Abstract
The protein tyrosine phosphatase PEST (PTP-PEST) is involved in the regulation of the actin cytoskeleton. Despite the emerging functions attributed to both PTPs and the actin cytoskeleton in apoptosis, the involvement of PTP-PEST in apoptotic cell death remains to be established. Using several cell-based assays, we showed that PTP-PEST participates in the regulation of apoptosis. As apoptosis progressed, a pool of PTP-PEST localized to the edge of retracting lamellipodia. Expression of PTP-PEST also sensitized cells to receptor-mediated apoptosis. Concertedly, specific degradation of PTP-PEST was observed during apoptosis. Pharmacological inhibitors, immunodepletion experiments, and in vitro cleavage assays identified caspase-3 as the primary regulator of PTP-PEST processing during apoptosis. Caspase-3 specifically cleaved PTP-PEST at the 549DSPD motif and generated fragments, some of which displayed increased catalytic activity. Moreover, caspase-3 regulated PTP-PEST interactions with paxillin, leupaxin, Shc, and PSTPIP. PTP-PEST acted as a scaffolding molecule connecting PSTPIP to additional partners: paxillin, Shc, Csk, and activation of caspase-3 correlated with the modulation of the PTP-PEST adaptor function. In addition, cleavage of PTP-PEST facilitated cellular detachment during apoptosis. Together, our data demonstrate that PTP-PEST actively contributes to the cellular apoptotic response and reveal the importance of caspases as regulators of PTPs in apoptosis.
Tyrosine phosphorylationis involved in signal transduction pathways that are necessary for multiple cellular phenomena such as growth and proliferation, differentiation, locomotion, and apoptosis. The protein tyrosine phosphatases (PTPs) are the predominant enzymes that mediate the removal of the phosphate moiety from tyrosine residues and, therefore, are indispensable regulators of the proper development and homeostasis of a wide variety of living organisms.
PTP-PEST is classified as a cytosolic PTP whose closest homologues are PTP-PEP and PTP-HSCF (2). Although PTP-PEST contains PEST regions previously proposed to stimulate protein degradation (67), pulse-chase analysis demonstrated that PTP-PEST is a stable protein with a half-life of more than 4 h (14). PTP-PEST is ubiquitously expressed, although it is found at higher levels in hemopoietic tissues (23, 28). Inactivation of the PTP-PEST gene results in early embryonic death and establishes PTP-PEST as an essential gene for mouse development (22, 74). Studies performed in fibroblast cells link PTP-PEST expression with regulation of cell migration and adhesion (3, 34). Indeed, Sastry et al. have shown that PTP-PEST inhibits Rac1-induced cytoskeletal changes, thereby preventing membrane ruffle formation (69). Furthermore, in aortic smooth muscle cells, nitric oxide inhibits cell migration by activating PTP-PEST (53). In Xenopus blastomeres, forced overexpression of PTP-PEST interferes with cell motility and results in a gastrulation defect (25). Concomitant with these reported phenotypes, PTP-PEST was found to directly interact or be associated in complexes with several signaling and cytoskeleton-associated molecules, including p130Cas, Sin, Hef-1, leupaxin, Hic-5, paxillin, FAK, Pyk2, Grb-2, Shc, Csk, PSTPIP, WASP, Abl, and gelsolin (12, 13, 16, 21, 23, 24, 28, 32, 38, 54, 62). Interestingly, PTP-PEST decreased WASP-promoted immunological synapse formation and actin polymerization in T cells (5). PTP-PEST reduces lymphocyte activation by inhibiting the Ras-mitogen-activated protein (MAP) kinase pathway (29). Finally, altered PTP-PEST interactions were associated with the human autoinflammatory disorder PAPA syndrome (82).
Apoptosis-mediated cell death is necessary for proper development, efficient immune function, and maintenance of tissue homeostasis (1). Two primary pathways activate apoptosis: the extrinsic pathway and the intrinsic pathway (9). The extrinsic pathway depends on activation of the death receptor members of the tumor necrosis factor (TNF) receptor family (27), whereas proteins sensing the diverse cellular stress trigger the intrinsic pathway (1). To amplify the apoptotic signal, these two pathways lead to the activation of the caspase cascade (9). Once activated, the executioner caspases commit cells to apoptosis by cleavage and alteration of function of their substrates (31).
Proper cellular adhesion is essential to mediate anchorage-dependent cell survival signals (75). In addition, to maintain physiological equilibrium, cells need to adopt a specific morphology. Apoptosis can result from the loss of cellular attachment, termed anoikis, or from a disturbance of the cytoskeleton leading to improper cellular morphology, known as amorphosis cell death (56). Importantly, cleavage of cytoskeletal proteins by caspases correlates with the morphological changes and cellular detachment that characterize apoptosis (19, 31). Interestingly, the caspase-mediated cleavage of p130Cas, a PTP-PEST-targeted protein, contributes to the dismantling of adhesion structures, whereas the cleavage of ROCK I induces formation of membrane blebbing (20, 50, 71).
Cumulative evidence indicates that phosphatases are critical regulators of apoptosis. For example, LAR-PTP (78), SHP-1 (84), YopH (11), PTP-1B (36), SAP-1 (77), TC-PTP (39), and PTP-PEP (41) were reported to promote apoptosis, whereas SHP-2 (45), FAP-1 (44), and MAP kinase phosphatase (83) attenuate the apoptotic response. Reactive oxygen species, which inhibit PTP activity (59), contribute to the induction of apoptosis (48). In addition, several kinases and phosphatases have been identified as important modulators of the apoptotic response by short interfering RNA screening (55). Some protein serine/threonine phosphatases are substrates of caspases, as exemplified by calcineurin and PP2A (31). Furthermore, caspase-3 regulates the protein stability of the dual-specific phosphatase PTEN (80). Nevertheless, there are not yet any reports of tyrosine-specific PTPs cleaved by caspases during apoptosis.
Through its numerous protein-protein interactions, PTP-PEST contributes to cytoskeletal organization, regulates immune cell activation, and plays a pivotal role in embryonic development. The function and regulation of PTP-PEST in apoptosis, however, have not yet been explored. Herein, we show that expression of PTP-PEST sensitizes cells to receptor-mediated apoptosis. In cells undergoing apoptosis, we observed a consistent correlation between the presence of PTP-PEST in membrane ruffles and lamellipodia retraction. We demonstrated that PTP-PEST integrity, activity, and adaptor function were all modulated specifically by caspase-3. Furthermore, the cleavage of PTP-PEST contributes to cellular detachment during apoptosis. This study links PTP-PEST to the regulation of apoptosis and implicates caspases in the regulation of PTPs.
MATERIALS AND METHODS
Reagents and antibodies used in this study.
The chemicals were from BioShop Canada, Inc.; Fisher Scientific; and Sigma. Recombinant active caspase-3 was from Upstate. The PTP-PEST polyclonal (2528 and 2530) and monoclonal (AG25) antibodies were previously described (23, 32). Monoclonal antibodies specific for Shc, p130Cas, and paxillin were from the BD Transduction Laboratory. Antibodies against Csk, GST, and SHP-2 were from Santa Cruz Biotechnology. TC-PTP monoclonal antibodies (3E2), kindly provided by Daniel Brunet, were described previously (43), and CF4-1D antibodies were from Calbiochem. Polyclonal rabbit antibodies against PTP-1B and the monoclonal anti-myc (9E10) antibody were from Upstate. Antibodies specific for caspase-3, caspase-8, and caspase-9 were from Cell Signaling Technology. Antibodies against green fluorescent protein (GFP), PTP-1B (FG6), caspase-6, and caspase-7 were, respectively, from Clonetech, Calbiochem, Upstate, and BD Biosciences.
Plasmids, cDNAs, and transfections.
Murine PTP-PEST expression vectors, pcDNA3.1/Zeo-PTP-PEST (wild type [WT] and C231S), and pEGFP-C2-PTP-PEST (WT), were described previously (23). PSTPIP cDNA, described by Côté et al. (23), was subcloned into pEBG, using BamH1 and Not1 restriction sites. The pcDNA3.1/Zeo-PTP-PEST 549ASP552A, 604ADS607A, 740AKK743A, and 549ASPA/604ADSA constructs were generated by site-directed mutagenesis (QuikChange site-directed mutagenesis kit; Stratagene). pEGFP-C2-PTP-PEST WT and 549ASP552A were generated by PCR cloning of the appropriate cDNA, using oligonucleotides containing BamH1 and Xho1, and were inserted between the BglII and Sal1 restriction sites. pEBG-PTP-PEST WT and 549ASP552A were generated by PCR cloning of the appropriate cDNA, using oligonucleotides containing the BamH1 and Not1 restriction sites. pCS2mycECFP-PTP-PEST-EYFP (WT, 549ASP552A, 604ADS607A, 740AKK743A, or 549ASPA/604ADSA) was obtained by replacing PTPαSpD2 with PTP-PEST in pCS2mycECFP-PTPαSpD2-EYFP (8), using Nco1/BglII with the corresponding PTP-PEST cDNA amplified by PCR. Human PTP-PEST (hPTP-PEST) cDNA (Open Biosystems) amplified by PCR was subcloned into pGEX-6P1 (Pharmacia), using the BamH1 and Sal1 restriction sites. Glutathione S-transferase (GST)-hPTP-PEST cDNA was further amplified by PCR and subcloned into pRK5, using the Xba1 and Sal1 restriction sites. All cDNAs were verified by sequencing. All transfections were performed using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions.
Cell lines.
All cell lines were maintained in Dulbecco modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 50 μg/ml gentamicin (Gibco) at 37°C in a 5% CO2 humid atmosphere. For 293T cells, the media was also supplemented with 1% L-glutamine. Primary mouse embryonic fibroblasts (P-MEFs) from BALB/c mice (Jackson) were generated as previously described (18). Mouse spontaneously immortalized fibroblasts were previously described (35). HeLa299 cells were a generous gift of N. Sonenberg (McGill University). PTP-PEST−/− parental cells were described by Côté et al. (22). Stable reexpression of PTP-PEST in the PTP-PEST-null cells was obtained as follows: subconfluent PTP-PEST−/− fibroblasts were transfected with linearized (Sca1) pcDNA3.1/Zeo (empty vector [EV]) or pcDNA3.1/Zeo-PTP-PEST (WT). After 48 h, the cells were split, and positive clones were selected, using 100 μg/ml of Zeocin (Invitrogen). The media was changed every 2 days for 10 days, and isolated colonies were picked and expanded in selection media. Clones expressing PTP-PEST were identified by immunoblotting. The clones were grown in media supplemented with 50 μg/ml Zeocin during routine culture and were maintained in the absence of Zeocin during the course of each experiment.
Induction of apoptosis and immunoblotting.
Cells were stimulated with 60 μM cisplatin (Calbiochem), 1 μM staurosporine (Sigma), or 10 ng/ml tumor necrosis factor alpha (TNF-α) (Sigma) in the presence of 10 μg/ml cycloheximide (Sigma) for the indicated time. Adherent and detached cells were rinsed on ice with phosphate-buffered saline (PBS) and lysed (1% NP-40, 150 mM NaCl, 50 mM Tris [pH 7.4], 0.1% sodium dodecyl sulfate [SDS], 50 mM NaF, 1 mM Na3VO4, and Complete protease inhibitors [Roche]). Cell extracts were centrifuged at 16,000 × g for 10 min at 4°C, and the protein concentration of each sample was calculated by a Bradford assay (Bio-Rad protein assay). Equal amounts (20 μg) of denatured protein were separated by SDS-polyacrylamide gel electrophoresis (PAGE) and transferred to polyvinylidene difluoride membranes (Immobilon-P; Millipore). Commercial antibodies were used according to the manufacturers' instructions, and noncommercial TC-PTP and PTP-PEST antibodies were used as described previously (23, 43). The blots were stripped using Re-Blot Plus Strong (Chemicon International) and reprobed according to the instructions from the manufacturer.
The effect of pharmacological protease inhibitors on PTP-PEST degradation during apoptosis was determined as follows. Caspase inhibitor (Z-VAD-FMK; Cedarlane) and calpain inhibitor (PD150606; Calbiochem) were added to cells 2 h prior to the addition of 1 μM staurosporine, and coincubation was continued for an additional 6 h. Adherent and floating cells were harvested, lysed, and analyzed as described above.
Analysis of PTP-PEST localization.
HeLa299 cells were transfected for 6 h with 6 μg of pEGFP-C2-PTP-PEST in 10-cm dishes. The cells were then replated at 2.2 × 105 cells per 35-mm fibronectin-coated (10 μg/ml fibronectin/PBS [Sigma]; 2 h, 37°C) glass bottom dish (Mat Tek Corporation) and incubated for 16 h. HeLa299 cells expressing EGFP-PTP-PEST were treated with 50 ng/ml anti-Fas (human activating; Upstate) and 10 μg/ml cycloheximide for the indicated period of time. The cells were maintained at 37°C in a 5% CO2 humid atmosphere, using a digital temperature controller 37-2 (Zeiss) and a digital CTI-controller 3700 (Zeiss) connected to an Inkubator S (Zeiss) installed on an Axiovert 200 M (Zeiss) confocal microscope. Random field images were acquired at the indicated times, using the LSM510 Meta confocal system and software. Profile analysis was performed using the same software. For time-lapse video confocal microscopy, images were acquired at 15-s intervals.
Measurement of apoptosis.
PTP-PEST−/− parental cells and reexpressing clones were seeded at 1.7 × 106 cells/60-mm dish. Eight hours following their adhesion, the cells were rinsed with PBS and starved in 0.1% FBS DMEM for 16 h. The cells were treated with 10 μg/ml cycloheximide in the presence or absence of 10 ng/ml TNF-α in 10% FBS DMEM for 2.5 h. Adherent and floating cells were rinsed with ice-cold PBS and lysed (20 mM HEPES [pH 7.4], 2 mM EDTA, 1% NP-40, 10 μg/ml aprotinin, 10 μM leupeptin, and 2 mM dithiothreitol [DTT]), and samples were centrifuged at 4°C for 10 min at 16,000 × g. The protein content was normalized, using the Bradford method, and caspase activation was quantified by DEVDase assay as described by Hardy and coworkers (40). HeLa299 cells were transfected with 6 μg DNA for 6 h in 10-cm dishes, trypsinized, harvested, and seeded at 5 × 105 cells/60-mm dish for 18 h. After 24 h of serum starvation (0.1% FBS DMEM), the cells were treated for 6 h with or without 50 ng/ml human anti-Fas (Upstate) in the presence of 10 μg/ml cycloheximide in 10% FBS DMEM. The cells were lysed, and caspase activity was measured as described above.
