Abstract
Yeast Kar4 is a putative transcription factor required for karyogamy (the fusion of haploid nuclei during mating) and possibly other functions. Previously known to be required only for the transcriptional induction of KAR3 and CIK1, microarray experiments identified many genes regulated by Kar4 in both mating and mitosis. Several gene clusters are positively or negatively regulated by mating pheromone in a Kar4-dependent manner. Chromatin immunoprecipitation and gel shift assays confirmed that Kar4 binds to regulatory DNA sequences upstream of KAR3. Together with one-hybrid experiments, these data support a model in which both Kar4 and Ste12 bind jointly to the KAR3 promoter. Analysis of the upstream regions of Kar4-induced genes identified a DNA sequence motif that may be a binding site for Kar4. Mutation within the motif upstream of KAR3 eliminated pheromone induction. Genes regulated by Kar4, on average, are delayed in their temporal expression and exhibit a more stringent dose response to pheromone. Furthermore, the induction of Kar4 by pheromone is necessary for the delayed temporal induction of KAR3 and PRM2, genes required for efficient nuclear fusion during mating. Accordingly, we propose that Kar4 plays a critical role in the choreography of the mating response.
In the budding yeast Saccharomyces cerevisiae, mating occurs between two haploid cells of different mating types, MATa and MATα (reviewed in references 9 and 31). Mating is initiated by the exchange of specific mating pheromones; MATa cells secrete a-factor and MATα cells secrete α-factor. Binding of the pheromone to its specific transmembrane receptor elicits a complex signal transduction pathway, resulting in cell cycle arrest and the transcriptional induction of mating-specific genes (reviewed in references 11 and 47). Mating-induced transcription requires activation of the transcription factor Ste12, which binds to pheromone response elements (PREs) in the upstream activating sequences of target genes (12, 16). Ste12 is also involved in the regulation of genes required for a distinct developmental program: filamentous growth in response to starvation (28).
Exposure of cells to mating pheromone leads to the induction of genes that are regulated by Ste12 alone (16), as well as genes that require a second transcription factor, Mcm1 (35, 50). Similarly, during filamentous growth, Ste12-dependent induction requires Tec1, with which it binds cooperatively at filamentation-responsive elements (2, 20, 27, 50). Thus, differential gene expression by Ste12 is achieved through cooperative interactions with other transcription factors such as Tec1 and Mcm1 and possibly others.
Mating consists of the fusion of two haploid cells, followed by the congression and eventual fusion of the two nuclei in an event called karyogamy (reviewed in reference 41). Preceding cell fusion, cytoplasmic microtubules stemming from the spindle pole body move each nucleus to the mating projection of each haploid cell (26, 34). Upon cell fusion, the cytoplasmic contents of each cell intermingle, facilitating the connection of cytoplasmic microtubules via Kar3, a kinesin-related protein, and Cik1, its associated light chain (33, 38). During nuclear congression, movement and microtubule shortening are produced by Kar3 and Cik1 acting on the cytoplasmic microtubules (25, 26, 33). Finally, the nuclear envelopes fuse at the spindle pole bodies, resulting in the formation of a single diploid nucleus (3).
The mating process must be precisely regulated both spatially and temporally. For example, premature cell wall degradation prior to cell adhesion, at ectopic or supernumerary sites, would be catastrophic. Moreover, many of the genes induced during mating, such as Kar3, have other functions during mitosis, and their premature overexpression may be deleterious during the cell cycle. The current model for the pathway of pheromone-induction suggests that all mating-induced genes are regulated by Ste12, providing no mechanism for the sequential expression of genes during mating.
Kar4 is a good candidate for controlling the temporal regulation of genes that show a delay during the pheromone response. Kar4 was identified previously as a putative transcription factor, which is required, together with Ste12, for the transcriptional induction of KAR3 and CIK1 by pheromone (14, 23). Previous work demonstrated that a 30-bp DNA sequence upstream of KAR3 is necessary for Kar4- and Ste12-dependent induction by pheromone (23). Overexpression of Ste12 is sufficient to induce the expression of KAR3, in the absence of both Kar4 and pheromone (23). In contrast, the overexpression of Kar4 does not bypass the requirement for Ste12, although it does partially alleviate the requirement for pheromone (23). These data suggest that Kar4 facilitates Ste12's transcriptional induction of genes such as KAR3.
Together with yeast Ime4, Kar4 is a member of the large MT-A70 superfamily of proteins, which includes the S-adenosylmethionine binding subunit of mRNA:m6A methyl transferases (MTase) and bacterial DNA:m6A MTases (5). Ime4 is required for the appearance of m6A in mRNA during yeast sporulation (7). In contrast, Kar4 is the prototypical member of highly divergent MT-A70 subfamily, conserved within fungi, in which the critical residues in the S-adenosylmethionine binding site have been altered (5). Kar4 and other members of this subfamily are unlikely to be MTases (5).
The regulation of Kar4 expression is highly complex, occurring at the level of transcription, translation, and protein turnover. There are two forms of Kar4: a constitutive, low-abundance 38.5-kDa protein (Kar4-long), which predominates during vegetative growth, and an abundant 35.5-kDa protein, which is induced by mating pheromone (Kar4-short) (14, 23). The two forms differ at their amino termini; Kar4-short is translated from a shorter mRNA and initiated from an AUG codon that is internally encoded in the Kar4-long message. In addition to differential transcriptional regulation, the two protein forms have different rates of protein synthesis and turnover (14).
The induction of KAR4 transcription by Ste12 after exposure to pheromone potentially provides an additional level of regulation for a subset of genes expressed during mating. In particular, the requirement that Kar4 act along with Ste12 for the induction of KAR3 and CIK1 might provide a mechanism for temporal regulation (23). Formally, Kar4 and Ste12 comprise a “feed-forward loop” regulatory circuit (29, 30). In this model, Kar4 and Ste12 jointly regulate the transcription of their target genes, providing a delayed response to the inducing signal.
The complexity of its regulation underscores the likelihood that Kar4 performs other regulatory functions beyond the mating-dependent regulation of KAR3 and CIK1. In support of this hypothesis, Kar4 is required for meiosis, its overexpression is inhibitory to vegetative growth, and kar4 deletion causes a cell cycle delay in G1 (23). Consistent with multiple functions for Kar4, we show here that Kar4 regulates a large array of genes in both mating and mitosis. Remarkably, the set of genes affected by Kar4 includes both positively and negatively regulated genes.
MATERIALS AND METHODS
Strains and microbial techniques.
The yeast strains and plasmids used in the present study are listed in Table 1. Standard yeast media and genetic techniques were as described previously (42). Synthetic alpha factor (Princeton Syn/Seq facility) was used at 3 to 10 μM. Microscopic analysis of karyogamy in zygotes by DAPI (4′,6′-diamidino-2-phenylindole) staining was performed as described previously (22). To analyze the defect of kar4C214G/C217G, matings of host kar4Δ strains MS3216 and MS3212 were performed. Each strain harbored either the wild-type KAR4 plasmid pMR2654, the kar4C214G/C217G plasmid pMR5511, or the vector pRS416 (45). Analysis of the prm2 mating defect was performed by crossing MATa (Y01430) and MATα (Y11430) deletion strains; wild-type control strains were MS3265 and MS3213 and the kar4Δ strains were MS3216 and MS3212. Matings were allowed to proceed for 90 min on yeast extract-peptone-dextrose (YEPD) plates. For each mating, >100 zygotes were scored (22).
TABLE 1.
Strains and plasmids used in the present studya
| Strain or plasmida | Genotype or relevant markersb | Source or reference |
|---|---|---|
| Strains | ||
| MS3212 | MATα leu2-3,112 his3Δ200 ura3-52 ade2-101 kar4Δ::HIS3 | |
| MS3213 | MATatrp1Δ1 his3Δ200 ura3-52 ade2-101 | |
| MS3215 | MATaleu2-3,112 his3Δ200 ura3-52 ade2-101 kar4Δ::HIS3 | |
| MS3216 | MATaleu2-3,112 his3Δ200 ura3-52 ade2-101 kar4Δ::HIS3 | |
| MY3265 | MATaura3-52 trp1 his4-519 leu2 can1-101 | |
| MY3375 | MATaura3-52 leu2Δ1 his3Δ200 | |
| MY4166 | MATakar4Δ ura3-52 leu2Δ1 his3Δ200 | |
| MY5100 | MATaste12Δ::leu2Δ1 ura3-52 trp1 his4-519 leu2 can1-101 | |
| MY5792 | MATaura3-52 leu2Δ1 his3Δ200 KAR4::HA | |
| MY10133 | MATaste12Δ::LEU2 trp1-901 leu2-3,112 ura3-52 his3Δ200 gal4Δ gal80Δ GAL2-ADE2 LYS2::GAL1-HIS3 met2::GAL7-lacZ | |
| PJ69-4A | MATatrp1-901 leu2-3,112 ura3-52 his3Δ200 gal4Δ gal80Δ GAL2-ADE2 LYS2::GAL1-HIS3 met2::GAL7-lacZ | 19 |
| BY4741 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | 49 |
| BY4742 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 | 49 |
| Y01430 | MATaprm2Δ::KanMX his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | 49 |
| Y11430 | MATα prm2Δ::KanMX his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 | 49 |
| Y03462 | MATakar4Δ::KanMX his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | 49 |
| Y13462 | MATα kar4Δ::KanMX his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 | 49 |
| Plasmids | ||
| pLG669-Z | lacZ 2μm URA3 | 15 |
| pMR2806 | PKAR3-L11-lacZ 2μm URA3 | |
| pMR2802 | PKAR3-L16-lacZ 2μm URA3 | |
| pMR2892 | PKAR3-L18-lacZ 2μm URA3 | |
| pMR2808 | PKAR3-L14-lacZ 2μm URA3 | |
| pMR2896 | PKAR3-L15-lacZ 2μm URA3 | |
| pMR5620 | PKAR3-min-lacZ 2μm URA3 | |
| pMR5621 | PKAR3-min31-2-lacZ 2μm URA3 | |
| pMR5622 | PKAR3-min41-2-lacZ 2μm URA3 | |
| pMR2654 | KAR4::HA CEN ARS URA3 | |
| pMR2516 | KAR4 CEN ARS URA3 | |
| pRS416 | CEN ARS URA3 | 45 |
| pMR5511 | kar4C214GC217G::HA CEN ARS URA3 | |
| pMR3356 | PGAL-KAR4::HA-short CEN ARS URA3 | |
| pMR3459 | PGAL-KAR4::HA-long CEN ARS URA3 | |
| pGBD-C2 | GBD TRP1 2μm | 19 |
| pMR4997 | GBD-KAR4 TRP1 2μm | |
| pGAD-C2 | GAD LEU2 2μm | 19 |
| pMR5000 | GAD-KAR4 LEU2 2μm | |
| pMR5665 | Ptet-6xHN-KAR4; Cmr | |
| pMR5669 | Ptet-6xHN-STE12; Cmr | |
| pPROTet.E133 | Ptet-6xHN; Cmr | Clontech |
| pSUL16 | ste12Δ::LEU2 | 11 |
Unless stated otherwise, all strains and plasmids were constructed in this laboratory.
