Abstract
Hypersuppressiveness, as observed in Saccharomyces cerevisiae, is an extremely biased inheritance of a small mitochondrial DNA (mtDNA) fragment that contains a replication origin (HS [rho−] mtDNA). Our previous studies showed that concatemers (linear head-to-tail multimers) are obligatory intermediates for mtDNA partitioning and are primarily formed by rolling-circle replication mediated by Mhr1, a protein required for homologous mtDNA recombination. In this study, we found that Mhr1 is required for the hypersuppressiveness of HS [ori5] [rho−] mtDNA harboring ori5, one of the replication origins of normal ([rho+]) mtDNA. In addition, we detected an Ntg1-stimulated double-strand break at the ori5 locus. Purified Ntg1, a base excision repair enzyme, introduced a double-stranded break by itself into HS [ori5] [rho−] mtDNA at ori5 isolated from yeast cells. Both hypersuppressiveness and concatemer formation of HS [ori5] [rho−] mtDNA are simultaneously suppressed by the ntg1 null mutation. These results support a model in which, like homologous recombination, rolling-circle HS [ori5] [rho−] mtDNA replication is initiated by double-stranded breakage in ori5, followed by Mhr1-mediated homologous pairing of the processed nascent DNA ends with circular mtDNA. The hypersuppressiveness of HS [ori5] [rho−] mtDNA depends on a replication advantage furnished by the higher density of ori5 sequences and on a segregation advantage furnished by the higher genome copy number on transmitted concatemers.
Eukaryotic cells gain most of the energy required for cellular functions by oxidative respiration in mitochondria. These organelles contain hundreds to thousands of copies of a mitochondrial DNA (mtDNA) genome that encodes components essential for respiration and protein synthesis. Generally, mitochondrial alleles segregate during vegetative cell growth (vegetative segregation), and all copies of mtDNA within a cell or an individual generally have the same sequence (homoplasmy). In patients with mitochondrial myopathies, the progressive accumulation of mtDNA with large deletions leads to a heteroplasmic state in specific tissues (20; for review, see references 43 and 54). This phenomenon may be caused by the selective replication and/or segregation of mtDNA bearing the deletion; however, genetic analysis of mtDNA processes in mammals has been hampered by the strict maternal inheritance of mitochondria. The yeast Saccharomyces cerevisiae is a suitable model system because of its biparental mtDNA inheritance (14).
The extremely biased inheritance of mtDNA with a large deletion is known in S. cerevisiae as “hypersuppressiveness.” Vegetative petite mutants ([rho−]) of S. cerevisiae are respiration-deficient cells containing mtDNA with a large deletion or tandem arrays of a mtDNA segment. Crosses of respiration-proficient haploids ([rho+] cells) with [rho−] haploids generally yield [rho+] progeny, but some segregants are [rho−] (a phenomenon termed “suppressiveness”). In contrast, when [rho+] cells are crossed with cells with hypersuppressive [rho−] mtDNA (HS [rho−] mtDNA), almost all progeny are respiration defective (7, 12). HS [rho−] mtDNA is a tandem array of a small region (1 kb or less) of mtDNA that has one of the several replication origins of [rho+] mtDNA (70 to 85 kbp) (7, 12). HS [rho−] mtDNA has a higher density of replication origins than [rho+] mtDNA, and this suggests that a replication advantage of HS [rho−] mtDNA over [rho+] mtDNA results in hypersuppressiveness.
Two mechanisms for the initiation of mtDNA replication in yeast have been proposed: mitochondrial transcriptional RNA polymerase (Rpo41)-primed initiation and recombination-mediated initiation. Rpo41-primed DNA replication is similar to that observed in mammalian mitochondria. In support of this mechanism, each mtDNA replication origin shares sequence similarity with the heavy-strand replication origin of mammalian mtDNA, including the presence of a transcription promoter and three GC-rich clusters (2, 13). RNA synthesized by Rpo41 from mtDNA replication-origin promoters has been detected, and an endoribonuclease that cleaves the synthesized RNA at sites that correspond to regions of transition from RNA to DNA synthesis has been detected (3, 49, 53). The intact replication-origin promoter is required for hypersuppressiveness (36). However, this transcription-dependent process is not the sole mechanism for the initiation of mtDNA replication, since both the replication origin and RPO41 are dispensable for the maintenance of [rho−] mtDNA (18, 53). In addition, it has been reported that the selective inheritance of hypersuppressive [rho−] mtDNA relative to nonhypersuppressive [rho−] mtDNA (lacking a replication origin) is independent of RPO41 (34).
Concatemers (linear head-to-tail multimers) are the major form of mtDNA in yeast and other organisms (1, 5, 37). Inspired by the late-stage replication of the Escherichia coli phage λ (16, 47), the recombination-mediated mechanism for the initiation of mtDNA replication in a rolling-circle mode has been proposed to explain mtDNA concatemer formation (37).
MHR1 was the first gene that was shown to be required for [rho+] mtDNA recombination of a gene conversion type (27) and for the recombinational repair of mtDNA (28). In addition, we found that the partitioning of [rho+] mtDNA and [rho−] mtDNA (lacking a replication origin) into daughter cells depends on the formation of concatemers and, although concatemers are the major mtDNA form in mother cells, monomers are the major form in growing buds (future daughter cells) (30). Concatemers are formed by either rolling-circle replication or crossover-type homologous recombination. Label-and-chase experiments and genetic analysis support the conclusion that the concatemers are formed primarily by an Mhr1-dependent rolling-circle mode of replication and that, during or immediately after their transmission into buds, the concatemers are processed into circular mtDNA monomers (29). In addition, Mhr1 plays an active role in vegetative segregation to generate homoplasmic cells, which can be explained by the selective inheritance of an mtDNA clone as concatemers formed by rolling-circle replication on a few selected templates (29). Since Mhr1-dependent rolling-circle replication is the major pathway of mtDNA inheritance, we supposed that Mhr1 functions in hypersuppressiveness.
The first genetic observation that connected hypersuppressiveness to mtDNA recombination was reported as an effect on the suppressiveness of a null mutation of CCE1 (MGT1), which encodes a mitochondrial protein that resolves junctions in Holliday intermediates, an essential intermediate for crossover-type homologous recombination (17, 24). The cce1 null mutation completely eliminates suppressiveness (57) and causes the formation of a large network of mtDNA molecules connected by Holliday junctions. cce1-null mutant cells exhibit normal [rho+] mtDNA inheritance and mtDNA recombination, but the transmission of [rho−] mtDNA from the mother cell to the bud is partially blocked. Thus, the effects of the cce1 null mutation on hypersuppressiveness might be due to preferential blockage of the transmission of [rho−] mtDNA to daughter cells (i.e., partitioning), rather than to defects in the replication of [rho−] mtDNA (31).
Meiotic homologous recombination is initiated by double-stranded breaks at specific sites on chromosomal DNA (52). Following break induction, a 3′-single-stranded tail derived from a nascent end invades intact double-stranded DNA bearing the same or a similar sequence (homologous DNA) to form an intermolecular double-stranded structure, called a heteroduplex joint, with the complementary strand (homologous DNA pairing). Homologous pairing is generally catalyzed by members of the RecA/Rad51 family of proteins in the presence of ATP (39, 46, 51). DNA synthesis initiated at the 3′ end of the paired single-stranded tail in the heteroduplex joint restores sequences lost by the breakage. These intermediates are processed into recombinant DNA molecules (52). Mhr1 catalyzes homologous DNA pairing in the absence of ATP (30) (see below), as in the case for the lambda phage β protein and the related protein RecT, which are involved in homologous recombination and the initiation of rolling-circle DNA replication (41, 44). Rolling-circle replication depends on the homologous DNA pairing activity of Mhr1, suggesting that the 3′ termini of single-stranded tails derived from a double-stranded break paired with circular mtDNA serve as primers to initiate mtDNA replication.
In this study, we address the following questions: is Mhr1 essential for hypersuppressiveness? If so, do double-stranded breaks induce Mhr1-dependent rolling-circle replication of mtDNA and does this mode of replication contribute to hypersuppressiveness? We find evidence to support possible roles for Mhr1-mediated homologous pairing and Ntg1-stimulated double-stranded cleavage in the ori5 region in rolling-circle replication and hypersuppressiveness.
MATERIALS AND METHODS
Yeast strains, media, and general genetic techniques.
The yeast strains used in this study are listed in Table 1. Media were as previously described (27, 30). General genetic techniques were used in this study (23). Yeast transformation was carried out using the lithium-acetate method (22).
TABLE 1.
Yeast strains used in this study
| Strain | Nuclear genotype | Mitochondrial genotype | Source or reference |
|---|---|---|---|
| OP11c-55R5 | aleu2 ura3 trp1 MHR1 NTG1 | [rho+] [omega+] Δens2 Oli2r | 27 |
| FL672c-55R5 | aleu2 trp1 can1 mhr1-1 | [rho+] [omega+] Δens2 Oli2r | 27 |
| OP11c-55R5/Δntg1 | aleu2 ura3 trp1 ntg1::LEU2 | [rho+] [omega+] Δens2 Oli2r | This study |
| YKN1423 | α leu2 ura3 met3 MHR1 NTG1 | [rho+] [omega+] Δens2 Oli2r | 30 |
| YKN1423A-3 | α leu2 ura3 met3 MHR1 NTG1 | Normal suppressive [rho−] | This study |
| YKN1423A-3/Δmhr1 | α leu2 ura3 met3 mhr1::LEU2 | Normal suppressive [rho−] | This study |
| YKN1423A-3/Δntg1 | α leu2 ura3 met3 ntg1::LEU2 | Normal suppressive [rho−] | This study |
| YKN1423C-1 | α leu2 ura3 met3 MHR1 NTG1 | HS [ori5] [rho−] mtDNA | This study |
| YKN1423C-1/Δmhr1 | α leu2 ura3 met3 mhr1::LEU2 | HS [ori5] [rho−] mtDNA | This study |
| YKN1423C-1/Δntg1 | α leu2 ura3 met3 ntg1::LEU2 | HS [ori5] [rho−] mtDNA | This study |
| YKN1423C-1GalNTG1 | α leu2 ura3 met3 GalNTG1 | HS [ori5] [rho−] mtDNA | This study |
| YKN1423C-1/Δnuc1 | α leu2 ura3 met3 nuc1::LEU2 | HS [ori5] [rho−] mtDNA | This study |
| YKN1423C-1/Δngl1 | α leu2 ura3 met3 ngl1::Kanr | HS [ori5] [rho−] mtDNA | This study |
| YKN1423C-1/Δogg1 | α leu2 ura3 met3 ogg1::Kanr | HS [ori5] [rho−] mtDNA | This study |
| YKN1423C-1/Δrpo41 | α leu2 ura3 met3 rpo41::Kanr | HS [ori5] [rho−] mtDNA | This study |
| YKN1423C-1/Δntg1Δrpo41 | α leu2 ura3 met3 ntg1::LEU2 rpo41::Kanr | HS [ori5] [rho−] mtDNA | This study |
| YKN1423C-1/Δntg1Δogg1 | α leu2 ura3 met3 ntg1::LEU2 ogg1::Kanr | HS [ori5] [rho−] mtDNA | This study |
| YKN1423C-1/Δntg1Δnuc1 | α leu2 ura3 met3 ntg1::LEU2 nuc1::Kanr | HS [ori5] [rho−] mtDNA | This study |
| YKN1423C-1 [rho0] | α leu2 ura3 met3 | [rho0] | This study |
Isolation of hypersuppressive [rho−] mutant cells.