Measurement of cellular attachment by flow cytometry.
HeLa299 cells were transfected with 13 μg of either pEGFP-PTP-PEST WT or pEGFP-PTP-PEST 549ASPA for 6 h in 10-cm dishes, trypsinized, harvested, and seeded at 310,000 cells/well (6-well plates) in 1% FBS DMEM for 16 h. The cells were treated for the indicated time with or without 50 ng/ml human anti-Fas (Upstate) in the presence of 10 μg/ml cycloheximide in 10% FBS DMEM. Following incubation, the media was removed, the cells were rinsed two times with PBS, and the attached cells were gently detached, using TrypLEExpress (Gibco) and harvested in PBS (supplemented with 2% FBS and 10 mM EDTA). The cells were transferred to polystyrene round-bottom tubes (BD Falcon) and centrifuged at 1,550 rpm at 4°C for 5 min (5810R centrifuge; Eppendorf). The cell pellets were resuspended in 500 μl PBS (supplemented with 2% FBS and 10 mM EDTA), and samples were kept on ice. The tubes were vortexed for 3 s and read, using FACScan (BD Bioscience), leaving 10 s for flow stabilization followed by a 30-s acquisition time for each reading. Each sample was analyzed three times using this method and vortexed for 3 s between each analysis. The results are representative of two independent experiments in which each condition was performed in triplicate. The cells were analyzed with Cell Quest Pro software (BD Bioscience) based on the forward and side scatter (eliminating doublets), and enhanced GFP (EGFP)-positive cells were read in the FL1 channel.
In vitro caspase-3 cleavage assay.
To monitor the cleavage of protein in vitro, clones from PTP-PEST−/− cells were rinsed with ice-cold PBS and lysed in cleavage assay buffer (20 mM HEPES [pH 7.4], 1% NP-40, 2 mM EDTA, 5 mM DTT, 10 μM leupeptin, 10 μg/ml aprotinin) as described by Kook et al. (50). Following determination of the protein concentration, 65 μg of protein lysate was incubated with 25 ng of recombinant caspase-3 (Upstate) for the indicated time period at 30°C. Control samples were incubated under the same conditions in the absence of caspase-3. Samples were immediately placed on ice, SDS sample buffer was added, the samples were boiled for 4 min, and the proteins were separated and detected using immunoblotting. To identify the caspase-3 cleavage site in PTP-PEST, HeLa299 cells transiently expressing either EGFP or a version of myc-ECFP-PTP-PEST-EYFP (WT, 549ASPA, 604ADSA, 740AKKA, or 549ASPA/604ADSA) were lysed, incubated with recombinant active caspase-3, and processed as described above. When isolated GST recombinant proteins were used as a substrate, PTP-PEST−/− cells expressing GST or GST-PTP-PEST were lysed in HMNETG (50 mM HEPES [pH 7.5], 1% Triton X-100, 150 mM NaCl, 1.5 mM MgCl2, 1 mM EGTA, 10% glycerol, and Complete protease inhibitors) as described above, and the lysate was incubated for 1 h at 4°C with glutathione Sepharose beads (Amersham Biosciences). The beads were then rinsed three times with HMNETG and two times with cleavage assay buffer. Samples were resuspended in cleavage assay buffer in the presence or absence of 50 ng of recombinant caspase-3, incubated at 30°C for the indicated time period, chilled on ice, rinsed once with lysis buffer (1% NP-40, 150 mM NaCl, 50 mM Tris [pH 7.4]), resuspended in SDS-sample buffer, and analyzed by immunoblotting.
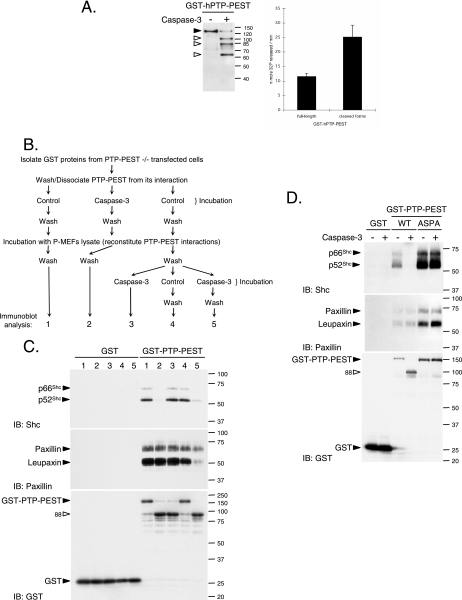
Analysis of PTP-PEST and PSTPIP protein complexes.
PTP-PEST−/− cells transiently expressing GST or GST-PTP-PEST were rinsed with ice-cold PBS and lysed in HMNETG, and protein extracts were centrifuged 10 min at 16,000 × g at 4°C. GST proteins were extracted on glutathione Sepharose beads at 4°C for 80 min. The beads were washed once with HMNETG, two times by inversion for 5 min at room temperatures in HMN300ETG (containing 300 mM NaCl), and twice with cleavage assay buffer. Beads bearing GST proteins were incubated for 30 min in caspase assay buffer with (sample number 2) or without (samples 1, 3, 4, and 5) 50 ng recombinant caspase-3 (Upstate) at 30°C (see Fig. 8B). The samples were chilled on ice and washed once with HMN300ETG and twice with the interaction buffer (1% NP-40, 150 mM NaCl, 50 mM Tris [pH 7.4], and Complete protease inhibitor). At this step, association of PTP-PEST with paxillin and Shc could not be detected (data not shown). Each sample was then incubated at 4°C for 2.5 h with P-MEF cell extract (2.5 mg of protein in 1 ml of interaction buffer) to recreate PTP-PEST interactions in vitro. Then the beads were washed three times with the interaction buffer and once with modified cleavage assay buffer (without DTT). Samples 3, 4, and 5 were then incubated in modified cleavage assay buffer with or without 250 ng caspase-3 for 30 min at 30°C (see Fig. 8B). Then interaction buffer was added to samples 4 and 5, and they were incubated for 10 min on ice and washed twice with interaction buffer. All samples were resuspended in SDS-sample buffer, and the protein content was analyzed by immunoblotting as described above.
FIG. 8.
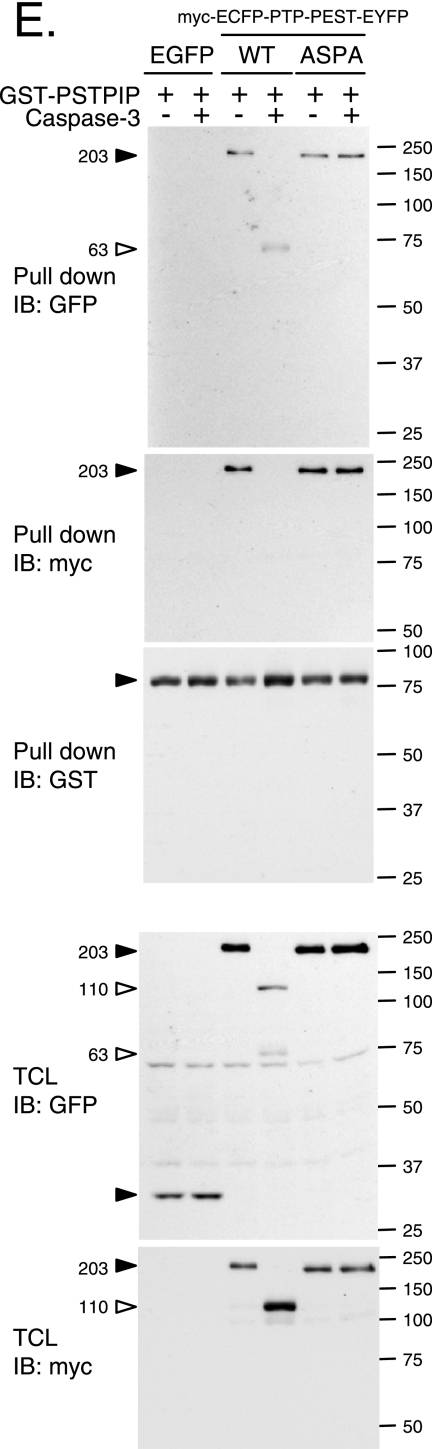
Caspase-3 regulates PTP-PEST activity and interactions. (A) GST-hPTP-PEST was isolated from transfected 293T cells, incubated with or without caspase-3, and subjected to in-solution phosphatase assays as described in Materials and Methods. GST, GST-PTP-PEST (C and D), and GST-PTP-PEST 549ASPA (D) were isolated from transfected PTP-PEST−/− cells. (B) Schematic representation of the approach used to investigate the effect of caspase-3 on the different protein-protein interactions. (C) Samples were processed (see numbers 1 to 5 outlined in panel B and Materials and Methods) and subjected to immunoblot analysis with antibodies directed against Shc, paxillin, and GST. (D) Samples were processed as numbers 1 and 2 described in panel B and then examined by immunoblotting with anti-Shc, anti-paxillin, and anti-GST antibodies. (E) PTP-PEST−/− cells transiently coexpressing GST-PSTPIP with either EGFP, myc-ECFP-PTP-PEST (WT)-EYFP, or myc-ECFP-PTP-PEST (549ASPA)-EYFP were lysed and treated with or without active recombinant caspase-3. GST-PSTPIP was isolated using glutathione Sepharose beads. Isolated GST-PSTPIP and bound PTP-PEST were detected by immunoblotting, using antibodies directed against GFP, GST, and myc. The expression of EGFP and PTP-PEST fusion proteins was also verified by immunoblotting the cell lysates. Filled arrowheads point to intact proteins, and empty arrowheads identify cleavage products. IB, immunoblot, TCL, total cell lysate. The values on the right correspond to molecular sizes in kDa.
To evaluate the importance of the 549DSPD site of PTP-PEST on the effect of caspase-3 interactions, GST proteins expressed in PTP-PEST−/− cells were isolated, washed, and incubated with or without recombinant caspase-3. After additional washes, they were incubated with P-MEF cell extract, washed again, and analyzed as described above for samples 1 and 2. To assess the ability of caspase-3 to regulate PSTPIP/PTP-PEST association, PTP-PEST−/− cells transiently expressing either EGFP or myc-ECFP-PTP-PEST (WT or 549ASPA)-EYFP with GST-PSTPIP were rinsed with PBS and lysed in modified cleavage assay buffer as described above. Aliquots of 1 mg of protein were incubated for 60 min at 30°C with or without 0.3 μg of recombinant caspase-3. Following the incubation, the samples were centrifuged at 16,000 × g for 2 min, chilled on ice, and diluted 10 times with interaction buffer. Proteins were recovered following incubation with glutathione Sepharose beads as described above. The beads were washed three times with interaction buffer, boiled in SDS-sample buffer, and analyzed by immunoblotting.
To analyze the PSTPIP protein complexes in vivo, HeLa299 cells transiently expressing either GST or GST-PSTPIP (WT or W232A) with myc-ECFP-PTP-PEST (WT or 549ASPA)-EYFP were treated with cycloheximide with or without anti-Fas for the indicated time. Both floating and adherent cells were colleted and lysed in interaction buffer for 10 min on ice, and the samples were then centrifuged at 16,000 × g for 10 min. Cleared lysates (2.5 mg) were then incubated at 4°C for 2.5 h with glutathione Sepharose beads as described above. The beads were washed three times with interaction buffer, boiled in SDS-sample buffer, and analyzed by immunoblotting.
In vitro caspase activation assay.
293T cells were trypsinized and collected by centrifugation. The cell pellets were washed with an equal volume of ice-cold NPM buffer {50 mM PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid).], 50 mM NaCl, 5 mM EGTA, 1 mM MgCl2, 5 mM DTT, 1 mM phenylmethylsulfonyl fluoride, 50 μg/ml pepstatin, 50 μg/ml leupeptin, 100 μg/ml aprotinin} and subsequently washed with five volumes of NPM buffer. The 293T cells were lysed with three cycles of snap-freeze-and-thaw, and the cell lysates were cleared by centrifugation at 16,000 × g for 5 min. Aliquots of the 293T lysates were stored in liquid nitrogen. The 293T cell lysates were stimulated with 2 mM dATP (Roche) at 37°C at the indicated time points. The reaction was terminated by mixing lysates with SDS-sample buffer and boiling for 3 min prior to analysis.
In-gel phosphatase assay.
[32P]ATP-labeled polypeptide (1 × 106 cpm) was incorporated into a 10% SDS-polyacrylamide gel prior to polymerization (60). Following electrophoresis, the gel was incubated for 1 h and then for 16 h in fixing buffer (20% isopropanol, 50 mM Tris [pH 8.0]). The gel was incubated in denaturation buffer (6 M guanidine-HCl, 50 mM Tris [pH 8.0], 0.3% β-mercaptoethanol) for 1.5 h and twice in renaturation buffer (50 mM Tris [pH 8.0], 1 mM EDTA, 0.04% Tween-40, 0.3% β-mercaptoethanol) for 1 h. The gel was subsequently incubated within the PTP reaction buffer (50 mM Tris [pH 8.0], 1 mM EDTA, 0.04% Tween-40, 0.3% β-mercaptoethanol, 3 mM DTT) for 1 h, followed by incubation with fresh reaction buffer for 16 h. Following Coomassie blue staining, the gel was dried and subjected to autoradiography for visualization of PTP activity.
Immunodepletion.
