the presence of recombinant GST-F402 or GST-F011 (Fox-2), and the cross-linked proteins were immunoprecipitated with the anti-U2AF65 antibody. Addition of GST-F011 (Fox-2) but not of GST-F402 significantly inhibited the binding of U2AF65 to the wild-type RNA (Fig. 5C, lanes 1 and 2). When the double-mutant RNA was used, U2AF65 protein was only slightly reduced when GST-F011 was added compared to GST-F402 (Fig. 5C, lanes 3 and 4). Addition of Fox-1 yielded the same results as did the addition of Fox-2 (data not shown). Note that although there is a slight reduction of U2AF65 cross-linking to the double-mutant RNA when Fox-2 is added, the reduction is consistently smaller than that observed with the wild-type RNA (compare lane 4 to lane 3 versus lane 2 to lane 1). This result is in agreement with the cotransfection experiment wherein the overexpression of Fox proteins led to a slight reduction of exon 4 inclusion of the reporter that contains double mutations at −34 and +45 UGCAUG (Fig. 3). In conclusion, Fox-1 and Fox-2 proteins reduce U2AF65 binding in a UGCAUG element-dependent manner.
To confirm that U2AF65 binding is in fact to the polypyrimidine sequence of the 3′ splice site of exon 4 in the RNA substrate, we mutated the TTTTCCCT element located five nucleotides upstream of the 3′-AG to AAAAGGGA and tested the U2AF65 binding of the resulting mutant substrate. The in vitro-transcribed wild-type or mutant RNA substrate was labeled with both [32P]UTP and [32P]ATP. The result of the UV cross-linking/IP assay showed dramatically reduced the U2AF65 binding to the mutant RNA compared to wild type, suggesting that U2AF65 specifically binds to bona fide polypyrimidine tract sequence (data not shown).
Exon 4 definition results from a balance of the function of UGCAUG silencer element and intronic enhancer element located downstream of exon 4.
Previous studies showed that positive regulation of exon 4 inclusion is mediated by the binding of U1 snRNP/TIAR/SRp20 to an intronic enhancer element, the key sequence motifs of which include a pseudo 5′ splice site (5′ss) and a U-tract, in the downstream of exon 4. Since these results demonstrate that the negative trans-acting factors Fox-1 and Fox-2 repress exon 4 inclusion by binding to the silencer UGCAUG motifs at the −34 and +45 positions, we wanted to determine how each element contributes to the overall outcome of exon 4 inclusion. Thus, we combined different mutations in the silencer and enhancer sequence motifs as illustrated in Fig. 6A and tested their effects on exon 4 inclusion in transient transfection experiments. The effects of the 5′ss and U-tract sequence mutations were described previously (25, 48). The results of both single and double mutants are shown in Fig. 6B and C. In HeLa cells, consistent with previous studies, mutation of the pseudo 5′ splice site (5′ss) and U-tract element (U Mut) resulted in decreased exon 4 inclusion to 15 and 35%, respectively. Intriguingly, combining the 5′ss mutation with the +45 or −34+45 mutation increased exon 4 inclusion significantly (Fig. 6B, compare lanes 8 and 9 to lane 5 in the top panel). Interestingly, the mutation of both silencer motifs +45 and the pseudo 5′ splice site resulted in the same level of inclusion of exon 4 as the wild-type reporter (Fig. 6B, compare lane 9 to lane 1 in the top panel). Combining the U-tract mutation with the +45 or −34+45 mutation resulted in a similar increase of exon 4 inclusion (Fig. 6B and 6C). These results indicate that the intronic enhancer element is required only when the +45 UGCAUG silencer element is present. In contrast, combining the −34 mutation with the 5′ss mutation or U-tract mutation did not change exon 4 inclusion compared to the mutation of 5′ss or U-tract alone (Fig. 6B, compare lanes 7 and 10 to lanes 5 and 6). The UGCAUG elements at −34 position has a weak effect on splicing of calcitonin/CGRP, so it is possible that mutation of the −34 element cannot override the effect of the mutation of 5′ss or U-tract. In addition, these results indicate that the UGCAUG silencer element and intronic enhancer element can independently regulate exon 4 splicing.
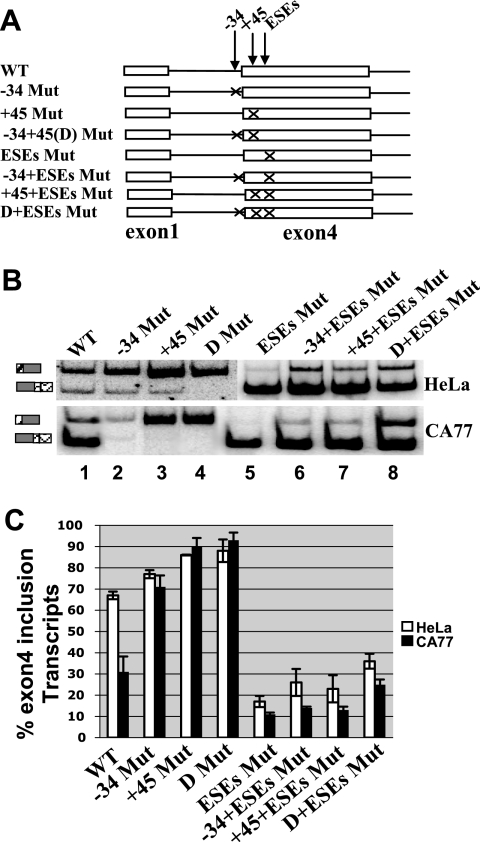
FIG. 6.
