Abstract
ATF6 is a membrane-bound transcription factor activated by proteolysis in response to endoplasmic reticulum (ER) stress to induce the transcription of ER chaperone genes. We show here that, owing to the presence of intra- and intermolecular disulfide bridges formed between the two conserved cysteine residues in the luminal domain, ATF6 occurs in unstressed ER in monomer, dimer, and oligomer forms. Disulfide-bonded ATF6 is reduced upon treatment of cells with not only the reducing reagent dithiothreitol but also the glycosylation inhibitor tunicamycin, and the extent of reduction correlates with that of activation. Although reduction is not sufficient for activation, fractionation studies show that only reduced monomer ATF6 reaches the Golgi apparatus, where it is cleaved by the sequential action of the two proteases S1P and S2P. Reduced monomer ATF6 is found to be a better substrate than disulfide-bonded forms for S1P. ER stress-induced reduction is specific to ATF6 as the oligomeric status of a second ER membrane-bound transcription factor, LZIP/Luman, is not changed upon tunicamycin treatment and LZIP/Luman is well cleaved by S1P in the absence of ER stress. This mechanism ensures the strictness of regulation, in that the cell can only process ATF6 which has experienced the changes in the ER.
Protein unfolding or misfolding causes serious problems to the cell and must therefore be immediately and appropriately countered by cellular activities. Unfolded proteins accumulated in the endoplasmic reticulum (ER) under ER stress activate a series of homeostatic responses collectively termed the unfolded protein response (UPR). The UPR consists of translational and transcriptional controls whose purposes are to decrease the burden on the ER and cope with the various consequences of ER stress, respectively (10, 16, 22, 29). The three types of UPR regulator identified to date are localized in the mammalian ER as transmembrane proteins which sense ER stress and transmit signals across the ER membrane. The first, the transmembrane protein kinase and endoribonuclease IRE1 (composed of ubiquitous α and gut-specific β), contributes to transcriptional control by initiating unconventional (frame switch) splicing of an mRNA encoding the basic leucine zipper (bZIP) type transcription factor XBP1, which activates transcription of a number of genes involved in the homeostasis of the ER (15, 21, 34).
The second, transmembrane protein kinase PERK, is responsible for translational control; activated PERK attenuates translation generally by phosphorylating the α subunit of eukaryotic initiation factor 2 (32). The luminal domains of IRE1 and PERK are to some extent similar, and both are thought to be maintained under normal conditions in an inactive state via binding to BiP, a major molecular chaperone in the ER. ER stress-induced release of BiP allows oligomerization and autophosphorylation of IRE1 or PERK, leading to activation of the respective downstream event (2, 28). Dissection studies of Saccharomyces cerevisiae Ire1p, however, have suggested that the role of BiP is not so simple (17) (see Discussion). Furthermore, a completely different model was presented recently based on the crystal structure, in which the luminal domain of yeast Ire1p binds directly to unfolded proteins to be oligomerized (6). Unfortunately, however, this model was not supported by the more recently reported crystal structure of the luminal domain of human IRE1 (48). Thus, clarification of the ER stress-sensing mechanism remains an important and challenging issue in this field.
The third molecule is the ER membrane-bound bZIP transcription factor ATF6, which is composed of both ubiquitous α and β. ATF6 is constitutively synthesized as a precursor form designated pATF6(P), a type II transmembrane protein with its bZIP domain facing the cytoplasm (12, 13). Upon ER stress, ATF6 is transported from the ER to the Golgi apparatus to be cleaved by the sequential actions of site 1 protease (S1P) and site 2 protease (S2P) (5, 26, 44), resulting in the production of a soluble and active transcription factor designated pATF6(N). pATF6(N) enters the nucleus where it mainly activates the transcription of genes encoding ER-localized molecular chaperones and folding enzymes (27, 45, 46). Interestingly, it has been shown that the binding and dissociation of BiP is also critical to the activation of ATF6 (36). The dissociation of BiP from ATF6 appears to be actively triggered upon ER stress rather than resulting from any simple competition between ATF6 and unfolded proteins for binding to BiP (38).
The results suggest a common control mechanism for all three regulators of UPR originally; however, the activation mechanism of ATF6 has been shown to include at least two steps additional to those for IRE1 or PERK. First, ATF6 must be discriminated from unfolded proteins in the ER to allow it to be packed into a COPII vesicle for transport from the ER to the Golgi apparatus (23). Second, ATF6 must be recognized as a substrate of S1P and S2P in the Golgi apparatus (44). In this report, we demonstrate that the luminal domain of ATF6 is disulfide bonded and that ER stress-induced reduction is involved in both of these two steps. These findings provide a novel insight into the regulatory mechanism of the UPR.
MATERIALS AND METHODS
Construction of plasmids.
Recombinant DNA techniques were performed according to standard procedures (33). The integrity of all constructed plasmids was confirmed by extensive sequencing analysis.
pCMVshort-EGFP-ATF6α(WT), to drive expression of human ATF6α (amino acids 6 to 670) fused to the C terminus of green fluorescent protein (GFP) from the shortened cytomegalovirus (CMV) promoter, was constructed previously (23). pCMVshort-EGFP-ATF6α(C467A), pCMVshort-EGFP-ATF6α(C618A), and pCMVshort-EGFP-ATF6α(C467&618A) were generated by mutagenizing pCMVshort-EGFP-ATF6α(WT) using the QuikChange site-directed mutagenesis kit (Stratagene). pCGN-HA-ATF6α to express full-length ATF6α tagged with the hemagglutinin (HA) epitope at the N terminus (43) was the kind gift of R. Prywes (Columbia University, New York, NY). pCGN-HA-ATF6α(C467A), pCGN-HA-ATF6α(C618A), and pCGN-HA-ATF6α(C467&618A) were created similarly to enhanced GFP (EGFP)-ATF6α mutants.
pcDNA-S1P(ΔTMD)-myc-KDEL, to express human S1P/subtilisin kexin isozyme 1 (amino acids 1 to 997) lacking C-terminal transmembrane and tail regions but carrying instead the myc epitope tag and the KDEL sequence at the C terminus, was constructed previously (26). pECFP-N1-tsVSVG, to express the temperature-sensitive mutant (ts045) of vesicular stomatitis virus G protein (VSV-G) fused to a cyan-emitting variant of GFP (CFP), was constructed previously (23). pcDNA-LZIP-V5-His, to express LZIP tagged with the V5 epitope at the C terminus, was kindly provided by D. Y. Jin (The University of Hong Kong, China).
Cell culture and transfection.
CHO cells were grown in a 1:1 mixture of Ham's F-12 and Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum, 2 mM glutamine, and antibiotics (100 U/ml penicillin and 100 μg/ml streptomycin sulfate) in a 5% CO2, 95% air incubator at 37°C. Cells were transfected with plasmid DNA as described previously (23) using Superfect (QIAGEN) basically according to the manufacturer's instructions and then incubated at 37°C for an appropriate time for expression of the transfected gene.
Treatment with thiol-reactive probes.
CHO cells were washed and harvested in phosphate-buffered saline (PBS) containing or not containing 10 mM N-ethylmaleimide (NEM). Cell pellets were incubated with 200 μl of 10% ice-cold trichloroacetic acid (TCA) for 30 min on ice, followed by centrifugation at 4°C for 30 min at 14,000 × g. Precipitated proteins were washed twice with 10% TCA and then washed twice with acetone. Air-dried precipitates were dissolved in 100 μl of reduction buffer (100 mM Tris-HCl, pH 9.5, containing 10 mM dithiothreitol [DTT], 10 mM EDTA, and 0.5% sodium dodecyl sulfate [SDS]). After incubation at 44°C for 1 h, 20 μl of the sample was incubated with or without 4 μl of 100 mM 4-acetamido-4′-maleimidylstilbene-2,2′-disulfonate (AMS) (Molecular Probes) for 90 min at room temperature in the dark. The reaction was stopped by the addition of an equal volume of 2× Laemmli's sample buffer and boiling in the absence of reducing agents.
Metabolic labeling.
CHO cells were incubated for 20 min in l-methionine- and l-cysteine-free Dulbecco's modified Eagle's medium supplemented with 2 mM glutamine, antibiotics, and 10% dialyzed fetal calf serum. Cells were then labeled for 1 h with 0.25 mCi (9.25 MBq)/plate EXPRE35S35S protein labeling mix (PerkinElmer) dissolved in 1 ml of the above medium. Cells expressing HA-tagged ATF6α(WT) or ATF6α(C618A) by transfection were starved for 30 min and then labeled for 1 h with 1 ml/plate of 0.19 mCi (7 MBq) EXPRE35S35S protein labeling mix in the above medium. Alternatively, cells were incubated for 30 min in the above medium containing 1/10 the normal concentrations of l-methionine and l-cysteine and then labeled for 4 h with 1 ml/plate of 0.25 mCi (9.25 MBq) EXPRE35S35S protein labeling mix dissolved in the above medium containing 1/10 the normal concentrations of l-methionine and l-cysteine. Four hours later, the medium was replaced with 1 ml/plate of 0.25 mCi (9.25 MBq) EXPRE35S35S protein labeling mix dissolved in the above medium containing 1/10 the normal concentrations of l-methionine and l-cysteine. Labeled cells were washed with ice-cold PBS, harvested, and suspended in 0.1 ml of 1% SDS lysis buffer (20 mM Tris-HCl, pH 7.4, containing 1% SDS, protease inhibitor cocktail [Nacalai Tesque], 10 μM MG132, and 10 mM NEM). Immunoprecipitation was carried out as described previously (13).
Immunological techniques.
Immunoblotting analysis was carried out according to the standard procedure (33) as described previously (27) using Western blotting luminol reagent (Santa Cruz Biotechnology). Chemiluminescence was detected using an LAS-1000 plus LuminoImage analyzer (Fuji Film). ATF6α and ATF6β were detected with rabbit anti-ATF6α (13) and anti-ATF6β (12) polyclonal antibodies, respectively. Rabbit anti-myc epitope polyclonal antibody and anti-V5 epitope polyclonal antibody were obtained from Medical and Biological Laboratories (MBL). Rabbit anti-HA epitope polyclonal antibody (Y-11) and goat anti-ribophorin I polyclonal antibody (C-15) were purchased from Santa Cruz. Mouse anti-GM130 monoclonal antibody and rabbit anti-P-PERK polyclonal antibody were obtained from BD Transduction Laboratories and Cell Signaling Technology, respectively.
Immunoprecipitation and indirect immunofluorescence were carried out essentially as described previously (23). Anti-GFP monoclonal antibody (mixture of clone 7.1 and 13.1) was purchased from Roche Molecular Biochemicals. Mouse anti-myc monoclonal antibody (clone 9E10) was purchased from Santa Cruz.
