Abstract
Ectopic expression of Cdc6p results in mitotic delay, and this has been attributed to Cdc6p-mediated inhibition of Cdc28 protein kinase and failure to activate the anaphase-promoting complex (APC). Here we show that endogenous Cdc6p delays a specific subset of mitotic events and that Cdc28 inhibition is not sufficient to account for it. The depletion of Cdc6p in G2/M cells reveals that Cdc6p is rate limiting for the degradation of the APC/Cdc20 substrates Pds1p and Clb2p. Conversely, the premature expression of Cdc6p delays the degradation of APC/Cdc20 substrates. Abolishing Cdc6p/Cdc28p interaction does not eliminate the Cdc6-dependent delay of these anaphase events. To identify additional Cdc6-mediated, APC-inhibitory mechanisms, we looked for mutants that reversed the mitotic delay. The deletion of SWE1, RAD24, MAD2, or BUB2 had no effect. However, disrupting CDC55, a PP2A regulatory subunit, suppressed the Cdc6p-dependent delay of Pds1 and Clb2 destruction. A specific role for CDC55 was supported by demonstrating that the lethality of Cdc6 ectopic expression in a cdc16-264 mutant is suppressed by the deletion of CDC55, that endogenous Cdc6p coimmunoprecipitates with the Cdc55 and Tpd3 subunits of PP2A, that Cdc6p/Cdc55p/Tpd3 interaction occurs only during mitosis, and that Cdc6 affects PP2A-Cdc55 activity during anaphase. This demonstrates that the levels and timing of accumulation of Cdc6p in mitosis are appropriate for mediating the modulation of APC/Cdc20.
In order to maintain genomic stability, cells must coordinate DNA replication such that every origin of replication fires once and only once per cell cycle (28, 69). The assembly of prereplicative complexes (pre-RCs) at origins of replication during G1 is therefore a tightly regulated process. In Saccharomyces cerevisiae, the main cyclin-dependent kinase (CDK), Cdc28p, promotes the initiation of DNA replication at the G1/S transition and also inhibits aberrant reinitiation by preventing the assembly of new pre-RCs before each of the progeny cells has its own complete genome (14, 15, 19, 41).
One of the components of the pre-RC is the Cdc6 protein (10, 13, 59). Both CDC6 transcription and protein levels are tightly regulated during the cell cycle. Cdc6 transcription occurs primarily in late M/early G1, but a second independently controlled wave of transcription has also been observed in late G1 (37, 45, 73, 74). The MCB box is responsible for CDC6 transcription in late G1, and the ECB box is responsible for the peak of CDC6 mRNA in late M/G1, 60 to 70 min after α-factor release, when 80% of the cells have metaphase or anaphase spindles (46). The ECB box is the primary regulator of CDC6 transcription in rapidly dividing cells, and it is also responsible for the timely expression of other genes involved in mitosis, such as CDC20 or SPO12, whose mRNA levels peak simultaneously with those of CDC6, and also for the replication genes MCM2 to MCM7 (46). In addition to transcriptional regulation, the levels of the Cdc6 protein are very accurately controlled by cell cycle-dependent proteolysis (16, 20, 43, 45, 52) and cellular localization (44). There is a peak of protein production in late M/early G1, and the protein is very stable during G1, when it accumulates to high levels in nuclei, but it becomes very unstable after the G1/S transition. It is also more evenly distributed between the nucleus and cytoplasm. Cdc6p contains several consensus CDK phosphorylation sites, and the phosphorylation of Cdc6p by Cdc28p is important for proteolysis, although this dependence varies with the phase of the cell cycle (7, 17, 20, 43).
The existence of distinctive active Cdc6p degradation mechanisms in S phase versus that in mitosis, where the rate of transcription and protein levels of Cdc6 are already very low, suggests that the regulation of Cdc6p is still necessary after the initiation of replication and that, as a consequence, Cdc6p might play a role in addition to its function in the initiation of replication.
In this study, in order to better understand the roles of Cdc6p during the cell cycle, we depleted Cdc6p from cells that had gone through complete DNA replication and observed a faster rate of Pds1p and Clb2p degradation in mitosis, suggesting a role of Cdc6p in modulating the activity of anaphase-promoting complex (APC)/Cdc20 in anaphase. We also observed that Cdc6 expressed under the control of its own promoter accumulates in mitosis to levels similar to those obtained by expression from the GAL1,10 promoter. Previous work showed that the overexpression of a dominant-negative Cdc6 mutant resulted in a delay of mitosis and that this correlated with the inhibition of Cdc28 protein kinase (43), which is required for the activation of APC/Cdc20. Although this previous data suggests that endogenous Cdc6p could affect APC/Cdc20 by inhibiting Cdc28 protein kinase activity, our results suggest that there is an additional Cdc6-mediated mechanism. We find that the expression of two Cdc6 phosphorylation site mutants interferes with the APC by a mechanism that requires neither interaction with nor inhibition of Cdc28p or the activation of mitotic checkpoints. The second pathway of APC inhibition may involve PP2A phosphatase, since the mitotic delay induced by mutant Cdc6 proteins can be reversed by the disruption of the CDC55 gene, a regulatory subunit of PP2A. In agreement with this hypothesis, we show that Cdc6p interacts with Cdc55p. In addition, the downregulation of PP2A-Cdc55, which occurs in normal cells, is not observed in cells devoid of Cdc6 in mitosis. We discuss a model that relates these results to the regulation of the APC by Cdc6 in a normal cell cycle.
MATERIALS AND METHODS
Strains and media.
Strains were derived from W303 (mata ura3 trp1 leu2 his3 ade2), with the exception of strains expressing hemagglutinin (HA)-tagged Cdc28p, which were derived from strain RJD635 (mata ura3 leu2 trp1 pep4::TRP1 cdc28::CDC28-HA-HIS3 bar1::LEU2), kindly provided by Ray Deshaies, California Institute of Technology, Pasadena, CA. A complete strain list with the relevant genotype is available upon request. Strain names used in individual experiments are provided in the figure legends.
W303 or RJD635 strains expressing myc-tagged Cdc6p under the GAL1,10 promoter were obtained by integrative transformation using previously described plasmids (20). swe1Δ, cdc55Δ, and Clb2-HA-, p86/91-HA-, and Cdc55-HA-tagged strains were obtained by the PCR gene replacement method using the plasmids pFA6a-His3MX6, pFA6a-3HA-His3MX6, and pFA6a-kanMX6 (36). PDS1-HA strains were obtained by transformation with a PDS1(HA)3::TRP1 fragment from plasmid pSB56, a plasmid derived from pOC52 (kindly provided by D. Koshland, Carnegie Institution, Baltimore, MD) by replacing the URA3 marker with the TRP1 marker. The disruption of the MIH1 gene was carried out by transformation with a mih1::LEU2 fragment from plasmid p119 (provided by P. Russell, Scripps Institute, La Jolla, CA). Gene-disrupted strains were detected by their specific phenotypes and/or PCR. Epitope-tagged strains were identified by Western blotting.
Strains with CDC6 under the control of the MET promoter (K4055; mata leu2 his3 ade2 GAL cdc6::hisG trp1::TRP1-MET-CDC6) were obtained from Kim Nasmyth and further modified by tagging the chromosomal copies of several genes as described above. CDC6 and mutant CDC6 expressed under its own promoter and myc tagged at the C terminus were introduced by transformation with a CEN plasmid or an empty vector.
Cells were routinely grown in 2% glucose selective media, YPRaf (2% raffinose) and YPRaf-Gal (2% raffinose plus 2% galactose) or yeast extract-peptone-dextrose (2% glucose). Cell plating dilution assays were carried out on selective plates containing raffinose plus either 2% galactose or 0.02% galactose, obtaining identical results (not shown). For experiments involving liquid cultures of cdc55Δ strains, YPSuc (2% sucrose) was used instead of YPRaf, since the latter does not support the growth of these strains.
Plasmids.
Plasmids expressing wild-type Cdc6 and Cdc6 phosphorylation site mutants under the control of the GAL promoter have been described previously (21). CDK phosphorylation sites were substituted for alanines as follows: CDC6 A-C (T7A, T23A, and S43A), CDC6 F (S372A), CDC6 E+F (S354A, T356A, T370A, T371A, and S372A), CDC6 A-C+F (T7A, T23A, S43A, T370A, T371A, and S372A), and CDC6 A-F (T7A, T23A, S43A, and S372A). CDC6 ΔN lacks the N-terminal amino acids 8 to 48. To express CDC6 and mutant CDC6 under the control of its own promoter, a cassette containing myc-tagged CDC6 and the CDC6 terminator was excised from the GAL plasmids and cloned in a CEN vector containing the CDC6 promoter. To construct plasmids expressing C-terminally deleted Cdc6, we obtained PCR fragments encompassing CDC6 from residue 1 to residue 401, 428, or 494 and having BamHI and SpeI restriction sites at their 5′ and 3′ ends, respectively. These PCR fragments were cloned into the BamHI-SpeI sites of plasmids derived from pRS306 and pRS315 containing the GAL1,10 and the CDC6 promoters, respectively, and eight myc tags.
Cell cycle experiments.
Early-exponential-phase cells were incubated with 10 μg/ml α-factor for 2.5 h. To release cells from the arrest, cells were harvested and washed once with twice the volume of supernatant and released in twice the original volume of fresh medium. For each time point, 2 ml of the culture was sampled; 1 ml was processed for flow cytometry, and the other was processed for immunoblotting as described previously (20). Samples were electrophoresed on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), transferred onto Hybond ECL membrane (Amersham) using a semidry transfer system (Bio-Rad), and blocked with 5% dry milk. Before detection with antibodies, all membranes were routinely stained with Ponceau S to check for equal loading and transfer. The detection of myc-tagged Cdc6 was performed using the monoclonal antibody 9E10, and for the detection of HA-tagged proteins, we used the monoclonal 12CA5 antibody. We used the enhanced chemiluminescence reagents according to manufacturer's instructions. All experiments were performed at least two times with results equivalent to those presented. The staining of microtubules was performed as described previously (18) using rat anti-tubulin antibody (clone YOL1/34). Fluorescence images were collected using a Carl Zeiss Axiophot FL microscope with a 63× objective and an Olympus DP70 camera. At least 200 cells were counted for each time point.
Immunoprecipitation and PP2A-Cdc55 phosphatase assay.
Yeast extracts were prepared as described previously (20), except that the resuspension buffer was 150 mM NaCl and phosphatase inhibitors (50 mM NaF, 5 mM Na4P2O7, and 0.1 mM Na3VO4) were included. A total of 2 mg of protein for strains expressing wild-type Cdc6p or an empty vector was typically incubated in the resuspension buffer in a final volume of 0.25 ml with ∼20 μg of the appropriate antibody. In experiments involving extracts from cells expressing endogenous Cdc6p, we used 5 mg of total extract. After 1 h at 4°C with rotation, 100 μl of a 50% slurry of protein A-Sepharose was added and incubated for one more hour at 4°C with rotation. Immunoprecipitates were washed six times in the same buffer (four times with the same buffer and two times with the same buffer but with 250 mM NaCl to immunoprecipitate Cdc55p) and released from protein A by boiling for 10 min in 1× SDS-PAGE loading buffer. Samples were subjected to SDS-PAGE and transferred to ECL membranes, and proteins were detected with the 12CA5 (for HA-tagged proteins), 9E10 (for Cdc6-myc), or Tpd3 antibody as described above.
