Abstract
Small GTPases of the Rho family are key regulators of phagocytic leukocyte function. Abr and Bcr are homologous, multidomain proteins. Their C-terminal domain has GTPase-activating protein (GAP) activity that, in vitro, is specific for Rac and Cdc42. To address the in vivo relevance of these entire proteins, of which little is known, the current study examined the effect of the genetic ablation of Abr and Bcr in murine macrophages. The concomitant loss of Abr and Bcr induced multiple alterations of macrophage cellular behavior known to be under the control of Rac. Macrophages lacking both Abr and Bcr exhibited an atypical, elongated morphology that was reproduced by the ectopic expression of GAP domain mutant Abr and Bcr in a macrophage cell line and of constitutively active Rac in primary macrophages. A robust increase in colony-stimulating factor 1 (CSF-1)-directed motility was observed in macrophages deficient for both proteins and, in response to CSF-1 stimulation, Abr and Bcr transiently translocated to the plasma membrane. Phagocytosis of opsonized particles was also increased in macrophages lacking both proteins and correlated with sustained Rac activation. Bcr and Abr GAP mutant proteins localized around phagosomes and induced distinct phagocytic cup formation. These results identify Abr and Bcr as the only GAPs to date that specifically negatively regulate Rac function in vivo in primary macrophages.
Small GTPases of the Rho family (Rac, Cdc42, and Rho) are key regulators of a variety of cellular responses in many different cell types and are best known for their role in controlling actin-cytoskeleton rearrangements (15). Phagocytic leukocytes are prominent examples of cell types of which the behavior is tightly regulated by Rho GTPases. It has been well established that Rho GTPases are critical for morphological changes of leukocytes, chemokine sensing and subsequent motility, phagocytosis, and reactive oxygen species generation through the NADPH oxidase (7, 26, 44).
The activity of Rho GTPases, like all other small GTPases, is determined by their cycling between active GTP-bound forms and inactive GDP-bound forms. In their GTP-bound form, Rho GTPases initiate signaling cascades involved in biological processes, and the subsequent hydrolysis of bound GTP to GDP terminates the signal. This GTP/GDP cycling is regulated by both guanine exchange factors (GEFs) and GTPase-activating proteins (GAPs). GEFs facilitate the exchange of the bound GDP for GTP, thereby activating Rho GTPases, and GAPs facilitate the intrinsic GTPase activity of Rho family proteins, thereby inactivating them. Many studies have focused on activation of Rho family proteins by GEFs, partly due to the fact that most GEFs were originally identified as oncogenes (47). However, there is accumulating evidence that negative regulation of Rho GTPase by GAPs is also critical for many cellular responses, such as neural morphogenesis, cell growth, and endocytosis (36).
The breakpoint cluster region (BCR) gene was originally identified due to its involvement in a specific chromosomal translocation that causes the development of chronic myeloid leukemia and a subset of acute lymphoblastic leukemia (19). Abr (active Bcr related) is the only protein known in humans and mice to share high homology with Bcr (68% amino acid identity) (21). Both Abr and Bcr contain a tandem Dbl homology (DH)-Pleckstrin homology (PH) domain and a C-terminal domain homologous to GAPs that are specific for Rho family GTPases. Bcr has an additional serine/threonine kinase domain and an oligomerization domain at its N terminus, both of which are absent in Abr (19). The isolated GAP domains of Abr and Bcr are catalytically active towards both Rac and Cdc42. Also, the isolated DH domains of Abr and Bcr were shown to be moderately active as GEFs toward Rac, Cdc42, and Rho (10, 20, 52). Thus, Abr and Bcr are unique in the sense that they have two potentially opposing regulatory domains toward small GTPases in one protein. However, it is still unclear whether both of the GEF and the GAP domains of Bcr and Abr are active in vivo and what the normal cellular functions of Abr and Bcr are.
To elucidate the in vivo function of Bcr, we previously generated Bcr null mutant mice and investigated their innate immune response, given the importance of Rho GTPases in regulating this and the potential role of Bcr in modulating Rho GTPase activity. In that study, mice deficient for Bcr were more sensitive to bacterial endotoxin (lipopolysaccharide) than the control mice, and neutrophils from those null mice produced significantly elevated levels of reactive oxygen species upon activation (56).
We subsequently generated mice deficient for Abr, which were fully viable and lacked any overt phenotype. If two highly homologous proteins have redundant functions in cells, it is possible that they compensate for each other and thus the effect of the loss of only one protein might not be well visible. To unmask common functions that could be regulated by Abr and Bcr, mice deficient for both were generated through breeding. These (abr × bcr)−/− double null mice are born with inner ear defects causing constant circling (28), and they also have abnormalities in cerebellar development, with highly activated astroglia (27). Interestingly, they exhibit a severe inflammatory response to lipopolysaccharide-induced endotoxemia, resulting in multiple organ damage and subsequent death, whereas Abr and Bcr single null mutant mice show a milder response and eventually recover (J. M. Cunnick et al., unpublished data).
The known involvement of Rho family GTPases in regulating cellular behavior of phagocytic leukocytes and the elevated innate immune response in (abr × bcr)−/− double null mice prompted us to examine their macrophages for possible abnormalities in cellular responses that are known to be under control of Rho family GTPases. Here we report support for the hypothesis that Abr and Bcr function as Rac GAPs in vivo, regulating multiple aspects of macrophage function, including morphology, colony-stimulating factor 1 (CSF-1)-induced motility, and phagocytosis.
MATERIALS AND METHODS
Mice.
The generation of bcr−/−, abr−/−, and (abr × bcr)−/− mice was reported previously (27, 56). All animals were maintained at the Animal Care Facility of the Research Institute of Childrens Hospital of Los Angeles, and all animal research was performed in accordance with institutional guidelines and with the National Research Council's Guide for the Care and Use of Laboratory Animals (36a).
Cell culture.
RAW 264.7 mouse macrophage (RAW) cells, Chinese hamster ovary (CHO) cells, and L929 cells were obtained from the ATCC (Rockville, MD). All tissue culture media and supplements were from Invitrogen (Carlsbad, CA). RAW cells were cultured in RPMI containing 10% fetal calf serum (FCS), penicillin (100 U/ml), and streptomycin (100 μg/ml), and CHO cells were cultured in Dulbecco's modified Eagle's medium (DMEM) with 10% FCS, penicillin, and streptomycin. L929 cells were maintained in minimal essential medium containing Earle's salts and l-glutamine and supplemented with 10% calf serum, penicillin, streptomycin, nonessential amino acids (0.1 mM), and sodium pyruvate (1 mM).
Isolation of BMMs and PEMs.
Bone marrow mononuclear phagocytic precursor cells were isolated from pelvises, femurs, and tibiae from mice as described by Stanley (50). These precursor cells were differentiated into adherent mature macrophages for 3 days in petri dishes (Kord-Valmark; PGC Scientifics Corporation, Frederick, MD) in complete macrophage medium containing 20% L929-conditioned medium as a source of CSF-1. For routine subculture of bone marrow-derived macrophages (BMMs), adherent BMMs were treated with Versene (Invitrogen) for 15 min, detached from the petri dish by pipetting, split 1:3, and maintained in 20% L929-conditioned medium. Thioglycolate medium (4% in distilled water; Difco, Detroit, MI) was prepared as described elsewhere (31) and administered to mice via intraperitoneal injection. Five days after injection, the peritoneal lavage was recovered from the injected mice by washing the peritoneal cavity with 8 ml ice-cold Hanks' balanced salt solution, from which peritoneal elicited macrophages (PEMs) were recovered by centrifugation at 800 × g at 4°C for 5 min.
Plasmids and antibodies.
An 0.7-kb enhanced green fluorescent protein (EGFP) cDNA fragment was obtained from pEGFP-C2 (Clontech, Mountain View, CA) digested with Eco47III and EcoRI. The BCR cDNA as a 3.2-kb EcoRI-BstEII fragment plus a 0.8-kb BstEII-BglII fragment was ligated with the EGFP fragment into pIRES Neo (Clontech) digested with EcoRV and BamHI to generate the EGFP-Bcr expression vector. An ABR cDNA as a 0.4-kb BamHI-BstEII fragment plus a 2.2-kb BstEII-EcoRI fragment was ligated with a 0.7-kb Eco47III-Bgl II EGFP cDNA fragment into pIRES Neo digested with EcoRV and EcoRI. Also, an ABR cDNA was cloned into pEYFP-C1 (Clontech) digested with Bgl II and XhoI. The entire fusion protein was excised by digestion with AgeI and XhoI and ligated with the lentiviral vector pCCL-cppt-MNDU3 digested with XmaI and XhoI. A BCR cDNA was cloned into pECFP-C1 (Clontech) digested with Bgl II and KpnI. The ECFP-BCR was excised as an AgeI and KpnI fragment and inserted into pCCLcpptMNDU3 digested with XmaI and KpnI.
Plasmids encoding Abr R683A (numbering of amino acid residues refers to human Abr variant 1/isoform a [NM_021962/NP_068781]) and Bcr R1090A in pEGFP-C2 (Clontech) were a kind gift from E. J. Kim and J. R. Lee (Korea Advanced Institute of Science and Technology, Daejon, Korea). The Abr coding region from plasmid EGFP-AbrR683A was subcloned as an EcoRI and XbaI fragment into pcDNA 3.1 HisB and used as a template for further mutagenesis (QuikChange II XL site-directed mutagenesis kit; Stratagene) to add the N795A mutation. The introduction of the mutations was confirmed by sequencing. Segments containing the wild-type, singly R683A mutated, and doubly mutated R683A, N795A GAP domains from residues 597 to 859 (until the stop codon and including 3′ untranslated sequences) were subcloned as 0.8-kb PvuII-EcoRI fragments into pGEX 3X digested with SmaI and EcoRI. The Bcr coding region from plasmid EGFP-BcrR1090A was subcloned into pcDNA 3.1 HisB as an EcoRI-XbaI fragment. This plasmid was used as a template to further generate an N1202A mutation. The mutations were confirmed by sequencing. Segments from the GAP domain including amino acid residues 1004 to 1271 containing the wild type or R1090A and R1090A/N1202A mutations were expressed in pGEX 3X. Plasmids encoding HA-Rac1, HA-Rac2, HA-Cdc42, and HA-RhoA were obtained from the UMR cDNA Resource Center (Rolla, MO). The construction of Xpress-Rac3 was reported previously (17). To generate EGFP-V14RhoC, a 0.7-kb HindIII-XhoI fragment from HA-V14RhoC (UMR cDNA Resources) was subcloned into pEGFP-C3 (Clontech) digested with HindIII-SalI. Myc-V12Cdc42 was a kind gift from A. Hall (Memorial Sloan-Kettering Cancer Center, New York). EGFP-V12mycCdc42 was constructed by subcloning an EcoRI fragment encoding V12mycCdc42 into pEGFP-C1. EGFP-V12Rac1 was constructed by subcloning a BamHI-EcoRI fragment from glutathione S-transferase (GST)-Rac1 into pEGFP-C1. Anti-GFP and anti-Bcr (N-20) antibodies were from Santa Cruz Biotechnology (Santa Cruz, CA). Anti-Abr antiserum was generated against the GAP domain of Abr as described elsewhere (27), and antihemagglutinin (anti-HA) antibodies were purchased from the USC Antibody Core (Los Angeles, CA). Anti-Xpress antibodies were purchased from Invitrogen, and anti-glyceraldehyde-3-phosphate dehydrogenase antibodies were from Chemicon (Temecula, CA).
