Abstract
The forkhead box M1 (FoxM1) transcription factor regulates expression of cell cycle genes essential for DNA replication and mitosis during organ repair and cancer progression. Here, we demonstrate that FoxM1-deficient (−/−) mouse embryonic fibroblasts and osteosarcoma U2OS cells depleted in FoxM1 levels by small interfering RNA transfection display increased DNA breaks, as evidenced by immunofluorescence focus staining for phosphospecific histone H2AX. FoxM1-deficient cells also exhibit stimulation of p53 transcriptional activity, as evidenced by increased expression of the p21cip1 gene. FoxM1-deficient cells display reduced expression of the base excision repair factor X-ray cross-complementing group 1 (XRCC1) and breast cancer-associated gene 2 (BRCA2), the latter of which is involved in homologous recombination repair of DNA double-strand breaks. Furthermore, FoxM1 protein is phosphorylated by checkpoint kinase 2 (Chk2) in response to DNA damage. This phosphorylation of FoxM1 on serine residue 361 caused increased stability of the FoxM1 protein with corresponding increased transcription of XRCC1 and BRCA2 genes, both of which are required for repair of DNA damage. These results identify a novel role for FoxM1 in the transcriptional response during DNA damage/checkpoint signaling and show a novel mechanism by which Chk2 protein regulates expression of DNA repair enzymes.
In response to DNA damage, activation of cell cycle checkpoint pathways arrests mammalian cell cycle progression to provide adequate time to repair DNA damage or to induce apoptosis damage is catastrophic (1, 48). These checkpoint pathways inhibit progression into DNA replication (S phase) by arresting cells in either G1 or S phase and may also stop the cells from entering mitosis (M phase) to prevent segregation of damaged chromosomes to daughter cells. Unrepaired DNA damage can result in permanent cell cycle arrest (senescence), induction of apoptosis, or mitotic cell death caused by genomic instability and loss of essential chromosomes (26). Unfaithful repair of DNA damage causes immortalization and transformation of cells leading to translocations, inversions, or deletions, which activate oncogenes and/or inactivate tumor suppressor genes that lead to the development of cancer (2, 12, 40). The checkpoint network consists of DNA damage-sensing kinases ataxia-telangiectasia mutated (ATM), ATM and Rad3 related (ATR) (1), and the downstream signaling effectors checkpoint kinase 1 (Chk1) and Chk2 (2). The damage signals are spread by the subsequent phosphorylation of downstream target proteins that mediate cell cycle arrest and repair in response to DNA damage. Although the p53 transcription factor plays an important role in mediating cell cycle arrest during DNA damage/checkpoint signaling, the transcriptional pathways that mediate DNA repair response are not completely understood.
The mammalian forkhead box (Fox) family of transcription factors consists of more than 50 mammalian proteins (13) that share homology in the winged helix DNA binding domain (5). FoxM1 (or the splicing FoxM1b isoform) is expressed in all proliferating mammalian cells and tumor-derived cell lines (6, 46), where it regulates transcription of cell cycle genes critical for progression into DNA replication and mitosis (41, 42, 45). Mouse hepatocytes with conditional deletion of the FoxM1 gene show an 80% reduction in DNA replication (S phase) and a complete inhibition of mitosis during liver regeneration (42). FoxM1-deficient hepatocytes display nuclear accumulation of cyclin-dependent kinase (Cdk) inhibitor proteins p21Cip1 and p27Kip1, which explains the significant reduction of S-phase progression in FoxM1-deficient cells (15, 41, 42). FoxM1 is essential for transcription of S-phase kinase-associated protein 2 and Cdk subunit 1, which are specificity subunits of the S-phase kinase-associated protein 1-Cullin1-F-box ubiquitin ligase complex (41) that targets the Cdk inhibitor proteins p21Cip1 and p27Kip1 for degradation during the G1/S transition. For G2/M and mitotic progression, FoxM1 is essential for transcription of the cyclin B1, Cdc25B phosphatase, polo-like kinase 1, aurora B kinase, survivin, and centromere protein A and B genes (18, 41-43). Moreover, mouse hepatocytes deficient in the FoxM1 transcription factor fail to progress into mitosis and are resistant to developing carcinogen-induced liver tumors (15).
FoxM1 transcriptional activity requires binding of the Cdk-cyclin complexes and subsequent phosphorylation at Thr596 of FoxM1 carboxyl-terminal region to recruit the CREB-binding protein coactivator protein (22). Despite the dependence of FoxM1 transcriptional activity on the cell cycle regulator Cdk-cyclin complexes, we provide evidence that FoxM1 is phosphorylated by DNA damage-induced Chk2, and this modification resulted in stabilization of the FoxM1 protein. We show that FoxM1 stimulates transcription of the base excision repair factor X-ray cross-complementing group 1 (XRCC1) (23) and breast cancer-associated gene 2 (BRCA2), the latter of which is involved in homologous recombination repair of DNA double-strand breaks (16, 37, 47). Furthermore, FoxM1-deficient (−/−) mouse embryonic fibroblasts (MEFs) and osteosarcoma U2OS cells depleted in FoxM1 levels by small interfering RNA (siRNA) transfection display increased DNA breaks, as evidenced by immunofluorescence focus staining for phosphospecific histone H2AX and terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL). Increased transcriptional activity of p53 tumor suppressor was found in FoxM1-deficient cells, with corresponding stimulation in expression of the p53 downstream target gene p21cip1. These results identify a novel role for FoxM1 in the transcriptional response during DNA damage/checkpoint signaling.
MATERIALS AND METHODS
Expression plasmids and recombinant proteins.
The following plasmids have been described previously: pCMV-T7FoxM1b (22), pGST-FoxM1b amino acids (aa) 1 to 138 (46), pGST-FoxM1b aa 365 to 748 (41). The pGST-huChk2(WT) and pGST-huChk2(D347A)(KD) fusion protein expression plasmids were gifts from S. Elledge (24). The pGST-FoxM1b aa 326 to 383 fusion protein expression plasmid was created through PCR amplification of the human FoxM1b cDNA with the following primers: glutathione S-transferase (GST) (EcoRI) FoxM1b 326 forward, 5′-TCT GAA TTC CAG CAG AAA CGA CCG AAT-3′; GST (XhoI) FoxM1b 383 reverse, 5′-TCT CTC GAG TGG CAC CTT CAC CGA GGG-3′. The site-directed FoxM1b mutants S360A, S361A, and S360:361A were introduced into the pGST-FoxM1b aa 326 to 383 plasmid by the QuikChange in vitro mutagenesis system (Stratagene, La Jolla, CA) using the following primers: S360A forward, 5′-CTG CTA CCA CGG GTC GCC TCA TAC CTG GTA CC-3′; S360A reverse, 5′-GGT ACC AGG TAT GAG GCG ACC CGT GGT AGC AG-3′; S361A forward. 5′-CTA CCA CGG GTC AGC GCA TAC CTG GTA CCT ATC-3′; S361A reverse, 5′-GAT AGG TAC CAG GTA TGC GCT GAC CCG TGG TAG-3′; S360:361A forward, 5′-CTG CTA CCA CGG GTC GCC GCA TAC CTG GTA CCT ATC-3′; S360:361A reverse, 5′-GAT AGG TAC CAG GTA TGC GGC GAC CCG TGG TAG CAG-3′. The S361A site-directed mutation was introduced into the pCMV-T7FoxM1b expression plasmid using the method described above with S361A forward and reverse primers. The pCMV-V5huChk2(WT) or pCMV-V5huChk2(D347A)(KD) expression plasmids were generated by PCR from pGST-huChk2(WT) or pGST-huChk2(D347A)(KD) DNA, respectively, with the following primers: V5(EcoRI) hChk2ATG, 5′-TCT GAA TTC ATG TCG GAT GTT GAG GCT CA-3′; V5(NotI) hChk2stop, 5′-TCT GCG GCC GCC ACA ACA CAG CAG CAC AC-3′.