The isolation of hypersuppressive [rho−] mutant cells and tests for the suppression of wild-type (WT) cells by [rho−] mutants were carried out as described previously (7), with some modifications. MATα [rho+] MHR1 NTG1 cells (YKN1423) were grown in yeast extract-peptone-glycerol (YPGly) medium, transferred into yeast extract-peptone-dextrose (YPD) medium containing 10 μg/ml of ethidium bromide, and cultivated at 30°C for 3 h. The cells were spread on YPD plates and allowed to form colonies at 30°C for 4 days. These colonies were crossed with a lawn of MATa [rho+] MHR1 NTG1cells (OP11c-55R5). HS [rho−] mutant-derived colonies formed only faint traces on YPGly plates in replica-plating tests. From about 10,000 ethidium bromide-treated cells, we obtained 5 HS [rho−] mutants which, when crossed with [rho+] cells, generated [rho−] cells at a frequency of 88 to 97%. One (YKN1423C-1) of these was used as the wild-type (with respect to nuclear genes) HS [rho−] strain.
The YKN1423C-1 cells carry HS [rho−] mtDNA called HSC-1. HSC-1 consists of tandem repeats (concatemers) of the 1.1-kb unit containing a region whose sequence is identical to that of the mtDNA replication origin ori5 of [rho+] mtDNA, which contains four ori's, and to that of HS3324, a previously isolated HS [rho−] mtDNA (6, 34). The 1.1-kb unit contains single BglII, EcoRV, and NdeI restriction sites.
Assay for hypersuppressiveness.
The [rho−] cells with the HS [ori5] [rho−] mtDNA and the disrupted genes (Δ) to be tested were crossed with parental (WT) [rho+] cells or with [rho+] cells bearing the same disruption. The diploid cells from these crosses were selectively grown in SD medium supplemented with required amino acids, cultivated for 18 to 22 generations, plated on SD, and allowed to form colonies at 30°C for 3 days. The colonies were replica plated to YPGly to test for respiration proficiency. Respiration-deficient ([rho−]) diploid cells from each cross formed colonies on SD, but not on YPGly.
Gene disruption.
Disruptions of the MHR1, NUC1, and NTG1 genes were performed as previously described (27, 30). RPO41, OGG1, and NGL1 were disrupted by the insertion of a Kanr cassette (32). The presence of the disrupted gene in each strain was confirmed by PCR with the primers that were used for amplification of the complete open reading frame (ORF) of the disrupted gene.
Southern blot analysis.
Whole yeast DNA and DNA digested with restriction endonucleases were analyzed by standard gel electrophoresis through a 1.5 or 2.0% agarose gel (FMC), at 2.0 V/cm in TAE buffer (40 mM Tris-acetate, 10 mM EDTA, pH 8.0) at room temperature. Unless otherwise stated, 6 to 15 μg of intact or digested DNA was loaded on gels. λ/HindIII-digested DNA (New England Biolabs) and a 100-bp ladder (Invitrogen) were used as size markers. The DNA in the gel was transferred to a Hybond-N+ membrane (Amersham Biosciences) in 10× SSC (1.5 M NaCl, 0.15 M sodium citrate) by capillary blotting. HS [ori5] [rho−] mtDNA was detected by Southern blot analysis using 32P-labeled HS [ori5] [rho−] mtDNA fragments as probes. 32P signals were qualitatively and quantitatively analyzed with a Fuji BAS2000 image analyzer.
Double-stranded break detection and quantification in the ori5 region of HS [ori5] [rho−] mtDNA.
Whole cellular DNA was isolated from early-log-phase cultures of wild-type and mutant cells grown in YPD at 30°C. To investigate the effects of increased amounts of Ntg1 protein on double-stranded break formation, NTG1 under the GAL1 promoter (PGAL1NTG1) control was constructed by integrating the Kanr-GAL1 cassette into the region upstream of the ATG codon of NTG1 (32). Early-log-phase cells of PGA1LNTG1 bearing HS [ori5] [rho−] mtDNA (YKN1423C-1GalNTG1) were grown in raffinose medium and transferred to galactose-containing medium for the induction of the GAL1 promoter. Whole cellular DNA (∼10 μg) was digested with BglII, fractionated by electrophoresis through a 2% agarose gel, and transferred to Hybond-N+ membranes for Southern blot analysis using the 32P-labeled HS [ori5] [rho−] mtDNA probe. 32P signals were detected and measured with a Fuji BAS2000 image analyzer. The background for each DNA fragment was corrected by subtracting an averaged signal from same-sized areas located just above and below the band of the fragment. The relative levels of double-stranded breaks were determined by calculating the ratio of the signals for the 0.8-kb fragment, derived by double-stranded cleavage in the ori5 region, to the signals for the unit-sized 1.1-kb HS [ori5] [rho−] mtDNA.
Production and purification of the Ntg1 and CΔntg1 proteins.
The ORF of NTG1 (1,200 bp) and its C-terminal deletion mutant (CΔntg1; the N-terminal 474 bp of the ORF from the initiation codon ATG) were each ligated into the pET14b expression vector (Novagen) to form six-His-tagged fusion proteins, which were overexpressed in Escherichia coli BL21(DE3)pLyS host cells (Stratagene). The E. coli cells were cultured at 37°C to early log phase (ca. 1.5 × 108 cells/ml) and cultured at 16°C for another 4 h in the presence of 1 mM isopropyl-β-d-thiogalactopyranoside. The Ntg1 and CΔntg1 proteins were purified using HisTrap resin (Pharmacia Biotech). The proteins were fractionated by 12.5% polyacrylamide gel electrophoresis in the presence of sodium dodecyl sulfate and detected by Coomassie brilliant blue staining.
In vitro assay for detection of double-stranded breaks in the ori5 region.
The standard reaction mixture (10 μl) consisted of 0.2 μg total DNA digested with BglII, 70 mM 3-(N-morpholino) propanesulfonic acid (pH 7.5), 1 mM dithiothreitol, 1 mM EDTA, 5% glycerol, and various concentrations of the Ntg1 or CΔntg1 protein. The reaction was carried out at 37°C for 30 min as described elsewhere (15). After removing the proteins from the reaction solution with proteinase K, the digests were analyzed by electrophoresis on a 2% agarose gel. The 0.8-kb DNA fragment derived from the unique double-stranded break in the ori5 region and the unit-sized 1.1-kb HS [ori5] [rho−] mtDNA were detected by Southern blot analysis as described above.
Two-dimensional gel electrophoresis.
Two-dimensional gel electrophoresis analysis was carried out as previously described (8, 29). The first dimension was run for 6 h at 1 V/cm on a 0.4% agarose gel in TBE buffer (45 mM Tris, 45 mM boric acid, 1 mM EDTA, pH 8.0) at 23°C. The λ/HindIII DNA size markers were used to determine the DNA electrophoresis positions. The second dimension was run at 5 V/cm on a 1.0% agarose gel in TBE buffer with 0.3 μg/ml ethidium bromide at 4°C. 32P signals from Southern blot analysis were detected and quantified with a Fuji BAS2000 image analyzer. The DNA species detected on the two-dimensional gel were assigned as described elsewhere (33). The gross 32P signals appearing in the framed area in Fig. 4A, below, corresponding to concatemers and supercoiled or nicked circular monomers in the two-dimensional gel, were measured and corrected by subtracting the background. The ratios of the net signals of concatemers to monomers were calculated to estimate the extent of concatemer formation by HS [ori5] [rho−] mtDNA.
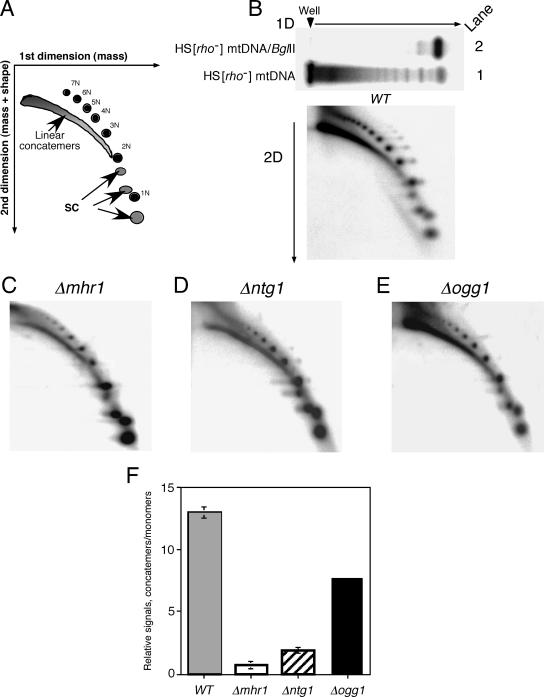
FIG. 4.