Cmr, chloramphenicol resistant.
Molecular techniques.
DNA and RNA manipulations, including plasmid DNA isolation and Northern blot analysis, were performed as described previously (43). Total RNA for microarray experiments and Northern blots was extracted from yeast cultures by using published procedures (44). Specific DNA fragments for Northern blot probes were prepared by PCR.
Plasmid construction was done by PCR-based in vivo recombination (37) and verified by DNA sequencing (Princeton Syn/Seq facility). The upstream KAR3 minimal sequence was recombined into the expression vector pLG670-Z (15) using the forward primer 5′-GGA AAG CAG GAA AGG AAA AAA TTT TTA GGC TCG AGC AAA ATG AAT CTC AAA CAA AAT CAA AAC AAA CAC-3′ and the reverse primer 5′-ACG CTA TAT ATA CAC GCC TGG CGG ATC TGC TCG AGG TGT TTG TTT TGA TTT TGT TTG AGA TTC ATT TTG-3′. Mutations in the putative Kar4 consensus sites were introduced into the same expression vector using the following primers: k3minmut31 forward, 5′-GGA AAG CAG GAA AGG AAA AAA TTT TTA GGC TCG AGC AAA ATG AAT CTC AAA CAA AAT CAG AAC AAA CAC-3′; k3minmut31reverse, 5′-ACG CTA TAT ATA CAC GCC TGG CGG ATC TGC TCG AGG TGT TTG TTC TGA TTT TGT TTG AGA TTC ATT TTG-3′; k3minmut41 forward, 5′-GGA AAG CAG GAA AGG AAA AAA TTT TTA GGC TCG AGC AAA ATG AAT CTC AAA CAG AAT CAA AAC AAA CAC-3′, and k3minmut41 reverse, 5′-ACG CTA TAT ATA CAC GCC TGG CGG ATC TGC TCG AGG TGT TTG TTT TGA TTC TGT TTG AGA TTC ATT TTG-3′.
The pGBD-KAR4 and pGAD-KAR4 constructs contain the ADH1 promoter for the constitutive expression of in-frame fusions between KAR4 and the GAL4 DNA-binding domain (GBD) or the GAL4 transcription activation domain (GAD). Constructs were made by PCR-based in vivo recombination using genomic DNA (37). The PCR primers were KAR4-FOR1 (AAAGAGATCGAATTCCCGGGGATCCATATGGCATTCCAAGATCCAAC) and KAR4-REV1 (TACTACGATTCATAGATCTCTGCAGGTTTATTGTGCTTTTTGTACTGGACTTT). Vectors pGBD-C2 and pGAD-C2 (19) were linearized with BamHI and PstI and introduced into yeast with PCR-amplified KAR4 DNA (6). Constructs were confirmed by restriction mapping and nucleotide sequencing (Princeton Syn/Seq facility). pGBD-C2 and pGBD-KAR4 were transformed into PJ69-4A, a MATa yeast two-hybrid reporter strain (19), and an isogenic ste12Δ strain (MY10133), created by using pSUL16 (10). Pheromone-dependent expression was assayed by spotting fivefold serial dilutions of transformants, pregrown to exponential phase in liquid SC−TRP, onto SC−TRP and SC−TRP,−HIS agar plates, with or without α-factor at 3 μM. Plates were incubated for 2 to 5 days at 30°C. pGAD-C2 and pGAD-KAR4 were transformed into S288C-derived isogenic strains MY3375 (KAR4 STE12) and MY4166 (kar4Δ STE12), as well as W303-derived isogenic strains MY3265 (KAR4 STE12) and MY5100 (KAR4 ste12Δ). All strains harbored a reporter construct with the KAR3 minimal promoter region upstream of lacZ (pMR5620). β-Galactosidase assays on the minimal and mutated upstream region of KAR3 were performed as previously described (23, 42).
Microarray and cluster analysis.
Strains MS3213 (wild type) and MS3215 (kar4Δ) were grown to mid-exponential phase in YEPD (pH 3.5), after which α-factor or methanol (mock treatment) was added. The cultures were incubated for 90 min to allow full expression of Kar4 and other late genes involved in mating. RNA was extracted as described above, and the preparation of biotinylated cRNA, hybridization, and microarray scanning were performed according to the manufacturer's instructions (Affymetrix, Santa Clara, CA). Hybridization to the yeast genome S98 array was done in a GeneChip hybridization oven 640 at 45°C for 16 h, washing steps were performed by using the Affymetrix GeneChip fluidics station 400, and the GeneChips were scanned by using an Agilent GeneArray scanner. Raw data were normalized and clustered using the k-means clustering algorithm to group genes based on the similarity of their normalized gene expression profiles (49). The fold induction or repression for each of the obtained clusters was determined from the ratio of the raw data for each of the genes in the cluster (wild type to kar4Δ mutant).
To identify upstream Kar4 binding sites, the AlignACE algorithm was used (18, 48). The statistical significance of the sequence motif obtained is expressed as the maximum a priori log likelihood (MAP) score and the specificity score (18). The MAP score measures the degree to which a motif is over-represented relative to the expectation for the random occurrence of such a motif in the sequence under consideration. The specificity score is a measure of the degree to which the distribution of sites is skewed toward the input set, and it takes into account the sequence of all of the 5′ upstream regulatory regions in the genome, highlighting motifs that are found preferentially in association with the genes under consideration (18).
Temporal regulation of Kar4-dependent genes.
The 29 Kar4-dependent and 34 Kar4-independent genes showing >2.5-fold induction by pheromone were selected for analysis. The data for the timing of induction in a bar1Δ strain for these genes (0, 15, 30, 45, and 60 min after induction) by 50 nM pheromone were obtained from previously published data (39). Similarly, data for the dose response of a bar1Δ strain to alpha factor exposure (0.15, 0.5, 1.5, 5, 15.8, 50, and 158 nM pheromone) after 30 min were previously published (39). A Stineman function was fit to all datum points to obtain a local smoothing (KaleidaGraph 4.01; Synergy Software). The datum points corresponding to the half-maximal value (t1/2) were interpolated, and the t1/2 for each gene was obtained. The statistical significance was tested by using a heteroscedastic Student t test.
Detection of Kar4.
Hemagglutinin (HA) epitope-tagged forms of Kar4 (14, 23) were detected with 12CA5 monoclonal antibody (Princeton monoclonal facility). Total yeast protein was extracted from cells as previously described (36). Proteins were run on a 10% polyacrylamide gel at 100 V and transferred to supported nitrocellulose membrane (Optitran; pore size, 0.2 μm; Schleicher & Schuell) at 100 V for 60 min. Western blotting was performed as previously described (14), and the Kar4::HA proteins were detected by using the ECL kit (Amersham, Arlington Heights, IL).
Chromatin immunoprecipitation assay.
Strain MY5792, which contains a chromosomal version of KAR4 with the HA epitope inserted at the carboxyl terminus of the coding region (14), and an untagged control strain MY3375, were grown to an optical density at 600 nm (OD600) of 0.7. Cells were we treated with α-factor dissolved in methanol or methanol alone for 90 min. Chromatin immunoprecipitation was performed as described previously (21), except that formaldehyde cross-linking was carried out for 1 h at room temperature. The primers used to amplify the upstream region of KAR3 were 5′-TTC GGC TTC TCT ATC GCT TT-3′ and 5′-TGC CAG AAT TTC TCT AAG TCC-3′.
Gel shift assay.
The open reading frames of KAR4 and STE12 were amplified from genomic DNA by PCR (6) and inserted into pPROTet.E133 vector (Clontech, Palo Alto, CA) to create in-frame fusions to a tag of six repeated His-Asn residues (6xHN). Fusion proteins were expressed in Escherichia coli (BL21Pro) by induction with 100 ng/ml of anhydrotetracycline (Clontech) for 4 h and partially purified by using NTA spin columns (QIAGEN, Valencia, CA). Cells containing the pPROTet.E133 vector were processed in parallel as a control for background DNA-binding activity. Protein concentrations were determined by using the Bradford protein assay (Bio-Rad Laboratories, Hercules, CA) with bovine serum albumin as the protein standard. Protein purity was assessed by using sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Coomassie blue staining (1).