For phosphatase assays, protein extracts from strains having a chromosomal copy of CDC55-HA and either expressing myc-tagged CDC6 from the MET promoter or carrying an empty vector (YSB393 or YSB534, respectively) were prepared as described above for immunoprecipitation, except that the extraction buffer did not contain phosphatase inhibitors. A total of 100 μg of protein was immunoprecipitated with the anti-HA antibody to purify Cdc55-HA, and beads were washed four times with extraction buffer and two times with phosphatase buffer (50 mM imidazole, pH 7.2, 0.2 mM EGTA, 0.02% β-mercaptoethanol, 0.1 mg/ml bovine serum albumin). The beads were then incubated with 45 μl of phosphatase buffer and 5 μl of a specific phosphopeptide, RRA(pT)VA, provided with a serine/threonine phosphatase assay kit (Promega, Madison, WI). Reactions were allowed to proceed for 30 min at 30°C and stopped by incubation on ice. Supernatants were collected, transferred to a 96-well plate, mixed with 50 μl of the molybdate dye provided with the kit, and incubated at room temperature for 15 min. The total amount of phosphate was determined by reading the absorbance at 600 nm of the molybdate-malachite green-phosphate complex using a Victor3 Wallac spectrofluorometer (PerkinElmer, Inc., Wellesley, MA). Cdc55 recovered from the beads was quantified by Western blotting using a Personal FX scanner (Bio-Rad) and used to normalize the PP2Ase enzymatic activity.
RESULTS
Cdc6 plays a role in mitosis before mitotic exit.
We reported several years ago that the ectopic expression of one Cdc6 phosphorylation site mutant (Cdc6 F) (see Fig. 2A) was lethal in a cdc16 background, and we suggested that Cdc6p might inhibit the APC (20). Two major events, the metaphase-to-anaphase transition and mitotic exit, occur sequentially in mitosis (reviewed in references 25, 30, and 72). In the metaphase-to-anaphase transition, chromatids are aligned and begin to separate to opposite poles of the cell. This event is triggered by APC/Cdc20-dependent degradation of the securin protein Pds1, which serves both as a marker for the onset of mitosis in our studies as well as a marker for APC/Cdc20 activity (11, 66, 67). APC/Cdc20 is also involved in the degradation of Clb5p (54) and in a first round of Clb2p degradation contributing to an initial decrease in Clb/Cdc28 protein kinase activity (34, 63, 71). The second mitotic event, exit from mitosis, involves the degradation of Clb2p in an APC/Cdh1-dependent manner (23, 26, 27, 58, 60). The resulting global decrease in Clb/Cdc28 activity finally leads to mitotic exit.
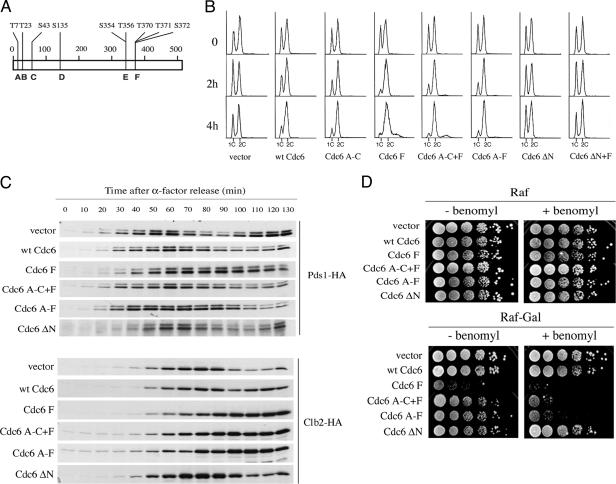
FIG. 2.
(A) Schematic diagram of Cdc6 protein. Vertical lines represent CDK sites A to F, and the amino acid numbers at the top of the diagram are given for consensus threonines and serines. (B). Cell cycle delay of cells expressing myc-tagged wild-type and mutant Cdc6p. Cells growing logarithmically in YPRaf medium were transferred to 2% galactose-containing medium, and samples were taken for flow cytometry analysis at the indicated times. The strains used were YSB57, empty vector; YSB58, GAL-CDC6; YSB117, GAL-CDC6 A-C; YSB116, GAL-Cdc6 F; YSB60, GAL-CDC6 A-C+F; YSB61, GAL-CDC6 A-F; YSB62, GAL-CDC6 ΔN8-48; and YSB196, GAL-CDC6 ΔN8-48+F. (C) The degradation of cell cycle-regulated Pds1p and Clb2p is delayed in strains overexpressing Cdc6p. W303 strains expressing either the indicated myc-tagged mutant Cdc6 proteins from the GAL1,10 promoter or an empty vector and expressing either HA-tagged Pds1p (upper panels) or HA-tagged Clb2p (lower panels) were grown in YPRaf to early log phase and arrested with α-factor. After arrest, Cdc6p expression was induced by the addition of galactose. Fifteen minutes later, cells were washed and released from the arrest into YPRaf-Gal. Samples were taken at the time of release and every 10 min thereafter during a 130-min time course to be processed for immunoblotting to detect Pds1-HA and Clb2-HA. The Pds1-HA strains were YSB169, empty vector; YSB170, GAL-CDC6; YSB171, GAL-Cdc6 F; YSB306, GAL-CDC6 A-C+F; YSB307, GAL-CDC6 A-F; and YSB308, GAL-CDC6 ΔN8-48. The Clb2-HA strains were YSB271, empty vector; YSB272, GAL-CDC6; YSB273, GAL-Cdc6 F; YSB274, GAL-CDC6 A-C+F; YSB275, GAL-CDC6 A-F; and YSB304, GAL-CDC6 ΔN8-48. wt, wild type. (D) Benomyl hypersensitivity of Cdc6 phosphorylation site mutants. W303 strains expressing either the indicated myc-tagged mutant Cdc6 proteins from the GAL1,10 promoter or an empty vector, described in the legend for panel B, were grown in glucose-containing selective medium to stationary phase, serially diluted 10-fold, spotted in raffinose (as a control) and raffinose plus galactose plates containing 5 μg/ml benomyl, and incubated at room temperature for 1 week. −, absence of; +, presence of.
In order to test the hypothesis that Cdc6p affected the APC, we first analyzed the steady-state level of Cdc6p through a single cell cycle and compared it to the profile of the two well-characterized mitotic markers: Pds1p and Clb2p. We constructed yeast strains that expressed C-terminally tagged myc-Cdc6p under the control of its own promoter and that contained a chromosomal copy of Pds1 or Clb2 tagged with HA. We monitored the levels of myc-Cdc6p, Pds1p-HA, and Clb2p-HA in cells released from α-factor arrest to monitor the metaphase-to-anaphase transition. We observed a peak of myc-Cdc6p at 20 min after release, after which the levels decreased until around 65 min. Levels again increased, peaking at 80 min after release (Fig. 1A). Interestingly, myc-Cdc6p levels began to increase at exactly the same time when Pds1p-HA degradation began to be apparent, about 65 min after release from α-factor and in the presence of high levels of Clb2p-HA. This pattern of expression of Cdc6 is entirely reproducible in replicate experiments and is in agreement with other recent data (2).
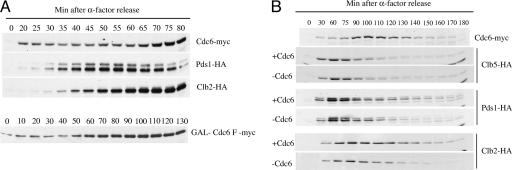
FIG. 1.
Steady-state levels of Cdc6p through the cell cycle. (A) Yeast cells with a copy of MET-CDC6 and another copy of myc-tagged CDC6 expressed under its own promoter (YSB75) and containing a chromosomal copy of Pds1-HA (YSB441) or Clb2-HA (YSB407) were grown in synthetic medium and arrested with α-factor. Cells were released in rich medium, and samples were taken at 20 min after release and, from then on, every 5 min to be processed for immunoblotting. α-Factor was readded 60 min after release to prevent entry into a new cell cycle. For a comparison of levels of Cdc6 expressed from its own promoter with those reached by GAL overexpression, we measured the levels of Cdc6 F in cells arrested with α-factor in YPRaf medium, treated with galactose for 15 min, and released in YPRaf-Gal. Note that four times less protein was loaded in gels from cells expressing GAL-myc-Cdc6 F than in endogenous myc-Cdc6. (B) Advancement of Pds1 and Clb2 degradation by reduction in Cdc6 levels. Yeast cells with a copy of MET-CDC6 and another copy of myc-tagged CDC6 expressed under its own promoter (YSB75) or an empty vector (YSB431) and containing a chromosomal copy of Pds1-HA (YSB441 and YSB442), Clb2-HA (YSB407 and YSB408), or Clb5-HA (YSB410 and YSB411) were grown in synthetic medium without methionine and arrested with α-factor. Cells were released in synthetic medium without methionine, and 30 min after release, methionine was added. At the indicated times, samples were taken to be processed for immunoblotting and flow cytometry. α-Factor was readded 70 min after release to prevent reentry into another cell cycle. Notice that for panel A, cells were released in rich yeast extract-peptone-dextrose medium and, in panel B, they were released in synthetic medium. Therefore the peak of accumulation of Cdc6p is slightly delayed in panel B.
The cell cycle profile of Cdc6p levels shown in Fig. 1A suggests that Cdc6p is present at significant levels at a time when it could inhibit the APC/Cdc20 form of the APC, i.e., when Pds1 levels begin to decline. The timing of peak levels of Cdc6 is 80 min (Fig. 1A), whereas Pds1 degradation is apparent at 65 min after release, suggesting that APC inhibition would occur after initial activation of APC/Cdc20 and is more consistent with Cdc6p contributing to the shutoff of APC/Cdc20 once it had functioned. According to this hypothesis, cells that are depleted of Cdc6p in mitosis should show a faster rate of degradation of APC/Cdc20 substrates compared to cells normally expressing Cdc6p. In order to test this prediction, we constructed yeast strains where CDC6 expression was regulated by the repressible MET promoter and that had chromosomal copies of Clb5, Pds1, or Clb2 tagged with HA. In addition, they contained either an additional copy of CDC6 under its own promoter or an empty vector. We grew these cells in the absence of methionine to allow growth of the cells with the empty vector and arrested them with α-factor. Cells were released, and methionine was added to interrupt the expression of Cdc6 during the ensuing cycle. Notice that methionine was added not immediately after release, but 30 min later in order to allow sufficient Cdc6p synthesis from the MET promoter to support late G1 pre-RC assembly and DNA replication. We analyzed the levels of Pds1p-HA, Clb2p-HA, and Clb5p-HA in these cells. As predicted by our model, the time of onset of Pds1p-HA degradation is the same time in the absence of Cdc6p as in the presence of Cdc6p. However, in the presence of Cdc6, the degradation of Pds1p-HA and Clb2p-HA fails to go to completion (Fig. 1B). This result is verified and extended in Fig. 2. Since the depletion of Cdc6p allows Pds1p degradation to go to completion, this suggests for the first time that endogenous Cdc6p may be rate limiting for that degradation.