Requirements for MHR1 and NTG1 in formation of major molecular species of HS [ori5] [rho−] mtDNA (probably rolling-circle replication products). The cells with disrupted genes were grown in YPD at 30°C. About 20 μg of whole cellular DNA was separated by two-dimensional gel electrophoresis and transferred to a Hybond-N+ membrane. The HS [ori5] [rho−] mtDNA on the membrane was detected by Southern blot analysis using the 32P-labeled HS [ori5] [rho−] mtDNA as a probe. (A) Schematic diagram. The first-dimension run separates DNA species according to mass, and the second-dimension run fractionates molecules according to both mass and shape. The positions of DNA species are indicated: concatemers, linear multimers of various continuous lengths; 1N, open circular monomers; 2-7N, open circular multimers [circular mtDNA with nick(s)] (2N, dimers; 3N, trimers; 4N, tetramers; 5N, pentamers; 6N, hexamers; 7N, heptamers); SC, supercoiled (natural closed circular) monomers, dimers, and trimers (33). The signals corresponding to concatemers, circular monomers, and multimers in the encircled areas after the two-dimensional run were measured and corrected by subtracting the background of the same-sized area lacking radioactivity. (B to E) Two-dimensional gel profiles of HS [ori5] [rho−] mtDNA isolated from parental cells (WT) and cells with the indicated disrupted gene. A one-dimensional gel profile of HS [ori5] [rho−] mtDNA from wild-type cells is shown above the two-dimensional gel profile in panel B. In this one-dimensional gel electrophoresis, lanes 1 and 2 contain undigested HS [ori5] [rho−] mtDNA and BglII-digested HS [ori5] [rho−] mtDNA, respectively. (F) Relative amounts of concatemers to circular monomers (nicked circular monomers plus supercoiled monomers). The relative signals are the net signals of concatemers divided by those of circular monomers. Each bar indicates the average of at least two independent experiments. Wild-type and disrupted strains containing HS [ori5] [rho−] mtDNA are labeled as follows: WT, the parental strain (YKN1423C-1); Δmhr1, YKN1423C-1/Δmhr1; Δntg1, YKN1423C-1/Δntg1; Δogg1, YKN1423C-1/Δogg1.
Nucleotide sequence accession number.
The nucleotide sequence of HSC-1 mtDNA was determined and submitted to DDB/EMBL/GenBank under accession no. AB182994.
RESULTS
The Mhr1 protein plays a role in hypersuppressiveness.
To test whether the Mhr1 protein is required for hypersuppressiveness, we used one of five newly isolated HS [rho−] mutants. The HS [rho−] mutant carries HSC-1 mtDNA (HS [ori5] [rho−] mtDNA; unit size, 1.1 kb) containing the ori5 of [rho+] mtDNA (rep2) (12). Most of the [rho−] cells isolated along with the HS [rho−] mutants showed moderate suppressiveness (normal [rho−] cells). Among the progeny of diploids generated by crossing randomly selected normal [rho−] strains with a [rho+] strain, the average proportion of [rho−] cells was 31 ± 6% (n = 9).
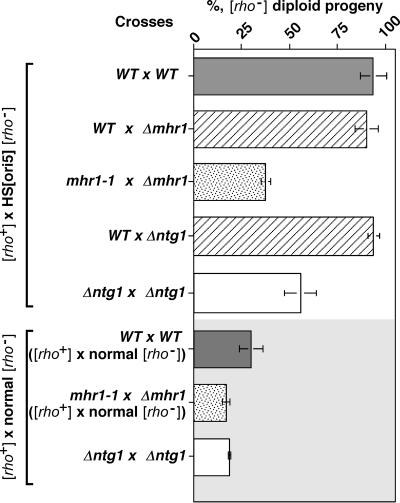
Cells containing HS [ori5] [rho−] mtDNA were crossed with [rho+] cells in the presence or absence of active Mhr1. In diploids generated by crosses between mhr1-1 and Δmhr1 haploids, the fraction of [rho−] cells among diploid progeny decreased from 87 to 100% to ca. 30% (Fig. 1). In the MHR1 and Δmhr1 cross, HS [ori5] [rho−] mtDNA showed hypersuppressiveness over [rho+] mtDNA to the same extent as the MHR1 and MHR1 cross (Fig. 1), indicating that the mutant mhr1 allele is recessive. It should be noted that a factor in the residual suppressiveness in the absence of active Mhr1 is the leakiness of mhr1-1 at 30°C, which is essential to these experiments since, in the complete absence of Mhr1, yeast cells do not maintain [rho+] mtDNA (27) and hypersuppressiveness cannot be tested.
FIG. 1.
MHR1 and NTG1 are required for hypersuppressiveness. The [rho−] cells containing HS [ori5] [rho−] mtDNA and the disrupted genes (Δ) to be tested were crossed with parental (WT) [rho+] cells and with [rho+] cells bearing the same disruption, as described in the text. For comparison, experiments using normal suppressive [rho−] cells were also carried out. The percentage of [rho−] zygotes from each cross was calculated and plotted. The strains used in crosses between [rho+] cells and [rho−] cells with the HS [ori5] [rho−] mtDNA were the following: WT × WT, [rho+] MHR1 NTG1 cells (OP11c-55R5) and MHR1 NTG1 cells containing HS [ori5] [rho−] mtDNA (YKN1423C-1); WT × Δmhr1, [rho+] MHR1 NTG1 cells (OP11c-55R5) and Δmhr1 cells containing HS [ori5] [rho−] mtDNA (YKN1423C-1/Δmhr1); mhr1-1 × Δmhr1, [rho+] mhr1-1 cells (FL672c-55R5) and Δmhr1 cells containing HS [ori5] [rho−] mtDNA (YKN1423C-1/Δmhr1); WT × Δntg1, [rho+] MHR1 NTG1 cells (OP11c-55R5) and Δntg1 cells containing the HS [ori5] [rho−] mtDNA (YKN1423C-1/Δntg1); Δntg1 × Δntg1, [rho+] Δntg1 cells (OP11c-55R5/Δntg1) and Δntg1 cells with HS [ori5] [rho−] mtDNA (YKN1423C-1/Δntg1). The strains used in crosses between [rho+] cells and the normal [rho−] cells were the following: WT × WT, [rho+] MHR1 NTG1 cells (OP11c-55R5), and normal suppressive [rho−] MHR1 NTG1 cells (YKN1423A-3); mhr1-1 × Δmhr1, [rho+] mhr1-1 cells (FL672c-55R5), and normal suppressive [rho−] Δmhr1 cells (YKN1423A-3/Δmhr1); Δntg1 × Δntg1, [rho+] Δntg1 cells (OP11c-55R5/Δntg1), with normal [rho−] Δntg1 cells (YKN1423A-3/Δntg1).
We repeated these experiments using a normal [rho−] strain, which was isolated along with the petite mutant containing HS [ori5] [rho−] mtDNA, and found that the suppressiveness of normal [rho−] was also reduced by one-third in the absence of MHR1 (Fig. 1). It has been reported that most isolated petite mutants have [rho−] mtDNA containing a replication-origin sequence (12). We confirmed that [rho−] cells containing mtDNA without replication origin (the mt-pMK2 DNA used in our previous studies [29]) are neutral suppressive petite mutants, i.e., few [rho−] progeny (1.8 ± 0.3%) were obtained after crossing [rho−] cells containing mt-pMK2 and [rho+] cells. Thus, it is likely that the suppressiveness of normal [rho−] also depends on an Mhr1 function at a replication origin.
It is known that a defective mutation in MHR1 affects the stability or copy number of mtDNA (27). The amount of HS [ori5] [rho−] mtDNA in Δmhr1 cells was about one-half of that in MHR1 cells under these experimental conditions (YPD medium, 30°C) (F. Ling, unpublished observation), but this decrease was about the same as that of [rho+] mtDNA in mhr1-1 cells (27). Thus, the extensive loss of hypersuppressiveness of HS [ori5] [rho−] mtDNA in mhr1/mhr1 zygotes is not due primarily to a change in mtDNA stability.
Based on these results, we conclude that Mhr1 is required for hypersuppressiveness, consistent with the proposed contribution of Mhr1-dependent rolling-circle mtDNA replication to hypersuppressiveness.
A double-stranded break was detected in the ori5 region of HS [ori5] [rho−] mtDNA (HSC-1).
To determine whether double-stranded breaks have a role in the initiation of Mhr1-dependent mtDNA replication and hypersuppressiveness, we subjected HS [ori5] [rho−] mtDNA to Southern analysis. We first examined mtDNA prepared from isolated mitochondria but found no improvement in the results obtained compared with whole cellular DNA. In addition, no mtDNA or background signal was detected in whole cellular DNA from [rho0] cells (cells devoid of mtDNA), even when more DNA was loaded and a longer exposure time was used (see below) (Fig. 2A). Thus, throughout this study we examined whole cellular DNA to minimize the risk of degradation.
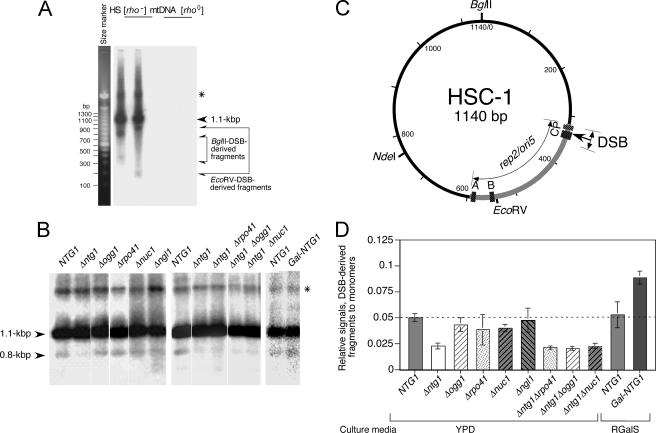
FIG. 2.
