Abstract
The Drosophila gypsy insulator contains binding sites for the Suppressor of Hairy-wing [Su(Hw)] protein. Enhancer and silencer blocking require Su(Hw) recruitment of Mod(mdg4)-67.2, a BTB/POZ domain protein that interacts with Su(Hw) through a carboxyl-terminal acidic domain. Here we conducted mutational analyses of the Mod(mdg4)-67.2 BTB domain. We demonstrate that this domain is essential for insulator function, in part through direction of protein dimerization. Our studies revealed the presence of a second domain (DD) that contributes to Mod(mdg4)-67.2 dimerization when the function of the BTB domain is compromised. Additionally, we demonstrate that mutations in amino acids of the charged pocket in the BTB domain that retain dimerization of the mutated protein cause a loss of insulator function. In these cases, the mutant proteins failed to localize to chromosomes, suggesting a role for the BTB domain in chromosome association. Interestingly, replacement of the Mod(mdg4)-67.2 BTB domain with the GAF BTB domain produced a nonfunctional protein. Taken together, these data suggest that the Mod(mdg4)-67.2 BTB domain confers novel activities to gypsy insulator function.
Enhancer-mediated promoter activation is a fundamental mechanism of gene regulation in eukaryotes (10, 44). Recently, sequences in different organisms have been identified that constrain enhancer action. These elements, known as insulators, block communication between an enhancer and promoter only when the insulator is positioned between these regulatory elements. Similarly, insulators prevent silencer interactions with promoters (6, 10, 30, 44, 45). The properties of insulators are exemplified by the gypsy insulator that originally was found in the gypsy transposable element (19, 24).
Genetic and molecular approaches have led to the identification and characterization of three proteins, Suppressor of Hairy wing [Su(Hw)], Mod(mdg4)-67.2, and CP190, that are required for the activity of the gypsy insulator (6, 36). Su(Hw) is a zinc finger protein that binds 12 directly repeated copies of a short sequence motif in the gypsy insulator (9, 42). In addition, Su(Hw) has two acidic domains located at the amino (N) and carboxyl (C) termini of the protein and a C-terminal enhancer-blocking region that is essential for insulation (23, 29). The mod(mdg4) gene, also known as E(var)3-93D, encodes a large set of protein isoforms with specific functions in regulating the chromatin structure of different genes (3). All isoforms encoded by mod(mdg4) contain a BTB/POZ domain and a glutamine-rich (Q) region in the N terminus (3, 7). The BTB (broad complex, tramtrack, bric-a-brac) or POZ (poxvirus and zinc finger) domain identifies a large family of proteins in organisms from yeast to humans (43, 47). This domain functions as a protein interaction domain that facilitates homodimer (2, 33, 34) and heterodimer formation as well as oligomerization (11, 28, 37). One of the mod(mdg4)-encoded protein isoforms, Mod(mdg4)-67.2, interacts with the enhancer-blocking domain of the Su(Hw) protein (12, 20) through a C-terminal acidic domain. This domain is affected in two viable mutations mod(mdg4)u1 and mod(mdg4)T6 (12, 15). The third component of the insulator complex, CP190, also contains a BTB domain (38). It was suggested that Mod(mdg4)-67.2 and CP190 interact through their BTB domains.
The mod(mdg4)u1 and mod(mdg4)T6 mutations have varying effects on insulator function, resulting in partial restoration of enhancer-promoter communication in some cases, while transforming the insulator into a silencer in others (4, 12, 13, 14, 15, 22). The domains of the Mod(mdg4)-67.2 protein required for the insulator and antirepression activity are not determined. Although the essential role of the BTB domain for Mod(mdg4)-67.2 activity was predicted in the previous studies, this postulate has not been proven (12, 20). Here we examined the role of the BTB domain in the functional activities of the Mod(mdg4)-67.2 protein.
The structure of the BTB domain has been examined for mammalian transcriptional repressors PLZF and Bcl-6 (33, 34). The high degree of sequence identity between the BTB domains of Mod(mdg4), Bcl-6, and PZLF (1) allowed us to predict key residues of the Mod(mdg4)-67.2 BTB domain to test for function. In these studies, we used a combination of in vitro and in vivo analyses to define the regions of Mod(mdg4)-67.2 required for insulator function. Using yeast two-hybrid analyses, we define a second homodimerization domain that substitutes for the BTB domain when its homodimerization capacity is reduced by mutation. We find that a complete absence of Mod(mdg4)-67.2 homodimerization results in the loss of insulator activity. Finally, our studies indicate that the BTB domain of Mod(mdg4)-67.2 makes specific contributions to the formation of a functional insulator complex, as a fusion protein containing the GAF BTB domain and the Mod(mdg4)-67.2 C terminus, while retaining the ability to dimerize, does not reconstitute enhancer blocking.
MATERIALS AND METHODS
Drosophila melanogaster strains, plasmid constructions, germ line transformation and genetic crosses.
All flies were maintained at 25°C on standard yeast medium. The plasmid constructions are described in the supplemental material. The transposon constructs, together with P25.7wc, a P element with defective inverted repeats used as a transposase source, were injected into y ac w1118 preblastoderm embryos (27). The generation of transgenic lines and introduction into the mod(mdg4)u1 or mod(mdg4)T6 background were done as described previously (14).
The effects of the various Mod(mdg4) proteins produced from homozygous expression vectors were scored independently by two authors. To express transgenes regulated by the hsp70 promoter, flies homozygous for the construct were heat shocked for 1 h every day from the second larval to middle pupal stages of fly development. To determine the yellow, cut, and sc phenotypes, we examined 3- to 5-day-old males developing at 25°C. For yellow phenotypes, wild-type expression in abdominal cuticle, wings, and bristles was assigned an arbitrary score of 5, while the absence of y expression was ranked 1. Flies with the previously characterized y allele were used as a reference to determine y pigmentation levels. The representative abdomens and wings displayed in the figures were selected by arranging several abdomens or wings in order of the severity of their mutant phenotype and selecting the average mutant phenotype to photograph.
Mutation of Mod(mdg4)-67.2.