Antagonistic effect of the UGCAUG silencer elements and intronic enhancer element on exon 4 definition. (A) Diagram illustrating the wild-type and mutant reporter minigene constructs used in this experiments. An “X” indicates mutations of either UGCAUG to UGACUG at −34 and+45 position, CAGGUAAGAC to CAGCAUAGAC at the pseudo 5′ splice site (5′ss), or UUUUUAUUUUC to GUGUGAUGGUC at U-tract, respectively. (B) RT-PCR assay of total RNA isolated from HeLa or CA77 Cells transfected with the calcitonin/CGRP reporter minigenes. (C) Quantification of the RT-PCR analysis shown in panel B.
When the same mutant reporters were transfected in CA77 cells and analyzed for exon 4 inclusion, similar results were observed (Fig. 6B, bottom panel, and Fig. 6C). Amazingly, combination of the 5′ss and the UGCAUG mutant resulted in the same level of exon 4 inclusion in CA77 cells as in HeLa cells (compare lane 9 in Fig. 6B in the bottom panel to lane 1 in the top panel). Loss of cell-specific splicing of calcitonin/CGRP pre-mRNA in these double mutants suggests that major tissue-specific elements for calcitonin/CGRP are located within the silencer or intronic enhancer elements. Interestingly, the simultaneous mutation of the −34 element and the U-tract appears to significantly increase exon 4 inclusion compared to the single U-tract mutant in CA77 cells, which is different from in HeLa cells (Fig. 5B and C, compare lane 10 to lane 6). This different effect in HeLa and CA77 cells suggests that an additional trans-acting factor(s) is expressed differentially in the two cell lines.
The UGACUG silencer element at the +45 position functions to suppress ESE-dependent calcitonin specific exon 4 inclusion.
Previous studies demonstrate that two exonic enhancer elements (ESEs), designated A and B, are present on exon 4, and that they are bound by Tra2β and SRP55, respectively (42, 43). The UGCAUG element at the +45 position is 18 and 62 nucleotides away from the A and B elements, respectively (Fig. 1B). We suspected that the silencer element and ESEs are antagonistic to each other and tested this hypothesis by combining the two mutants (Fig. 7A). Consistent with previous studies, mutation of the ESEs decreased exon 4 inclusion drastically from 70 to 17% (Fig. 7B and C, compare lane 5 to lane 1 in the top panel). In contrast to the intronic enhancer element mutant, reduced exon 4 inclusion in the ESE mutant cannot be fully rescued by mutation of the UGCAUG silencer element at the +45 position (Fig. 7B, compare lanes 6 and 7 to lane 1). This is particularly obvious with the double mutants +45-ESEs and D+ESEs. Interestingly, although the effect of the +45 mutation is consistently stronger than that of the −34 mutation in the wild-type ESE background (exon 4 inclusion increased by 18% versus 9% increase over wild-type for the +45 and −34 mutants, respectively) (Fig. 7B and C, compare lanes 2 to 3 to lane 1; similar results are also shown in Fig. 1 and 6), the effect of the +45 mutation is very similar to that of the −34 mutation in the ESE mutant background (exon 4 inclusion increased by 6% versus 7% increase over the ESE mutant for the +45 and −34 mutants, respectively) (Fig. 7B and 7C, compare lanes 6 and 7 to lane 5). These results demonstrate that mutation of the ESEs decreased the effect of the +45 silencer element on exon 4 inclusion. Transfection with these different reporter minigenes into CA77 cells demonstrated similar results as in HeLa cells (Fig. 7B and C). Thus, these results suggest that the exonic UGACUG silencer element at the +45 position functions to suppress the ESE-dependent calcitonin specific exon 4 inclusion and that the intronic UGACUG silencer element at the −34 position regulates exon 4 splicing independent of the ESEs.
FIG. 7.
The UGACUG silencer element at the +45 position functions to suppress ESE-dependent calcitonin-specific exon 4 inclusion. (A) Diagram illustrating the wild-type and mutant calcitonin/CGRP reporter minigene constructs used to test the role of UGCAUG silencer elements and the ESE A and B enhancer elements in exon 4 definition. An “X” indicates mutations of either UGCAUG to UGACUG at −34 and +45 position or ESEs (both A and B elements), respectively. (B) RT-PCR assay of HeLa and CA77 cells transfections. (C) Quantification of RT-PCR analysis shown in panel B.