Electroporation of BMMs.
BMMs were electroporated as described previously (53) with modifications; BMMs were detached with Versene (Invitrogen) and washed once with and then resuspended in DMEM containing 15% FBS. Plasmid DNA (10 μg; endotoxin free) was incubated with BMMs (1.5 × 106 cells; 300-μl volume) for 15 min and electroporated with a pulse of 200 V, 960 μF three times, using a Gene Pulser (Bio-Rad). Electroporated cells were immediately transferred to 5 ml of DMEM containing 15% FBS and rested for 15 min at room temperature. Cells were pelleted and plated in a well of a six-well plate with a glass coverslip insert. After 3 to 5 h of incubation, the medium was replaced with 20% L929-conditioned macrophage medium. Cells were fixed 24 to 72 h after electroporation and stained for F-actin for morphological analysis.
Rho, Rac, and Cdc42 activation assay.
The GST-Pak1 Rac binding domain (RBD) and GST-Rhotekin Rho binding domain (RBD) were isolated and precoupled with glutathione agarose (Sigma, St. Louis, MO) as described elsewhere (4, 43). CHO cells (4 × 106) were transfected with Lipofectamine and Plus reagent (Invitrogen) according to the manufacturer's instructions and grown for 48 h prior to the assay. Rac and Cdc42 activation assays were performed as described elsewhere (4) with some modification; cells were washed twice with ice-cold phosphate-buffered saline (PBS) and lysed in 0.5 to 1 ml MLB (25 mM HEPES, pH 7.5, 150 mM NaCl, 10 mM MgCl2, 1 mM EDTA, pH 8.0, 1% Igepal, and 10% glycerol) and precleared with glutathione-agarose beads (15 min, 4°C). Lysate, 500 μg to 1 mg, was incubated with 10 μg of GST-Pak RBD precoupled with glutathione-agarose beads in a 500-μl volume (1 h, 4°C). For the positive and negative controls of each assay, cell lysates were first incubated with either 0.1 mM GTPγS or 100 mM GDP (Upstate, Lake Placid, NY) (15 min, 30°C) prior to the addition of GST-Pak RBD. Rho activation assays were performed as described elsewhere (43) with 500 μg of lysate. Affinity-precipitated proteins as well as 10 μl of the supernatant were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, followed by immunoblotting with anti-HA antibodies or anti-Xpress antibodies (Invitrogen). The resulting blots were scanned and analyzed with Un-Scan-It software (Silk Scientific, Orem, UT). GTPase activation is reported as the ratio of GTP-bound GTPase to total GTPase after normalization to a control.
To measure Rac activation during phagocytosis of opsonized Escherichia coli, BMMs were starved of CSF-1 and serum in DMEM containing 1% FBS for 24 h. Starved cells were detached with EDTA and washed once with Hanks' balanced salt solution without magnesium and calcium. Cells (1.5 × 106; 250-μl volume) were incubated with opsonized fluorescein isothiocyanate (FITC)-E. coli (50 bacteria per cell) for 30 min on ice, and phagocytosis was initiated by placing cells at 37°C. At the indicated time points, 250 μl of ice-cold 2× MLB was added to the cell suspension to stop the phagocytosis reaction. The entire cell lysate was used for the Rac activation assay.
GAP activity assay.
Wild-type and mutant GAP domains of Bcr and Abr as bacterially expressed GST fusions were purified as described previously (10). GAP activity was measured using a RhoGAP assay kit (Cytoskeleton, Denver, CO) according to the manufacturer's specifications. Briefly, 0.34 μM GST or GST fusion protein was added to an assay mixture containing Rac1 GTPase and GTP. After 20 min at 37°C, developing reagent was added and phosphate production was measured at 650 nm.
Immunofluorescence microscopy for morphology analysis.
For morphology analysis on glass, PEMs or BMMs (2.5 × 104 cells) were cultured on Lab-Tec glass chamber slides (Nunc-Nalgene, Naperville, IL) for 24 h in complete macrophage medium without CSF-1 for PEMs and with 2.22 nM CSF-1 for BMMs. RAW cells were plated in a six-well plate containing coverglass slides (Fisher Scientific), followed by transfection using FuGene HD (Roche, Indianapolis, IN) according to the manufacturer's instructions and cultured for 24 to 48 h prior to fixation. For microscopy, cells were washed twice with PBS and fixed in 4% paraformaldehyde (Electronic Scientific Co., Hatfield, PA) (15 min at room temperature [RT]), followed by permeabilization in 0.2% Triton X-100 (5 min, RT). To localize filamentous actin (F-actin), cells were stained with FITC or tetramethyl rhodamine isocyanate-phalloidin (Sigma) (1 μg/ml; 30 min; 10% normal goat serum) and mounted in Vectashield containing 4′,6′-diamidino-2-phenylindole (DAPI; Vector Laboratories, Burlingame, CA). Cell images were acquired with a Leica DM RA upright microscope and Easy Fish software.
Intracellular translocation of EYFP-Abr and ECFP-Bcr with CSF-1 stimulation.
Lentiviral transduction of BMMs was previously reported (12). BMMs expressing EGFP, enhanced yellow fluorescent protein (EYFP)-Abr, or enhanced cyan fluorescent protein (ECFP)-Bcr were starved of CSF-1 for 24 h. BMMs were maintained alive in DMEM supplemented with 25 mM HEPES and 0.1% bovine serum albumin and dropped onto glass-bottom petri dishes (MatTek Corp., Ashland, MA) that had been coated with fibronectin (10 ng/ml). After allowing the BMMs to attach to the glass slides for 10 min, serial images of cells expressing fluorescent proteins were recorded every 30 seconds. To stimulate cells, CSF-1 (15 ng/ml) was directly added to the medium in the vicinity of the cells under study. Images were obtained with a Leica TCS SP1 spectral confocal laser scanning microscope mounted on a Leica DM IRBE inverted microscope (oil immersion, 40× objective; 458-nm excitation with argon laser).
Migration assay.
Migration assays using Transwells were performed as described elsewhere (22) with modification. BMMs (5 × 104 cells, in 100 μl macrophage medium) were placed in the upper compartment of the Transwell chamber membrane (8-μm pore size, polycarbonate membrane; Corning Costar, Cambridge, MA) and allowed to migrate for 3 h toward the lower compartment of the chamber containing 2.22 nM CSF-1. To measure random motility, 2.22 nM CSF-1 was added to both compartments of the Transwell chamber. After incubation, the migrated cells on the Transwell membrane were stained with crystal violet (Sigma). The images of the stained cells were obtained with a Leica fluorescence MZ FL III stereomicroscope, and the cells in a field that corresponded to half of the total area of the Transwell membrane were counted. Each assay was performed in duplicate, and two independent sets of BMMs were tested.
Fluorescence-activated cell sorting (FACS) and microscopic analysis of phagocytosis.
BMMs were starved of CSF-1 for 24 h in DMEM containing 1% FCS, resuspended in 0.5 ml DMEM, and prechilled on ice. FITC-conjugated E. coli cells (Invitrogen-Molecular Probes) were serum opsonized according to the manufacturer's instruction and mixed with the prechilled BMMs at a ratio of 25 bacteria per cell on ice for 30 min. Phagocytosis was initiated by incubating the cell and bacteria mixture at 37°C for the indicated time period and stopped by placing the mixture on ice immediately after incubation. Cells were washed with cold PBS five times and incubated in 0.25% trypan blue (pH 2.5, in acetate-buffered saline; EMD-Merck, Darmstadt, Germany) for 15 min on ice to quench the fluorescence from externally bound bacteria. After washing off the quenching solution, cells were fixed in 2% paraformaldehyde (15 min, RT) and analyzed with a Coulter ELITE flow cytometer. Mean fluorescence intensities (MFIs) for FITC-positive populations were compared. For microscopic analysis of phagocytosis, RAW cells were transfected directly on a coverglass slide. At 24 h posttransfection, cells were starved in RPMI medium containing 1% FCS. Texas Red-coupled zymosan was opsonized according to the manufacturer's instructions (Invitrogen-Molecular Probes). Twenty-five particles of zymosan per cell in 0.5 ml cold RPMI were applied to washed cells, and these were incubated on ice for 30 min. After unbound zymosan particles were washed away, cells were allowed to phagocytose for 1 h and fixed immediately. To observe phagosome formation in BMMs, cells (5 × 104) were cultured on Lab-Tec chamber slides (Nunc) in macrophage medium for 24 h. AlexaFluor 594- or Texas Red-coupled zymosan particles were opsonized according to the manufacturer's instructions (Invitrogen-Molecular Probes). Forty particles of opsonized zymosan per cell in 0.5 ml of cold DMEM with 15% FBS were centrifuged onto the adherent BMMs (400 × g; 5 min; 4°C). After washing twice with cold DMEM, phagocytosis was initiated by adding prewarmed DMEM with 15% FBS. After 15 min of incubation, the medium was removed and the BMMs were washed twice with cold PBS, fixed, and stained for F-actin. To quantitate phagocytosis, the number of phagosomes of 100 wild-type or double null BMMs was determined as the phagocytic index.
RESULTS
Abr and Bcr function as specific GAPs for Rac in vivo.