DNA gel shift assays were conducted according to the method of Dolan et al. (10) with minor modifications. DNA probes were prepared by annealing complementary oligonucleotides encoding the minimal Kar4-dependent region contained in the KAR3 promoter plus five downstream nucleotides to include the full Kar4-responsive consensus sequence. The 5′ overhangs were filled with Klenow fragment (New England Biolabs), dCTP, dTTP, dGTP, and 32P-α-dATP (Amersham) according to the enzyme manufacturer's specification. For binding experiments, poly(dI-dC) competitor was added to a final concentration of 100 μM. Protein extracts were added to a constant final concentration in each experiment (typically between 3 and 6 μg per reaction). Where Ste12 or Kar4 concentrations were varied, compensatory amounts of equivalent protein from cells bearing the vector plasmid were added.
Site-directed mutagenesis.
Site-directed mutagenesis of the zinc-finger domain in the 3X HA-tagged version of KAR4 (pMR2654) was performed by using the QuikChange II XL kit (Stratagene, La Jolla, CA). The primers used to mutate the putative zinc-finger domain of KAR4 were 5′-AAA AAT GCA ATG GCA CGG TTG GAT GGG TAT CAC AGG TAC A-3′ (sense) and 5′-TGT ACC TGT GAT ACC CAT CCA ACC GTG CCA TTG CAT TTT T-3′ (antisense). The pMR5511 construct was sequenced to confirm the change (Princeton Syn/Seq facility).
Galactose induction of Kar4.
For galactose induction experiments, the kar4Δ strain MS3216 harboring the plasmids pMR3356 (PGAL-KAR4-short::HA), pMR3459 (PGAL-KAR4-long::HA), and pMR2654 (KAR4::HA) were grown in synthetic complete media without uracil, with 2% raffinose, to mid-exponential phase. Cultures were induced by the addition of 2% galactose plus 0.35% glucose (for KAR4-long::HA) and 2% galactose plus 0.2% glucose (for KAR4-short::HA). Galactose induction was for 3 h, after which pheromone was added. Aliquots of cells were harvested at each time point (0, 5, 10, 20, 30, 45, 60, 75, and 90 min after addition of pheromone) and snap-frozen in liquid nitrogen. Northern blotting was performed as previously described.
RESULTS
Kar4 binds to DNA upstream of the KAR3 gene.
Although Kar4 was implicated as a transcriptional activator of KAR3 and CIK1, it was not determined whether Kar4 binds to DNA along with Ste12 or acts indirectly to facilitate Ste12's association or activity. As a first test, we used chromatin immunoprecipitation to detect the association of epitope-tagged Kar4 with the upstream regulatory region of KAR3. Strains expressing Kar4::HAp or untagged Kar4 were treated with α-factor or mock treated with methanol, the DNA was cross-linked, and the chromatin containing Kar4 was immunoprecipitated (Fig. 1A). Enrichment of KAR3-specific DNA sequences was detected by PCR and compared to a nonspecific DNA sequence from the telomere of chromosome 1 (TEL1).
FIG. 1.
(A) Kar4 functions at the promoter region of KAR3. Chromatin immunoprecipitation of Kar4::HA-bound upstream sequence of KAR3. Strain MY5792 harboring a chromosomal version of KAR4::HA (tagged Kar4) and the untagged control strain MY3375 were either treated with pheromone for 90 min or mock treated with MeOH. Immunoprecipitated (ChIP) or whole-cell extract (WCE) DNA was amplified with primers specific to upstream sequences of KAR3 and TEL1 (nonspecific control). PCR fragments were fractionated and visualized by using conventional agarose gel electrophoresis. Lane 1, MY5792 no-cross-link control; lane 2, MY5792 whole-extract control; lane 3, cross-linked tagged strain MY5792, not induced by pheromone; lane 4, pheromone-induced cross-linked MY5792; lane 5, cross-linked, pheromone-induced untagged strain MY3375. (B) Kar4 and Ste12 bind to the KAR3 promoter. The results of a gel shift assay using radioactively labeled KAR3 minimal promoter region (KAR3-min) in the presence of various amounts of E. coli expressed Ste12 and Kar4 is shown. The relative amount of each protein is indicated. (C) Kar4 binding confers pheromone inducible expression. The wild-type yeast two-hybrid reporter strain PJ69-4A (WT) or an isogenic strain containing a ste12Δ were transformed with either the GBD vector plasmid or with GBD-Kar4. Strains were grown to exponential phase, and serial dilutions were spotted onto selective plates with or without 3 μM pheromone (α-factor) dissolved in MeOH. All cells can grow on −TRP plates (GROWTH). Only cells expressing the GAL-HIS3 reporter gene can grow on −TRP,−HIS plates (EXPRESSION). (D) Fusing an activation domain to Kar4 does not remove the requirement for Ste12. S288C-derived strains MY3375 (KAR4 STE12) and MY4166 (kar4Δ STE12), as well as W303-derived strains MY3265 (KAR4 STE12) and MY5100 (KAR4 ste12Δ), were transformed with either the GAD vector plasmid or with the GAD-Kar4 plasmid. Each strain also harbored the reporter construct with the KAR3 minimal promoter region fused upstream of lacZ (pMR5620). Expression from the promoter is presented in β-galactosidase (β-gal) units from cultures grown in the presence (+) or absence (−) of 6 μM α-factor (pheromone).
A small amount of KAR3 upstream DNA, but not TEL1, was precipitated from vegetative cells. The addition of pheromone caused a significant enrichment of the KAR3 DNA above the level observed in vegetative cells, a finding consistent with increased levels of Kar4. Immunoprecipitation by the HA antibody was dependent both on the presence of the HA epitope on Kar4 and on the addition of cross-linker. These data support the hypothesis that Kar4 plays a direct role in the induction of KAR3.
We next used a gel shift assay to determine whether Kar4 can bind directly to the regulatory DNA sequences upstream of KAR3. 6xHN-tagged Kar4 and Ste12 were expressed in E. coli, partially purified, and tested for binding to a 35-bp DNA segment, KAR3-min (Fig. 1B). High levels of Ste12 alone were sufficient to retard the electrophoretic mobility of 42% of KAR3-min, indicating that Ste12 can bind to the sole PRE contained within the DNA. Threefold reduction in the amount of Ste12 reduced the amount of DNA retarded by more than fourfold (10%). In the absence of Ste12, Kar4 retarded a smaller amount of KAR3-min (10 to 17%). When both Kar4 and Ste12 were present, more KAR3-min was bound than by either protein alone (34%). The amount bound was somewhat greater than the sum for each protein alone, and the specific pattern of bands was different from that observed with each protein alone. These data confirm that both Ste12 and Kar4 can bind to the KAR3-min sequence and suggests that they may bind cooperatively.
To determine whether Kar4 binding could confer pheromone-dependent transcriptional activation on an exogenous promoter, Kar4 was fused to the GBD, constitutively expressed from the ADH1 promoter, and tested for activation of a HIS3 reporter construct fused to the GAL1 regulatory region (Fig. 1C). In the absence of pheromone, GBD-Kar4 did not activate the HIS3 reporter, indicating that Kar4 does not contain a constitutive activation domain. In the presence of pheromone, HIS3 expression was strongly activated by GBD-Kar4; expression was dependent on the presence of Ste12. These results indicate that Kar4 localization to a promoter is sufficient to confer pheromone regulation. The requirement for Ste12 may reflect either an indirect Ste12-dependent activation of Kar4 or the direct activation of transcription by Ste12 via an association with Kar4.
In a complementary approach, we next determined whether fusion of the GAD to Kar4 could obviate the requirement for Ste12 (Fig. 1D). Constitutively expressed GAD-Kar4 was functional and able to replace Kar4 for both the basal level and the pheromone-induced activation of the lacZ reporter via the KAR3-min sequence (Fig. 1D, line 4). However, transcriptional activation by GAD-Kar4 was still dependent on Ste12 (Fig. 1D, line 8). One interpretation of these data is that Ste12 may facilitate association of Kar4 with the KAR3 promoter, which is consistent with the results from the in vitro gel shift assay.
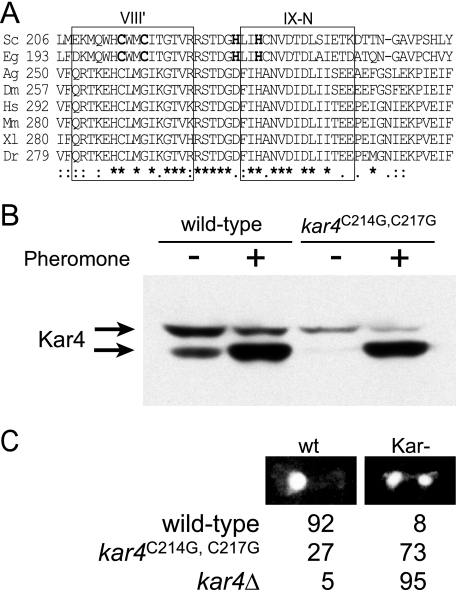
The sequence of Kar4 contains a region similar to a zinc-finger motif of the Cys2-His2 or Cys2-His-Cys type (4, 8). However, the loop region in Kar4 is one residue smaller than that of canonical Zn-finger domains, and critical hydrophobic residues important for packing of the hydrophobic core are not present (4). Nevertheless, this region of the protein is highly conserved and corresponds to motifs VIII′ and IX-N of the MTase family. Although all of the putative histidine and cysteine Zn ligands are conserved within the orthologues of Kar4 in the fungal lineage (cf. E. gossypii, Fig. 2A), only two of four residues are conserved in the larger class B lineage found in higher eukaryotes (5). To determine whether the conserved cysteines are required for Kar4 function, positions 214 and 217 were mutated to glycine. The levels of the mutated protein in pheromone-induced cells were similar to in the wild type, indicating that the mutations had little effect on the steady-state level of Kar4 (Fig. 2B). The effect of the mutations was examined by microscopic examination of zygotes (Fig. 2C). In crosses between wild-type cells, more than 90% of the zygotes have completed nuclear fusion. In contrast, in matings between kar4Δ strains, 95% of the nuclei failed to fuse. In matings between the mutants (kar4C214GC217G × kar4C214GC217G), 73% of the zygotes contained two nuclei, a ninefold increase in the failure of nuclear fusion. Although the defect is not as severe as that caused by complete deletion of KAR4, this domain is clearly important for Kar4 function.