Mod(mdg)-67.2 cDNA cloned in pGEX2T was kindly provided by D. Dorsett. To make mutations in BTB, Mod(mdg)-67.2 BTB cDNA was subcloned in pBluescript II SK+ digested with EcoRI and DraII. One half of the BTB domain (first 122 bp of BTB domain) was PCR amplified with a primer containing one substitution and M13 reverse sequencing primer. The second half of the BTB domain was amplified with a primer complementary to the BTB domain (in case of one amino acid substitution) or with a primer containing the second substitution (in case of double substitutions) and M13 forward sequencing primer. PCR products were digested with DraII and EcoRI and cloned in pBluescript II SK+ digested with EcoRI and DraII. The following primers were used to produce mutations: S25A, 5′-CATAGCGCCTCGTGG-3′; D33N, 5′-GGCCCTCGGCGGCCAGCGAGACGTTCACC-3′; H46D, 5′-AAATAGTGAAGGCCGACCG-3′; R47Q, 5′-AAATAGTGAAGGCCCACCAATTG-3′. To prepare Mod(mdg4)ΔQ, Mod(mdg)-67.2 cDNA in pGEX2T was digested with BlpI, filled in with Klenow fragment, and self-ligated. The BTB domain of GAF was PCR amplified from GAF cDNA in pET3 with the primer 5′-AATACGACTCACTATAG-3′ and 5′-CCGCGGCGGTGCCAGTCCCTGAATG-3′ containing a SacII site. The PCR product was digested with SacII and ligated in vector pSK containing Mod(mdg4)-67.2 cDNA, which was first digested with EcoRI, blunted, and digested with SacII.
Construction of plasmids expressing Mod(mdg4)-67.2 and its mutant derivatives in flies.
The Su(Hw) promoter (29) was kindly provided by D. Dorsett. To construct transposons, Mod(mdg4)-containing mutant BTB domains were cloned in pCaSpeR4 under control of the hsp70 promoter in the case of the H46D, H46D/D33N, and D33N/S25A mutants or under control of the Su(Hw) promoter in the case of the R47Q, D33N, R47Q/D33N, and Mod(mdg4)Gaf mutants. Vectors were digested with EcoRI and BamHI and ligated with the 1.8-kb EcoRI-BamHI fragment of Mod(mdg)-67.2 containing mutations described above.
Construction of plasmids for in vitro experiments.
For protein expression, Mod(mdg4) was cloned in frame with a six-His tag in pET23a (Novagene). pGEX2TMod was digested with BamHI, and the end was filled in with Klenow fragment. After EcoRI digestion, the gene fragment was cloned in pET23a that was first digested with EagI and then filled in and digested with EcoRI, producing pET23mod. To make an expression vector with mutant Mod(mdg4) proteins, we replaced the EcoRI-Eco72I fragment of pET23mod with the same fragment of the pCaSpeR4 construct containing mutations.
Construction of plasmids for yeast two-hybrid system.
For yeast two-hybrid assays, the coding regions described above were cloned in vector pGBT9 and pGAD424 (Clontech) using EcoRI and BamHI sites. Su(Hw) was PCR amplified from pGEM3ZfSu(Hw) plasmid with primers 5′-AATGAGTGCCTCCAAGGAGGGC-3′ (upstream) and 5′-CCGTCGACTCAAGCTTTCTCTTGTTC-3′ (downstream) containing the SalI site. The PCR product was digested with SalI and cloned in vectors pGBT9 and pGAD424 digested with SmaI and SalI. Su(Hw) lacking the C-terminal end was PCR amplified as previously described, but the next downstream primer used was 5′-TTTGTCGACTTCGCCTGTGAC-3′ (also with the SalI site). The PCR product was digested with SalI and cloned in vector pGBT9 digested with SmaI and SalI, so we have pGBT9Su(Hw). To clone only the domain of SuHw interacting with Mod(mdg4), pGBT9Su(Hw) was digested with EcoRI and SalI and ligated with pGBT9 and pGAD424 vectors digested with the same enzymes. To make a plasmid carrying the Gal4-activating domain at the C-terminal end of the fusion protein, all pGAD plasmids with mutated or wild-type Mod(mdg4) were digested with EcoRI and then HindIII, filled in by Klenow, and self-ligated [the resulting plasmid was called pGAD(−)]. The activation domain was PCR amplified from pGAD424 with primers 5′-AGCGGATCCATGGATAAAGCGG-3′, containing a BamHI site, and 5′-GACAGATCTCTCTTTTTTTGGGTTTGGT-3′, containing a BglII site, digested with BamHI and BglII, and ligated with pGAD(−) digested with the same endonuclease. These operations produced plasmids containing fusions of all mutant forms and the wild type of Mod(mdg4) with the activation domain of Gal4 on the C-terminal end, designated pGDA. CP-1901-765 was PCR amplified with primers 5′-CATGGGTGAAGTCAAGTC-3′ and 5′-TTCAGATCTTTCCAGGTTGTCAATGG-3′, containing the BglII site. This PCR product was cloned in pGDA vector digested with EcoRI, filled in by Klenow and then BamHI. To prepare Mod(mdg4)1-273, PCR amplification from pGBT Mod 67.2 with the help of 5′-ATAGGATCCTTGCGGCACAAGTTG-3′, containing the BamHI site, and GAL DNA binding primers was done. The PCR product was digested with EcoRI and BamHI enzymes and cloned in either a pGBT or pGDA vector. PCR amplification with one primer 5′-TATGGATCCCTTCTTCTTGTTCTG-3′ containing a BamHI site and another primer containing EcoRI were used to prepare Mod(mdg4)234-610 (5′-TATGAATTCATGTCCTCGAGCGCC-3′), Mod(mdg4)317-610 (5′-ACCGAATTCATGTACTCTGAAGAC-3′), and Mod(mdg4)390-610 (5′-ATAGAATTCATGGTCGACACCAGCGGG-3′) mutants. PCR products were cloned in either pGBT or pGDA vector as previously described. Deletion of DD was done by PCR amplification (primers 5′-ATAAGGCCTGGGCAATTCCATGGGGAG-3′ and 5′-ATAAGGCCTGTCGACACCAGCGGG-3′) following StuI digestion and self-ligation of the resulting plasmid.
Two-hybrid and in vitro interactions.
Two-hybrid assays were carried out using yeast strain pJ694A, plasmids, and protocols obtained from Clontech (Palo Alto, CA). For growth assays, plasmids were transformed into yeast strain pJ694A by the lithium acetate method as described by the manufacturer and plated on media lacking tryptophan and leucine. After 2 days of growth at 30°C, the cells were plated on selective media lacking tryptophan, leucine, histidine, and adenine, and growth was compared. Liquid culture assays were performed according to protocols described in the yeast protocols handbook (Clontech).