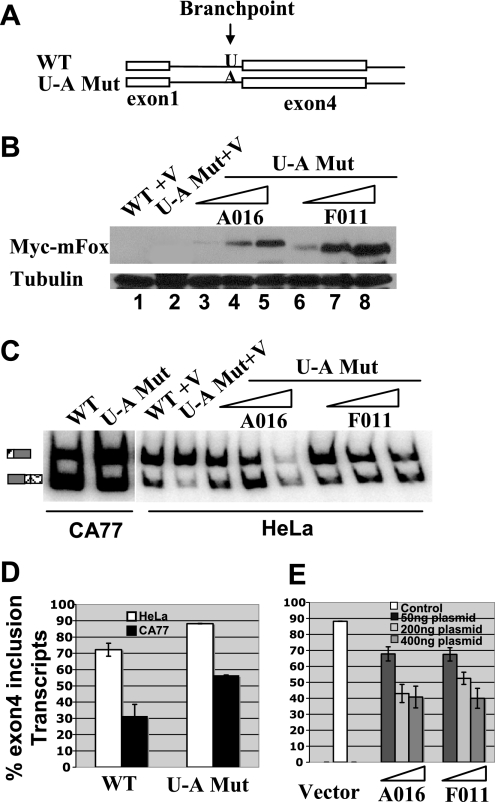
Overexpression of Fox represses the exon 4 inclusion of a reporter containing a mutation in the branchpoint sequence.
Repression of calcitonin specific exon 4 is mediated by the presence of a weak uridine branch acceptor at the 3′ splice site of exon 4. Mutation of this uridine residue into an adenosine residue resulted in a significant increase in calcitonin-specific splicing in several cell lines tested (1, 2). We wanted to determine whether Fox-1 or Fox-2 protein can still repress exon 4 inclusion after the weak uridine branch acceptor was changed to the canonical adenosine branch acceptor. When HeLa cells were transfected with the reporter construct that contains U-A point mutation at the branchpoint, exon 4 inclusion increased from 71% for the wild type to 88%, whereas in CA77 cells exon 4 inclusion increased from 31% for the wild type to 57% (Fig. 8D). This result is consistent with the previous studies (1, 2). Interestingly, 57% exon 4 inclusion in CA77 cells with this mutant suggests that some other silencer elements in calcitonin/CGRP reporter may still be at work repressing exon 4 inclusion. We next cotransfected the wild-type reporter construct with increasing amounts of the Fox-1 or Fox-2 expression plasmid in HeLa cells. Indeed, in the presence of the U-A branchpoint mutation, Fox proteins can still suppress exon 4 inclusion in a dose-dependent manner. For example, with 50 ng of transfected plasmid, exon 4 inclusion decreased from 87 to 67%, and when 400 ng of cDNA was transfected, exon 4 inclusion decreased to 40% (Fig. 8E). These results suggest that the branchpoint and UGCAUG silencer elements function independently in regulating exon 4 inclusion in the calcitonin/CGRP pre-mRNA.
FIG. 8.
Overexpression of either Fox-1 or Fox-2 represses the calcitonin-specific exon 4 inclusion of a reporter minigene containing a U-A branchpoint mutation at the 3′ splice site upstream of exon 4. (A) Diagram indicating the branchpoint mutation in the calcitonin/CGRP reporter. (B) Western blot analysis showing overexpression of Fox-1 or Fox-2 proteins. The blot was probed with anti-tubulin (control) and anti-Myc antibodies. (C) RT-PCR assay of HeLa and CA77 cells transfections. (D) Quantification of RT-PCR analysis with RNA isolated from both HeLa and CA77 cells transfected with the wild-type or branchpoint U-A mutant reporter minigene. (E) Quantification of RT-PCR analysis with RNA isolated from HeLa cells cotransfected with branchpoint U-A mutant calcitonin/CGRP reporter gene and a vector plasmid or increasing amounts of A016 (Fox-1) or F011 (Fox-2) expression plasmid (50, 200, or 400 ng).
DISCUSSION
Previous studies have shown that the (U)GCAUG element functions as both a silencer and enhancer of alternative splicing (9, 12, 13, 17, 19, 29). The evolutionarily conserved (U)GCAUG element is widely associated with neuron-specific and muscle-specific exons (28). Recently, Fox proteins have been shown to regulate splicing via binding to (U)GCAUG elements (5, 15, 31, 44). The mouse Fox-1 and Fox-2 are both specifically expressed in the brain in addition to the muscle and heart (30). In the brain, these proteins are specifically expressed in neurons and not glia (44). We report here that Fox-1- and Fox-2-mediated splicing regulation also plays an important role in controlling tissue-specific alternative RNA processing of human calcitonin/CGRP pre-mRNA. We demonstrate that two UGCAUG elements, one located 34 nucleotides upstream of and the other 45 nucleotides downstream of the beginning of exon 4, function as splicing silencers for exon 4 inclusion. Furthermore, Fox-1 and Fox-2 proteins specifically bind these silencers and negatively regulate exon 4 inclusion. These results indicate that Fox-1 and Fox-2 are important for the neuron-specific CGRP-specific pathway, and they function by blocking the calcitonin-specific exon 4 inclusion. These results appear to be contradictory to a previous study on the role of the (U)GCAUG elements in regulating the rat calcitonin/CGRP alternative RNA processing (12). The previous study concluded that these elements function as enhancer elements for the calcitonin-specific exon 4 inclusion (12). However, in that study, the results appear to be very complex when deletion mutants and mutants with inserted multimerized repeats were tested. For example, an intronic deletion removing two (U)GCAUG repeats did cause a slight increase in exon 4 inclusion in a CGRP-specific cell line. In addition, when multimerized UGCAUG elements were inserted upstream of exon 4, calcitonin-specific splicing was substantially reduced (12). The discrepancy between the two studies may also reflect differences between the rat and human calcitonin/CGRP gene.