Double-stranded breaks in the replication origin of HS [ori5] [rho−] mtDNA (HSC-1) and effects of NTG1 disruption and Ntg1 protein overproduction on double-stranded cleavage. (A) Detection of double-stranded breaks in HS [ori5] [rho−] mtDNA. Whole cellular DNA from MHR1 NTG1 cells with HS [ori5] [rho−] mtDNA and from [rho0] MHR1 NTG1 cells was digested with BglII or EcoRV and separated by electrophoresis on a 1.5% agarose gel. HS [ori5] [rho−] mtDNA signals were detected by Southern blot analysis using 32P-labeled HS [ori5] [rho−] mtDNA as a probe. Under standard conditions for electrophoresis (ca. 10 μg DNA loaded), unit-size HS [ori5] [rho−] mtDNA (1.1 kb) and a 0.8-kb DNA fragment were observed in BglII digests (lane 1 in panel C). To detect less-abundant restriction fragments, a much larger amount of each DNA sample (ca. 75 μg) was loaded and the Southern blot was exposed to the imaging plate for a longer period. Under these conditions, a pair of fragments (0.8 kb and 0.3 kb as for BglII digests and 0.9 kb and 0.2 kb as for EcoRV digests; indicated with pairs of arrows) were detected in addition to the 1.1-kb unit-size DNA, the background signals were higher, and the 1.1-kb signal was saturated and broadened. No signals were detected from DNA prepared from [rho0] cells. mtDNA containing single-stranded regions resistant to digestion with restriction enzymes is indicated by an asterisk. (B) Effects of disruption of various mtDNA-related genes on double-stranded cleavage. Total DNA (ca. 10 μg), isolated from early-log-phase cells grown in YPD at 30°C, was digested with BglII and subjected to gel electrophoresis through a 2% agarose gel and Southern analysis. A unit-size HS [ori5] [rho−] mtDNA, derived from the BglII digestion, and a 0.8-kb DNA fragment, derived from the unique double-stranded break in the ori5 region from the BglII digest, are indicated with arrowheads. mtDNA containing single-stranded regions resistant to digestion with BglII is indicated by an asterisk. Disrupted genes are indicated above the panel. Strains were as follows: NTG1, YKN1423C-1; Δntg1, YKN1423C-1/Δntg1; Δogg1, YKN1423C-1/Δogg1; Δrpo41, YKN1423C-1/Δrpo41; Δnuc1, YKN1423C-1/Δnuc1; Δngl1, YKN1423C-1/Δngl1; Δntg1 Δrpo41, YKN1423C-1/Δntg1Δrpo41; Δntg1 Δogg1, YKN1423C-1/Δntg1Δogg1; Δntg1 Δnuc1, YKN1423C-1/Δntg1Δnuc1. To investigate the effects of Ntg1 overproduction, the expression of NTG1 under the GAL1 promoter was induced in YKN1423C-1GalNTG1 cells in which the GAL1 promoter was integrated 5′ to the NTG1-coding region and mitochondria containing HS [ori5] [rho−] mtDNA. YKN1423C-1 and YKN1423C-1GalNTG1 were cultured under conditions for the induction of the GAL1 promoter. Then, their mtDNA was analyzed as described. NTG1, YKN1423C-1; Gal-NTG1, YKN1423C-1GalNTG1. (C) Physical map of HSC-1 (HS [ori5] [rho−] mtDNA) showing the in vivo double-stranded breakage site. The 282-bp ori5 sequence is indicated by a thick gray line (nucleotides 313 to 594 with respect to the BglII site). The three GC clusters within ori5 are indicated by black boxes with white stippling (C, nucleotides 329 to 345; B, nucleotides 545 to 553; A, nucleotides 582 to 594). The transcriptional promoter (P, nucleotides 316 to 325) is indicated by a black box with white hatching. Restriction sites are indicated. The in vivo double-stranded breakage site is indicated by an arrow, and the standard deviation is shown by a doubleheaded arrow (nucleotides 334 ± 16 [n = 12]). (D) Quantitative analysis of double-stranded breaks in mtDNA. The extent of double-stranded breakage in the HS [ori5] [rho−] mtDNA in each sample shown in panel B was calculated from the signals of the 0.8-kb and the unit-sized 1.1-kb DNA fragments (indicated with arrowheads in panel B). After correcting band intensities by subtracting the background, the ratios of the signals specific to the 0.8- and 1.1-kb fragments were determined and plotted as “relative signals.”
Total DNA isolated from yeast cells was digested with BglII or EcoRV into unit lengths and fractionated by electrophoresis. The gel was examined by Southern blotting using the HS [ori5] [rho−] mtDNA-specific probe. We detected a clear major signal corresponding to the intact HS [ori5] [rho−] mtDNA unit (1.1 kb), indicating that the mtDNA was not affected by uncontrolled degradation during preparation and restriction (Fig. 2B, NTG1; see also Fig. 4B and related discussion, below). A weak signal was detected above the major signal (Fig. 2 and 3). Since these signals disappeared with single-strand-specific nuclease S1 treatment, they were caused by single-stranded regions that conferred resistance to restriction enzyme digestion at the cleavage site.
FIG.3.
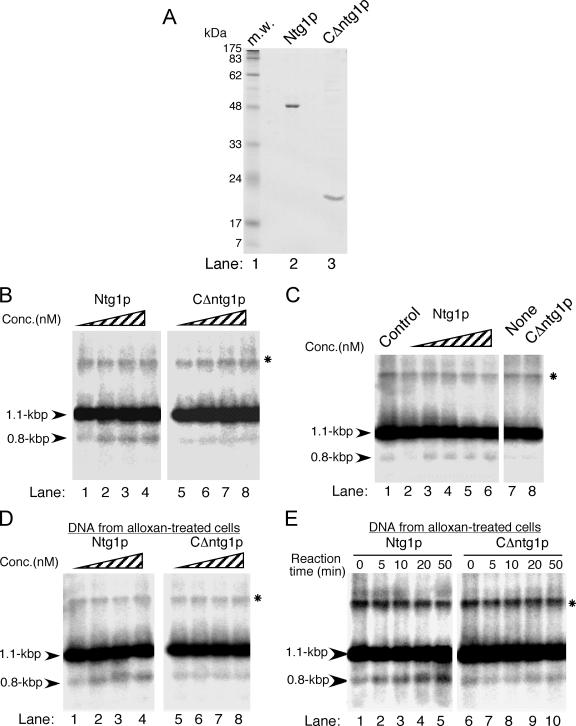
The Ntg1 protein introduces a double-stranded break in vitro by recognizing oxidative damage in ori5. (A) Purification of the Ntg1 protein (Ntg1p) and a C-terminal truncation mutant (CΔntg1p). Purified Ntg1 (lane 2) and CΔntg1 (lane 3) proteins (ca. 3.2 μg) were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Lane 1, prestained protein size markers. (B) In vitro cleavage by Ntg1 of HS [ori5] [rho−] mtDNA isolated from wild-type cells. Whole cellular DNA was extracted from the MHR1 NTG1 cells containing HS [ori5] [rho−] mtDNA (YKN1423C-1) grown in YPD medium at 30°C to mid-log phase. About 15 μg of BglII-digested DNA was incubated with 0, 0.8, 1.6, and 3.2 nM of Ntg1 (lanes 1 to 4) or CΔntg1 (lanes 5 to 8) at 37°C for 30 min. After the proteins were removed by proteinase K treatment, the digests were analyzed by electrophoresis on a 2% agarose gel. The 0.8-kb DNA fragment derived from the unique double-stranded break in the ori5 region and BglII digestion and the unit-size 1.1-kb fragment corresponding to HS [ori5] [rho−] mtDNA (indicated with arrowheads) were detected by Southern blot analysis using 32P-labeled HS [ori5] [rho−] mtDNA as a probe. (C) In vitro cleavage by Ntg1 of HS [ori5] [rho−] mtDNA isolated from ntg1Δ cells. Whole cellular DNA was extracted from the Δntg1 cells containing HS [ori5] [rho−] mtDNA (YKN1423C-1/Δntg1), digested with BglII, and incubated with 0, 0.8, 1.6, 2.4, and 3.2 nM of Ntg1 (lanes 2 to 6) or 0 and 3.2 nM of CΔntg1 protein (lanes 7 and 8) at 37°C for 30 min. An equal amount of BglII-treated DNA extracted from NTG1 cells containing HS [ori5] [rho−] mtDNA (YKN1423C-1) was used as a control (lane1). Electrophoretic analysis was carried out as described for panel B.
We detected a second weak signal that migrated faster than the major signal (Fig. 2B, NTG1). This faster-migrating signal would be expected if HS [ori5] [rho−] mtDNA had a site-specific double-stranded break. Next, we selected a set of conditions (ca. eightfold more DNA loaded and longer exposure time) to detect small amounts of other fragments. These conditions resulted in an apparent broadening of the major signal derived from unit-length HS [ori5] [rho−] mtDNA, and we detected a pair of signals corresponding to 0.8- and 0.3-kb DNA fragments produced by BglII digestion and a pair of signals corresponding to 0.9- and 0.2-kb fragments produced by EcoRV digestion (Fig. 2A). Since the sum of the lengths of the fragments in each pair was equal to unit-size HS [ori5] [rho−] mtDNA, these fragments are likely derived from double-stranded cleavage at a site that was mapped near the promoter end of the ori5 sequence of HS [ori5] [rho−] mtDNA (about nucleotide 334 ± 16 [n = 12]) (Fig. 2C). The mapped cleavage site was further confirmed by cleavage of the isolated 0.8-kb BglII fragment with NdeI, which produced two fragments of about 0.4 kb each (Fig. 2C and 3F, lanes 2 and 3).
Figure 3b.