Confocal microscopy.
GFP in CHO cells cultured in 35-mm glass-base dishes was visualized using a Leica TCS SP2 confocal microscope system (400-mW Ar/Kr 457- to 675-nm laser, Leica DM IRE2 fluorescence microscope, 63× 1.40- to 0.60-numerical aperture HCXPLAPO oil objective).
Discontinuous sucrose gradient centrifugation.
CHO cells unstressed or ER stressed were scraped after being washed once with 1 ml of ice-cold PBS containing protease inhibitor cocktail (Nacalai Tesque) and 10 mM MG132. Cells from 8 dishes were pooled, suspended into 200 μl of ice-cold sucrose buffer (10 mM HEPES/KOH, pH 7.4, containing 0.25 M sucrose, 1 mM EDTA, protease inhibitor cocktail, and 10 mM NEM), and homogenized with 30 strokes using a Dounce tissue homogenizer (1 ml size; Wheaton). After the addition of 200 μl of ice-cold sucrose buffer, cells were further homogenized with 20 strokes and then centrifuged at 1,000 × g for 5 min at 4°C. Two hundred microliter aliquots of the resulting supernatant were laid on the top of a discontinuous sucrose gradient, consisting of five layers of 200 μl each with distinct sucrose concentrations (20%, 30%, 40%, 50%, and 60% from top to bottom), and centrifuged at 4°C at 50,000 rpm for 30 min in a S55S-1096 rotor (Hitachi) using a Himac CS 150 GXL microultracentrifuge (Hitachi), with deceleration performed without braking. Six fractions of 200 μl each were then collected from the top.
RESULTS
ATF6 is disulfide bonded in the luminal domain.
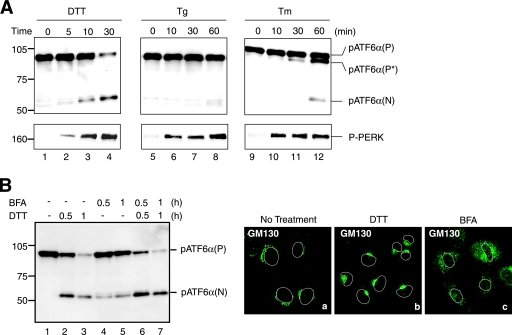
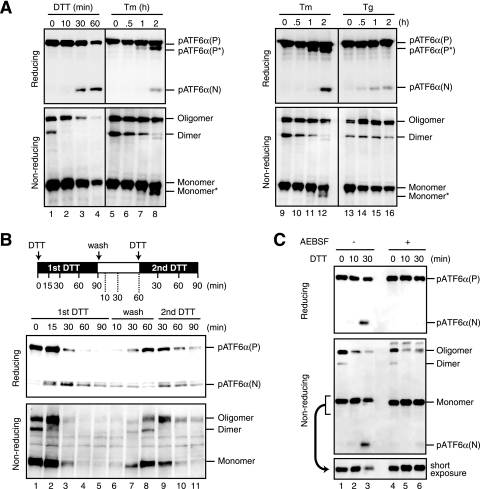
We previously found that the extent of ER stress-induced cleavage of ATF6 varied depending on inducers added to HeLa cells, with cleavage much more extensive in cells treated with DTT (a reducing reagent) than in those treated with tunicamycin (Tm, an inhibitor of protein N-glycosylation) or thapsigargin (Tg, an inhibitor of Ca2+-ATPase in the ER) (12, 13). These observations were well reproduced in Chinese hamster ovary (CHO) cells employed for the analyses in the present study, except that Tg was unexpectedly found to be a weaker inducer of ATF6 cleavage than Tm in these cells (Fig. 1A, upper panel), unlike their similar effects in HeLa cells. In contrast, PERK was activated similarly in response to treatment with any of the three reagents (Fig. 1A, lower panel), indicating a potential difference in activation mechanisms between ATF6 and PERK.
FIG. 1.
Effects of various ER stress inducers on activation of ATF6. (A) CHO cells were treated with 1 mM DTT, 600 nM Tg, or 4 μg/ml Tm for the indicated periods, and then cell lysates were prepared and analyzed by immunoblotting using anti-ATF6α or anti-phosphorylated PERK (P-PERK) antibodies. Migration positions of pATF6α(P), pATF6α(N), and P-PERK as well as full-range rainbow molecular weight markers (GE Healthcare) are indicated. pATF6α(P*) denotes the nonglycosylated form of pATF6α(P). (B) CHO cells were untreated (lane 1) or treated with 1 mM DTT alone (lanes 2 and 3), 5 μg/ml BFA alone (lanes 4 and 5), or 1 mM DTT and 5 μg/ml BFA simultaneously (lanes 6 and 7) for the indicated periods, and then cell lysates were prepared and analyzed by immunoblotting using anti-ATF6α antibody (left). CHO cells treated with 1 mM DTT or 5 μg/ml BFA for 1 h were analyzed by immunofluorescence using anti-GM130 antibody (right). An outline of the nucleus is indicated by a white line in each cell.
We also found that brefeldin A (BFA), occasionally used as an ER stress inducer in the literature (19), was weak in activating ATF6 (Fig. 1B, left panel). BFA is known to block anterograde transport from the ER to the Golgi apparatus without affecting retrograde transport from the Golgi apparatus to the ER (18), an effect which may result in the accumulation of unfolded proteins in the ER. In addition, S1P and S2P, responsible for the ER stress-induced cleavage of ATF6, became colocalized with ATF6 in BFA-treated cells, as BFA treatment caused a merging of the Golgi apparatus into the ER (Fig. 1B, right panels) (GM130 is a marker for the Golgi apparatus) (35). pATF6α(P) was indeed cleaved to produce pATF6α(N) in cells treated with BFA (Fig. 1B, left panel, lanes 4 and 5); however, the extent of cleavage was markedly less than that in cells treated with DTT (lanes 2 and 3). Here, it is the levels of pATF6α(P) which should be compared because cleaved products are highly unstable, and thus, the level of pATF6α(N) is often less quantitative. Double treatment of cells with BFA and DTT showed additive effects on the cleavage (lanes 6 and 7), suggesting that DTT treatment has stimulatory effects on the cleavage of ATF6 by S1P and S2P.
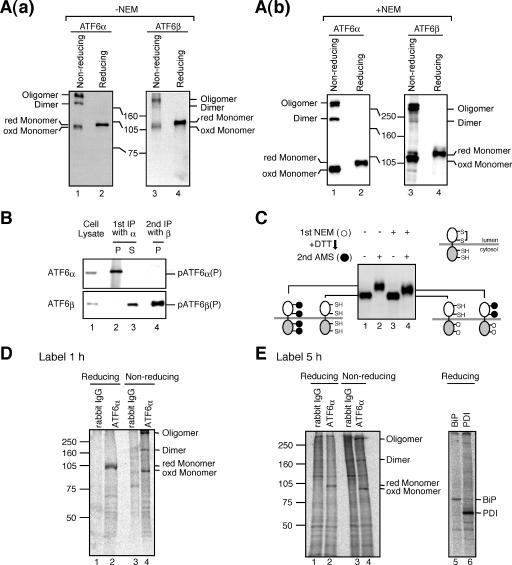
The above two results prompted us to look into the luminal domain of ATF6, particularly with regard to the presence of DTT-sensitive disulfide bridges. Alignment of ATF6α and ATF6β revealed significant amino acid sequence homology (45% identity) in their luminal domains, which were shown to contain two cysteine residues at similar positions (Fig. 2). These cysteines are well conserved between humans and mice for both ATF6α and ATF6β (Fig. 2). Caenorhabditis elegans possesses a single ATF6 gene (39), and the luminal domain of worm ATF6 also contains two cysteine residues at similar positions, although sequences around them are not well conserved (Fig. 2). Conservation during evolution suggests the importance of these two cysteine residues to the function or regulation of ATF6. We therefore examined whether ATF6 is disulfide-bonded. Laemmli's SDS sample buffer without reducing agent was added directly to unstressed CHO cells that had been scraped in PBS and recovered by centrifugation, followed by boiling for 10 min. When an aliquot of these cell lysates was boiled again for 5 min after the addition of β-mercaptoethanol (5% vol/vol) and subjected to SDS-polyacrylamide gel electrophoresis (PAGE) (reducing conditions), ATF6α and ATF6β were detected as a single band at the molecular mass bands of 90 kDa and 110 kDa, respectively [Fig. 3A (a), lanes 2 and 4, respectively], as we reported previously (12, 13). When an aliquot of cell lysates was boiled for 5 min in the absence of β-mercaptoethanol and subjected to SDS-PAGE (nonreducing conditions), however, both ATF6α and ATF6β were detected as three bands [Fig. 3A(a), lanes 1 and 3, respectively], which were assigned as monomer, dimer, and oligomer forms on the basis of the results of mutational analysis (see Fig. 7). The putative monomeric form of ATF6α or ATF6β obtained under nonreducing conditions migrated faster than the respective reduced form of ATF6α (compare lane 1 with lane 2) or ATF6β (compare lane 3 with lane 4), suggesting that even monomer ATF6α and ATF6β are disulfide bonded intramolecularly. Given the possibility that the observed disulfide bond formation may have represented a postlysis artifact, we repeated the experiments by including 10 mM NEM, a sulfhydryl alkylating reagent, in PBS and Laemmli's SDS sample buffer to harvest and lyse CHO cells, respectively, to quench free cysteines. Although the monomer tended to be higher with than without NEM treatment, three bands were always obtained, and the putative monomeric form migrated faster than the respective reduced form [Fig. 3A(b)]. We also noticed that the relative ratio of these three forms varied between experiments, which might reflect their dynamic status: all had short half-lives and were interchangeable (Fig. 4). These results indicated that endogenous ATF6α and ATF6β are indeed disulfide bonded.
FIG. 2.
Alignment of the transmembrane and luminal domains of human and mouse ATF6α and ATF6β as well as worm ATF6. Amino acid sequences of the transmembrane (TM, red square) and luminal domains of human (hs) and mouse (ms) ATF6α and ATF6β as well as C. elegans (ce) ATF6 are aligned. Amino acids identical to those of ATF6α are marked by white letters in black boxes, whereas amino acids identical to those of ATF6β are shaded with gray. The two highly conserved cysteine residues are highlighted by red boxes. Asparagine residues marked by red and asterisks denote putative N-glycosylation sites.
FIG. 3.
