Abstract
Transposons have contributed protein coding sequences to a unexpectedly large number of human genes. Except for the V(D)J recombinase and telomerase, all remain of unknown function. Here we investigate the activity of the human SETMAR protein, a highly expressed fusion between a histone H3 methylase and a mariner family transposase. Although SETMAR has demonstrated methylase activity and a DNA repair phenotype, its mode of action and the role of the transposase domain remain obscure. As a starting point to address this problem, we have dissected the activity of the transposase domain in the context of the full-length protein and the isolated transposase domain. Complete transposition of an engineered Hsmar1 transposon by the transposase domain was detected, although the extent of the reaction was limited by a severe defect for cleavage at the 3′ ends of the element. Despite this problem, SETMAR retains robust activity for the other stages of the Hsmar1 transposition reaction, namely, site-specific DNA binding to the transposon ends, assembly of a paired-ends complex, cleavage of the 5′ end of the element in Mn2+, and integration at a TA dinucleotide target site. SETMAR is unlikely to catalyze transposition in the human genome, although the nicking activity may have a role in the DNA repair phenotype. The key activity for the mariner domain is therefore the robust DNA-binding and looping activity which has a high potential for targeting the histone methylase domain to the many thousands of specific binding sites in the human genome provided by copies of the Hsmar1 transposon.
DNA transposons are genomic parasites that exist purely at the molecular level. Although ubiquitous, they are short-lived in any given eukaryotic genome and rely on frequent horizontal transfer to new hosts (19, 24). At present, DNA transposons are accepted as extinct in humans, and the youngest family identified thus far appeared some 50 million years ago, after the divergence of the prosimians from the anthropoid lineage (19).
DNA transposons contribute less than 3% of the human genome, compared to 41% contributed by retroelements that transpose via an RNA intermediate. Nevertheless, the DNA-based elements have provided 43 of the 47 human genes derived from transposons (19). Of these 43 genes, only the immune system V(D)J recombinase RAG1 has been systematically analyzed for transposition. Transposition mediated by RAG1 was first detected at a low frequency in vitro and then in vivo (4, 14, 27). Since several steps in V(D)J recombination are identical to transposition, RAG1-mediated transposition was, perhaps, not unexpected. However, the activities of the other human domesticated transposases are far from certain because they are of unknown function, and only two have documented phenotypes. SETMAR is involved in nonhomologous end joining (NHEJ) and in promoting resistance to ionizing radiation (20), while mutations in the human homolog of mouse JERKY are associated with idiopathic generalized epilepsy (29). The only other known phenotype for a domesticated transposase in higher eukaryotes is for the plant DAYSLEEPER gene, which is essential for development in Arabidopsis thaliana (2).
The human SETMAR protein is a fusion between an active histone H3 methylase and a mariner family transposase now encoded by exon 3 of the gene (Fig. 1A) (5, 20, 32). The fusion event occurred about 50 million years ago and is therefore present in all anthropoid primates but not in other mammals (5). We chose to investigate the potential activities of this protein because it is highly expressed in many different human tissues and cancers and has a documented DNA repair phenotype (5, 20, 32).
FIG. 1.
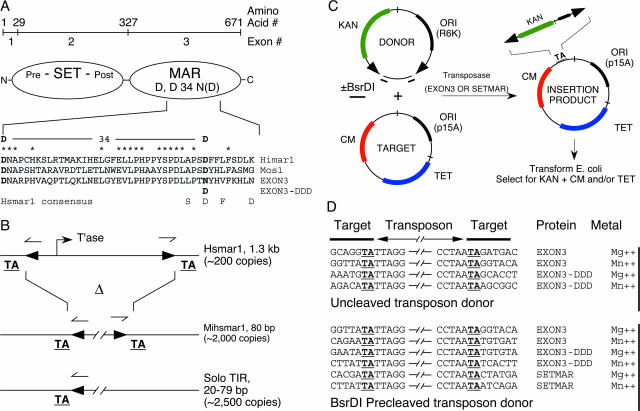
The domain structure of SETMAR and a genetic assay for transposition. (A) SETMAR exons 1 and 2 encode the histone methylase domain. The transposase domain is encoded by exon 3. The structure illustrated is the most common splice isoform (5) and yields numerous cDNA hits in a Web-based search of the NCBI database. The SETMAR residues surrounding the active site D34N region are shown aligned with the transposase sequences for Himar1 and Mos1. Himar1 and Mos1 are closely related transposons that have been shown to be fully active in vitro and in vivo in a wide range of cell types. The DDN motif corresponds to SETMAR residues D483, D575, and N610. (B) Exon 3 of SETMAR was derived from the transposase gene of the Hsmar1 element. The TIRs flanking the transposon are represented by solid black arrow heads. There are about 200 defective copies of Hsmar1 in the human genome. Hsmar1 is associated with a family of MITES which we refer to as mini-Hsmar1, or Mihsmar1. Superficially, Mihsmar1 resembles an internal deletion derivative of the parental transposon. However, although it shares the TIRs of the parental element, the central region may have a different origin. MITES are rare in bacteria (1) but common in eukaryotes where the nuclear membrane mandates a trans-acting transposase. Upon insertion, mariner transposons cause the duplication of a TA dinucleotide derived from the target site. (C) Schematic representation of the genetic transposition assay showing a random transposition event. Solid black arrow heads represent the transposon TIRs. The drug resistance markers are as follows: KAN, kanamycin; CM, chloramphenicol; TET, tetracycline; ori, origin of plasmid replication. The BsrDI restriction endonuclease sites placed adjacent to the transposon ends can be used to artificially cleave the transposon ends to generate a linear transposon donor. (D) An example of the transposon integration sites recovered under each of the conditions tested. DNA sequencing reactions were initiated from primer sites within the transposon, out across the transposon ends into the flanking target DNA. Transposon insertions were precise and flanked by the TA dinucleotide (marked in boldface type) duplication diagnostic of mariner family elements.
Exon 3 of the SETMAR gene is derived from the Hsmar1 transposon (32). The human genome contains approximately 200 copies of this element. In addition, there exists a large family of miniature inverted-repeat transposable elements (MITES) associated with Hsmar1. MITES do not encode transposases but, as in this case, can sometimes be identified with a full-length parental transposon with which they share closely related terminal inverted repeats (TIRs). Although MITES are probably more sophisticated than simple internal-deletion derivatives of the parental transposon (11), we propose to refer to these elements as mini-Hsmar1, or Mihsmar1. There are about 2,000 copies of Mihsmar1 in the human genome that, together with the solo TIRs, represent almost 7,000 potential binding sites for SETMAR (Fig. 1B).