Although the presence or absence of Cdc6p did not affect the rate of degradation of Clb5p-HA, the lack of effect on Clb5p-HA degradation is easily explained when comparing its levels with those of myc-Cdc6p. By the time that Cdc6p levels peak (90 to 100 min after α-factor release, under the growth conditions for Fig. 1B), most of Clb5p-HA has already been degraded. These results show not only that Cdc6p is expressed significantly before exit from mitosis (Fig. 1A) but also that it functions in an earlier step than exit from mitosis (Fig. 1B). Cdc6p does not appear to inhibit the triggering of mitosis since Clb5p degradation is normal and the onset of Pds1p degradation occurs with normal timing. Rather, Cdc6p appears to downregulate APC/Cdc20 after its execution point.
Cell cycle delay in cells expressing stabilized Cdc6p.
In order to gain more insight into the mechanism by which Cdc6p could modulate APC/Cdc20 activity, we tested the ability of different Cdc6p mutants to inhibit APC/Cdc20. There are abundant data in the literature showing that ectopic expression of Cdc6p results in mitotic delay (3, 5, 43), and this delay depends on the time of expression of Cdc6p, but not on the levels of Cdc6p (5). We reasoned that we could induce the ectopic expression of Cdc6p and its mutants at the metaphase-to-anaphase transition by using the GAL1,10 promoter. We could then monitor the effects on the APC/Cdc20, measured as a mitotic delay by flow cytometry or by the time of onset and extent of degradation of Pds1p and Clb2p. To validate our approach, we wanted to show that, in a situation of ectopic expression of Cdc6p in metaphase-to-anaphase cells, the levels of Cdc6p attained were similar to the maximum levels of Cdc6p expressed from its own promoter as shown in Fig. 1A. To that end, we measured the levels of a stabilized mutant, myc-Cdc6p F (21). We expressed myc-Cdc6 F from the GAL1,10 promoter in cells treated with galactose for 15 min during α-factor arrest and released them from the arrest in the continuous presence of galactose. We observed that at 60 min after release, i.e., at the metaphase-to-anaphase transition (Fig. 1A), the levels of myc-Cdc6p F were similar to the peak of myc-Cdc6p expressed from its own promoter (Fig. 1A, compare the 70-to-80-min time points in the upper panel with the 60-min time point in the lower panel). Similar results comparing the levels of Cdc6p expressed from its own promoter with those of other stabilized Cdc6 mutant proteins have been described in the literature (40). Thus, GAL-induced expression at metaphase to anaphase is at the same level eventually reached by wild-type Cdc6p somewhat later in mitosis. This level of GAL-induced Cdc6p is sufficient to lead to mitotic delay, as we will show below. These results suggest that expressing Cdc6p from the GAL1,10 promoter is a valid approach to studying a physiologically significant mechanism by which the Cdc6p might downregulate APC/Cdc20 activity.
Cdc6p contains at least six SP or TP motifs that could potentially serve as CDK phosphorylation sites (A to F), three at the N terminus and three at the C terminus (Fig. 2A). The mutation of all these sites, a subset of them, or just one site at the C terminus (Cdc6 F) into unphosphorylatable sites results in the stabilization of Cdc6p (7, 17, 20, 43). We tested the effect of expressing these stabilized mutant proteins under the GAL1,10 promoter on cell growth. Cells containing the endogenous CDC6 gene and C-terminal, myc-tagged derivatives of either wild-type CDC6 or mutant cdc6 under the control of the GAL1,10 promoter were grown in the presence of galactose for 4 h. Flow cytometry shows that the expression of wild-type myc-Cdc6p and myc-Cdc6p sites A to C (myc-Cdc6p A-C) resulted in an accumulation of cells with a 2C content of DNA after 4 h in galactose (Fig. 2B), suggesting a delay in mitosis, as previously reported (5, 43). When myc-Cdc6p F, myc-Cdc6p A-C+F, or myc-Cdc6p A-F were expressed, the mitotic delay was stronger than that in the wild type or Cdc6p A-C since it was readily observed as early as 2 h after the addition of galactose and the remaining population of G1 cells was smaller than that observed for cells expressing wild-type Cdc6p. In conjunction with this strong mitotic delay, cells had abnormal cell morphology with hyperpolarized buds (not shown). The expression of myc-Cdc6p ΔN (lacking the N-terminal amino acids 8 to 48 containing three Cdk phosphorylation sites [20]) or myc-Cdc6p ΔN+F did not have an observable effect on the cell cycle or on cell morphology. These results suggest that the N terminus is an important component for the mitotic delay produced by the expression of C-terminal phosphorylation site mutants. This result does not contradict the reported effect of expressing Cdc6 ΔN as the sole source of Cdc6p in the cell, in which case, Cdc6 ΔN actually caused a significant mitotic delay (6) since wild-type endogenous Cdc6p is present in our experiments. Furthermore, that work addressed an event in mitotic exit and we will show below that the dominant phosphorylation site mutants are monitoring an earlier event.
To further characterize the effect of the different Cdc6p mutants in mitosis, we monitored the levels of Pds1p-HA and Clb2p-HA as cells progressed through the cell cycle after a G1 arrest and release. Compared to control cells carrying an empty vector, where the minimum levels of Pds1p-HA occurred at 90 min after release, cells expressing wild-type myc-Cdc6p showed a delay of around 10 min in Pds1p-HA degradation and Pds1p-HA levels remained low for a longer time than those in the control cells (Fig. 2C, upper panel). The degradation of Clb2p-HA in cells expressing wild-type myc-Cdc6p was diminished compared to that in control cells. Clb2p-HA began to decay at about the same time as in cells with an empty vector, 100 min after release (Fig. 2C, lower panel), but levels did not continue to decline as they did in the control. Cells expressing the phosphorylation site mutants A-C+F and A-F showed an even greater delay in Pds1p-HA degradation than did cells expressing wild-type myc-Cdc6p. In cells expressing myc-Cdc6p A-C+F and myc-Cdc6p A-F, minimum levels of Pds1p-HA were not attained until 110 and 120 min after release. These mutations also affected Clb2p-HA levels, which did not decrease during the entire time course of the experiment. Cells expressing myc-Cdc6p F showed extremely slow kinetics of Pds1p degradation, as Pds1p-HA levels did not decrease during the entire time course. In a similar manner, the levels of Clb2p remained undegraded and constant in cells expressing myc-Cdc6p F. As expected since there was no cell cycle delay, cells expressing myc-Cdc6p ΔN showed normal oscillation in the timing of degradation and in the levels of both Pds1p-HA and Clb2p-HA. The delay and/or failure in Pds1p and Clb2p degradation indicates that the expression of Cdc6 phosphorylation site mutants results in a delay during both the metaphase-to-anaphase transition and the exit of mitosis. Since Clb2p degradation by the APC/Cdc20 is required for complete Clb2 proteolysis by APC/Cdh1 at the exit of mitosis, we cannot be certain from our results which step of the degradation of Clb2 is affected by the Cdc6 protein (34, 67, 71).
To gain more evidence that Cdc6 ectopic expression was affecting early mitosis, we tested the ability of cells expressing phosphorylation site mutants to grow in the presence of sublethal concentrations of the microtubule-depolymerizing drug benomyl, which triggers the spindle assembly checkpoint and transient inhibition of APC/Cdc20.
We found that benomyl did not have any effect in cells expressing wild-type myc-Cdc6p (Fig. 2D). However, it had a very strong inhibitory effect on the growth of cells expressing myc-Cdc6p A-C+F, myc-Cdc6p A-F, and myc-Cdc6p F. While the myc-Cdc6p F cells were compromised for growth in even the absence of benomyl, they are further inhibited by benomyl and the Cdc6p A-C+F and Cdc6p A-F are clearly supersensitive to benomyl. Our interpretation of these results is that benomyl treatment and ectopic expression of phosphorylation site mutants have additive effects in the inhibition of the APC/Cdc20, resulting in a permanent arrest.
Pds1p is a primary target of the cell cycle checkpoint pathways that survey DNA integrity and the proper assembly and orientation of the mitotic spindle (9). Therefore, one possible explanation for the Pds1p degradation delay in cells expressing Cdc6p is that ectopic expression of Cdc6p triggers one of these checkpoints and arrests cells at the metaphase-to-anaphase transition. To investigate this possibility, we constructed null strains in genes involved in the DNA damage and spindle checkpoint pathways and tested whether they could suppress the cell cycle defects. We did not observe the suppression of the delay in Pds1p-HA degradation or the extremely elongated cell morphology (not shown) in any of the strains tested (rad24Δ, mad1Δ, and bub2Δ). These results suggest that the G2/M delay caused by ectopic expression of Cdc6p is not mediated by the DNA replication or the spindle checkpoint.
Cdc28 activity in cells expressing different Cdc6p mutants.
The full activation of the APC at the metaphase-to-anaphase transition requires active Cdc28 protein kinase (49, 50). Since Cdc6p interacts with Cdc28 protein kinase through the N terminus (7, 21, 44, 64) and very high levels of Cdc6p may inhibit Cdc28 protein kinase activity (3, 21, 43), we hypothesized that stable Cdc6p mutants could result in an increased inhibition of the Cdc28 protein kinase activity and this would result in decreased APC activity. Such inhibition has been observed for another dominant-negative C-terminal mutant, cdc6-d2 (T368M), which maps near to the site F (S372A) used in our studies (43). In addition, previous work from other groups showed that the overexpression of wild-type Cdc6p late in the cell cycle resulted in an accumulation of cells with a 2C content of DNA (5) and this delay in cell cycle progression was significantly increased when Cdc6p was expressed in a mih1Δ background. Mih1p is a phosphatase that removes an inhibitory phosphate group at Tyr 19 in Cdc28p and an activator of Cdc28p (4, 51). Cells lacking MIH1 have compromised Cdc28 activity. We wanted to know whether the disruption of the MIH1 gene would also have an effect when the Cdc6 phosphorylation site mutants were expressed in the yeast background used in our studies. Figure 3A shows that the expression of wild-type myc-Cdc6p in cells defective for the MIH1 gene results in a clear growth defect, and the expression of the C-terminal phosphorylation site mutant (myc-Cdc6p E+F) results in lethality. However, the expression of the other phosphorylation site mutants does not have a greater effect than that observed in a wild-type background. Interestingly, the expression of wild-type Cdc6p in a mih1Δ background produces the same elongated bud cell morphology observed in wild-type cells expressing mutant Cdc6p F (not shown).
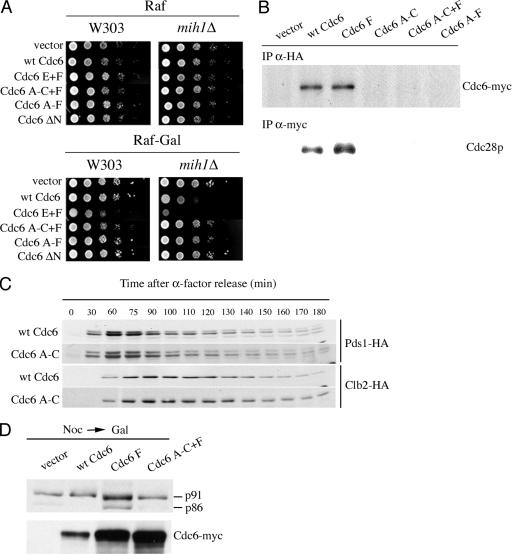
FIG. 3.