Presence of disulfide bridges in ATF6. (A) (a) CHO cells cultured in 60-mm dishes were harvested, lysed in 100 μl of 2× Laemmli's SDS sample buffer without reducing reagent, and boiled for 10 min. After boiling again for 5 min in the presence (reducing) or absence (nonreducing) of 5% (vol/vol) 2-mercaptoethanol, 10-μl aliquots of each sample were subjected to SDS-PAGE and analyzed by immunoblotting with anti-ATF6α (left panel) or anti-ATF6β (right panel) antibodies. Migration positions of the oxidized (oxd) and reduced (red) monomers, dimer, and oligomer are indicated. (b) Analysis was done as for panel a, except that PBS and Laemmli's SDS sample buffer used to harvest and lyse CHO cells, respectively, contained 10 mM NEM. (B) CHO cells were lysed in 1% octylglucopyranoside lysis buffer and immunoprecipitated first with anti-ATF6α antibody (1st IP with α). Proteins remaining in the supernatant were then immunoprecipitated with anti-ATF6β antibody (2nd IP with β). Proteins recovered in the precipitates (P) and supernatant (S) as well as cell lysates were analyzed by immunoblotting under reducing conditions using anti-ATF6α (upper panel) or anti-ATF6β (lower panel) antibodies. (C) CHO cells were washed and harvested with PBS containing or not containing 10 mM NEM [1st NEM (+) or (−), respectively]. Cellular proteins were precipitated with TCA. Precipitated proteins were reduced with 10 mM DTT, followed by incubation with or without an excess amount of AMS [2nd AMS (+) or (−), respectively]. Proteins were then subjected to SDS-PAGE under reducing conditions and analyzed by immunoblotting using anti-ATF6α antibody. Cysteines with free SH or disulfide bonded as well as those covalently modified with NEM (○) or AMS (•) are illustrated. (D) CHO cells pulse-labeled for 1 h with [35S]methionine and cysteine were lysed in 1% SDS lysis buffer, boiled for 5 min, and then after dilution with 2% Triton X-100 buffer, immunoprecipitated with control rabbit IgG or anti-ATF6α antibody. All buffer contained 10 mM NEM. The immunoprecipitates were subjected to SDS-PAGE under reducing and nonreducing conditions. (E) CHO cells labeled for 5 h with [35S]methionine and cysteine (radioisotope was added to the culture at 5 and 1 h prior to harvest) were lysed in 1% SDS lysis buffer, boiled for 5 min, diluted with 2% Triton X-100 buffer, and then split into two halves. All buffer contained 10 mM NEM. The first half was subjected to immunoprecipitation with control rabbit IgG or anti-ATF6α antibody. The immunoprecipitates were analyzed by SDS-PAGE under reducing and nonreducing conditions (left panel). The second half was subjected to immunoprecipitation with anti-BiP or anti-protein disulfide isomerase (PDI) antibody to show that proteins with long half-lives were also labeled during the 5-h period. The immunoprecipitates were analyzed by SDS-PAGE under reducing conditions (right panel). Note that neither BiP nor PDI was coimmunoprecipitated with ATF6α under these condition (lane 2) even though they were metabolically labeled (lane 5 or 6).
FIG. 7.
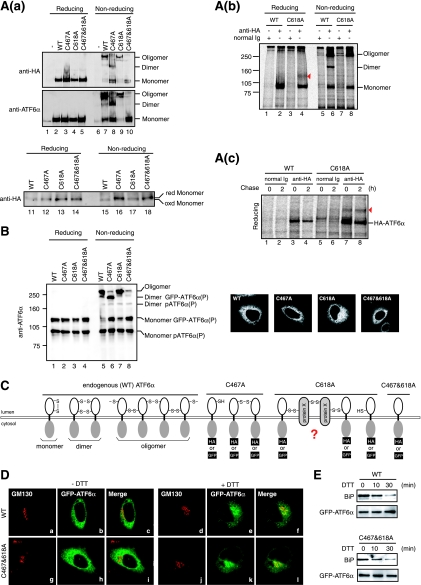
Effects of Cys-to-Ala mutations on oligomeric status and localization of ATF6. (A) (a) CHO cells were transfected with pCGN-ATF6α(WT), pCGN-ATF6α(C467A), pCGN-ATF6α(C618A), or pCGN-ATF6α(C467&618A) to overexpress WT or mutant versions of full-length ATF6α tagged with the HA epitope at the N terminus. Cell lysates were prepared and analyzed as described for Fig. 3A(a) (without NEM) using anti-HA (upper panel) or anti-ATF6α (lower panel) antibody. The bottom panel shows the difference in the migration positions of monomer forms derived from WT and mutant versions of HA-tagged ATF6α after longer electrophoresis. (b) CHO cells expressing pCGN-ATF6α(WT) or pCGN-ATF6α(C618A) by transfection were pulse-labeled for 1 h with [35S]methionine and cysteine, lysed in 1% SDS lysis buffer, boiled for 5 min, and then immunoprecipitated with control rabbit IgG or anti-HA antibody as described for Fig. 3D. All buffer contained 10 mM NEM. The immunoprecipitates were subjected to SDS-PAGE under reducing and nonreducing conditions. The red arrowhead marks protein X, which was disulfide bonded to ATF6α(C618A). (c) CHO cells expressing pCGN-ATF6α(WT) or pCGN-ATF6α(C618A) by transfection were pulse-labeled for 1 h with [35S]methionine and cysteine, chased for the indicated periods, and analyzed as described for panel b. The immunoprecipitates were subjected to SDS-PAGE under reducing and nonreducing conditions. The red arrowhead marks protein X, which was disulfide bonded to ATF6α(C618A). (c) CHO cells expressing pCGN-ATF6α(WT) or pCGN-ATF6α(C618A) by transfection were pulse-labeled for 1 h with [35S]methionine and cysteine, chased for the indicated periods, and analyzed as described for panel b. The immunoprecipitates were subjected to SDS-PAGE under reducing conditions. Migration positions of HA-ATF6α and protein X are indicated. (B) CHO cells were transfected with pCMVshort-EGFP-ATF6α(WT), pCMVshort-EGFP-ATF6α(C467A), pCMVshort-EGFP-ATF6α(C618A), and pCMVshort-EGFP-ATF6α(C467&618A) to express WT or mutant versions of GFP-ATF6α fusion protein at physiological levels. Cell lysates were prepared and analyzed as described for Fig. 3A(b) (with NEM) using anti-ATF6α antibody. Transfected cells were also analyzed by fluorescence microscopy to visualize the localization of each GFP-ATF6α fusion protein (panels a to d). (C) Redox and oligomeric status of endogenous ATF6α as well as the WT and mutant (C467A, C618A, and C467&618A) ATF6α are schematically shown. (D) CHO cells transfected with pCMVshort-EGFP-ATF6α(WT) or pCMVshort-EGFP-ATF6α(C467&618A) were treated with (+) or without (−) 1 mM DTT for 30 min and then fixed, immunostained with anti-GM130 antibody, and analyzed by fluorescence microscopy. (E) CHO cells transfected with pCMVshort-EGFP-ATF6α(WT) or pCMVshort-EGFP-ATF6α(C467&618A) were treated with 1 mM DTT for the indicated periods, then lysed with 1% Triton X-100 lysis buffer, and immunoprecipitated with anti-GFP antibody. The precipitates were analyzed by immunoblotting using anti-BiP or anti-ATF6α antibodies.
FIG. 4.
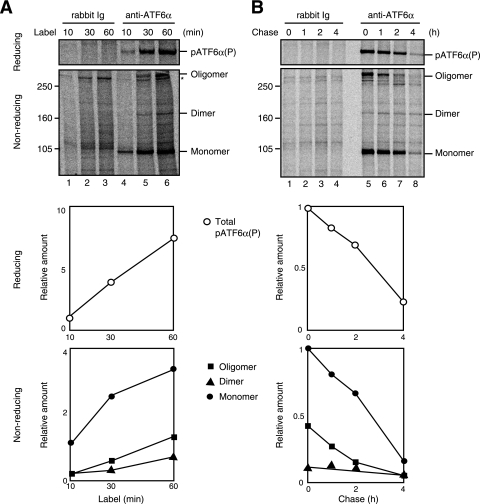
Synthesis and degradation of various forms of ATF6. (A) CHO cells were labeled with [35S]methionine and cysteine for the indicated periods, followed by cell lysis and immunoprecipitation with control rabbit IgG or anti-ATF6α antibody as described for Fig. 3D. The immunoprecipitates were subjected to SDS-PAGE under reducing and nonreducing conditions. (Bottom panels) The radioactive intensity of each band was determined and is presented against labeling period after normalization to the value of pATF6α(P) (reducing) or monomer ATF6α (nonreducing). (B) CHO cells pulsed labeled with [35S]methionine and cysteine for 1 h and then chased for the indicated periods were analyzed as described for panel A.
Sequential immunoprecipitation was carried out to determine whether ATF6α and ATF6β are homo- or hetero-oligomerized. Unstressed cells were lysed with 1% octylglucopyranoside lysis buffer and subjected to first immunoprecipitation with anti-ATF6α antibody, and then both precipitate and supernatant were immunoblotted with anti-ATF6α and -ATF6β antibodies. As shown in Fig. 3B, all of the pATF6α(P) was immunoprecipitated with anti-ATF6α antibody, whereas pATF6β(P) was not immunoprecipitated with it at all (compare lane 2 with lane 3). pATF6β(P) remaining in the supernatant was successfully immunoprecipitated with anti-ATF6β antibody (lane 4). Thus, ATF6 is disulfide bonded homogeneously.