In this paper we show for the first time that the transposase domain of human SETMAR retains full transposase activity, although limited by a defect in one of the intermediate stages of the reaction. Our studies reveal that SETMAR retains robust site-specific DNA-looping, nicking, and integration activities. We discuss the contribution of these activities to the DNA repair functions reported by others (20) and the possibility that SETMAR has a novel role in chromatin modification and gene regulation.
MATERIALS AND METHODS
Media.
All strains were grown in Luria-Bertani (LB) media at 37°C, unless otherwise stated. The following antibiotics were used at the indicated concentrations: ampicillin, 50 or 100 μg/ml; kanamycin, 50 μg/ml; chloramphenicol, 50 μg/ml.
Strains.
The following Escherichia coli strains were used in the experiments: RC5024 (identical to DH5α) {endA1 hsdR17 glnV44 thi-1 recA1 gyrA relA1 Δ(lacIZYA-argF)U169 deoR [φ80dlacΔ(lacZ)M15]}, RC5081 (identical to S17.1λ pir; a gift from Judy Armitage), and RC5082 (identical to Rosetta 2; Novagen).
Plasmids.
Exon 3 was amplified by PCR from human genomic DNA with primers TACAGTTACTGTGGAGGTTTATCAGACA and TGTAAGCCACCATGCCAGGCCACAT, followed by a second round of PCR with primers CGGGATCCATGAAAATGATGTTAGACAA and CGGGATCCTTAATCAAAATAGGAACCAT. The resulting PCR product was digested with BamHI and inserted into the unique BamHI site of pMAL-c2X (New England Biolabs) to create pRC802.
The SETMAR gene was constructed as follows: exon 1 and exon 2 were amplified by PCR from human genomic DNA with primers CCTTGGCACAAACCCTATCACATGTCAGTCAAC and TTCAGGGAATGGACTGAAAAGGACAACACCC for exon 1 and AGAATGGGTCCCAGTCAGAAGCTACTTGG and AGCATAACTTCACAAGATTTGGAGTACAGCC for exon 2, followed by a second round of PCR amplification with CGGGATCCATGGCGGAGTTTAAGGAGAAG (Exon1-F), CTGGAAGGGCGCCGG (Exon1-R), TACACTCCTGATCATGTAGTTGGACC (Exon2-F), and CTCAAGGGTCAATCGCTTGCAGG (Exon2-R). The PCR products were phosphorylated, ligated together, and reamplified with primers Exon1-F and Exon2-R. Exon3 was amplified with primers ACTATGAAAATGATGTTAGACAAAAAGCAA (Exon3-F) and CGGGATCCTTAATCAAAATAGGAACCATTACAATCAAC (Exon3-R). The PCR product was phosphorylated and ligated to the exon 1 + 2 PCR product and then reamplified with primers Exon1-F and Exon3-R. This was digested with BamHI and inserted into BamHI-digested pMAL-c2X to generate pRC813.
Plasmids pRC656 and pRC657 were constructed by PCR amplification of a kanamycin resistance cassette with primer GCGAATTCTATTAGGTTGGTGCAAAAGTAATTGCGGTTTTGGATCCCTGTAATCCGGGC and ligation of the EcoRI-digested PCR product to pBluescript (Stratagene) digested with EcoRI. BamHI digestion of the plasmid resulted in the excision of the kanamycin resistance marker, which was replaced with a BamHI fragment containing a kanamycin resistance gene and an R6K plasmid origin of replication (lacking the R6K pir gene that is required for replication) to generate pRC656 and pRC657 (opposite orientation of BamHI fragment).
pRC707 was constructed by phosphorylation and ligation of two PCR products spanning the length of exon 3, generated by PCR amplification of exon 3 with primers GCAGGAGGAATTCCATATGAAAATGATGTTAGAC (EXON3-DDD-F) and GTGGTAGTCGGTTGGCAAGAGGTCAGGTG and GTCTTTAAGCATCTCAACAACTTTTTGCAGGG and CAGCCAAGCTTGCCTGCAGGTCGACTC (EXON3-DDD-R). This was followed by reamplification with primers EXON3-DDD-F and EXON3-DDD-R, digestion of the PCR product with NdeI and HindIII, and ligation to a NdeI- and HindIII-digested derivative of pBAD24.
Other plasmids were constructed as follows: pRC901, by digestion of pRC707 with XbaI and ClaI and ligation of the EXON3-DDD-containing fragment to XbaI- and ClaI-digested pRC802; pRC816, by PCR amplification of an Hsmar1 transposon end-containing fragment from pRC657 with primers CGGGATCCGCAATGTTAGGTTGGTGCAAAAGTAATTGC and CCACATGTGGAATTGTGAGCGG and insertion of the PCR product into pCR-Blunt II-TOPO (Invitrogen); pRC824, by PCR amplification of a kanamycin resistance marker with primers AGGAATTCGCAATGTTAGGTTGGTGCAAAAGTAATTGCGGTTTTGGATCCGCACAAGAAAAAAATATATCATCATG and AGGAATTCGCAATGTTAGGTTGGTGCAAAAGTAATTGCGGTTTTGGATCCTTTGTTGTAGGTGG and insertion of the EcoRI-digested PCR product into the EcoRI site of pBluescript; pRC902 and pRC903, by digestion of pRC657 with EcoRI and insertion of the Hsmar1 transposon end-containing fragment into EcoRI-digested pBluescript (the two plasmids contain the EcoRI fragment in opposite orientations); pRC704, by SapI- and XmnI-mediated digestion of pRC656, followed by the filling in of the staggered end with Klenow polymerase and self-ligation of the vector. This step removes the pBluescript origin of replication, resulting in a plasmid wholly dependent on the R6K pir gene product for replication. Therefore, the plasmid was propagated in E. coli S17.1λ pir which carries a chromosomal copy of the pir gene.
Molecular procedures.