To express His-tagged proteins in vitro, the vector pET23mod was transformed in Escherichia coli strain BL21(DE3), grown in LB with ampicillin at 37°C to an optical density at 600 nm (OD600) of 0.5, and then induced with 1 mmol of isopropyl-β-d-thiogalactopyranoside (IPTG), followed by growth at 18°C for 6 h. Protein purification was done with Talon superflow resin (Clontech) under native conditions according to the manufacturer's instructions. To express N-terminally glutathione S-transferase (GST)-tagged Mod(mdg4) protein, the pGEX2Tmod plasmid was transferred to E. coli strain BL21(DE3) and induced as described before. Purification was performed on glutathione-Sepharose 4B (Amersham) according to the manufacturer's instructions.
To study homodimerization in vitro, we performed polyacrylamide gel electrophoresis (PAGE) of six-His-tagged protein under native conditions. Gel electrophoresis in 7% gels was done in Tris-glycine buffer for 3 h at 20 V/cm. The proteins were blotted on a polyvinylidene difluoride membrane, incubated with primary antibody specific for the Mod(mdg4) 67.2 isoform, and developed with the ECL-plus kit (Amersham).
To investigate the possibility of heteromultimerization, we performed GST pull-down experiments. GST-Mod(mdg4) protein was incubated with glutathione-Sepharose 4B beads in binding buffer (20 mM HEPES-KOH, pH 7.6, 200 mM KCl, 2.5 mM MgCl, 10% glycerol, and 0.05% NP-40) for 2 h. Beads were blocked in 5% bovine serum albumin for 1 h and incubated with six-His-tagged protein for 3 h. After incubation, the beads were washed three times in wash buffer (10 mM Tris-HCl, pH 7.5, 1 mM EDTA, 0.2% NP-40, 400 mM NaCl). Washed beads were boiled in Laemmli buffer and separated on an 8% sodium dodecyl sulfate-polyacrylamide gel. The proteins were blotted on a polyvinylidene difluoride membrane, which was then incubated with anti-six-His antibody (Amersham).
Immunofluorescence analyses.
Antibodies against residues 403 to 610 of the Mod(mdg4)-67.2 were generated in chickens. Cy3-conjugated anti-chicken antibody (1:500; Chemicon) was used as a secondary antibody. Fixation and squashing of salivary glands and antibody staining were performed as originally described by Platero (40). Antibodies to lamin were generated in rabbits or mice and detected by secondary Cy5-conjugated antibody. Antibodies to Su(Hw) protein were generated in rabbits and detected by secondary fluorescein isothiocyanate-conjugated antibody. Imaginal disk staining was performed as described previously (31) with the same antibody.
RESULTS
Design of mutations in the BTB domain of the Mod(mdg4)-67.2 protein.
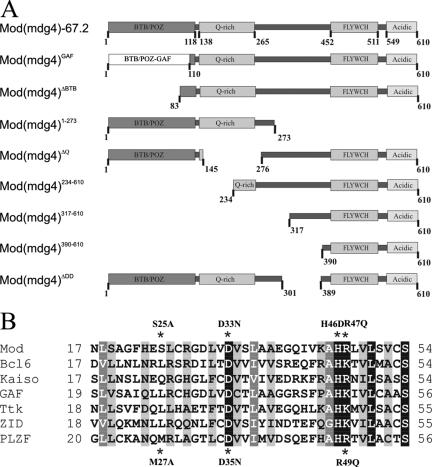
To examine the role of the BTB domain in Mod(mdg4)-67.2 activity, we made several derivatives and tested the function of the resulting protein. First, we deleted 83 amino acids from the BTB domain [Mod(mdg4)ΔBTB]. These amino acids represent the most conserved and functionally important region of the BTB domain (33). Second, we made point mutations in critical amino acids within the charge pocket (Fig. 1). Melnick et al. (33, 34) identified two conserved charged residues in the BTB domain, an aspartate at position 35 and arginine at position 49 (Fig. 1B). Each monomer of the BTB dimer contributes a wall of the pocket containing the D35 (negatively charged) and R49 (positively charged) residues, leading to the formation of a coordinately charged pocket containing two positive and two negative charges. Since Mod(mdg4)-67.2 has both conserved residues, we made the same single, ModD33N (aspartate to asparagine at position 33) and ModR47Q (arginine to glutamine at position 47), and double, ModD33N/R47Q, substitutions. Third, we changed histidine 46, which is the most conserved residue among BTB domains (42), to an acidic aspartate in the ModH46D mutant. Histidine 46 is not involved in formation of the charge pocket. We also made the double mutant ModD33N/H46D in which the alteration in charge by the H46D substitution is compensated by the neutralization of a negative charge in the D33N substitution. Finally, we constructed a BTB swap derivative. Read et al. (41) demonstrated that the BTB domain of the Mod(mdg4) protein substituted for that of GAF in transcription stimulation. To determine whether the GAF BTB domain is functionally equivalent to the Mod(mdg4) BTB domain, we replaced the first 108 residues of Mod(mdg4) with the first 122 residues of the GAF protein, retaining the position of the BTB domain with respect to the GAF protein, to form Mod(mdg4)Gaf (Fig. 1A).
FIG. 1.
Mod(mdg4)-67.2 protein domains and sequence comparisons. (A) Schematic representation of the Mod(mdg4)-67.2 protein. Mod(mdg4)-67.2 contains discrete functional domains, including the N-terminal BTB domain, glutamine-rich domain (Q-rich), conserved Cys2His2 motif, named FLYWCH motif (8), and the C-terminal acidic domain that interacts with the Su(Hw) protein. The structure of the Mod(mdg4)-67.2, Mod(mdg4)Gaf, Mod(mdg4) ΔBTB, Mod(mdg4) ΔQ, Mod(mdg4) ΔDD, Mod(mdg4)1-273, Mod(mdg4)234-610, Mod(mdg4)317-610, and Mod(mdg4)390-610 proteins are shown. (B) Sequence alignment of the N terminus of the Mod(mdg4)-67.2 BTB domain with other BTB domains. This region of the BTB corresponds mainly to the charged pocket. Darker shading indicates more highly conserved residues. Conserved residues selected for mutational analysis are marked with an asterisk. The boxes indicate the charged residues in the pocket.
Study of dimerization of the Mod(mdg4) mutants.
Each of the Mod(mdg4) mutants was tested in the yeast two-hybrid system for its ability to interact with Su(Hw). Recently we found that the acidic domains and DNA binding region of Su(Hw) partially repress transcription in yeast (32), complicating the interpretation of results obtained using this system. For this reason, we used a truncated version of the Su(Hw) protein that contains only the Mod(mdg4)-interacting domain [Su(Hw)MID], Su(Hw)MID and Mod(mdg4)-67.2 were fused in frame with either the yeast GAL4 DNA binding domain (GAL4BD) or activation domain (GAL4AD). As expected, Su(Hw)MID interacts strongly with Mod(mdg4)-67.2 in both reciprocal two-hybrid tests (see Table S1 in the supplemental material).