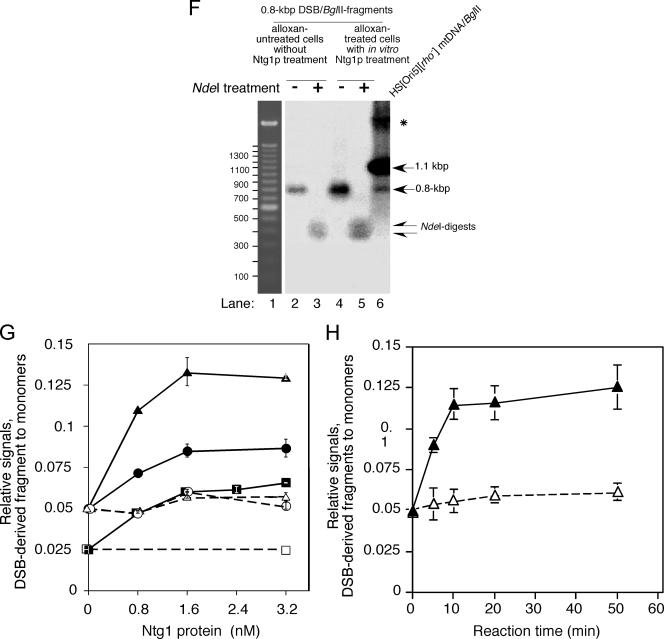
(D) In vitro cleavage by Ntg1 of HS [ori5] [rho−] mtDNA isolated from alloxan-treated cells. NTG1 cells containing HS [ori5] [rho−] mtDNA (YKN1423C-1) were grown in YPD medium at 30°C to mid-log phase and treated with 10 mM alloxan at 30°C for 10 min. Whole cellular DNA was extracted, digested with BglII, and incubated with 0, 0.8, 1.6, and 3.2 nM Ntg1 (lanes 1 to 4) or CΔntg1 (lanes 5 to 8) at 37°C for 30 min. Analysis was carried out as described for panel B. (E) Time-dependent in vitro cleavage by Ntg1 of HS [ori5] [rho−] mtDNA isolated from alloxan-treated cells. NTG1 cells containing HS [ori5] [rho−] mtDNA (YKN1423C-1) were grown in YPD medium at 30°C to mid-log phase and treated with 10 mM alloxan at 30°C for 10 min. Whole cellular DNA was extracted, digested with BglII, and incubated with 1.6 nM Ntg1 (lanes 1 to 5) or CΔntg1 (lanes 6 to 10) at 37°C. Aliquots were withdrawn from the reaction mixtures at the indicated time points and analyzed as described for panel B. (F) NdeI digestion of the 0.8-kb fragment. BglII-digested whole cellular DNA from alloxan-treated cells was incubated with 1.2 nM Ntg1 at 37°C for 30 min, and the resulting 0.8-kb fragment was purified (lane 4). As a control, the 0.8-kb fragment was also purified from BglII digests (without Ntg1 treatment) of HS [ori5] [rho−] mtDNA from untreated cells (lane 2). These fragments were further digested with NdeI (lanes 5 and 3, respectively). If Ntg1 cuts the BglII fragment of HS [ori5] [rho−] mtDNA in the ori5 region, NdeI generates two fragments of about 0.4 kb each (indicated with arrows in lanes 3 and 5; see the restriction map in Fig. 2C). Lane 6, BglII digests of HS [ori5] [rho−] mtDNA from untreated cells. All signals were detected by Southern blot analysis using 32P-labeled 1.1-kb HS [ori5] [rho−] mtDNA as a probe. Lane 1, 100-bp size ladder. mtDNA containing single-stranded regions resistant to digestion with BglII is indicated by an asterisk. (G and H) Quantification of Ntg1-generated double-stranded breaks in the ori5 region of HS [ori5] [rho−] mtDNA isolated from alloxan-treated or untreated cells. The frequency of double-stranded breaks, as represented by the signals corresponding to the 0.8-kb DNA fragment, was calculated for each concentration of Ntg1 and CΔntg1. The ratios of the 0.8- and the 1.1-kb signals are plotted as relative signals. •, DNA from NTG1 cells incubated with Ntg1; ○, DNA from NTG1 cells incubated with CΔntg1; ▪, DNA from ntg1-disrupted cells incubated with Ntg1; □, DNA from ntg1-disrupted cells incubated with CΔntg1; ▴, DNA from alloxan-treated cells incubated with Ntg1; ▵, DNA from alloxan-treated cells incubated with CΔntg1.
These results suggest that, in vivo, a double-stranded break occurs in ori5 near its promoter end in a small population of the HS [ori5] [rho−] mtDNA.
The Ntg1 protein by itself functions in ori5-specific double-stranded cleavage of HS [ori5] [rho−] mtDNA.
We next tested the assumption that the double-stranded break functions in the hypersuppressiveness of the HS [ori5] [rho−] mtDNA by using a genetic approach to identify a gene(s) responsible for double-stranded break induction.
By monitoring the amount of the 0.8-kb DNA fragment derived from the double-stranded cleavage in the ori5 region of the BglII-generated 1.1-kb unit relative to the amount of the uncleaved 1.1-kb HS [ori5] [rho−] mtDNA, we tested several gene disruptions for direct or indirect effects on double-stranded cleavage. These genes included those encoding proteins that function directly or potentially in repair, recombination, and/or replication of mtDNA: DNA N-glycosylase/lyases (NTG1 and OGG1), nucleases (NUC1 and NGL1), and the mitochondrial RNA polymerase RPO41. We found that disruption of the NTG1 gene repressed the double-stranded cleavage in ori5 to one-third of the wild-type level (Fig. 2B and D). As described in detail below, the Ntg1 protein is a base excision enzyme that is active on oxidized bases. The disruption of OGG1, a gene encoding another base excision enzyme, had little effect on the extent of double-stranded cleavage (Fig. 2B and D), suggesting that the substrate specificity of the Ntg1 protein is an important factor for double-stranded breakage in the ori5 region. The disruptions of RPO41, NUC1, and NGL1 had little, if any, effect on double-stranded cleavage (Fig. 2B and D). To further examine the effects of the disruptions of OGG1, RPO41, and NUC1, we constructed double mutants with disruptions of one of these genes and Δntg1. We found no further decrease in the amount of double-stranded cleavage in the ori5 region above that caused by Δntg1 alone (Fig. 2B and D). Thus, it is very unlikely that RPO41, OGG1, or NUC1 is involved in double-stranded cleavage in the ori5 region.
The Ntg1 protein exists primarily in mitochondria, although some is found in the nucleus as well (55). Ntg1 is required for the repair of oxidative DNA damage in vivo, and it shares amino acid sequence similarity with E. coli endonuclease III and the human Nth1 protein (21). Ntg1 has DNA repair glycosylase and apurinic/apyrimidinic (AP) lyase activities. It recognizes various oxidatively modified bases and causes single-stranded breakage (nicking) in double-stranded DNA (15, 45, 55). Thus, the effect of NTG1 disruption on ori5-specific double-stranded cleavage is not readily explainable by the known biochemical activities of the Ntg1 protein. Therefore, we further investigated the role of Ntg1 in ori5-specific double-stranded breakage.
We overexpressed Ntg1 and found that it enhanced double-stranded cleavage of ori5 by almost twofold, as estimated by the relative signal strength of the 0.8-kb and unit-sized fragments (Fig. 2B and D). This result supports an important role for Ntg1 in the ori5-specific double-stranded cleavage of HS [ori5] [rho−] mtDNA.
To determine whether Ntg1 introduces double-stranded breaks into ori5 directly, indirectly, or with the aid of other proteins, we treated isolated DNA with purified Ntg1 and, as a negative control, with an inactive derivative lacking a portion of the C terminus (CΔntg1, consisting of only a 40% region of the wild-type protein) (Fig. 3A). We confirmed that purified Ntg1 had DNA repair N-glycosylase/AP lyase activity. Since alloxan treatment generates reactive oxidative species in vivo (9, 19), closed-circular double-stranded DNA (pUC18), isolated from alloxan-treated E. coli cells, was nicked with increasing amounts of purified Ntg1 to generate the open circular form (see Fig. S1B and C in the supplemental material). However, the same DNA isolated from untreated cells generated only a small amount of the open circular form (Fig. A1, compare lane 2 in A with lane 5 in B). No double-stranded cleavage was detected under these conditions. Thus, the Ntg1 protein purified from E. coli has the specific activity described previously (15).
We treated BglII-digested DNA isolated from NTG1-proficient (Fig. 3B and G) and -deficient (Fig. 3C and G) cells with purified Ntg1 and found that signals specific for the 0.8-kb fragment were increased by Ntg1 treatment. The untreated levels were similar to those in mtDNA isolated from wild-type and Δngt1 cells (Fig. 2B and D), but the level of the 0.8-kb fragment after treatment of Δntg1 mtDNA with sufficient purified Ntg1 was close to that obtained with mtDNA isolated from NTG1 cells (Fig. 3G). The in vitro double-stranded cleavage of isolated HS [ori5] [rho−] mtDNA to generate the 0.8-kb fragment was significantly enhanced by treating the cells with alloxan before extraction of HS [ori5] [rho−] mtDNA (Fig. 3D and G). The appearance of the signal for the 0.8-kb fragment was dependent on the length of incubation with Ntg1 (Fig. 3E and H). When Ntg1 was replaced by its inactive derivative, CΔntg1, there was no increase in cleavage (Fig. 3B to E, G, and H), indicating that cleavage was due to the activity of Ntg1 itself, not to contaminating DNases from E. coli.
We sought further confirmation that the 0.8-kb fragment produced by the in vitro treatment of isolated DNA with purified Ntg1 resulted from double-stranded cleavage of ori5. The 0.8-kb fragments produced by in vivo double-stranded cleavage of ori5 (in BglII-digested total DNA) and by in vitro treatment with purified Ntg1 of DNA isolated from alloxan-treated cells (followed by treatment with BglII) were extracted from a gel. These fragments were digested with NdeI, which would generate a pair of 0.4-kb fragments if the 0.8-kb fragment were derived from double-stranded cleavage of ori5 (Fig. 1C). As shown in Fig. 3F, digestion of these 0.8-kb fragments yielded only the predicted pair of 0.4-kb fragments and no other signals.
These results indicate that the isolated HS [ori5] [rho−] mtDNA is preferentially modified near the promoter end of ori5, that Ntg1 alone introduces a double-stranded break in vitro by recognizing this modification at ori5, and that the modification is enhanced in vivo by subjecting the cells to oxidative stress. These conclusions, in turn, suggest that the ori5 region is modified by oxidative base damage (see Discussion, below).
The Ntg1 protein is required for the hypersuppressiveness of HS [ori5] [rho−] mtDNA.