Materials and reagents were generally of the best quality available. Chemicals were obtained from Sigma or BDH Laboratory Supplies. Enzymes were from New England Biolabs or Roche Applied Science. DNA manipulations using commercially available enzymes were done according to manufacturer's recommendations. All cloned PCR products were confirmed by nucleotide sequencing.
Transposase expression and purification.
E. coli Rosetta 2 cells harboring pRC802 or pRC901 were grown overnight at 37°C in LB medium containing selection for the plasmid. The following day, the culture was diluted 1:100 in fresh LB medium with selection and grown to mid-log phase (optical density at 600 nm reading of ∼0.5). The culture was shifted from 37°C to 30°C and incubated with 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 2.5 h. Cells were harvested by centrifugation and resuspended in HSG buffer medium (50 mM HEPES [pH 7.5], 200 mM NaCl, 2 mM dithiothreitol [DTT], 5 mM EDTA, 10% glycerol) supplemented with a protease inhibitor cocktail (Roche Applied Science). The cells were lysed by French press, and the soluble fraction, retrieved by centrifugation (25,000 × g for 30 min), was loaded onto an amylose resin column (New England Biolabs). The column was washed several times with HSG buffer and the protein eluted with 10 mM maltose. The fraction containing MBP-EXON3 (or MBP-EXON3-DDD) was diluted fourfold in buffer A (50 mM HEPES [pH 7.5], 50 mM NaCl, 2 mM DTT, 5 mM EDTA, 5% glycerol), loaded on to a MonoS HR5/5 column (Amersham Pharmacia), and eluted with a 20-ml gradient of 0.05 to 1 M NaCl. Glycerol (15%) was added to the fractions containing MBP-EXON3 or MBP-EXON3-DDD, and 5- to 10-μl aliquots were stored at −80°C. Induction of protein expression in the cell extract and elution fractions of the final column are shown in Fig. S5 in the supplemental material.
MBP-SETMAR expression and purification were done in a similar manner, except that 500 mM NaCl and 0.1% Triton were included in all the buffers and a MonoQ HR5/5 column (Amersham Pharmacia) was used for the last purification step. Induction of protein expression in the cell extract and elution fractions of the final column are shown in Fig. S6 in the supplemental material.
Paired-end complex assembly.
DNA fragments containing Hsmar1 transposon ends with flanking DNA were generated by digestion of pRC902 with AccI and SacII (162-bp fragment) and pRC903 with XbaI (83-bp fragment). The former was 3′ radiolabeled with [α-P32]dCTP (Amersham Pharmacia) at the AccI site, and the latter was 3′ radiolabeled at both ends. Different combinations of cold and labeled transposon ends (100 fmol each) were incubated with 2 nM transposase in a 20-μl reaction mix containing 20 mM HEPES (pH 7.5), 100 mM NaCl, 250 μg/ml bovine serum albumin, 2 mM DTT, 10% glycerol, 5% dimethyl sulfoxide (DMSO), 5 mM CaCl2 and 250 ng nonspecific DNA at room temperature for 10 min. Only one DNA fragment was radiolabeled in the mixed-probe reactions, and cold DNA was used in half the quantity of the labeled probe.
Precleaved transposon ends were generated by digestion of pRC816 with BsrDI and XbaI (48-bp fragment) or with BsrDI and XhoI (164-bp fragment) and 3′ radiolabeled with [α-P32]dCTP at the XbaI or XhoI end. Various combinations of cold and labeled transposon ends (50 fmol each) were incubated with 1.25 nM transposase in buffer B (20 mM HEPES [pH 7.5], 100 mM NaCl, 2 mM DTT, 240 μg/ml bovine serum albumin, 10% glycerol) in a 20-μl reaction mix for 2 or 4 h at 37°C. The quantity of transposase was doubled in the mixed-probe reactions.
Products were separated on a 5% Tris-acetic acid (TAE)-polyacrylamide gel. Gels were dried, exposed to PhosphorImager plates, and analyzed by Image Reader software (Fujifilm).
DNA cleavage assay.
A 109-bp Hsmar1 transposon end-containing DNA fragment was generated by HindIII- and Acc65I-mediated digestion of pRC816. The resulting DNA fragment was 3′ radiolabeled at the HindIII end, and 50 fmol was incubated with 1.25 nM transposase in buffer B (20-μl reaction volume) for 2 or 4 h at 37°C. Complexes were separated on a 5% TAE-polyacrylamide gel, excised, and soaked overnight at 37°C in buffer B supplemented with 5% DMSO and 5 mM MgCl2 or MnCl2. The DNA was purified by phenol-chloroform extraction and ethanol precipitation and electrophoresed on a 5% TAE-polyacrylamide gel.
To determine the exact site of cleavage, pRC816 was digested with HindIII and 3′ radiolabeled at both ends with [α-P32]dCTP, and 50 fmol was incubated in buffer B supplemented with 5% DMSO and 5 mM MgCl2 or MnCl2 (20 μl-reaction volume) for 4 h at 37°C. The DNA was purified as described above and run on a 10% polyacrylamide sequencing gel.
Integration assay.
A precleaved Hsmar1 transposon end was generated by digestion of pRC816 with BsrDI and XbaI. The DNA was 3′ radiolabeled at the XbaI end with [α-P32]dCTP, and 50 fmol was incubated with 1 μg pBluescript and 1.25 nM transposase in buffer B supplemented with 5% DMSO and 5 mM MgCl2 or MnCl2 (20-μl reaction volume) for 2 or 4 h at 37°C. Products were separated by agarose gel electrophoresis and viewed by ethidium bromide staining and audioradiography.
In vitro transposition.
pRC704 contains a kanamycin resistance marker and an R6K plasmid origin of replication flanked by 30-bp Hsmar1 terminal inverted repeats. To assay for transposition in vitro, 1 to 5 nM pRC704 was incubated with a similar quantity of transposase and 5 to 10 nM pACYC184 (New England Biolabs) in buffer B supplemented with 5% DMSO and 5 mM MgCl2 or MnCl2 (10- or 20-μl reaction volume) for 3 or 6 h at 37°C. The reaction was used to transform E. coli DH5α cells, transformants were selected for on media containing selection for both plasmids, and potential transposition products were sequenced with primers directed to each end of the transposon.
Transposition with precleaved substrate was performed as described above, except that the substrate (a precleaved Hsmar1 transposon harboring a kanamycin resistance gene) was generated by digestion of pRC824 with BsrDI.