The human −2 KB BRCA2 promoter region or −1 KB XRCC1 promoter region was PCR amplified from human U2OS genomic DNA with the following primers: human BRCA2 (hBRCA2) −2kbXhoI, 5′-TCT CTC GAG GTT AGG GAG GAA GGA GCA-3′; hBRCA2 −58bpXhoI, 5′-TCT CTC GAG AAT CTG ATG ATG GAC GCC-3′; hXRCC1 −1kbXhoI, 5′-TCT CTC GAG GTT TTC TCA CCT TCC CAC; hXRCC1 +22bpHindIII, 5′-TCT AAG CTT CTG TCT CCT GGG GCC AAG. The resulting PCR human promoter fragments were cloned into the XhoI site or the corresponding XhoI and HindIII sites of the pGL3 basic Luciferase vector (Promega).
GST-tagged proteins were purified from crude Escherichia coli lysates with glutathione beads (Amersham Pharmacia Biotech, Piscataway, NJ) as described previously (46). The Dual-Luciferase assay system (Promega) was used to measure firefly luciferase enzyme activity and normalized with the Renilla luciferase internal control as described previously (22).
Cell culture and transient transfection.
U2OS and MEFs were grown in Dulbecco's modified Eagle's medium (Cellgro) supplemented with 10% fetal calf serum. A U2OS clone C3 cell line (U2OS C3 cell) (15) that allowed doxycycline (Dox)-inducible expression of the green fluorescent protein (GFP)-FoxM1b fusion protein was maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum plus 50 μg/ml of hygromycin B (Invitrogen). Transient transfections were carried out using Fugene 6 (Roche) as described previously (22).
Western blotting.
Cell lysates were resolved by denaturing gel electrophoresis before electrotransfer to a Protran nitrocellulose membrane. The membrane was subjected to Western blot analysis with antibodies against proteins of interest as described previously (41). The signals from the primary antibody were amplified by horseradish peroxidase-conjugated anti-mouse immunoglobulin G (IgG; Bio-Rad, Hercules, CA), and detected with Enhanced Chemiluminescence Plus (ECL-plus; Amersham Pharmacia Biotech, Piscataway, NJ). The following antibodies and dilutions were used for Western blotting: rabbit anti-FoxM1 (1:5,000) (41), mouse anti-Chk2 (clone 7, 1:1,000; Upstate), mouse anti-β-actin (AC-15, 1:20,000; Sigma), mouse anti-V5 (1:5,000; Invitrogen), mouse anti-T7 (1:10,000; Novagen), mouse anti-BRCA2 (clone 5.23, 1:500; Upstate), rabbit anti-XRCC1 (1:1,000; Serotec), phospho-p53Ser15 antibody (1:1,000; Cell Signaling), p53 antibody (1:200; Santa Cruz Biotechnology), and mouse anti-p21cip (1:1,000; Upstate). The anti-phosphoserine 361 FoxM1 peptide antibody (FoxM1 pS361) was generated and affinity purified by Genemed Synthesis (South San Francisco, CA). Briefly, two peptides were generated on the basis of the sequence around Ser 361, one of which was chemically phosphorylated at Ser 361. After immunization, antibodies were affinity purified over a column containing the immobilized phosphorylated peptide. Next, the bound material was passed over a second column containing immobilized nonphosphorylated peptide. Phospho-Ser 361-specific antibodies were eluted in the flowthrough of the second peptide column.
γH2AX and TUNEL immunostaining.
Passage 3 FoxM1+/+ or FoxM1−/− MEFs were seeded on an eight-well chamber slide (Nalge Nunc International, Naperville, IL) at a density of 2 × 104 cells per well. After 24 h, cells were subjected to TUNEL staining with a commercially available TUNEL assay kit (Chemicon international) by following the manufacturer's protocol or to anti-γH2AX (Ser139) staining. Briefly, cells were fixed with 10% formalin (Sigma), permeabilized with 0.2% Triton X-100, and blocked with phosphate-buffered saline containing 1% bovine serum albumin. The rabbit anti-γH2AX (Ser139) (1:50; Upstate) was used to detect phosphorylated H2AX at Ser 139. Secondary chicken anti-rabbit IgG-fluorescein isothiocyanate (1:100) was from Santa Cruz Biotechnology. Cells were counterstained with 4′,6′-diamidino-2-phenylindole (DAPI). Images were captured using a microscope Axioplan2 (Carl Zeiss).
In vitro kinase assays.
Purified recombinant wild-type or mutant FoxM1b GST protein fragments (2 μg) were incubated with purified GST-Chk2 wild type (WT) or GST-Chk2 kinase-dead (KD) proteins (1 μg) for 30 min in kinase buffer (10 mM HEPES at pH 7.5, 75 mM KCl, 5 mM MgCl2, 0.5 mM EDTA, 2 mM dithiothreitol, 100 μM ATP) in the presence of 5 μCi [γ-32P]ATP. Reactions were stopped by the addition of 5× sodium dodecyl sulfate (SDS) loading buffer, and the protein mixtures were resolved by denaturing gel electrophoresis. Gels were subjected to autoradiography to monitor 32P incorporation. Equal levels of FoxM1 GST protein fragments were confirmed by Coomassie blue staining.
Real-time PCR and quantitative chromatin immunoprecipitation (ChIP) assay.
The mRNA levels of FoxM1, p21cip, BRCA2, and XRCC1 were monitored by real-time PCR as described previously (41). The following sense (S) and antisense (AS) primer sequences and annealing temperature (Ta) were used for human mRNA: FoxM1-S, 5′-GGA GGA AAT GCC ACA CTT AGC G-3′; FoxM1-AS, 5′-TAG GAC TTC TTG GGT CTT GGG GTG-3′ (Ta, 55.7°C); p21cip-S, 5′-TTT GAT TAG CAG CGG AAC AAG G-3′; p21cip-AS, 5′-AAA GAC AAC TAC TCC CAG CCC C-3′ (Ta, 55.7°C); BRCA2-S, 5′-GCC TTG GAT TTC TTG AGT AGA CTG C-3′; BRCA2-AS, 5′-GTG TTT CGT ATT TGG TGC CAC AAC-3′ (Ta, 55.7°C); XRCC1-S, 5′-ACG GAT GAG AAC ACG GAC AGT G-3′; XRCC1-AS, 5′-CGT AAA GAA AGA AGT GCT TGC CC-3′ (Ta, 55.7°C). The following S and AS primer sequences and Ta were used for mouse mRNA: FoxM1-S, 5′-CAC TTG GAT TGA GGA CCA CTT-3′; FoxM1-AS, 5′-GTC GTT TCT GCT GTG ATT CC-3′ (Ta, 57.5°C); BRCA2-S, 5′-GTG GCA CGA AAT ACG CAA CAC-3′; BRCA2-AS, 5′-GCA AGG CAA GTT CTT CAT CAG C-3′ (Ta, 57.5°C); XRCC1-S, 5′-GCT CCA CAG ATG AGA ACA CAG ACA G-3′; XRCC1-AS, 5′-GAA CTC GCC ATA CAG GAA GAA GTG-3′ (Ta, 57.5°C). These real-time reverse transcription (RT)-PCR RNA levels were normalized to human cyclophilin or mouse cyclophilin (mcyclophilin) mRNA levels, and these primers are as follows: hcyclophilin-S, 5′-GCA GAC AAG GTC CCA AAG ACA G-3′; hcyclophilin-AS, 5′-CAC CCT GAC ACA TAA ACC CTG G-3′ (Ta, 55.7°C); mcyclophilin-S, 5′-GGC AAA TGC TGG ACC AAA CAC-3′; mcyclophilin-AS, 5′-TTC CTG GAC CCA AAA CGC TC-3′ (Ta, 57.5°C).