It has been shown that many factors can bind to the calcitonin/CGRP pre-mRNA and regulate exon 4 inclusion. U1 snRNP, SRp20, PTB, and TIAR bind to the intronic enhancer elements located downstream of exon 4 and promote exon 4 inclusion (21-23, 25, 48). Tra2β and SRp55 are recruited to ESE A and B and activate exon 4 inclusion (42, 43, 46). However, all of these regulators are expressed in many tissues and are unlikely to be responsible for neuron-specific regulation of calcitonin/CGRP alternative processing. Therefore, how the neuronal pattern of processing in the calcitonin/CGRP transcript is achieved remained poorly understood. Recently, our group showed that the neuron-specific Hu proteins can bind to the U-tract of the intronic enhancer element located downstream of exon 4, thereby blocking access of TIAR and as a result, promoting the CGRP-specific pathway (H. Zhu, R. A. Hasman, and H. Lou, unpublished data). Interestingly, Hu proteins are not sufficient to block exon 4 inclusion because overexpression of Hu proteins in HeLa cells did not change the ratio of RNA with exon 4 included (Zhu et al., unpublished). Studies in the present report identify Fox-1 and Fox-2 proteins as major contributors to neuron-specific exon 4 exclusion for calcitonin/CGRP pre-mRNA. However, it is also clear that Fox-1 and Fox-2 are not the only neuron-specific regulators responsible for the CGRP pathway. For example, as shown in Fig. 7B and C, when both ESE and UGCAUG silencer elements are mutated, exon 4 inclusion is consistently lower in CA77 cells than in HeLa cells. In this case, Hu proteins may be responsible for the reduced exon 4 inclusion.
Hints that sequences located near the 3′ splice site of exon 4 contain neuron-specific splicing silencers came from a few previous studies. Analyzing a number of deletion and substitution mutant surrounding the 3′ splice site of exon 4 in the rat calcitonin/CGRP reporter constructs, Emeson et al. suggested that a cis-acting element at the calcitonin-specific 3′ splice junction plays an important role in regulating tissue-specific processing of the calcitonin/CGRP pre-mRNA (10). Consistent with this idea, two calcitonin/CGRP splice regulator proteins (CSRs) were suggested to function as neuron-specific negative regulators in alternative calcitonin/CGRP splicing (8, 33). These CSRs were purified through biochemical approaches from rat brain nuclear extracts based on their ability to bind an RNA substrate containing a sequence of 380 nucleotides surrounding the calcitonin-specific exon 4 (8, 33). However, to date, the identity of these two CSR proteins has not been reported. It is likely that Fox-1 and Fox-2 proteins are the two CSR proteins based on the following considerations. First, the two CSR proteins show approximate molecular masses of 43 and 41 kDa, respectively, similar to Fox-1 and Fox-2. Second, previous studies demonstrated that both proteins have the ability to form a weak complex with a 21-nucleotide target sequence containing the −34 UGCAUG silencer elements (33). Finally, Fox-1 and Fox-2 proteins are expressed in the brain, specifically in neurons (44).
Although it is clear that Fox-1 and Fox-2 proteins regulate alternative splicing of a handful of pre-mRNAs through binding to (U)GCAUG elements, the mechanism of such regulation has not been explored. Our results demonstrate for the first time that the Fox-1 and Fox-2 proteins reduce U2AF65 binding to the polypyrimidine tract of the 3′ splice site of the calcitonin/CGRP exon 4 in a UGCAUG element-dependent manner (Fig. 5). U2AF plays an essential role in promoting the ATP-dependent binding of U2 snRNP to the pre-mRNA branchpoint (36). It has been shown that a number of splicing regulatory factors affect U2AF binding to the polypyrimidine tract upstream from the 3′ splice site, thereby regulating splicing. For example, in female fruit flies, SXL protein binds to the polypyrimidine tract of the male-specific 3′ splice site of the transformer pre-mRNA to block access of U2AF, thereby promoting usage of the downstream female-specific 3′ splice site (45). Repression of splicing by PTB can also occur by competing with U2AF65 binding (20, 39). It has also been shown that ESE binding proteins such as SR proteins promote splicing by increasing U2AF65 binding (16, 50). It remains to be determined how Fox-1 and Fox-2 interfere with U2AF65 binding. Since the UGCAUG at −34 is only six nucleotides upstream from the branchpoint sequence, it is possible that Fox-1 or Fox-2 bound at this element physically interferes with U2AF or U2 snRNP interaction with their cognate binding sites. It is also possible that Fox-1 or Fox 2 interacts with a component(s) of the splicing complex, thereby affecting its assembly at the 3′ splice site upstream of exon 4, which may explain why the small increase in U2AF binding does not appear to correlate with the large decrease in Fox binding in the mutant. Additional experiments are necessary to distinguish these possibilities.