RESULTS
Construction of SETMAR.
The SETMAR gene was cloned directly from human genomic DNA. Several rounds of PCR were used to remove the introns, and the uninterrupted gene was cloned in a vector for expression and purification from a bacterial host. The transposase domain encoded by exon 3 of the gene was also cloned, expressed, and purified separately.
The amino acid sequence of the exon 3 product has been well conserved with respect to the consensus sequence for the Hsmar1 transposase, with which it shares 94% identity (32). One important difference is that the DD34D catalytic triad, typical of mariner elements, is mutated to DDN. Since we were also interested in investigating the potential effects of this substitution, we constructed a derivative of the transposase domain encoded by exon 3 in which the DDN motif had been changed to the consensus DDD. Henceforth, we will refer to the protein product of wild-type exon 3 as EXON3 and to the derivative with the consensus DDD motif as EXON3-DDD.
SETMAR and EXON3 mobilize an Hsmar1 transposon.
Full-length SETMAR, EXON3, and EXON3-DDD were assayed for transposition using a genetic in vitro hop assay (Fig. 1C). In this assay, the protein is incubated with an artificial Hsmar1 transposon and a target plasmid. Transposition events are recovered by genetic transformation of bacteria and selection for drug resistance markers present on the transposon plus the target plasmid. Transposon insertions were precise and flanked by the TA dinucleotide duplication diagnostic of mariner family elements (Fig. 1D).
Transposition events were recovered using EXON3 and EXON3-DDD. Restoration of the consensus DDD catalytic triad in the EXON3-DDD protein increased the frequency of events slightly, as did the use of Mn2+ as the catalytic metal ion (see Fig. 4A below for relative frequencies). The absolute frequency was extremely low but comparable to V(D)J transposition (4, 14).
FIG. 4.
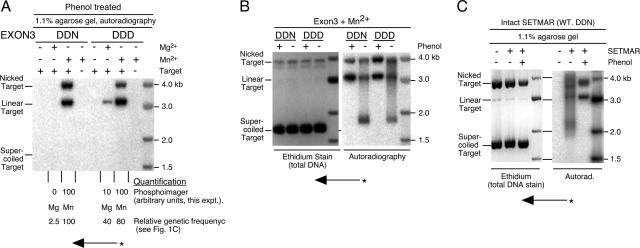
The integration product is topologically constrained. (A) Insertion of radioactively labeled linear transposon ends into a supercoiled plasmid target. The transposon ends were precleaved with BsrDI and incubated with the target plasmid and EXON3 protein, as indicated. Reactions were stopped with EDTA and SDS, treated with phenol to disrupt the integration complex, and electrophoresed on a standard TAE-buffered 1.1% agarose gel. After electrophoresis, the gel was dried and recorded by autoradiography. Concerted integration of transposon ends at the target site yield a linear product. Single-ended integration events yield a lariat structure which will migrate close to the position of the nicked topoisomer of the target plasmid (for an example, see reference 3). The bands in the gel were quantified using a PhosphorImager, as indicated below the panel. Also given is the relative frequency of transposition measured in the genetic assay for which results are shown in Fig. 1 under the same conditions. (B) The effect of phenol on the integration complex. Insertion reactions were performed with a precleaved transposon end as described for paned A. The reactions were stopped with the addition of EDTA and SDS and visualized either before or after treatment with phenol to disrupt the integration complex. After electrophoresis, the gel was stained for total DNA with ethidium bromide. It was then dried, and the radioactive products were recorded on a PhosphorImager. (C) Insertion reactions as described for panel B but using intact SETMAR protein. After electrophoresis, the gel was stained for total DNA with ethidium bromide, dried, and recorded by autoradiography.
For the cut-and-paste transposons, which include the members of the mariner family, an important mechanistic transition occurs between the pre- and postcleavage steps. During the postcleavage steps, the active site must accommodate a target site in place of the flanking DNA, and perform integration instead of excision. In some systems, the efficiency of integration is increased if an artificially precleaved transposon is provided as a substrate. To test this possibility for SETMAR, the ends of the Hsmar1 transposon were cleaved using BsrDI restriction endonuclease sites located precisely at either end of the element (Fig. 1C). This increased transposition >100-fold under all conditions tested, reaching levels comparable to other mariner family elements, such as Himar1 and Mos1. Under this regime, transposition events were easily recovered using intact SETMAR (Fig. 1D).
The increased efficiency of integration with the precleaved transposon substrate suggests that SETMAR and EXON3 may be partially defective for one or more of the preceding steps of the reaction. To identify the defective step(s), we therefore decided to examine each of the individual biochemical stages of the transposition reaction.
Assembly of the paired-end complex.
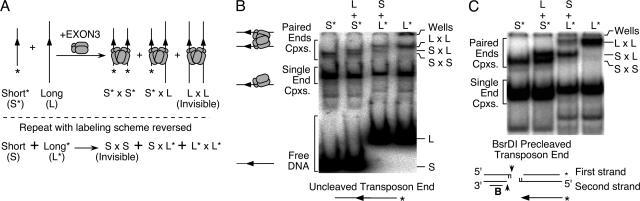
Concerted integration of the transposon ends on either side of the TA dinucleotide at the target site implies the existence of a paired-end complex (PEC). Indeed, synapsis of transposon ends into a PEC has been found to be an early intermediate step in every DNA-transposition reaction studied in molecular detail (6). The ability of EXON3 to organize the assembly of a PEC was examined in a variation of the classical electrophoretic mobility shift assay (Fig. 2A). In this experiment, the PEC is revealed by mixing the protein with labeled and unlabeled transposon ends of different lengths. Using this strategy, PEC assembly with EXON3 was detected with either uncleaved or precleaved transposon ends (Fig. 2B and C). Indeed, PEC assembly was extremely efficient, reaching 10% of the complexes in some reactions. This efficiency is much better than previously demonstrated for any other mariner family element where the PEC is undetectable or barely detectable under the most favorable conditions (8, 24).
FIG. 2.