ChIP assays were used to measure FoxM1 binding on endogenous promoter regions of BRCA2, XRCC1, and Cdc25B as described previously (41). The primers used to amplify the following human gene promoter fragments are annotated with the binding position upstream of the transcription start site, Ta, and whether they are in the S or AS orientation: Cdc25B −92S, 5′-AAG AGC CCA TCA GTT CCG CTT G-3′; Cdc25B + 120AS, 5′-CCC ATT TTA CAG ACC TGG ACG C-3′ (Ta, 62°C); BRCA2 −1648S, 5′-CAT AAG GGG GCA GAA TAA GAG TTG-3′; BRCA2 −1545AS, 5′-TGA TAG AAG GTG GAA ATG AGG TCC-3′ (Ta, 54°C); XRCC1 −1055S, 5′-TGG GAA GGC AGA ACC AGA ATC-3′; XRCC1 −912AS, 5′-GGG AAG GTG AGA AAA CAG GGT G-3′ (Ta, 58.2°C). Normalization was carried out using the ΔΔCT method. Briefly, immunoprecipitation (IP) samples and total input threshold cycles (CT) for each treatment were subtracted from the CT of the corresponding serum control IP (rabbit serum). The resulting corrected value for the total input was then subtracted from the corrected experimental IP value (ΔΔCT), and these values were raised to the power of two (2ΔΔCT). These values were then expressed as relative promoter binding ± standard deviations.
Transfection of dominant-negative Chk2 and Chk2 siRNA.
U2OS cells were transfected with 10 μg of plasmid expressing V5-tagged mutant Chk2 by Fugene 6 (Roche) or with 100 nM Chk2 siRNA (33) by Lipofectamine 2000 (Invitrogen). Forty-eight hours after transfection, the transfected cells were exposed to 10 Gy γ-irradiation (IR), 20 μM etoposide, or 20 J m−2 UV. Whole-cell extracts were prepared 6 h after exposure by lysis in extraction buffer. The endogenous protein levels of FoxM1 or Chk2 and expression of V5-tagged Chk2d/n protein were determined by Western blotting with corresponding specific antibodies.
Endogenous FoxM1 Ser361 phosphorylation.
Cell extracts were made from U2OS cells treated with 10 Gy IR 3 h later or from nontreated cells. The lysates (500 μg) were immunoprecipitated with rabbit anti-FoxM1 serum (1 μl) as described previously (15, 41). Phosphorylation of endogenous FoxM1 on Ser 361 was assayed by Western blotting with anti-pS361 FoxM1 peptide antibody (1:200). The amounts of extracts used had similar FoxM1 levels, as shown by Western blotting with rabbit anti-FoxM1 serum.
Statistical analysis.
We used Microsoft Excel to calculate standard deviations and statistically significant differences between samples using the Student t test. The asterisks in each graph indicate statistically significant changes, with P values calculated by the Student t test as follows: *, P < 0.05; **, P ≤ 0.01; ***, P ≤ 0.001. P values of <0.05 were considered statistically significant.
RESULTS AND DISCUSSION
FoxM1-deficient cells exhibit DNA breaks and increased activation of the p53 tumor suppressor.
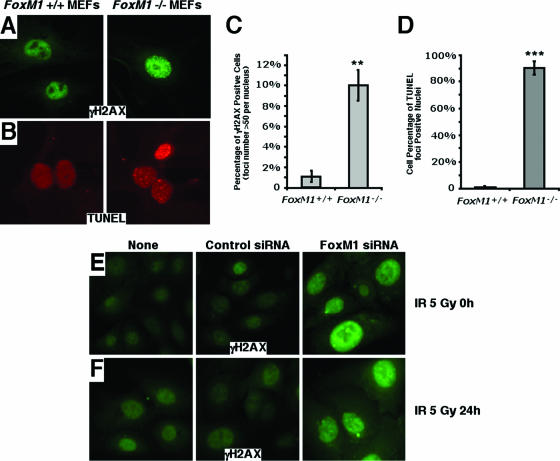
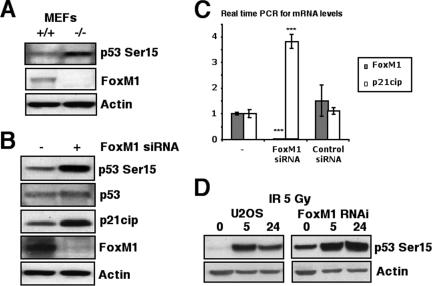
Growth of MEFs in culture, under normal oxygen concentrations, causes cell senescence through oxidative stress-induced DNA damage, which requires activation of the DNA repair pathways (3). Consistent with an important role of FoxM1 in DNA repair, we previously reported that early passage FoxM1-deficient (−/−) MEFs fail to proliferate in culture and undergo premature cellular senescence, as evidenced by increased expression of senescence-associated β-galactosidase gene, p19ARF, and p16Ink4A (41). To examine whether FoxM1-deficient cells are susceptible to DNA breaks, FoxM1−/− and wild type (+/+) MEFs were immunostained for phosphospecific histone H2AX foci (γH2AX), which is a known marker for the presence of DNA breaks (29). The FoxM1−/− MEFs exhibited high levels of spontaneous γH2AX focus formation compared to the wild type (+/+) MEFs (Fig. 1A and 1C). In addition, the FoxM1−/− MEFs displayed high levels of TUNEL foci (Fig. 1B and 1D), which efficiently end label sites of DNA breaks (38). The TUNEL focus pattern is distinct from that observed in apoptotic cells. Also, we have shown that the FoxM1−/− cells are refractory to apoptosis (9, 41). The presence of both TUNEL foci and γH2AX foci indicates a deficiency in DNA repair in the FoxM1−/− cells. Furthermore, osteosarcoma U2OS cells depleted in FoxM1 levels by siRNA transfection showed significant increases in γH2AX focus formation (Fig. 1E). The depletion of FoxM1 also increased cell sensitivity to the DNA-damaging agent γ irradiation (5 Gy) and delayed the DNA repair posttreatment, evidenced by higher levels of γH2AX foci in the FoxM1-deficient cells 24 h after γ irradiation (5 Gy)-induced DNA damage (Fig. 1F). The increased number of DNA breaks in FoxM1−/− MEFs and FoxM1-deficient U2OS cells was correlated with increased phosphorylation of the Ser 15 residue on the p53 protein (Fig. 2A and B), which is specifically phosphorylated by ATM, ATR, and DNA-PK in response to DNA damage and stimulates p53 transcriptional activity (31, 36). These results suggest that increased kinase activity of ATM, ATR, and DNA-PK were found in FoxM1-deficient cells. Consistent with increased p53 transcriptional activity, FoxM1-deficient cells displayed increased mRNA and protein levels of p21Cip1/Waf1 (Fig. 2B and C), which is a transcriptional downstream target of the p53 tumor suppressor protein (7, 10). Furthermore, U2OS cells depleted in FoxM1 levels by siRNA transfection exhibited persistent phosphorylation of the Ser 15 residue on the p53 protein at 24 h after γ irradiation (5 Gy)-induced DNA damage, whereas control cells showed diminished phosphorylation of p53 at Ser15 (Fig. 2D). These results suggest that diminished FoxM1 levels caused reduced DNA repair leading to increased DNA breaks with corresponding increases in p53 transcriptional activity and downstream target gene expression.
FIG. 1.
FoxM1-deficient cells exhibit increased DNA breaks. (A to D) To examine for DNA breaks, wild type (+/+) or FoxM1−/− (−/−) MEFs were immunostained for phosphospecific histone H2AX (γH2AX) foci or for TUNEL foci. FoxM1−/− MEFs exhibited high levels of spontaneous γH2AX foci (A and C) or TUNEL-positive foci (B and D) compared to wild-type MEFs. (E and F) Osteosarcoma U2OS cells depleted in FoxM1 levels by siRNA transfection exhibited high levels of spontaneous γH2AX foci. U2OS cells were transfected with FoxM1 siRNA or control siRNA or received no treatment, as indicated. Cells were then exposed to 5 Gy of IR and immunostained for γH2AX foci at 0 h (E) or 24 h (F) post-IR. The asterisks indicate statistically significant changes: **, P ≤ 0.01; ***, P ≤ 0.001.