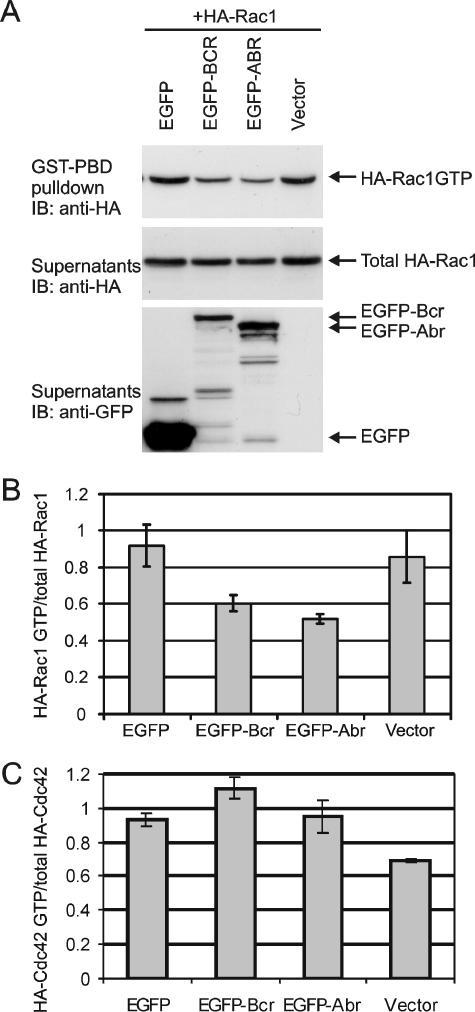
The bacterially expressed GAP domains of Abr and Bcr have been shown to be catalytically active toward both Rac and Cdc42 (10, 20). In addition, we have previously detected elevated levels of Rac GTP in (abr × bcr)−/− PEMs that were activated in vivo by injection of mice with thioglycolate (27). To confirm the in vivo GAP activity of Abr and Bcr as whole proteins and to determine the in vivo specificity of the GAP activity towards different Rho family GTPases, EGFP, EGFP-Abr, or EGFP-Bcr was transiently expressed in CHO cells together with either HA-Rac1, HA-Rac2, Xpress-Rac3, HA-Cdc42, or HA-RhoA. Intracellular levels of Rac GTP/Cdc42 GTP and Rho GTP were analyzed using GST-Pak and GST-Rhotekin pull-down assays, respectively. Each pull-down assay included cell lysates incubated with GTPγS as a positive control and cell lysates with GDP as a negative control. As shown in Fig. 1A, upper panel, the amount of Rac1 GTP precipitated with GST-Pak RBD was clearly lower in cells expressing EGFP-Bcr or EGFP-Abr than in cells expressing EGFP alone, whereas the amount of total Rac1 protein was equal (Fig. 1A, middle panel). The quantitation showed that the overexpression of EGFP-Abr and EGFP-Bcr without any further treatment led to a 50% reduction of Rac1 GTP levels compared to cells expressing EGFP alone (Fig. 1B). Similarly, the GST-Pak pull-down assay on CHO cells expressing EGFP-Abr and EGFP-Bcr with either HA-Rac2 or Xpr-Rac3 showed up to a 50% decrease of Rac2 GTP and Rac3 GTP levels, respectively (data not shown). In contrast, EGFP-Abr and EGFP-Bcr had no effect on Cdc42 GTP levels (Fig. 1C) or on RhoA GTP levels (data not shown). Using similar pull-down methods with GST-ELMO1 as the detection tool to affinity precipitate RhoG GTP (41), we were unable to find evidence for activity of Abr or Bcr as GAPs for the RhoG protein, which is closely related to Rac (62, 63) (results not shown). These data strongly suggest that, in vivo, Abr and Bcr only act as Rac-specific GAPs.
FIG. 1.
Abr and Bcr reduce levels of activated Rac but not activated Cdc42. (A) Plasmids encoding Bcr and Abr as EGFP fusion proteins as well as EGFP alone were coexpressed with HA-tagged Rac1 in CHO cells. HA-Rac1 GTP was affinity precipitated with GST-Pak RBD. Total Rac levels and Abr and Bcr expression were analyzed from the supernatants after GST-Pak RBD precipitation. Top panel, HA-Rac1 GTP; middle panel, total HA-Rac1 protein; bottom panel, expression of EGF-Bcr, EGFP-Abr, and EGFP. One representative image of three independent experiments is shown. (B) The ratio of HA-Rac1 GTP levels to total HA-Rac1 levels was determined and then normalized to that of EGFP-expressing cells. Bars represent the mean ± standard deviation of at least three independent experiments. (C) EGFP-Bcr, EGFP-Abr, and EGFP were coexpressed with HA-Cdc42, and the relative activation of HA-Cdc42 was determined. Bars represent the mean ± standard deviation of two independent experiments.
Loss of Abr and Bcr leads to a highly elongated morphology in primary macrophages.
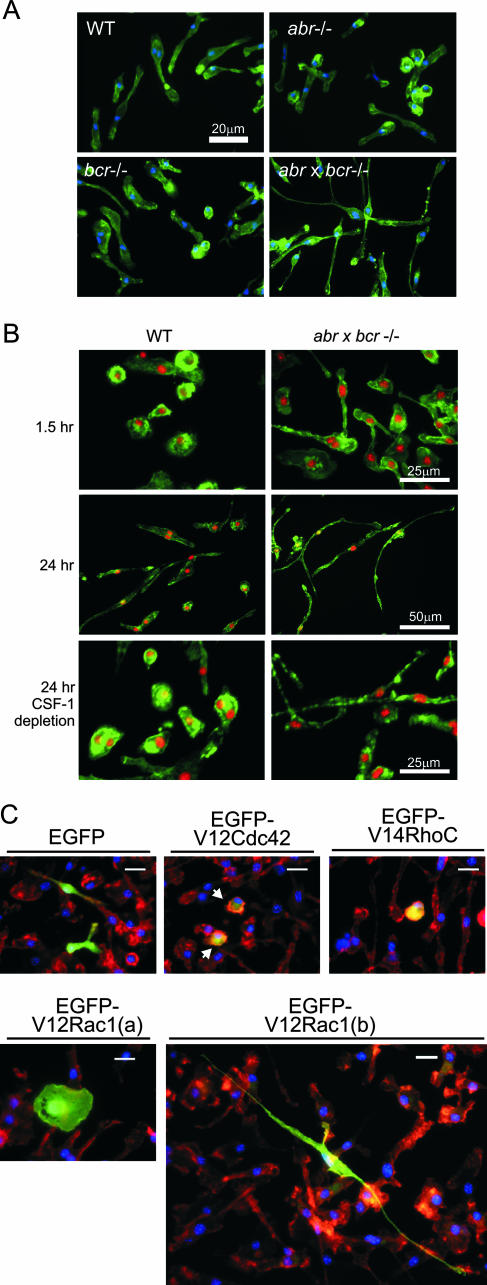
Rho GTPases and their regulators are known to govern actin cytoskeletal organization and, thus, regulate the morphology and polarity of phagocytic leukocytes, such as the macrophage and neutrophil (2, 26, 60, 61). Both Abr and Bcr are expressed across different types of phagocytic leukocytes, including primary BMMs, PEMs, and peritoneal elicited neutrophils (Cunnick et al., unpublished). Given the role of Abr and Bcr in regulating Rac GTP levels and the known involvement of Rac in regulating the actin cytoskeleton, the overall morphology of PEMs and BMMs from wild-type, abr−/−, bcr−/−, and (abr × bcr)−/− mice was first explored. After activation with thioglycolate in vivo, PEMs were isolated and cultured on glass slides for 24 h in the absence of CSF-1 and stained for F-actin. Microscopic analysis of their morphology showed that there was no overt difference among wild-type and Abr and Bcr single null PEMs, which all showed a mixture of round and elongated shapes (Fig. 2A). In sharp contrast, the double null mutant PEMs had developed a remarkably elongated and polarized shape with a clear leading edge and a retraction tail. This markedly elongated morphology was also prominent in (abr × bcr)−/− BMMs when they were plated on glass slides in the presence of CSF-1. Within 1.5 h the double null mutant BMMs initiated long protrusions and developed a polarized morphology (Fig. 2B, upper panel), whereas the wild-type BMMs remained round and unspread. Twenty-four hours later, the double null mutant BMMs had developed an almost neurite-like, long, thin morphology and some of the cells reached a length of 170 μm, while the control wild-type BMMs displayed a mixture of round and elongated shapes (Fig. 2B, middle panel). With subsequent depletion of CSF-1, the double null mutant BMMs maintained their highly elongated morphology, in contrast to the control wild-type BMMs, which started to round up (Fig. 2B, bottom panel). There was no noticeable difference in morphology on glass surfaces among wild-type and abr−/− and bcr−/− single null BMMs (data not shown). When we reintroduced EGFP-Bcr into wild-type or (abr × bcr)−/− BMMs using electroporation, both types of cells failed to develop long thin extensions and remained rounded. Reintroduction of EGFP-Abr only partly reduced elongation of the cells (see Fig. S1A in the supplemental material).
FIG. 2.
Abnormally elongated morphology of macrophages lacking both Abr and Bcr is phenocopied by activated Rac. PEMs of the indicated genotypes were plated on glass chamber slides and incubated for 24 h in the absence of CSF-1 (A), or BMMs of the wild type and double null mutants were cultured on glass chamber slides for 1.5 h (B, upper panel) or 24 h (B, bottom panels) in the presence of 120 ng/ml CSF-1. F-actin was stained with FITC-phalloidin (green), and nuclei were stained with DAPI (blue in panel A, red in panel B). Note that the double null BMMs already started showing the elongated morphology after 1.5 h in culture. Bars, 20 μm (A and C), 25 μm (B, top and bottom panels), or 50 μm (B, middle panel). (C) Wild-type BMMs were electroporated with the indicated constructs. Arrows point to cells expressing V12Cdc42. Cells were stained for nuclei (blue) and F-actin (red); EGFP-expressing cells appear green.
To investigate if the highly elongated morphology of cells lacking both Abr and Bcr was correlated with Rac activity, we introduced fluorescently labeled, constitutively active Rac1, Cdc42, or RhoC into the primary wild-type BMMs and analyzed their morphology. As shown in Fig. 2C, expression of EGFP alone did not alter the morphology of these cells significantly, whereas expression of both constitutively active Cdc42 and RhoC caused cells to become rounder and less spread. Expression of V12Rac had a very clear phenotypic effect on the primary BMMs, which depended on the expression levels as judged by the EGFP fluorescence intensity. Cells that expressed the highest levels of V12Rac were very round and spread [Fig. 2C, EGFP-V12Rac1(a)]. Interestingly, cells that had lower V12Rac expression showed a very extended, long, thin morphology [Fig. 3C, EGFP-V12Rac1(b)] that phenocopies the morphology seen in (abr × bcr)−/− BMMs. These data showed that the concomitant loss of Abr and Bcr affects cellular morphology and causes a highly polarized shape in macrophages which is correlated with Rac activity.
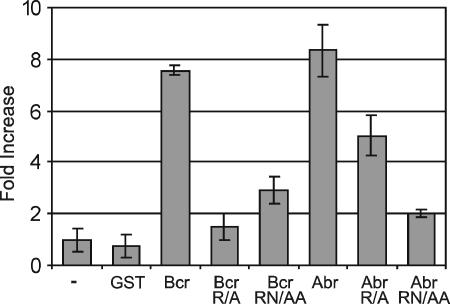
FIG. 3.