The dATP-treated 293T cell lysate (240 μg) was diluted to 0.8 μg/μl with lysis buffer (20 mM HEPES [pH 7.5], 50 mM NaCl, 1% NP-40, 10 mM NaF, 10 mM Na4P2O7, 10% glycerol, 100 μM phenylmethylsulfonyl fluoride, 5 μg/ml pepstatin, 5 μg/ml leupeptin, 10 μg/ml aprotinin). Following precleaning with 50 μl protein-G Sepharose beads (Amersham), the lysate was incubated with 25 μg PTP-PEST antibody (AG25), which had been conjugated with protein-G Sepharose, at 4°C for 2.5 h. Samples were analyzed by immunoblotting and in-gel PTP assay. For immunodepletion of capase-9, caspase-3, and caspase-7, 1.4 mg of 293T lysates were incubated with 12.5 μg of the appropriate antibody conjugated with protein-A/G Sepharose beads (Amersham) at 4°C for 2.5 h. The samples were then centrifuged, and the supernatants were collected and subjected to an additional round of immunodepletion in order to completely remove the specific caspases. Subsequently, each caspase-deprived lysate was subjected to treatment with 2 mM dATP and was then examined by immunoblotting or in-gel PTP assay.
In-solution phosphatase activity assay.
Five hundred twenty nanograms of GST-hPTP-PEST bound to glutathione Sepharose beads was incubated for 20 h at 37°C in the presence or absence of active caspase-3. Treated and control beads coupled with GST-hPTP-PEST were mixed with 3 μM 32P-labeled PTP substrates [polypeptide (Glu:Tyr)4:1] in 60 μl reaction buffer (25 mM HEPES [pH 7.5], 1 mM EDTA, 1 mM DTT) and then incubated at 30°C for 10 min with vigorous shaking. The reaction was terminated by the addition of 180 μl 20% trichloroacetic acid. Twenty microliters of bovine serum albumin (25 mg/ml) was added into the solution as a carrier protein to facilitate substrate precipitation. Following centrifugation (16,000 × g, 5 min), the supernatant was collected and counted, using a Multi-Purpose Scintillation counter (LS6500; Beckman) to detect the amount of released 32P. The catalytic activity of PTP was expressed as picomoles of 32P released per minute.
RESULTS
PTP-PEST localizes to retracting membrane ruffles of apoptotic cells.
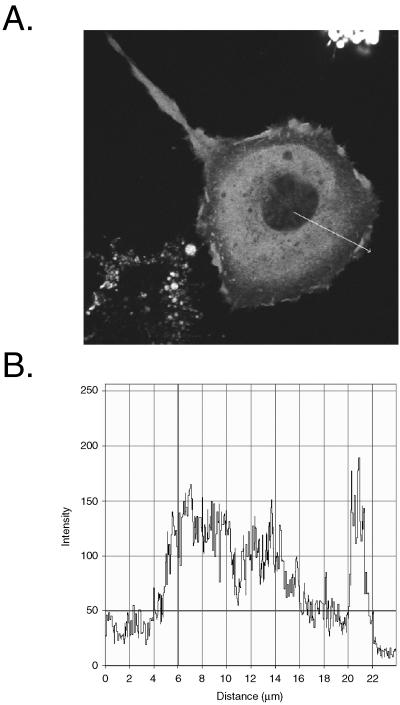
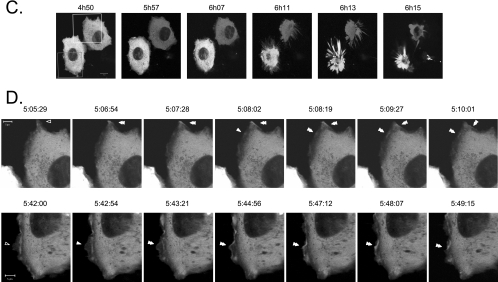
Progression of apoptosis is characterized by important changes in cellular morphology, such as membrane blebbing, cell rounding, and detachment, that are intimately associated with modifications of the actin cytoskeleton (19). Several reports link PTP-PEST to the regulation of the architecture of the actin cytoskeleton (3-5, 52, 69). To explore whether PTP-PEST physiologically contributes to apoptosis, we first examined its cellular localization during this phenomenon. HeLa299 cells expressing EGFP-PTP-PEST were incubated with anti-Fas and observed live using confocal microscopy. Prior to cellular detachment, PTP-PEST was localized predominantly in the cytosol. Interestingly, there was also a pronounced signal at the periphery of the cell (Fig. 1A). Profile analysis showed that the signal corresponding to PTP-PEST detected in membrane ruffles on the cell periphery was at least as intense as that present in the cytosol (Fig. 1B). This suggests substantial localization of PTP-PEST to the cytosol and likely the plasma membrane. To obtain more insight into the dynamic localization of PTP-PEST during apoptosis, time-lapse confocal video microscopy analysis was performed on anti-Fas-treated cells. Figure 1C illustrates that incubation with anti-Fas induced an apoptotic cell phenotype. In lamellipodia that either expanded or remained static, a low amount of PTP-PEST was observed (Fig. 1D). Preceding apoptotic cell death, PTP-PEST appeared at the edge of the lamellipodias that immediately retracted (Fig. 1D and Movies S1 and S2 in the supplemental material). Moreover, PTP-PEST was maintained at the edge of the contracting membrane ruffles during their movement inward (Fig. 1D and Movies S1 and S2 in the supplemental material). The presence of PTP-PEST in the retracting lamellipodia was observed in 91% of the cells analyzed (55 cells). Similar results were obtained when caspase-3 was microinjected into COS-7 cells expressing EGFP-PTP-PEST (data not shown). PTP-PEST was also concentrated at the base of membrane blebs in some cells (data not shown). Two other nonreceptor PTPs, PTP-1B and TC-PTP, as well as the adaptor protein Nck, did not significantly localize to the retracting membrane ruffle of anti-Fas-treated cells (data not shown). The presence of PTP-PEST in retracting lamellipodia implies that this enzyme could participate in the morphological changes and membrane detachment of cells undergoing apoptosis.
FIG. 1.
Localization of EGFP-PTP-PEST in live cells during apoptosis. HeLa299 cells expressing EGFP-PTP-PEST were stimulated with 50 ng/ml anti-Fas in the presence of 10 μg/ml cycloheximide and analyzed using confocal microscopy. (A) Cellular distribution of EGFP-PTP-PEST following 5 h and 50 min of treatment. (B) Profile of the intensity (arbitrary unit) of the fluorescence against the distance (μm) traversed by the arrow (drawn on panel A) representing the quantity of signal detected in different areas of the cell. (C) Pictures taken at the indicated time of incubation. Scale bar, 10 μm. (D) Time-lapse analysis of the area delimited by the squares drawn on panel C (see Movies S1 and S2 in the supplemental material). Images captured between 5:05:29 and 5:10:01 were acquired inside the white square, whereas those from 5:42:00 to 5:49:15 correspond to the gray square. Scale bar, 5 μm. Empty arrowheads point to sites where low amounts of EGFP-PTP-PEST were detected, filled arrowheads indicate the accumulation of signals, and double-filled arrowheads identify sites of inward movement on the cell periphery.
Expression of PTP-PEST sensitizes cells to receptor-mediated apoptosis.
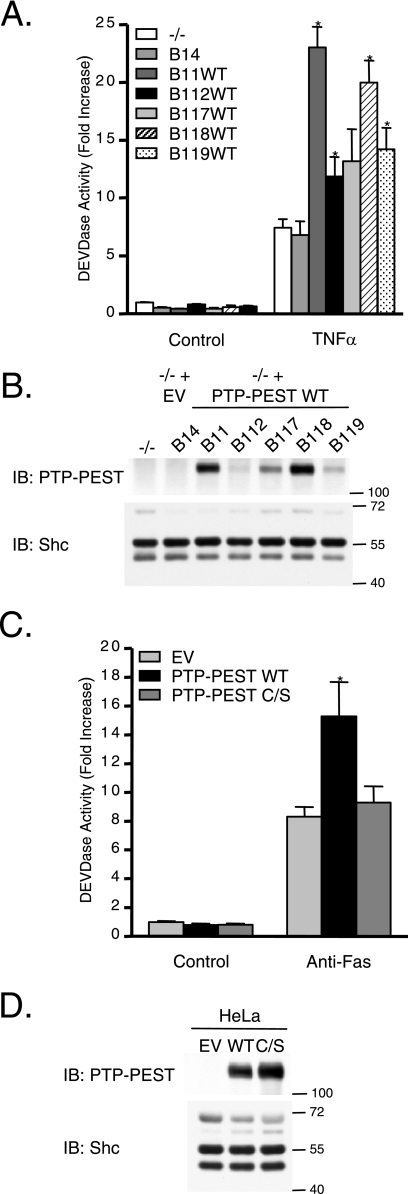
The specific presence of PTP-PEST in retracting membrane ruffles of apoptotic cells suggests that this enzyme potentially plays important functions in this type of cell death. Consequently, we evaluated the possibility that PTP-PEST influences the cellular response to apoptotic stimuli. For this purpose, PTP-PEST cDNA was reintroduced into PTP-PEST−/− parental cells, and single clones were isolated. Each clone expressed a different, yet stable, level of PTP-PEST (Fig. 2B). Clones expressing PTP-PEST that were treated with TNF-α exhibited enhanced induction of apoptosis compared to PTP-PEST−/− parental cells and PTP-PEST−/− EV control clones (Fig. 2A). In addition, the level of DEVDase induction appeared to correlate with the level of PTP-PEST expression: clones expressing higher levels of PTP-PEST exhibited greater sensitivity to TNF-α than the clones with lower PTP-PEST expression.
FIG. 2.
Expression of PTP-PEST sensitizes cells to receptor-mediated apoptosis. (A) PTP-PEST−/− parental cells and rescued clones were treated with 10 μg/ml cycloheximide in the absence (control) or presence of 10 ng/ml TNF-α for 2.5 h, and apoptosis was measured by DEVDase assay as described in Materials and Methods. Values are n-fold increases over those for the control PTP-PEST−/− parental cells (−/−) and represent means ± the standard errors (SE) of three independent experiments. Asterisks denote values significantly different from those of −/− cells ( , P < 0.05). (C) DEVDase activity measured in cell lysates obtained from HeLa299 cells transfected with either pcDNA3 (EV), pcDNA3-PTP-PEST-WT (WT), or pcDNA3-PTP-PEST-C231S (C/S) incubated for 6 h with 10 μg/ml cycloheximide in the absence (control) or presence of 50 ng/ml anti-Fas. Values are n-fold increases over those for the control EV and represent means ± SE of three independent experiments performed in duplicate. Asterisks denote values significantly different from those for control EV cell lysates (
, P < 0.05). (C) DEVDase activity measured in cell lysates obtained from HeLa299 cells transfected with either pcDNA3 (EV), pcDNA3-PTP-PEST-WT (WT), or pcDNA3-PTP-PEST-C231S (C/S) incubated for 6 h with 10 μg/ml cycloheximide in the absence (control) or presence of 50 ng/ml anti-Fas. Values are n-fold increases over those for the control EV and represent means ± SE of three independent experiments performed in duplicate. Asterisks denote values significantly different from those for control EV cell lysates ( , P < 0.01). Expression of PTP-PEST in the parental cells and in the various clones (B) and in transfected HeLa299 cells (D) was measured by SDS-PAGE and immunoblotting for PTP-PEST (2528 rabbit antibody). Clones derived from parental cells, which harbor the empty vector (B14), do not express PTP-PEST, whereas clones in which wild-type PTP-PEST was reintroduced (B11WT, B112WT, B117WT, B118WT, and B119WT) express different levels of PTP-PEST. The amount of protein on the immunoblot was normalized with the expression of Shc, and the values on the right correspond to molecular sizes in kDa. IB, immunoblot.
, P < 0.01). Expression of PTP-PEST in the parental cells and in the various clones (B) and in transfected HeLa299 cells (D) was measured by SDS-PAGE and immunoblotting for PTP-PEST (2528 rabbit antibody). Clones derived from parental cells, which harbor the empty vector (B14), do not express PTP-PEST, whereas clones in which wild-type PTP-PEST was reintroduced (B11WT, B112WT, B117WT, B118WT, and B119WT) express different levels of PTP-PEST. The amount of protein on the immunoblot was normalized with the expression of Shc, and the values on the right correspond to molecular sizes in kDa. IB, immunoblot.
These results were also validated in transient expression experiments using a different cell line (HeLa299) and a different apoptotic stimulus (anti-Fas) (Fig. 2C). Cells expressing PTP-PEST displayed a greater increase in apoptosis than the empty vector-transfected controls. In contrast, the catalytically dead PTP-PEST mutant (C231S) failed to induce a significant increase in apoptosis compared to that for the empty vector control, indicating that the catalytic activity of PTP-PEST is essential for its effect on the Fas-mediated apoptotic response. Efficiency of the transfection is shown in Fig. 2D. In both the rescued fibroblast and HeLa models, PTP-PEST expression did not spontaneously induce apoptosis in cells growing under control conditions (Fig. 2A and C). Together, these results suggest that the expression and catalytic activity of PTP-PEST increase sensitivity to receptor-mediated apoptosis.
PTP-PEST is cleaved during apoptosis.