FIG. 2.
A highly conserved region among Kar4 protein family. (A) Pileup sequences of Kar4-related proteins. Family members shown are from Saccharomyces cerevisiae (Sc), Eremothecium gossypii (Eg), Anopheles gambiae (Ag), Drosophila melanogaster (Dm), Homo sapiens (Hs), Mus musculus (Mm), Xenopus laevis (Xl), and Danio rerio (Dr). Identical residues ( ) and similar residues (: or .) are indicated below the pileup. Boxed regions highlight the regions similar to methytransferase domains VIII′ and IX-N (5). Putative zinc-finger ligand residues are indicated in boldface. (B) Mutations in conserved cysteines do not decrease protein levels of Kar4. Western blot showing levels of MS3216 kar4Δ::HIS strain expressing wild-type HA epitope-tagged Kar4 (KAR4::HA on pMR2654) and mutagenized HA epitope-tagged Kar4 (kar4C214G C217G::HA on pMR5511). The cells were grown in the absence (−) or presence (+) of pheromone. The arrows indicate the two forms of Kar4::HAp. (C) Microscopic analysis of mating. Homozygous matings (wild-type × wild-type, kar4C214GC217G × kar4C214GC217G, and kar4Δ × kar4Δ) were mated for 75 min before being fixed in methanol-acetic acid, stained with DAPI to reveal the positions of the nuclei, and examined by fluorescence microscopy. Zygotes were scored as being wild type (nuclei have fused [wt]) or karyogamy defective (nuclei separate and unfused [Kar−]). The percentages of each class are listed.
) and similar residues (: or .) are indicated below the pileup. Boxed regions highlight the regions similar to methytransferase domains VIII′ and IX-N (5). Putative zinc-finger ligand residues are indicated in boldface. (B) Mutations in conserved cysteines do not decrease protein levels of Kar4. Western blot showing levels of MS3216 kar4Δ::HIS strain expressing wild-type HA epitope-tagged Kar4 (KAR4::HA on pMR2654) and mutagenized HA epitope-tagged Kar4 (kar4C214G C217G::HA on pMR5511). The cells were grown in the absence (−) or presence (+) of pheromone. The arrows indicate the two forms of Kar4::HAp. (C) Microscopic analysis of mating. Homozygous matings (wild-type × wild-type, kar4C214GC217G × kar4C214GC217G, and kar4Δ × kar4Δ) were mated for 75 min before being fixed in methanol-acetic acid, stained with DAPI to reveal the positions of the nuclei, and examined by fluorescence microscopy. Zygotes were scored as being wild type (nuclei have fused [wt]) or karyogamy defective (nuclei separate and unfused [Kar−]). The percentages of each class are listed.
Kar4 regulates the expression of many genes.
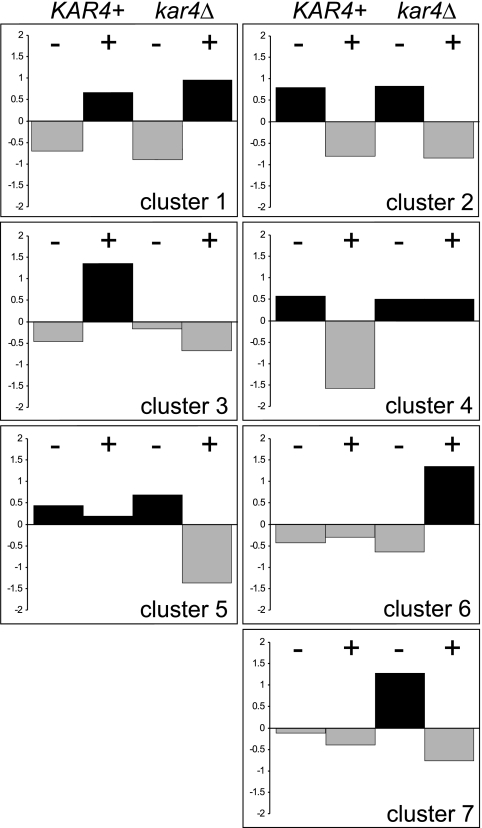
We performed microarray experiments to determine whether Kar4 has a more global effect on the transcriptional regulation of genes involved in the yeast pheromone response. cRNA prepared from total RNA of pheromone or mock-treated wild-type and kar4Δ cells and used to probe Affymatrix yeast microarrays in duplicate. Raw hybridization data were collected, normalized, and clustered into 20 original clusters according to a k-means clustering algorithm, grouping the genes based on the similarity of their normalized gene expression profiles (48). Clusters containing genes that showed neither pheromone-dependent regulation nor evidence of Kar4-dependent expression were not examined further. Clusters with similar patterns of expression were pooled together into seven new clusters (Fig. 3). Clusters 1 (414 genes) and 2 (850 genes) are comprised of pheromone-regulated genes whose induction or repression was independent of Kar4. The remaining clusters were filtered by the ratio of gene expression in the kar4Δ mutant relative to the wild type; only genes showing a >2-fold difference were further analyzed. Cluster 3 contains 70 genes whose expression was reduced in the kar4Δ mutant after pheromone treatment. Two expected genes within this cluster are KAR3 and CIK1. In addition to these three anticipated gene clusters, four other clusters were unexpected. Cluster 4 contains 225 genes that were expressed more strongly after pheromone induction in the kar4Δ mutant, indicating a possible role for Kar4 in the repression of genes after exposure to pheromone. Two clusters contained genes whose expression is constitutive in wild-type cells but which are either repressed (cluster 5) or induced (cluster 6) by pheromone in the kar4Δ mutant. These clusters suggest that Kar4 plays a specific role in maintaining the expression of these genes at normal levels during the pheromone response. Cluster 7 contained 55 genes whose vegetative expression was elevated in the kar4Δ mutant but which showed constitutive expression in wild-type cells, a finding consistent with Kar4 having a significant regulatory function during vegetative growth.
FIG. 3.
Kar4-dependent clusters after pheromone treatment. Strains MS3213 (wild type) and MS3215 (kar4Δ) were exposed to pheromone (+) or mock treated with methanol (−) for 90 min. RNA was extracted and prepared for microarray analysis as described in Materials and Methods. Genes were clustered according to their normalized expression profiles. Specifically, we identified seven clusters that show Kar4-independent pheromone induction (cluster 1), Kar4-independent repression in pheromone (cluster 2), Kar4-dependent pheromone induction (cluster 3), Kar4-dependent repression in pheromone (cluster 4), constitutive expression in wild-type cells but repression by pheromone in the kar4Δ strain (cluster 5), constitutive expression in wild-type cells but induction by pheromone in the kar4Δ strain (cluster 6), and constitutive expression in wild-type cells but higher levels of mitotic expression in the kar4Δ strain (cluster 7).
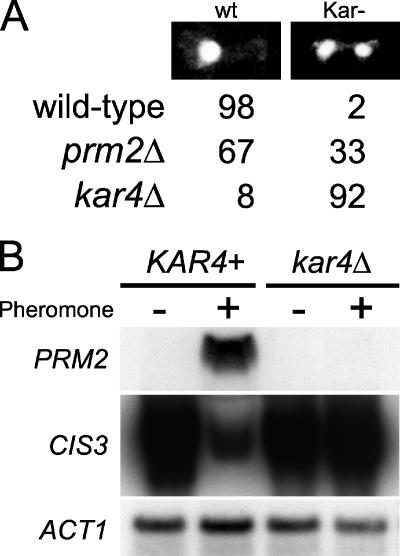
To determine whether the clustering of genes based on their pattern of Kar4 dependence reflected a deeper functional relationship, we examined the gene annotations for significant correlations by using FunSpec (40). Cluster 3 was significantly enriched in genes previously shown to be involved in nuclear congression (e.g., KAR3 and CIK1) or known to affect microtubule function (e.g., SPC25) or else implicated in the structure of the cytoskeleton (e.g., YOR129C), a finding consistent with Kar4's function in regulating genes involved in nuclear congression (P < 0.0004 [FunSpec]). Other genes within the pheromone-induced, Kar4-dependent cluster may have roles in mating that are not directly related to microtubule function, such as PRM2, which encodes a putative pheromone-induced membrane protein (17). Although no function has been previously reported for PRM2, we found that karyogamy failed in 30% of prm2Δ/prm2Δ zygotes, a finding consistent with a role in mating (Fig. 4A). Many of the remaining genes in the Kar4-dependent cluster are not characterized. Kar4 most likely regulates genes in which both mating partners must be mutant to express the mating defect, as is the case for both KAR3 and CIK1. Such “bilateral” mutants and mutants with less severe defects would have been missed in previous screens for mutations affecting mating (22, 41).
FIG. 4.
Kar4-dependent induction and repression of genes. (A) PRM2 is required for efficient nuclear fusion. Three different mating mixtures (wild-type × wild-type, n = 55; kar4Δ × kar4Δ, n = 148; and prm2Δ × prm2Δ, n = 195) were examined for karyogamy defects. Mating mixtures were stained with DAPI, and the phenotypes were observed and scored as fused (wt) or unfused (Kar−) nuclei. The percentages of each class are shown. (B) Northern blot analysis of wild type (MS3213) and kar4Δ (MS3215) mRNA in the absence (−) or presence (+) of pheromone. The blot was hybridized to a probe for PRM2 (upper panel) or CIS3 (middle panel). A duplicate Northern blot was hybridized with an ACT1 probe (bottom panel).