(A) Lethality of Cdc6 phosphorylation site mutants in a mih1Δ background. W303 or isogenic mih1Δ (W303 mih1Δ) strains expressing either the indicated myc-tagged mutant Cdc6 proteins from the GAL1,10 promoter or an empty vector were grown in glucose-containing selective medium to stationary phase, serially diluted 10-fold, spotted in either raffinose (as a control) or raffinose plus galactose selective plates, and incubated at 30°C for 2 days. The mih1Δ strains were YSB198, empty vector; YSB199, GAL-CDC6; YSB157, GAL-CDC6 E+F; YSB201, GAL-CDC6 A-C+F; YSB202, GAL-CDC6 A-F; and YSB203, GAL-CDC6 ΔN8-48. (B) Interaction between Cdc6p and Cdc28p. Strains (YSB161, empty vector; YSB162, GAL-CDC6; YSB163, GAL-Cdc6 F; YSB164, GAL-CDC6 A-C; YSB166, GAL-CDC6 A-C+F; and YSB167, GAL-CDC6 A-F) containing HA-tagged Cdc28p under the control of its natural promoter and expressing either the indicated myc-tagged mutant Cdc6 proteins from the GAL1,10 promoter or an empty vector were grown in YPR to mid-log phase, and galactose was added to induce the expression of Cdc6p. After 2 h, total yeast extracts were prepared and subjected to immunoprecipitation (IP). The top panel corresponds to HA immunoprecipitates blotted with the 9E10 anti-myc antibody. The bottom panel corresponds to myc immunoprecipitates blotted with the 12CA5 anti-HA antibody. (C) Yeast cells with a copy of MET-CDC6 and another copy of myc-tagged wild-type CDC6 (YSB75) or CDC6 A-C (YSB133) expressed under its own promoter and containing a chromosomal copy of Pds1-HA or Clb2-HA were grown in synthetic medium without methionine and arrested with α-factor (the Pds1-HA strains were YSB441 and YSB478, and the Clb2-HA strains were YSB407 and YSB472). The cells were further treated as described in the legend for Fig. 1B. (D) Effect of wild-type and mutant Cdc6p overexpression on Cdc28 kinase activity. W303 strains (YSB359, empty vector; YSB360, GAL-CDC6; YSB361, GAL-CDC6 F; and YSB364, GAL-CDC6 A-C+F) containing HA-tagged p86/91 and expressing the indicated forms of myc-tagged Cdc6p under the GAL1,10 promoter were grown to early log phase in YPRaf and arrested with nocodazole (Noc). After nocodazole arrest, galactose was added for 2.5 h and samples were taken to be processed for Western blotting to detect p86/91. wt, wild type.
To study the inhibition of Cdc28p by Cdc6p in more detail, we looked at the ability of Cdc6 phosphorylation site mutants to interact with Cdc28p and we performed coimmunoprecipitation assays. We used yeast strains that express Cdc28p tagged with the HA epitope from its natural promoter and induced the expression of myc-tagged Cdc6p with galactose. As shown in Fig. 3B, only wild-type myc-Cdc6p and myc-Cdc6p F were capable of interacting with endogenous Cdc28p. These are the same myc-Cdc6 proteins to which mih1Δ strains are supersensitive, that is, strains with reduced Cdc28 activity (Fig. 3A). myc-Cdc6p A-C, myc-Cdc6p A-C+F, and myc-Cdc6p A-F, lacking three N-terminal phosphorylation sites, failed to coimmunoprecipitate with detectable levels of Cdc28p (Fig. 3B). Similar conclusions about the effects of these mutations on Cdc6p/Cdc28p interaction have been reached in in vitro studies using Cdc6 mutant proteins expressed in insect cells (64). The fact that phosphorylation site mutants were capable of producing a mitotic delay (Fig. 2) but only those with wild-type N-terminal sites were capable of binding Cdc28p and were lethal in a mih1Δ background is the key evidence that the inhibitory interaction between Cdc6p with Cdc28p is not the exclusive mechanism for the mitotic delay.
More evidence was obtained by studying the Pds1p degradation kinetics in cells expressing myc-Cdc6p A-C from its own promoter and as the only source of Cdc6p (Fig. 3C), rather than from the GAL1,10 promoter (Fig. 2B). Despite the fact that this mutant does not interact with Cdc28p, the degradation kinetics of both Pds1p-HA and Clb2p-HA are identical to those in cells expressing wild-type Cdc6p. These results suggest that there may be different pathways for the mitotic delay produced by different phosphorylation site mutants.
To directly analyze Cdc28p activity in vivo, we measured the phosphorylation state of the B subunit of DNA polymerase α (p86/91; POL12), an in vivo substrate of the Cdc28 protein kinase (22). The inhibition of Cdc28p by the overproduction of Sic1p, a CDK inhibitor, results in the complete inhibition of the phosphorylation of p86 to p91 in nocodazole-arrested cells (12, 22). We constructed yeast strains that expressed myc-tagged CDC6 under the control of the GAL1,10 promoter and that contained a single chromosomally integrated copy of HA-tagged POL12. We induced the expression of wild-type or mutant Cdc6p with galactose in nocodazole-arrested cells. As expected from results of investigating the overexpression of cdc6-d2 with a mutation in a nearby amino acid and which specifically prevents late G2/M degradation of Cdc6p (43), myc-Cdc6p F had a small inhibitory effect on Cdc28 protein kinase activity since a band of p86 could be detected (Fig. 3D). Importantly, the expression of myc-Cdc6p A-C+F, which, like myc-Cdc6p F, results in Pds1p degradation delay and the accumulation of cells with a 2C content of DNA, did not result in a detectable inhibition of Cdc28p, as nonphosphorylated p86 was not present. This result indicates that, in nocodazole-arrested cells, only Cdc6 mutants that interact with Cdc28p are capable of inhibiting it, suggesting either that Cdc6p A-C+F does not inhibit Cdc28p at all or that it inhibits a different form of Cdc28p than the one present in nocodazole-arrested cells. More important, this shows that Cdc6 mutants that neither interact with nor inhibit Cdc28p can nevertheless cause a G2 delay.
As described above, we and others have observed that the mitotic delay due to Cdc6p ectopic expression in G2 is enhanced in a mih1Δ background. Although, in S. cerevisiae, the phosphorylation state of Tyr 19 on Cdc28p is not important in an unperturbed cell cycle (1, 57), it becomes important in conditions in which the actin cytoskeleton is perturbed (33, 38, 56). Under these conditions, the Swe1 protein kinase phosphorylates Cdc28p at Tyr 19 and the resulting inhibition of Cdc28 protein kinase prevents the entry into mitosis. Dephosphorylation by Mih1 phosphatase is then essential for entry into mitosis. Therefore, it is possible that Cdc28p inhibition by Cdc6p ectopic expression, if any, results from the stimulation of inhibitory phosphorylation of Tyr 19 by Swe1 protein kinase. If so, swe1Δ yeast strains would be expected to suppress the mitotic delay due to GAL-induced Cdc6p expression.
To study this possibility, we performed cell cycle kinetic experiments monitoring the levels of Pds1p-HA in cells ectopically expressing myc-Cdc6p F in a swe1Δ background. These experiments were performed simultaneously with those shown in Fig. 2C and 4C, where a wild-type, isogenic strain carrying an empty vector was used as control for normal Pds1 degradation kinetics. swe1Δ does not suppress the delay of Pds1p degradation (shown in Fig. 4C so that important controls are in the same figure). In addition, swe1Δ did not suppress the extreme elongated bud morphology of cells expressing myc-Cdc6p F, myc-Cdc6p A-C+F, or myc-Cdc6p A-F (not shown). However, swe1Δ did suppress the growth defects and the elongated phenotype of mih1Δ strains expressing wild-type GAL-myc-Cdc6p and also the lethality of expressing GAL-myc-Cdc6p F in a mih1Δ background (not shown). In summary, swe1Δ is able to overcome only the effects of deleting MIH1, as expected, not those of expressing mutant Cdc6p from the GAL1,10 promoter.
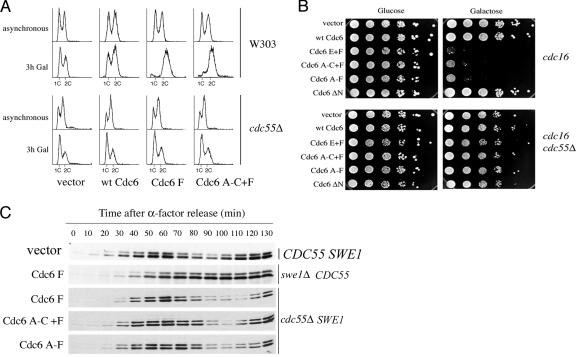
FIG. 4.
Suppression of mitotic delay. (A) W303 and W303 cdc55Δ cells expressing the indicated myc-tagged mutant Cdc6 proteins from the GAL1,10 promoter and growing logarithmically in either YPRaf or YPSuc were transferred to 2% galactose-containing medium, and samples were taken for flow cytometry analysis at the indicated times. Flow cytometry profiles of normally cycling cultures (asynchronous; no galactose added) were also obtained. The cdc55Δ strains were YSB335, empty vector; YSB336, GAL-CDC6; YSB337, GAL-CDC6 F; and YSB340, GAL-CDC6 A-C+F. (B) cdc16-264 and cdc16-264 cdc55Δ strains expressing either the indicated myc-tagged mutant Cdc6 proteins from the GAL1,10 promoter or an empty vector were grown in glucose-containing selective medium to stationary phase, serially diluted 10-fold, spotted in either glucose (as a control) or raffinose plus galactose selective plates, and incubated at 30°C for 2 days. The cdc16-264 strains were YSB172, empty vector; YSB173, GAL-CDC6; YSB176, GAL-CDC6 E+F; YSB177, GAL-CDC6 A-C+F; YSB178 GAL-CDC6 A-F; and YSB323 GAL-CDC6 ΔN8-48. The cdc16-264 cdc55Δ strains were YSB416, empty vector; YSB417, GAL-CDC6; YSB418, GAL-CDC6 E+F; YSB419, GAL-CDC6 A-C+F; YSB420, GAL-CDC6 A-F; and YSB421, GAL-CDC6 ΔN8-48. (C) Wild-type (YSB169), cdc55Δ (strains described in the legend for panel A), and swe1Δ strains (YSB248) expressing the indicated myc-tagged mutant Cdc6 proteins from the GAL1,10 promoter and expressing HA-tagged Pds1p were grown to early log phase in YPSuc (cdc55Δ) or YPRaf (swe1Δ) and arrested with α-factor. After arrest, Cdc6p expression was induced by the addition of galactose. Fifteen minutes later, cells were washed and released from the arrest into YPSuc-Gal or YPRaf-Gal. Samples were taken at the time of release and every 10 min thereafter during a 130-min time course to be processed for immunoblotting.
Altogether, these results show that the mitotic delay produced by ectopic Cdc6p expression might result from Cdc28p inhibition. However, the mechanism for this inhibition would be only partially explained by inhibitory interaction with Cdc6p as not all mutants that show the delay interact with Cdc28p. This also does not appear to be an effect of Cdc6p on stimulating Swe1-mediated inhibitory phosphorylation of Cdc28p since swe1Δ does not reverse the mitotic delay in any mutant. Thus, there must be an additional, Cdc6-mediated mechanism of inhibiting the APC/Cdc20.
Suppression of mitotic delay by deletion of CDC55.