GAP activity of Bcr or Abr is inhibited by mutation of conserved arginine and asparagines residues. GST fusion proteins tested include the wild-type Bcr and Abr GAP domains (bars labeled Bcr and Abr), Bcr R1090A (Bcr R/A), Abr R683A (Abr R/A), Bcr R1090A/N1202A (Bcr RN/AA), and Abr R683A/N795A (Abr RN/AA). Purified Bcr or Abr GAP domains were tested for Rac1 GAP activity. Results are expressed as the change with respect to Rac GTPase alone. Each GAP assay was performed in duplicate, and bars represent the mean ± standard deviation of two independent experiments.
Expression of GAP-defective mutants of Abr and Bcr results in elongated morphology in RAW cells.
Apart from the GAP domain, Abr and Bcr also contain a DH domain that, in vitro, is modestly active as a GEF for Rho family GTPases (10, 20). Therefore, it was possible that the abnormal morphology of macrophages lacking Abr and Bcr could be partly attributed to loss of the DH domains of Abr and Bcr. In Dbl, mutation of residues N673 and D674 completely abolished its activity as a GEF towards Rac (1). These residues are highly conserved among DH domains and are also conserved as residues N689/E690 in Bcr and N282/E283 in Abr. We therefore mutated these residues in both Abr and Bcr and introduced the mutants into the RAW 264.7 macrophage cell line. However, neither construct's expression caused morphological changes in these cells (results not shown).
We next examined whether the loss of GAP activity is directly linked to the abnormal morphology observed in (abr × bcr)−/− macrophages. To investigate this, we similarly made point mutations in the GAP domain to knock out its activity. GAP domains containing a mutation in the highly conserved arginine residue (Abr R683A and Bcr R1090A) were compared to wild-type GAP domains. In some GAPs such a single arginine mutation is not sufficient to completely abrogate the GAP function. Therefore, we introduced an additional mutation in conserved asparagine residues, making double mutants Bcr R1090A/N1202A and Abr R683A/N795A, and tested these for GAP activity in vitro. As shown in Fig. 3, both Abr and Bcr wild-type GAP domains were highly active against Rac. The R1090A mutation in Bcr was sufficient to abrogate its activity, whereas the analogous mutation in Abr, R683A, substantially reduced but did not entirely eliminate the GAP activity. However, the addition of the N795A mutation essentially abolished the GAP activity. To test whether such mutants are still able to bind activated Rac, we cotransfected wild-type and mutant, full-length Abr and Bcr with GST-V12Rac. Pull-down experiments confirmed that V12Rac was still able to form a stable complex with these mutants that was detectable in a pull-down assay (see Fig. S2 in the supplemental material).
To further verify that these mutants were inactive as GAPs in vivo, we compared the ability of wild-type EGFP-Bcr and EGFP-Bcr R1090A to complement the loss of bcr in bcr−/− BMMs spreading on fibronectin. Similar experiments were performed with EGFP-Abr and EGFP-AbrR683A in abr−/− BMMs. The results (see Fig. S1B in the supplemental material) showed that the BcrR1090A mutant could not prevent cell spreading, whereas AbrR683A showed a partial inhibition, consistent with this mutant still having residual GAP activity.
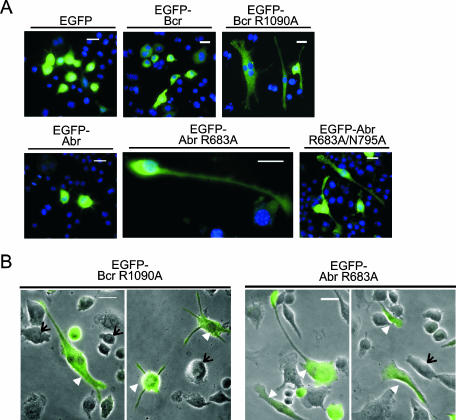
We next cultured RAW cells on glass coverslips and transiently transfected them with EGFP, EGFP-Bcr, EGFP-Abr, EGFP-Bcr R1090A, EGFP-Abr R683A, or EGFP-Abr R683A/N795A. RAW 264.7 cells express moderate amounts of endogenous Abr and Bcr (data not shown). At 24 h after transfection, cells were fixed and analyzed under a fluorescence microscope. In Fig. 4A, the effects of each of these on overall morphology are shown. All proteins except EGFP-Bcr R1090A showed an even, diffuse staining throughout the cytoplasm, whereas EGFP-Bcr R1090A also exhibited punctate staining in some cells (not visible in Fig. 4A). RAW cells expressing EGFP-Bcr R1090A, EGFP-Abr R683A, or EGFP-Abr R683A/N795A developed a prominent long and thin morphology, whereas cells expressing EGFP, EGFP-Abr, or EGFP-Bcr remained round and showed few cellular protrusions. To visualize the cell periphery more clearly and to compare the morphology of transfected cells to that of untransfected cells, EGFP fluorescence images (green) were superimposed with phase-contrast images (black and white) taken from the identical microscopic field. As shown in Fig. 4B, cells that were expressing EGFP-Abr R683A developed a highly polarized cell shape without any stimulation, which mimicked that of (abr × bcr)−/− macrophages. The expression of EGFP-Bcr R1090A also caused an elongated shape in some cells as well as multiple spike-like cellular protrusions in others. In great contrast, nontransfected cells maintained a mixture of a round or moderately elongated morphology (Fig. 4B). These data indicate that the GAP-defective mutants of Abr and Bcr act as dominant negative proteins, profoundly affecting macrophage cellular morphology by inducing an abnormally elongated shape in the macrophage cell line.
FIG. 4.
GAP-defective mutants of Abr and Bcr induce abnormally elongated morphology in a macrophage-derived cell line, RAW 264.7. Cells were transfected with the indicated plasmids. After 24 h, cells were fixed, stained for nuclei, and analyzed with a fluorescence microscope. (A) Cells were visualized for EGFP (green) and nuclei (blue). Bars, 20 μm. EGFP-Abr R683A-expressing cells are shown at a higher magnification. (B) EGFP images (green) of cells were superimposed on those of phase-contrast images (black and white) to illustrate overall cell shape. Note that some of the cells expressing GAP mutants of Abr and Bcr (arrowheads) developed a highly elongated shape or multiple protrusions, while untransfected cells (arrows) remained round.
Combined loss of Abr and Bcr results in a synergistic increase in directed cell migration both in vivo and in vitro.
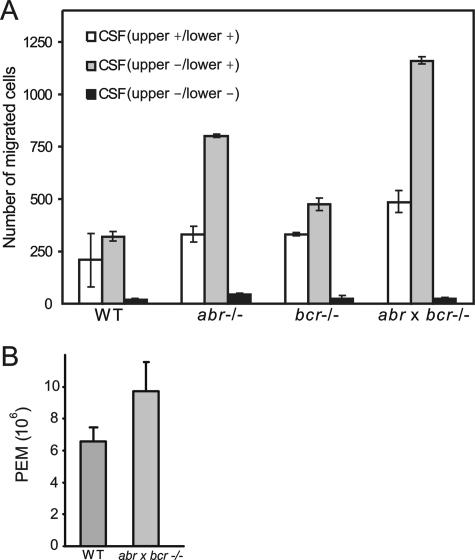
During migration, cells protrude lamellipodia to form new adhesion sites at the leading edge in the direction of movement and retract the other opposite end (46). Therefore, a polarized shape is closely correlated with migration of a cell (48, 59), and both the polarization and migration of macrophages and neutrophils are known to be regulated by Rac (2, 9, 26). Given that the combined loss of Abr and Bcr resulted in a polarized morphology in BMMs in the presence of CSF-1, it was of interest to test if the polarized elongated shape developed in macrophages lacking both Bcr and Abr is linked to changes in their ability to migrate toward chemokines. To test this possibility, CSF-1, a well-known attractant of macrophages, was used to induce directed migration in macrophages. Random migration was measured by adding CSF-1 to both compartments of the Transwell and allowing cells to move in the presence of a uniform concentration of CSF-1. Directed migration was measured by adding CSF-1 to only the lower compartment. As shown in Fig. 5A, there was no statistically significant difference in random motility among wild type, abr−/−, bcr−/−, and (abr × bcr)−/− BMMs (P > 0.05). However, in the presence of a CSF-1 gradient (Fig. 5A), Abr single null BMMs showed a 3-fold increase in their motility and Bcr single null BMMs showed a modest increase of 1.5-fold compared to the control wild type. Interestingly, the double null mutant BMMs displayed a robust 3.7-fold increase in directed migration toward CSF-1 compared to that of the wild-type BMMs.
FIG. 5.
(abr × bcr)−/− macrophages have increased motility both in vitro and in vivo. (A) BMMs were added to the upper chamber of Transwells and allowed to migrate for 3 h. Random motility was measured in the presence of CSF-1 in both the upper and lower chambers of Transwells (white bars), and directed migration was measured in the presence of CSF-1 in the lower chamber (gray bars). Black bars represent motility in the absence of CSF-1 in both the upper and lower chambers. Values shown are the mean ± standard deviation (n = 2 per genotype), and each experiment was performed in duplicate. The differences in CSF-1-directed migration between the wild-type and abr−/− BMMs and between the wild-type and (abr × bcr)−/− BMMs were statistically significant (P < 0.05; two-tailed Student t test, Bonferroni adjustment). (B) The number of macrophages in peritoneal exudates from wild-type or double null mutant mice (n = 3) was compared 4 days after intraperitoneal thioglycolate injection and was greater in double null mutant mice (P < 0.05).
In vivo, circulating monocyte/macrophages are attracted to cytokines and chemokines that are generated at the site of inflammation. The peritoneum after thioglycolate injection represents an experimentally induced site of inflammation, and the number of macrophages recruited to the peritoneum has been used to assess the in vivo motility of macrophages (64). To examine whether loss of Abr and Bcr affects the in vivo motility of macrophages, we compared the number of macrophages in peritoneal exudates recovered from wild-type, abr−/−, bcr−/−, and (abr × bcr)−/− mice 4 days after thioglycolate injection. There was no significant difference in the number of recovered PEMs between wild-type and Abr and Bcr single null mice (data not shown). In contrast, as shown in Fig. 5B, the number of macrophages in peritoneal exudates from the double null mutant mice was greatly increased (P < 0.05) compared to that of the control wild-type mice. These data indicated that, both in vivo and in vitro, the combined loss of Abr and Bcr leads to a synergistic increase in directed motility of macrophages.
Abr and Bcr translocate to the plasma membrane upon stimulation with CSF-1.