Cluster 4 contained genes whose expression was reduced during mating in a Kar4-dependent manner. Cluster 4 was enriched in genes involved in cell wall functions (e.g., CIS3/PIR2, CWP1, SKG1, MHP1, and YDL222C) or which act as molecular chaperones (e.g., SSE2, HSP30, HSP42, SIS1, YNL077W, HSP78, SSA4, HSP104, and HSP82). Downregulation of cell wall proteins would be consistent with the requisite removal of cell wall at the zone of cell fusion during mating (13).
To confirm the microarray data, Northern blots were performed on two representative genes from clusters 3 and 4 (Fig. 4B). PRM2 was observed to be strongly induced by pheromone, in a Kar4-dependent way. Similarly, CIS3 was strongly repressed by the addition of pheromone, but levels remained high in the kar4Δ mutant.
Identification of a consensus Kar4 regulatory motif.
To determine whether the KAR3-min region is both necessary and sufficient for Kar4-mediated pheromone induction of KAR3, the 30-bp segment was synthesized and placed upstream of a lacZ reporter gene (Fig. 5A). In the presence of pheromone, the 30-bp region drove the expression of the reporter gene to the same extent as the full-length KAR3 promoter (L11) (23), indicating that this 30-bp minimal sequence contained all of the necessary regulatory information. It was previously noted that the 30 bp contains a repeated sequence motif (CAAA), which was suggested to be a regulatory element for Kar4, in addition to a weak Ste12-binding site (23).
FIG. 5.
Defining Kar4 activation parameters. (A) KAR4-dependent minimal regulatory region. The DNA region upstream of KAR3 was fused upstream of the lacZ gene, lacking upstream activator sequences. Expression is presented as units of β-galactosidase specific activity in the presence of mating pheromone. Assays were performed with MY3265 (KAR4) or MY4166 (kar4Δ) harboring plasmids with deletions in the KAR3 promoter regions. L11 (pMR2806), L16 (pMR2802) and L18 (pMR2892) delimit the KAR4-dependent region to a 30-bp region upstream of the KAR3 gene. Flanking the lines are numbers that correspond to the endpoints of the deletions within the original 560-bp region (23). K3 min (pMR5620) contains the minimal 30-bp KAR4-dependent region indicated by the shaded box. The nucleotide sequence is shown below K3 min. The vertical line at position 485 indicates the endpoints for deletions L14 (pMR2808) and L15 (pMR2896). The results for L11, L14, L15 L16, and L18 were published previously (23). (B) Sequence alignment of upstream DNA from Kar4-regulated genes as identified by AlignAce analysis. Upstream regulatory regions of Kar4-dependent pheromone-induced genes showing a fourfold or greater induction were analyzed for sequence enrichment. See the Materials and Methods for descriptions of the MAP score and the specificity score. (C) Mutational analysis of the Kar4-dependent region. The results of β-galactosidase assays represented as a percentage of the KAR3-minimal region promoter activity (% K3 min) in the presence of mating pheromone are shown. MY3265 harboring plasmids containing mutated (31-2, pMR5621; 41-2, pMR5622) or wild-type (pMR5620) minimal KAR3 upstream regulatory region, as well as the vector control (pLG669-Z, vector), were analyzed for promoter activity. Mutated nucleotides are underlined. The consensus pheromone response element (PRE) is boxed.
Two KAR3 promoter deletions, L14 (pMR2802) and L15 (pMR2896), with endpoints within one of the CAAA repeats, had previously been found to destroy pheromone induction, which is consistent with an essential role for this element in Kar4-dependent pheromone regulation (Fig. 5A). Mutation of one of the A residues within this CAAA repeat eliminated pheromone induction of the lacZ reporter gene (Fig. 5C). In contrast, mutation of an equivalent A, adjacent to the PRE (TCAAACA), within an overlapping CAAA, had no effect on induction (Fig. 5C).
To determine whether the CAAA sequence is also upstream of other genes regulated by Kar4, we compared the DNA sequences of the 15 genes in cluster 3 whose expression after pheromone induction was reduced at least fourfold or more in the kar4Δ mutant relative to the wild type. These genes included KAR3, CIK1, SPC25, PRM2, ISC1, IME4, CDA2, JJJ2, ECM23 YPL033C, YDR281C, YOR129C, YNL033W, YJL218W, and YNL105W. Using AlignACE (18) to identify DNA sequence motifs that are overrepresented upstream of the input genes, we found significant enrichment of one motif, C/TAANNCAAANNCNGNYT (Fig. 5B, MAP score of 7.45104 and specificity score of 1.6 × 10−10). This motif was found upstream of 9 of the input genes, including KAR3 (CAAAACAAACACTGTTT). As discussed above, mutation of the second A of the conserved motif destroyed promoter activity from the KAR3 minimal activating sequence (Fig. 5C). Two other putative motifs encompassed the same CAAA sequence but exhibited low specificity scores due to the high TA content (motif 1, MAP score of 28.9849, specificity score of 7.2 × 10−1; motif 2, MAP score of 18.691, specificity score of 1.1 × 10−1).
Temporal regulation by Kar4.
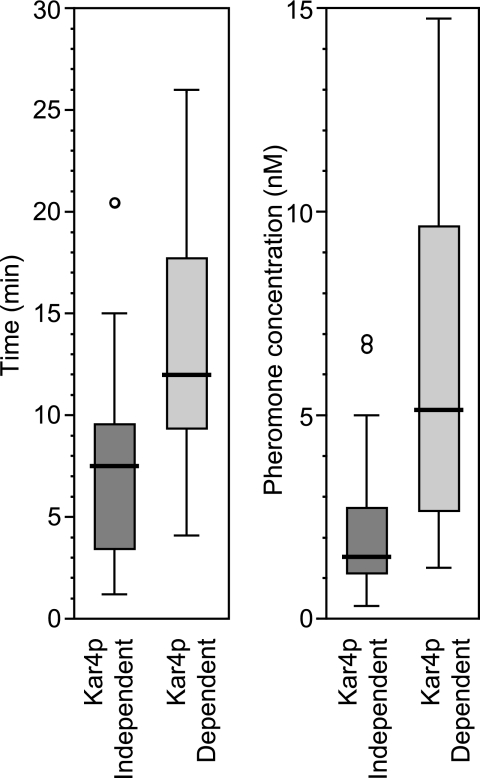
The additional requirement of Kar4 along with Ste12 for the regulation of genes during mating provides a potential mechanism for temporal regulation during the pheromone response. From the microarray data, we generated lists of the genes that were induced by pheromone at least 2.5-fold in both the Kar4-independent cluster and the Kar4-dependent cluster (Table 2). To compare the two sets of genes, published gene expression profiles were compared with respect to both the kinetics of induction and the dose response to pheromone (39). For each gene we determined the time required for half-maximal induction (t1/2). For the set of 34 Kar4-independent genes, ca. 25% showed half-maximal inductions of 3 min or less after addition of pheromone and ca. 75% of genes showed t1/2's of 9 min or less, with a median t1/2 of 7.5 min (Fig. 6A). The rapid induction of this class of genes is consistent with previous data showing that the prototype pheromone-induced gene, FUS1, does not require protein synthesis for its induction (32). In contrast, the set of 29 Kar4-dependent genes were delayed in their response to pheromone. Only 25% showed half-maximal induction by 10 min after pheromone addition and 75% required as long as 16 min, with a median t1/2 of 12 min. Thus, the requirement for Kar4 imposed a significant delay in pheromone induction to this class of genes (P = 3.3 × 10−7).
TABLE 2.
Kar4-dependent and -independent genes induced ≥2.5-fold based on microarray data
| Gene type | No. of genes | Genes |
|---|---|---|
| Kar4-dependent genes | 29 | KAR3, CIK1, PRM2, IME4, SPC25, YOR129C, YNL105W, YPL033C, YJL218W, ECM23, JJJ2, YNL033W, CDA2, ISC1, FUI1, MMP1, PCL2/CLN4, RPO31, MHT1, HXT9, CLN3, YGL204C, PGM1, YDR340W, SCW11, CUP9, RCL1, HOS2, YOR225W |
| Kar4-independent genes | 34 | PRM6, FUS2, YDR124W, PRM1, ASG7, YOR343C, FIG1, YIL080W, FIG2, FUS1, KAR5, RRN11, PRM5, SST2, HYM1, DIG2, FAR1, YJL169W, SUT1, HMI1, YNL324W, BOS1, BOP3, YML048W-A, ORT1, ECM13, AGA1, MFA1, GPA1, KTR2, PRP39, YMR304C-A, AGA2, STE2 |
FIG. 6.
Temporal and pheromone concentration regulation of Kar4-dependent genes. (A) Genes showing greater than 2.5-fold induction by pheromone from cluster 1 (Kar4-independent [34]) and cluster 2 (Kar4 dependent [29]) were selected, and datum points were taken representing the log2 (ratio) of normalized expression at 0, 15, 30, 45, and 60 min after induction by pheromone (39). A pheromone response curve was generated for each gene, and the t1/2 was extracted from the interpolated data. Primary data were from Roberts et al. (39). (B) Box plot representing the concentration of pheromone corresponding to the half-maximal induction of each of the Kar4-dependent and Kar4-independent pheromone response genes. The data determined from the log2 (ratio) of normalized expression at 30 min after exposure of a bar1Δ strain to various concentrations of pheromone (0.15, 0.5, 1.5, 5, 15.8, 50, and 158 nM α-factor) Primary data were from Roberts et al. (39).
The two sets of genes also showed significant differences with respect to the pheromone dose response. In the bar1Δ strain used in the study, Kar4-independent genes required a median concentration of only 1.25 nM pheromone to achieve half-maximal expression (Fig. 6B). In contrast, the Kar4-dependent genes required a significantly higher median concentration of pheromone (5.2 nM) to reach half-maximal expression (P = 1.59 × 10−5).