FIG. 2.
FoxM1-deficient cells exhibit increased activation of the p53 tumor suppressor. (A and B) FoxM1-deficient cells exhibited increased phosphorylation of serine 15 of the p53 tumor suppressor protein. (A) Cell extracts were prepared from wild type (+/+) or FoxM1−/− (−/−) MEFs, and levels of p53 Ser15 phosphorylation (top), FoxM1 (middle), and β-actin (bottom) were analyzed by Western blotting. (B) Also, cell extracts prepared from FoxM1 siRNA-transfected U2OS cells (+) or control cells (−) were analyzed by Western blotting for levels of p53 Ser15 phosphorylation, p53, p21Cip1 (downstream target gene of p53), FoxM1, and β-actin. (C) Increased levels of p21Cip1 mRNA were found in FoxM1-deficient U2OS cells. U2OS cells were transfected with FoxM1 siRNA or control siRNA, and then 72 h later, RNA was prepared to examine for expression of p21Cip1 mRNA by qRT-PCR. The asterisks indicate statistically significant changes: ***, P ≤ 0.001. (D) FoxM1-deficient U2OS cells show increased phosphorylation of serine 15 of the p53 tumor suppressor protein in response to γ-irradiation. U2OS cells were transfected with or without FoxM1 siRNA and then exposed 5 Gy of IR. Cell extracts were prepared at 0 h, 5 h, or 24 h post-IR and analyzed by Western blotting for levels of p53 Ser15 phosphorylation (top) and β-actin (bottom).
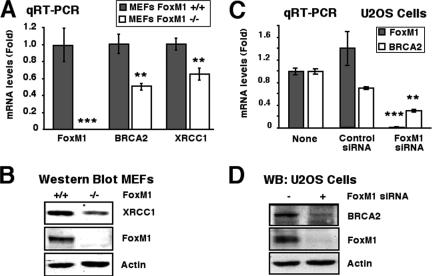
FoxM1-deficient cells exhibit reduced expression of the DNA repair genes XRCC1 and BRCA2.
We previously used microarray analysis to demonstrate that premature expression of FoxM1b during mouse liver regeneration caused earlier expression of the base excision repair factor XRCC1, suggesting a role for FoxM1 in regulating transcription of DNA repair genes. We also scanned −5-kb promoter regions of a number of DNA repair genes with the FoxM1 DNA binding consensus sequence (46), and several tandem FoxM1 putative binding sites (−1,171 to −1,148 bp) were found in the promoter of BRCA2, which is involved in homologous recombination repair of DNA double-strand breaks (16, 37, 47). We showed that the increased DNA breaks in FoxM1−/− MEFs correlated with decreased expression of both XRCC1 and BRCA2 genes (Fig. 3A and B). Furthermore, U2OS cells depleted in FoxM1 levels show significant decreases in expression of BRCA2 mRNA and protein (Fig. 3C and D). These results suggest that FoxM1 is critical for transcription of these DNA repair genes.
FIG. 3.
FoxM1-deficient cells exhibit reduced expression of the DNA repair genes XRCC1 and BRCA2. (A) FoxM1−/− MEFs display reduced mRNA levels of BRCA2 and XRCC1 DNA repair genes, as determined by qRT-PCR. (B) FoxM1−/− MEFs display reduced XRCC1 protein, as determined by Western blot analysis. (C and D) FoxM1-depleted U2OS cells exhibit reduced expression of BRCA2 mRNA and protein. U2OS cells were transfected with FoxM1 siRNA or control siRNA, and then 72 h later, protein and RNA were prepared to examine for expression of BRCA2 mRNA by qRT-PCR and BRCA2 protein by Western blot analysis. The asterisks indicate statistically significant changes: **, P ≤ 0.01; ***, P ≤ 0.001.
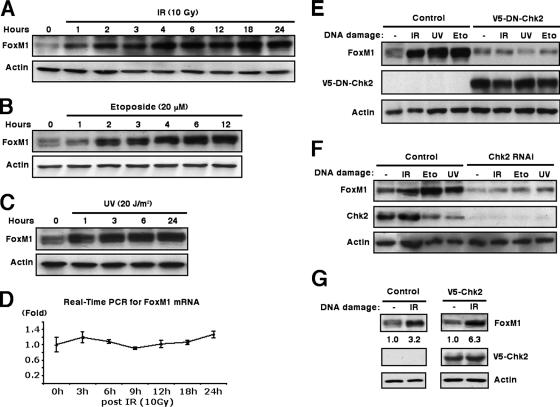
Phosphorylation of FoxM1 protein by Chk2 stabilizes FoxM1 protein in response to DNA damage.
To determine the mechanism of activation of FoxM1 in response to DNA damage, we treated human U2OS cells with DNA-damaging agents, IR, or UV irradiation and monitored the levels of FoxM1 protein by Western blot analysis. All of these treatments caused a gradual increase in levels of FoxM1 protein (Fig. 4A, B, and C) without affecting FoxM1 mRNA expression (Fig. 4D and data not shown). These results suggest that the DNA damage-signaling pathway stabilizes the FoxM1 protein. Chk2 has previously been shown to phosphorylate and stimulate transcriptional activity of the p53 (4, 11, 30) and E2F1 (34) proteins. Given that FoxM1 was induced with kinetics similar to those of E2F1 and p53 in response to DNA damage, we reasoned that FoxM1 protein levels might be regulated by Chk2 phosphorylation. To examine this hypothesis, we transfected a dominant-negative Chk2 mutant (4) to inhibit Chk2 function and determine whether endogenous FoxM1 protein is still induced in response to DNA damage. The Chk2 dominant-negative mutant blocked induction of the FoxM1 protein in response to all three types of DNA-damaging agents (Fig. 4E), suggesting that Chk2 is involved in phosphorylation and stabilization of FoxM1 protein. To confirm this result, we depleted U2OS cells of Chk2 levels by siRNA transfection (Fig. 4F) and showed that expression of the FoxM1 protein was not induced by IR, etoposide, or UV in Chk2-depleted U2OS cells (Fig. 4F). This was in marked contrast to the clear induction of FoxM1 protein levels in control experiments (Fig. 4F). Transfection of U2OS cells with the V5-tagged wild-type Chk2 expression vector further stimulated the relative induction of FoxM1 protein induced by IR treatment (Fig. 4G, from 3.2-fold in control to 6.3-fold with V5-Chk2). Despite the possibility of protein nuclear export mediated by Chk2 phosphorylation (32), the FoxM1 protein remained nuclear at all time points (hours) following IR treatment (data not shown). Taken together, these results suggest a physiological role for Chk2 in regulating increased levels of FoxM1 protein in response to DNA damage.
FIG. 4.
FoxM1 protein is stabilized after DNA damage and requires Chk2 kinase. (A to C) FoxM1 protein is stabilized in response to IR, etoposide, or UV-induced DNA damage. Extracts were prepared from U2OS cells at the indicated time points following exposure to 10 Gy of IR (A), 20 μM etoposide (B), or 20 J/m2 of UV light (UV) (C) and were immunoblotted for endogenous FoxM1 or β-actin protein levels. (D) No change was observed in FoxM1 mRNA levels from U2OS cells exposed to 10 Gy IR as determined by real-time quantitative RT-PCR. (E) Transfection of dominant-negative Chk2 expression vector in U2OS cells inhibits stabilization of FoxM1 protein in response to DNA damage. U2OS cells were transfected with V5-tagged dominant-negative Chk2 (V5-DN-Chk2) expression vector or vector alone, as indicated. Cells were then exposed to IR (10 Gy), UV (20 J/m2), or etoposide (20 μM), and cell extracts were prepared 6 h later. Levels of endogenous FoxM1 (top), V5-tagged dominant-negative Chk2 (middle), and β-actin (bottom) were analyzed by Western blot analysis. (F) Depletion of Chk2 levels in U2OS cells by siRNA transfection inhibits stabilization of FoxM1 protein in response to DNA damage. U2OS cells were transfected with Chk2 siRNA (100 nM) and then exposed to IR (10 Gy), etoposide (20 μM), or UV (20 J/m2), and cell extracts were prepared 6 h later. Levels of endogenous FoxM1 (top), Chk2 (middle), and β-actin (bottom) were analyzed by Western blot analysis. (G) Increased expression of Chk2 protein increases stabilization of FoxM1 protein in response to IR. U2OS cells were transfected with V5-tagged Chk2 expression vector and then exposed to IR (10 Gy). Cell extracts were prepared 6 h later. Levels of endogenous FoxM1 (top), V5-tagged Chk2 (middle), and β-actin (bottom) were analyzed by immunoblotting.