The increased CSF-1-induced motility of macrophages that are lacking both Abr and Bcr suggested to us that Abr and Bcr might regulate the pool of Rac that is activated by CSF-1 stimulation. Activated Rac and its GEFs are known to localize at the plasma membrane after different cellular stimuli (13, 54, 65) and would be expected to be removed from that location upon GTP hydrolysis. Indeed, in a time course following CSF-1 stimulation, increased levels of endogenous Rac were recovered in the cytosolic fraction of BMMs, whereas those of Cdc42 and RhoA remained constant (see Fig. S3A in the supplemental material). Also, in order to inactivate RacGTP, Abr and Bcr would be expected to transiently associate with the plasma membrane, the site of activated Rac accumulation. However, in RAW cells, EGFP-Bcr and EGFP-Abr are mainly present in the cytosol (Fig. 4A).
Recent studies have suggested that Bcr localizes to the plasma membrane at the cellular junctions of epithelial cells (34, 42). Therefore, possible transient changes in Bcr and Abr localization could occur depending on the cell type and stimulus. To investigate a possible membrane association of Abr, CHO cells were transiently transfected with wild-type Abr and a mutant Abr carrying an N-terminal myristoylation sequence. A similar construct could not be made for Bcr since it has an N-terminal oligomerization domain thought to be important for its normal cellular function. Subcellular fractionation showed that a small amount of Abr protein is present in the membrane fraction in unstimulated cells and that myristoylation increases this (see Fig. S3B in the supplemental material). Interestingly, under steady-state levels, less HA-RacGTP was measured in the samples expressing myristoylated Abr (see Fig. S3C and D in the supplemental material).
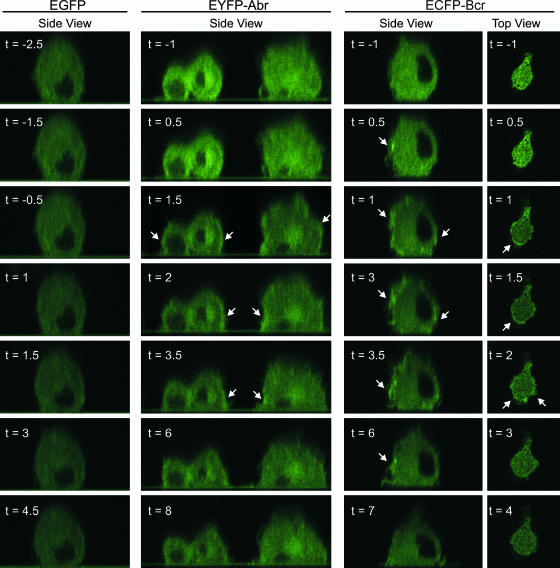
To further determine the subcellular localization of Abr and Bcr in live quiescent and CSF-1-stimulated cells and to be able to detect transient movement, BMMs were transduced with lentivirus encoding EYFP-Abr or ECFP-Bcr and stimulated with CSF-1. In Fig. 6 and Movies S1 though S4 in the supplemental material, serial confocal microscope images of live BMM in the x-y (top view) or x-z (side view) plane are shown. Prior to the addition of CSF-1 (Fig. 6, time with negative numbers), the fluorescent signal from cells expressing EYFP-Abr or ECFP-Bcr appeared to be distributed evenly through the cytoplasm of each cell. In sharp contrast, within 1 to 2 minutes after the addition of CSF-1 (Fig. 6, time with plus numbers), an intensification of the fluorescent signal from EYFP-Abr or ECFP-Bcr could be seen at the plasma membrane of the cell. This signal was transient in nature but remained distinguishable for up to 6 min. In Fig. 6, areas of the cell periphery that display a noticeable brightening after stimulation are evident. Some of these signals appear to be localized to ruffling processes induced by CSF-1 (ECFP-Bcr; side view, t = 3 and 3.5). However, this type of transient intensification at the plasma membrane was entirely absent in cells expressing EGFP after stimulation, which confirmed that the translocation of Bcr and Abr was not an artifact caused by a general concentration of the fluorescent signal at the cell periphery.
FIG. 6.
Abr and Bcr translocate to the plasma membrane upon stimulation with CSF-1. Wild-type BMMs were transduced with lentivirus encoding the indicated proteins. The transduced BMMs were starved of CSF-1 overnight and allowed to attach to the fibronectin-coated surface of a glass-bottom petri dish. Serial confocal images in the x-z mode (side view) and x-y mode (top view), before and after the addition of CSF-1 to the cells, were collected every 30 seconds. Each number represents a time point in minutes before (designated with a minus sign) and after (designated with a plus sign) CSF-1 addition. CSF-1 was added between t = −1 and t = 1 for each series of images. The nucleus of each cell was identified by the dark area in the center of the cell, which lacks expression of the fusion proteins. The plane of focus for images captured in the x-z mode was through the nucleus of the cell, while that of the images captured in the x-y mode was near the attachment of the cell to the glass. The arrows mark areas of fluorescence intensification that occurred at the periphery of the cells.
To complement these results, we captured images of cells stimulated with CSF-1 in the x-y mode (Fig. 6, top view; see also Movie S4 in the supplemental material). These also displayed an enrichment of the fluorescent signal at the plasma membrane. Interestingly, in the top view series of Fig. 6, the fluorescent signal from ECFP-Bcr became concentrated along the plasma membrane of what appears to be the leading edge of a polarized BMM, displaying a migrating morphology after stimulation with CSF-1. Although it was technically challenging to locate migrating cells during these studies, a migrating BMM expressing ECFP-Bcr was captured and is shown in Movie S5 of the supplemental material. Similar to the cell in the top view series of ECFP-Bcr in Fig. 6, an enrichment of the signal from ECFP-Bcr was observed near or at the leading edge of a clearly migrating BMM in a gradient of CSF-1. These images further showed that Abr and Bcr dynamically change their localization, transiently moving to the leading edge of cells as well as to the plasma membrane in response to chemokine stimulation.
Phagocytosis is enhanced in the absence of Abr and Bcr.
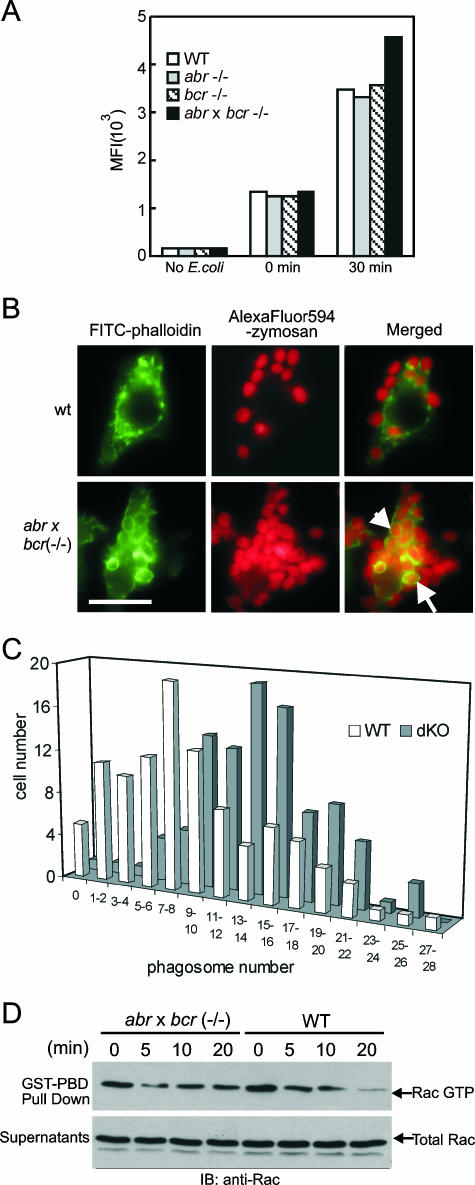
As with cell morphology and directed cell migration, phagocytosis is also an important cellular response of macrophages that is under the control of Rho GTPases. In particular, Rac is known to reorganize the actin cytoskeleton necessary for phagocytic cup formation and closure (8, 35). To explore the potential involvement of Bcr and Abr in phagocytosis, the effect of the loss of Abr and Bcr on phagocytosis in BMMs was examined with fluorescently labeled E. coli. The level of phagocytosis was determined by MFIs from the internalized FITC-E. coli using FACS analysis. As shown in Fig. 7A, after 30 min of incubation at 37°C, all BMMs of the different genotypes showed a noticeable increase in MFI, indicating that these cells were able to ingest FITC-E. coli under these experimental conditions. Importantly, phagocytosis was substantially enhanced in (abr × bcr)−/− BMMs, whereas Abr and Bcr single null BMMs showed no difference compared to wild type. In BMMs that were kept at 4°C (Fig. 7A, 0 min), no difference was observed in MFIs, indicating that the internalization rather than the binding of E. coli was affected by loss of Abr and Bcr.
FIG. 7.
Combined loss of Abr and Bcr results in increased phagocytosis by macrophages and is associated with sustained Rac activation. (A) BMMs were incubated with FITC-E. coli at a ratio of 1:25 in serum-free medium and analyzed by FACS. MFIs of FITC fluorescence from FACS were plotted on the y axis. (B) Representative images of wild-type and double null mutant cells phagocytosing opsonized zymosan particles. F-actin was stained with FITC-phalloidin (green), and zymosan was coupled with AlexaFluor (red). Red and green images are merged to identify the internalized zymosan particles. The external particles appear bright red, whereas the internalized zymosan particles appear orange and are surrounded by F-actin-rich phagocytic cups (arrows point to examples). Bar, 20 μm. (C) Number of phagosomes containing opsonized zymosan. The amount of internalized particles in each cell was counted and plotted against the number of cells containing that amount of particles. The difference in phagosome number distribution between wild-type and the double null BMMs was statistically significant (P < 0.01). (D) BMMs of the indicated genotypes were preincubated with opsonized FITC-E. coli, allowed to phagocytose for the indicated time, and assayed for activated and total Rac as described for Fig. 1.