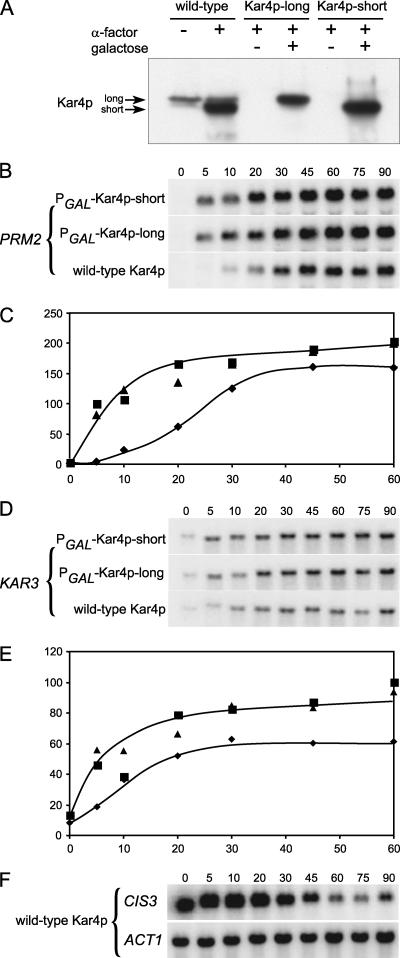
These observations suggest that Kar4 specifically imparts a delayed response to genes regulated by mating pheromone. To test this hypothesis, we examined the effect of precocious expression of Kar4 on the temporal regulation of genes that are either induced (KAR3 and PRM2) or repressed (CIS3) by pheromone, depending on Kar4. To perturb the kinetics of induction, Kar4 was exogenously regulated using the galactose-inducible GAL1 promoter. We used constructs expressing one of the two forms of Kar4 from the GAL1 promoter (PGAL). The shorter, pheromone-induced form was expressed from PGAL-KAR4-short::HA, and the longer, constitutively expressed form was expressed from PGAL-KAR4-long::HA. In both cases the concentrations of glucose and galactose were adjusted to achieve levels of the PGAL-expressed proteins comparable to those observed for the endogenous short form after pheromone induction (14). Protein levels of the endogenous (wild-type) and galactose-induced forms of Kar4 were verified to be similar (Fig. 7A). All strains were grown in galactose and then exposed to mating pheromone. At various time intervals, RNA was extracted, blotted, and probed for expression of PRM2 (Fig. 7B and C), KAR3 (Fig. 7D and E), and CIS3 (Fig. 7F).
FIG. 7.
Kar4-dependent temporal regulation of PRM2, KAR3, and CIS3. (A) Western blot comparison of pheromone- and galactose-induced Kar4::HA. The MS3216 kar4Δ::HIS3 strain harbored either the wild-type KAR4::HA (pMR2654), PGAL-KAR4-short::HA (pMR3356), or PGAL-KAR4-long::HA (pMR3459) plasmid constructs. The cells were grown in the absence (−) or presence (+) of galactose and in the absence (−) or presence (+) of pheromone (α-factor). The arrows indicate the two forms of Kar4::HA. (B to F) Time course of kar4Δ cells (MS3216) harboring plasmid pMR2654 (⧫, wild-type Kar4) or the galactose-induced plasmids pMR3356 (▪, galactose-induced short form of Kar4, PGAL-Kar4-short) or pMR3459 (▴, galactose-induced long form of Kar4 predominant, PGAL-Kar4-long). Cells were grown in medium containing galactose, and pheromone was added at t = 0. Cells were harvested at the indicated times, and RNA was prepared. Northern blots were hybridized with probes to either PRM2 (B), KAR3 (D), CIS3 (F), or ACT1 (F). The graphs in panels C and E show the levels of mRNA in arbitrary intensity units.
In wild-type cells, PRM2 mRNA levels were not detected prior to the pheromone treatment. After pheromone addition, PRM2 mRNA levels increased gradually, reaching half-maximal levels after approximately 25 min (Fig. 7B and C). In both strains in which Kar4 was expressed from PGAL, initial mRNA levels were not detected but increased very rapidly after pheromone addition, reaching half-maximal levels within 5 min (Fig. 7B and C). Plateau levels of PRM2 were somewhat elevated relative to the wild type.
In wild-type cells, a basal mitotic level of KAR3 mRNA was observed prior to pheromone addition (Fig. 7D and E). The KAR3 mRNA levels increased more quickly than PRM2, reaching half-maximal levels approximately 10 min after pheromone addition. In both strains in which Kar4 was expressed from PGAL, the initial levels of KAR3 mRNA were similar to that of the wild type but increased more rapidly, reaching half maximal levels with 5 min after pheromone addition (Fig. 7D and E). In addition, the plateau levels of KAR3 mRNA were significantly elevated compared to cells with endogenously expressed Kar4. Thus, the kinetics of both PRM2 and KAR3 induction were significantly accelerated by the premature expression of Kar4, albeit to different extents.
Either form of Kar4 when prematurely produced was able similarly to induce and alter the kinetics of PRM2 and KAR3 expression. These observations confirm that the two proteins are functionally equivalent but differ mainly in their patterns of expression and stability (14).
In contrast to KAR3 and PRM2, CIS3 mRNA levels were high in mitotic cells and began to decline after 20 to 30 min after pheromone addition, reaching levels three- to fourfold lower (Fig. 7F). Premature expression of Kar4 from PGAL had no significant effect on the rate of mRNA decline (not shown), suggesting that the half-life of the mRNA is longer than any potential shift in the timing of repression. ACT1 mRNA levels were essentially constant throughout the time of the experiment, indicating that equal concentrations of RNA were analyzed at each time point (Fig. 7F).
DISCUSSION
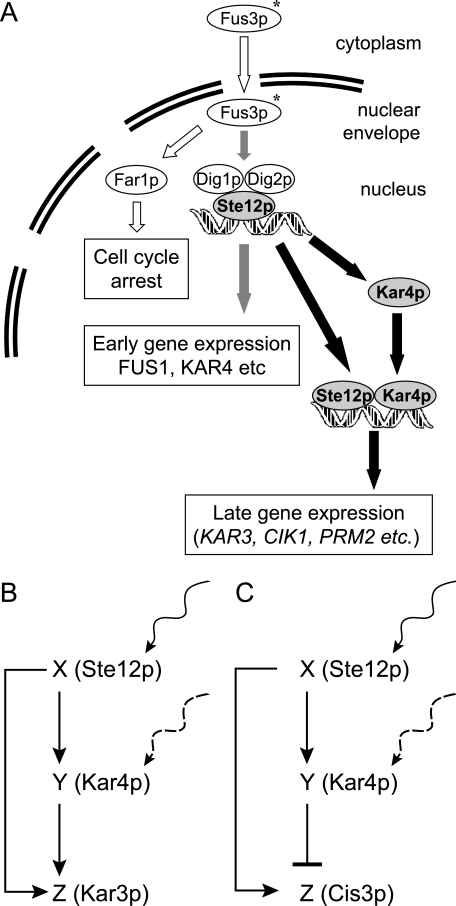
Originally identified as a regulator of two specific genes required for karyogamy, we find that Kar4 has a more widespread role in the yeast mating response. In addition to being required for the pheromone-dependent induction of KAR3 and CIK1, Kar4 is required for both the positive and the negative regulation of a number of genes during both mating and mitosis (Fig. 8A).
FIG. 8.
(A) Proposed model of Kar4 action in the pheromone response. During the pheromone response, cytoplasmic Fus3 (Fus3-C) enters the nucleus (Fus3-N), where it phosphorylates Far1 and Dig1 and Dig2, leading to cell cycle arrest, and the activation of Ste12, which in turn induces the early pheromone response genes. Ste12 also induces the expression of the pheromone-specific short form of Kar4, which begins to accumulate in the cell. After sufficient Kar4 has accumulated, it facilitates Ste12 binding to the promoter regions of genes involved in functions required late in the pheromone response. (B) Representation of the feed-forward loop. The diagram was modified from that of Mangan and Alon (29). Coherent type I feed forward loop (AND gate logic) in which transcription factor X is represented by Ste12 and transcription factor Y is represented by Kar4 and the target gene, Z, is represented by KAR3. The inducer of X and Y is pheromone, although other inducers of Kar4 are possible. This type of feed-forward loop is suggested to act as a persistence detector where only persistent stimulus of both X and Y will lead to activation of Z (29). (C) Incoherent type I feed-forward loop (AND gate logic) in which X is represented by Ste12, Y is represented by Kar4, and Z is represented by CIS3. This type of feed-forward loop is characterized by the combined induction of X and Y by a stimulus (pheromone) in order to cause Z to be repressed by Y.
Among the genes positively regulated by Kar4 during mating is PRM2, encoding a member of the pheromone-regulated putative integral membrane proteins (PRM). We have found that Prm2 is required for efficient karyogamy, and it is likely that other proteins in this cluster will also have roles in mating.
An unexpected cluster contained genes repressed by pheromone in a Kar4-dependent manner. The observation that a gene is repressed by pheromone may reflect either a specific regulatory response to pheromone or downregulation of the gene in the G1 phase of the cell cycle. However, because the kar4Δ strain arrests in G1 like the wild-type strain, genes dependent on Kar4 for pheromone-dependent downregulation are showing a specific response to pheromone. Many genes in this cluster encode proteins that are either predicted to be membrane localized or are involved in cell wall function, including CIS3/PIR2, HOC1, CWP1, YBR209W, CRC1, PHO5, PDR15, YGL146C, IZH4, YOL013W-A, FMP45, HBT1, RSN1, YSR3, HXT16, EDE1, ECM30, OPT1, MAL11, ECI1, EMP70, PST1, TGL1, SIM1, JSN1, LST8, YKL008C, SNX4, STD1, SKG1, and YNL181W. One interpretation of this cluster of genes is that cell wall biosynthesis may be negatively regulated at the shmoo tip to allow efficient cell fusion during mating. Consistent with this hypothesis, the shmoo tip has reduced levels of β-1-3 glucan that are correlated with the efficiency of mating (13).