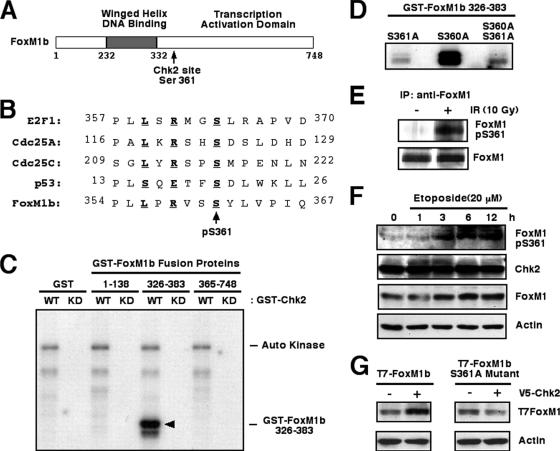
Comparison of the Chk2 phosphorylation sequences from several known target proteins (8, 20, 30, 34) allowed development of a Chk1/Chk2 phosphorylation consensus sequence (27) (Fig. 5B). A Chk2 consensus phosphorylation site centered on human FoxM1 Ser 361, which is completely conserved between mouse and human FoxM1 proteins (Fig. 5A and B) and more closely resembles the Chk2 phosphorylation sites found in Cdc25 and E2F1 proteins than that found in the p53 protein (Fig. 6B). To explore the importance of this potential Chk2 phosphorylation site in the FoxM1b protein at Ser361, we assessed whether this protein region functions as a substrate for Chk2 phosphorylation using an in vitro kinase assay. First, we prepared three GST-FoxM1b fusion proteins containing aa 1 to 138, 326 to 383, or 365 to 748 from the FoxM1b protein and found that recombinant Chk2 protein efficiently phosphorylated only the GST-FoxM1b aa 326 to 383 fusion protein in vitro, which contains the potential Chk2 phosphorylation site at Ser 361 (Fig. 5C). Site-directed mutagenesis that converted the Ser 361 residue to an Ala residue (S361A) in the GST-FoxM1b aa 326 to 383 fusion protein prevented its phosphorylation by Chk2 in vitro (Fig. 5D). In contrast, mutation of the adjacent Ser 360 to an Ala residue (S360A) in the GST-FoxM1b aa 326 to 383 fusion protein did not affect its phosphorylation by Chk2 (Fig. 5D), demonstrating that FoxM1b Ser361 residue is phosphorylated by recombinant Chk2 protein in vitro. To establish whether the FoxM1 Ser 361 residue is phosphorylated by Chk2 in vivo, we prepared an anti-phosphospecific FoxM1 peptide antibody directed against the FoxM1b sequence situated around the phosphorylated Ser 361 residue (Fig. 5E, FoxM1 pS361). We used affinity-purified anti-pS361 FoxM1 peptide antibody to determine that Chk2 phosphorylated the Ser 361 residue of the endogenous FoxM1 protein in response to IR-induced DNA damage compared to untreated cells (Fig. 5E). We next determined that Chk2-mediated phosphorylation of endogenous FoxM1 protein correlated with stabilization of FoxM1 protein. We used Western blot analysis with antibodies specific to either FoxM1 or FoxM1 pS361 peptide antibody at different time points following etoposide treatment of U2OS cells. Under these experimental conditions, FoxM1 protein levels began to increase at 3 h after treatment (Fig. 5F), and this correlated with detectable phosphorylation of FoxM1 protein at Ser residue 361, as determined by Western blot analysis (Fig. 5F, top). Furthermore, transfection of V5-Chk2 expression vector stimulated levels of the T7-tagged FoxM1 protein, whereas Chk2 was unable to influence levels of the FoxM1 S361A mutant protein that was disrupted in the Chk2 phosphorylation site (Fig. 5G). These results suggest that stabilization of endogenous FoxM1 protein correlates with Chk2-mediated phosphorylation of FoxM1 Ser residue 361 in response to DNA damage in vivo.
FIG. 5.
Chk2 phosphorylates FoxM1 on Ser 361 in vitro and in vivo. (A) Schematic representation of the FoxM1 transcription factor. The location of the Chk2 phosphorylation site is indicated between the winged helix DNA binding domain and the transcription activation domain. (B) Comparison of the Chk2 phosphorylation sites in E2F1, Cdc25A, Cdc25C, and p53 protein sequences with the potential Chk2 phosphorylation site in the FoxM1 protein. Residues in boldface type fit the consensus phosphorylation motif, and the arrow indicates the phosphorylated serine residue. (C) The GST-FoxM1b aa 326 to 383 fusion protein is phosphorylated by recombinant GST-Chk2 fusion protein. Recombinant GST-Chk2 (WT) or the GST-Chk2 KD form (KD) (1 μg) was incubated with [32P]ATP in the presence of 2 μg recombinant GST protein or GST-FoxM1b fusion proteins aa 1 to 138, aa 326 to 383, and aa 365 to 748, respectively. Reaction products were resolved on denaturing SDS-polyacrylamide gels and visualized by autoradiography. (D) GST-FoxM1 S361A mutant fusion protein is no longer phosphorylated by recombinant GST-Chk2 fusion protein. A Ser-to-Ala mutation was generated in GST-FoxM1 aa 326 to 383 fusion protein on Ser 361, Ser 360, or both. The mutated GST-FoxM1 aa 326 to 383 recombinant proteins (2 μg) were incubated with [γ-32P]ATP in the presence of recombinant GST-Chk2 (1 μg). Reaction products were resolved on denaturing SDS-polyacrylamide gels and visualized by autoradiography. (E) FoxM1 protein is phosphorylated at Ser residue 361 following IR-induced DNA damage. U2OS cells were left untreated or exposed to IR (10 Gy). Cell extracts were prepared 3 h later and immunoprecipitated with anti-FoxM1 antibody and protein A bead incubation. Phosphorylation of endogenous FoxM1 on Ser 361 was monitored by Western blot analysis with the anti-pS361 FoxM1 peptide antibody (top). The level of immunoprecipitated FoxM1 (bottom) is also shown. (F) The Chk2 protein in vivo phosphorylates FoxM1 protein 3 h following etoposide treatment. Extracts from U2OS cells exposed to 20 μM etoposide were harvested at the indicated hours after treatment and immunoblotted with the anti-pS361 FoxM1 peptide antibody. Levels of endogenous Chk2, FoxM1, and β-actin were also analyzed by Western blot analysis. The modest change in the signal for FoxM1 was a result of uneven loading (compare the β-actin signals). (G) Mutation of the Chk2 phosphorylation site in FoxM1 protein at serine residue 361 prevents stabilization of FoxM1 protein in response to increased expression of Chk2 protein. U2OS cells were transfected with expression vectors containing either T7-tagged WT FoxM1 protein or FoxM1 S361A mutant protein with or without the V5-Chk2 expression vector. Protein extracts were prepared 24 h later and examined for FoxM1 protein levels by Western blot analysis using the T7-tagged antibody.