Since the activation of APC/Cdc20 involves the phosphorylation of the APC core subunits (49, 50) and dephosphorylation should occur to inhibit APC/Cdc20 in the process of shifting to APC/Cdh1 for exit from mitosis, a role for phosphatases in APC/Cdc20 regulation is attractive. Actually, a role for PP2A phosphatase in the inhibition of the APC/Cdc20 has been described in Xenopus and clams (32, 55). In addition, there is also documented genetic and biochemical interaction between PP2A and the APC (31, 61). In yeast, there are only two regulatory subunits, Cdc55p and Rts1p. The function of Cdc55p has been related to the mitotic checkpoint, although the precise role is controversial (39, 61). Cdc55p has also been related to Cdc28p phosphorylation by Swe1p because cells disrupted for CDC55 have increased levels of Swe1p (39, 61, 68).
To determine whether PP2A-Cdc55 played a role in the mitotic delay produced by Cdc6p, we first looked at the cell cycle profile of cdc55Δ cells expressing Cdc6 mutants. As shown in Fig. 4A, the deletion of CDC55 suppressed the mitotic cell cycle delay seen in wild-type strains due to expressing myc-Cdc6p F and myc-Cdc6p A-C+F under the GAL1,10 promoter. Flow cytometry showed identical cell cycle distribution in cdc55Δ cells expressing an empty vector, wild-type myc-Cdc6p, or the mutants (Fig. 4A). cdc55Δ cells had two peaks corresponding to content of 1C and 2C DNA as opposed to wild-type W303 cells expressing mutant Cdc6, which showed an accumulation of 2C DNA, as is also shown in Fig. 2B. To further confirm that cdc55Δ suppressed the mitotic delay, we tested whether the deletion of CDC55 would suppress the lethality of the expression of the Cdc6 mutants in an APC-defective background. In fact, cdc55Δ suppressed the lethality of expressing all stable Cdc6 phosphorylation site mutants in the cdc16-264 strain (Fig. 4B). Finally, we compared the kinetics of Pds1p-HA degradation of cdc55Δ cells expressing Cdc6p phosphorylation site mutants to the kinetics of degradation in wild-type strains expressing the same Cdc6 mutants. cdc55Δ cells containing HA-PDS1 under the control of its natural promoter and myc-CDC6 mutants under the GAL1,10 promoter were arrested with α-factor and released as described in the legend for Fig. 2C. As described for experiments with the swe1Δ strain, these experiments were performed simultaneously with those shown in Fig. 2C, where a wild-type isogenic strain carrying an empty vector was used as control for normal Pds1 degradation kinetics and where the effects of the Cdc6 mutants in the wild-type (CDC55) background are shown. Figure 4C shows a return to normal kinetics of Pds1p-HA degradation in the cdc55Δ strain expressing myc-Cdc6p F, myc-Cdc6p A-C+F, and myc-Cdc6p A-F as opposed to swe1Δ CDC55 strains expressing myc-Cdc6p F. These results demonstrate that Cdc55p is involved in the delays caused by ectopic Cdc6p expression (39, 61, 68).
Cdc6 interacts physically and functionally with Cdc55, but only during mitosis.
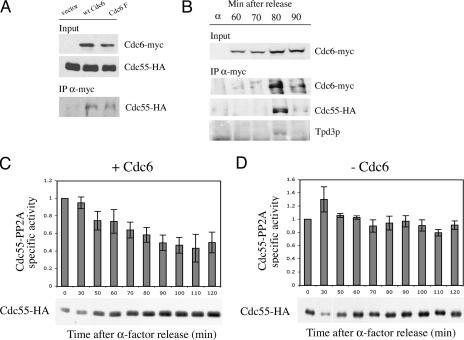
To obtain evidence for a direct role for Cdc6 in Cdc55 activity, we determined whether Cdc6p and Cdc55p could interact. We HA tagged the chromosomal copy of CDC55 in cells expressing wild-type and myc-Cdc6 F under the GAL1,10 promoter and performed coimmunoprecipitation assays. As shown in Fig. 5A, Cdc55p immunoprecipitated with both wild-type myc-Cdc6p and myc-Cdc6p F. It also interacted with other Cdk-phosphorylation site mutants of Cdc6 (not shown).
FIG. 5.
(A) Interaction between Cdc6p and Cdc55p. W303 strains expressing HA-tagged Cdc55p (YSB399) and either the indicated myc-tagged mutant Cdc6p from the GAL1,10 promoter or an empty vector were grown to mid-log phase in YPRaf, galactose was added to induce the expression of Cdc6p for 3 h, and total extracts were prepared. The top and middle panels represent a fraction (1/500) of the total input material in the immunoprecipitation (IP) blotted with the anti-myc-Cdc6 (9E10) and anti-Cdc55-HA (12CA5) antibodies, respectively. The bottom panel represents one-third of the Cdc6-myc immunoprecipitates blotted with the anti-HA antibody 12CA5. The Cdc55-HA strains used were YSB289, empty vector; YSB290, GAL-CDC6; and YSB291, GAL-CDC6 F. wt, wild type. (B) Cell cycle-specific interaction between Cdc6, Cdc55, and Tpd3. Yeast cells with a copy of MET-CDC6 and another copy of myc-tagged CDC6 expressed under its own promoter and containing a chromosomal copy of Cdc55-HA (YSB393) was grown in synthetic medium and arrested with α-factor and released in rich medium. Samples were taken, and total cell extracts were prepared from arrested cells and from cells at the indicated times after release. Cdc6-myc immunoprecipitates prepared with the 9E10 antibody were blotted and probed with 9E10, 12CA5, and α-Tpd3 for Cdc6-myc, Cdc55-HA, and Tpd3, respectively. (C and D) Cdc55-PP2A activity during the cell cycle in wild-type cells (C) or in cells depleted of Cdc6 in mitosis (D). Yeast cells with a copy of MET-CDC6 and another copy of myc-tagged CDC6 expressed under its own promoter (YSB393) or an empty vector (YSB534) and containing a chromosomal copy of Cdc55-HA were grown in synthetic medium without methionine and arrested with α-factor. They were further treated as described in the legend for Fig. 1B. Extracts, immunoprecipitations, and phosphatase assays were carried out at the indicated time points. Histograms represent the mean values and standard errors of three experiments. Immunoprecipitated HA-Cdc55 from both yeast strains that was used to normalize the phosphatase levels is shown below the graphs. −, absence of; +, presence of.
To rule out the possibility that the interaction was an artifact of GAL induction, we performed immunoprecipitation assays with extracts from cells expressing myc-Cdc6 from its own promoter. Furthermore, we assessed the interaction at various points in the cell cycle in cells growing synchronously after α-factor release. We observed a clear, cell cycle-specific interaction between myc-Cdc6p and Cdc55p-HA that occurred at about 80 min after release and was somewhat diminished by 90 min (Fig. 5B). The narrow interval of interaction may explain why the asynchronous cells used in Fig. 5A showed less Cdc55 in the Cdc6 immunoprecipitates than did the synchronized cells in Fig. 5B. The interaction occurred when the levels of myc-Cdc6p were maximum, which is in agreement with our hypothesis that Cdc6p is present at sufficient levels to inhibit APC/Cdc20 after the metaphase-to-anaphase transition. We also probed the blot carrying proteins immunoprecipitated by the anti-myc-Cdc6 antibody with an antibody against a structural subunit of the PP2A, Tpd3p, and we found that Tpd3p was present in the immunoprecipitate, suggesting that Cdc6p interacted with the PP2A holoenzyme and not just with the regulatory subunit Cdc55p (Fig. 5B). We obtained identical results with a strain where Cdc55 was not tagged (not shown).
The suppression of Cdc6-mediated cell cycle delay in cdc55Δ mutants and the Cdc6/Cdc55 interaction suggest the possibility that Cdc6p might affect the kinetics of dephosphorylation or the ratio of phosphorylation/dephosphorylation of a Cdc55 substrate and, in turn, affect the passage through mitosis. Phosphatase PP2A associated with Ccd55p has recently been described as a negative regulator of mitosis (62, 70) by preventing Net1p phosphorylation and Cdc14p nucleolar release (47). We reasoned that Cdc6p could affect the cell cycle-regulated activity of PP2A-Cdc55. To study this possibility, we measured the Cdc55-associated phosphatase activity in wild-type cells released from α-factor arrest and in cells treated similarly but deprived of Cdc6p. We observed that PP2A-Cdc55-specific activity gradually decreased in wild-type cells, reaching 50% of the initial activity at 80 min after release (Fig. 5C). In contrast, in cells that proceeded through mitosis without Cdc6p, the specific activity of PP2A-Cdc55 was maintained at a level similar to that observed in G1 phase (Fig. 5D). Note the change in ordinate scales in the two graphs shown in Fig. 5C and D. As an independent control for cell cycle progression, we monitored spindle elongation. Long anaphase spindles started to appear at the same time in both strains, namely, by 70 min after release, and reached a maximum in both strains at 90 min, which was in agreement with numerous previous studies (13, 58, 74). This indicates that the reduction in PP2A-Cdc55 activity in cells expressing Cdc6 occurred during anaphase and that the sustained PP2A-Cdc55 activity in cells deprived of mitotic Cdc6 also occurred during anaphase and was not due to failure to enter mitosis. The phosphatase assay shows a functional interaction between Cdc6 and PP2A-Cdc55 at the same point in the cell cycle as where physical interaction is observed. The functional interaction supports the conclusion that the physical interaction is physiologically significant (see Discussion).
Ability of Cdc6 mutants to interact with Cdc55 and cause delay.
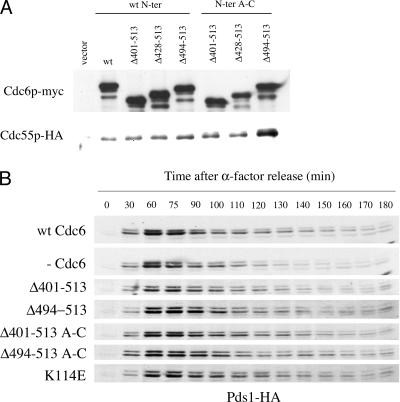
To gain more insight into the interaction between Cdc6p and Cdc55p and into its physiological significance, we analyzed the strength of interactions between Cdc55p and various Cdc6p deletion mutants. According to crystallographic data obtained with archaeal Cdc6 and upon alignment of archaeal Cdc6 with the Schizosaccharomyces pombe Cdc6p ortholog, Cdc18, Liu and colleagues proposed that Cdc6p is structured in three domains: the two more N-terminal domains form an AAA+-type nucleotide binding fold, and the third domain, at the C terminus, adopts a winged-helix fold similar to that of known DNA binding modules (35). Since domains I and II are involved in the assembly of pre-RC and the formation of active replication complexes (42, 53, 65, 48) and since Cdc6p with a deletion in the amino acids 8 to 48 can still interact with Cdc55p (not shown), we reasoned that domain III in Cdc6p could be important for Cdc55p interaction. By alignment of S. cerevisiae Cdc6p with S. pombe Cdc18, we determined that domain III in S. cerevisiae encompassed the last 113 amino acids, from 401 to 513. We constructed yeast strains that expressed Cdc6p mutants lacking different portions of the C-terminal domain under the GAL1,10 promoter, and we carried out Cdc6p/Cdc55p coimmunoprecipitation assays. A series of mutants was also constructed in which, in addition to various deletions at the Cdc6p C terminus, the N-terminal domain had the Cdc28 phosphorylation site mutations A to C. The A to C mutants failed to interact with Cdc28p. As shown in Fig. 6A, Cdc55p-HA can interact with myc-Cdc6p lacking amino acids 401 to 513, suggesting that the interaction involves amino acids 49 to 401. Interestingly, the deletion of the last 20 amino acids (494 to 513) seemed to increase the interaction with Cdc55p. This effect was especially evident when Cdc6p had the A to C Cdk site mutations in the N-terminal domain, suggesting a competition between Cdc55p and Cdc28p for binding Cdc6p. This competition may account for the fact that the cdc6ΔN mutant binds Cdc55 but does not lead to detectable attenuation of APC/Cdc20 Pds1 degradation. The results also show that the last 20 amino acids of Cdc6p (494 to 513) may have an inhibitory role in the interaction between Cdc6p and Cdc55p.