The cluster of genes exhibiting Kar4-dependent pheromone repression was also enriched in molecular chaperone genes, such as HSP42, SSA4, HSP78, HSP82, HSP104, and SSE2. Although a specific role for these chaperones in mating is not known, it is possible that elevated levels would interfere with mating. Assessing chaperone regulation during mating has been complicated by the difficulty of ensuring constant stress over the course of the experiment. Indeed, different studies have yielded conflicting data (39, 46). However, the Kar4 dependence of their regulation suggests that repression is real.
Cluster 6 is comprised of genes showing similar levels of expression during vegetative growth and after pheromone treatment in wild type but which showed significant induction by pheromone in the kar4Δ strain. Cluster 6 was enriched in genes known to play a role in the mating response, encoding proteins involved in pheromone synthesis (AXL1, MFα1, and MFα2), pheromone receptors (STE3), agglutination (SAG1 and MUC1), and signaling (FUS3). Most of these genes show relatively little change during an extended time course of pheromone response (39). Because most of these genes act in the earliest stages of the mating response, delayed Kar4-dependent negative regulation may allow for a pulse of expression preparatory to mating.
Genes in cluster 5 were also constitutively expressed in the wild-type strain but, in contrast to cluster 6, were downregulated by pheromone in the kar4Δ mutant. One hypothesis to explain cluster 5 is that it is comprised of genes whose expression is cell cycle regulated and not normally expressed during the G1 phase. However, expression of these genes may be required during mating and therefore induced by pheromone. Consistent with this, cluster 5 is enriched in genes that are involved in RNA processing and ribosome biogenesis, and may facilitate high levels of protein synthesis during mating. Several genes within this cluster (RPB11, MAK16, TRM1, SRP40, NSR1, RRP8, and HCA4) have been shown to be expressed in pheromone-arrested cells, repressed as the cells reenter the cell cycle at G1, and then induced later in the cell cycle (46).
Cluster 7 was comprised of genes that are expressed constitutively in wild-type cells but showed strongly elevated levels of expression during mitotic growth in the kar4Δ mutant. The behavior of these genes suggests that Kar4 may negatively regulate the expression of some genes during vegetative growth. Because their levels of gene expression are not elevated in the kar4Δ mutant after pheromone treatment, these genes must not be expressed at G1 but would be expressed at other times in the cell cycle. Consistent with this hypothesis, cluster 7 is enriched in genes involved in cell cycle regulation (CDC5 and CDC6), as well as in microtubule and spindle pole body function (SPC42, VIK1, and SPC24).
Temporal regulation of Kar4-dependent pheromone induced genes.
The Kar4-dependent pheromone-induced genes in cluster 3 were enriched in genes induced later in the pheromone response. Because Kar4 is itself induced by pheromone, it was proposed that Kar4 may function as a temporal regulator of a subset of mating-specific proteins (23).
In support of this hypothesis, premature expression of Kar4 significantly accelerated the expression of KAR3 and PRM2, causing both genes to become half-maximally induced within 5 min after pheromone addition. The accelerated kinetics match those observed for the bulk of the genes whose expression is independent of Kar4 (Fig. 6). In wild-type cells, KAR3 expression was induced more quickly than PRM2, indicating that the various Kar4-dependent promoters may respond to different levels of Kar4.
These results suggest that, with respect to pheromone-dependent transcriptional induction, Kar4 acts like a type I coherent feed-forward loop (Fig. 8B) (29, 30). Briefly, a feed-forward loop is composed of a transcription factor X (e.g., Ste12) that regulates a second transcription factor Y (e.g., Kar4), such that both regulate downstream genes, Z (e.g., KAR3 and other cluster 2 genes induced later in the pheromone response). In the AND-gate version of the type I coherent feed-forward loop, the requirement for the second transcription factor reaching a concentration threshold results in a sigmoid-shaped delayed response. This model predicts that constitutive expression of activating levels of Kar4 would transform the system into one exhibiting a simple dependence on the activating stimulus, erasing both the delay and the sigmoid behavior, as we observed (Fig. 7C).
With respect to genes showing Kar4-dependent repression on pheromone (clusters 4 and 6), a second class of a feed-forward loop may be more relevant. In the type I incoherent feed-forward loop with AND-gate logic (Fig. 8C), a transcription factor X (i.e., Ste12) activates both gene Z (e.g., Cis3) and a second transcription factor Y (i.e., Kar4), which in turn represses gene Z. CIS3 expression is repressed during pheromone response in both a Ste12 and Kar4-dependent manner (our data and reference 39), although the dependence on Ste12 may simply reflect the requirement for Kar4 expression. One characteristic of incoherent type I feed-forward loops is that they show a modest pulse of Z expression upon stimulation of X, since it takes time for Y to accumulate. Cluster 6 included several genes that act early in mating and so might be expected to be expressed at a high level early, but not later, in the mating response.
Kar4 regulatory motif.
Previous experiments demonstrated the Kar4-dependent transcriptional regulation of KAR3 and CIK1 but did not establish that Kar4 binds to DNA. Given the sequence homology between Kar4 and mRNA methyltransferases, it was critical to reexamine whether Kar4 interacts with DNA. In this work, we showed that Kar4 is associated with the chromatin upstream of KAR3 and that purified Kar4 binds to DNA regulatory sequences upstream of KAR3 in vitro. Furthermore, Kar4-dependent regulation was observed using a heterologous reporter construct (CYC1-lacZ) containing the upstream regulatory elements but no mRNA sequences from KAR3 (23; the present study). Thus, Kar4-dependent regulation during mating most likely acts via regulatory DNA sequences rather than by elements present within the mRNA.
To characterize the Kar4 regulatory sequence, we used AlignAce (18) to identify DNA motifs enriched upstream of the genes showing strong Kar4-dependent pheromone induction. This analysis identified a motif, C/TAANNCAAANNCNGNYT, contained within the minimal region of KAR3, which was necessary for pheromone induction. Interestingly, this motif was not enriched in the other clusters showing Kar4-dependent regulation. Kar4 may either (i) bind to more than one motif, (ii) bind via interactions with a second DNA-binding protein, or (iii) act indirectly through the induction of other regulatory proteins. Motif analysis yielded a large set of DNA motifs enriched in the upstream regulatory regions of the other clusters, but it remains to be determined whether any are required for Kar4-dependent regulation.
In sum, we have shown that Kar4 plays a more extensive and complex role in yeast mating than previously thought. Interestingly, the human pathogen Candida albicans contains a homolog of Kar4 that is transcriptionally regulated both by mating pheromone and by whether the cell is in the alternate “opaque” phase of growth (24). Similar to our observations with budding yeast, several opaque-phase genes, including gene associated with skin colonization are downregulated during the pheromone response. These results suggest that in Candida, as in Saccharomyces, Kar4 may help provide part of the complex pattern of gene regulation during different cell developmental phases of growth.
Acknowledgments
We thank the members of the Rose lab for many helpful discussions. We thank Donna Storton for help with use of the microarray facility. This study was supported by grant GM37739 to M.D.R.
Footnotes
Published ahead of print on 13 November 2006.
REFERENCES
- 1.Ausubel, F. M. 1987. Current protocols in molecular biology. Greene Publishing Associates, Media, PA.
- 2.Baur, M., R. K. Esch, and B. Errede. 1997. Cooperative binding interactions required for function of the Ty1 sterile responsive element. Mol. Cell. Biol. 17:4330-4337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beh, C. T., V. Brizzio, and M. D. Rose. 1997. KAR5 encodes a novel pheromone-inducible protein required for homotypic nuclear fusion. J. Cell Biol. 139:1063-1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bohm, S., D. Frishman, and H. W. Mewes. 1997. Variations of the C2H2 zinc finger motif in the yeast genome and classification of yeast zinc finger proteins. Nucleic Acids Res. 25:2464-2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bujnicki, J. M., M. Feder, M. Radlinska, and R. M. Blumenthal. 2002. Structure prediction and phylogenetic analysis of a functionally diverse family of proteins homologous to the MT-A70 subunit of the human mRNA:m6A methyltransferase. J. Mol. Evol. 55:431-444. [DOI] [PubMed] [Google Scholar]
- 6.Burke, D., D. Dawson, T. Stearns, et al. 2000. Methods in yeast genetics. Cold Spring Harbor Laboratory Press, Plainview, NY.