The exonic UGCAUG silencer element at+45 position on exon 4 is very close to the ESEs mediated by Tra2β and SRp55 protein (18 nucleotides from ESE A and 62 nucleotides from ESE B) (Fig. 1B). Thus, repression of splicing by Fox-1 and Fox-2 can also occur by interfering with Tra2β and SRp55 proteins binding to these ESEs. Our results indicate that the +45 UGACUG silencer element functions to suppress ESE-dependent calcitonin-specific exon 4 splicing. When the ESEs are mutated, the second mutation at the +45 UGCAUG silencer did not increase exon 4 inclusion to the same extent as it did with the 5′ss mutation-containing reporter (Fig. 6 and 7). A number of studies have demonstrated that the ESE-dependent 3′ splice site activation by SR proteins involves the recruitment of U2AF65 to weak polypyrimidine tracts (16, 50). It is therefore possible that Fox-1 and Fox-2 antagonize the binding of Tra2β and SRp55 to ESEs through interacting with the +45 UGCAUG silencer element. As a result, the Fox proteins inhibit U2AF65 binding, leading to repression of the exon 4 inclusion.
Our results also indicate an interesting coordinated regulation of exon 4 inclusion. In HeLa cells, the mutant reporter minigene containing a mutation at the intronic pseudo 5′ splice site produced little exon 4 inclusion (15%). Intriguingly, combining the pseudo 5′ splice site mutation with the −34+45 silencer mutation resulted in 63% exon 4 inclusion, which is very close to the 70% exon 4 inclusion for the wild-type reporter minigene (Fig. 6). This result indicates that a delicate balance between the hexanucleotide silencer and intronic enhancer elements has evolved to control exon 4 definition. The silencer elements and intronic enhancer elements contribute individually to exon 4 recognition. However, the intronic enhancer element is only required when the UGCAUG silencer element is present. When the same combined mutant reporter was introduced into CA77 cells, the level of exon 4 inclusion approaches that in HeLa cells, indicating that this mutant combination results in a loss of cell-specific regulation of exon 4 inclusion. This result suggests that neuron-specific regulatory elements for calcitonin/CGRP pre-mRNA are located within the silencer elements and/or intronic enhancer elements, which is consistent with the identification of Fox-1/Fox-2 and Hu proteins as neuron-specific regulators of the calcitonin/CGRP system. A number of alternatively included exons have been demonstrated to undergo complex control through a regulatory network involving both positive and negative factors (11, 14, 34, 49).
Like many other alternatively included exons, the calcitonin-specific exon 4 is flanked by suboptimal splicing signals, which is typified by the presence of a uridine (cytosine in the rat calcitonin/CGRP pre-mRNA) instead of the canonical adenosine branchpoint. It was demonstrated previously that exon 4 inclusion is significantly increased when the noncanonical branchpoint is mutated to adenosine in a cell type-independent fashion (47). Our results indicate that Fox-mediated repression of exon 4 inclusion can override the presence of a strong branchpoint. Addition of Fox proteins decreased exon 4 inclusion of the reporter that contains the U-A branchpoint mutation (Fig. 8D). This result is consistent with the fact that exon 4 inclusion increases from 31 to 57% (compare to 88% in HeLa cells) when the branchpoint is changed from U to A in CA77 cells (Fig. 8C), suggesting that the repressors in these cells such as Fox proteins are still at work. Therefore, the suboptimal uridine branchpoint and UGCAUG silencer elements may coregulate exon 4 inclusion in an additive but not synergistic manner.
Based on our results, a model for how tissue-specific regulation of calcitonin/CGRP is achieved is depicted in Fig. 9. Identification of a novel neuron-specific regulatory factor in this system represents a significant step forward toward fully understanding the regulatory mechanism controlling this specific alternative splicing event. It will be important to decipher how Fox proteins interact with the spliceosome to repress splicing of specific exons.
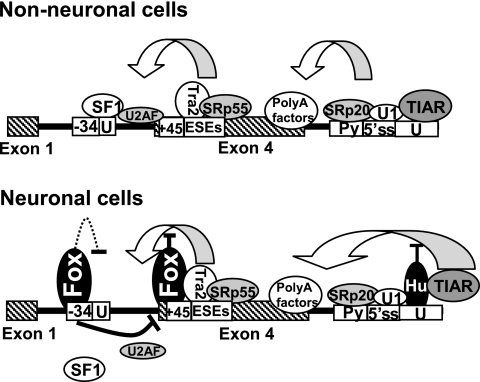
FIG. 9.