A detailed microscopic analysis of phagocytosis was performed to further examine the difference between wild-type and the double null BMMs during this process. After BMMs were allowed to engulf AlexaFluor 594-zymosan particles (red), cells were fixed and stained for actin using FITC-phalloidin (green). The internalized zymosan particles were microscopically identified by F-actin-rich phagocytic cups completely surrounding the engulfed zymosan. As shown in Fig. 7B, the double null BMMs appeared to ingest more zymosan particles than the wild type and formed more distinctive F-actin-rich phagocytic cups around the zymosan. To quantitate this difference in phagocytosis, the number of phagosomes was plotted against the number of cells that developed that number of phagosomes. In addition, the number of phagosomes in each of 100 wild-type and double null mutant cells was determined as the phagocytic index. This analysis showed the remarkable difference in the distribution of phagosome number per cell between the two genotypes (Fig. 7C), with many more double null mutant cells ingesting large numbers of particles than the wild-type cells. The phagocytic index from this analysis showed that the double null BMMs ingested a total of 1,337 particles per 100 cells, in comparison to 915 in the wild-type BMMs. This difference in phagosome distribution was statistically significant (two-tailed Student t test; P < 0.01). We also compared wild-type and (abr × bcr)−/− BMMs for the kinetics of Rac deactivation following initiation of phagocytosis of opsonized E. coli. As shown in Fig. 7D, cells that were in the initial steps of uptake of E. coli had a measurable amount of endogenous activated Rac at t = 0. Over the course of 20 min, this level decreased in the wild-type cells. Interestingly, and consistent with the lack of proteins that would downregulate RacGTP levels, we found sustained Rac activation in the (abr × bcr)−/− BMMs under these conditions. These data show that loss of Abr and Bcr profoundly affects phagosome formation and leads to an increase in phagocytosis in the (abr × bcr)−/− macrophages through decreased deactivation of Rac.
GAP-defective mutants of Abr and Bcr localize to the phagocytic cup during phagocytosis.
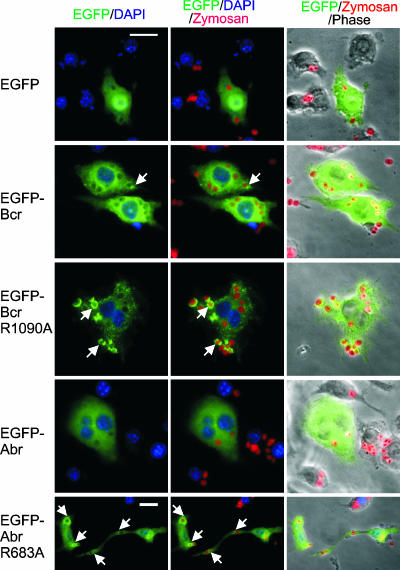
The enhanced phagocytosis as well as the pronounced cup formation in the double null BMMs prompted us to further examine the subcellular localization of Abr and Bcr during phagosome formation, because the specific translocation of Rho GTPase and their regulators to nascent phagosomes had been previously suggested (11, 39). To examine the localization of Abr and Bcr during phagocytosis, RAW cells were transiently transfected with EGFP, EGFP-Bcr, EGFP-Abr, EGFP-Bcr R1090A, or EGFP-Abr R683A. After 24 h of serum starvation, the transfected RAW cells were allowed to phagocytose opsonized Texas Red-zymosan particles. Engulfed zymosan particles were distinguished from those that were bound on the surface by using phase-contrast microscopy, since the ingestion of particles by a cell is known to cause a contrast change of the particle from phase bright to dimmer, flatter, and phase dark (14). As shown in Fig. 8, there was no clear local enrichment of EGFP-Abr around phagosomes, partly due to the fact that it was difficult to find cells that formed a complete phagosome. There was no apparent accumulation of EGFP-Bcr around phagosomes either, with few, rare exceptions, as shown in Fig. 8. In contrast, there was a striking recruitment of EGFP-Bcr R1090A to phagosomes that had both started forming and that had been completely formed. In the mature phagosomes, the EGFP-Bcr R1090A signal completely surrounded the engulfed zymosan particles (Fig. 8). A similar observation was made with EGFP-Abr R683A, although the enrichment was moderate compared to that of EGFP-Bcr R1090A. These data indicated that Bcr and Abr, in their mutant GAP forms, were brought to the actively forming phagosome, and EGFP-Bcr R1090A, in particular, could induce strong phagocytic cup formation.
FIG. 8.
GAP-defective mutant Bcr and Abr localize to phagocytic cups during phagocytosis of opsonized zymosan particles. After transfection with the indicated plasmids, RAW cells were serum starved and then incubated with Texas Red-coupled zymosan particles at a ratio of 1:25. EGFP-expressing cells appear green, zymosan is red, and nuclei are blue. Images with different fluorescences were merged, as indicated above the panels, to identify EGFP signal localizing to phagosomes (arrowheads point to examples). Bars, 20 μm. Note that the images of the EGFP-Abr R683A-transfected cells are shown at a lower magnification than the others, in order to capture one elongated cell.
DISCUSSION
Recent reviews have reported that the human genome contains more than 69 genes encoding RhoGAPs (5, 36), far exceeding the 22 genes encoding Rho family GTPases (25). The excess of RhoGAPs over their target Rho GTPases suggests that these RhoGAPs perform very specific functions and might be critical for the regulation of small GTPases during different cellular events (6, 36). Abr and Bcr are highly homologous multidomain proteins that contain such GAP domains with in vitro specificity for the Rac and Cdc42 members of the Rho family. The in vivo function and specificity of the GAP domain of Abr and Bcr was unclear. In this study, using a mouse model that lacks both proteins, we addressed this issue and report the synergistic regulation of macrophage cellular behavior by these two highly homologous proteins. We found the concomitant loss of Abr and Bcr in macrophages caused dramatic changes in cell morphology and led to robust increases in directed migration and phagocytosis, which were unequivocally linked to the loss of their Rac GAP activity. This study is the first to show such profound effects of the genetic ablation of GAPs specific for Rac in macrophages.
Abr and Bcr contain a domain with homology to those that are found in Rho GEFs (DH domains) in addition to their GAP domains. The isolated DH domains of Abr and Bcr exhibited catalytic GEF activity toward bacterially expressed Rac, Cdc42, and RhoA (10). However, in the current study, we did not find positive evidence supporting an activity of these domains either in vitro or in vivo. Recently, others found that Bcr can also act as a GAP for Rho in vitro if RhoA is expressed in baculovirus and thus is properly prenylated (33). Therefore, it was of importance to further investigate the specificity of the GAP activity of Abr and Bcr. We confirmed that the isolated GAP domains of Abr and Bcr are highly active as GAPs in vitro towards Rac and that mutations of residues that, in other Rho GAPs, are critical for their catalytic activity completely abrogate Abr and Bcr GAP activity. In this study, we also found biochemical evidence supporting in vivo activity of the entire Abr and Bcr proteins only as Rac GAPs. The expression of Bcr and Abr as EGFP fusion proteins in a mammalian cell line specifically reduced the amount of active Rac (Rac1, Rac2, and Rac3) only, and not of active Cdc42, RhoG, or RhoA. These results, showing that in vivo Abr and Bcr function as Rac GAPs, are entirely consistent with our previous studies in which loss of both Abr and Bcr was associated with increased Rac activation of PEMs (27).
Rac is a well-known regulator of morphology. The introduction of constitutively active Rac (caRac) via microinjection of bacterially produced proteins induces immediate ruffling and lamellipodia in monocyte/macrophage cell lines (2) and causes actin reorganization in many different cell types. The final morphological effect of the actin reorganization induced by Rac activation was suggested to vary depending on the cell type and on the surface to which the cells under study were attached. For example, fibroblasts microinjected with caRac developed a uniform, very round cell spreading pattern on fibronectin (38). However, in a neuronal cell line, caRac induced transient uniform spreading that was immediately followed by long and thin neurite formation on laminin-coated surfaces, which was not observed on fibronectin (30). In this study, we observed increased ruffle formation (data not shown) as well as pronounced morphological changes in the double null macrophages that are entirely consistent with an activity of Abr and Bcr in the specific downregulation of Rac. Interestingly, the loss of Abr and Bcr in macrophages led to markedly increased cell elongation rather than uniform spreading of the cells and this phenotype was prominent on a glass surface, which was reminiscent of the effect of caRac in neuronal cell types. By expressing caRac1 in primary macrophages via DNA electroporation, allowing expression of a properly prenylated protein, we noted that it can cause two different morphologies depending on the expression level. Importantly, one morphology phenocopies that seen in BMMs lacking Abr and Bcr. Since neither caCdc42 nor caRhoC produced this morphology in the same cells, this further underscores that the (abr × bcr)−/− macrophages develop this phenotype because they lack negative regulation of activated Rac by Bcr and Abr.
The dramatically elongated cell morphology was also observed in a macrophage cell line transiently overexpressing GAP-defective mutants of Abr and Bcr. Several GAP-defective Rho GAPs act in a dominant negative manner by sequestering substrate GTPases, preventing their inactivation but still allowing their interaction with downstream effectors (23, 29, 32). We showed that the GAP mutants of Abr and Bcr could still bind to caRac, providing evidence that these act as dominant negative proteins. It has been also reported that inhibition of RhoGAP activity is sufficient to activate Rho (55). Therefore, the elongated morphology induced by these Abr and Bcr GAP dead mutants was likely caused by increased levels of activated Rac at specific locations where Rac would normally be inactivated by Abr and Bcr. The data obtained with the mutants are consistent with the concept that the morphological abnormalities of the double null macrophages are directly caused by the specific loss of the GAP activity of Abr and Bcr. Together, our results demonstrate that actin reorganization in a defined direction is enhanced in the macrophages as a result of the loss of Abr and Bcr GAP activity.
Rac is required for cell migration, including CSF-1-induced macrophage motility, to polarize and to form protrusions toward the direction of movement (26, 37). It is believed that localized activation/deactivation of Rac is critical for CSF-1-induced motility, because the microinjection of both dominant negative Rac and constitutively active Rac prevented cells from developing the proper polarization required for migration, thereby inhibiting motility (3, 26). This view is further supported by the finding of Gardiner et al. (16) showing that Rac is periodically activated at the leading edge of neutrophils migrating toward chemokines. Many studies have focused on the positive regulators of Rac, such as phosphatidylinositol 3-kinase and Vav that link CSF-1 stimulation to the localized Rac activation at the cell membrane and to motility (45, 54, 60). However, little is known about the negative regulators of Rac that deactivate it during the process. Our study qualifies Abr and Bcr as important negative regulators of Rac in CSF-1-induced macrophage migration. Firstly, the loss of Abr and Bcr led to a dramatic increase in chemotaxis of primary macrophages toward CSF-1 that was accompanied by a highly polarized morphology. Secondly, Abr and Bcr exhibited dynamic changes in their subcellular location in response to CSF-1 stimulation within a time frame that corresponds to the movement of a fraction of the total Rac to the cytosol upon CSF-1 stimulation. Whereas Abr and Bcr were normally localized throughout the cytoplasm and/or at the tip of cellular protrusions, CSF-1 triggered a transient and robust translocation of Abr and Bcr to the membrane in macrophages. Thirdly, we also found Bcr enriched at the leading edge of migrating cells, where periodic Rac activation normally occurs. Fourthly, addition of an N-terminal myristoylation sequence to Abr resulted in a mutant protein that had an increased localization in the membrane fraction of CHO cells and further decreased levels of activated Rac. These combined results strongly suggest the involvement of Abr and Bcr in the negative regulation of localized Rac activity in directed macrophage migration. These results also reillustrate that actin rearrangement in a defined direction is taking place in these cells, which is in accordance with the abnormally polarized morphology that we observed in the double null macrophages.