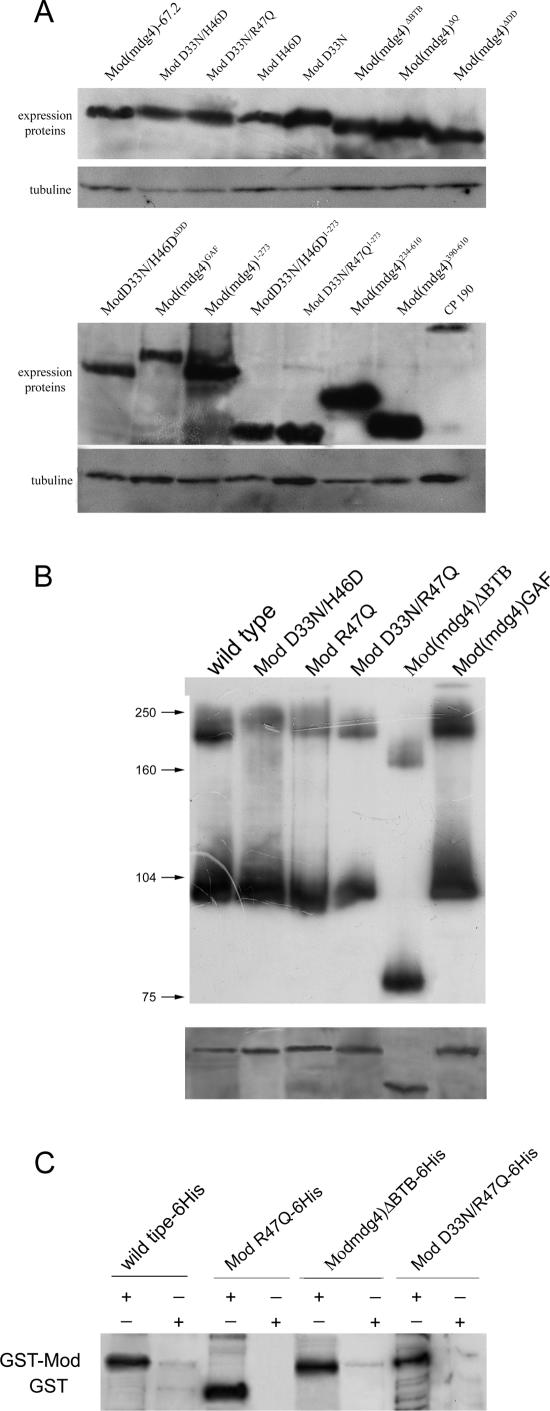
As a first step in our studies, we tested interactions between the Mod(mdg4) BTB mutants and Su(Hw)MID (Table 1; also see Table S1 in the supplemental material). Unexpectedly, we found that most of the mutant forms of Mod(mdg4) displayed a polar effect in the two-hybrid system, as the interaction was strong only when the Mod(mdg4) derivative was fused to GAL4BD. Fusion of the Mod(mdg4) derivatives to GAL4AD produced only weak colony growth on selective plates. To overcome this problem, we fused the GAL4AD domain to the C-terminal part of the Mod(mdg4) mutants. In this case, all Mod(mdg4) mutants showed robust interactions with Su(Hw)MID in the reciprocal two-hybrid tests, suggesting that the Mod(mdg4) mutants accumulate stably in yeast. These data were supported by immunoblot analysis of Mod(mdg4) proteins demonstrating a comparable level of accumulation in yeast cells (Fig. 2A).
TABLE 1.
Summary of interactions between Mod(mdg4) mutants and Mod(mdg4)-67.2a
| Interacting proteins (GAL4BD, GALAD) | Strength of interaction with AD fused:
|
|
|---|---|---|
| Before protein | Behind protein | |
| Mod(mdg4), Mod(mdg4) | +++ | +++ |
| Mod(mdg4), ModD33N/H46D | − | +++ |
| ModD33N/H46D, Mod(mdg4) | − | +++ |
| ModD33N/H46D, ModD33N/H46D | − | + |
| Mod(mdg4), ModH46D | − | ++ |
| ModH46D, Mod(mdg4) | − | +++ |
| ModH46D, ModH46D | − | + |
| Mod(mdg4), ModD33N | − | ++ |
| ModD33N, Mod(mdg4) | − | ++ |
| ModD33N, ModD33N | − | − |
| Mod(mdg4), ModR47Q | +++ | +++ |
| ModR47Q, Mod(mdg4) | ++ | +++ |
| ModR47Q, ModR47Q | ++ | +++ |
| Mod(mdg4), ModD33N/R47Q | + | +++ |
| ModD33N/R47Q, Mod(mdg4) | + | +++ |
| ModD33N/R47Q, ModD33N/R47Q | − | ++ |
| Mod(mdg4), Mod(mdg4)GAF | − | +++ |
| Mod(mdg4)GAF, Mod(mdg4) | − | +++ |
| Mod(mdg4)GAF, Mod(mdg4)GAF | − | +++ |
| Mod(mdg4), Mod(mdg4)ΔBTB | − | ++ |
| Mod(mdg4)ΔBTB, Mod(mdg4) | − | ++ |
| Mod(mdg4)ΔBTB, Mod(mdg4)ΔBTB | − | ++ |
No growth occurred after transformation with single plasmids, indicating that interactions between the proteins are required for expression of the reporter genes (data not shown). The GAL4AD is fused in front of or behind the tested protein. The + symbol refers to the relative strength of the two-hybrid interaction. The − symbol indicates the absence of interaction. Equivalent expression of the chimeric proteins in yeast was confirmed by immunoblotting with GAL4BD or AD monoclonal antibodies (Fig. 2A and data not shown).
FIG. 2.