Models to explain how nonneuronal and neuron-specific alternative RNA processing of the human calcitonin/CGRP pre-mRNA are achieved. In nonneuronal cells, a number of trans-acting factors promote exon 4 inclusion at both ends of this exon. In neuronal cells, Fox proteins bind to UGCAUG elements and interfere with binding of splicing factors at the 3′ splice site, whereas Hu proteins antagonize the activity of TIAR protein leading to decreased exon 4 inclusion. The dashed line indicates a hypothesized role of Fox proteins in interfering with SF1 binding at the branchpoint sequence.
Acknowledgments
We thank the following individuals for providing antibody and plasmids: Doug Black at UCLA (anti-Fox-1 and anti-Fox-2 antibodies), Sachiyo Kawamoto at the NIH (F011, F411, F402, A016, and A713 Fox expression plasmids), and Andrew Lieberman at the University of Michigan (Fox-1 cDNA plasmid). We thank Helen Salz and Jo Ann Wise for critical reading of the manuscript.
This study was supported by an NIH grant to H.L. (NS-049103-01). A.P.B. was supported by NIH grant RO1 GM63090 to M. A. Garcia-Blanco.
Footnotes
Published ahead of print on 13 November 2006.
REFERENCES
- 1.Adema, G. J., R. A. Bovenberg, H. S. Jansz, and P. D. Baas. 1988. Unusual branch point selection involved in splicing of the alternatively processed Calcitonin/CGRP-I pre-mRNA. Nucleic Acids Res. 16:9513-9526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adema, G. J., K. L. van Hulst, and P. D. Baas. 1990. Uridine branch acceptor is a cis-acting element involved in regulation of the alternative processing of calcitonin/CGRP-l pre-mRNA. Nucleic Acids Res. 18:5365-5373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amara, S. G., V. Jonas, M. G. Rosenfeld, E. S. Ong, and R. M. Evans. 1982. Alternative RNA processing in calcitonin gene expression generates mRNAs encoding different polypeptide products. Nature 298:240-244. [DOI] [PubMed] [Google Scholar]
- 4.Auweter, S. D., R. Fasan, L. Reymond, J. G. Underwood, D. L. Black, S. Pitsch, and F. H. Allain. 2006. Molecular basis of RNA recognition by the human alternative splicing factor Fox-1. EMBO J. 25:163-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baraniak, A. P., J. R. Chen, and M. A. Garcia-Blanco. 2006. Fox-2 mediates epithelial cell-specific fibroblast growth factor receptor 2 exon choice. Mol. Cell. Biol. 26:1209-1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Black, D. L. 2003. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 72:291-336. [DOI] [PubMed] [Google Scholar]
- 7.Brudno, M., M. S. Gelfand, S. Spengler, M. Zorn, I. Dubchak, and J. G. Conboy. 2001. Computational analysis of candidate intron regulatory elements for tissue-specific alternative pre-mRNA splicing. Nucleic Acids Res. 29:2338-2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coleman, T. P., Q. Tran, and J. R. Roesser. 2003. Binding of a candidate splice regulator to a calcitonin-specific splice enhancer regulates calcitonin/CGRP pre-mRNA splicing. Biochim. Biophys. Acta 1625:153-164. [DOI] [PubMed] [Google Scholar]
- 9.Deguillien, M., S. C. Huang, M. Moriniere, N. Dreumont, E. J. Benz, Jr., and F. Baklouti. 2001. Multiple cis elements regulate an alternative splicing event at 4.1R pre-mRNA during erythroid differentiation. Blood 98:3809-3816. [DOI] [PubMed] [Google Scholar]
- 10.Emeson, R. B., F. Hedjran, J. M. Yeakley, J. W. Guise, and M. G. Rosenfeld. 1989. Alternative production of calcitonin and CGRP mRNA is regulated at the calcitonin-specific splice acceptor. Nature 341:76-80. [DOI] [PubMed] [Google Scholar]
- 11.Eperon, I. C., O. V. Makarova, A. Mayeda, S. H. Munroe, J. F. Caceres, D. G. Hayward, and A. R. Krainer. 2000. Selection of alternative 5′ splice sites: role of U1 snRNP and models for the antagonistic effects of SF2/ASF and hnRNP A1. Mol. Cell. Biol. 20:8303-8318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hedjran, F., J. M. Yeakley, G. S. Huh, R. O. Hynes, and M. G. Rosenfeld. 1997. Control of alternative pre-mRNA splicing by distributed pentameric repeats. Proc. Natl. Acad. Sci. USA 94:12343-12347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huh, G. S., and R. O. Hynes. 1994. Regulation of alternative pre-mRNA splicing by a novel repeated hexanucleotide element. Genes Dev. 8:1561-1574. [DOI] [PubMed] [Google Scholar]