Assembly of a paired-end complex. (A) Schematic representation of the “long by short” assay for PEC assembly. Synaptic complexes are revealed by mixing combinations of long and short transposon ends. These are either unlabeled or radioactively labeled, as indicated by the asterisk. Only two of the three possible complexes are detected in each lane because any complexes containing unlabeled DNA fragments are invisible in the experiment. EXON3 is arbitrarily illustrated as dimeric and tetrameric based on previous work with Himar1 (24). (B) PEC assembly with uncleaved transposon ends. Complexes (Cpxs) were assembled with EXON3 and uncleaved linear transposon ends, as indicated below the panel. Products were analyzed on a 5% polyacrylamide gel. Homogeneous complexes containing two transposon ends of the same length are present in the outermost pair of lanes. Mixed complexes containing one long and one short transposon end are present in the central pair of lanes. (C) Experiment as described for panel B, but the transposon ends are precleaved with the restriction endonuclease BsrDI, as indicated below the panel. The lack of flanking DNA improves the resolution of the bands in the gel.
Similar electrophoretic mobility shift assays performed with SETMAR revealed specific binding to the Hsmar1 transposon end (see Fig. S7 in the supplemental material). Visualization of a PEC was precluded by the high molecular weight of intact SETMAR and by solubility problems at higher concentrations that caused aggregation in the wells. However, as noted above, the integration of precleaved Hsmar1 transposon ends on either side of a TA dinucleotide target (Fig. 1D) demonstrates that SETMAR, like EXON3, supports PEC assembly. Further evidence for this is also presented below.
Cleavage at the Hsmar1 transposon ends.
In the DDE(D) family of transposases, integration of the transposon ends at the target site is via a direct transesterification mechanism (28). This reaction requires that the transposase cleave the donor DNA at a specific location, generating a free 3′-hydroxyl group precisely at each end of the transposon. The direct transesterification mechanism for integration does not, however, constrain the position of cleavage on the opposite strand of the DNA. Consequently, variations in this step have been detected in different systems. For example, the closely related Himar1 and Mos1 transposons cleave the 5′ phosphate imprecisely, predominantly at positions 2 or 3 bp within the element (8, 18). In the more distantly related Sleeping Beauty transposon, in vivo excision footprints suggest that 5′ cleavage is likewise predominantly 3 bp within the element (16, 25).
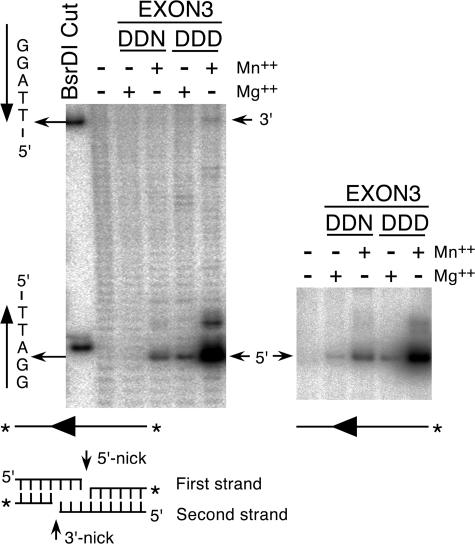
The positions at which EXON3 nicks the transposon ends during the cleavage step of the reaction were determined by analysis on a denaturing DNA-sequencing gel (Fig. 3). Cleavage of the substrate with the BsrDI restriction endonuclease provided DNA markers for nicks at the 3′ transposon end and at a 5′ phosphate 2 bp inside the element on the opposite strand. This experiment revealed that the 5′ nick is located 3 bp within the element. The 3′ nick was precisely at the end of the transposon, as expected. However, it was very inefficient compared to the nicking at the 5′ end. The inefficiency of the 3′ nick identifies at least one partial defect in the transposition reaction, as we had postulated above. It also explains the increased recovery of transposition events in the genetic assay when the transposon ends were precleaved with BsrDI (above) (Fig. 1C and D).
FIG. 3.
In vitro cleavage assay. DNA cleavage reactions were performed with EXON3 and EXON3-DDD using an uncleaved transposon end as a substrate. The positions of the first and second strand cleavage sites were determined by analysis on a DNA sequencing gel. A BsrDI restriction endonuclease site placed at the transposon end was used to generate markers for cleavage at the 3′-end of the transposon and at a position 2 bp within the element on the opposite strand. The actual positions of transposon end cleavage are illustrated below and to the left of the gels. In the left panel, the DNA is labeled on both 3′ ends, as indicated by the asterisk. In the right panel, only the 3′ end of the transposon arm is labeled. Using this labeling strategy, only the 5′ nick can be visualized.
Detection of integration products.
The final event in transposition is the concerted integration of the 3′ ends of the element at a target site. This was assayed using radioactively labeled BsrDI-precleaved linear transposon ends as a substrate and a supercoiled plasmid as a target (Fig. 4A). Concerted integration of a pair of transposon ends will yield a radioactively labeled linear product slightly larger than the target plasmid. A product of the correct size was detected in the presence of the EXON3 protein. Single-end integrations were also detected at the position expected for the nicked topoisomer of the target. Integration in Mn2+ was efficient and little changed by restoration of the DDD motif. The quantification of the products using a PhosphorImager is presented below the gel, together with the relative frequencies of the transposition events detected genetically in Fig. 1D.
SETMAR and EXON3 integration products are topologically constrained.
Transposon integration typically yields an extremely stable complex that constrains the topology of the transposon and the target (23). This is a further practical demonstration of the role of the PEC at this stage of the reaction. To test whether this is true for EXON3, the integration reaction was performed with precleaved transposon ends as described above (Fig. 4B). The reactions were first treated with sodium dodecyl sulfate (SDS) to disrupt any weak nonspecific protein-DNA interactions. Reaction products were then analyzed either before or after treatment with phenol, which is a strong denaturant and is required to disrupt the integration complex in the Himar1 system (23). To help establish the location and identity of the complexes, the gel was first stained for total DNA with ethidium bromide and then dried and recorded by autoradiography (Fig. 4B). A phenol-sensitive complex was detected just above the position of the supercoiled target, indicating that the integration complex is stable and topologically constrained.
Finally, a similar experiment was performed with the intact SETMAR protein (Fig. 4C). In the absence of phenol, a radioactively labeled smear was detected, together with a product just above the position of the supercoiled target plasmid. Treatment of the reaction mixture with phenol yielded radioactively labeled products at the positions of the linear and nicked isomers of the target plasmid, diagnostic of concerted and single-end integration events, respectively. The concerted integration products detected in this experiment are further confirmation that intact SETMAR has the same DNA binding and looping activities shown for EXON3 in the previous experiments.