FIG. 6.
(A) Structure/function analysis of the effect of various Cd6p deletion mutations on the extent of interaction with Cdc55p. W303 strains expressing HA-tagged Cdc55p (YSB399) and carrying either an empty vector or vectors expressing the indicated myc-tagged Cdc6p C-terminal deletion mutant from the GAL1,10 promoter were grown to mid-log phase in YPRaf, galactose was added to induce the expression of Cdc6p for 3 h, and total extracts were prepared and subjected to immunoprecipitation. The top panel corresponds to myc immunoprecipitates blotted with the 9E10 anti-myc antibody. The bottom panel corresponds to myc immunoprecipitates blotted with the 12CA5 anti-HA antibody. The Cdc55-HA strains were YSB289, empty vector; YSB290, GAL-CDC6; YSB423, GAL-CDC6 Δ401-513; YSB424, GAL-CDC6 Δ428-513; YSB425, GAL-CDC6 Δ494-513; YSB427, GAL-CDC6 Δ401-513 A-C; YSB428, GAL-CDC6 Δ428-513 A-C; and YSB429, GAL-CDC6 Δ494-513 A-C. (B) Pds1-HA degradation kinetics in cells expressing endogenous levels of either wild-type (wt) Cdc6p or different Cdc6 deletion mutants, each of which is indicated to the left of each blot. As a control of nonfunctional Cdc6p, the effect of the expression of Cdc6 K114E is also included. Yeast cells with a copy of MET-CDC6 (K4055) and a plasmid-borne copy of the indicated myc-tagged CDC6 mutant expressed under the control of its own promoter or an empty vector and also containing a chromosomal copy of Pds1-HA were subjected to the same experimental conditions as those described in the legend for Fig. 1B. The strains used were YSB441, CDC6; YSB442, empty vector; YSB488, CDC6 Δ401-513; YSB490, Δ494-513; YSB482, Δ401-513 A-C; YSB483 Δ494-513 A-C; and YSB479, CDC6 K114E.
We also tested the ability of these Cdc6p mutant proteins to carry out various Cdc6p functions. When these mutants were expressed as the only source of Cdc6p, none of them could support growth (not shown). We next tested the effect of expressing these C-terminal deletion mutants on the Pds1p degradation in a normal cell cycle. The expression of all C-terminal deletion mutants resulted in a wild-type profile of Pds1p accumulation and degradation, which was expected, as all of them could interact with HA-Cdc55p. Mutant Cdc6pΔ401-513 A-C shows an enhanced interaction with HA-Cdc55p but an identical Pds1-HA degradation profile, suggesting that a limited quantity of Cdc55-HA interacting with Cdc6 may suffice to exert a physiological effect.
We have thus found that C-terminal deletion mutants that interact with Cdc55p have an effect on Pds1p accumulation which is like that of wild-type Cdc6p. The fact that none of these mutants can support growth and yet most of them inhibit the APC to levels similar to those of wild-type Cdc6p indicates that the faster Pds1p degradation kinetics in cells not expressing any source of Cdc6p is not due to lethality effects associated with the lack of Cdc6p replication function, but rather to another function in the Cdc6p itself. In a similar manner, the expression of myc-Cdc6pK114E as the only source of Cdc6p, a mutant defective in pre-RC formation that does not support growth (42, 65), also results in normal kinetics of Pds1p degradation (Fig. 6B). This result also suggests that preventing the assembly of the pre-RC in mitosis is not an essential step for the role of Cdc6p in modulating mitosis.
DISCUSSION
These studies were initiated in an attempt to explain the inhibition of mitosis observed by Bueno and Russell (5) due to the ectopic expression of Cdc6 in G2 phase, but not in other phases of the cell cycle. A partial explanation for Cdc6-induced mitotic delay was previously found in the fact that a highly overproduced Cdc6 mutant protein inhibited Cdc28 activity, which is needed to activate the APC through phosphorylation of the Cdc20 subunit. However, it was not clear whether this was the only mechanism by which Cdc6 inhibited mitosis or whether it could occur with endogenous levels of Cdc6. Therefore, we decided to study the effects of various Cdc6 mutants, particularly but not exclusively those that fail to interact with Cdc28, to try to identify other, potentially more direct, Cdc6-mediated mechanisms for modulating the APC (20). Four lines of evidence support the existence of two Cdc6-dependent mitotic inhibitory mechanisms, one dependent on Cdc28 and one not. First, we previously showed that ectopic expression of one Cdc6 phosphorylation site mutant (Cdc6 F) was lethal in a mutant affecting a core subunit of the APC, cdc16. Second, here we find that the depletion of Cdc6 causes an advancement of the time of complete degradation of APC substrates. Third, the manipulation of levels of Cdc6 through GAL induction causes a delay of complete degradation of two APC substrates, Pds1 and Clb2, but some Cdc6 mutants that neither interact with nor inhibit Cdc28 cause even greater delay than does wild-type Cdc6. Fourth, these latter mutants have an additive effect on cell viability with benomyl, a drug that leads to the inhibition of APC/Cdc20.
The Cdc6 depletion experiment seems to confirm that endogenous levels of Cdc6 could inhibit the APC and that the high levels used in previous studies and required to inhibit Cdc28 were not essential. The cell cycle expression profile of Cdc6p (Fig. 1A) and the effect of turning off Cdc6p expression (Fig. 1B) suggest that Cdc6p would inhibit the APC/Cdc20 after its activation, perhaps contributing to its shutoff once it had functioned.
One novel observation made during control experiments was that, unexpectedly, the levels of Cdc6p in late mitosis are at least as high as those reached upon the induction of the GAL1,10 promoter (Fig. 1A). This reduced the concern that effects observed after galactose induction could be artifacts of overproduction and allowed us to use GAL induction to study possible Cdc28-independent effects of Cdc6. By flow cytometry, we showed that Cdc6 mutants produce a strong delay of cells in G2/M and that this cell cycle delay correlates with the stabilization of Pds1p and Clb2 (Fig. 2C). The stronger effect of various Cdc6 mutants versus that of the wild type is probably due to the fact that Cdc6 is an intrinsically unstable protein and that the mutations we used lead to stabilization (7, 17, 21, 43).
Several further findings suggested that, while Cdc28 interaction can contribute to these effects of Cdc6, they cannot explain the whole cell cycle delay. An indirect argument is that Pds1 and Clb2 degradation starts at the same time in all situations, suggesting that there is sufficient Cdc28 to activate the APC. A more direct argument derives from the difference in the ability of Cdc6p A-C+F and Cdc6p F to inhibit Cdc28p, which implies the existence of two mechanisms relevant to the G2/M delay. One involves inhibitory interaction between Cdc6p and Cdc28p at very high levels of Cdc6p and is represented by Cdc6p F, which interacts with Cdc28p in vitro and partially inhibits Cdc28p in nocodazole-arrested cells in vivo (Fig. 3). The second and new mode of inhibition is highlighted by Cdc6p A-C+F, which does not bind Cdc28p in vitro or inhibit Cdc28p in vivo detectably (Fig. 3) but nevertheless extends the half life of APC targets and blocks the cell cycle in mitosis (Fig. 2 and 4). The Cdc28-independent mechanism could operate in the presence of lower levels of Cdc6 protein and would be in agreement with the fact that we observed a delay in Pds1p degradation very shortly after Cdc6 induction with galactose, when Cdc6 levels were still quite low and would not be expected to inhibit Cdc28, and with the fact that we observed the same growth defects in medium containing either 2% galactose or 0.02% galactose, which induces levels of Cdc6p expression much lower than 2% (not shown). It may also be significant that, although we did observe some in vivo inhibition of Cdc28p by Cdc6p F, the level of inhibition was much lower than that previously described by others studying the Cdc6-d2 mutant protein (43). This difference most likely arises from the fact that Cdc6p F mutants are less stable and do not accumulate to levels as high as those of Cdc6-d2 (43). The presence of active Cdc28p in Cdc6-F mutants demonstrated in Fig. 3, however, was unexpected, even though the APC seems to be inhibited, as evidenced by Pds1p stabilization and cell cycle delay.
Several potential Cdc6-mediated mechanisms that might explain Cdc6-dependent, Cdc28-independent cell cycle delays were considered and ruled out. The stabilization of the Sic1p Clb/Cdc28 repressor was not at work since sic1Δ does not reduce Cdc6-induced mitotic delay (43). Stimulating inhibitory phosphorylation of Cdc28p Tyr 19 by the Swe1 kinase was not the explanation, since swe1Δ did not reverse either the inhibition of Pds1p degradation in cells expressing Cdc6 F (Fig. 4C) or the morphological defects of the strains expressing other phosphorylation site mutants. The inhibition of Mih1 seemed unlikely since the disruption of MIH1 affected only strains overexpressing wild-type Cdc6 or C-terminal mutants, which, interestingly, are those that are capable of interacting with Cdc28p, but not strains expressing other phosphorylation site mutants (Fig. 3A and B). These data are very similar to previous reports describing cdc34 sic1Δ mutants. The disruption of SWE1 was not capable of rescuing the G2/M arrest of cdc34 sic1Δ strains grown at the restrictive temperature, whereas these cell cycle defects were increased in a mih1Δ background (29), which is what we see with cell cycle delay during the expression of stable Cdc6 mutants. Our results are in agreement with this Cdc34 substrate being Cdc6. Cell cycle checkpoints were not involved.
Adenovirus E4orf4 protein induces Cdc55-PP2A-dependent growth arrest in yeast and interacts with the APC. Also, as with Cdc6, adenovirus protein E4orf4 expression in yeast results in a mitotic delay increased in a mih1Δ background but not suppressed by the disruption of SWE1 (31). We therefore investigated cdc55Δ mutants defective in a regulatory subunit of PP2A and found that cdc55Δ suppresses the Cdc6-mediated delay of APC substrate destruction, the Cdc6-mediated cell cycle delay, and the lethality of the overexpression of Cdc6p in a cdc16 (APC mutant) background (Fig. 4). Normal Pds1 degradation (Fig. 4C) suggests that cdc55Δ does not just lead to a bypass of a mitotic exit block related to the inhibition of separase, which is required to downregulate Cdc55-phosphatase 2A activity (47), as a result of the increased levels of Pds1 (securin), a separase inhibitor. Instead, the reversal of Cdc6-induced APC target stabilization suggests that CDC55 may be involved at a step prior to the separase-PP2A interaction step (47). Furthermore, the premature release of Cdc14 into the nucleus, which might occur in cdc55Δ cells (47), would not be expected to enhance Pds1 degradation since Cdc14 is not an activator of APC-Cdc20. We also do not think that the cdc55Δ is acting to decrease the inhibition of Cdc28p, since Swe1p protein kinase levels are increased in cdc55Δ cells which should result in increased Cdc28p inhibition (39, 61, 68). Furthermore, Cdc55-PP2A is not the phosphatase involved in removing the activating phosphorylation of Cdc28p at Thr 169 (8). Despite the fact that cdc55Δ does not suppress the growth defects of cdc23 or cdc16 strains grown at the restrictive temperature (61), it can suppress the temperature sensitivity of cdc20-1 mutants (61), maybe by indirectly activating APC/Cdh1, as suggested by Wang and Ng (62). A more likely possibility is that ectopic expression of Cdc6p could be leading to changes in the phosphorylation status of a common substrate of both Cdc28 and Cdc55 required for APC/Cdc20 activity or even in the phosphorylation status of the APC/Cdc20 itself. This could occur by Cdc6 targeting Cdc55-PP2A to this substrate. CDC55 deletion would result in the inability of Cdc6 to target Cdc55-PP2A to this substrate and thus restore APC/Cdc20 activity. In other words, Cdc6 may affect a substrate whose phosphorylation state is kept within a functional range by an appropriate balance of Cdc28 and Cdc55 activities.