FIG. 6.
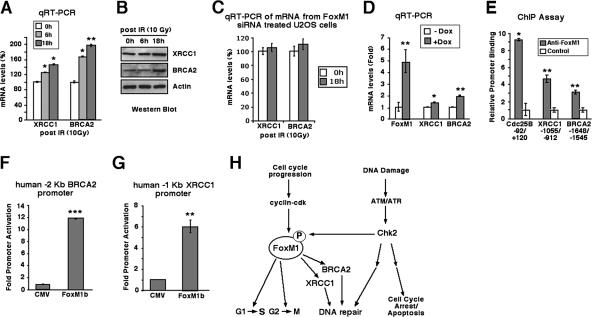
FoxM1 stimulates transcription of the DNA repair genes XRCC1 and BRCA2. (A and B) Expression of BRCA2 and XRCC1 mRNA is increased in response to IR DNA damage. U2OS cells were left untreated or exposed to IR (10 Gy), and then RNA was harvested at 6 or 18 h after treatment and analyzed for BRCA2 and XRCC1 mRNA levels by qRT-PCR (A) and for BRCA2 and XRCC1 protein levels by Western blotting (B). (C) The induction of XRCC1 and BRCA2 mRNAs following DNA damage depends on FoxM1. U2OS cells were transfected with FoxM1 siRNA and left untreated or exposed to IR (10 Gy), and then RNA was harvested at 18 h after treatment and analyzed for BRCA2 and XRCC1 mRNA levels by qRT-PCR. (D) Conditional expression of FoxM1b protein in U2OS cells stimulates BRCA2 and XRCC1 mRNA levels. U2OS C3 cells were induced for GFP-FoxM1b expression by Dox treatment, and then RNA was isolated 48 h later and analyzed for FoxM1, BRCA2, and XRCC1 mRNA levels by qRT-PCR. (E) ChIP assays show direct binding of FoxM1 to the endogenous human Cdc25B, XRCC1, and BRCA2 promoter regions. The U2OS cell chromatin was cross-linked, sonicated to DNA fragments of 500 to 1,000 nucleotides in length, and then IP with antibodies specific to either FoxM1 or rabbit serum (control), and the amount of promoter DNA associated with the IP chromatin was quantitated by qRT-PCR with primers specific to the human Cdc25B, XRCC1, and BRCA2 promoter regions. (F and G) Transcription of the human XRCC1 and BRCA2 promoter regions is stimulated in cotransfection assays with FoxM1b expression vector. We performed cotransfection assays with the CMV-FoxM1b expression vector and luciferase plasmids containing either −2 kb of the human BRCA2 promoter region or −1 kb of the human XRCC1 promoter region, prepared protein extracts from U2OS cells at 24 h following transfection, and used them to measure dual luciferase enzyme activity. The asterisks indicate statistically significant changes: *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001. (H) Model of DNA damage induction of FoxM1 protein. The FoxM1 transcription factor regulates expression of cell cycle genes essential for progression into DNA replication and mitosis. In response to DNA damage signaling, we show that Chk2 phosphorylates and stabilizes FoxM1 protein and increased levels of FoxM1 activate transcription of the DNA repair genes XRCC1 and BRCA2. DNA damage signaling also induces DNA repair enzymes and cell cycle arrest independent of the FoxM1 transcription factor.
FoxM1 stimulates transcription of the DNA repair genes XRCC1 and BRCA2.
We showed that FoxM1-deficient cells displayed increased DNA breaks and reduced expression of XRCC1 and BRCA2 (Fig. 1 and 3), suggesting that FoxM1 plays a critical role in regulating transcription of these DNA repair genes. Increased levels of FoxM1 protein were also found in response to DNA damage (Fig. 4). Consistent with these findings, IR-treated U2OS cells displayed increased expression of XRCC1 and BRCA2 both on mRNA and protein levels at 6 and 18 h after treatment (Fig. 6A and B). The induction of XRCC1 and BRCA2 mRNAs observed after ionizing radiation depended on FoxM1 because siRNA-mediated depletion of FoxM1 in the U2OS cells prevented the increased expression of XRCC1 and BRCA2 after ionizing radiation (Fig. 6C). Furthermore, Dox induction of GFP-FoxM1b fusion protein in U2OS C3 cells (15) demonstrated that increased FoxM1b levels stimulated expression of endogenous XRCC1 and BRCA2 genes, as determined by quantitative real-time RT-PCR (qRT-PCR) (Fig. 6D).
We next used quantitative ChIP assays (41) to determine whether FoxM1 bound to the endogenous human XRCC1 and BRCA2 promoter regions. The U2OS cell chromatin was cross-linked, sonicated to DNA fragments of 500 to 1,000 nucleotides in length, and then IP with antibodies specific to either FoxM1 or rabbit serum (control), and the amount of promoter DNA associated with the IP chromatin was quantitated by qRT-PCR with primers specific to the human XRCC1 and BRCA2 promoter regions. These ChIP PCR primers were made to DNA sequences situated near several tandem potential FoxM1 binding sites in the human BRCA2 (bp −1171 to −1148) and XRCC1 (bp −900 to −893) promoter regions. This quantitative ChIP assay with FoxM1 antibody showed that the FoxM1 protein binds to the endogenous human XRCC1 and BRCA2 promoter regions compared to the rabbit serum control (Fig. 6E). As a positive control, we performed ChIP assays with the human Cdc25B promoter region (Fig. 6E), a known FoxM1 transcriptional target gene (41). We also performed control ChIP assays with cross-linked extracts prepared from U2OS cells and FoxM1 antibody or control mouse IgG serum, and the IP genomic DNA was analyzed for the presence of the liver-specific human transthyretin (TTR) promoter region by real-time PCR. Consistent with the specificity of our ChIP assays, this control ChIP experiment demonstrated that neither the FoxM1 antibody nor IgG serum immunoprecipitated significant levels of this proximal TTR promoter region from either untransfected or FoxM1-depleted U2OS cell extracts (data not shown). To determine whether FoxM1 regulates transcription of the human XRCC1 and BRCA2 genes, the luciferase reporter gene was linked to either the −2 kb of the human BRCA2 promoter region or −1 kb of the human XRCC1 promoter region. We performed cotransfection assays with the cytomegalovirus (CMV)-FoxM1b expression vector and these human promoter luciferase plasmids, prepared protein extracts from U2OS cells at 24 h following transfection, and used them to measure dual luciferase enzyme activity. Cotransfection of FoxM1b expression vector caused a 12-fold increase in BRCA2 promoter activity (Fig. 6F) and a 6-fold induction of XRCC1 promoter expression (Fig. 6G). These results demonstrate that the XRCC1 and BRCA2 genes are direct transcriptional targets of the FoxM1 protein.
FoxM1 protein is critical for regulated transcription of DNA repair genes in response to Chk2 signaling.
In the current study, we showed that FoxM1 protein is phosphorylated by Chk2, causing increased stability of the FoxM1 protein in response to DNA damage (Fig. 6H). Stabilization of FoxM1 protein in response to DNA damage correlates with the increased expression of DNA repair genes XRCC1 and BRCA2 (Fig. 6G), which we demonstrated by cotransfection and ChIP assays as direct transcriptional targets of FoxM1. We demonstrated that DNA damage signaling increased FoxM1 protein stability, which in turn stimulates transcription of genes encoding DNA repair enzymes (Fig. 6H). These results identify a novel role for FoxM1 in the transcriptional regulation of DNA repair genes. Furthermore, the FoxM1 transcription factor is overexpressed in a number of aggressive human tumors (19, 25, 28, 35, 39, 44). In published studies, reduced expression of FoxM1 significantly diminished DNA replication and mitosis of tumor cells and reduced development of mouse tumors in response to carcinogens or oncogenes (14, 15, 17, 41). Our current study suggests that inhibition of FoxM1 levels will also lead to defective DNA repair and that this defect may result in cell cycle arrest of tumor cells (41).