- 14.Izquierdo, J. M., N. Majos, S. Bonnal, C. Martinez, R. Castelo, R. Guigo, D. Bilbao, and J. Valcarcel. 2005. Regulation of Fas alternative splicing by antagonistic effects of TIA-1 and PTB on exon definition. Mol. Cell 19:475-484. [DOI] [PubMed] [Google Scholar]
- 15.Jin, Y., H. Suzuki, S. Maegawa, H. Endo, S. Sugano, K. Hashimoto, K. Yasuda, and K. Inoue. 2003. A vertebrate RNA-binding protein Fox-1 regulates tissue-specific splicing via the pentanucleotide GCAUG. EMBO J. 22:905-912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kan, J. L., and M. R. Green. 1999. Pre-mRNA splicing of IgM exons M1 and M2 is directed by a juxtaposed splicing enhancer and inhibitor. Genes Dev. 13:462-471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kawamoto, S. 1996. Neuron-specific alternative splicing of nonmuscle myosin II heavy chain-B pre-mRNA requires a cis-acting intron sequence. J. Biol. Chem. 271:17613-17616. [PubMed] [Google Scholar]
- 18.Lieberman, A. P., D. L. Friedlich, G. Harmison, B. W. Howell, C. L. Jordan, S. M. Breedlove, and K. H. Fischbeck. 2001. Androgens regulate the mammalian homologues of invertebrate sex determination genes tra-2 and fox-1. Biochem. Biophys. Res. Commun. 282:499-506. [DOI] [PubMed] [Google Scholar]
- 19.Lim, L. P., and P. A. Sharp. 1998. Alternative splicing of the fibronectin EIIIB exon depends on specific TGCATG repeats. Mol. Cell. Biol. 18:3900-3906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin, C. H., and J. G. Patton. 1995. Regulation of alternative 3′ splice site selection by constitutive splicing factors. RNA 1:234-245. [PMC free article] [PubMed] [Google Scholar]
- 21.Lou, H., G. J. Cote, and R. F. Gagel. 1994. The calcitonin exon and its flanking intronic sequences are sufficient for the regulation of human calcitonin/calcitonin gene-related peptide alternative RNA splicing. Mol. Endocrinol. 8:1618-1626. [DOI] [PubMed] [Google Scholar]
- 22.Lou, H., and R. F. Gagel. 1998. Alternative RNA processing: its role in regulating expression of calcitonin/calcitonin gene-related peptide. J. Endocrinol. 156:401-405. [DOI] [PubMed] [Google Scholar]
- 23.Lou, H., and R. F. Gagel. 1999. Mechanism of tissue-specific alternative RNA processing of the calcitonin CGRP gene. Front. Horm. Res. 25:18-33. [DOI] [PubMed] [Google Scholar]
- 24.Lou, H., R. F. Gagel, and S. M. Berget. 1996. An intron enhancer recognized by splicing factors activates polyadenylation. Genes Dev. 10:208-219. [DOI] [PubMed] [Google Scholar]
- 25.Lou, H., Y. Yang, G. J. Cote, S. M. Berget, and R. F. Gagel. 1995. An intron enhancer containing a 5′ splice site sequence in the human calcitonin/calcitonin gene-related peptide gene. Mol. Cell. Biol. 15:7135-7142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Massiello, A., A. Salas, R. L. Pinkerman, P. Roddy, J. R. Roesser, and C. E. Chalfant. 2004. Identification of two RNA cis-elements that function to regulate the 5′ splice site selection of Bcl-x pre-mRNA in response to ceramide. J. Biol. Chem. 279:15799-15804. [DOI] [PubMed] [Google Scholar]
- 27.Matlin, A. J., F. Clark, and C. W. Smith. 2005. Understanding alternative splicing: toward a cellular code. Nat. Rev. Mol. Cell. Biol. 6:386-398. [DOI] [PubMed] [Google Scholar]
- 28.Minovitsky, S., S. L. Gee, S. Schokrpur, I. Dubchak, and J. G. Conboy. 2005. The splicing regulatory element, UGCAUG, is phylogenetically and spatially conserved in introns that flank tissue-specific alternative exons. Nucleic Acids Res. 33:714-724.15691898 [Google Scholar]
- 29.Modafferi, E. F., and D. L. Black. 1997. A complex intronic splicing enhancer from the c-src pre-mRNA activates inclusion of a heterologous exon. Mol. Cell. Biol. 17:6537-6545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nakahata, S., and S. Kawamoto. 2005. Tissue-dependent isoforms of mammalian Fox-1 homologs are associated with tissue-specific splicing activities. Nucleic Acids Res. 33:2078-2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ponthier, J. L., C. Schluepen, W. Chen, R. A. Lersch, S. L. Gee, V. C. Hou, A. J. Lo, S. A. Short, J. A. Chasis, J. C. Winkelmann, and J. G. Conboy. 2006. Fox-2 splicing factor binds to a conserved intron motif to promote inclusion of protein 4.1R alternative exon 16. J. Biol. Chem. 281:12468-12474. [DOI] [PubMed] [Google Scholar]