The mechanism through which Abr and Bcr are brought to the membrane upon CSF-1 stimulation is not known and requires further investigation. Abr and Bcr contain a PH domain that can potentially mediate the membrane translocation of both proteins. CSF-1 is known to activate phosphatidylinositol 3-kinase to generate phospholipid and therefore could provide a ligand for the PH domain-containing proteins (45). Recent work by Vedham et al. (54) showed that the PH domain of Vav, a GEF for Rac, accounts for specific recruitment of Vav to the plasma membrane upon CSF-1 stimulation. This finding further implicates the PH domain of Abr and Bcr as a potential candidate to be involved in the translocation of Abr and Bcr in CSF-1 stimulation.
The loss of Abr and Bcr also profoundly affected phagocytosis of opsonized particles by macrophages, which is known to be dependent on Rac. Rac is recruited to the phagosome and activated during FcγR-mediated phagocytosis in macrophage cell lines (39). Using knockout mouse models that are deficient for Rac2 or for both Rac1 and Rac2, others have shown that the loss of Rac prevents phagocytosis in primary murine macrophages (18, 64). The recent study by Hoope et al. (24), which examined the dynamics of Rac activation during phagocytosis, showed that rapid deactivation of Rac occurs after phagocytic cup closure. To date, the only GAP implicated in phagosome formation is the Rho-specific myosin IXb (40, 51). Our study provides Bcr and Abr as the first example of GAPs specific for Rac to negatively regulate phagocytosis in macrophages. The combined loss of Abr and Bcr resulted in the increased phagocytosis of opsonized bacteria and zymosan particles with distinct actin-rich phagocytic cup formations. In addition, we found the clear local enrichment of GAP mutants of Bcr and Abr along phagocytic cups in RAW cells. Although we did not see the clear localization of wild-type Bcr and Abr to those structures, this would not be unexpected, since the overexpression of Bcr and Abr in RAW cells is predicted to inhibit the Rac activation required to form distinctly visible phagocytic cups.
Very few other studies have addressed the role of GAPs specific for Rho family GTPases by genetically deleting the GAP under study in the mouse. Interestingly, those few studies commonly reported that the ablation of a specific GAP resulted in the global elevation of the activity of the target small GTPase (49, 57, 58); Rho GTP levels were highly elevated in P190B RhoGAP null murine embryonic fibroblasts (49), as was the Cdc42 GTP level in Cdc42GAP null murine embryonic fibroblasts and hematopoietic stem cells (57, 58). Those studies also reported changes in the overall size of the cells or the whole animal as well as in the growth of cells (49, 57, 58). However, we did not see any difference in basal Rac GTP levels among bcr−/−, abr−/−, and (abr × bcr)−/− BMMs, nor did we observe any change in the overall cell size of BMMs or the whole animal (data not shown). Therefore, it is more likely that, at least in BMMs, Abr and Bcr downregulate localized pools of Rac GTP that were generated by specific cellular stimuli in a defined location rather than control the general pool of Rac GTP. This is also consistent with the fact that (abr × bcr)−/− mice are viable. Thus, among the 69 genes encoding RhoGAPs, Abr and Bcr occupy a specific niche in the regulation of defined Rac functions in macrophages. In view of the increasing number of pathological states which involve the innate immune system and, specifically, macrophages, ranging from atherosclerosis, chronic inflammation, and cancer to sepsis, our identification of Abr and Bcr as negative regulators of Rac may have a medical application in the treatment of these conditions.
Supplementary Material
Acknowledgments
We thank George McNamara for help with microscopy, Ana Romero for help with isolation of mouse tail DNAs, and Donna Foster for care of the mice. Gary Bokoch and Ulla Knaus (The Scripps Research Institute, La Jolla, CA) are acknowledged for GST-Pak-RBD, and JaeRan Lee and EunJoon Kim (Korea Advanced Institute of Science and Technology, Daejon, Korea) are acknowledged for the generous gift of the EGFP-Bcr R1090A- and EGFP-Abr R683A-encoding plasmids. We thank Allan Hall (Memorial Sloan-Kettering Cancer Center, New York) for Myc-V12Cdc42, Manabu Negishi (University of Kyoto, Kyoto, Japan) for Myc-RhoG and Myc-RhoG G12V, and Kodi Ravichandran (University of Virginia) for GST-ELMO1.
This work was supported by National Institutes of Health Public Health Service grants HL071945 and HL060231.
Footnotes
Published ahead of print on 20 November 2006.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Aghazadeh, B., K. Zhu, T. J. Kubiseski, G. A. Liu, T. Pawson, Y. Zheng, and M. K. Rosen. 1998. Structure and mutagenesis of the Dbl homology domain. Nat. Struct. Biol. 5:1098-1107. [DOI] [PubMed] [Google Scholar]
- 2.Allen, W. E., G. E. Jones, J. W. Pollard, and A. J. Ridley. 1997. Rho, Rac and Cdc42 regulate actin organization and cell adhesion in macrophages. J. Cell Sci. 110:707-720. [DOI] [PubMed] [Google Scholar]
- 3.Allen, W. E., D. Zicha, A. J. Ridley, and G. E. Jones. 1998. A role for Cdc42 in macrophage chemotaxis. J. Cell Biol. 141:1147-1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benard, V., and G. M. Bokoch. 2002. Assay of Cdc42, Rac, and Rho GTPase activation by affinity methods. Methods Enzymol. 345:349-359. [DOI] [PubMed] [Google Scholar]
- 5.Bernards, A. 2003. GAPs galore! A survey of putative Ras superfamily GTPase activating proteins in man and Drosophila. Biochim. Biophys. Acta 1603:47-82. [DOI] [PubMed] [Google Scholar]
- 6.Bernards, A., and J. Settleman. 2004. GAP control: regulating the regulators of small GTPases. Trends Cell Biol. 14:377-385. [DOI] [PubMed] [Google Scholar]
- 7.Bokoch, G. M. 2005. Regulation of innate immunity by Rho GTPases. Trends Cell Biol. 15:163-171. [DOI] [PubMed] [Google Scholar]
- 8.Castellano, F., P. Montcourrier, and P. Chavrier. 2000. Membrane recruitment of Rac1 triggers phagocytosis. J. Cell Sci. 113:2955-2961. [DOI] [PubMed] [Google Scholar]
- 9.Cho, S. Y., and R. L. Klemke. 2002. Purification of pseudopodia from polarized cells reveals redistribution and activation of Rac through assembly of a CAS/Crk scaffold. J. Cell Biol. 156:725-736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chuang, T. H., X. Xu, V. Kaartinen, N. Heisterkamp, J. Groffen, and G. M. Bokoch. 1995. Abr and Bcr are multifunctional regulators of the Rho GTP-binding protein family. Proc. Natl. Acad. Sci. USA 92:10282-10286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cougoule, C., S. Hoshino, A. Dart, J. Lim, and E. Caron. 2006. Dissociation of recruitment and activation of the small G-protein Rac during Fcγ receptor-mediated phagocytosis. J. Biol. Chem. 281:8756-8764. [DOI] [PubMed] [Google Scholar]
- 12.Cunnick, J., P. Kaur, Y. Cho, J. Groffen, and N. Heisterkamp. 2006. Use of bone marrow-derived macrophages to model murine innate immune responses. J. Immunol. Methods 311:96-105. [DOI] [PubMed] [Google Scholar]
- 13.del Pozo, M. A., L. S. Price, N. B. Alderson, X. D. Ren, and M. A. Schwartz. 2000. Adhesion to the extracellular matrix regulates the coupling of the small GTPase Rac to its effector PAK. EMBO J. 19:2008-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Diakonova, M., G. Bokoch, and J. A. Swanson. 2002. Dynamics of cytoskeletal proteins during Fcγ receptor-mediated phagocytosis in macrophages. Mol. Biol. Cell 13:402-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Etienne-Manneville, S., and A. Hall. 2002. Rho GTPases in cell biology. Nature 420:629-635. [DOI] [PubMed] [Google Scholar]
- 16.Gardiner, E. M., K. N. Pestonjamasp, B. P. Bohl, C. Chamberlain, K. M. Hahn, and G. M. Bokoch. 2002. Spatial and temporal analysis of Rac activation during live neutrophil chemotaxis. Curr. Biol. 12:2029-2034. [DOI] [PubMed] [Google Scholar]
- 17.Haataja, L., V. Kaartinen, J. Groffen, and N. Heisterkamp. 2002. The small GTPase Rac3 interacts with the integrin-binding protein CIB and promotes integrin α(IIb)β(3)-mediated adhesion and spreading. J. Biol. Chem. 277:8321-8328. [DOI] [PubMed] [Google Scholar]
- 18.Hall, A. B., M. A. Gakidis, M. Glogauer, J. L. Wilsbacher, S. Gao, W. Swat, and J. S. Brugge. 2006. Requirements for Vav guanine nucleotide exchange factors and Rho GTPases in FcγR- and complement-mediated phagocytosis. Immunity 24:305-316. [DOI] [PubMed] [Google Scholar]
- 19.Heisterkamp, N., and J. Groffen. 2002. Philadelphia-positive leukemia: a personal perspective. Oncogene 21:8536-8540. [DOI] [PubMed] [Google Scholar]
- 20.Heisterkamp, N., V. Kaartinen, S. van Soest, G. M. Bokoch, and J. Groffen. 1993. Human ABR encodes a protein with GAPrac activity and homology to the DBL nucleotide exchange factor domain. J. Biol. Chem. 268:16903-16906. [PubMed] [Google Scholar]
- 21.Heisterkamp, N., C. Morris, and J. Groffen. 1989. ABR, an active BCR-related gene. Nucleic Acids Res. 17:8821-8831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hemmeryckx, B., A. van Wijk, A. Reichert, V. Kaartinen, R. de Jong, P. K. Pattengale, I. Gonzalez-Gomez, J. Groffen, and N. Heisterkamp. 2001. Crkl enhances leukemogenesis in BCR/ABL P190 transgenic mice. Cancer Res. 61:1398-1405. [PubMed] [Google Scholar]