Reduced cellular proliferation during aging is associated with a progressive decline in FoxM1 expression (21, 43). In published studies, we used the −3 kb TTR promoter to maintain hepatocyte expression of the FoxM1b transgene in 12-month-old (old-aged) transgenic (TG) mice during liver regeneration (43). We showed that maintaining hepatocyte expression of FoxM1b alone in old-aged TTR-FoxM1b TG mice is sufficient to restore regenerating hepatocyte DNA synthesis and mitosis and expression of cell cycle genes to levels found in young regenerating mouse liver2 (43). Based on our current studies, reduction of FoxM1 levels during aging may also cause defects in DNA repair, which will lead to increases in the incidence of cancer during aging resulting from gene mutations and chromosome abnormalities.
Acknowledgments
We thank the members of Robert Costa's laboratory for critically reviewing the manuscript and S. Elledge for providing us with the pGST-Chk2(WT) and pGST-Chk2(D347A)(KD) fusion protein expression plasmids.
This work was supported by U.S. Public Health Service grants DK 54687 from NIDDK and RO1 AG 21842 from NIA (to R.H.C.) and by the “985” Project of the China Education Ministry (to Y.T.). P.R. is supported by grants from the NIH (CA 77637 and CA 100035).
Footnotes
Published ahead of print on 13 November 2006.
We dedicate this work to the memory of Robert H. Costa.
REFERENCES
- 1.Abraham, R. T. 2001. Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes Dev. 15:2177-2196. [DOI] [PubMed] [Google Scholar]
- 2.Bartek, J., and J. Lukas. 2003. Chk1 and Chk2 kinases in checkpoint control and cancer. Cancer Cell 3:421-429. [DOI] [PubMed] [Google Scholar]
- 3.Busuttil, R. A., M. Rubio, M. E. Dolle, J. Campisi, and J. Vijg. 2003. Oxygen accelerates the accumulation of mutations during the senescence and immortalization of murine cells in culture. Aging Cell 2:287-294. [DOI] [PubMed] [Google Scholar]
- 4.Chehab, N. H., A. Malikzay, M. Appel, and T. D. Halazonetis. 2000. Chk2/hCds1 functions as a DNA damage checkpoint in G(1) by stabilizing p53. Genes Dev. 14:278-288. [PMC free article] [PubMed] [Google Scholar]
- 5.Clark, K. L., E. D. Halay, E. Lai, and S. K. Burley. 1993. Co-crystal structure of the HNF-3/fork head DNA-recognition motif resembles histone H5. Nature 364:412-420. [DOI] [PubMed] [Google Scholar]
- 6.Costa, R. H., V. V. Kalinichenko, A. X. Holterman, and X. Wang. 2003. Transcription factors in liver development, differentiation, and regeneration. Hepatology 38:1331-1347. [DOI] [PubMed] [Google Scholar]
- 7.el-Deiry, W. S., T. Tokino, V. E. Velculescu, D. B. Levy, R. Parsons, J. M. Trent, D. Lin, W. E. Mercer, K. W. Kinzler, and B. Vogelstein. 1993. WAF1, a potential mediator of p53 tumor suppression. Cell 75:817-825. [DOI] [PubMed] [Google Scholar]
- 8.Falck, J., N. Mailand, R. G. Syljuasen, J. Bartek, and J. Lukas. 2001. The ATM-Chk2-Cdc25A checkpoint pathway guards against radioresistant DNA synthesis. Nature 410:842-847. [DOI] [PubMed] [Google Scholar]
- 9.Gusarova, G. A., I.-C. Wang, M. L. Majors, V. V. Kalinichenko, T. Ackerson, V. Petrovic, and R. H. Costa. A cell penetrating ARF peptide inhibitor of FoxM1 in mouse hepatocellular carinoma treatment. J. Clin. Investig., in press. [DOI] [PMC free article] [PubMed]
- 10.Harper, J. W., G. R. Adami, N. Wei, K. Keyomarsi, and S. J. Elledge. 1993. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell 75:805-816. [DOI] [PubMed] [Google Scholar]
- 11.Hirao, A., Y. Y. Kong, S. Matsuoka, A. Wakeham, J. Ruland, H. Yoshida, D. Liu, S. J. Elledge, and T. W. Mak. 2000. DNA damage-induced activation of p53 by the checkpoint kinase Chk2. Science 287:1824-1827. [DOI] [PubMed] [Google Scholar]
- 12.Hoeijmakers, J. H. 2001. Genome maintenance mechanisms for preventing cancer. Nature 411:366-374. [DOI] [PubMed] [Google Scholar]
- 13.Kaestner, K. H., W. Knochel, and D. E. Martinez. 2000. Unified nomenclature for the winged helix/forkhead transcription factors. Genes Dev. 14:142-146. [PubMed] [Google Scholar]
- 14.Kalin, T. V., I. C. Wang, T. J. Ackerson, M. L. Major, C. J. Detrisac, V. V. Kalinichenko, A. Lyubimov, and R. H. Costa. 2006. Increased levels of the FoxM1 transcription factor accelerate development and progression of prostate carcinomas in both TRAMP and LADY transgenic mice. Cancer Res. 66:1712-1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kalinichenko, V. V., M. Major, X. Wang, V. Petrovic, J. Kuechle, H. M. Yoder, B. Shin, A. Datta, P. Raychaudhuri, and R. H. Costa. 2004. Forkhead Box m1b transcription factor is essential for development of hepatocellular carcinomas and is negatively regulated by the p19ARF tumor suppressor. Genes Dev. 18:830-850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karran, P. 2000. DNA double strand break repair in mammalian cells. Curr. Opin. Genet. Dev. 10:144-150. [DOI] [PubMed] [Google Scholar]
- 17.Kim, I. M., T. Ackerson, S. Ramakrishna, M. Tretiakova, I. C. Wang, T. V. Kalin, M. L. Major, G. A. Gusarova, H. M. Yoder, R. H. Costa, and V. V. Kalinichenko. 2006. The Forkhead Box m1 transcription factor stimulates the proliferation of tumor cells during development of lung cancer. Cancer Res. 66:2153-2161. [DOI] [PubMed] [Google Scholar]
- 18.Krupczak-Hollis, K., X. Wang, V. V. Kalinichenko, G. A. Gusarova, I.-C. Wang, M. B. Dennewitz, H. M. Yoder, H. Kiyokawa, K. H. Kaestner, and R. H. Costa. 2004. The mouse forkhead box m1 transcription factor is essential for hepatoblast mitosis and development of intrahepatic bile ducts and vessels during liver morphogenesis. Dev. Biol. 276:74-88. [DOI] [PubMed] [Google Scholar]
- 19.Lee, J. S., I. S. Chu, J. Heo, D. F. Calvisi, Z. Sun, T. Roskams, A. Durnez, A. J. Demetris, and S. S. Thorgeirsson. 2004. Classification and prediction of survival in hepatocellular carcinoma by gene expression profiling. Hepatology 40:667-676. [DOI] [PubMed] [Google Scholar]
- 20.Lee, J. S., K. M. Collins, A. L. Brown, C. H. Lee, and J. H. Chung. 2000. hCds1-mediated phosphorylation of BRCA1 regulates the DNA damage response. Nature 404:201-204. [DOI] [PubMed] [Google Scholar]
- 21.Ly, D. H., D. J. Lockhart, R. A. Lerner, and P. G. Schultz. 2000. Mitotic misregulation and human aging. Science 287:2486-2492. [DOI] [PubMed] [Google Scholar]