To substantiate disulfide bond formation in ATF6 unambiguously, we determined the accessibility of ATF6α to the thiol-reactive probe 4-acetamido-4′-maleimidylstilbene-2,2′-disulfonate (AMS; molecular weight, 536). ATF6α contains four cysteine residues, two in the luminal domain and two in the cytosolic domain, as depicted in Fig. 3C. Unstressed cells were harvested in PBS not containing 10 mM NEM [1st NEM (−)], and then cellular proteins were precipitated with TCA. Precipitated proteins were reduced with 10 mM DTT such that all four cysteine residues in ATF6α became accessible to AMS, followed by incubation with or without an excessive amount of AMS [2nd AMS (+) or (-), respectively]. ATF6α with 1st NEM (−) and 2nd AMS (−) (lane 1), in which four cysteine were reduced, migrated faster than ATF6α with 1st NEM (−) and 2nd AMS (+) (lane 2), in which four cysteines were covalently modified with AMS, as expected. Unstressed cells were also harvested in PBS containing 10 mM NEM [1st NEM (+)] to covalently modify preexisting free thiols. Cellular proteins were precipitated with TCA to remove unreacted NEM. Precipitated proteins were then reduced with 10 mM DTT, followed by incubation with or without an excessive amount of AMS [2nd AMS (+) or (−), respectively]. Importantly, ATF6α with 1st NEM (+) and 2nd AMS (+) (lane 4) migrated between ATF6α with 1st NEM (−) and 2nd AMS (−) (lane 1) and ATF6α with 1st NEM (−) and 2nd AMS (+) (lane 2), indicating that only two cysteine residues were modified with AMS when cells were pretreated with NEM. This means that two cysteine residues in the cytosolic domain of ATF6α were reduced in unstressed cells and thus easily modified by the 1st NEM treatment, whereas two cysteine residues in the luminal domain of ATF6α were disulfide bonded and protected from the 1st NEM attack and became accessible only after reduction with DTT. ATF6α with the 1st NEM (+) and 2nd AMS (−) (lane 3) migrated to a position similar to that of ATF6α with 1st NEM (−) and 2nd AMS (−) (lane 1) even though two cysteine residues in the cytosolic domain were thought to have been covalently modified with NEM, which was explained by the smallness of the NEM molecule (molecular weight 125) compared with AMS. These results demonstrate the presence of a disulfide bond in ATF6α.
Immunoprecipitation with anti-ATF6α antibody was performed in lysates of 35S-labeled cells to determine whether ATF6α is disulfide bonded to other cellular proteins. As shown in Fig. 3D, one and three bands were obtained under reducing and nonreducing conditions, respectively (lanes 2 and 4), from unstressed cells which had been labeled with [35S]methionine and cysteine for 1 h and then lysed with 1% SDS lysis buffer, followed by boiling for 5 min and dilution with 2% Triton X-100 buffer. The labeling period was further extended to 5 h, and the radiolabeled isotope was added to the culture at 5 h and 1 h before harvest to efficiently label both proteins with short half-lives, such as ATF6α (Fig. 4B), and those with long half-lives, such as BiP and protein disulfide isomerase (Fig. 3E, lanes 5 and 6). Again, the only bands obtained specifically with anti-ATF6α antibody were ATF6α compared with those obtained with rabbit normal immunoglobulin G (IgG) (Fig. 3E, lanes 1 to 4). It should be noted that lysis with 1% SDS lysis buffer followed by boiling eliminated the coimmunoprecipitation of BiP with ATF6α, which was observed when cells were lysed with 1% Triton X-100 lysis buffer (see Fig. 7E). We also noticed that inclusion of 10 mM NEM in lysis and dilution buffers as well as incubation and wash buffers for immunoprecipitation was essential in this experiment to obtain monomer, dimer, and oligomer forms of ATF6α after SDS-PAGE under nonreducing conditions. Without NEM, most of ATF6α was converted to oligomer during the procedure, which required many hours in a low-SDS concentration environment. As mentioned above, the cytoplasmic region of ATF6α also contains two cysteines. These results clearly indicate that ATF6α is disulfide bonded homogeneously.
To characterize the nature of the monomer, dimer, and oligomer forms of ATF6α, we labeled cells with [35S]methionine and cysteine for increasing periods from 10 min to 60 min, followed by cell lysis and immunoprecipitation with anti-ATF6α antibody (Fig. 4A). SDS-PAGE analysis under reducing conditions showed that the amount of 35S incorporated into ATF6α increased linearly with time (upper panel, lanes 4 to 6; see also figure at bottom for quantitative data). SDS-PAGE analysis under nonreducing conditions revealed that only the monomer was labeled during the initial 10 min (lower panel, lane 4) and then the dimer and oligomer were labeled from 30 min (lower panel, lanes 5 and 6), indicating that newly synthesized monomer is converted to dimer and oligomer. We then conducted a pulse-chase experiment (Fig. 4B). SDS-PAGE analysis under reducing conditions showed that the half-life of ATF6α was <3 h in CHO cells (upper panel, lanes 5 to 8; see also figure at bottom for quantitative data), with this value being comparable to the approximately 2 h determined with HeLa cells (13). SDS-PAGE analysis under nonreducing conditions revealed that the half-life of the monomer was <3 h and that the oligomer had a half-life of <2 h (lower panel, lanes 5 to 8), indicating that oligomerized ATF6α is not a dead-end product formed during the biosynthesis of ATF6α. Dimer ATF6α remained relatively constant during the chase periods (lower panel, lanes 5 to 8), probably because the oligomer was converted to the dimer and then monomer during degradation, and therefore, the dimer was in equilibrium between a decrease (conversion of dimer to monomer) and increase (conversion of oligomer to dimer). These results support the notion that all forms of ATF6α are highly dynamic.
ATF6 is reduced in response to ER stress, and reduced monomer ATF6 is transported to the Golgi apparatus.
We next investigated the changes in oligomeric status of ATF6α in cells treated with various ER stress inducers. In DTT-treated cells, dimer ATF6α disappeared 10 min after the addition of DTT (Fig. 5A, lanes 1 and 2), followed from 30 min after addition by the disappearance of oligomer ATF6α and decrease in monomer (lanes 3 and 4), producing pATF6α(N). We noticed that the extent of DTT-induced ATF6α cleavage varied depending on experiments, possibly reflecting a difference in the confluence of cell cultures. In the results shown in Fig. 5B, dimer ATF6α disappeared 15 min after the addition of DTT, producing pATF6α(N) (Fig. 5B, lanes 1 and 2), while oligomer and monomer ATF6α disappeared from 30 min after addition (lanes 3 to 5), resulting in a severe decrease in the level of pATF6α(P) and the detection of higher levels of pATF6α(N) [as mentioned above, the summation of the levels of pATF6α(P) and pATF6α(N) was not constant due to the rapid degradation of pATF6α(N)]. When DTT was washed out, all forms of ATF6α were restored within 60 min (lanes 6 to 8). In response to a second DTT treatment, the immediate disappearance of dimer ATF6α (lane 9), followed by that of oligomer ATF6α and monomer ATF6α, was again observed (lanes 10 and 11), revealing that the oligomeric status of ATF6 is highly dynamic and that all forms can be activated when cells are treated with reducing reagent.
FIG. 5.
Effects of ER stress inducers on oligomeric status of ATF6. (A) CHO cells were treated with 1 mM DTT, 2 μg/ml Tm, or 1 μM Tg for the indicated periods, and then cell lysates were prepared and analyzed as described for Fig. 3A(b) (with NEM). Monomer* denotes the nonglycosylated form of monomer. The same set of data for Tm treatment is shown in the left and right panels with different exposure times to emphasize the difference in the extent of ATF6α cleavage between DTT and Tg. (B) CHO cells were treated with 1 mM DTT for the indicated periods (1st DTT, lanes 1 to 5). Cells treated with 1 mM DTT for 90 min were washed and then incubated in the absence of DTT for the indicated periods (wash, lanes 6 to 8). Cells incubated without DTT for 60 min were treated again with 1 mM DTT for the indicated periods (2nd DTT, lanes 9 to 11). Cell lysates were prepared and analyzed as described for Fig. 3A(a) (without NEM). (C) CHO cells were pretreated with (+) or without (−) 300 μM AEBSF for 5 min and then treated with 1 mM DTT for the indicated periods without removing AEBSF. Cell lysates were prepared and analyzed as described for Fig. 3A(b) (with NEM). The bottom panel shows the result of shorter exposure of the region around the monomer.
Importantly, the disappearance of dimer ATF6α was also observed in cells treated with Tm. Further, monomer ATF6α was decreased when pATF6α(N) was detected (Fig. 5A, lanes 5 to 8 and lanes 9 to 12), whereas oligomer ATF6α was maintained at a constant level, probably because Tm is not a direct reducing reagent for ATF6α. Accordingly, the level of pATF6α(N) detected in Tm-treated cells was much lower than that in DTT-treated cells (compare lane 8 with lanes 3 and 4). In cells treated with Tg, the levels of dimer and oligomer ATF6α remained constant, but a decrease in monomer ATF6α was observed when pATF6α(N) was detected (Fig. 5A, lanes 13 to 16), probably because Tg is not a direct reducing reagent for ATF6α either. A very low level of pATF6α(N) produced in Tg-treated cells was consistent with the results shown in Fig. 1A. We previously showed that the serine protease inhibitor 4-(2-aminoethyl)benzenesulfonyl fluoride (AEBSF) was able to block ER stress-induced cleavage of ATF6α via inhibition of S1P (25). When CHO cells were pretreated with AEBSF and then treated with DTT without removal of the AEBSF, pATF6α(N) was not produced (Fig. 5C, lanes 4 to 6). Importantly, dimer pATF6α disappeared and oligomer ATF6α decreased, whereas monomer ATF6α was accumulated under these conditions. Thus, ATF6 changes its oligomeric status in response to ER stress in the direction of oligomer to monomer, and the extent of reduction is correlated with activation status.
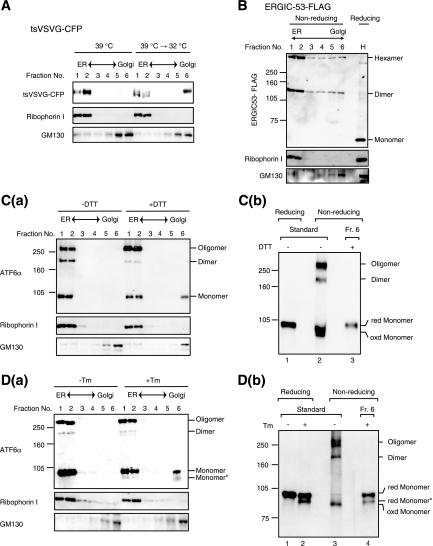
To determine the role of ER stress-induced reduction in the activation process of ATF6, we fractionated cell lysates in a sucrose density gradient, as ATF6 is transported from the ER to the Golgi apparatus when activated. As a control, we used the temperature-sensitive mutant (ts045) of VSV-G (9) fused to CFP (the resulting fusion protein is referred to as tsVSVG-CFP). At the nonpermissive temperature 39°C, tsVSVG-CFP was misfolded due to a point mutation in its luminal domain and was thus retained in the ER. Accordingly, tsVSVG-CFP was recovered upon fractionation in heavy fractions together with the ER marker ribophorin I (Fig. 6A, 39°C, fractions 1 and 2). In contrast, most of the tsVSVG-CFP was found in the lightest fraction together with the Golgi marker GM130 15 min after a temperature shift to the permissive temperature of 32°C (Fig. 6A, 39°C → 32°C, fraction 6), indicating that tsVSVG-CFP was properly folded and reached the Golgi apparatus. The band detected in fraction 6 migrated more slowly than that in fraction 1 or 2, reflecting the conversion of its carbohydrate moieties from an endo-β-N-acetylglucosaminidase H (endoH)-sensitive to an endoH-resistant form, which we confirmed experimentally (data not shown). Thus, proteins in the Golgi apparatus were discriminated from proteins in the ER after this fractionation procedure.