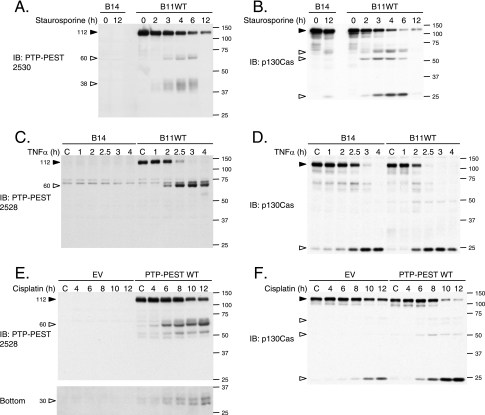
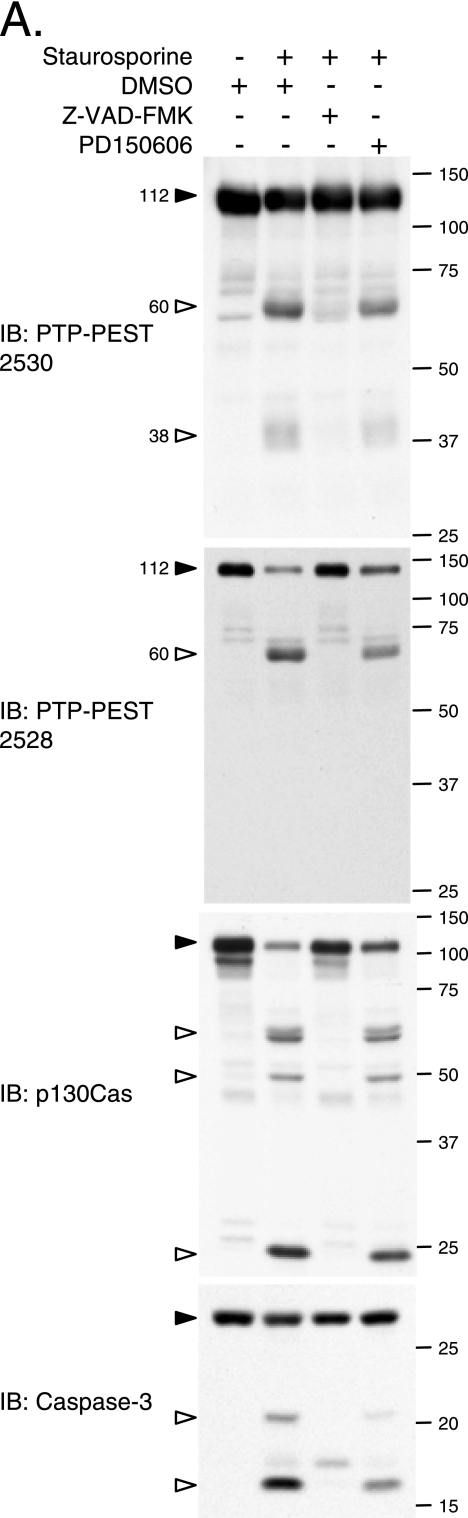
Several cytoskeletal proteins are degraded during apoptosis (31). Since PTP-PEST seems to play an important role in programmed cell death, we sought more insight into its regulation during apoptosis. We examined the consequences of activating the intrinsic or extrinsic apoptotic pathways on the integrity of PTP-PEST. Following stimulation of the PTP-PEST−/−-expressing WT clone with staurosporine or TNF-α and treatment of transfected HeLa299 cells with cisplatin, the amount of full-length PTP-PEST decreased in a time-dependent fashion (Fig. 3A, C, and E). PTP-PEST antibodies detected a protein fragment of approximately 60 kDa. Also, PTP-PEST degradation induced by staurosporine and cisplatin treatment resulted in diffuse bands detectable by SDS-PAGE at ∼38 and ∼30 kDa (Fig. 3A and E). Treatment with tunicamycin induced similar PTP-PEST degradation (data not shown). Interestingly, under the same apoptotic conditions, p130Cas was also degraded and gave the typical band pattern described previously (50) (Fig. 3B, D, and F). We observed that PTP-PEST expression accelerated TNF-α- and cisplatin-induced p130Cas degradation in rescued PTP-PEST−/− cells and in transfected HeLa cells (Fig. 3D and F). The enhanced p130Cas cleavage correlated with the increased DEVDase activity in PTP-PEST-WT-expressing cells (Fig. 2). Taken together, these results show that PTP-PEST is cleaved in a time-dependent manner during apoptosis triggered by intrinsic cellular stress, DNA damage, and extrinsic pathways.
FIG. 3.
PTP-PEST is cleaved during apoptosis. PTP-PEST−/− EV and PTP-PEST−/−-expressing WT cells (clones B14 and B11WT), described in Fig. 2, were treated with 1 μM staurosporine (A and B), or 10 ng/ml TNF-α in the presence of 10 μg/ml cycloheximide for the indicated times (C and D; note that the control [C] received only 10 μg/ml cycloheximide). HeLa299 cells transfected with either empty vector or PTP-PEST WT cDNA were treated under control conditions (dimethyl sulfoxide) or with 60 μM cisplatin for different periods of time (E and F). (E) A longer exposure of the anti-PTP-PEST immunoblot is shown in the bottom panel. Protein extracts were analyzed by immunoblotting for PTP-PEST, using 2530 rabbit antibody (A) or 2528 rabbit antibody (C and E), as well as for p130Cas (B, D, and F). Filled arrowheads point to intact proteins, and empty arrowheads identify degradation products. IB, immunoblot. The values on the right correspond to molecular sizes in kDa.
Degradation of PTP-PEST during apoptosis is specific.
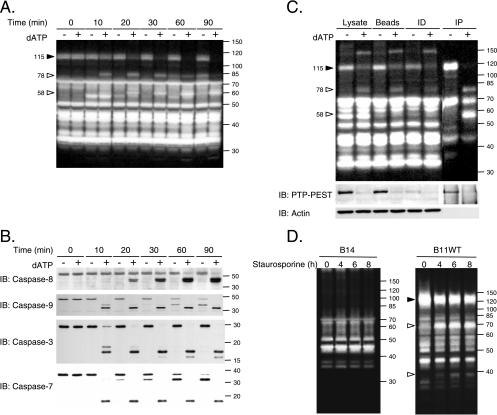
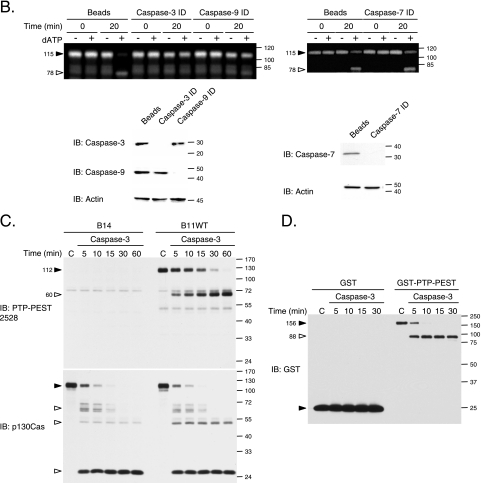
Increasing evidence in the literature implicates PTPs in the regulation of apoptotic signal transduction, but the mechanism of PTP regulation is not largely understood, and none of the classical PTPs have been reported to be cleaved during apoptosis. Therefore, we examined whether the cleavage of PTP-PEST during apoptosis was specific or reflected general processing of PTP family members. For this purpose, we employed a cell-free system in which dATP stimulates caspase activity (66). Control and dATP-treated 293T cell extracts were analyzed by in-gel PTP assay in order to visualize the population of active PTPs (Fig. 4A). Following in vitro caspase activation, a PTP of approximately 115 kDa became less abundant, and the phosphatase activity of proteins migrating at ∼78 and ∼58 kDa increased. Furthermore, the effect of dATP-mediated caspase activation seemed to be specific for these three bands, since the signal corresponding to PTPs at other molecular masses remained unchanged. Under these conditions, we observed that the initiator caspases, caspase-8 and caspase-9, and the effector caspases, caspase-3 and caspase-7, were activated (Fig. 4B). To confirm the identity of the modulated PTPs (Fig. 4A), dATP-stimulated 293T cell extracts were subjected to immunodepletion with anti-PTP-PEST antibody prior to an in-gel PTP assay. The efficiency of the immunodepletion was assessed by immunoblotting (Fig. 4C). Our results suggest that the ∼115-kDa band corresponds to the noncleaved form of PTP-PEST, while both the ∼78- and ∼58-kDa signals represent truncated forms of the protein (Fig. 4C). To confirm these data in vivo, we took advantage of our PTP-PEST−/− cells in which either an empty vector or PTP-PEST was reintroduced and expressed in a stable manner. Following staurosporine stimulation, no significant changes could be observed in tyrosine phosphatase activity patterns among the PTP-PEST−/− EV (B14, B15) clones analyzed (Fig. 4D and data not shown). In contrast, treatment of cells expressing PTP-PEST WT (B11) (as well as the B118 clone [data not shown]) led to a decrease in the 120-kDa band and increased activity detected near the 70- and 40-kDa markers. PTP-PEST expression and modulation were confirmed by immunoblotting (data not shown). As expected, PTP-PEST catalytic activity is not required for its cleavage, since the PTP-PEST C231S mutant was also efficiently degraded during staurosporine treatment (data not shown). Among the enzymes detected by in-gel PTP assay, PTP-PEST is the predominant phosphatase showing variation during apoptosis. The generation of smaller bands with phosphatase activity, seen with expression of an active form of PTP-PEST, suggests that some cleaved products of this enzyme are potentially catalytically active.
FIG. 4.
PTP-PEST is specifically cleaved during apoptosis. (A) Proteins were extracted from 293T cells, treated with or without 2 mM dATP for various times, and subjected to in-gel PTP assays. (B) Activation of caspases in dATP-treated 293T cell extracts was confirmed by immunoblotting with anti caspase-8, -9, -3, and -7. (C) 293T cell lysates treated with or without dATP were incubated with beads in the presence or absence of AG25 PTP-PEST antibody. Supernatants from bead control and PTP-PEST immunodepleted (ID) samples and PTP-PEST immunoprecipitate (IP) samples were subjected to in-gel PTP assay analysis. The control lysate, beads, and immunodepleted and immunoprecipitated protein samples were analyzed for their actin and PTP-PEST contents by immunoblotting (bottom). (D) PTP-PEST−/− EV and PTP-PEST−/−-expressing WT cells (clones B14 and B11WT), described in Fig. 2, were treated with 1 μM staurosporine for different periods of time, lysed, and subjected to in gel-PTP assays. PTP-PEST expression in these samples was also confirmed by immunoblotting (data not shown). Filled arrowheads indicate decreasing signals, whereas empty arrowheads point to increasing signals observed during the different treatments. IB, immunoblot. The values on the right correspond to molecular sizes in kDa.
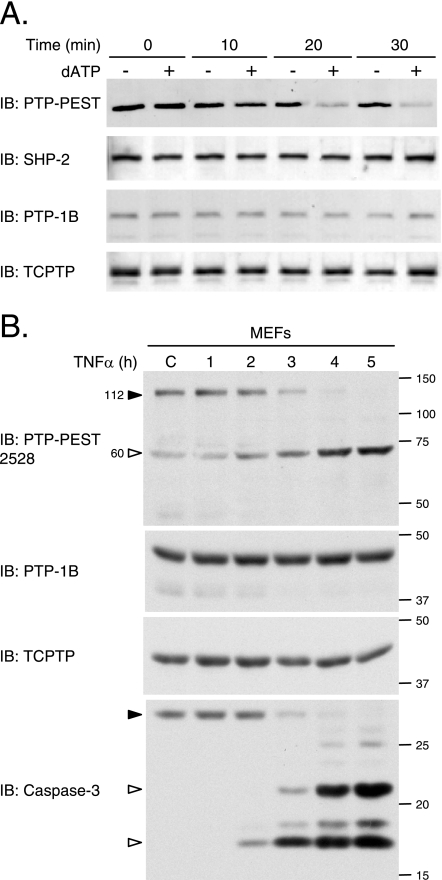
We then investigated the integrity of particular members of the PTP family during apoptosis. Following in vitro treatment with dATP, immunoblot analysis showed that only the PTP-PEST signal, but not that of SHP-2, PTP-1B, or TC-PTP, was reduced (Fig. 5A). The stability of endogenous phosphatases was then confirmed in cell culture, using spontaneously immortalized mouse embryonic fibroblasts. In cells exposed to TNF-α, PTP-1B and TC-PTP remained intact, whereas PTP-PEST was degraded as activation of caspase-3 occurred (Fig. 5B). Together, these results demonstrate that the cleavage of PTPs during apoptosis is not a general phenomenon among the family but is likely limited to specific members, including PTP-PEST.
FIG. 5.
PTP-PEST, but not SHP-2, PTP-1B, and TC-PTP, is cleaved during apoptosis. (A) Caspases were activated in vitro in 293T cell lysates, and the stability of different phosphatases was investigated by immunoblotting, using antibodies specific for SHP-2, PTP-1B (FG6), TC-PTP (CF4-1D), and PTP-PEST (AG25). (B) Spontaneously immortalized MEFs were incubated with 10 μg/ml cycloheximide in the absence (control [C]) or presence of 10 ng/ml TNF-α for the indicated times. Samples were analyzed by immunoblotting for PTP-PEST (2528 rabbit polyclonal antibody), TC-PTP (3E2), PTP-1B (rabbit polyclonal antibody), and caspase-3. IB, immunoblot. Filled arrowheads point to intact proteins, and empty arrowheads identify cleavage products. The values on the right correspond to molecular sizes in kDa.
PTP-PEST is a substrate of caspase-3.
Degradation of PTPs caused by UV irradiation was recently shown to be dependent on activation of calpain, a group of proteases activated during apoptosis (37, 73). In addition, in apoptotic cells, several proteins, including molecules involved in cell adhesion and cytoskeletal network organization, are substrates of caspases (31). In order to identify whether calpain or caspase is responsible for PTP-PEST degradation in vivo, apoptosis was induced in cells expressing PTP-PEST (clone B11) in the presence of specific pharmacological inhibitors. Degradation of PTP-PEST induced by staurosporine was prevented by the caspase inhibitor Z-VAD, but not by the calpain inhibitor PD150606 (Fig. 6A). In Rat-1 cells, inhibition of caspase by Z-VAD also blocked degradation of endogenous PTP-PEST (data not shown). The proteasome inhibitor lactacystin did not impair efficient PTP-PEST proteolysis during apoptosis (data not shown). The cleavage of PTP-PEST occurred in parallel with that of p130Cas and with the activation of caspase-3 (Fig. 6A). This experiment reveals that caspases are the proteases responsible for PTP-PEST cleavage during apoptosis.
FIG. 6.
Caspase-3 cleaves PTP-PEST. (A) To inhibit caspases or calpain proteases, PTP-PEST−/−-expressing WT cells (clone B11WT) were preincubated with either 100 μM Z-VAD-FMK or 100 μM PD150606 for 2 h and subsequently treated with 1 μM staurosporine in the presence of the indicated inhibitor for an additional 6 h. Protein lysates were then subjected to immunoblot analysis for PTP-PEST (2528 and 2530 rabbit polyclonal antibodies), p130Cas, and caspase-3. (B) 293T cell extracts were immunodepleted (ID) for either caspase-3, caspase-9, or caspase-7, treated with dATP for the indicated time, and subjected to in-gel PTP assays. The presence of caspase-9, caspase-3, and caspase-7 in the immunodepleted samples was verified by immunoblotting (bottom). (C) Protein lysates from PTP-PEST−/− EV (B14) and WT rescued (B11WT) cells were incubated with active recombinant caspase-3 or under control conditions (C) for the indicated times. Samples were further analyzed by immunoblotting for PTP-PEST (2528 rabbit polyclonal antibody) and p130Cas. (D) GST or GST-PTP-PEST was isolated from transfected PTP-PEST−/− cells, incubated with active recombinant caspase-3 or under control conditions (C), and analyzed by immunoblotting, using anti-GST antibody. Filled arrowheads point to intact proteins, and empty arrowheads identify cleavage products. The values on the right correspond to molecular sizes in kDa. IB, immunoblot.