Analysis of the mutant Mod(mdg4) proteins in vitro. (A) Western analyses of yeast extracts carrying different Mod(mdg4) mutants. The panel shows the expression Mod(mdg4) proteins fused to GAL4 binding domain detected with GAL4 antibodies. In the lower panel, the same filter was stripped and reprobed with anti-tubuline antibodies. (B) Western blot analyses of E. coli-expressed and purified mutant Mod(mdg4) proteins. The native PAGE and sodium dodecyl sulfate-PAGE of the same proteins are shown in the upper and lower panels, respectively. Experimental details are described in Materials and Methods. (C) Interaction of the Mod(mdg4) mutants with Mod(mdg4)-67.2 by GST pull-down assay. The interactions of mutant Mod(mdg4) proteins with wild-type Mod(mdg4)-67.2 were visualized by Western blot analysis using the anti-His tag monoclonal antibodies. All results were reproduced in three independent experiments.
We examined whether the mutated forms of the Mod(mdg4) protein were able to self-associate (Table 2). Note that the yeast two-hybrid assay does not discriminate between homodimerization and multimerization. According to previous observations (33, 34), we expected that alterations in the BTB domain would compromise self-association. We tested this in two ways. First, we tested for interactions with full-length Mod(mdg4)-67.2. Second, we tested for homologous interactions between BTB domain mutants. Surprisingly, in these tests, we found that deletion or alteration of the BTB domain did not eliminate homodimerization, as previously reported (20). This difference may result from our use of the C-terminal fusion proteins, as, for example, we also failed to observe interactions when GAL4AD was fused to the N-terminal part of the Mod(mdg4)ΔBTB protein. As in the case of Mod(mdg4)ΔBTB, most Mod(mdg4) mutants supported yeast growth on selective plates when the GAL4AD domain was on the C terminus of the fusion protein. For ModH46D, ModD33N/H46D, and ModD33N/R47Q, growth on the selective plates was weaker, suggesting that these proteins were able to self-associate with lower efficiency. Only ModD33N lost the ability to self-associate in the two hybrid assay. Thus, the properties of ModD33N contrast with those of Mod(mdg4)ΔBTB. These results might be explained by unfolding of the mutant BTB domain that reduces the ability of the ModD33N protein to self-associate. Indeed, a similar D33N mutation in PLZF and BCL-6 BTB domains led to unfolding of the proteins (33, 34).
TABLE 2.
Identification of the second domain required for dimerization of Mod(mdg4)-67.2a
| Interacting proteins | Strength of interaction |
|---|---|
| Mod(mdg4)ΔBTB, Mod(mdg4)ΔQ | +++ |
| Mod(mdg4)ΔBTB, Mod(mdg4)1-273 | − |
| Mod(mdg4)1-273, Mod(mdg4)1-273 | +++ |
| ModD33N, Mod(mdg4)1-273 | − |
| ModH46D, Mod(mdg4)1-273 | − |
| ModD33N/H46D, Mod(mdg4)1-273 | + |
| ModD33N/R47Q, Mod(mdg4)1-273 | + |
| Mod(mdg4)GAF, Mod(mdg4)1-273 | ++ |
| ModR47Q, Mod(mdg4)1-273 | +++ |
| ModR47Q1-273, ModR47Q1-273 | +++ |
| ModR47Q1-273, Mod(mdg4)1-273 | +++ |
| ModD33N/H46D1-273, ModD33N/H46D1-273 | − |
| ModD33N/H46D1-273, Mod(mdg4)1-273 | + |
| ModD33N/R47Q1-273, ModD33N/R47Q1-273 | − |
| ModD33N/R47Q1-273, Mod(mdg4)1-273 | + |
| Mod(mdg4)234-610, Mod(mdg4)234-610 | ++ |
| Mod(mdg4)317-610, Mod(mdg4)317-610 | ++ |
| Mod(mdg4)390-610, Mod(mdg4)390-610 | − |
| Mod(mdg4)ΔDD, Su(Hw)MID | +++ |
| ModD33N/H46DΔDD, Su(Hw)MID | +++ |
| Mod(mdg4)ΔDD, Mod(mdg4)ΔDD | +++ |
| ModD33N/H46DΔDD, Mod(mdg4)ΔDD | + |
| ModD33N/H46DΔDD, Mod(mdg4) | + |
| ModD33N/H46DΔDD, ModD33N/H46DΔDD | − |
The relative strength of the two-hybrid interaction was similar in both directions. The GAL4AD was on the C terminus of the fused proteins. No growth occurred after transformation with single plasmids, indicating that interactions between the proteins are required for expression of the reporter genes (data not shown). Equivalent expression of the chimeric proteins in yeast was confirmed by immunoblotting with GAL4 BD or AD monoclonal antibodies (Fig. 2A and data not shown). Designations are as defined for Table 1.
The ability of several Mod(mdg4) proteins to self-associate was confirmed in vitro by native PAGE (Fig. 2B). Mod(mdg4)-67.2 was found both as monomer and dimer. The Mod(mdg4)ΔBTB, ModD33N/H46D, ModR47Q, ModD33N/R47Q, and Mod(mdg4)Gaf proteins were detected in the same two forms. Thus, all tested Mod(mdg4) mutants preserved the ability to self-associate in vitro, confirming the results obtained by the yeast two-hybrid method.
Finally, we confirmed, by the GST pull-down assay, interactions of Mod(mdg4)-67.2 with Mod(mdg4)ΔBTB, ModD33N/R47Q, and ModR47Q mutants (Fig. 2C). Equal amounts of purified GST or GST fused to full-length Mod(mdg4)-67.2 were mixed with purified recombinant mutant Mod(mdg4) proteins fused to a histidine tag. All mutant forms of Mod(mdg4) were retained on the glutathione beads when incubated with the GST-Mod(mdg4)-67.2. The lack of binding to GST alone confirmed the specificity of the interaction with Mod(mdg4)-67.2.
Mod(mdg4)-67.2 contains a second homodimerization domain.
As Mod(mdg4)ΔBTB is able to self-associate, we tried next to identify a second interaction domain in the Mod(mdg4)-67.2 protein. Mod(mdg4)-67.2 contains a glutamine-rich (Q-rich) region (between 138 amino acids [aa] and 265 aa) in common with other Mod(mdg4) isoforms (3, 7). Glutamine-rich (Q) domains are frequently involved in homodimerization (46). To examine an involvement of the Q domain in self-association of the Mod(mdg4) protein, we made Mod(mdg4)ΔQ that contains a deletion extending from 145 aa to 277 aa and Mod(mdg4)1-273 that includes both the BTB and Q domains (Fig. 1A). In the yeast two-hybrid system, Mod(mdg4)ΔBTB interacted with Mod(mdg4)ΔQ but not with Mod(mdg4)1-273 (Table 2). At the same time, Mod(mdg4)1-273 was able to self-interact. These results suggest that the Q domain is not involved in self-association of the Mod(mdg4)-67.2 protein.