If Cdc6 targets Cdc55 to substrates in mitosis, then we predict that the two proteins would interact. In fact, we presented evidence for both physical and functional interaction between Cdc6 and Cdc55 during mitosis (Fig. 5). Cdc6 and Cdc55 coimmunoprecipitate during mitosis, and Cdc6 has an effect on PP2A-Cdc55 phosphatase since cells expressing Cdc6 show downregulation of PP2A-Cdc55 activity in anaphase and cells lacking Cdc6 show no decrease in PP2A-Cdc55 activity (Fig. 5C and D). The sustained levels of PP2A-Cdc55 activity might be expected to inhibit mitosis, but this is difficult to sort out given that the absence of Cdc6 will also lead to an increase in Cdc28 protein kinase activity. The fact that Cdc6p interacts with Cdc55p in mitosis but not other parts of the cell cycle and the fact that, in the absence of Cdc6p, Pds1p degradation is increased suggest that Cdc6p-Cdc55p interaction should have an inhibitory effect on the protease activity of the APC. This APC inhibitory effect might occur in the context of decreasing levels of Cdc55 phosphatase activity, as our results suggest, by increased phosphorylation of APC regulators. This might seem to contradict the observed reduced inhibition of APC in elevated PP2A-Cdc55 in cells lacking Cdc6, but in the absence of Cdc6, the higher levels of Cdc55 could not be targeted to mitotic substrates, and as mentioned above, an increase in Cdc28 protein kinase activity would counteract higher Cdc55 phosphatase activity.
The idea that Cdc6 may play a role in mitosis in addition to its function as a component of the pre-RC has been repeatedly proposed since it was shown that the ectopic expression of wild-type Cdc6 resulted in a delay in the G2/M stage of the cell cycle (5), and other groups have consistently reported similar results both for budding and fission yeast (24, 43). In addition to Cdc6 inhibiting entry into mitosis, Clb/Cdc28 inhibition by Cdc6 has been implicated in promoting timely exit from the cell cycle (46, 63, 6, 2). Newly synthesized and stabilized Cdc6, which is present at significant levels (Fig. 1), could also serve in a normal cell cycle to fully inhibit APC/Cdc20 before Cdc20 degradation in late mitosis. At this point, Cdc6 could interact with CDC55-PP2A and perhaps contribute to its downregulation, as results in Fig. 5 suggest. An interesting possibility is that Cdc28-bound Cdc6 could affect Cdc55 phosphatase activity. Despite the decrease in Cdc55-PP2A activity, the remaining activity could be targeted to a mitotic substrate that would then contribute to the deactivation of the APC/Cdc20. Alternatively, increased phosphorylation of APC regulators would result in APC/Cdc20 inhibition. Cdc6-mediated inhibition of Clb/Cdc28 would provide an additional means to inactivate APC/Cdc20. In fact, we have observed that Cdc6 ΔN interacts with Cdc55, but does not interact with Cdc28 and does not inhibit the cell cycle, suggesting that both Cdc28 inhibition and Cdc55 inhibition may be required for proper APC regulation.
Acknowledgments
We thank Doug Koshland and Paul Russell for plasmids, Karen Heichmann, Ray Deshaies, and Kim Nasmyth for strains, and J. R. Broach for antibodies against Tpd3. We also thank Benjamin Pina for the use of his laboratory for some of the work.
This work was supported by TRDRP 9RT-0179 and NIH GM25508.
Footnotes
Published ahead of print on 27 November 2006.
REFERENCES
- 1.Amon, A., U. Surana, I. Muroff, and K. Nasmyth. 1992. Regulation of p34CDC28 tyrosine phosphorylation is not required for entry into mitosis in S. cerevisiae. Nature 355:368-371. [DOI] [PubMed] [Google Scholar]
- 2.Archambault, V., C. X. Li, A. J. Tackett, R. Wasch, B. T. Chait, M. P. Rout, and F. R. Cross. 2003. Genetic and biochemical evaluation of the importance of Cdc6 in regulating mitotic exit. Mol. Biol. Cell 14:4592-4604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Basco, R. D., M. D. Segal, and S. I. Reed. 1995. Negative regulation of G1 and G2 by S-phase cyclins of Saccharomyces cerevisiae. Mol. Cell. Biol. 15:5030-5042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Booher, R. N., R. J. Deshaies, and M. W. Kirschner. 1993. Properties of Saccharomyces cerevisiae wee1 and its differential regulation of p34CDC28 in response to G1 and G2 cyclins. EMBO J. 12:3417-3426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bueno, A., and P. Russell. 1992. Dual functions of CDC6: a yeast protein required for DNA replication also inhibits nuclear division. EMBO J. 11:2167-2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Calzada, A., M. Sacristan, E. Sanchez, and A. Bueno. 2001. Cdc6 cooperates with Sic1 and Hct1 to inactivate mitotic cyclin-dependent kinases. Nature 412:355-358. [DOI] [PubMed] [Google Scholar]
- 7.Calzada, A., M. Sanchez, E. Sanchez, and A. Bueno. 2000. The stability of the Cdc6 protein is regulated by cyclin-dependent kinase/cyclin B complexes in Saccharomyces cerevisiae. J. Biol. Chem. 275:9734-9741. [DOI] [PubMed] [Google Scholar]
- 8.Cheng, A., K. E. Ross, P. Kaldis, and M. J. Solomon. 1999. Dephosphorylation of cyclin-dependent kinases by type 2C protein phosphatases. Genes Dev. 13:2946-2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clarke, D. J., and J. F. Gimenez-Abian. 2000. Checkpoints controlling mitosis. Bioessays 22:351-363. [DOI] [PubMed] [Google Scholar]
- 10.Cocker, J. H., S. Piatti, C. Santocanale, K. Nasmyth, and J. F. Diffley. 1996. An essential role for the Cdc6 protein in forming the pre-replicative complexes of budding yeast. Nature 379:180-182. [DOI] [PubMed] [Google Scholar]
- 11.Cohen-Fix, O., J. M. Peters, M. W. Kirschner, and D. Koshland. 1996. Anaphase initiation in Saccharomyces cerevisiae is controlled by the APC-dependent degradation of the anaphase inhibitor Pds1p. Genes Dev. 10:3081-3093. [DOI] [PubMed] [Google Scholar]
- 12.Desdouets, C., C. Santocanale, L. S. Drury, G. Perkins, M. Foiani, P. Plevani, and J. F. Diffley. 1998. Evidence for a Cdc6p-independent mitotic resetting event involving DNA polymerase alpha. EMBO J. 17:4139-4146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Detweiler, C. S., and J. J. Li. 1997. Cdc6p establishes and maintains a state of replication competence during G1 phase. J. Cell Sci. 110:753-763. [DOI] [PubMed] [Google Scholar]
- 14.Diffley, J. F. 2001. DNA replication: building the perfect switch. Curr. Biol. 11:R367-R370. [DOI] [PubMed] [Google Scholar]
- 15.Donaldson, A. D., and J. J. Blow. 1999. The regulation of replication origin activation. Curr. Opin. Genet. Dev. 9:62-68. [DOI] [PubMed] [Google Scholar]
- 16.Drury, L. S., G. Perkins, and J. F. Diffley. 1997. The Cdc4/34/53 pathway targets Cdc6p for proteolysis in budding yeast. EMBO J. 16:5966-5976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Drury, L. S., G. Perkins, and J. F. Diffley. 2000. The cyclin-dependent kinase Cdc28p regulates distinct modes of Cdc6p proteolysis during the budding yeast cell cycle. Curr. Biol. 10:231-240. [DOI] [PubMed] [Google Scholar]
- 18.Dua, R., D. L. Levy, and J. L. Campbell. 1998. Role of the putative zinc finger domain of Saccharomyces cerevisiae DNA polymerase ɛ in DNA replication and the S/M checkpoint pathway. J. Biol. Chem. 273:30046-30055. [DOI] [PubMed] [Google Scholar]
- 19.Dutta, A., and S. P. Bell. 1997. Initiation of DNA replication in eukaryotic cells. Annu. Rev. Cell Dev. Biol. 13:293-332. [DOI] [PubMed] [Google Scholar]
- 20.Elsasser, S., Y. Chi, P. Yang, and J. L. Campbell. 1999. Phosphorylation controls timing of Cdc6p destruction: a biochemical analysis. Mol. Biol. Cell 10:3263-3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Elsasser, S., F. Lou, B. Wang, J. L. Campbell, and A. Jong. 1996. Interaction between yeast Cdc6 protein and B-type cyclin/Cdc28 kinases. Mol. Biol. Cell 7:1723-1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Foiani, M., G. Liberi, G. Lucchini, and P. Plevani. 1995. Cell cycle-dependent phosphorylation and dephosphorylation of the yeast DNA polymerase α-primase B subunit. Mol. Cell. Biol. 15:883-891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Glotzer, M., A. W. Murray, and M. W. Kirschner. 1991. Cyclin is degraded by the ubiquitin pathway. Nature 349:132-138. [DOI] [PubMed] [Google Scholar]
- 24.Greenwood, E., H. Nishitani, and P. Nurse. 1998. Cdc18p can block mitosis by two independent mechanisms. J. Cell Sci. 111:3101-3108. [DOI] [PubMed] [Google Scholar]
- 25.Harper, J. W., J. L. Burton, and M. J. Solomon. 2002. The anaphase-promoting complex: it's not just for mitosis any more. Genes Dev. 16:2179-2206. [DOI] [PubMed] [Google Scholar]
- 26.Holloway, S. L., M. Glotzer, R. W. King, and A. W. Murray. 1993. Anaphase is initiated by proteolysis rather than by the inactivation of maturation-promoting factor. Cell 73:1393-1402. [DOI] [PubMed] [Google Scholar]
- 27.Irniger, S., S. Piatti, C. Michaelis, and K. Nasmyth. 1995. Genes involved in sister chromatid separation are needed for B-type cyclin proteolysis in budding yeast. Cell 81:269-278. [DOI] [PubMed] [Google Scholar]
- 28.Jensen, S., M. Segal, D. J. Clarke, and S. I. Reed. 2001. A novel role of the budding yeast separin Esp1 in anaphase spindle elongation: evidence that proper spindle association of Esp1 is regulated by Pds1. J. Cell Biol. 152:27-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kaiser, P., R. A. Sia, E. G. Bardes, D. J. Lew, and S. I. Reed. 1998. Cdc34 and the F-box protein Met30 are required for degradation of the Cdk-inhibitory kinase Swe1. Genes Dev. 12:2587-2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koepp, D. M., J. W. Harper, and S. J. Elledge. 1999. How the cyclin became a cyclin: regulated proteolysis in the cell cycle. Cell 97:431-434. [DOI] [PubMed] [Google Scholar]