DISCUSSION
Transposition activity.
Human SETMAR is a chimeric protein, created by the fusion of a SET-domain histone H3 methylase and a mariner family transposase. A previous study provided evidence of histone methyltransferase activity for SETMAR and implicated the protein in the nonhomologous end-joining of double-stranded DNA breaks (20). In the present work, we have investigated the properties of the transposase domain of SETMAR. We show that while SETMAR is largely defective for transposition in vitro, many of the specific activities required for transposition have been preserved. The primary defect appears to be a deficiency in the nicking reaction that generates the crucial 3′-hydroxyl group at the end of the transposon.
The transposase domain, encoded by exon 3 of SETMAR, contains 19 single amino acid differences relative to the consensus sequence for the Hsmar1 transposase (32). Any one or more of these differences could be responsible for the low level of transposase activity. The most obvious a priori candidate is the substitution which changes the canonical DDD catalytic motif to DDN (Fig. 1A). However, this need not necessarily abolish the catalytic activity. In the two-metal ion catalytic mechanism employed by the DDE(D) superfamily of enzymes, coordination of the A metal ion is somewhat flexible and the final E(D) residue of the motif is not absolutely required (10, 12, 38). Furthermore, the D-to-N substitution in SETMAR is structurally conservative and retains the physical presence of the oxygen atom that normally coordinates metal ion A in the active site.
In light of these considerations, it should be noted that restoration of the consensus DDD motif in our EXON3-DDD construct did not substantially rescue cleavage at the 3′ end of the transposon (Fig. 3) nor did it substantially increase the frequency of integration with the precleaved transposon end (Fig. 4A). The only significant enhancement in activity is observed in the 5′-nicking assay. Thus, the DDN substitution is probably not important in limiting the transposition activity of SETMAR. Indeed, we have evidence that when Mn2+ is provided as the catalytic metal ion, the defect in 3′-strand cleavage can be rescued by restoration of the consensus S residue located two positions before the active site N610 in SETMAR (Fig. 1A; result to be presented elsewhere).
The in vitro transposition assays were heavily influenced by the choice of catalytic metal ion. In particular, the 5′-nicking reaction was much more efficient in Mn2+ than in Mg2+. This could be attributed to limitations of the protein purification protocol or to the in vitro reaction conditions, in which potential host factors are lacking. Similar limitations have been encountered with human immunodeficiency virus integrase and V(D)J recombination (12, 14, 21, 26). In the case of the human immunodeficiency virus integrase protein, the physiological Mg2+ dependence was restored by changes in the purification protocol. Another example is provided by the bacterial NHEJ protein LigD, in which the polymerase and nuclease activities are absolutely dependent on Mn2+ in vitro but presumably not in vivo (40, 41).
The rarity of transposition events in the in vitro hop assay suggest that SETMAR is unlikely to catalyze transposition in the human genome. Of course, exceptional cases may be identified, such as the rare examples of unforced V(D)J transposition in vivo (27). The present results make a better case for a SETMAR nicking activity (Fig. 3). Indeed, residual nicking activity has been proposed as a mechanism by which the supposedly defunct DNA transposons can form hotspots for meiotic crossovers and gene conversion events, as suggested for the generation of the inherited peripheral neuropathies Charcot-Marie tooth disease type 1A and hereditary neuropathy with liability to pressure palsies in humans (30).
Site-specific DNA-binding and -looping activity.
Most of the domesticated transposons in the human genome are derived from DNA transposons, as opposed to retroelements that transpose via an RNA intermediate (19). One possibility is that this bias arose as a result of their utility as site-specific DNA-binding proteins. In this view, they would be expected to perform a structural role, perhaps in the organization of specialized chromatin features, as transcription factors or as mediators that target the activity of fusion proteins (2, 5, 17, 19). This mode of action for SETMAR is highly feasible, in light of the strong site-specific DNA-binding activity reported here and previous work demonstrating strong selective pressure on the 5′ portion of exon 3 encoding the DNA-binding domain of SETMAR (5).
In bacteria, DNA looping is a well-established mechanism for the regulation of transcription and DNA recombination (references 6, 7, and 35 and references therein). In eukaryotes, the best known DNA-looping factors are transcriptional enhancers which typically operate over large distances (for examples, see reference 34). However, recent work has shown that long-range chromosomal interactions are important for a much wider range of regulatory circuitry than had been previously realized. The chromosome conformation capture technique has provided evidence for the specific spatial organization of chromosomes within the nucleus (9, 34). Superimposed upon this global level of organization are site-specific DNA-looping events and protein-mediated interactions between distant loci on the same or even different chromosomes. Such interactions are involved in a diverse collection of important biological processes, such as genetic imprinting (22), and in the regulation of genes, such as the globins (34), cytokines (33), and the polycomb genes that direct development (13).
Assembly of the transposon ends into a PEC is a specific type of DNA-looping event. To date, the only mariner family elements reconstituted in vitro are Himar1 and Mos1 (8, 23, 24), and in both cases, the PEC is unstable and extremely difficult to detect. Our present results, which indicate robust PEC formation by SETMAR, suggest that the protein possesses an unusually stable DNA-looping activity. We propose that a primary function of SETMAR involves DNA looping between distant chromosomal loci. The numerous Hsmar1 transposon ends in the genome would provide the requisite DNA-binding sites, while the histone methyltransferase activity, previously demonstrated for SETMAR, would serve to open the chromatin structure and increase its flexibility, thereby facilitating the synapsis of distant SETMAR binding sites. The DNA structures thus created might be important for gene regulation. Future investigation will address the question of whether SETMAR functions as one of the many agents mediating the histone code or epigenetic regulation.
A role for the transposase catalytic domain?