To define the region of the putative dimerization domain in Mod(mdg4)-67.2, we tested interactions between Mod(mdg4)1-273 and various mutant Mod(mdg4) proteins (Table 2). Mod(mdg4)1-273 interacted strongly with ModR47Q and Mod(mdg4)Gaf, interacted weakly with ModD33N/H46D and ModD33N/R47Q, and did not interact with ModD33N and ModH/46D. Next, we determined whether BTB mutants that were truncated would retain the ability to self-associate. We constructed derivatives of ModD33N/H46D, ModD33N/R47Q, and ModR47Q that contain 273 aa (Table 2). As expected, ModR47Q1-273 showed self-association, as this mutation retained a functional BTB. In contrast, ModD33N/H46D1-273 and ModD33N/R47Q1-273 lost the ability to self-associate while retaining the ability to weakly interact with Mod(mdg4)1-273. These results suggest that the C-terminal part of Mod(mdg4)-67.2 is essential for the interaction with Mod(mdg4) mutants.
To refine the C-terminal domain required for self-association, we made three N-terminally truncated versions of the Mod(mdg4)-67.2 protein: Mod(mdg4)234-610, Mod(mdg4)317-610, and Mod(mdg4)390-610 (Fig. 1). In the yeast two-hybrid system, only Mod(mdg4)234-610 and Mod(mdg4)317-610 proteins show growth on the selective media, indicating that these proteins are able to self-associate (Table 3). Mod(mdg4)390-610 was unable to self-associate, implying that at least a part of a putative dimerization domain, named DD, might be located between positions 317 and 390.
TABLE 3.
Summary of results obtained with the Mod(mdg4) mutantsa
| Protein | Dimerization | Enhancer blocking | Transcription stimulation | Binding to chromosomes | Speckle pattern | Interaction with CP190 |
|---|---|---|---|---|---|---|
| Mod(mdg4)-67.2 | + | + | + | + | + | + |
| Mod(mdg4)ΔBTB | +/− | − | − | − | − | − |
| Mod(mdg4)ΔDD | + | + | + | + | + | + |
| Mod(mdg4)GAF | + | − | − | − | +/− | + |
| ModD33N | − | − | − | − | − | − |
| ModH46D | +/− | − | − | − | − | − |
| ModR47Q | + | + | + | + | + | + |
| ModD33N/R47Q | +/− | +/− | +/− | + | +/− | +/− |
| ModD33N/H46D | +/− | + | + | + | + | +/− |
| ModD33N/H46DΔDD | − | − | − | − | − | +/− |
+, wild-type level of functionality [like Mod(mdg4)-67.2]; +/−, weak functionality; −, almost no functionality. The enhancer blocking mediated by the Mod(mdg4) mutants was examined with the aid of the y2 scD1, scms, ct6, and ctk alleles (Fig. 4; see also Table S1 in the supplemental material). Transcription stimulation was examined with aid of the y2 (yellow expression in bristles) (Fig. 4) and scv2 (see Table S1 in the supplemental material) alleles.
To test the role of the DD domain in self-association of Mod(mdg4)-67.2, we made the Mod(mdg4)ΔDD (Fig. 1A) and D33N/H46DΔDD mutants carrying a deletion between positions 301 and 389. In the yeast two-hybrid system, both proteins efficiently interacted with Su(Hw)MID (Table 2). Mod(mdg4)ΔDD was able to self-associate, most likely because the BTB domain is intact (Table 2). In contrast, the D33N/H46DΔDD protein does not show interaction. Like ModD33N/H46D1-273, D33N/H46DΔDD showed poor growth on selective plates, which indicated a weak interaction with Mod(mdg4) and Mod(mdg4)ΔDD. These results support the proposal that the DD plays a critical role in self-association of D33N/H46D.
Genetic systems used to study the Mod(mdg4) activities.
To determine the in vivo effects of mutations in the BTB domain, we used three gypsy-induced alleles in the yellow, scute, and cut loci. Two activities were demonstrated for the Mod(mdg4)-67.2 protein interacting with Su(Hw): participation in enhancer blocking and transcription stimulation. The features of these model systems are detailed below.
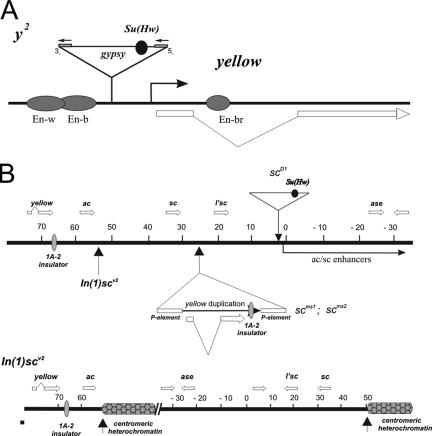
The first system we used involved the yellow gene. In the y2 mutation (Fig. 3A), a gypsy element is inserted between the yellow promoter and the enhancers controlling yellow expression in the wing and body (17). As a result, males hemizygous for the y2 allele show brown abdominal pigmentation in the fifth and sixth abdominal segments instead of the black pigmentation observed in wild-type males (Fig. 4). These effects are caused by the gypsy insulator that blocks the wing and body enhancers but not the bristle enhancer that is located in the yellow intron (17, 18). The mod(mdg4)u1 and mod(mdg4)T6 mutations enhance the mutant y2 phenotype by repressing yellow expression in bristles and other derivative cuticle structures (14, 15). At the same time, mod(mdg4)u1 causes a partial loss of the enhancer-blocking activity of the gypsy insulator. In the mod(mdg4)u1 background, y2 males display a variegated cuticular phenotype resulting from different expression levels of the yellow gene in adjacent groups of cells in the abdominal segments (16). In some cuticle cells, the effect of the gypsy insulator is reversed, resulting in normal expression of the yellow gene; in other cells, the effect of the gypsy insulator is enhanced due to direct repression of yellow. Thus, in the y2 allele, binding of the Mod(mdg4)-67.2 protein strengthens the enhancer-blocking activity of the gypsy insulator and prevents repression of the yellow promoter.
FIG. 3.