To determine which caspase cleaves PTP-PEST, caspase-3, -7, and -9 were individually immunodepleted from 293T lysates prior to addition of dATP. Samples were then subjected to in-gel PTP assays. Immunodepletion of caspase-7 did not interfere with the cleavage of PTP-PEST (Fig. 6B). Conversely, immunodepletion of either caspase-3 or caspase-9 could block the cleavage of PTP-PEST. Under these conditions, caspase-3 requires active caspase-9 for its induction (30). Since caspase-9 was still present following caspase-3 immunodepletion, these results suggest that only caspase-3 was responsible for this cleavage. Immunoblot analysis performed on the supernatants demonstrated that specific immunodepletion of caspase-3, -7, and -9 was obtained (Fig. 6B). Caspase-8 was excluded from the immunodepletion analysis, since its activation occurred following the rapid cleavage of PTP-PEST (Fig. 4A and B). In contrast, the activation of caspase-3 in vitro and in vivo (Fig. 4, 5, and 6) was concomitant with the proteolysis of PTP-PEST. In order to verify that active caspase-3 could directly cleave PTP-PEST, we incubated cell extracts (Fig. 6C) and purified GST-PTP-PEST (Fig. 6D) with the recombinant active caspase-3. As depicted in Fig. 6C, addition of active purified caspase-3 to lysates from PTP-PEST-expressing cells led to a degradation pattern similar to that previously observed in vivo (Fig. 3 and 6C). p130Cas was also cleaved under these conditions, and the cleavage products typically generated during apoptosis were obtained (Fig. 6C) (50). Importantly, caspase-3 could cleave purified GST-PTP-PEST, suggesting that other proteins were not required for the proteolysis (Fig. 6D). Together, these observations indicate that caspase-3 cleaves PTP-PEST during apoptosis. To our knowledge, this is the first example of a classical PTP directly cleaved by a caspase.
Caspase-3 cleaves PTP-PEST on its 549DSPD site.
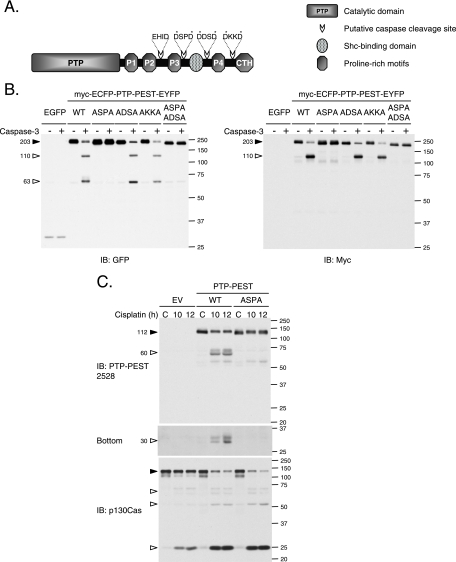
Analysis of the murine PTP-PEST sequence revealed the presence of several potential caspase cleavage sites (Fig. 7A). We mutated each DXXD motif individually in PTP-PEST cDNA (549DSPD to 549ASPA, 604DDSD to 604ADSA, 740DKKD to 740AKKA) and inserted it between sequences corresponding to myc-ECFP and EYFP tags.
FIG. 7.
Caspase-3 cleaves PTP-PEST on its 549DSPD site. (A) Schematic views of murine PTP-PEST showing the catalytic domain (PTP), the five regions rich in proline (Pro1, Pro2, Pro3, Pro4, and CTH) and the Shc binding domain (SBD). Four potential caspase cleavage sites (423EHID, 549DSPD, 604DDSD, and 740DKKD) are indicated. Asterisks ( ) identify the amino acids targeted by mutagenesis. (B) Lysates from HeLa299 cells expressing EGFP or different versions of myc-ECFP-PTP-PEST-EYFP were prepared, incubated with or without active recombinant caspase-3, and analyzed by immunoblot, using antibodies directed against GFP and myc (9E10). (C) HeLa299 cells transfected with a nontagged version of PTP-PEST in pcDNA3 (EV, WT, or 549ASPA) were incubated with 60 μM cisplatin for the indicated times and lysed as described in Materials and Methods. Protein lysates were resolved by SDS-PAGE and analyzed by immunoblotting for PTP-PEST (2528 rabbit polyclonal antibody) and p130Cas. (C, bottom) A longer exposure of the anti-PTP-PEST immunoblot is shown. Filled arrowheads point to intact proteins, and empty arrowheads identify cleavage products. IB, immunoblot. The values on the right correspond to molecular sizes in kDa.
) identify the amino acids targeted by mutagenesis. (B) Lysates from HeLa299 cells expressing EGFP or different versions of myc-ECFP-PTP-PEST-EYFP were prepared, incubated with or without active recombinant caspase-3, and analyzed by immunoblot, using antibodies directed against GFP and myc (9E10). (C) HeLa299 cells transfected with a nontagged version of PTP-PEST in pcDNA3 (EV, WT, or 549ASPA) were incubated with 60 μM cisplatin for the indicated times and lysed as described in Materials and Methods. Protein lysates were resolved by SDS-PAGE and analyzed by immunoblotting for PTP-PEST (2528 rabbit polyclonal antibody) and p130Cas. (C, bottom) A longer exposure of the anti-PTP-PEST immunoblot is shown. Filled arrowheads point to intact proteins, and empty arrowheads identify cleavage products. IB, immunoblot. The values on the right correspond to molecular sizes in kDa.
Protein extracts from HeLa299 cells expressing the different mutants of PTP-PEST were incubated with or without caspase-3 and subjected to immunoblot analysis. An anti-GFP antibody, which recognizes both ECFP and EYFP, revealed that following incubation with caspase-3, two fragments of approximately 110 kDa and 63 kDa appeared when WT, 604ADSA, and 740AKKA forms of PTP-PEST were expressed (Fig. 7B). The 549ASPA mutation was sufficient to completely prevent cleavage of PTP-PEST by caspase-3. These observations establish that caspase-3 cleaves PTP-PEST into two main fragments by targeting its 549DSPD motif. Detection of the myc-epitope tag identified the 110-kDa cleavage product as the N-terminal portion of PTP-PEST that contains its catalytic domain, whereas the 63-kDa fragment revealed by anti-GFP immunoblotting corresponded to the C-terminal segment. With regard to the in vivo situation, PTP-PEST was cleaved during cisplatin-induced apoptosis of HeLa cells. Efficient proteolysis of PTP-PEST depended on the 549DSPD site, since its mutation to 549ASPA significantly reduced its degradation in vivo (Fig. 7C). Importantly, this mutation abolished the generation of the ∼60-kDa product and of the smaller fragments near 30 kDa. The degradation of p130Cas occurred in a similar fashion in both WT- and 549ASPA PTP-PEST-expressing cells (Fig. 7C). Moreover, as previously observed in Fig. 3, the rate of p130Cas degradation was more apparent in PTP-PEST-expressing cells than in the empty vector controls. This demonstrates that caspase-3 cleaves PTP-PEST at 549DSPD while apoptotic cell death progresses.
Caspase-3 regulates PTP-PEST activity, interactions, and adaptor function.
Cleavage of proteins involved in organizing the actin cytoskeleton affects their properties, redistributes them to different protein complexes, and modifies their functions (31, 49). We observed that the catalytic activity of PTP-PEST was important for its ability to increase sensitivity to receptor-mediated apoptosis. To evaluate the effect of caspase-3 on PTP-PEST catalytic activity, purified GST-hPTP-PEST from 293T-transfected cells was incubated in the presence or absence of caspase-3, followed by an in-solution phosphatase assay. Figure 8A shows that after caspase-3 proteolysis, PTP-PEST exhibited an approximately twofold increase in catalytic activity. Immunoblotting with anti-GST showed the efficiency of the treatment with caspase-3. These observations indicate that caspase-3 regulates PTP-PEST catalytic activity in a cleavage-dependent manner.
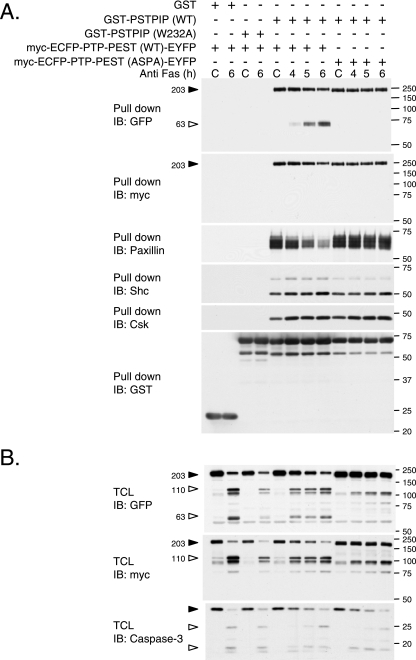
To understand how caspase-3 could affect PTP-PEST function, we investigated whether the cleavage of PTP-PEST could regulate its ability to interact with signaling molecules. Located at the amino terminus, the catalytic domain of PTP-PEST is followed by a carboxy-terminal (C-terminal) tail. This segment contains a Shc binding domain (SBD) and five proline-rich segments, among which the second (P2) interacts with paxillin family members (paxillin, Hic-5, and leupaxin), the fourth (P4) recruits Csk, and the fifth, termed CTH, associates with PSTPIP (12, 23, 24, 28, 38, 62). We used an in vitro approach to characterize the effect of caspase-3 on PTP-PEST interactions, as described in Fig. 8B. GST-PTP-PEST was purified from PTP-PEST−/−-transfected cells and incubated with P-MEF cell lysates in order to reconstitute PTP-PEST protein complexes. As expected, intact PTP-PEST can interact with Shc (p52 and p66), paxillin, and leupaxin (Fig. 8C, lanes 1 and 4). When PTP-PEST was cleaved before being incubated with the lysates, the N-terminal fragment failed to bind Shc, whereas paxillin and leupaxin interactions were maintained (Fig. 8C, lane 2). We also investigated the effect of caspase-3 cleavage on PTP-PEST protein complexes following in vitro reconstitution (Fig. 8C, lanes 3 and 5). Under these conditions, caspase-3 drastically decreased the association of PTP-PEST with Shc and leupaxin but only slightly affected recruitment of paxillin (Fig. 8C, compare lane 4 with lane 5). Paxillin and leupaxin have been previously identified as caspase-3 substrates, which explains their reduced affinity for PTP-PEST following treatment with active caspase-3 in vitro (15). The interactions observed in that experiment were specific to PTP-PEST, as they were not detected with the GST tag alone (Fig. 8C). The impact of active caspase-3 on PTP-PEST interactions was then measured using the 549ASPA mutant. Samples were processed as condition 1 and 2 depicted in Fig. 8B. Prior incubation with caspase-3 was sufficient to abolish the interaction between the PTP-PEST (WT) N-terminal fragment and Shc without affecting the interaction with paxillin and leupaxin (Fig. 8D). The 549ASPA mutations of PTP-PEST efficiently reverted the caspase-3-mediated dissociation of Shc. Finally, we examined the effect of caspase-3 on the C-terminal region of PTP-PEST (Fig. 8E), using lysates of cells coexpressing myc-ECFP-PTP-PEST-EYFP (WT or 549ASPA) with GST-PSTPIP. Whole-cell extracts were incubated with or without active caspase-3, and PSTPIP was pulled down using glutathione Sepharose beads. We noticed that following incubation with caspase-3, PSTPIP could interact with a shorter form of PTP-PEST-WT (Fig. 8E, top panel). This fragment was identified as the PTP-PEST C-terminal cleavage product because it could not be detected by anti-myc antibody (Fig. 8E). Caspase-3 cleavage of PTP-PEST did not inhibit the interaction with PSTPIP, as binding was seen to both the intact protein and the C-terminal fragment (Fig. 8E). In addition, no detectable caspase-3 cleavage of PSTPIP was observed under these conditions. These results support the model that the action of caspase-3 on PTP-PEST prevents its catalytic domain from associating with leupaxin, Shc (p52 and p66), PSTPIP, and, to a lesser extent, paxillin. This is due to the action of caspase-3 on both PTP-PEST, at the 549DSPD site, and leupaxin/paxillin. In addition, caspase-3 cleavage of PTP-PEST liberated a C-terminal fragment that was still able to interact with PSTPIP. Together, these results implicate active caspase-3 in the modulation of both the catalytic activity and the interactions of PTP-PEST.
The numerous partners reported to interact with the carboxy-terminal end of PTP-PEST are indicative of an adaptor function. To support this premise, we investigated the capability of PTP-PEST to bridge PSTPIP to new interacting partners. As observed in Fig. 9, PSTPIP associated with PTP-PEST was also recruited to paxillin, Shc, and Csk. In contrast, a mutant version of PSTPIP (W232A), which is unable to bind PTP-PEST (23), failed to associate with paxillin, Shc, and Csk (Fig. 9A). These complexes were also specifically observed when the WT but not the W232A form of PSTPIP was expressed in the PTP-PEST−/−-reexpressing WT cells (clone B11) (data not shown). These results suggest that PTP-PEST has the ability to perform adaptor functions that link PSTPIP toward paxillin, Shc, or Csk. To define the consequences of caspase activation on PTP-PEST scaffolding functions, we examined the proteins associated with PSTPIP in cells coexpressing GST-PSTPIP with either WT or the 549ASPA mutant of myc-ECFP-PTP-PEST-EYFP during treatment with anti-Fas. As apoptosis progressed, the association of PSTPIP with the intact form of PTP-PEST WT was diminished, while the interaction with the PTP-PEST C-terminal fragment increased (Fig. 9A). This paralleled a decreased association of PSTPIP with paxillin. In contrast, the associations of PSTPIP with Shc and Csk were maintained during apoptosis. This modulation did not occur when PTP-PEST 549ASPA was expressed, since paxillin, Shc, Csk, and intact PTP-PEST coprecipitated with PSTPIP during apoptotic progression. The isolation of each GST fusion protein was confirmed by immunoblotting. Figure 9B shows the expression of PTP-PEST in the different conditions and the activation of caspase-3 following treatment with anti-Fas. The reduction in paxillin interacting with PSTPIP indicates that the scaffolding function of PTP-PEST is modulated by caspase cleavage on its 549DSPD site during apoptosis. In addition, the preserved association of PSTPIP with Shc and Csk suggests that the PTP-PEST C-terminal cleavage product is competent to perform these associations. Together, these data support that PTP-PEST adaptor functions are modulated in cells undergoing apoptosis.