- 23.Hirose, K., T. Kawashima, I. Iwamoto, T. Nosaka, and T. Kitamura. 2001. MgcRacGAP is involved in cytokinesis through associating with mitotic spindle and midbody. J. Biol. Chem. 276:5821-5828. [DOI] [PubMed] [Google Scholar]
- 24.Hoppe, A. D., and J. A. Swanson. 2004. Cdc42, Rac1, and Rac2 display distinct patterns of activation during phagocytosis. Mol. Biol. Cell 15:3509-3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jaffe, A. B., and A. Hall. 2005. Rho GTPases: biochemistry and biology. Annu. Rev. Cell Dev. Biol. 21:247-269. [DOI] [PubMed] [Google Scholar]
- 26.Jones, G. E., W. E. Allen, and A. J. Ridley. 1998. The Rho GTPases in macrophage motility and chemotaxis. Cell Adhes. Commun. 6:237-245. [DOI] [PubMed] [Google Scholar]
- 27.Kaartinen, V., I. Gonzalez-Gomez, J. W. Voncken, L. Haataja, E. Faure, A. Nagy, J. Groffen, and N. Heisterkamp. 2001. Abnormal function of astroglia lacking Abr and Bcr RacGAPs. Development 128:4217-4227. [DOI] [PubMed] [Google Scholar]
- 28.Kaartinen, V., A. Nagy, I. Gonzalez-Gomez, J. Groffen, and N. Heisterkamp. 2002. Vestibular dysgenesis in mice lacking Abr and Bcr Cdc42/RacGAPs. Dev. Dyn. 223:517-525. [DOI] [PubMed] [Google Scholar]
- 29.Lee, J. S., K. Kamijo, N. Ohara, T. Kitamura, and T. Miki. 2004. MgcRacGAP regulates cortical activity through RhoA during cytokinesis. Exp. Cell Res. 293:275-282. [DOI] [PubMed] [Google Scholar]
- 30.Leeuwen, F. N., H. E. Kain, R. A. Kammen, F. Michiels, O. W. Kranenburg, and J. G. Collard. 1997. The guanine nucleotide exchange factor Tiam1 affects neuronal morphology; opposing roles for the small GTPases Rac and Rho. J. Cell Biol. 139:797-807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li, Y. M., G. Baviello, H. Vlassara, and T. Mitsuhashi. 1997. Glycation products in aged thioglycollate medium enhance the elicitation of peritoneal macrophages. J. Immunol. Methods 201:183-188. [DOI] [PubMed] [Google Scholar]
- 32.Liang, X., N. A. Draghi, and M. D. Resh. 2004. Signaling from integrins to Fyn to Rho family GTPases regulates morphologic differentiation of oligodendrocytes. J. Neurosci. 24:7140-7149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ligeti, E., and J. Settleman. 2006. Regulation of RhoGAP specificity by phospholipids and prenylation. Methods Enzymol. 406:104-117. [DOI] [PubMed] [Google Scholar]
- 34.Malmberg, E. K., C. X. Andersson, M. Gentzsch, J. H. Chen, A. Mengos, L. Cui, G. C. Hansson, and J. R. Riordan. 2004. Bcr (breakpoint cluster region) protein binds to PDZ-domains of scaffold protein PDZK1 and vesicle coat protein Mint3. J. Cell Sci. 117:5535-5541. [DOI] [PubMed] [Google Scholar]
- 35.Massol, P., P. Montcourrier, J. C. Guillemot, and P. Chavrier. 1998. Fc receptor-mediated phagocytosis requires CDC42 and Rac1. EMBO J. 17:6219-6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moon, S. Y., and Y. Zheng. 2003. Rho GTPase-activating proteins in cell regulation. Trends Cell Biol. 13:13-22. [DOI] [PubMed] [Google Scholar]
- 36a.National Research Council. 1996. Guide for the care and use of laboratory animals. National Academy Press, Washington, D.C.
- 37.Nobes, C. D., and A. Hall. 1999. Rho GTPases control polarity, protrusion, and adhesion during cell movement. J. Cell Biol. 144:1235-1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nobes, C. D., and A. Hall. 1995. Rho, Rac, and Cdc42 GTPases regulate the assembly of multimolecular focal complexes associated with actin stress fibers, lamellipodia, and filopodia. Cell 81:53-62. [DOI] [PubMed] [Google Scholar]
- 39.Patel, J. C., A. Hall, and E. Caron. 2002. Vav regulates activation of Rac but not Cdc42 during FcγR-mediated phagocytosis. Mol. Biol. Cell 13:1215-1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Post, P. L., G. M. Bokoch, and M. S. Mooseker. 1998. Human myosin-IXb is a mechanochemically active motor and a GAP for rho. J. Cell Sci. 111:941-950. [DOI] [PubMed] [Google Scholar]
- 41.Prieto-Sanchez, R. M., I. M. Berenjeno, and X. R. Bustelo. 2006. Involvement of the Rho/Rac family member RhoG in caveolar endocytosis. Oncogene 25:2961-2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Radziwill, G., R. A. Erdmann, U. Margelisch, and K. Moelling. 2003. The Bcr kinase downregulates Ras signaling by phosphorylating AF-6 and binding to its PDZ domain. Mol. Cell. Biol. 23:4663-4672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ren, X. D., and M. A. Schwartz. 2000. Determination of GTP loading on Rho. Methods Enzymol. 325:264-272. [DOI] [PubMed] [Google Scholar]
- 44.Ridley, A. J. 2001. Rho GTPases and cell migration. J. Cell Sci. 114:2713-2722. [DOI] [PubMed] [Google Scholar]
- 45.Ridley, A. J. 2001. Rho proteins, PI 3-kinases, and monocyte/macrophage motility. FEBS Lett. 498:168-171. [DOI] [PubMed] [Google Scholar]
- 46.Ridley, A. J. 2004. Pulling back to move forward. Cell 116:357-358. [DOI] [PubMed] [Google Scholar]
- 47.Schmidt, A., and A. Hall. 2002. Guanine nucleotide exchange factors for Rho GTPases: turning on the switch. Genes Dev. 16:1587-1609. [DOI] [PubMed] [Google Scholar]
- 48.Servant, G., O. D. Weiner, P. Herzmark, T. Balla, J. W. Sedat, and H. R. Bourne. 2000. Polarization of chemoattractant receptor signaling during neutrophil chemotaxis. Science 287:1037-1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sordella, R., M. Classon, K. Q. Hu, S. F. Matheson, M. R. Brouns, B. Fine, L. Zhang, H. Takami, Y. Yamada, and J. Settleman. 2002. Modulation of CREB activity by the Rho GTPase regulates cell and organism size during mouse embryonic development. Dev. Cell 2:553-565. [DOI] [PubMed] [Google Scholar]
- 50.Stanley, E. R. 1997. Murine bone marrow-derived macrophages. Methods Mol. Biol. 75:301-304. [DOI] [PubMed] [Google Scholar]
- 51.Swanson, J. A., M. T. Johnson, K. Beningo, P. Post, M. Mooseker, and N. Araki. 1999. A contractile activity that closes phagosomes in macrophages. J. Cell Sci. 112:307-316. [DOI] [PubMed] [Google Scholar]
- 52.Tan, E. C., T. Leung, E. Manser, and L. Lim. 1993. The human active breakpoint cluster region-related gene encodes a brain protein with homology to guanine nucleotide exchange proteins and GTPase-activating proteins. J. Biol. Chem. 268:27291-27298. [PubMed] [Google Scholar]
- 53.Thompson, C. D., M. R. Frazier-Jessen, R. Rawat, R. P. Nordan, and R. T. Brown. 1999. Evaluation of methods for transient transfection of a murine macrophage cell line, RAW 264.7. BioTechniques 27:824-830, 832. [DOI] [PubMed] [Google Scholar]
- 54.Vedham, V., H. Phee, and K. M. Coggeshall. 2005. Vav activation and function as a rac guanine nucleotide exchange factor in macrophage colony-stimulating factor-induced macrophage chemotaxis. Mol. Cell. Biol. 25:4211-4220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vincent, S., and J. Settleman. 1999. Inhibition of RhoGAP activity is sufficient for the induction of Rho-mediated actin reorganization. Eur. J. Cell Biol. 78:539-548. [DOI] [PubMed] [Google Scholar]
- 56.Voncken, J. W., H. van Schaick, V. Kaartinen, K. Deemer, T. Coates, B. Landing, P. Pattengale, O. Dorseuil, G. M. Bokoch, J. Groffen, et al. 1995. Increased neutrophil respiratory burst in bcr-null mutants. Cell 80:719-728. [DOI] [PubMed] [Google Scholar]
- 57.Wang, L., L. Yang, K. Burns, C. Y. Kuan, and Y. Zheng. 2005. Cdc42GAP regulates c-Jun N-terminal kinase (JNK)-mediated apoptosis and cell number during mammalian perinatal growth. Proc. Natl. Acad. Sci. USA 102:13484-13489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang, L., L. Yang, M. D. Filippi, D. A. Williams, and Y. Zheng. 2006. Genetic deletion of Cdc42GAP reveals a role of Cdc42 in erythropoiesis and hematopoietic stem/progenitor cell survival, adhesion, and engraftment. Blood 107:98-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Watanabe, T., J. Noritake, and K. Kaibuchi. 2005. Regulation of microtubules in cell migration. Trends Cell Biol. 15:76-83. [DOI] [PubMed] [Google Scholar]
- 60.Wells, C. M., P. J. Bhavsar, I. R. Evans, E. Vigorito, M. Turner, V. Tybulewicz, and A. J. Ridley. 2005. Vav1 and Vav2 play different roles in macrophage migration and cytoskeletal organization. Exp. Cell Res. 310:303-310. [DOI] [PubMed] [Google Scholar]
- 61.Wells, C. M., M. Walmsley, S. Ooi, V. Tybulewicz, and A. J. Ridley. 2004. Rac1-deficient macrophages exhibit defects in cell spreading and membrane ruffling but not migration. J. Cell Sci. 117:1259-1268. [DOI] [PubMed] [Google Scholar]
- 62.Wennerberg, K., and C. J. Der. 2004. Rho-family GTPases: it's not only Rac and Rho (and I like it). J. Cell Sci. 117:1301-1312. [DOI] [PubMed] [Google Scholar]
- 63.Wennerberg, K., S. M. Ellerbroek, R. Y. Liu, A. E. Karnoub, K. Burridge, and C. J. Der. 2002. RhoG signals in parallel with Rac1 and Cdc42. J. Biol. Chem. 277:47810-47817. [DOI] [PubMed] [Google Scholar]
- 64.Yamauchi, A., C. Kim, S. Li, C. C. Marchal, J. Towe, S. J. Atkinson, and M. C. Dinauer. 2004. Rac2-deficient murine macrophages have selective defects in superoxide production and phagocytosis of opsonized particles. J. Immunol. 173:5971-5979. [DOI] [PubMed] [Google Scholar]
- 65.Zhang, Q., J. Calafat, H. Janssen, and S. Greenberg. 1999. ARF6 is required for growth factor- and Rac-mediated membrane ruffling in macrophages at a stage distal to rac membrane targeting. Mol. Cell. Biol. 19:8158-8168. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.