FIG. 6.
Effects of ER stress inducers on localization of ATF6. (A) CHO cells expressing tsVSVG-CFP by transfection were cultured at the nonpermissive temperature of 39°C and then shifted to the permissive temperature of 32°C for 0 min (39°C) and 15 min (39°C → 32°C). Cells were homogenized and fractionated through a sucrose gradient. Aliquots of the six resulting fractions were subjected to SDS-PAGE under reducing conditions and analyzed by immunoblotting with anti-GFP, anti-ribophorin I (ER marker), and anti-GM130 (Golgi marker) antibodies. (B) CHO cells expressing ERGIC-53 tagged with the FLAG epitope by transfection were fractionated as described for panel A. Aliquots of the six resulting fractions as well as the homogenate (H) were subjected to SDS-PAGE under reducing and nonreducing conditions as indicated and analyzed by immunoblotting with anti-FLAG antibody. Aliquots of each fraction were also analyzed under reducing conditions using anti-ribophorin I and anti-GM130 antibodies. (C) (a) CHO cells were treated with (+) or without (−) 1 mM DTT for 15 min and then fractionated as described for panel A. All buffer contained 10 mM NEM. Aliquots of the six resulting fractions were subjected to SDS-PAGE under nonreducing conditions and analyzed by immunoblotting with anti-ATF6α antibody. Aliquots of each fraction were also analyzed under reducing conditions using anti-ribophorin I and anti-GM130 antibodies. (b) An aliquot of fraction 6 of DTT-treated cells was applied to centrifugation-coupled membrane filter (molecular weight cut at 10 kDa, Amicon Ultra-4; Millipore) to replace buffer with 10 mM Tris-HCl buffer, pH 7.4, containing 1% SDS, and then subjected to SDS-PAGE under nonreducing conditions together with reduced or nonreduced lysates of unstressed cells. (D) (a) CHO cells were treated with (+) or without (−) 2 μg/ml Tm for 2 h and then fractionated as described for panel A. All buffer contained 10 mM NEM. Aliquots of the six resulting fractions were analyzed as described for panel C. (b) An aliquot of fraction 6 of Tm-treated cells was analyzed as described for panel C together with reduced or nonreduced lysates of unstressed and Tm-treated cells.
ERGIC-53, which functions as a receptor molecule for some cargo proteins (11), was also used as a control for this experiment. Consistent with the notion that ERGIC-53, containing two cysteine residues, circulates between the ER and Golgi apparatus as a dimer and hexamer (25), FLAG-tagged ERGIC-53 was present in all 6 fractions (Fig. 6B), indicating that changes in oligomeric status are not prerequisite for trafficking from the ER to the Golgi apparatus. Under the conditions used, all forms of ATF6α were recovered in heavy fractions in unstressed cells [Fig. 6C(a), −DTT, and D(a), −Tm, fractions 1 and 2]. In contrast, only monomer ATF6α was recovered in the lightest fraction in cells treated with DTT for 15 min [Fig. 6C(a), +DTT, fraction 6] and in cells treated with Tm for 2 h [Fig. 6D(a), +Tm, fraction 6]. These incubation periods were selected as the earliest time points which would allow detection of pATF6α(N) in respective treatment. To determine whether the band(s) detected in fraction 6 of DTT- or Tm-treated cells represented reduced or oxidized monomer ATF6α, an aliquot of fraction 6 was applied to centrifugation-coupled membrane filtration to replace the buffer and then subjected to SDS-PAGE under nonreducing conditions, on the basis that high concentrations of sucrose somewhat disturbed migration of ATF6α during SDS-PAGE. Comparison with the migration positions of standards containing reduced and oxidized monomer ATF6α revealed that the band in fraction 6 of DTT-treated cells was reduced monomer ATF6α [Fig. 6C(b), lane 3] and that the bands in fraction 6 of Tm-treated cells were reduced monomer ATF6α with and without the carbohydrate moiety [Fig. 6D(b), lane 4]. These results indicate that ATF6 is reduced in response to ER stress and that reduced monomer ATF6 is transported from the ER to the Golgi apparatus, in contrast to the case of ERGIC-53.
Reduction is not sufficient for activation of ATF6.
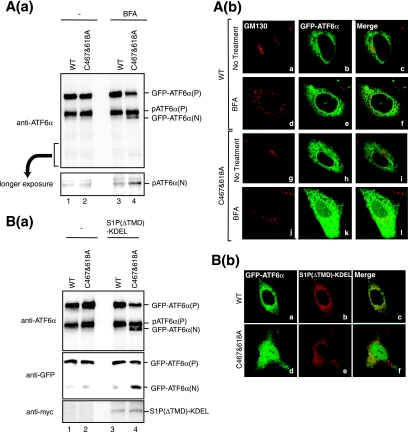
We next mutated two conserved cysteines in the luminal domain to alanine and examined their effects on the oligomeric status of ATF6α. Wild-type (WT) ATF6α or ATF6α carrying a mutation of C467A, C618A or both (C467&618A) were expressed by transfection from a plasmid encoding ATF6α tagged with the HA epitope at the N terminus, whose expression was under the control of a strong CMV promoter (43). Although exogenous ATF6α was expressed at an extremely high level over endogenous ATF6α [Fig. 7A(a), middle panel, compare lane 1 with lanes 2 to 5], HA-tagged ATF6α(WT) showed three bands under nonreducing conditions (lane 7) similar to endogenous ATF6α. HA-tagged ATF6α(C467A) exhibited two of three bands (lane 8) while HA-tagged ATF6α(C467&618A) exhibited only one (lane 10), as expected. Monomer ATF6α derived from HA-tagged ATF6α(WT) migrated faster than that from HA-tagged ATF6α(C467A), ATF6α(C618A), or ATF6α(C467&618A) when they were run for a longer time on SDS-PAGE under nonreducing conditions [Fig. 7A(a), bottom panels, lanes 15 to 18]. These results demonstrated that C467 and C618 are indeed engaged in the disulfide bonding. Thus, the three bands observed for WT and endogenous ATF6α were assigned from the bottom as monomer, dimer, and oligomer forms, respectively, as schematically shown in Fig. 7C. Unexpectedly, HA-tagged ATF6α(C618A) formed an oligomer in addition to a monomer [Fig. 7A(a), lane 9], indicating that ATF6α(C618A) is able to be disulfide bonded with endogenous ATF6α or other cellular proteins. To confirm this notion, immunoprecipitation was performed in lysates of 35S-labeled cells expressing HA-tagged ATF6α(WT) or ATF6α(C618A) using anti-HA antibody. Results showed that, consistent with the results of immunoblotting analysis shown in Fig. 7A(a), ATF6α(WT) exhibited monomer, dimer, and oligomer forms [Fig. 7A(b), lane 6], whereas ATF6α(C618A) exhibited monomer and oligomer forms (lane 8) under nonreducing conditions. In this experiment, transfected cells were harvested in PBS containing 10 mM NEM, lysed in 1% SDS lysis buffer, boiled for 5 min, and diluted with 2% Triton X-100 buffer for immunoprecipitation. Cell lysis in buffer containing 2% SDS and 6 M urea followed by boiling for 10 min did not alter the oligomeric status of ATF6α(C618A) (data not shown), suggesting that ATF6α(C618A) was oligomerized with some other molecules through disulfide bonding. Under reducing conditions, only monomer ATF6α was recovered from cells expressing HA-tagged ATF6α(WT) (lane 2), whereas a band migrating between monomer and dimer ATF6α was obtained in addition to monomer ATF6α from cells expressing HA-tagged ATF6α(C618A), suggesting that ATF6α(C618A) was disulfide bonded to this protein X (Fig. 7C). However, the marked difference in intensity between band X and ATF6α(C618A) casts doubt on the stoichiometry of the complex formed between them. We therefore conducted a pulse-chase experiment. SDS-PAGE analysis under reducing conditions showed reproducible coimmunoprecipitation of band X with ATF6α(C618A) [Fig. 7A(c), lane 7] but not with ATF6α(WT) (lane 3). Both ATF6α(WT) and ATF6α(C618A) decreased after chase (lanes 4 and 8, respectively), as was shown for endogenous ATF6α (Fig. 4B), whereas band X remained unchanged after chase [Fig. 7A(c), lane 8], suggesting that band X is a stable protein. This might explain the apparently unstoichiometric association between ATF6α(C618A) and band X observed after pulse-labeling for 1 h. Nonetheless, a complete understanding of the unusual oligomeric nature of ATF6α(C618A) must await identification of band X (see Discussion), which is in progress in our laboratory.
We have succeeded in expressing GFP-ATF6α fusion protein at a level comparable to that of endogenous ATF6α using a truncated CMV promoter, and we showed that the GFP-ATF6α behaved quite similarly to endogenous ATF6α (23). Introduction of cysteine mutation(s) into GFP-ATF6α produced essentially the same effects on oligomeric status as those observed for HA-tagged ATF6α (Fig. 7B). GFP-ATF6α(C467A) formed a monomer and dimer (lane 6), GFP-ATF6α(C618A) formed a monomer and oligomer (lane 7), and GFP-ATF6α(C467&618A) formed a monomer (lane 8). These results demonstrate that ATF6α is disulfide bonded through Cys467 and Cys618.
The observation that reduced monomer ATF6 reaches the Golgi apparatus (Fig. 6) raised the possibility that ATF6α devoid of disulfide bridges might be constitutively active. However, we found that WT and three mutant GFP-ATF6α were localized in the ER in unstressed cells similarly (Fig. 7B, right panels). Indeed, like GFP-ATF6α(WT), GFP-ATF6α(C467&618A) required ER stress to move to the Golgi apparatus (Fig. 7D); in both cases, GFP was colocalized with the Golgi marker GM130 in cells treated with DTT for 30 min but not in unstressed cells. We did not observe a significant difference in kinetics for transport between GFP-ATF6α(WT) and GFP-ATF6α(C467&618A). The ER chaperone BiP constitutively bound to GFP-ATF6α(C467&618A) and dissociated from GFP-ATF6α(C467&618A) immediately in response to DTT treatment, again similarly to GFP-ATF6α(WT) (Fig. 7E). Thus, ER retention of ATF6α was accounted for by BiP binding but not by disulfide bonding, at least in part, and reduction is not sufficient for activation of ATF6.