(A) Structure of the yellow locus in wild-type and y2 alleles. Exons of the yellow gene are represented by white rectangles, and filled ovals represent various tissue-specific transcriptional enhancers that control yellow gene expression in the respective tissues. The gypsy insertion responsible for the y2 allele is represented by a triangle. Closed boxes flanking gypsy represent the long terminal repeats. The Su(Hw) insulator is represented by a closed circle located in the 5′ transcribed untranslated region of gypsy. (B) Schematic representation of the yellow/ac/sc region in the scD1, scms, and In(I)scV2 mutations (5, 21). The coordinates of the AS-C region are as defined in Campuzano et al. (5). Vertical arrows indicate the position of chromosomal breakpoints associated with the In(I)scv2 mutation. Arrows with a triangle show insertions of gypsy (scD1) and of P elements (scms) associated with duplication of the yellow sequences. Thick horizontal white arrows show the positions and direction of yellow and AS-C genes transcripts. The filled oval indicates the endogenous 1A-2 insulator and the Su(Hw) binding sites in gypsy.
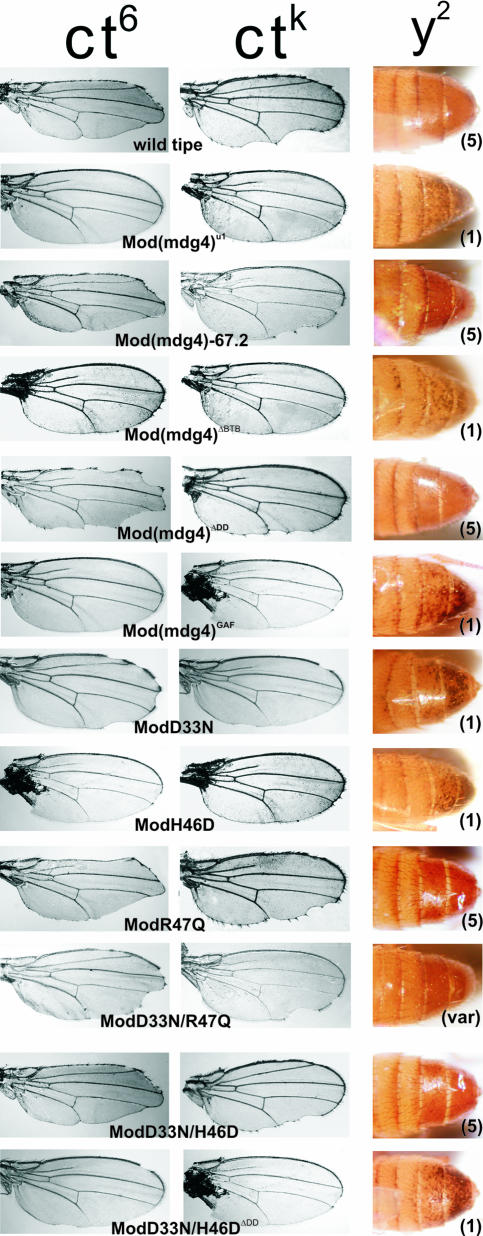
FIG. 4.
Phenotypic effects of the mutant Mod(mdg4) proteins. Effects of the Mod(mdg4) mutants on the cut wing phenotype in the ctK and ct6 alleles and on abdomens of 3-day-old males. Mod (mdg4)-67.2 and the mutant Mod(mdg4) proteins were expressed in ct6 or ctK or y2; mod(mdg4)u1/mod(mdg4)u1 flies. Figures in parentheses indicate the levels of yellow expression in bristles, as follows: 1, loss of pigmentation in all bristles; var, parts of bristles are pigmented; 5, pigmentation of all bristles like in wild-type flies.
The second model system (Fig. 3B) utilized in our studies involved genes in the Achaete-Scute complex (AS-C), located adjacent to the yellow gene (5). The expression of the ac and sc genes is confined to the proneural clusters that determine the precise positions of macrochaetae (36). A complex pattern of ac and sc expression is mediated by the action of site-specific enhancer-like elements distributed over about 90 kb of the AS-C cluster (5, 36). Several AS-C alleles were tested. The scD1 mutation is caused by an insertion of gypsy 20 kb downstream of the sc gene (5) that blocks the communication between many bristle-specific enhancers and the sc promoter. The mod(mdg4)u1 mutation only partially suppresses the sc mutant phenotype, suggesting that Mod(mdg4)-67.2 is not critical for the block of the sc enhancers by the gypsy insulator. Recently, the first endogenous insulator 1A-2, containing two Su(Hw) binding sites, was found to separate the yellow gene from the AS-C (21, 39). In the scms1 and scms2 mutants (Fig. 3B), the 1A-2 insulator was duplicated between the sc gene and its enhancers (21). In contrast to its effects on the scD1 allele, mod(mdg4)u1 almost completely suppresses the mutant phenotype of the scms alleles. Finally, we used the In(1)scv2 mutation (Fig. 3B), which carries an inversion with one breakpoint located very close to the 3′ end of the ac coding region and a second in centric heterochromatin (5). Despite the close proximity to centric heterochromatin, In(1)scv2 causes only a weak mutant phenotype (5). However, in the mod(mdg4)u1 background, this inversion strongly enhances ac and sc phenotypes, suggesting that the Mod(mdg4)-67.2 protein blocks heterochromatin-mediated repression. Thus, the AS-C mutations allow us to test the ability of mutant Mod(mdg4) proteins to function at the gypsy insulator as well as an endogenous Su(Hw) insulator.
The third genetic model system is the cut locus, where ct6 and ctK are two gypsy-induced alleles. In the ct6 allele, a gypsy element is inserted close to and completely blocks a wing margin enhancer located nearly 85 kb upstream of the cut promoter (12, 26), producing a cut wing phenotype (Fig. 4). The mod(mdg4)u1 and mod(mdg4)T6 mutations almost completely suppress the ct6 mutant phenotype, suggesting that Mod(mdg4)-67.2 is essential for blocking the wing enhancer (Fig. 4). In the ctK allele, the wing margin enhancer of cut is only partially blocked, resulting in an intermediate cut wing phenotype (Fig. 4), presumably because the inserted gypsy contains fewer Su(Hw)-binding sites than most gypsy elements (25). The ctK mutant phenotype is more sensitive to the levels of Su(Hw) and Mod(mdg4)-67.2 activity than most gypsy insertions and is almost completely suppressed by heterozygous su(Hw) mutations or mod(mdg4)u1 (12). Thus, the ctK allele provides a sensitive assay to examine the ability of the Mod(mdg4) mutant proteins to support the blocking of the wing enhancer.
Role of the BTB and DD domains in insulation and transcription stimulation mediated by the Mod(mdg4)-67.2 protein.