Site-specific DNA binding in mariner family transposases is determined by an N-terminal domain located within the first 60 to 120 amino acids (15, 31, 36, 37, 39). Statistical analysis of the synonymous-versus-nonsynonymous substitution ratio among the copies of SETMAR in the anthropoid lineage suggests that amino acids 1 to 171 are under purifying selection (5), while the remaining portion of the protein, including a large part of the catalytic domain, may be evolving neutrally. On the basis of this evidence, it has been suggested that the physiological role of SETMAR requires only the SET and DNA-binding domains of the protein. However, the presence of a full-length catalytic domain in many ancient domesticated transposons suggests that additional constraints are operating besides DNA binding. This notion is supported by the catalytic activities demonstrated here and by examples of domesticated transposases, such as the human BUSTER genes which are derived from an hAT family transposon and maintain a perfect DDD catalytic triad of amino acid residues (19).
In a previous study, expression of SETMAR was associated with increased levels of NHEJ in human embryonic kidney cells and enhanced resistance to ionizing radiation (20). The authors envisaged a role for SETMAR in DNA repair and postulated a nicking activity mediated by the transposase domain of the protein. We have observed 5′-nicking activity for EXON3, and it is tempting to consider the possibility that the DNA binding, looping, and nicking activities contribute to a DNA repair pathway involving NHEJ. Interestingly, a mutation introduced at residue D490 of SETMAR, seven residues after the start of the DDN motif, abolished NHEJ activity and had a dominant-negative effect when coexpressed with wild-type SETMAR (20). This is further evidence that the transposase active site is required for the physiological function of SETMAR in vivo. Future work is expected to focus on analyzing the DNA repair function of SETMAR with mutations that specifically abolish one defined aspect of the DNA binding, looping, or nicking activities of this protein. The work presented here provides a solid base from which to assess the properties of the desired mutations.
Supplementary Material
Acknowledgments
We thank Malcolm Robertson for technical help with PCR and cloning steps. We also thank Paul Crellin and Maria-Joao Gravato-Nobre for editorial suggestions.
This work was funded by grants from The Wellcome Trust to R.C.
Footnotes
Published ahead of print on 27 November 2006.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Buisine, N., C. M. Tang, and R. Chalmers. 2002. Transposon-like Correia elements: structure, distribution and genetic exchange between pathogenic Neisseria sp. FEBS Lett. 522:52-58. [DOI] [PubMed] [Google Scholar]
- 2.Bundock, P., and P. Hooykaas. 2005. An Arabidopsis hAT-like transposase is essential for plant development. Nature 436:282-284. [DOI] [PubMed] [Google Scholar]
- 3.Chalmers, R. M., and N. Kleckner. 1996. IS10/Tn10 transposition efficiently accommodates diverse transposon end configurations. EMBO J. 15:5112-5122. [PMC free article] [PubMed] [Google Scholar]
- 4.Chatterji, M., C. L. Tsai, and D. G. Schatz. 2006. Mobilization of RAG-generated signal ends by transposition and insertion in vivo. Mol. Cell. Biol. 26:1558-1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cordaux, R., S. Udit, M. A. Batzer, and C. Feschotte. 2006. Birth of a chimeric primate gene by capture of the transposase gene from a mobile element. Proc. Natl. Acad. Sci. USA 103:8101-8106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Craig, N. L., R. Craigie, M. Gellert, and A. M. Lambowitz. 2002. Mobile DNA II. American Society for Microbiology, Washington, DC.
- 7.Crellin, P., S. Sewitz, and R. Chalmers. 2004. DNA looping and catalysis; the IHF-folded arm of Tn10 promotes conformational changes and hairpin resolution. Mol. Cell 13:537-547. [DOI] [PubMed] [Google Scholar]
- 8.Dawson, A., and D. J. Finnegan. 2003. Excision of the Drosophila mariner transposon mos1. Comparison with bacterial transposition and v(d)j recombination. Mol. Cell 11:225-235. [DOI] [PubMed] [Google Scholar]
- 9.Dekker, J., K. Rippe, M. Dekker, and N. Kleckner. 2002. Capturing chromosome conformation. Science 295:1306-1311. [DOI] [PubMed] [Google Scholar]
- 10.Derbyshire, V., N. D. Grindley, and C. M. Joyce. 1991. The 3′-5′ exonuclease of DNA polymerase I of Escherichia coli: contribution of each amino acid at the active site to the reaction. EMBO J. 10:17-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feschotte, C., N. Jiang, and S. R. Wessler. 2002. Plant transposable elements: where genetics meets genomics. Nat. Rev. Genet. 3:329-341. [DOI] [PubMed] [Google Scholar]
- 12.Goldgur, Y., F. Dyda, A. B. Hickman, T. M. Jenkins, R. Craigie, and D. R. Davies. 1998. Three new structures of the core domain of HIV-1 integrase: an active site that binds magnesium. Proc. Natl. Acad. Sci. USA 95:9150-9154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grimaud, C., F. Bantignies, M. Pal-Bhadra, P. Ghana, U. Bhadra, and G. Cavalli. 2006. RNAi components are required for nuclear clustering of Polycomb group response elements. Cell 124:957-971. [DOI] [PubMed] [Google Scholar]
- 14.Hiom, K., M. Melek, and M. Gellert. 1998. DNA transposition by the RAG1 and RAG2 proteins: a possible source of oncogenic translocations. Cell 94:463-470. [DOI] [PubMed] [Google Scholar]
- 15.Izsvak, Z., D. Khare, J. Behlke, U. Heinemann, R. H. Plasterk, and Z. Ivics. 2002. Involvement of a bifunctional, paired-like DNA-binding domain and a transpositional enhancer in Sleeping Beauty transposition. J. Biol. Chem. 277:34581-34588. [DOI] [PubMed] [Google Scholar]
- 16.Izsvak, Z., E. E. Stuwe, D. Fiedler, A. Katzer, P. A. Jeggo, and Z. Ivics. 2004. Healing the wounds inflicted by sleeping beauty transposition by double-strand break repair in mammalian somatic cells. Mol. Cell 13:279-290. [DOI] [PubMed] [Google Scholar]