FIG. 9.
The scaffolding function of PTP-PEST is modulated during apoptosis. (A) HeLa299 cells cotransfected with cDNA encoding either GST or GST-PSTPIP (WT or W232A) with myc-ECFP-PTP-PEST (WT or 549ASPA)-EYFP were treated with 10 μg/ml cycloheximide in the absence (control [C]; 6 h) or presence of 50 ng/ml anti-Fas for the indicated times. Lysates extracted from the cells were incubated with glutathione Sepharose beads, and the ability of the expressed GST fusion protein to interact with other partners was determined by immunoblotting. The coprecipitated paxillin, Shc, Csk, and PTP-PEST were analyzed by immunoblotting for the indicated proteins and epitopes. Filled arrowheads point to intact PTP-PEST, and empty arrowheads indicate cleavage products. (B) Total cell extracts were subjected to immunoblotting with anti-GFP, 9E10 (anti-myc), and anti-caspase-3. Filled arrowheads point to intact PTP-PEST or caspase-3, and empty arrowheads indicate cleavage products. IB, immunoblot, TCL, total cell lysate. The values on the right correspond to molecular sizes in kDa.
Caspase cleavage of PTP-PEST facilitates cellular detachment during apoptosis.
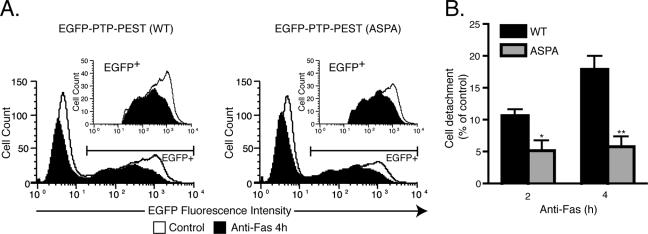
Caspase-mediated proteolysis of cytoskeletal protein causes membrane blebbing, cell rounding, and adhesion lost as apoptosis progress (19, 31). Localization of PTP-PEST to the retracting membrane ruffles and its caspase-3-dependent modulation implies that regulation of this enzyme could participate in cellular detachment during apoptosis. To determine whether the cleavage of PTP-PEST during apoptosis plays a role in loss of adhesion, we used flow cytometry to count the number of cells remaining attached to the culture dish following treatment with anti-Fas. Incubation of HeLa299 cells expressing EGFP-PTP-PEST (WT or 549ASPA) with anti-Fas decreased the number of cells attached to the culture dish (Fig. 10A).Interestingly, the proportion of cells that detached during the progression of apoptosis was significantly diminished in the 549ASPA mutant compared to that in WT PTP-PEST-expressing cells (Fig. 10B). Two hours following anti-Fas treatment, the 549ASPA PTP-PEST mutant-expressing cells exhibited a twofold decrease in the loss of adhesion versus those expressing WT PTP-PEST. Moreover, at 4 h, detachment of cells harboring the 549ASPA mutant was delayed compared to that for PTP-PEST WT-expressing cells, as revealed by a threefold decrease in adhesion lost. These results indicate that cleavage of PTP-PEST by caspase-3 contributes to cellular detachment during apoptosis.
FIG. 10.
Caspase cleavage of PTP-PEST facilitates cellular detachment during apoptosis. (A) HeLa299 cells expressing either EGFP-PTP-PEST WT or EGFP-PTP-PEST 549ASPA were incubated with 10 μg/ml cycloheximide in the absence (control) or presence of 50 ng/ml anti-Fas. Following treatment, adherent cells were harvested and counted, using flow cytometry as described in Materials and Methods. Cells expressing EGFP-PTP-PEST (WT or 549ASPA) were selected based on fluorescence intensity (inset). The area between the control curve and the anti-Fas-stimulated curve corresponds to the number of cells that detached during the treatment. (B) The values given are the percentages of cellular detachment in EGFP-PTP-PEST WT- or EGFP-PTP-PEST 549ASPA-expressing cells compared to that in the controls and represent means ± SE of two independent experiments in which each condition was performed in triplicate and each sample was analyzed three times by flow cytometry. Asterisks denote values significantly different from those of EGFP-PTP-PEST WT-expressing cells ( , P < 0.05;
, P < 0.05; 
 , P < 0.01).
, P < 0.01).
DISCUSSION
Ubiquitous expression of PTP-PEST maintained throughout development and in adult animals points toward a fundamental biologic role for this enzyme (14). Indeed, mice lacking the PTP-PEST gene exhibit severe developmental defects leading to embryonic death (22, 74). PTP-PEST accomplishes several of its functions through the regulation of the actin cytoskeleton (4, 52). Nevertheless, the contribution and regulation of PTP-PEST have not been investigated in apoptosis, a physiological phenomenon essential for proper development involving major cytoskeletal rearrangement. Here, we provide the first evidence that PTP-PEST plays a role in the progression of apoptotic cell death. Importantly, we have identified PTP-PEST as a new caspase substrate and linked its caspase-mediated cleavage and modulation to the process of cell detachment during apoptosis.
PTP-PEST was previously reported to regulate membrane ruffling, cell adhesion, and spreading; proper balance of these physiological processes is crucial for cell migration (3, 34, 69). Functions involving p130Cas, Rac, WASP, paxillin, paxillin kinase linker (PKL), and VAV2 are suggested to couple PTP-PEST to the modulation of the cytoskeleton organization in migrating cells (3, 5, 23, 24, 34, 46, 69, 70). Interestingly, several proteins that govern cell migration are modulated during apoptosis in order to remodel cell shape (19). The effects of PTP-PEST on signaling cascades that orchestrate actin dynamics led us to explore its possible modulation in apoptotic cell death. In growing cells, PTP-PEST is reported to be primarily localized in the cytosol (3, 23). However, in cells undergoing apoptosis, we observed concurrent localization of PTP-PEST to retracting lamellipodia as well as to the cytosol. The exact mechanism by which PTP-PEST is specifically targeted to the edges of retracting lamellipodia is still unknown, but it is possible that some of the mechanisms involved in detachment of apoptotic cells are conserved with those controlling tail retraction in cells undergoing polarized migration. Indeed, in cells stimulated with fibronectin, PTP-PEST is recruited to the plasma membrane and has been detected in adhesion structures (3, 69). Moreover, PTP-PEST promotes tail retraction by regulating p190RhoGAP (70). Numerous regulators of actin cytoskeleton organization are also associated with the morphological conversion occurring during apoptosis (19, 20, 26, 71). The presence of PTP-PEST in retracting lamellipodia during apoptosis indicates a novel function for this enzyme in the progression of cell death and potentially in cellular detachment.
Since PTP-PEST sensitizes cells to both anti-Fas- and TNF-α-induced cell death, our results identify PTP-PEST as a modulator of receptor-mediated apoptosis. Observations showing that orthovanadate, a general PTP inhibitor, could protect cells against the cytotoxic effect of TNF-α provided the first evidence for a function of these enzymes in receptor-mediated programmed cell death (81). In hematopoietic cells, SHP-1 is required for efficient Fas-induced apoptosis (76, 84). On the other hand, FAP-1 associates with and attenuates Fas (Apo-1) trafficking to the cell surface, thereby inhibiting Fas-induced apoptosis (44). Recently, PTP-PEST was shown to be involved in the trafficking of the Fas ligand (7). Inhibition by reactive oxygen species of MAP kinase phosphatase induced by TNF-α increases Jun N-terminal protein kinase-mediated cell death (48), whereas PTP1B-null mice exhibited an increased resistance to Fas-dependent hepatic apoptosis (68). Together, these reports underline the importance of PTP regulation in receptor-mediated apoptosis. Interestingly, the effect of PTP-PEST is dependent on its catalytic activity, which suggests that an unidentified substrate is targeted in this phenomenon. In addition, PTP-PEST-expressing cells display accelerated cleavage of p130Cas, suggesting that PTP-PEST can act as a regulator of the apoptotic response. Interestingly, the proteolysis of p130Cas, a substrate of PTP-PEST, has been proposed to be highly dependent on its phosphorylation status and to contribute to the morphological characteristics of apoptotic cells (32, 42, 50). FAK, a reported substrate of PTP-PEST (3, 29, 72), was recently proposed to bind RIP and suppress its proapoptotic signal in death receptor-induced apoptosis (51).
Posttranslational modifications of PTPs, such as nitrosylation (6), ubiquitination (47, 64), phosphorylation (33), calpain degradation (37), and oxidation (79), are important for their regulation. Here, we report the caspase-mediated cleavage of a classical PTP, PTP-PEST, during apoptosis. In vitro and in vivo experiments identified caspase-3 as the protease executing PTP-PEST cleavage during apoptosis. Despite the presence of four putative caspase cleavage sites, mutation of the 549DSPD site was sufficient to block caspase-3 cleavage of PTP-PEST in vitro and precluded its degradation in vivo. The DSPD sequence is processed by caspases in other proteins, including p130Cas and RAD21 (17, 50), and its corresponding site (580DLVD) found in human PTP-PEST has been described as a caspase cleavage site in Hef1 (63). In addition, our observations indicate that the cleavage of PTP-PEST during apoptosis is conserved between different cell types in several mammalian species and is a common characteristic of different apoptotic pathways. This degradation of PTPs appears to be specific to PTP-PEST, since TC-PTP, PTP-1B, SHP-2, and most candidates highlighted by in-gel PTP assays were resistant to caspase activity.
The noncleavable mutant of PTP-PEST contributed to caspase activation to the same extent as the WT form (data not shown). Consequently, we also observed equivalent, accelerated p130Cas cleavage in both WT- and 549ASPA PTP-PEST- expressing cells (Fig. 7C). This implies that the cleavage of PTP-PEST is not involved in amplifying the caspase cascade. As caspase activation precedes PTP-PEST cleavage, the cleaved products may have an impact on the apoptotic process downstream of caspase induction. Interestingly, in vitro analysis indicates that caspase cleavage of PTP-PEST liberates fragments that display increased catalytic activity compared to that of the intact form. Furthermore, proteolysis of PTP-PEST would prevent its catalytic domain from being associated with its targets, Shc and PSTPIP (23, 29), and, therefore, would change its substrate specificity. Intramolecular interaction has been characterized as an autoinhibition mechanism for other PTPs (61). Thus, in addition to its adaptor function, the C terminus of PTP-PEST could inhibit its catalytic domain or may bind a protein that modulates PTP-PEST enzymatic activity. As the process of apoptotic cell death involves tightly regulated molecular mechanisms, the caspase-dependent modulation of the interactions and catalytic activity of PTP-PEST suggests that it participates in the progression of apoptosis.
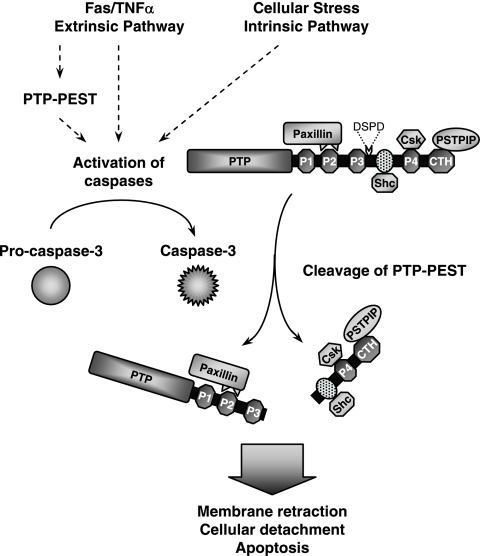
Cellular detachment is a major feature of the execution phase of apoptosis. The detailed mechanisms that coordinate this complex event remain unclear. In this study, we show that the cleavage of PTP-PEST during apoptotic cell death promotes cellular detachment. Our experiments indicate that cleavage of PTP-PEST produces an N-terminal fragment that maintains the ability to recruit the multidomain focal adhesion adaptor protein paxillin. Paxillin plays a central role in regulating cell attachment and migration (10). The association of PTP-PEST with paxillin was reported to target the enzyme to its substrate PKL and is essential for inhibition of cell adhesion and membrane protrusion (46). Therefore, the recognition of an altered subset of substrates, such as paxillin-linked focal adhesion proteins, by the active PTP-PEST fragment might stimulate cellular detachment during apoptosis. Another binding partner of PTP-PEST is PSTPIP, through which PTP-PEST regulates the phosphorylation status and actin polymerization activity of WASP (5, 23). Mutations in the gene encoding WASP were reported to accelerate lymphocyte apoptotic cell death (65). Importantly, we observed that active caspase-3 uncouples PSTPIP from the PTP-PEST catalytic domain. Because modifications to the structure of the actin network cause the morphological conversions associated with apoptosis (19, 56), deregulation of WASP activity during programmed cell death may also contribute to this phenomenon. We demonstrated that PTP-PEST is additionally able to act as a scaffolding molecule bridging PSTPIP to paxillin, providing a link between adhesion signaling and actin polymerization. During apoptosis, caspase cleavage of PTP-PEST uncouples PSTPIP from the PTP-PEST catalytic domain and from paxillin, consequently interrupting their potential communication. In addition, we observed that proteolysis of PTP-PEST in cells undergoing apoptosis generates a fragment still competent to couple PSTPIP to Shc or Csk. Notably, Shc and Csk have both been implicated in cell adhesion (57, 58). Through these multiple protein interactions, PTP-PEST may also function as a platform, bringing together different signaling modules in close proximity to control cellular adhesion and migration. Caspase-dependent cleavage of PTP-PEST rearranges these complexes, which results in unstable cytoskeletal architecture and reduced cell adhesion. In summary, we propose that the cleavage of PTP-PEST during apoptosis enhances its catalytic activity, alters its substrate specificity, and disrupts its scaffolding properties, thereby facilitating cellular detachment (Fig. 11).