We assessed whether Ntg1-dependent double-stranded cleavage at ori5 is responsible for the hypersuppressiveness of HS [ori5] [rho−] mtDNA. When [rho+] cells were crossed with cells bearing HS [ori5] [rho−] mtDNA in the absence of active Ntg1, the fraction of [rho−] cells among the diploid progeny was reduced to about 50% and, thus, HS [ori5] [rho−] mtDNA failed to exhibit hypersuppressiveness under these conditions (Fig. 1). As in the absence of active Mhr1, the disruption of NTG1 caused repression of the suppressiveness of normal [rho−] mtDNA (Fig. 1). The extent of the decrease in the suppressiveness of HS [ori5] [rho−] mtDNA and normal [rho−] mtDNA (both ca. 50%) caused by disruption of NTG1 qualitatively correlated with the reduction of double-stranded cleavage (30 to 50% of the wild-type level; see the preceding section). The disruption of NTG1 mildly affects the copy number of HS [ori5] [rho−] mtDNA (ca. 60% of the wild-type level), while the disruption of NTG1 did not significantly affect the amount of [rho+] mtDNA (F. Ling, preliminary unpublished observation). Thus, in this case, the decrease in the copy number can be a factor in preventing hypersuppressiveness. However, as described in the Discussion section below, it is likely that the specific action of Ntg1 at ori5 plays an important role in the inheritance of HS [ori5] [rho−] mtDNA. In the absence of active Mhr1, differential stability of HS [ori5] [rho−] mtDNA and [rho+] mtDNA is not a factor to prevent hypersuppressiveness.
In yeast mitochondria, a large portion of mtDNA molecules are present in a linear form (concatemers) (5, 37). One might wonder whether a minor increase in the free ends caused by Ntg1-mediated double-stranded breakage (2.5 to 3% [of total mtDNA units] above the level in Δntg1 cells) (Fig. 2D) is sufficient to confer an extreme advantage for HS [ori5] [rho−] mtDNA inheritance, resulting in hypersuppressiveness. The fraction of concatemers in the entire mtDNA population of wild-type cells was calculated to be 78.9 ± 0.7%, and the mean length of the concatemers was >15 mtDNA genomic units (Fig. 4B). Thus, the termini of the concatemers were calculated to be less than the value equivalent to that 5% (78.9% of total mtDNA/15 units [in each concatemer]) of mtDNA unit has a double-stranded break. Then, the 2.5 to 3% increase in the fraction of mtDNA units containing a double-stranded break is not a minor increase. Moreover, it is well known that, in the absence of protection at its termini, linear DNA is subjected to extensive degradation. Thus, the nascent termini would immediately undergo DNA repair. Therefore, the 2.5 to 3% increase in the monomeric units containing a double-stranded break would be an apparent increase in the steady-state level, and the actual double-stranded breakage mediated by Ntg1 would occur at a much higher rate. Another possibility is that the termini of concatemers are protected by some means, such as the binding of specific proteins (42), and only nascent termini produced by Ntg1 are used in the initiation of mtDNA replication.
Thus, the double-stranded cleavage of ori5 mediated by Ntg1 is responsible, at least partially, for the full expression of hypersuppressiveness of HS [ori5] [rho−] mtDNA.
Ntg1 and Mhr1 proteins promote HS [ori5] [rho−] mtDNA concatemer formation.
Finally, we tested the assumption that the observed double-stranded breakage of ori5 is responsible for the initiation of rolling-circle replication of HS [ori5] [rho−] mtDNA. We analyzed the effects of mutations in NTG1, MHR1, and other genes at the level of concatemers, the product of rolling-circle replication, relative to monomers. In a one-dimensional electrophoretic analysis of whole cellular DNA from wild-type cells containing HS [ori5] [rho−] mtDNA, a dense, smeared band (representing concatemers) and a set of discrete bands (representing mtDNA circular monomers, dimers, and multimers) overlapped (Fig. 4B, top, lane 1). Resolution of these molecules by two-dimensional electrophoresis allowed concatemers and circular forms to be visualized as an arc and a set of discrete spots, respectively (Fig. 4A and B).
One concern with this analysis is that the majority of mtDNA might not enter the gel, perhaps because of the much larger size of the multimeric form and, thus, the signals would represent only a portion of the mtDNA in the sample. To assess this possibility, we used gel electrophoresis and Southern analysis to compare the signals from untreated mtDNA and mtDNA treated with a restriction endonuclease that cuts HS [ori5] [rho−] mtDNA once. Equal amounts of DNA from wild-type cells were loaded into the gel with and without digestion with BglII. After one-dimensional electrophoresis, the ratio of the recovered signals from two independent experiments was 10:6 ± 1 for undigested and BglII-digested DNA (Fig. 4B, top panel, lanes 1 and 2, respectively). On the other hand, in two independent experiments, 8 ± 5% of the mtDNA from the undigested DNA remained in the well (Fig. 4B, top panel, lane 1). These results suggest that some of the mtDNA was lost during BglII digestion but that most of the undigested mtDNA entered the gel and was recovered.
Another concern was that the dense, smeared band and the arc of mtDNA observed after one- and two-dimensional electrophoresis, respectively, represented the products of uncontrolled degradation of a much larger mtDNA species during preparation of the cellular DNA. Although this possibility cannot be ruled out, we are confident that the majority of the signals represent concatemers, rather than degradation products, as described below. In our previous studies, we showed by both one- and two-dimensional electrophoresis that the levels of the dense, smeared bands and the arcs correlated positively with the intracellular activity of the Mhr1 protein, which has no DNase activity, indicating that the signals reflected the homologous pairing activity of Mhr1. A straightforward explanation of these previous observations is that the smeared bands and arcs represent concatemers that are formed by Mhr1-dependent rolling-circle replication and this explanation was supported by label-and-chase experiments (29). Our previous studies revealed the reproducibility of the relative amounts of mtDNA concatemers and monomers, and these results were independent of one- or two-dimensional gel electrophoresis (29). Therefore, these results are not a consequence of the uncontrolled degradation of mtDNA species during preparation, and mtDNA species are not lost during the second phase of two-dimensional electrophoresis.
The signals from HS [ori5] [rho−] mtDNA species after two-dimensional gel electrophoresis indicated that the disruption of NTG1, as well as that of MHR1, greatly reduced the relative amounts of rolling-circle replication products (concatemers) and circular multimers (crossover homologous recombination products) to monomers, compared with samples from wild-type cells (compare Fig. 4C and D with B; quantification is in panel F). From these results, we conclude that double-stranded breakage of ori5 and homologous DNA pairing play a role, at least partially, in rolling-circle replication, as well as in crossover homologous recombination.
DISCUSSION
The requirement for replication-origin sequences and associated promoters for hypersuppressiveness has been attributed to the replication advantage conferred by promoter-dependent RNA priming, rather than to the segregation advantage ( 36) (see also the introduction). In contrast, it has been reported that RPO41 is dispensable for the biased inheritance of HS [rho−] mtDNA bearing ori5 or another replication origin (ori3) (33). Our findings provide a new explanation as to why a replication origin, but not RPO41, is required for hypersuppressiveness.
We previously showed that mtDNA concatemers are obligatory intermediates of the partitioning of [rho+] and [rho−] mtDNA and that concatemer formation is carried out primarily by Mhr1-dependent rolling-circle replication (29, 30). In this study, we have shown that Ntg1, in addition to Mhr1, is required for the full expression of hypersuppressiveness by HS [ori5] [rho−] mtDNA (Fig. 1). We have detected an ori5-specific double-stranded break in HS [ori5] [rho−] mtDNA (Fig. 2). The extent of breakage was decreased or enhanced by disruption or overexpression, respectively, of NTG1 (Fig. 2). The double-stranded break was still detected, at one-half to one-third of the wild-type level, in HS [ori5] [rho−] mtDNA isolated from cells lacking NTG1. However, the extent of the decrease in suppressiveness conferred by disruption of NTG1 correlates qualitatively with the reduction in double-stranded cleavage (Fig. 1 and 2). Finally, both Ntg1 and Mhr1 are required for maximal concatemer formation of HS [ori5] [rho−] mtDNA (Fig. 4).
As described in Results, it remains a possibility that the decrease in the copy number of HS [ori5] [rho−] mtDNA is a factor preventing hypersuppressiveness. While the disruption of NTG1 extensively decreased the amount of HS [ori5] [rho−] mtDNA concatemers (Fig. 4D and F), the disruption of NTG1 caused little effect on the amount of concatemers of [rho−] mtDNA without replication origin (mt-pMK2 DNA) (F. Ling, unpublished observation). This supports the explanation that the effect of the disruption of NTG1 on hypersuppressiveness is a result of a specific interaction of Ntg1 with ori5 that is required for concatemer formation.
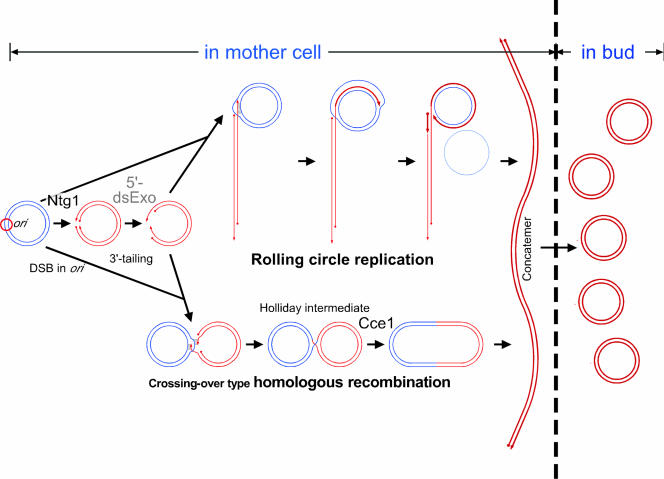
The observations in this study and previous ones are consistent with the model in which, as in the case of homologous recombination, a 3′ terminus of a single-stranded tail, derived from a double-stranded break at ori, is paired by the Mhr1 protein to circular mtDNA and used as a primer terminus to initiate rolling-circle mtDNA replication (Fig. 5).
FIG. 5.