- 17.Kipling, D., and P. E. Warburton. 1997. Centromeres, CENP-B and Tigger too. Trends Genet. 13:141-145. [DOI] [PubMed] [Google Scholar]
- 18.Lampe, D. J., M. E. Churchill, and H. M. Robertson. 1996. A purified mariner transposase is sufficient to mediate transposition in vitro. EMBO J. 15:5470-5479. [PMC free article] [PubMed] [Google Scholar]
- 19.Lander, E. S., et al. 2001. Initial sequencing and analysis of the human genome. Nature 409:860-921. [DOI] [PubMed] [Google Scholar]
- 20.Lee, S. H., M. Oshige, S. T. Durant, K. K. Rasila, E. A. Williamson, H. Ramsey, L. Kwan, J. A. Nickoloff, and R. Hromas. 2005. The SET domain protein Metnase mediates foreign DNA integration and links integration to nonhomologous end-joining repair. Proc. Natl. Acad. Sci. USA 102:18075-18080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leh, H., P. Brodin, J. Bischerour, E. Deprez, P. Tauc, J. C. Brochon, E. LeCam, D. Coulaud, C. Auclair, and J. F. Mouscadet. 2000. Determinants of Mg2+-dependent activities of recombinant human immunodeficiency virus type 1 integrase. Biochemistry 39:9285-9294. [DOI] [PubMed] [Google Scholar]
- 22.Ling, J. Q., T. Li, J. F. Hu, T. H. Vu, H. L. Chen, X. W. Qiu, A. M. Cherry, and A. R. Hoffman. 2006. CTCF mediates interchromosomal colocalization between Igf2/H19 and Wsb1/Nf1. Science 312:269-272. [DOI] [PubMed] [Google Scholar]
- 23.Lipkow, K., N. Buisine, and R. Chalmers. 2004. Promiscuous target interactions in the mariner transposon Himar1. J. Biol. Chem. 279:48569-48575. [DOI] [PubMed] [Google Scholar]
- 24.Lipkow, K., N. Buisine, D. J. Lampe, and R. Chalmers. 2004. Early intermediates of mariner transposition: catalysis without synapsis of the transposon ends suggests a novel architecture of the synaptic complex. Mol. Cell. Biol. 24:8301-8311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luo, G., Z. Ivics, Z. Izsvak, and A. Bradley. 1998. Chromosomal transposition of a Tc1/mariner-like element in mouse embryonic stem cells. Proc. Natl. Acad. Sci. USA 95:10769-10773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McBlane, J. F., D. C. van-Gent, D. A. Ramsden, C. Romeo, C. A. Cuomo, M. Gellert, and M. A. Oettinger. 1995. Cleavage at a V(D)J recombination signal requires only RAG1 and RAG2 proteins and occurs in two steps. Cell 83:387-395. [DOI] [PubMed] [Google Scholar]
- 27.Messier, T. L., J. P. O'Neill, S. M. Hou, J. A. Nicklas, and B. A. Finette. 2003. In vivo transposition mediated by V(D)J recombinase in human T lymphocytes. EMBO J. 22:1381-1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mizuuchi, K. 1992. Polynucleotidyl transfer reactions in transpositional DNA recombination. J. Biol. Chem. 267:21273-21276. [PubMed] [Google Scholar]
- 29.Moore, T., S. Hecquet, A. McLellann, D. Ville, D. Grid, F. Picard, B. Moulard, P. Asherson, A. J. Makoff, D. McCormick, L. Nashef, P. Froguel, A. Arzimanoglou, E. LeGuern, and B. Bailleul. 2001. Polymorphism analysis of JRK/JH8, the human homologue of mouse jerky, and description of a rare mutation in a case of CAE evolving to JME. Epilepsy Res. 46:157-167. [DOI] [PubMed] [Google Scholar]
- 30.Reiter, L. T., T. Murakami, T. Koeuth, L. Pentao, D. M. Muzny, R. A. Gibbs, and J. R. Lupski. 1996. A recombination hotspot responsible for two inherited peripheral neuropathies is located near a mariner transposon-like element. Nat. Genet. 12:288-297. [DOI] [PubMed] [Google Scholar]
- 31.Richardson, J. M., A. Dawson, N. O'Hagan, P. Taylor, D. J. Finnegan, and M. D. Walkinshaw. 2006. Mechanism of Mos1 transposition: insights from structural analysis. EMBO J. 25:1324-1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Robertson, H. M., and K. L. Zumpano. 1997. Molecular evolution of an ancient mariner transposon, Hsmar1, in the human genome. Gene 205:203-217. [DOI] [PubMed] [Google Scholar]
- 33.Spilianakis, C. G., M. D. Lalioti, T. Town, G. R. Lee, and R. A. Flavell. 2005. Interchromosomal associations between alternatively expressed loci. Nature 435:637-645. [DOI] [PubMed] [Google Scholar]
- 34.Tolhuis, B., R. J. Palstra, E. Splinter, F. Grosveld, and W. de Laat. 2002. Looping and interaction between hypersensitive sites in the active beta-globin locus. Mol. Cell 10:1453-1465. [DOI] [PubMed] [Google Scholar]
- 35.Travers, A., and G. Muskhelishvili. 1998. DNA microloops and microdomains: a general mechanism for transcription activation by torsional transmission. J. Mol. Biol. 279:1027-1043. [DOI] [PubMed] [Google Scholar]
- 36.Van Pouderoyen, G., R. F. Ketting, A. Perrakis, R. H. Plasterk, and T. K. Sixma. 1997. Crystal structure of the specific DNA-binding domain of Tc3 transposase of C. elegans in complex with transposon DNA. EMBO J. 16:6044-6054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Watkins, S., G. van Pouderoyen, and T. K. Sixma. 2004. Structural analysis of the bipartite DNA-binding domain of Tc3 transposase bound to transposon DNA. Nucleic Acids Res. 32:4306-4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang, W., J. Y. Lee, and M. Nowontny. 2006. Making and breaking nucleic acids: two-Mg2+-ion catalysis and substrate specificity. Mol. Cell 22:5-13. [DOI] [PubMed] [Google Scholar]
- 39.Zhang, L., A. Dawson, and D. J. Finnegan. 2001. DNA-binding activity and subunit interaction of the mariner transposase. Nucleic Acids Res. 29:3566-3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhu, H., J. Nandakumar, J. Aniukwu, L. K. Wang, M. S. Glickman, C. D. Lima, and S. Shuman. 2006. Atomic structure and nonhomologous end-joining function of the polymerase component of bacterial DNA ligase D. Proc. Natl. Acad. Sci. USA 103:1711-1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhu, H., and S. Shuman. 2005. Novel 3′-ribonuclease and 3′-phosphatase activities of the bacterial non-homologous end-joining protein, DNA ligase D. J. Biol. Chem. 280:25973-25981. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.