Reduced monomer ATF6 is a better substrate of site 1 protease.
To clarify the biological significance of disulfide bonding of ATF6 further, we focused on another aspect of the ATF6 activation process, namely its susceptibility to proteases. As already shown in Fig. 1B, BFA-mediated relocation of S1P and S2P from the Golgi apparatus to the ER resulted in inefficient cleavage of ATF6α compared with a strong cleavage inducer such as DTT. When CHO cells expressing GFP-ATF6α(WT) by transfection were treated with BFA, very weak cleavage of endogenous pATF6α(P) occurred to produce pATF6α(N), whereas exogenous GFP-ATF6α(WT) was somewhat more resistant to cleavage [Fig. 8A (a), compare lane 3 with lane 1]. Thus, colocalization of substrate with protease is not sufficient for full cleavage. In contrast, when CHO cells expressing GFP-ATF6α(C467&618A) by transfection were treated with BFA, the precursor form of GFP-ATF6α(C467&618A) was efficiently cleaved to produce its nuclear form, which migrated slightly faster than pATF6α(P) (compare lane 4 with lane 2). Accordingly, GFP-ATF6α(WT) was localized exclusively in the ER of BFA-treated cells [Fig. 8A(b), panel e], whereas GFP derived from GFP-ATF6α(C467&618A) was found in both the ER and nucleus (panel k). These findings indicated that reduced monomer ATF6α is a better substrate of S1P than disulfide-bonded ATF6α.
FIG. 8.
Effects of enforced expression of S1P in the ER on cleavage of ATF6. (A) (a) CHO cells transfected with pCMVshort-EGFP-ATF6α(WT) or pCMVshort-EGFP-ATF6α(C467&618A) were treated with or without 5 μg/ml BFA for 1 h. Cell lysates were prepared and analyzed under reducing conditions using anti-ATF6α antibody. The bottom panel shows the result of longer exposure of the region around pATF6α(N). (b) CHO cells transfected and treated as described for panel a were fixed and immunostained with anti-GM130 antibody (panels a, d, g, and j). GFP-ATF6α was visualized by its own fluorescence (panels b, e, h, and k). (B) (a) CHO cells were transfected with pCMVshort-EGFP-ATF6α(WT) or pCMVshort-EGFP-ATF6α(C467&618A) together with or without a plasmid to express S1P(ΔTMD)-KDEL tagged with the c-myc epitope at the C terminus. Cell lysates were prepared and analyzed under reducing conditions using anti-ATF6α (upper panel), anti-GFP (middle panel), or anti-myc (lower panel) antibodies. (b) CHO cells cotransfected as described for panel a were fixed and immunostained with anti-myc antibody (panels b and e). GFP-ATF6α was visualized by its own fluorescence (panels a and d).
We further confirmed this notion by utilizing mutant S1P, termed S1P(ΔTMD)-KDEL, which lacks S1P's transmembrane domain and instead carries the ER retention sequence KDEL at its C terminus (7). We previously showed that overexpression of S1P(ΔTMD)-KDEL resulted in constitutive production of pATF6α(N), albeit with markedly low cleavage efficiency (26). GFP-ATF6α(WT) and GFP-ATF6α(C467&618A) were expressed by transfection with or without S1P(ΔTMD)-KDEL tagged with the c-myc epitope. Results of immunoblotting analysis using anti-ATF6α or anti-GFP antibody showed that the coexpression of S1P(ΔTMD)-KDEL had little effect on GFP-ATF6α(WT) [Fig. 8B(a), compare lane 3 with lane 1] but did specifically induce the cleavage of GFP-ATF6α(C467&618A) (compare lane 4 with lane 2). Consistent with the results of biochemical analysis, GFP-ATF6α(WT) in the presence of S1P(ΔTMD)-KDEL was exclusively localized in the ER [Fig. 8B(b), panel a], whereas GFP derived from GFP-ATF6α(C467&618A) was localized in both the ER and nucleus (panel d).
ER stress-induced reduction is specific to ATF6.
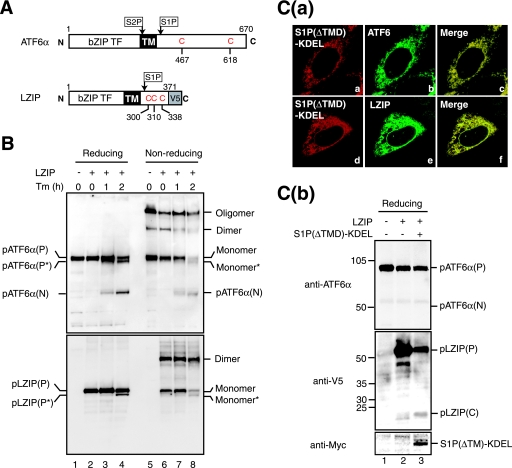
To determine whether ER stress-induced changes in oligomeric status are specific to ATF6, LZIP/Luman was employed as a control. LZIP is a bZIP-type transcription factor localized in the ER as a type II transmembrane protein similar to ATF6 (Fig. 9A) (31). Although it was shown that LZIP was cleavable by S1P, LZIP was not activated in response to ER stress. Thus, both the trigger for proteolysis and the consequence of the cleavage remain unknown. LZIP was tagged with the V5 epitope at the C terminus and expressed by transfection. Unlike ATF6α, Tm treatment of transfected cells did not result in cleavage of LZIP, although the deglycosylated form of LZIP, pLZIP(P*), was detected (Fig. 9B, reducing, lanes 2 to 4). Analysis under nonreducing conditions revealed that LZIP, which contains three cysteine residues in its luminal domain, as depicted in Fig. 9A, existed mainly as a monomer and dimer (lane 6). Importantly, the level of dimer LZIP was not changed in response to Tm treatment, in marked contrast to the case of dimer ATF6α which disappeared reproducibly (lanes 7 and 8). Essentially identical results were obtained when 10 mM NEM was included in the lysis buffer (data not shown). These results indicate that ER stress-induced reduction is not a general feature of ER membrane-bound transcription factors.
FIG. 9.
Effects of tunicamycin treatment and enforced expression of S1P in the ER on oligomeric status and cleavage of LZIP. (A) Schematic structures of ATF6α and LZIP tagged with the V5 epitope at the C terminus are shown. Both contain a single transmembrane domain (TM) in the middle of the respective protein, and their N-terminal bZIP transcription factor (TF) domains face the cytoplasm. Cleavage of ATF6α by S1P and S2P as well as that of LZIP by S1P was previously demonstrated. The positions of cysteine residues in their luminal regions are also indicated. (B) CHO cells transfected with (+) or without (−) a plasmid to express V5-tagged LZIP were treated with 2 μg/ml Tm for the indicated periods, and then cell lysates were prepared and analyzed as described for Fig. 3A(a) (without NEM) using anti-ATF6α (upper panel) or anti-V5 (lower panel) antibodies. Proteins marked by asterisks lack a carbohydrate moiety because of the action of Tm. (C) (a) CHO cells were cotransfected with plasmids to express myc-tagged S1P(ΔTMD)-KDEL and V5-tagged LZIP, and then immunostained with anti-myc (panels a and d), anti-ATF6α (panel b), or anti-V5 (panel e) antibodies. (b) CHO cells were mock transfected (lane 1), transfected with a plasmid to express V5-tagged LZIP alone (lane 2), or cotransfected with plasmids to express V5-tagged LZIP and myc-tagged S1P(ΔTMD)-KDEL (lane 3), and then cell lysates were prepared and analyzed by immunoblotting under reducing conditions using anti-ATF6α (upper panel), anti-V5 (middle panel), or anti-myc (lower panel) antibodies.
We then compared the extent of cleavage of ATF6α with that of LZIP induced by overproduction of S1P(ΔTMD)-KDEL. LZIP tagged with the V5 epitope was expressed by transfection with or without S1P(ΔTMD)-KDEL tagged with the c-myc epitope. Colocalization of S1P(ΔTMD)-KDEL with endogenous ATF6α or exogenous LZIP was confirmed by double immunostaining [Fig. 9C(a)]. Coexpression of S1P(ΔTMD)-KDEL resulted in efficient cleavage of pLZIP(P) and, thereby, the production of the luminal fragment of LZIP, pLZIP(C), as shown in Fig. 9C(b) (middle panel, compare lane 3 with lane 2). In contrast, endogenous pATF6α(P) was hardly cleaved under these conditions (top panel, compare lane 3 with lane 2), probably because a much better substrate for S1P(ΔTMD)-KDEL was present, namely pLZIP(P). These findings show that disulfide bridges formed in the luminal domain make ATF6α but not LZIP resistant to digestion by S1P.
DISCUSSION
Mammalian ER expresses three types of transmembrane proteins whose cytoplasmic domains achieve their function of countering ER stress through highly characteristic properties, namely endoribonuclease activity in IRE1, protein kinase activity in PERK, and transcriptional activator activity in ATF6. In contrast, their luminal domains are postulated to sense ER stress and transmit a signal across the membrane. It was initially reported that they are inactive when bound to the major ER chaperone BiP and that the dissociation of BiP is a trigger for their activation, providing a common mechanism for the activation of all three molecules. IRE1 or PERK freed from BiP becomes oligomerized and autophosphorylated, a process considered sufficient for the activation of their respective downstream events (2, 28), while ATF6 freed from BiP is transported to the Golgi apparatus for cleavage (36).
However, more recent analyses indicate that the story is not so simple. In the case of yeast Ire1p, the BiP binding site was mapped to a region close to the transmembrane domain and, unexpectedly, was separated from the two subregions indispensable to ER stress-induced activation. Thus, the deletion mutant of yeast Ire1p lacking the BiP-binding site fully responded to ER stress but became hypersensitive to temperature upshift and ethanol compared with the wild-type Ire1p, leading to the authors' proposal that BiP is not the principal determinant of yeast Ire1p activity but rather an adjuster for sensitivity to various stresses (17). In the case of ATF6, binding of BiP to ATF6 was found to be stable but not in a dynamic state, contradictory to the general assumption that BiP is dissociated from ATF6 as a result of competitive binding to unfolded proteins produced under ER stress. This finding led the authors to propose that the dissociation of the BiP-ATF6 complex, which is essential to the regulation of ATF6, operates under an active regulatory mechanism (38).