- 22.Major, M. L., R. Lepe, and R. H. Costa. 2004. Forkhead box M1B (FoxM1B) transcriptional activity requires binding of Cdk/cyclin complexes for phosphorylation-dependent recruitment of p300/CBP coactivators. Mol. Cell. Biol. 24:2649-2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marintchev, A., A. Robertson, E. K. Dimitriadis, R. Prasad, S. H. Wilson, and G. P. Mullen. 2000. Domain specific interaction in the XRCC1-DNA polymerase beta complex. Nucleic Acids Res. 28:2049-2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matsuoka, S., G. Rotman, A. Ogawa, Y. Shiloh, K. Tamai, and S. J. Elledge. 2000. Ataxia telangiectasia-mutated phosphorylates Chk2 in vivo and in vitro. Proc. Natl. Acad. Sci. USA 97:10389-10394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Obama, K., K. Ura, M. Li, T. Katagiri, T. Tsunoda, A. Nomura, S. Satoh, Y. Nakamura, and Y. Furukawa. 2005. Genome-wide analysis of gene expression in human intrahepatic cholangiocarcinoma. Hepatology 41:1339-1348. [DOI] [PubMed] [Google Scholar]
- 26.Olive, P. L. 1998. The role of DNA single- and double-strand breaks in cell killing by ionizing radiation. Radiat. Res. 150:S42-S51. [PubMed] [Google Scholar]
- 27.O'Neill, T., L. Giarratani, P. Chen, L. Iyer, C. H. Lee, M. Bobiak, F. Kanai, B. B. Zhou, J. H. Chung, and G. A. Rathbun. 2002. Determination of substrate motifs for human Chk1 and hCds1/Chk2 by the oriented peptide library approach. J. Biol. Chem. 277:16102-16115. [DOI] [PubMed] [Google Scholar]
- 28.Pilarsky, C., M. Wenzig, T. Specht, H. D. Saeger, and R. Grutzmann. 2004. Identification and validation of commonly overexpressed genes in solid tumors by comparison of microarray data. Neoplasia 6:744-750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rogakou, E. P., D. R. Pilch, A. H. Orr, V. S. Ivanova, and W. M. Bonner. 1998. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 273:5858-5868. [DOI] [PubMed] [Google Scholar]
- 30.Shieh, S. Y., J. Ahn, K. Tamai, Y. Taya, and C. Prives. 2000. The human homologs of checkpoint kinases Chk1 and Cds1 (Chk2) phosphorylate p53 at multiple DNA damage-inducible sites. Genes Dev. 14:289-300. [PMC free article] [PubMed] [Google Scholar]
- 31.Shieh, S. Y., M. Ikeda, Y. Taya, and C. Prives. 1997. DNA damage-induced phosphorylation of p53 alleviates inhibition by MDM2. Cell 91:325-334. [DOI] [PubMed] [Google Scholar]
- 32.Singh, S. V., A. Herman-Antosiewicz, A. V. Singh, K. L. Lew, S. K. Srivastava, R. Kamath, K. D. Brown, L. Zhang, and R. Baskaran. 2004. Sulforaphane-induced G2/M phase cell cycle arrest involves checkpoint kinase 2-mediated phosphorylation of cell division cycle 25C. J. Biol. Chem. 279:25813-25822. [DOI] [PubMed] [Google Scholar]
- 33.Sorensen, C. S., R. G. Syljuasen, J. Falck, T. Schroeder, L. Ronnstrand, K. K. Khanna, B. B. Zhou, J. Bartek, and J. Lukas. 2003. Chk1 regulates the S phase checkpoint by coupling the physiological turnover and ionizing radiation-induced accelerated proteolysis of Cdc25A. Cancer Cell 3:247-258. [DOI] [PubMed] [Google Scholar]
- 34.Stevens, C., L. Smith, and N. B. La Thangue. 2003. Chk2 activates E2F-1 in response to DNA damage. Nat. Cell Biol. 5:401-409. [DOI] [PubMed] [Google Scholar]
- 35.Teh, M. T., S. T. Wong, G. W. Neill, L. R. Ghali, M. P. Philpott, and A. G. Quinn. 2002. FOXM1 is a downstream target of Gli1 in basal cell carcinomas. Cancer Res. 62:4773-4780. [PubMed] [Google Scholar]
- 36.Tibbetts, R. S., K. M. Brumbaugh, J. M. Williams, J. N. Sarkaria, W. A. Cliby, S. Y. Shieh, Y. Taya, C. Prives, and R. T. Abraham. 1999. A role for ATR in the DNA damage-induced phosphorylation of p53. Genes Dev. 13:152-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tutt, A., D. Bertwistle, J. Valentine, A. Gabriel, S. Swift, G. Ross, C. Griffin, J. Thacker, and A. Ashworth. 2001. Mutation in Brca2 stimulates error-prone homology-directed repair of DNA double-strand breaks occurring between repeated sequences. EMBO J. 20:4704-4716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vafa, O., M. Wade, S. Kern, M. Beeche, T. K. Pandita, G. M. Hampton, and G. M. Wahl. 2002. c-Myc can induce DNA damage, increase reactive oxygen species, and mitigate p53 function: a mechanism for oncogene-induced genetic instability. Mol. Cell 9:1031-1044. [DOI] [PubMed] [Google Scholar]
- 39.van den Boom, J., M. Wolter, R. Kuick, D. E. Misek, A. S. Youkilis, D. S. Wechsler, C. Sommer, G. Reifenberger, and S. M. Hanash. 2003. Characterization of gene expression profiles associated with glioma progression using oligonucleotide-based microarray analysis and real-time reverse transcription-polymerase chain reaction. Am. J. Pathol. 163:1033-1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van Gent, D. C., J. H. Hoeijmakers, and R. Kanaar. 2001. Chromosomal stability and the DNA double-stranded break connection. Nat. Rev. Genet. 2:196-206. [DOI] [PubMed] [Google Scholar]
- 41.Wang, I.-C., Y.-J. Chen, D. E. Hughes, V. Petrovic, M. L. Major, H. J. Park, Y. Tan, T. Ackerson, and R. H. Costa. 2005. Forkhead box M1 regulates the transcriptional network of genes essential for mitotic progression and genes encoding the SCF (Skp2-Cks1) ubiquitin ligase. Mol. Cell. Biol. 25:10875-10894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang, X., H. Kiyokawa, M. B. Dennewitz, and R. H. Costa. 2002. The forkhead box m1b transcription factor is essential for hepatocyte DNA replication and mitosis during mouse liver regeneration. Proc. Natl. Acad. Sci. USA 99:16881-16886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang, X., E. Quail, N.-J. Hung, Y. Tan, H. Ye, and R. H. Costa. 2001. Increased levels of forkhead box M1B transcription factor in transgenic mouse hepatocytes prevents age-related proliferation defects in regenerating liver. Proc. Natl. Acad. Sci. USA 98:11468-11473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wonsey, D. R., and M. T. Follettie. 2005. Loss of the forkhead transcription factor FoxM1 causes centrosome amplification and mitotic catastrophe. Cancer Res. 65:5181-5189. [DOI] [PubMed] [Google Scholar]
- 45.Ye, H., A. Holterman, K. W. Yoo, R. R. Franks, and R. H. Costa. 1999. Premature expression of the winged helix transcription factor HFH-11B in regenerating mouse liver accelerates hepatocyte entry into S phase. Mol. Cell. Biol. 19:8570-8580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ye, H., T. F. Kelly, U. Samadani, L. Lim, S. Rubio, D. G. Overdier, K. A. Roebuck, and R. H. Costa. 1997. Hepatocyte nuclear factor 3/fork head homolog 11 is expressed in proliferating epithelial and mesenchymal cells of embryonic and adult tissues. Mol. Cell. Biol. 17:1626-1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yuan, S. S., S. Y. Lee, G. Chen, M. Song, G. E. Tomlinson, and E. Y. Lee. 1999. BRCA2 is required for ionizing radiation-induced assembly of Rad51 complex in vivo. Cancer Res. 59:3547-3551. [PubMed] [Google Scholar]
- 48.Zhou, B. B., and S. J. Elledge. 2000. The DNA damage response: putting checkpoints in perspective. Nature 408:433-439. [DOI] [PubMed] [Google Scholar]