- 31.Kornitzer, D., R. Sharf, and T. Kleinberger. 2001. Adenovirus E4orf4 protein induces PP2A-dependent growth arrest in Saccharomyces cerevisiae and interacts with the anaphase-promoting complex/cyclosome. J. Cell Biol. 154:331-344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lahav-Baratz, S., V. Sudakin, J. V. Ruderman, and A. Hershko. 1995. Reversible phosphorylation controls the activity of cyclosome-associated cyclin-ubiquitin ligase. Proc. Natl. Acad. Sci. USA 92:9303-9307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lew, D. J., and S. I. Reed. 1995. A cell cycle checkpoint monitors cell morphogenesis in budding yeast. J. Cell Biol. 129:739-749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lim, H. H., P. Y. Goh, and U. Surana. 1998. Cdc20 is essential for the cyclosome-mediated proteolysis of both Pds1 and Clb2 during M phase in budding yeast. Curr. Biol. 8:231-234. [DOI] [PubMed] [Google Scholar]
- 35.Liu, J., C. L. Smith, D. DeRyckere, K. DeAngelis, G. S. Martin, and J. M. Berger. 2000. Structure and function of Cdc6/Cdc18: implications for origin recognition and checkpoint control. Mol. Cell 6:637-648. [DOI] [PubMed] [Google Scholar]
- 36.Longtine, M. S., A. McKenzie III, D. J. Demarini, N. G. Shah, A. Wach, A. Brachat, P. Philippsen, and J. R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953-961. [DOI] [PubMed] [Google Scholar]
- 37.McInerny, C. J., J. F. Partridge, G. E. Mikesell, D. P. Creemer, and L. L. Breeden. 1997. A novel Mcm1-dependent element in the SWI4, CLN3, CDC6, and CDC47 promoters activates M/G1-specific transcription. Genes Dev. 11:1277-1288. [DOI] [PubMed] [Google Scholar]
- 38.McMillan, J. N., R. A. Sia, and D. J. Lew. 1998. A morphogenesis checkpoint monitors the actin cytoskeleton in yeast. J. Cell Biol. 142:1487-1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Minshull, J., A. Straight, A. D. Rudner, A. F. Dernburg, A. Belmont, and A. W. Murray. 1996. Protein phosphatase 2A regulates MPF activity and sister chromatid cohesion in budding yeast. Curr. Biol. 6:1609-1620. [DOI] [PubMed] [Google Scholar]
- 40.Nguyen, V. Q., C. Co, and J. J. Li. 2001. Cyclin-dependent kinases prevent DNA re-replication through multiple mechanisms. Nature 411:1068-1073. [DOI] [PubMed] [Google Scholar]
- 41.Pasero, P., and E. Schwob. 2000. Think global, act local—how to regulate S phase from individual replication origins. Curr. Opin. Genet. Dev. 10:178-186. [DOI] [PubMed] [Google Scholar]
- 42.Perkins, G., and J. F. Diffley. 1998. Nucleotide-dependent prereplicative complex assembly by Cdc6p, a homolog of eukaryotic and prokaryotic clamp-loaders. Mol. Cell 2:23-32. [DOI] [PubMed] [Google Scholar]
- 43.Perkins, G., L. S. Drury, and J. F. Diffley. 2001. Separate SCFCDC4 recognition elements target Cdc6 for proteolysis in S phase and mitosis. EMBO J. 20:4836-4845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Piatti, S., T. Bohm, J. H. Cocker, J. F. Diffley, and K. Nasmyth. 1996. Activation of S-phase-promoting CDKs in late G1 defines a “point of no return” after which Cdc6 synthesis cannot promote DNA replication in yeast. Genes Dev. 10:1516-1531. [DOI] [PubMed] [Google Scholar]
- 45.Piatti, S., C. Lengauer, and K. Nasmyth. 1995. Cdc6 is an unstable protein whose de novo synthesis in G1 is important for the onset of S phase and for preventing a ‘reductional’ anaphase in the budding yeast Saccharomyces cerevisiae. EMBO J. 14:3788-3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pramila, T., S. Miles, D. GuhaThakurta, D. Jemiolo, and L. L. Breeden. 2002. Conserved homeodomain proteins interact with MADS box protein Mcm1 to restrict ECB-dependent transcription to the M/G1 phase of the cell cycle. Genes Dev. 16:3034-3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Queralt, E., C. Lehane, B. Novak, and F. Uhlmann. 2006. Downregulation of PP2ACdc55 phosphatase by separase initiates mitotic exit in budding yeast. Cell 125:719-732. [DOI] [PubMed] [Google Scholar]
- 48.Randell, J. C., J. L. Bowers, H. K. Rodriguez, and S. P. Bell. 2006. Sequential ATP hydrolysis by Cdc6 and ORC directs loading of the Mcm2-7 helicase. Mol. Cell 21:29-39. [DOI] [PubMed] [Google Scholar]
- 49.Rudner, A. D., K. G. Hardwick, and A. W. Murray. 2000. Cdc28 activates exit from mitosis in budding yeast. J. Cell Biol. 149:1361-1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rudner, A. D., and A. W. Murray. 2000. Phosphorylation by Cdc28 activates the Cdc20-dependent activity of the anaphase-promoting complex. J. Cell Biol. 149:1377-1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Russell, P., S. Moreno, and S. I. Reed. 1989. Conservation of mitotic controls in fission and budding yeasts. Cell 57:295-303. [DOI] [PubMed] [Google Scholar]
- 52.Sánchez, M., A. Calzada, and A. Bueno. 1999. The Cdc6 protein is ubiquitinated in vivo for proteolysis in Saccharomyces cerevisiae. J. Biol. Chem. 274:9092-9097. [DOI] [PubMed] [Google Scholar]
- 53.Schepers, A., and J. F. Diffley. 2001. Mutational analysis of conserved sequence motifs in the budding yeast Cdc6 protein. J. Mol. Biol. 308:597-608. [DOI] [PubMed] [Google Scholar]
- 54.Shirayama, M., A. Toth, M. Galova, and K. Nasmyth. 1999. APCCdc20 promotes exit from mitosis by destroying the anaphase inhibitor Pds1 and cyclin Clb5. Nature 402:203-207. [DOI] [PubMed] [Google Scholar]
- 55.Shteinberg, M., Y. Protopopov, T. Listovsky, M. Brandeis, and A. Hershko. 1999. Phosphorylation of the cyclosome is required for its stimulation by Fizzy/cdc20. Biochem. Biophys. Res. Commun. 260:193-198. [DOI] [PubMed] [Google Scholar]
- 56.Sia, R. A., H. A. Herald, and D. J. Lew. 1996. Cdc28 tyrosine phosphorylation and the morphogenesis checkpoint in budding yeast. Mol. Biol. Cell 7:1657-1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sorger, P. K., and A. W. Murray. 1992. S-phase feedback control in budding yeast independent of tyrosine phosphorylation of p34cdc28. Nature 355:365-368. [DOI] [PubMed] [Google Scholar]
- 58.Surana, U., A. Amon, C. Dowzer, J. McGrew, B. Byers, and K. Nasmyth. 1993. Destruction of the CDC28/CLB mitotic kinase is not required for the metaphase to anaphase transition in budding yeast. EMBO J. 12:1969-1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tanaka, T., D. Knapp, and K. Nasmyth. 1997. Loading of an Mcm protein onto DNA replication origins is regulated by Cdc6p and CDKs. Cell 90:649-660. [DOI] [PubMed] [Google Scholar]
- 60.Visintin, R., S. Prinz, and A. Amon. 1997. CDC20 and CDH1: a family of substrate-specific activators of APC-dependent proteolysis. Science 278:460-463. [DOI] [PubMed] [Google Scholar]
- 61.Wang, Y., and D. J. Burke. 1997. Cdc55p, the B-type regulatory subunit of protein phosphatase 2A, has multiple functions in mitosis and is required for the kinetochore/spindle checkpoint in Saccharomyces cerevisiae. Mol. Cell. Biol. 17:620-626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang, Y., and T. Y. Ng. 2006. Phosphatase 2A negatively regulates mitotic exit in Saccharomyces cerevisiae. Mol. Biol. Cell 17:80-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wäsch, R., and F. R. Cross. 2002. APC-dependent proteolysis of the mitotic cyclin Clb2 is essential for mitotic exit. Nature 418:556-562. [DOI] [PubMed] [Google Scholar]
- 64.Weinreich, M., C. Liang, H. H. Chen, and B. Stillman. 2001. Binding of cyclin-dependent kinases to ORC and Cdc6p regulates the chromosome replication cycle. Proc. Natl. Acad. Sci. USA 98:11211-11217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Weinreich, M., C. Liang, and B. Stillman. 1999. The Cdc6p nucleotide-binding motif is required for loading Mcm proteins onto chromatin. Proc. Natl. Acad. Sci. USA 96:441-446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yamamoto, A., V. Guacci, and D. Koshland. 1996. Pds1p, an inhibitor of anaphase in budding yeast, plays a critical role in the APC and checkpoint pathway(s). J. Cell Biol. 133:99-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yamamoto, A., V. Guacci, and D. Koshland. 1996. Pds1p is required for faithful execution of anaphase in the yeast, Saccharomyces cerevisiae. J. Cell Biol. 133:85-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yang, H., W. Jiang, M. Gentry, and R. L. Hallberg. 2000. Loss of a protein phosphatase 2A regulatory subunit (Cdc55p) elicits improper regulation of Swe1p degradation. Mol. Cell. Biol. 20:8143-8156.11027284 [Google Scholar]
- 69.Yang, S. S., E. Yeh, E. D. Salmon, and K. Bloom. 1997. Identification of a mid-anaphase checkpoint in budding yeast. J. Cell Biol. 136:345-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yellman, C. M., and D. J. Burke. 2006. The role of Cdc55 in the spindle checkpoint is through regulation of mitotic exit in Saccharomyces cerevisiae. Mol. Biol. Cell 17:658-666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yeong, F. M., H. H. Lim, C. G. Padmashree, and U. Surana. 2000. Exit from mitosis in budding yeast: biphasic inactivation of the Cdc28-Clb2 mitotic kinase and the role of Cdc20. Mol. Cell 5:501-511. [DOI] [PubMed] [Google Scholar]
- 72.Zachariae, W., and K. Nasmyth. 1999. Whose end is destruction: cell division and the anaphase-promoting complex. Genes Dev. 13:2039-2058. [DOI] [PubMed] [Google Scholar]
- 73.Zhou, C., and A. Jong. 1990. CDC6 mRNA fluctuates periodically in the yeast cell cycle. J. Biol. Chem. 265:19904-19909. [PubMed] [Google Scholar]
- 74.Zwerschke, W., H. W. Rottjakob, and H. Kuntzel. 1994. The Saccharomyces cerevisiae CDC6 gene is transcribed at late mitosis and encodes a ATP/GTPase controlling S phase initiation. J. Biol. Chem. 269:23351-23356. [PubMed] [Google Scholar]