Model for double-stranded break-induced and homologous DNA pairing-dependent rolling-circle replication and crossover-type homologous recombination of mtDNA. Ntg1 introduces a double-stranded break in a replication origin region of mtDNA. The double-stranded break is processed into two 3′-single-stranded tails by a putative 5′-double-stranded DNA exonuclease (5′-dsExo). If the Mhr1 protein pairs only one of the 3′-single-stranded tails with a homologous sequence of intact circular mtDNA to form a heteroduplex joint, and then rolling-circle replication is initiated and produces a concatemer (a linear head-to-tail array of multiple mitochondrial genomic units), as shown at the top. If Mhr1 pairs both 3′-single-stranded tails with a homologous sequence on the same intact circular mtDNA to form heteroduplex DNA joints, then a double-stranded break-repair process (52) is initiated to form crossover products (circular mtDNA multimers of discrete sizes), as shown at the bottom via Holliday intermediates as well as gene conversion products (not shown). The resolution of Holliday intermediates to crossover products requires the Cce1 protein. Double-stranded cleavage of multimeric circular DNA also produces a concatemer. Concatemers are essential intermediates for mtDNA partitioning and, upon transmission into a growing bud, the concatemer is processed into a monomeric circular form (29, 30). Complete loss of mtDNA requires the disruption of both MHR1 and CCE1, indicating that yeast mitochondria have another minor homologous-pairing activity (30). hpp, homologous pairing protein.
In this study we sequenced two newly isolated hypersuppressive [rho−] mtDNAs and found that both contain ori5. It was reported that among 17 hypersuppressive [rho−] mtDNA isolated, 7 had ori5, 5 had ori3, 4 had ori2, and none had ori1 (48). This may reflect the sensitivity of ori5 to the in vivo double-stranded cleavage. A critical difference between rolling-circle replication and homologous recombination lies in the manner of homologous DNA pairing: in rolling-circle replication, pairing involves one of the tails derived from a double-stranded break, whereas homologous recombination involves pairing of both of the tails with a common intact circular mtDNA (Fig. 5). It is generally observed at hot spots for meiotic homologous recombination that double-stranded cleavage is detected at the initiation site in only a tiny fraction of double-stranded DNA (50). The small amount of double-stranded cleavage within ori5 is reminiscent of the initiation of meiotic homologous recombination.
Ntg1, which excises base-derived lesions from oxidatively damaged DNA, has AP-lyase (previously described as endonuclease) activity (15, 45; for review, see reference 26). From its known activity, Ntg1 is expected to produce single-stranded breaks (nicks) rather than double-stranded breaks. However, in vitro experiments with purified Ntg1 or its inactive derivative (CΔntg1) as a negative control and DNA from alloxan-treated cells (Fig. 3) indicate that Ntg1 alone can introduce a double-stranded break near the promoter end of ori5 by recognizing specific oxidative damage at the site.
The Ntg1 protein purified from NTG1-overexpressing E. coli cells recognizes oxidative damage and generates single-stranded breaks, but not double-stranded breaks, in plasmid DNA (lacking an mtDNA replication origin) isolated from E. coli cells, as previously reported (see Fig. S1 in the supplemental material). Thus, the Ntg1 preparation used in this study was not contaminated with an unidentified endonuclease that causes double-stranded breaks.
Disruption of OGG1, which encodes another mtDNA repair enzyme, had no effect on double-stranded cleavage of ori5 (Fig. 2). Whereas Ntg1 recognizes various oxidized bases, Ogg1 primarily recognizes products derived from guanine (see reference 26 for review). The region identified as the in vivo double-stranded break site consists of mostly A and T except for the GC cluster C. Thus, the substrate specificities of Ntg1 and Ogg1 and their differential effects at ori5 suggest that cleavage occurs outside the GC cluster C (Fig. 1C).
The site-specific (or preferential) sensitization to oxidation at the mtDNA ori5 locus, including the promoter, would be conferred by the specific melting or disruption of the normal double-stranded structure of ori5. DNA often melts at promoters (38, 56) or replication origins (11, 40) through specific interactions with protein factors, and single-stranded DNA is generally much more sensitive to DNA-damaging agents than is double-stranded DNA (4, 35). Both strands would be oxidized simultaneously at the melted region, which would make them susceptible to Ntg1 protein cleavage, resulting in a double-stranded break. Further analyses of both the structure of mtDNA at ori5 and of the factors interacting with the replication origin will be required to understand specific cleavage at ori5.
The higher density of replication origins in HS [rho−] mtDNA has been used to explain the replication advantage of HS [rho−] mtDNA over [rho+] mtDNA in diploid cells. Hypersuppressiveness is completely eliminated by a defective mutation in another recombination-related gene, CCE1 (MGT1) (57). The Cce1 protein is a recombination junction (Holliday junction)-resolving endo-DNase (17). Lockshon et al. proposed that the apparent dependence of hypersuppressiveness on CCE1 results from the marked segregation disadvantage of HS [rho−] mtDNA in cce1 cells, caused by the larger proportion of molecules linked together by recombination junctions in the absence of Cce1 (31).
Our previous studies supported a model in which mtDNA is inherited as concatemers that are immediately processed into circular mtDNA monomers after transmission into buds (29, 30). It is likely that randomly selected concatemers are transmitted and that the amount of mtDNA concatemer transmitted to daughter cells over a fixed time is independent of genome unit size. The transmitted copy number of HS [ori5] [rho−] mtDNA (unit size, 1.1 kb) within a given time is almost 80-fold more than that of the [rho+] mtDNA (unit size, 80 kb) (Fig. 2), which results in a supplementary segregation advantage for HS [rho−] mtDNA over [rho+] mtDNA.
Recently, evidence of recombination between human paternal and maternal mtDNAs has been reported, and recombination hot spots have been identified. It is noteworthy that the recombination hot spots are located near or overlap with mtDNA replication origins (10, 25). Together, these results and our findings suggest the novel possibility that, in both human and yeast cells, mtDNA replication is initiated by a common initiation event for mtDNA recombination. Homoplasmy and the biased inheritance of deleted mtDNA (enhancing heteroplasmy) both depend on the recombination-related initiation of mtDNA replication in yeast, as we have shown previously (29) and in this report.
Supplementary Material
Acknowledgments
This study was supported in part by a grant from the Bioarchitect Research Program of RIKEN to F.L. and T.S., by a grant from the Life Science Foundation of Japan to F.L., by a grant for CREST from JST, a Grant-in-Aid from the Ministry of Education, Sports, Culture, Science and Technology of Japan to T.S., and a Grant-in-Aid (18570168) from the Ministry of Education, Sports, Culture, Science and Technology of Japan to F.L.
Footnotes
Published ahead of print on 20 November 2006.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Backert, S., P. Dorfel, R. Lurz, and T. Borner. 1996. Rolling-circle replication of mitochondrial DNA in the higher plant Chenopodium album (L.). Mol. Cell. Biol. 16:6285-6294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baldacci, G., and G. Bernardi. 1982. Replication origins are associated with transcription initiation sequences in the mitochondrial genome of yeast. EMBO J. 1:987-994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baldacci, G., B. Cherif-Zahar, and G. Bernardi. 1984. The initiation of DNA replication in the mitochondrial genome of yeast. EMBO J. 3:2115-2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beletskii, A., and A. S. Bhagwat. 1996. Transcription-induced mutations: increase in C to T mutations in the nontranscribed strand during transcription in Escherichia coli. Proc. Natl. Acad. Sci. USA 93:13919-13924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bendich, A. J. 1996. Structural analysis of mitochondrial DNA molecules from fungi and plants using moving pictures and pulsed-field gel electrophoresis. J. Mol. Biol. 255:564-588. [DOI] [PubMed] [Google Scholar]
- 6.Blanc, H. 1984. Two modules from the hypersuppressive rho− mitochondrial DNA are required for plasmid replication in yeast. Gene 30:47-61. [DOI] [PubMed] [Google Scholar]
- 7.Blanc, H., and B. Dujon. 1980. Replicator regions of the yeast mitochondrial DNA responsible for suppressiveness. Proc. Natl. Acad. Sci. USA 77:3942-3946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brewer, B. J., and W. L. Fangman. 1987. The localization of replication origins on ARS plasmids in S. cerevisiae. Cell 51:463-471. [DOI] [PubMed] [Google Scholar]
- 9.Bromme, H. J., H. Ebelt, D. Peschke, and E. Peschke. 1999. Alloxan acts as a prooxidant only under reducing conditions: influence of melatonin. Cell Mol. Life Sci. 55:487-493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.D'Aurelio, M., C. D. Gajewski, M. T. Lin, W. M. Mauck, L. Z. Shao, G. Lenaz, C. T. Moraes, and G. Manfredi. 2004. Heterologous mitochondrial DNA recombination in human cells. Hum. Mol. Genet. 13:3171-3179. [DOI] [PubMed] [Google Scholar]
- 11.Dean, F. B., J. A. Borowiec, T. Eki, and J. Hurwitz. 1992. The simian virus-40 T-Antigen double hexamer assembles around the DNA at the replication origin. J. Biol. Chem. 267:14129-14137. [PubMed] [Google Scholar]
- 12.de Zamaroczy, M., R. Marotta, G. Faugeron-Fonty, R. Goursot, M. Mangin, G. Baldacci, and G. Bernardi. 1981. The origins of replication of the yeast mitochondrial genome and the phenomenon of suppressivity. Nature 292:75-78. [DOI] [PubMed] [Google Scholar]
- 13.de Zamaroczy, M., G. Faugeron-Fonty, G. Baldacci, R. Goursot, and G. Bernardi. 1984. The ori sequences of the mitochondrial genome of a wild-type yeast strain: number, location, orientation and structure. Gene 32:439-457. [DOI] [PubMed] [Google Scholar]
- 14.Dujon, B. 1981. Mitochondrial genetics and function, p. 505-635. In J. N. Strathern, E. W. Jones, and J. R. Broach (ed.), The molecular biology of the yeast Saccharomyces: life cycle and inheritance. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 15.Eide, L., M. Bjoras, M. Pirovano, I. Alseth, K. G. Berdal, and E. Seeberg. 1996. Base excision of oxidative purine and pyrimidine DNA damage in Saccharomyces cerevisiae by a DNA glycosylase with sequence similarity to endonuclease III from Escherichia coli. Proc. Natl. Acad. Sci. USA 93:10735-10740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Enquist, L. W., and A. Skalka. 1973. Replication of bacteriophage lambda DNA dependent on the function of host and viral genes. I. Interaction of red, gam and rec. J. Mol. Biol. 75:185-212. [DOI] [PubMed] [Google Scholar]
- 17.Ezekiel, U. R., and H. P. Zassenhaus. 1993. Localization of a cruciform cutting endonuclease to yeast mitochondria. Mol. Gen. Genet. 240:414-418. [DOI] [PubMed] [Google Scholar]
- 18.Fangman, W. L., J. W. Henly, and B. J. Brewer. 1990. RPO41-independent maintenance of [rho−] mitochondrial DNA in Saccharomyces cerevisiae. Mol. Cell. Biol. 10:10-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frei, B., K. H. Winterhalter, and C. Richter. 1985. Mechanism of alloxan-induced calcium release from rat liver mitochondria. J. Biol. Chem. 260:7394-7401. [PubMed] [Google Scholar]
- 20.Holt, I. J., A. E. Harding, and J. A. Morgan-Hughes. 1988. Deletions of muscle mitochondrial DNA in patients with mitochondrial myopathies. Nature 331:717-719. [DOI] [PubMed] [Google Scholar]
- 21.Ikeda, S., T. Biswas, R. Roy, T. Izumi, I. Boldogh, A. Kurosky, A. H. Sarker, S. Seki, and S. Mitra. 1998. Purification and characterization of human NTH1, a homolog of Escherichia coli endonuclease III. Direct identification of Lys-212 as the active nucleophilic residue. J. Biol. Chem. 273:21585-21593. [DOI] [PubMed] [Google Scholar]
- 22.Ito, H., Y. Fukuda, K. Murata, and A. Kimura. 1983. Transformation of intact yeast cells treated with alkali cations. J. Bacteriol. 153:163-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaiser, C., S. Michaelis, and A. Mitchell. 1994. Methods in yeast genetics: a Cold Spring Harbor Laboratory course manual. Cold Spring Harbor Laboratory Press, Plainview, N.Y.