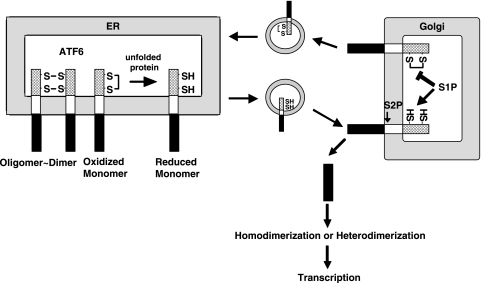
In the present report, we show that ATF6 occurs as a monomer, dimer, and oligomer in unstressed ER through the presence of inter- and intramolecular disulfide bridges formed between two conserved cysteine residues in the luminal domain (Fig. 2, 3, and 7) and that ATF6 is reduced in response to ER stress (Fig. 5), allowing reduced monomer ATF6 to relocate to the Golgi apparatus to be cleaved by the sequential actions of S1P and S2P (Fig. 6), as schematically drawn in Fig. 10. These findings thus reveal an unambiguous difference between the activation of IRE1 or PERK and of ATF6: when activated in response to ER stress, IRE1 and PERK are oligomerized, whereas ATF6 is deoligomerized. These differences may explain the different kinetics in the responses of IRE1, PERK, and ATF6 to various ER stress inducers, especially to thapsigargin, as reported recently (8). Differential mechanisms in sensing ER stress among the three UPR regulators are likely to be advantageous for the cell. The cell can selectively activate one or two branches of the UPR instead of always activating all three to supply the most desirable outcome according to the circumstances or needs under more-physiological ER stress conditions than those triggered by chemical ER stress inducers.
FIG. 10.
Model for regulation of ATF6 activation. ATF6 occurs in unstressed ER as the oxidized monomer, dimer, and oligomer due to intra- and intermolecular disulfide bridges formed in the luminal domain. Upon ER stress, ATF6 is reduced, and the reduced monomer ATF6 is transported to the Golgi apparatus to be cleaved by S1P and S2P. The N-terminal transcription factor domain liberated is homo- or heterodimerized and translocated into the nucleus to activate transcription of ER chaperone genes. ATF6 may move to the Golgi apparatus in the absence of ER stress but will return to the ER because oxidized ATF6 is resistant to cleavage by S1P. Thus, the cell processes only ATF6 which has experienced stress in the ER.
ATF6 is considered to be transported from the ER to the Golgi apparatus by vesicles coated with COP-II coat proteins (COP-II vesicles) on the basis that the transport of ATF6 was blocked by a dominant-negative form of Sar1 (23), a cytoplasmic GTPase, which plays a key role in the formation of COP-II vesicles (1). The smaller ATF6 may be more easily packed into COP-II vesicles of 60 to 80 nm; it has been reported that the transport of excessively large proteins, for example, procollagen of 300 nm, is not mediated by canonical COP-II vesicles (3, 40). However, because ERGIC-53 can traverse between the ER and Golgi apparatus as a dimer or hexamer (Fig. 6), the size of cargo proteins is unlikely to be a primary determinant for packaging. The relationship between reduction and packaging is not presently clear, since the reduction of ATF6 was not sufficient for transport (Fig. 7). Nonetheless, it is still possible that ATF6 with two free cysteine residues is preferably packed into COP-II vesicles because we could not produce such a molecule by mutating conserved cysteine residues to alanine. We are now analyzing the budding step from the ER.
The activation mechanism of ATF6 is categorized as regulated intramembrane proteolysis (Rip), a novel concept for protein activity regulation, which is conserved from bacteria to humans, in which a functional protein is synthesized as a part of a large transmembrane protein and excised when required by the cell (4). A substrate of Rip is cleaved sequentially by two proteases; the second cleavage is thought to occur in the transmembrane domain, and the second protease can only process a substrate which has been cleaved by the first protease. Thus, the first cleavage is the most important step in Rip. In the case of ATF6, cleavage of ATF6 by S1P reduced the size of the luminal domain from ∼270 amino acids to ∼20 amino acids (44). It was recently reported that ATF6 cleaved by S1P but not full-length ATF6 is a good substrate of S2P, leading to the authors' proposal that the role of S1P is to reduce the size of the luminal domain to prepare ATF6 to be an optimal S2P substrate (37).
In the present study, we show that reduced monomer ATF6 is a better substrate of S1P than disulfide-bonded ATF6 (Fig. 8), and this is a clear advantage for ER stress-induced reduction of ATF6. ATF6 may traverse between the ER and Golgi apparatus even in the absence of ER stress. Namely, as BiP sometimes leaves the ER to reach the cis-Golgi compartment and is then retrieved to the ER via interaction with the KDEL receptor (30), ATF6 might be mislocalized to the Golgi apparatus by accompanying BiP. We also show that all forms of ATF6 turn over rapidly due to proteasome-mediated degradation (Fig. 4 and data not shown). It is generally considered that a protein is retrotranslocated from the ER to the cytoplasm through a pore and, thus, that an oligomeric protein needs to be deoligomerized to obtain access to the proteasome (41). During the deoligomerization process, ATF6 may be packed into COP-II vesicles by mistake and transported to the Golgi apparatus. Even in these cases, however, unnecessary activation of the UPR is avoided because disulfide-bonded ATF6 is a poor substrate of S1P and is able to return to the ER, as schematically shown in Fig. 10. Thus, we propose that ER stress-induced reduction of ATF6 provides a basis for strict regulation: the cell can only process ATF6 which has undergone processing in the ER, or in other words, ATF6 is activated only when cellular UPR activity is required. One might argue that reduced ATF6 is easily cleaved by S1P because it is not properly folded; however, we consider this unlikely because the double-cysteine mutant GFP-ATF6α(C467&618A), found to be a better substrate of S1P than GFP-ATF6α(WT) (Fig. 8), is still transport competent from the ER to the Golgi apparatus upon ER stress (Fig. 7). BiP is released from GFP-ATF6α(WT) and GFP-ATF6α(C467&618A) with a similar time course (Fig. 7), and a putative ATF6 escort protein suggested in the literature (35) would not be able to discriminate GFP-ATF6α(C467&618A) from GFP-ATF6α(WT). We also emphasize that ER stress-induced reduction of ATF6 provides the opportunity for ATF6 to heterodimerize with other transcription factors on the way from the ER to the nucleus (24, 47).
What reduces ATF6 in response to ER stress is a completely open question. Although the lumen of ER is highly oxidized under normal conditions compared with the cytosol (42), and appears to undergo even further oxidization under ER stress conditions (20), ATF6 can be reduced in cells treated with not only the reducing reagent DTT but also the nonreducing reagent Tm. Reduction is specific to ATF6, as a second ER membrane-bound transcription factor unresponsive to ER stress, namely, LZIP, was not reduced in response to ER stress (Fig. 9). We thus consider that reduction is mediated by a specific enzyme. A number of oxidoreductases are present in mammalian ER (42). Indeed, it was recently reported that ERp44, one such luminal oxidoreductase, modulates the activity of a subtype of inositol 1,4,5-triphoshate receptor, depending on pH, Ca2+ concentration, and redox state, via direct interaction through free cysteine residues on the receptor molecule (14). An important next step is to identify a protein that binds to ATF6 in an ER stress-dependent manner. In this connection, protein X is interesting, as it forms a mixed disulfide with ATF6α(C618A) (Fig. 7). Results of the pulse-chase experiment suggested that band X was associated with pulse-labeled ATF6α(C618A), dissociated when pulse-labeled ATF6α(C618A) was degraded, and reassociated with ATF6α(C618A) newly synthesized during the chase period [Fig. 7A(c)]. Thus, an attractive candidate for band X would be a protein disulfide isomerase-like protein involved in the oxidative folding of ATF6 or an oxidoreductase that reduces the ATF6 luminal domain when ATF6 is degraded by the proteasome via an ER-associated degradation-related mechanism (41). As depicted in Fig. 7C, protein X might be a membrane protein capable of forming a mixed disulfide with Cys467 with free SH in the ATF6α(C618A) mutant but not with Cys618 with free SH in the ATF6α(C467A) mutant simply because Cys467 is sufficiently close to the catalytic site in protein X.
In conclusion, we demonstrate here that the luminal domain of ATF6 is disulfide bonded and that ER stress-induced reduction is involved in both transportation and subsequent recognition by the protease, both of which are unique to the ATF6 activation process compared with those of IRE1 and PERK.
Acknowledgments
We thank Kaoru Miyagawa for technical and secretarial assistance.
This work was supported, in part, by grants from the Ministry of Education, Culture, Sports, Science and Technology of Japan (14037233 and 15GS0310 to K.M. and 17770158 to T.O.).
Footnotes
Published ahead of print on 13 November 2006.