FIG. 11.
Proposed functions of PTP-PEST in apoptosis. By bridging PSTPIP to an additional partner (paxillin, Shc, or Csk), PTP-PEST acts as a scaffold involved in regulating cell adhesion and migration. During initiation of apoptosis, PTP-PEST expression enhances caspase activation in cells exposed to TNF-α and anti-Fas. The extrinsic and intrinsic apoptotic pathways both lead to activation of the executioner caspase-3, which cleaves PTP-PEST on its 549DSPD motif. This produces an N-terminal fragment that displays increased catalytic activity and retains the ability to bind paxillin. The C-terminal fragment remains competent to link PSTPIP exclusively to Shc or Csk. The targeting of active PTP-PEST fragments to paxillin-associated focal adhesion proteins and the remodeling of PTP-PEST protein complexes following caspase activation may destabilize the actin network and stimulate cellular loss of adhesion. Therefore, cleavage of PTP-PEST facilitates cellular detachment during the execution phase of apoptosis.
PTPs are major modulators of signal transduction and also function as integral regulators of the apoptotic response. Cellular progression through apoptosis correlates with the redistribution of PTP-PEST toward the site of cellular detachment, suggesting an active participation of PTP-PEST in the apoptotic process. PTP-PEST expression results in enhanced caspase activity and an accelerated cleavage of p130Cas subsequent to induction of apoptosis. This indicates that PTP-PEST operates upstream of the initiation of the caspase cascade. Interestingly, caspase-3 cleavage of PTP-PEST, which modulates its catalytic activity and scaffolding properties, facilitates cellular detachment, a hallmark of programmed cell death. In this manner, proteolysis would permit a functional transition of PTP-PEST during apoptosis. Apoptosis plays an important role in embryonic development and in lymphocyte selection during their maturation. Since PTP-PEST is essential for proper embryonic development and is highly expressed in T and B lymphocytes (22, 29, 74), our current characterization of its function during caspase-mediated cell death could contribute to a better understanding of these phenomena. Finally, our data reveal the importance of caspases in the regulation of PTPs during apoptosis.
Supplementary Material
Acknowledgments
We are grateful for support from the National Science Council of Taiwan (grants NSC 94-2311-B-011-032 and NSC 94-2311-B-001-020 to T.-C.M.). M.H. is a recipient of a Rolande and Marcel Gosselin Graduate Studentship; S.H. was supported by a postdoctoral fellowship from the Canadian Institute of Health Research (CIHR); C.B. is a recipient of a Human Frontier Science Program postdoctoral fellowship; and A.B. is a recipient of a fellowship from the Lymphoma Research Foundation. M.L.T. is the recipient of the Jeanne et Jean-Louis Lévesque Chair in Cancer Research, a chercheur-Nationaux of the Fonds de la Rercherche en Santé du Québec, and recipient of a James McGill Professor fellowship. This work was supported by a CIHR operating grant (MT-12466) to M.L.T.
We thank Ryan Petrie and Jacynthe Laliberté for their advice on live imaging and John W. Hanrahan for access to the confocal microscope. PTP-PEST polyclonal antibodies were provided by Jean-François Côté. We are grateful to Serge Morin and Martin Calille for their assistance with computer software and to Josée N. Lavoie for helpful discussions. We thank Yves Boiclair and Matthew Stuible for critical reviews of the manuscript.
Footnotes
Published ahead of print on 27 November 2006.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Adams, J. M. 2003. Ways of dying: multiple pathways to apoptosis. Genes Dev. 17:2481-2495. [DOI] [PubMed] [Google Scholar]
- 2.Alonso, A., J. Sasin, N. Bottini, I. Friedberg, A. Osterman, A. Godzik, T. Hunter, J. Dixon, and T. Mustelin. 2004. Protein tyrosine phosphatases in the human genome. Cell 117:699-711. [DOI] [PubMed] [Google Scholar]
- 3.Angers-Loustau, A., J. F. Côté, A. Charest, D. Dowbenko, S. Spencer, L. A. Lasky, and M. L. Tremblay. 1999. Protein tyrosine phosphatase-PEST regulates focal adhesion disassembly, migration, and cytokinesis in fibroblasts. J. Cell Biol. 144:1019-1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Angers-Loustau, A., J. F. Côté, and M. L. Tremblay. 1999. Roles of protein tyrosine phosphatases in cell migration and adhesion. Biochem. Cell Biol. 77:493-505. [PubMed] [Google Scholar]
- 5.Badour, K., J. Zhang, F. Shi, Y. Leng, M. Collins, and K. A. Siminovitch. 2004. Fyn and PTP-PEST-mediated regulation of Wiskott-Aldrich syndrome protein (WASp) tyrosine phosphorylation is required for coupling T cell antigen receptor engagement to WASp effector function and T cell activation. J. Exp. Med. 199:99-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barrett, D. M., S. M. Black, H. Todor, R. K. Schmidt-Ullrich, K. S. Dawson, and R. B. Mikkelsen. 2005. Inhibition of protein-tyrosine phosphatases by mild oxidative stresses is dependent on S-nitrosylation. J. Biol. Chem. 280:14453-14461. [DOI] [PubMed] [Google Scholar]
- 7.Baum, W., V. Kirkin, S. B. Fernandez, R. Pick, M. Lettau, O. Janssen, and M. Zornig. 2005. Binding of the intracellular Fas ligand (FasL) domain to the adaptor protein PSTPIP results in a cytoplasmic localization of FasL. J. Biol. Chem. 280:40012-40024. [DOI] [PubMed] [Google Scholar]
- 8.Blanchetot, C., L. G. Tertoolen, and J. den Hertog. 2002. Regulation of receptor protein-tyrosine phosphatase alpha by oxidative stress. EMBO J. 21:493-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boatright, K. M., and G. S. Salvesen. 2003. Mechanisms of caspase activation. Curr. Opin. Cell Biol. 15:725-731. [DOI] [PubMed] [Google Scholar]
- 10.Brown, M. C., and C. E. Turner. 2004. Paxillin: adapting to change. Physiol. Rev. 84:1315-1339. [DOI] [PubMed] [Google Scholar]
- 11.Bruckner, S., S. Rhamouni, L. Tautz, J. B. Denault, A. Alonso, B. Becattini, G. S. Salvesen, and T. Mustelin. 2005. Yersinia phosphatase induces mitochondrially dependent apoptosis of T cells. J. Biol. Chem. 280:10388-10394. [DOI] [PubMed] [Google Scholar]
- 12.Charest, A., J. Wagner, S. Jacob, C. J. McGlade, and M. L. Tremblay. 1996. Phosphotyrosine-independent binding of SHC to the NPLH sequence of murine protein-tyrosine phosphatase-PEST. Evidence for extended phosphotyrosine binding/phosphotyrosine interaction domain recognition specificity. J. Biol. Chem. 271:8424-8429. [DOI] [PubMed] [Google Scholar]
- 13.Charest, A., J. Wagner, M. Kwan, and M. L. Tremblay. 1997. Coupling of the murine protein tyrosine phosphatase PEST to the epidermal growth factor (EGF) receptor through a Src homology 3 (SH3) domain-mediated association with Grb2. Oncogene 14:1643-1651. [DOI] [PubMed] [Google Scholar]
- 14.Charest, A., J. Wagner, S. H. Shen, and M. L. Tremblay. 1995. Murine protein tyrosine phosphatase-PEST, a stable cytosolic protein tyrosine phosphatase. Biochem. J. 308:425-432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chay, K. O., S. S. Park, and J. F. Mushinski. 2002. Linkage of caspase-mediated degradation of paxillin to apoptosis in Ba/F3 murine pro-B lymphocytes. J. Biol. Chem. 277:14521-14529. [DOI] [PubMed] [Google Scholar]
- 16.Chellaiah, M. A., R. S. Biswas, D. Yuen, U. M. Alvarez, and K. A. Hruska. 2001. Phosphatidylinositol 3,4,5-trisphosphate directs association of Src homology 2-containing signaling proteins with gelsolin. J. Biol. Chem. 276:47434-47444. [DOI] [PubMed] [Google Scholar]
- 17.Chen, F., M. Kamradt, M. Mulcahy, Y. Byun, H. Xu, M. J. McKay, and V. L. Cryns. 2002. Caspase proteolysis of the cohesin component RAD21 promotes apoptosis. J. Biol. Chem. 277:16775-16781. [DOI] [PubMed] [Google Scholar]
- 18.Cheng, A., G. S. Bal, B. P. Kennedy, and M. L. Tremblay. 2001. Attenuation of adhesion-dependent signaling and cell spreading in transformed fibroblasts lacking protein tyrosine phosphatase-1B. J. Biol. Chem. 276:25848-25855. [DOI] [PubMed] [Google Scholar]
- 19.Coleman, M. L., and M. F. Olson. 2002. Rho GTPase signalling pathways in the morphological changes associated with apoptosis. Cell Death Differ. 9:493-504. [DOI] [PubMed] [Google Scholar]
- 20.Coleman, M. L., E. A. Sahai, M. Yeo, M. Bosch, A. Dewar, and M. F. Olson. 2001. Membrane blebbing during apoptosis results from caspase-mediated activation of ROCK I. Nat. Cell Biol. 3:339-345. [DOI] [PubMed] [Google Scholar]
- 21.Cong, F., S. Spencer, J. F. Côté, Y. Wu, M. L. Tremblay, L. A. Lasky, and S. P. Goff. 2000. Cytoskeletal protein PSTPIP1 directs the PEST-type protein tyrosine phosphatase to the c-Abl kinase to mediate Abl dephosphorylation. Mol. Cell 6:1413-1423. [DOI] [PubMed] [Google Scholar]
- 22.Côté, J. F., A. Charest, J. Wagner, and M. L. Tremblay. 1998. Combination of gene targeting and substrate trapping to identify substrates of protein tyrosine phosphatases using PTP-PEST as a model. Biochemistry 37:13128-13137. [DOI] [PubMed] [Google Scholar]
- 23.Côté, J. F., P. L. Chung, J. F. Théberge, M. Hallé, S. Spencer, L. A. Lasky, and M. L. Tremblay. 2002. PSTPIP is a substrate of PTP-PEST and serves as a scaffold guiding PTP-PEST toward a specific dephosphorylation of WASP. J. Biol. Chem. 277:2973-2986. [DOI] [PubMed] [Google Scholar]
- 24.Côté, J. F., C. E. Turner, and M. L. Tremblay. 1999. Intact LIM 3 and LIM 4 domains of paxillin are required for the association to a novel polyproline region (Pro 2) of protein-tyrosine phosphatase-PEST. J. Biol. Chem. 274:20550-20560. [DOI] [PubMed] [Google Scholar]
- 25.Cousin, H., and D. Alfandari. 2004. A PTP-PEST-like protein affects alpha5beta1-integrin-dependent matrix assembly, cell adhesion, and migration in Xenopus gastrula. Dev. Biol. 265:416-432. [DOI] [PubMed] [Google Scholar]
- 26.Croft, D. R., M. L. Coleman, S. Li, D. Robertson, T. Sullivan, C. L. Stewart, and M. F. Olson. 2005. Actin-myosin-based contraction is responsible for apoptotic nuclear disintegration. J. Cell Biol. 168:245-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Curtin, J. F., and T. G. Cotter. 2003. Live and let die: regulatory mechanisms in Fas-mediated apoptosis. Cell. Signal. 15:983-992. [DOI] [PubMed] [Google Scholar]
- 28.Davidson, D., J. F. Cloutier, A. Gregorieff, and A. Veillette. 1997. Inhibitory tyrosine protein kinase p50csk is associated with protein-tyrosine phosphatase PTP-PEST in hemopoietic and non-hemopoietic cells. J. Biol. Chem. 272:23455-23462. [DOI] [PubMed] [Google Scholar]
- 29.Davidson, D., and A. Veillette. 2001. PTP-PEST, a scaffold protein tyrosine phosphatase, negatively regulates lymphocyte activation by targeting a unique set of substrates. EMBO J. 20:3414-3426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fearnhead, H. O., J. Rodriguez, E. E. Govek, W. Guo, R. Kobayashi, G. Hannon, and Y. A. Lazebnik. 1998. Oncogene-dependent apoptosis is mediated by caspase-9. Proc. Natl. Acad. Sci. USA 95:13664-13669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fischer, U., R. U. Janicke, and K. Schulze-Osthoff. 2003. Many cuts to ruin: a comprehensive update of caspase substrates. Cell Death Differ. 10:76-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Garton, A. J., A. J. Flint, and N. K. Tonks. 1996. Identification of p130(cas) as a substrate for the cytosolic protein tyrosine phosphatase PTP-PEST. Mol. Cell. Biol. 16:6408-6418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Garton, A. J., and N. K. Tonks. 1994. PTP-PEST: a protein tyrosine phosphatase regulated by serine phosphorylation. EMBO J. 13:3763-3771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Garton, A. J., and N. K. Tonks. 1999. Regulation of fibroblast motility by the protein tyrosine phosphatase PTP-PEST. J. Biol. Chem. 274:3811-3818. [DOI] [PubMed] [Google Scholar]