- 24.Kleff, S., B. Kemper, and R. Sternberg. 1992. Identification and characterization of yeast mutants and the gene for a cruciform cutting endonuclease. EMBO J. 11:699-704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kraytsberg, Y., M. Schwartz, T. A. Brown, K. Ebralidse, W. S. Kunz, D. A. Clayton, J. Vissing, and K. Khrapko. 2004. Recombination of human mitochondrial DNA. Science 304:981. [DOI] [PubMed] [Google Scholar]
- 26.Krokan, H. E., R. Standal, and G. Slupphaug. 1997. DNA glycosylases in the base excision repair of DNA. Biochem. J. 325:1-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ling, F., F. Makishima, N. Morishima, and T. Shibata. 1995. A nuclear mutation defective in mitochondrial recombination in yeast. EMBO J. 14:4090-4101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ling, F., H. Morioka, E. Ohtsuka, and T. Shibata. 2000. A role for MHR1, a gene required for mitochondrial genetic recombination, in the repair of damage spontaneously introduced in yeast mtDNA. Nucleic Acids Res. 28:4956-4963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ling, F., and T. Shibata. 2004. Mhr1p-dependent concatemeric mitochondrial DNA formation for generating yeast mitochondrial homoplasmic cells. Mol. Biol. Cell 15:310-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ling, F., and T. Shibata. 2002. Recombination-dependent mtDNA partitioning. In vivo role of Mhr1p to promote pairing of homologous DNA. EMBO J. 21:4730-4740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lockshon, D., S. G. Zweifel, L. L. Freeman-Cook, H. E. Lorimer, B. J. Brewer, and W. L. Fangman. 1995. A role for recombination junctions in the segregation of mitochondrial DNA in yeast. Cell 81:947-955. [DOI] [PubMed] [Google Scholar]
- 32.Longtine, M. S., A. McKenzie III, D. J. Demarini, N. G. Shah, A. Wach, A. Brachat, P. Philippsen, and J. R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953-961. [DOI] [PubMed] [Google Scholar]
- 33.Lorimer, H. E. 2002. 2D gel electrophoresis of mtDNA. Methods Mol. Biol. 197:187-196. [DOI] [PubMed] [Google Scholar]
- 34.Lorimer, H. E., B. J. Brewer, and W. L. Fangman. 1995. A test of the transcription model for biased inheritance of yeast mitochondrial DNA. Mol. Cell. Biol. 15:4803-4809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lutgerink, J. T., E. van den Akker, D. Pachen, E. J. Smeets, P. van Dijk, J. M. Aubry, H. Joenje, M. V. Lafleur, and J. Retel. 1993. Singlet oxygen-induced DNA damage: product analysis, studies of biological consequences and characterization of mutations. IARC Sci. Publ. 124:115-125. [PubMed] [Google Scholar]
- 36.MacAlpine, D. M., J. Kolesar, K. Okamoto, R. A. Butow, and P. S. Perlman. 2001. Replication and preferential inheritance of hypersuppressive petite mitochondrial DNA. EMBO J. 20:1807-1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maleszka, R., P. J. Skelly, and G. D. Clark-Walker. 1991. Rolling circle replication of DNA in yeast mitochondria. EMBO J. 10:3923-3929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Matsunaga, M., and J. A. Jaehning. 2004. Intrinsic promoter recognition by a “core” RNA polymerase. J. Biol. Chem. 279:44239-44242. [DOI] [PubMed] [Google Scholar]
- 39.McEntee, K., G. M. Weinstock, and I. R. Lehman. 1979. Initiation of general recombination catalyzed in vitro by the recA protein of Escherichia coli. Proc. Natl. Acad. Sci. USA 76:2615-2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mukherjee, S., I. Patel, and D. Bastia. 1985. Conformational changes in a replication origin induced by an initiator protein. Cell 43:189-197. [DOI] [PubMed] [Google Scholar]
- 41.Noirot, P., and R. D. Kolodner. 1998. DNA strand invasion promoted by Escherichia coli RecT protein. J. Biol. Chem. 273:12274-12280. [DOI] [PubMed] [Google Scholar]
- 42.Oldenburg, D. J., and A. J. Bendich. 2001. Mitochondrial DNA from the liverwort Marchantia polymorpha: circularly permuted linear molecules, head-to-tail concatemers, and a 5′ protein. J. Mol. Biol. 310:549-562. [DOI] [PubMed] [Google Scholar]
- 43.Ozawa, T. 1997. Genetic and functional changes in mitochondria associated with aging. Physiol. Rev. 77:425-464. [DOI] [PubMed] [Google Scholar]
- 44.Rybalchenko, N., E. I. Golub, B. Bi, and C. M. Radding. 2004. Strand invasion promoted by recombination protein beta of coliphage lambda. Proc. Natl. Acad. Sci. USA 101:17056-17060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Senturker, S., P. Auffret van der Kemp, H. J. You, P. W. Doetsch, M. Dizdaroglu, and S. Boiteux. 1998. Substrate specificities of the ntg1 and ntg2 proteins of Saccharomyces cerevisiae for oxidized DNA bases are not identical. Nucleic Acids Res. 26:5270-5276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shibata, T., C. DasGupta, R. P. Cunningham, and C. M. Radding. 1979. Purified Escherichia coli recA protein catalyzes homologous pairing of superhelical DNA and single-stranded fragments. Proc. Natl. Acad. Sci. USA 76:1638-1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Silberstein, Z., S. Maor, I. Berger, and A. Cohen. 1990. Lambda red-mediated synthesis of plasmid linear multimers in Escherichia coli K12. Mol. Gen. Genet. 223:496-507. [DOI] [PubMed] [Google Scholar]
- 48.Skelly, P. J., and G. D. Clark-Walker. 1990. Conversion at large intergenic regions of mitochondrial DNA in Saccharomyces cerevisiae. Mol. Cell. Biol. 10:1530-1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stohl, L. L., and D. A. Clayton. 1992. Saccharomyces cerevisiae contains an RNase MRP that cleaves at a conserved mitochondrial RNA sequence implicated in replication priming. Mol. Cell. Biol. 12:2561-2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sun, H., D. Treco, N. P. Schultes, and J. W. Szostak. 1989. Double-strand breaks at an initiation site for meiotic gene conversion. Nature (London) 338:87-90. [DOI] [PubMed] [Google Scholar]
- 51.Sung, P. 1994. Catalysis of ATP-dependent homologous DNA pairing and strand exchange by yeast RAD51 protein. Science 265:1241-1243. [DOI] [PubMed] [Google Scholar]
- 52.Szostak, J. W., T. L. Orr-Weaver, R. J. Rothstein, and F. W. Stahl. 1983. The double-strand-break repair model for recombination. Cell 33:25-35. [DOI] [PubMed] [Google Scholar]
- 53.Van Dyck, E., and D. A. Clayton. 1998. Transcription-dependent DNA transactions in the mitochondrial genome of a yeast hypersuppressive petite mutant. Mol. Cell. Biol. 18:2976-2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wallace, D. C. 1999. Mitochondrial diseases in man and mouse. Science 283:1482-1488. [DOI] [PubMed] [Google Scholar]
- 55.You, H. J., R. L. Swanson, C. Harrington, A. H. Corbett, S. Jinks-Robertson, S. Senturker, S. S. Wallace, S. Boiteux, M. Dizdaroglu, and P. W. Doetsch. 1999. Saccharomyces cerevisiae Ntg1p and Ntg2p: broad specificity N-glycosylases for the repair of oxidative DNA damage in the nucleus and mitochondria. Biochemistry 38:11298-11306. [DOI] [PubMed] [Google Scholar]
- 56.Young, B. A., T. M. Gruber, and C. A. Gross. 2004. Minimal machinery of RNA polymerase holoenzyme sufficient for promoter melting. Science 303:1382-1384. [DOI] [PubMed] [Google Scholar]
- 57.Zweifel, S. G., and W. L. Fangman. 1991. A nuclear mutation reversing a biased transmission of yeast mitochondrial DNA. Genetics 128:241-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.