- 7.Clancy, M. J., M. E. Shambaugh, C. S. Timpte, and J. A. Bokar. 2002. Induction of sporulation in Saccharomyces cerevisiae leads to the formation of N6-methyladenosine in mRNA: a potential mechanism for the activity of the IME4 gene. Nucleic Acids Res. 30:4509-4518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Desjarlais, J. R., and J. M. Berg. 1992. Toward rules relating zinc finger protein sequences and DNA binding site preferences. Proc. Natl. Acad. Sci. USA 89:7345-7349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dohlman, H. G., and J. W. Thorner. 2001. Regulation of G protein-initiated signal transduction in yeast: paradigms and principles. Annu. Rev. Biochem. 70:703-754. [DOI] [PubMed] [Google Scholar]
- 10.Dolan, J. W., C. Kirkman, and S. Fields. 1989. The yeast STE12 protein binds to the DNA sequence mediating pheromone induction. Proc. Natl. Acad. Sci. USA 86:5703-5707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elion, E. A., M. Qi, and W. Chen. 2005. Signal transduction: signaling specificity in yeast. Science 307:687-688. [DOI] [PubMed] [Google Scholar]
- 12.Errede, B., and G. Ammerer. 1989. STE12, a protein involved in cell-type-specific transcription and signal transduction in yeast, is part of protein-DNA complexes. Genes Dev. 3:1349-1361. [DOI] [PubMed] [Google Scholar]
- 13.Fitch, P. G., A. E. Gammie, D. J. Lee, V. B. de Candal, and M. D. Rose. 2004. Lrg1p Is a Rho1 GTPase-activating protein required for efficient cell fusion in yeast. Genetics 168:733-746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gammie, A. E., B. G. Stewart, C. F. Scott, and M. D. Rose. 1999. The two forms of karyogamy transcription factor Kar4p are regulated by differential initiation of transcription, translation, and protein turnover. Mol. Cell. Biol. 19:817-825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guarente, L., and M. Ptashne. 1981. Fusion of Escherichia coli lacZ to the cytochrome c gene of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 78:2199-2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hagen, D. C., G. McCaffrey, and G. F. Sprague, Jr. 1991. Pheromone response elements are necessary and sufficient for basal and pheromone-induced transcription of the FUS1 gene of Saccharomyces cerevisiae. Mol. Cell. Biol. 11:2952-2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heiman, M. G., and P. Walter. 2000. Prm1p, a pheromone-regulated multispanning membrane protein, facilitates plasma membrane fusion during yeast mating. J. Cell Biol. 151:719-730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hughes, J. D., P. W. Estep, S. Tavazoie, and G. M. Church. 2000. Computational identification of cis-regulatory elements associated with groups of functionally related genes in Saccharomyces cerevisiae. J. Mol. Biol. 296:1205-1214. [DOI] [PubMed] [Google Scholar]
- 19.James, P., J. Halladay, and E. A. Craig. 1996. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144:1425-1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim, T. S., H. Y. Kim, J. H. Yoon, and H. S. Kang. 2004. Recruitment of the Swi/Snf complex by Ste12-Tec1 promotes Flo8-Mss11-mediated activation of STA1 expression. Mol. Cell. Biol. 24:9542-9556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kurdistani, S. K., D. Robyr, S. Tavazoie, and M. Grunstein. 2002. Genome-wide binding map of the histone deacetylase Rpd3 in yeast. Nat. Genet. 31:248-254. [DOI] [PubMed] [Google Scholar]
- 22.Kurihara, L. J., C. T. Beh, M. Latterich, R. Schekman, and M. D. Rose. 1994. Nuclear congression and membrane fusion: two distinct events in the yeast karyogamy pathway. J. Cell Biol. 126:911-923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kurihara, L. J., B. G. Stewart, A. E. Gammie, and M. D. Rose. 1996. Kar4p, a karyogamy-specific component of the yeast pheromone response pathway. Mol. Cell. Biol. 16:3990-4002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lockhart, S. R., R. Zhao, K. J. Daniels, and D. R. Soll. 2003. Alpha-pheromone-induced “shmooing” and gene regulation require white-opaque switching during Candida albicans mating. Eukaryot. Cell 2:847-855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mackey, A. T., L. R. Sproul, C. A. Sontag, L. L. Satterwhite, J. J. Correia, and S. P. Gilbert. 2004. Mechanistic analysis of the Saccharomyces cerevisiae kinesin Kar3. J. Biol. Chem. 279:51354-51361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maddox, P., E. Chin, A. Mallavarapu, E. Yeh, E. D. Salmon, and K. Bloom. 1999. Microtubule dynamics from mating through the first zygotic division in the budding yeast Saccharomyces cerevisiae. J. Cell Biol. 144:977-987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Madhani, H. D., and G. R. Fink. 1997. Combinatorial control required for the specificity of yeast MAPK signaling. Science 275:1314-1317. [DOI] [PubMed] [Google Scholar]
- 28.Madhani, H. D., T. Galitski, E. S. Lander, and G. R. Fink. 1999. Effectors of a developmental mitogen-activated protein kinase cascade revealed by expression signatures of signaling mutants. Proc. Natl. Acad. Sci. USA 96:12530-12535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mangan, S., and U. Alon. 2003. Structure and function of the feed-forward loop network motif. Proc. Natl. Acad. Sci. USA 100:11980-11985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mangan, S., A. Zaslaver, and U. Alon. 2003. The coherent feed-forward loop serves as a sign-sensitive delay element in transcription networks. J. Mol. Biol. 334:197-204. [DOI] [PubMed] [Google Scholar]
- 31.Marsh, L., and M. D. Rose. 1997. The pathway of cell and nuclear fusion during mating in Saccharomyces cerevisiae, p. 827-888. In J. Pringle, J. R. Broach, and E. W. Jones (ed.), The molecular and cellular biology of the yeast Saccharomyces, vol. 3. Cell cycle and cell biology. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 32.McCaffrey, G., F. J. Clay, K. Kelsay, and G. F. Sprague, Jr. 1987. Identification and regulation of a gene required for cell fusion during mating of the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 7:2680-2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meluh, P. B., and M. D. Rose. 1990. KAR3, a kinesin-related gene required for yeast nuclear fusion. Cell 60:1029-1041. [DOI] [PubMed] [Google Scholar]
- 34.Miller, R. K., and M. D. Rose. 1998. Kar9p is a novel cortical protein required for cytoplasmic microtubule orientation in yeast. J. Cell Biol. 140:377-390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oehlen, L. J., J. D. McKinney, and F. R. Cross. 1996. Ste12 and Mcm1 regulate cell cycle-dependent transcription of FAR1. Mol. Cell. Biol. 16:2830-2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ohashi, A., J. Gibson, I. Gregor, and G. Schatz. 1982. Import of proteins into mitochondria: the precursor of cytochrome c1 is processed in two steps, one of them heme-dependent. J. Biol. Chem. 257:13042-13047. [PubMed] [Google Scholar]
- 37.Oldenburg, K. R., K. T. Vo, S. Michaelis, and C. Paddon. 1997. Recombination-mediated PCR-directed plasmid construction in vivo in yeast. Nucleic Acids Res. 25:451-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Page, B. D., L. L. Satterwhite, M. D. Rose, and M. Snyder. 1994. Localization of the Kar3 kinesin heavy chain-related protein requires the Cik1 interacting protein. J. Cell Biol. 124:507-519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roberts, C. J., B. Nelson, M. J. Marton, R. Stoughton, M. R. Meyer, H. A. Bennett, Y. D. He, H. Dai, W. L. Walker, T. R. Hughes, M. Tyers, C. Boone, and S. H. Friend. 2000. Signaling and circuitry of multiple MAPK pathways revealed by a matrix of global gene expression profiles. Science 287:873-880. [DOI] [PubMed] [Google Scholar]
- 40.Robinson, M. D., J. Grigull, N. Mohammad, and T. R. Hughes. 2002. FunSpec: a web-based cluster interpreter for yeast. BMC Bioinformatics 3:35-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rose, M. D. 1996. Nuclear fusion in the yeast Saccharomyces cerevisiae. Annu. Rev. Cell Dev. Biol. 12:663-695. [DOI] [PubMed] [Google Scholar]
- 42.Rose, M. D., F. M. Winston, P. Hieter, et al. 1990. Methods in yeast genetics: a laboratory course manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 43.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 44.Schmitt, M. E., T. A. Brown, and B. L. Trumpower. 1990. A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res. 18:3091-3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Spellman, P. T., G. Sherlock, M. Q. Zhang, V. R. Iyer, K. Anders, M. B. Eisen, P. O. Brown, D. Botstein, and B. Futcher. 1998. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol. Biol. Cell 9:3273-3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sprague, G. F., and J. W. Thorner. 1992. Pheromone response and signal transduction during the mating process of Saccharomyces cerevisiae, p. 657-744. In J. R. Broach, J. Pringle, and E. W. Jones (ed.), The molecular and cellular biology of the yeast Saccharomyces, vol. 2. Gene expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 48.Tavazoie, S., J. D. Hughes, M. J. Campbell, R. J. Cho, and G. M. Church. 1999. Systematic determination of genetic network architecture. Nat. Genet. 22:281-285. [DOI] [PubMed] [Google Scholar]
- 49.Winzeler, E. A., D. D. Shoemaker, A. Astromoff, H. Liang, K. Anderson, B. Andre, R. Bangham, R. Benito, J. D. Boeke, H. Bussey, A. M. Chu, C. Connelly, K. Davis, F. Dietrich, S. W. Dow, M. El Bakkoury, F. Foury, S. H. Friend, E. Gentalen, G. Giaever, J. H. Hegemann, T. Jones, M. Laub, H. Liao, N. Liebundguth, D. J. Lockhart, A. Lucau-Danila, M. Lussier, N. M'Rabet, P. Menard, M. Mittmann, C. Pai, C. Rebischung, J. L. Revuelta, L. Riles, C. J. Roberts, P. Ross-MacDonald, B. Scherens, M. Snyder, S. Sookhai-Mahadeo, R. K. Storms, S. Veronneau, M. Voet, G. Volckaert, T. R. Ward, R. Wysocki, G. S. Yen, K. Yu, K. Zimmermann, P. Philippsen, M. Johnston, and R. W. Davis. 1999. Functional characterization of the Saccharomyces cerevisiae genome by gene deletion and parallel analysis. Science 285:901-906. [DOI] [PubMed] [Google Scholar]
- 50.Zeitlinger, J., I. Simon, C. T. Harbison, N. M. Hannett, T. L. Volkert, G. R. Fink, and R. A. Young. 2003. Program-specific distribution of a transcription factor dependent on partner transcription factor and MAPK signaling. Cell 113:395-404. [DOI] [PubMed] [Google Scholar]