- 32.Roesser, J. R. 2004. Both U2 snRNA and U12 snRNA are required for accurate splicing of exon 5 of the rat calcitonin/CGRP gene. RNA 10:1243-1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roesser, J. R., K. Liittschwager, and S. E. Leff. 1993. Regulation of tissue-specific splicing of the calcitonin/calcitonin gene-related peptide gene by RNA-binding proteins. J. Biol. Chem. 268:8366-8375. [PubMed] [Google Scholar]
- 34.Rooke, N., V. Markovtsov, E. Cagavi, and D. L. Black. 2003. Roles for SR proteins and hnRNP A1 in the regulation of c-src exon N1. Mol. Cell. Biol. 23:1874-1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rosenfeld, M. G., S. G. Amara, and R. M. Evans. 1984. Alternative RNA processing: determining neuronal phenotype. Science 225:1315-1320. [DOI] [PubMed] [Google Scholar]
- 36.Ruskin, B., P. D. Zamore, and M. R. Green. 1988. A factor, U2AF, is required for U2 snRNP binding and splicing complex assembly. Cell 52:207-219. [DOI] [PubMed] [Google Scholar]
- 37.Russo, A. F., T. M. Lanigan, and B. E. Sullivan. 1992. Neuronal properties of a thyroid C-cell line: partial repression by dexamethasone and retinoic acid. Mol. Endocrinol. 6:207-218. [DOI] [PubMed] [Google Scholar]
- 38.Sabate, M. I., L. S. Stolarsky, J. M. Polak, S. R. Bloom, I. M. Varndell, M. A. Ghatei, R. M. Evans, and M. G. Rosenfeld. 1985. Regulation of neuroendocrine gene expression by alternative RNA processing. Colocalization of calcitonin and calcitonin gene-related peptide in thyroid C-cells. J. Biol. Chem. 260:2589-2592. [PubMed] [Google Scholar]
- 39.Singh, R., J. Valcarcel, and M. R. Green. 1995. Distinct binding specificities and functions of higher eukaryotic polypyrimidine tract-binding proteins. Science 268:1173-1176. [DOI] [PubMed] [Google Scholar]
- 40.Stamm, S., S. Ben-Ari, I. Rafalska, Y. Tang, Z. Zhang, D. Toiber, T. A. Thanaraj, and H. Soreq. 2005. Function of alternative splicing. Gene 344:1-20. [DOI] [PubMed] [Google Scholar]
- 41.Sune, C., and M. A. Garcia-Blanco. 1995. Transcriptional trans activation by human immunodeficiency virus type 1 Tat requires specific coactivators that are not basal factors. J. Virol. 69:3098-3107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tran, Q., T. P. Coleman, and J. R. Roesser. 2003. Human transformer 2beta and SRp55 interact with a calcitonin-specific splice enhancer. Biochim. Biophys. Acta 1625:141-152. [DOI] [PubMed] [Google Scholar]
- 43.Tran, Q., and J. R. Roesser. 2003. SRp55 is a regulator of calcitonin/CGRP alternative RNA splicing. Biochemistry 42:951-957. [DOI] [PubMed] [Google Scholar]
- 44.Underwood, J. G., P. L. Boutz, J. D. Dougherty, P. Stoilov, and D. L. Black. 2005. Homologues of the Caenorhabditis elegans Fox-1 protein are neuronal splicing regulators in mammals. Mol. Cell. Biol. 25:10005-10016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Valcarcel, J., R. Singh, P. D. Zamore, and M. R. Green. 1993. The protein Sex-lethal antagonizes the splicing factor U2AF to regulate alternative splicing of transformer pre-mRNA. Nature 362:171-175. [DOI] [PubMed] [Google Scholar]
- 46.van Oers, C. C., G. J. Adema, H. Zandberg, T. C. Moen, and P. D. Baas. 1994. Two different sequence elements within exon 4 are necessary for calcitonin-specific splicing of the human calcitonin/calcitonin gene-related peptide I pre-mRNA. Mol. Cell. Biol. 14:951-960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zandberg, H., T. C. Moen, and P. D. Baas. 1995. Cooperation of 5′ and 3′ processing sites as well as intron and exon sequences in calcitonin exon recognition. Nucleic Acids Res. 23:248-255. [PMC free article] [PubMed] [Google Scholar]
- 48.Zhu, H., R. A. Hasman, K. M. Young, N. L. Kedersha, and H. Lou. 2003. U1 snRNP-dependent function of TIAR in the regulation of alternative RNA processing of the human calcitonin/CGRP pre-mRNA. Mol. Cell. Biol. 23:5959-5971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhu, J., A. Mayeda, and A. R. Krainer. 2001. Exon identity established through differential antagonism between exonic splicing silencer-bound hnRNP A1 and enhancer-bound SR proteins. Mol. Cell 8:1351-1361. [DOI] [PubMed] [Google Scholar]
- 50.Zuo, P., and T. Maniatis. 1996. The splicing factor U2AF35 mediates critical protein-protein interactions in constitutive and enhancer-dependent splicing. Genes Dev. 10:1356-1368. [DOI] [PubMed] [Google Scholar]