REFERENCES
- 1.Antonny, B., and R. Schekman. 2001. ER export: public transportation by the COPII coach. Curr. Opin. Cell Biol. 13:438-443. [DOI] [PubMed] [Google Scholar]
- 2.Bertolotti, A., Y. Zhang, L. M. Hendershot, H. P. Harding, and D. Ron. 2000. Dynamic interaction of BiP and ER stress transducers in the unfolded-protein response. Nat. Cell Biol. 2:326-332. [DOI] [PubMed] [Google Scholar]
- 3.Bonfanti, L., A. A. Mironov, Jr., J. A. Martinez-Menarguez, O. Martella, A. Fusella, M. Baldassarre, R. Buccione, H. J. Geuze, A. A. Mironov, and A. Luini. 1998. Procollagen traverses the Golgi stack without leaving the lumen of cisternae: evidence for cisternal maturation. Cell 95:993-1003. [DOI] [PubMed] [Google Scholar]
- 4.Brown, M. S., J. Ye, R. B. Rawson, and J. L. Goldstein. 2000. Regulated intramembrane proteolysis: a control mechanism conserved from bacteria to humans. Cell 100:391-398. [DOI] [PubMed] [Google Scholar]
- 5.Chen, X., J. Shen, and R. Prywes. 2002. The luminal domain of ATF6 senses endoplasmic reticulum (ER) stress and causes translocation of ATF6 from the ER to the Golgi. J. Biol. Chem. 277:13045-13052. [DOI] [PubMed] [Google Scholar]
- 6.Credle, J. J., J. S. Finer-Moore, F. R. Papa, R. M. Stroud, and P. Walter. 2005. On the mechanism of sensing unfolded protein in the endoplasmic reticulum. Proc. Natl. Acad. Sci. USA 102:18773-18784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DeBose-Boyd, R. A., M. S. Brown, W. P. Li, A. Nohturfft, J. L. Goldstein, and P. J. Espenshade. 1999. Transport-dependent proteolysis of SREBP: relocation of site-1 protease from Golgi to ER obviates the need for SREBP transport to Golgi. Cell 99:703-712. [DOI] [PubMed] [Google Scholar]
- 8.DuRose, J. B., A. B. Tam, and M. Niwa. 2006. Intrinsic capacities of molecular sensors of the unfolded protein response to sense alternate forms of endoplasmic reticulum stress. Mol. Biol. Cell. 17:3095-3107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gallione, C. J., and J. K. Rose. 1985. A single amino acid substitution in a hydrophobic domain causes temperature-sensitive cell-surface transport of a mutant viral glycoprotein. J. Virol. 54:374-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harding, H. P., M. Calfon, F. Urano, I. Novoa, and D. Ron. 2002. Transcriptional and translational control in the Mammalian unfolded protein response. Annu. Rev. Cell Dev. Biol. 18:575-599. [DOI] [PubMed] [Google Scholar]
- 11.Hauri, H. P., F. Kappeler, H. Andersson, and C. Appenzeller. 2000. ERGIC-53 and traffic in the secretory pathway. J. Cell Sci. 113:587-596. [DOI] [PubMed] [Google Scholar]
- 12.Haze, K., T. Okada, H. Yoshida, H. Yanagi, T. Yura, M. Negishi, and K. Mori. 2001. Identification of the G13 (cAMP-response-element-binding protein-related protein) gene product related to activating transcription factor 6 as a transcriptional activator of the mammalian unfolded protein response. Biochem. J. 355:19-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haze, K., H. Yoshida, H. Yanagi, T. Yura, and K. Mori. 1999. Mammalian transcription factor ATF6 is synthesized as a transmembrane protein and activated by proteolysis in response to endoplasmic reticulum stress. Mol. Biol. Cell 10:3787-3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Higo, T., M. Hattori, T. Nakamura, T. Natsume, T. Michikawa, and K. Mikoshiba. 2005. Subtype-specific and ER luminal environment-dependent regulation of inositol 1,4,5-trisphosphate receptor type 1 by ERp44. Cell 120:85-98. [DOI] [PubMed] [Google Scholar]
- 15.Iwakoshi, N. N., A. H. Lee, and L. H. Glimcher. 2003. The X-box binding protein-1 transcription factor is required for plasma cell differentiation and the unfolded protein response. Immunol. Rev. 194:29-38. [DOI] [PubMed] [Google Scholar]
- 16.Kaufman, R. J. 1999. Stress signaling from the lumen of the endoplasmic reticulum: coordination of gene transcriptional and translational controls. Genes Dev. 13:1211-1233. [DOI] [PubMed] [Google Scholar]
- 17.Kimata, Y., D. Oikawa, Y. Shimizu, Y. Ishiwata-Kimata, and K. Kohno. 2004. A role for BiP as an adjustor for the endoplasmic reticulum stress-sensing protein Ire1. J. Cell Biol. 167:445-456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lippincott-Schwartz, J. 1993. Bidirectional membrane traffic between the endoplasmic reticulum and Golgi apparatus. Trends Cell Biol. 3:81-88. [DOI] [PubMed] [Google Scholar]
- 19.Liu, E. S., J. H. Ou, and A. S. Lee. 1992. Brefeldin A as a regulator of grp78 gene expression in mammalian cells. J. Biol. Chem. 267:7128-7133. [PubMed] [Google Scholar]
- 20.Marciniak, S. J., C. Y. Yun, S. Oyadomari, I. Novoa, Y. Zhang, R. Jungreis, K. Nagata, H. P. Harding, and D. Ron. 2004. CHOP induces death by promoting protein synthesis and oxidation in the stressed endoplasmic reticulum. Genes Dev. 18:3066-3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mori, K. 2003. Frame switch splicing and regulated intramembrane proteolysis: key words to understand the unfolded protein response. Traffic 4:519-528. [DOI] [PubMed] [Google Scholar]
- 22.Mori, K. 2000. Tripartite management of unfolded proteins in the endoplasmic reticulum. Cell 101:451-454. [DOI] [PubMed] [Google Scholar]
- 23.Nadanaka, S., H. Yoshida, F. Kano, M. Murata, and K. Mori. 2004. Activation of mammalian unfolded protein response is compatible with the quality control system operating in the endoplasmic reticulum. Mol. Biol. Cell 15:2537-2548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Newman, J. R., and A. E. Keating. 2003. Comprehensive identification of human bZIP interactions with coiled-coil arrays. Science 300:2097-2101. [DOI] [PubMed] [Google Scholar]
- 25.Nufer, O., F. Kappeler, S. Guldbrandsen, and H. P. Hauri. 2003. ER export of ERGIC-53 is controlled by cooperation of targeting determinants in all three of its domains. J. Cell Sci. 116:4429-4440. [DOI] [PubMed] [Google Scholar]
- 26.Okada, T., K. Haze, S. Nadanaka, H. Yoshida, N. G. Seidah, Y. Hirano, R. Sato, M. Negishi, and K. Mori. 2003. A serine protease inhibitor prevents endoplasmic reticulum stress-induced cleavage but not transport of the membrane-bound transcription factor ATF6. J. Biol. Chem. 278:31024-31032. [DOI] [PubMed] [Google Scholar]
- 27.Okada, T., H. Yoshida, R. Akazawa, M. Negishi, and K. Mori. 2002. Distinct roles of activating transcription factor 6 (ATF6) and double-stranded RNA-activated protein kinase-like endoplasmic reticulum kinase (PERK) in transcription during the mammalian unfolded protein response. Biochem. J. 366:585-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Okamura, K., Y. Kimata, H. Higashio, A. Tsuru, and K. Kohno. 2000. Dissociation of Kar2p/BiP from an ER sensory molecule, Ire1p, triggers the unfolded protein response in yeast. Biochem. Biophys. Res. Commun. 279:445-450. [DOI] [PubMed] [Google Scholar]
- 29.Patil, C., and P. Walter. 2001. Intracellular signaling from the endoplasmic reticulum to the nucleus: the unfolded protein response in yeast and mammals. Curr. Opin. Cell Biol. 13:349-356. [DOI] [PubMed] [Google Scholar]
- 30.Pelham, H. R. 1991. Recycling of proteins between the endoplasmic reticulum and Golgi complex. Curr. Opin. Cell Biol. 3:585-591. [DOI] [PubMed] [Google Scholar]
- 31.Raggo, C., N. Rapin, J. Stirling, P. Gobeil, E. Smith-Windsor, P. O'Hare, and V. Misra. 2002. Luman, the cellular counterpart of herpes simplex virus VP16, is processed by regulated intramembrane proteolysis. Mol. Cell. Biol. 22:5639-5649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ron, D. 2002. Translational control in the endoplasmic reticulum stress response. J. Clin. Investig. 110:1383-1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 34.Schroder, M., and R. J. Kaufman. 2005. The mammalian unfolded protein response. Annu. Rev. Biochem. 74:739-789. [DOI] [PubMed] [Google Scholar]
- 35.Seemann, J., E. Jokitalo, M. Pypaert, and G. Warren. 2000. Matrix proteins can generate the higher order architecture of the Golgi apparatus. Nature 407:1022-1026. [DOI] [PubMed] [Google Scholar]
- 36.Shen, J., X. Chen, L. Hendershot, and R. Prywes. 2002. ER stress regulation of ATF6 localization by dissociation of BiP/GRP78 binding and unmasking of Golgi localization signals. Dev. Cell 3:99-111. [DOI] [PubMed] [Google Scholar]
- 37.Shen, J., and R. Prywes. 2004. Dependence of site-2 protease cleavage of ATF6 on prior site-1 protease digestion is determined by the size of the luminal domain of ATF6. J. Biol. Chem. 279:43046-43051. [DOI] [PubMed] [Google Scholar]
- 38.Shen, J., E. L. Snapp, J. Lippincott-Schwartz, and R. Prywes. 2005. Stable binding of ATF6 to BiP in the endoplasmic reticulum stress response. Mol. Cell. Biol. 25:921-932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shen, X., R. E. Ellis, K. Sakaki, and R. J. Kaufman. 2005. Genetic interactions due to constitutive and inducible gene regulation mediated by the unfolded protein response in C. elegans. PLoS Genet. 1:355-368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stephens, D. J., and R. Pepperkok. 2002. Imaging of procollagen transport reveals COPI-dependent cargo sorting during ER-to-Golgi transport in mammalian cells. J. Cell Sci. 115:1149-1160. [DOI] [PubMed] [Google Scholar]
- 41.Tsai, B., Y. Ye, and T. A. Rapoport. 2002. Retro-translocation of proteins from the endoplasmic reticulum into the cytosol. Nat. Rev. Mol. Cell. Biol. 3:246-255. [DOI] [PubMed] [Google Scholar]
- 42.Tu, B. P., and J. S. Weissman. 2004. Oxidative protein folding in eukaryotes: mechanisms and consequences. J. Cell Biol. 164:341-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang, Y., J. Shen, N. Arenzana, W. Tirasophon, R. J. Kaufman, and R. Prywes. 2000. Activation of ATF6 and an ATF6 DNA binding site by the endoplasmic reticulum stress response. J. Biol. Chem. 275:27013-27020. [DOI] [PubMed] [Google Scholar]
- 44.Ye, J., R. B. Rawson, R. Komuro, X. Chen, U. P. Dave, R. Prywes, M. S. Brown, and J. L. Goldstein. 2000. ER stress induces cleavage of membrane-bound ATF6 by the same proteases that process SREBPs. Mol. Cell 6:1355-1364. [DOI] [PubMed] [Google Scholar]
- 45.Yoshida, H., T. Okada, K. Haze, H. Yanagi, T. Yura, and K. Mori. 2000. ATF6 activated by proteolysis directly binds in the presence of NF-Y (CBF) to the cis-acting element responsible for the mammalian unfolded protein response. Mol. Cell. Biol. 20:6755-6767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yoshida, H., T. Okada, K. Haze, H. Yanagi, T. Yura, M. Negishi, and K. Mori. 2001. Endoplasmic reticulum stress-induced formation of transcription factor complex ERSF including NF-Y (CBF) and activating transcription factors 6a and 6b that activates the mammalian unfolded protein response. Mol. Cell. Biol. 21:1239-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang, K., X. Shen, J. Wu, K. Sakaki, T. Saunders, D. T. Rutkowski, S. H. Back, and R. J. Kaufman. 2006. Endoplasmic reticulum stress activates cleavage of CREBH to induce a systemic inflammatory response. Cell 124:587-599. [DOI] [PubMed] [Google Scholar]
- 48.Zhou, J., C. Y. Liu, S. H. Back, R. L. Clark, D. Peisach, Z. Xu, and R. J. Kaufman. 2006. The crystal structure of human IRE1 luminal domain reveals a conserved dimerization interface required for activation of the unfolded protein response. Proc. Natl. Acad. Sci. USA 103:14343-14348. [DOI] [PMC free article] [PubMed] [Google Scholar]