- 35.Gu, F., N. Dubé, J. W. Kim, A. Cheng, J. Ibarra-Sanchez Mde, M. L. Tremblay, and Y. R. Boisclair. 2003. Protein tyrosine phosphatase 1B attenuates growth hormone-mediated JAK2-STAT signaling. Mol. Cell. Biol. 23:3753-3762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gu, F., D. T. Nguyen, M. Stuible, N. Dubé, M. L. Tremblay, and E. Chevet. 2004. Protein-tyrosine phosphatase 1B potentiates IRE1 signaling during endoplasmic reticulum stress. J. Biol. Chem. 279:49689-49693. [DOI] [PubMed] [Google Scholar]
- 37.Gulati, P., B. Markova, M. Gottlicher, F. D. Bohmer, and P. A. Herrlich. 2004. UVA inactivates protein tyrosine phosphatases by calpain-mediated degradation. EMBO Rep. 5:812-817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gupta, A., B. S. Lee, M. A. Khadeer, Z. Tang, M. Chellaiah, Y. Abu-Amer, J. Goldknopf, and K. A. Hruska. 2003. Leupaxin is a critical adaptor protein in the adhesion zone of the osteoclast. J. Bone Miner. Res. 18:669-685. [DOI] [PubMed] [Google Scholar]
- 39.Gupta, S., V. Radha, C. Sudhakar, and G. Swarup. 2002. A nuclear protein tyrosine phosphatase activates p53 and induces caspase-1-dependent apoptosis. FEBS Lett. 532:61-66. [DOI] [PubMed] [Google Scholar]
- 40.Hardy, S., W. El-Assaad, E. Przybytkowski, E. Joly, M. Prentki, and Y. Langelier. 2003. Saturated fatty acid-induced apoptosis in MDA-MB-231 breast cancer cells. A role for cardiolipin. J. Biol. Chem. 278:31861-31870. [DOI] [PubMed] [Google Scholar]
- 41.Hasegawa, K., H. Yajima, T. Katagiri, M. Ogimoto, Y. Arimura, K. Mitomo, K. Mashima, K. Mizuno, and H. Yakura. 1999. Requirement of PEST domain tyrosine phosphatase PEP in B cell antigen receptor-induced growth arrest and apoptosis. Eur. J. Immunol. 29:887-896. [DOI] [PubMed] [Google Scholar]
- 42.Hoon Kim, D., S. Jeon Choi, S. Kook, W. Kim, and W. Keun Song. 2003. Phosphorylation-dependent cleavage of p130cas in apoptotic rat-1 cells. Biochem. Biophys. Res. Commun. 300:141-148. [DOI] [PubMed] [Google Scholar]
- 43.Ibarra-Sanchez, M. J., J. Wagner, M. T. Ong, C. Lampron, and M. L. Tremblay. 2001. Murine embryonic fibroblasts lacking TC-PTP display delayed G1 phase through defective NF-kappaB activation. Oncogene 20:4728-4739. [DOI] [PubMed] [Google Scholar]
- 44.Ivanov, V. N., P. Lopez Bergami, G. Maulit, T. A. Sato, D. Sassoon, and Z. Ronai. 2003. FAP-1 association with Fas (Apo-1) inhibits Fas expression on the cell surface. Mol. Cell. Biol. 23:3623-3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ivins Zito, C., M. I. Kontaridis, M. Fornaro, G. S. Feng, and A. M. Bennett. 2004. SHP-2 regulates the phosphatidylinositide 3′-kinase/Akt pathway and suppresses caspase 3-mediated apoptosis. J. Cell. Physiol. 199:227-236. [DOI] [PubMed] [Google Scholar]
- 46.Jamieson, J. S., D. A. Tumbarello, M. Hallé, M. C. Brown, M. L. Tremblay, and C. E. Turner. 2005. Paxillin is essential for PTP-PEST-dependent regulation of cell spreading and motility: a role for paxillin kinase linker. J. Cell Sci. 118:5835-5847. [DOI] [PubMed] [Google Scholar]
- 47.Jin, J., T. Shirogane, L. Xu, G. Nalepa, J. Qin, S. J. Elledge, and J. W. Harper. 2003. SCFbeta-TRCP links Chk1 signaling to degradation of the Cdc25A protein phosphatase. Genes Dev. 17:3062-3074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kamata, H., S. Honda, S. Maeda, L. Chang, H. Hirata, and M. Karin. 2005. Reactive oxygen species promote TNFalpha-induced death and sustained JNK activation by inhibiting MAP kinase phosphatases. Cell 120:649-661. [DOI] [PubMed] [Google Scholar]
- 49.Kim, W., S. Kook, D. J. Kim, C. Teodorof, and W. K. Song. 2004. The 31-kDa caspase-generated cleavage product of p130cas functions as a transcriptional repressor of E2A in apoptotic cells. J. Biol. Chem. 279:8333-8342. [DOI] [PubMed] [Google Scholar]
- 50.Kook, S., S. R. Shim, S. J. Choi, J. Ahnn, J. I. Kim, S. H. Eom, Y. K. Jung, S. G. Paik, and W. K. Song. 2000. Caspase-mediated cleavage of p130cas in etoposide-induced apoptotic Rat-1 cells. Mol. Biol. Cell 11:929-939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kurenova, E., L. H. Xu, X. Yang, A. S. Baldwin, Jr., R. J. Craven, S. K. Hanks, Z. G. Liu, and W. G. Cance. 2004. Focal adhesion kinase suppresses apoptosis by binding to the death domain of receptor-interacting protein. Mol. Cell. Biol. 24:4361-4371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Larsen, M., M. L. Tremblay, and K. M. Yamada. 2003. Phosphatases in cell-matrix adhesion and migration. Nat. Rev. Mol. Cell. Biol. 4:700-711. [DOI] [PubMed] [Google Scholar]
- 53.Lin, Y., A. C. Ceacareanu, and A. Hassid. 2003. Nitric oxide-induced inhibition of aortic smooth muscle cell motility: role of PTP-PEST and adaptor proteins p130cas and Crk. Am. J. Physiol. Heart Circ. Physiol. 285:H710-H721. [DOI] [PubMed] [Google Scholar]
- 54.Lyons, P. D., J. M. Dunty, E. M. Schaefer, and M. D. Schaller. 2001. Inhibition of the catalytic activity of cell adhesion kinase beta by protein-tyrosine phosphatase-PEST-mediated dephosphorylation. J. Biol. Chem. 276:24422-24431. [DOI] [PubMed] [Google Scholar]
- 55.MacKeigan, J. P., L. O. Murphy, and J. Blenis. 2005. Sensitized RNAi screen of human kinases and phosphatases identifies new regulators of apoptosis and chemoresistance. Nat. Cell Biol. 7:591-600. [DOI] [PubMed] [Google Scholar]
- 56.Martin, S. S., and K. Vuori. 2004. Regulation of Bcl-2 proteins during anoikis and amorphosis. Biochim. Biophys. Acta 1692:145-157. [DOI] [PubMed] [Google Scholar]
- 57.Mauro, L., D. Sisci, M. Bartucci, M. Salerno, J. Kim, T. Tam, M. A. Guvakova, S. Ando, and E. Surmacz. 1999. SHC-alpha5beta1 integrin interactions regulate breast cancer cell adhesion and motility. Exp. Cell Res. 252:439-448. [DOI] [PubMed] [Google Scholar]
- 58.McGarrigle, D., D. Shan, S. Yang, and X. Y. Huang. 2006. Role of tyrosine kinase Csk in G protein-coupled receptor- and receptor tyrosine kinase-induced fibroblast cell migration. J. Biol. Chem. 281:10583-10588. [DOI] [PubMed] [Google Scholar]
- 59.Meng, T. C., T. Fukada, and N. K. Tonks. 2002. Reversible oxidation and inactivation of protein tyrosine phosphatases in vivo. Mol. Cell 9:387-399. [DOI] [PubMed] [Google Scholar]
- 60.Meng, T. C., S. F. Hsu, and N. K. Tonks. 2005. Development of a modified in-gel assay to identify protein tyrosine phosphatases that are oxidized and inactivated in vivo. Methods 35:28-36. [DOI] [PubMed] [Google Scholar]
- 61.Neel, B. G., H. Gu, and L. Pao. 2003. The ′Shp'ing news: SH2 domain-containing tyrosine phosphatases in cell signaling. Trends Biochem. Sci. 28:284-293. [DOI] [PubMed] [Google Scholar]
- 62.Nishiya, N., Y. Iwabuchi, M. Shibanuma, J. F. Côté, M. L. Tremblay, and K. Nose. 1999. Hic-5, a paxillin homologue, binds to the protein-tyrosine phosphatase PEST (PTP-PEST) through its LIM 3 domain. J. Biol. Chem. 274:9847-9853. [DOI] [PubMed] [Google Scholar]
- 63.O'Neill, G. M., and E. A. Golemis. 2001. Proteolysis of the docking protein HEF1 and implications for focal adhesion dynamics. Mol. Cell. Biol. 21:5094-5108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ray, D., Y. Terao, D. Nimbalkar, L. H. Chu, M. Donzelli, T. Tsutsui, X. Zou, A. K. Ghosh, J. Varga, G. F. Draetta, and H. Kiyokawa. 2005. Transforming growth factor beta facilitates beta-TrCP-mediated degradation of Cdc25A in a Smad3-dependent manner. Mol. Cell. Biol. 25:3338-3347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rengan, R., H. D. Ochs, L. I. Sweet, M. L. Keil, W. T. Gunning, N. A. Lachant, L. A. Boxer, and G. M. Omann. 2000. Actin cytoskeletal function is spared, but apoptosis is increased, in WAS patient hematopoietic cells. Blood 95:1283-1292. [PubMed] [Google Scholar]
- 66.Rodriguez, J., and Y. Lazebnik. 1999. Caspase-9 and APAF-1 form an active holoenzyme. Genes Dev. 13:3179-3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rogers, S., R. Wells, and M. Rechsteiner. 1986. Amino acid sequences common to rapidly degraded proteins: the PEST hypothesis. Science 234:364-368. [DOI] [PubMed] [Google Scholar]
- 68.Sangwan, V., G. N. Paliouras, A. Cheng, N. Dubé, M. L. Tremblay, and M. Park. 2005. Protein tyrosine phosphatase 1B deficiency protects against fas-induced hepatic failure. J. Biol. Chem. [DOI] [PubMed]
- 69.Sastry, S. K., P. D. Lyons, M. D. Schaller, and K. Burridge. 2002. PTP-PEST controls motility through regulation of Rac1. J. Cell Sci. 115:4305-4316. [DOI] [PubMed] [Google Scholar]
- 70.Sastry, S. K., Z. Rajfur, B. P. Liu, J. F. Cote, M. L. Tremblay, and K. Burridge. 2006. PTP-PEST couples membrane protrusion and tail retraction via VAV2 and p190RhoGAP. J. Biol. Chem. 281:11627-11636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sebbagh, M., C. Renvoize, J. Hamelin, N. Riche, J. Bertoglio, and J. Breard. 2001. Caspase-3-mediated cleavage of ROCK I induces MLC phosphorylation and apoptotic membrane blebbing. Nat. Cell Biol. 3:346-352. [DOI] [PubMed] [Google Scholar]
- 72.Shen, Y., G. Schneider, J. F. Cloutier, A. Veillette, and M. D. Schaller. 1998. Direct association of protein-tyrosine phosphatase PTP-PEST with paxillin. J. Biol. Chem. 273:6474-6481. [DOI] [PubMed] [Google Scholar]
- 73.Shim, S. R., S. Kook, J. I. Kim, and W. K. Song. 2001. Degradation of focal adhesion proteins paxillin and p130cas by caspases or calpains in apoptotic rat-1 and L929 cells. Biochem. Biophys. Res. Commun. 286:601-608. [DOI] [PubMed] [Google Scholar]
- 74.Sirois, J., J.-F. Côté, A. Charest, N. Uetani, A. Bourdeau, S. A. Duncan, E. Daniels, and M. L. Tremblay. Essential function of PTP-PEST during mouse embryonic vascularization, mesenchyme formation, neurogenesis and early liver development. Mech. Dev., in press. [DOI] [PMC free article] [PubMed]
- 75.Stupack, D. G., and D. A. Cheresh. 2002. Get a ligand, get a life: integrins, signaling and cell survival. J. Cell Sci. 115:3729-3738. [DOI] [PubMed] [Google Scholar]
- 76.Su, X., T. Zhou, Z. Wang, P. Yang, R. S. Jope, and J. D. Mountz. 1995. Defective expression of hematopoietic cell protein tyrosine phosphatase (HCP) in lymphoid cells blocks Fas-mediated apoptosis. Immunity 2:353-362. [DOI] [PubMed] [Google Scholar]
- 77.Takada, T., T. Noguchi, K. Inagaki, T. Hosooka, K. Fukunaga, T. Yamao, W. Ogawa, T. Matozaki, and M. Kasuga. 2002. Induction of apoptosis by stomach cancer-associated protein-tyrosine phosphatase-1. J. Biol. Chem. 277:34359-34366. [DOI] [PubMed] [Google Scholar]
- 78.Tisi, M. A., Y. Xie, T. T. Yeo, and F. M. Longo. 2000. Downregulation of LAR tyrosine phosphatase prevents apoptosis and augments NGF-induced neurite outgrowth. J. Neurobiol. 42:477-486. [PubMed] [Google Scholar]
- 79.Tonks, N. K. 2005. Redox redux: revisiting PTPs and the control of cell signaling. Cell 121:667-670. [DOI] [PubMed] [Google Scholar]
- 80.Torres, J., J. Rodriguez, M. P. Myers, M. Valiente, J. D. Graves, N. K. Tonks, and R. Pulido. 2003. Phosphorylation-regulated cleavage of the tumor suppressor PTEN by caspase-3: implications for the control of protein stability and PTEN-protein interactions. J. Biol. Chem. 278:30652-30660. [DOI] [PubMed] [Google Scholar]
- 81.Totpal, K., S. Agarwal, and B. B. Aggarwal. 1992. Phosphatase inhibitors modulate the growth-regulatory effects of human tumor necrosis factor on tumor and normal cells. Cancer Res. 52:2557-2562. [PubMed] [Google Scholar]
- 82.Wise, C. A., J. D. Gillum, C. E. Seidman, N. M. Lindor, R. Veile, S. Bashiardes, and M. Lovett. 2002. Mutations in CD2BP1 disrupt binding to PTP PEST and are responsible for PAPA syndrome, an autoinflammatory disorder. Hum. Mol. Genet. 11:961-969. [DOI] [PubMed] [Google Scholar]
- 83.Wu, J. J., and A. M. Bennett. 2005. Essential role for mitogen-activated protein (MAP) kinase phosphatase-1 in stress-responsive MAP kinase and cell survival signaling. J. Biol. Chem. 280:16461-16466. [DOI] [PubMed] [Google Scholar]
- 84.Yousefi, S., and H. U. Simon. 2003. SHP-1: a regulator of neutrophil apoptosis. Semin. Immunol. 15:195-199. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.