We constructed transgenic flies bearing the mutant Mod(mdg4) transgenes under the control of two promoters, the inducible hsp70 promoter [Mod(mdg4)-67.2; ModH46D and ModD33N/H46D] or the ubiquitously expressed su(Hw) promoter [Mod(mdg4)-67.2; Mod(mdg4)ΔBTB, Mod(mdg4)ΔDD ModD33N/H46D, ModD33N/H46DΔDD, ModR47Q, ModD33N, ModD33N/R47Q, and Mod(mdg4)Gaf]. The activities of these mutant Mod(mdg4) proteins were assessed in mod(mdg4)u1 and mod(mdg4)T6 mutant backgrounds. At least eight independent transgenic lines were examined for rescue in both backgrounds by each Mod(mdg4) mutant to exclude position effects. If expression of the transgene did not rescue the mutant mod(mdg4)u1 phenotype, then we established lines that contained either two or three copies of the homozygous transgenes to reveal weak phenotypic effects. These studies revealed similar results with both mutant backgrounds. For this reason, we refer only to the mod(mdg4)u1 mutation in the subsequent text.
In 14 lines, expression of the Mod(mdg4)67-2 transgene under control of either the hsp70 or Su(Hw) promoter completely reverted the phenotypes associated with the mod(mdg4)u1 mutation (Fig. 4 and Table 3; see also Fig. S1 in the supplemental material). Thus, either promoter produces sufficient levels of functional protein to complement the loss of the Mod(mdg4)-67.2 protein. Interestingly, expression of Mod(mdg4) carrying a deletion of the BTB domain failed to rescue the mutant phenotypes, indicating a complete inactivation of the protein. These data confirm the essential role of Mod(mdg4)67.2 BTB domain.
We examined the function of proteins bearing point mutations that affect the integrity of the charge pocket of the BTB domain (Fig. 4 and Table 3; see also Fig. S1 in the supplemental material). The ModH46D protein has a substitution in the most conserved residue of the BTB domain that changes the uncharged histidine (47) to the negatively charged aspartic acid (H46D). This mutation almost completely inactivated all insulator functions. In contrast, the ModR47Q mutant manifested the same levels of activity as the wild-type Mod(mdg4)-67.2 protein. Neutralization of a negative charge in the BTB pocket of ModD33N resulted in almost complete loss of the insulating and antirepression activities. ModD33N only slightly rescued the enhancer-blocking activity of the gypsy insulator in the ct6 allele but was unable to restore enhancer blocking in scms, scD1, and In(1)scV2 mutants and ctK mutations. However, the loss of negative charge at one position in the pocket can be compensated by creation of a negative charge elsewhere in the pocket. For example, the ModD33N/H46D mutant had near wild-type function. These results suggest that inversion of the two most conserved residues preserves the function of BTB.
Additionally, the double mutant ModD33N/R47Q showed moderate levels of insulator and antirepression activities. ModD33N/R47Q restored the enhancer blocking activity of the gypsy insulator in the ct6 allele but did not significantly affect the mutant phenotype of the ctK, scD1, and scms alleles. Interestingly, scD1 and scms depend upon an endogenous insulator whose enhancer blocking effects are only partially dependent on the Su(Hw) and Mod(mdg4) proteins. ModD33N/R47Q reversed the variegation of abdominal pigmentation in the y2 flies but only partially suppressed inhibition of yellow expression in bristles. Finally, ModD33N/R47Q only slightly suppressed the heterochromatin-mediated repression in In(I)scV2 flies.
Taken together, our studies suggest that the charge pocket is important in Mod(mdg4) function. However, the absolute charge may not be the critical factor determining activity. The most severe effects on insulator function were observed when a conserved negative amino acid (D33N) was changed, whereas neutralizing a positive charge (R47Q) had little effect. Combining the D33N mutation with changes in positively charged residues restored function to a degree dependent upon the identity of the amino acid. Based on these observations, we propose that residues in the charge pocket may make specific contributions to the function of the BTB domain.
We were interested in determining the role of the newly discovered DD in gypsy insulator activities mediated by Mod(mdg4)-67.2 (Fig. 4 and Table 3; see also Fig. S1 in the supplemental material). Expression of Mod(mdg4)ΔDD completely reverted the phenotypes associated with the mod(mdg4)u1 mutation, suggesting that DD is not essential for the Mod(mdg4)-67.2 function. In contrast, if the DD was deleted in mutants that affect the BTB domain, such as ModD33N/H46DΔDD, then there was a complete loss of function. These results suggest that the DD domain is essential when the mutant BTB domain partially loses the ability to dimerize.
Models of insulator function propose that the Mod(mdg4) BTB domain plays an important role in directing protein interactions with other BTB proteins to form looped chromatin domains (6). Based on this postulate, we predicted that the BTB domain of Mod(mdg4) could be replaced with another BTB domain and retain insulator function. To test this prediction, we chose the related BTB domain of GAF (41). We found that the Mod(mdg4)Gaf protein was completely inactive in all assays, suggesting that the GAF BTB domain does not substitute for the Mod(mdg4) BTB (Fig. 4 and Table 3; see also Fig. S1 in the supplemental material). We propose that these effects are not due to the inability of the BTB swap protein to fold properly, as Mod(mdg4)Gaf formed homodimers and interacted with the Su(Hw) protein in the yeast two-hybrid system. These data imply that the Mod(mdg4) BTB domain mediates specific interactions with unidentified protein complexes required for the functional potency of Mod(mdg4)-67.2.
Localization of the Mod(mdg4) mutants on polytene chromosomes and in diploid cells.
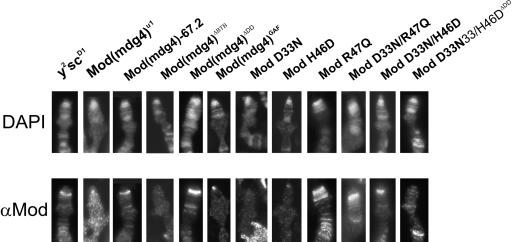
To further investigate the mechanism associated with the loss of insulator function among the BTB domain mutants, we examined the chromosome association of these proteins. First, we determined whether these mutant proteins were bound to the gypsy transposon present in the y2 and scD1 genes using a polytene chromosome assay. We used antibodies raised against the unique C-terminal domain of Mod(mdg4)-67.2 to specifically detect the insulator protein. In flies wild type for Mod(mdg4)-67.2, the sites corresponding to gypsy insertions in the y2 and scD1 mutations bind this protein, as demonstrated by strong bands of protein localization at the tip of the X chromosome (Fig. 5). In mod(mdg4)u1 flies expressing Mod(mdg4)-67.2, ModR47Q, ModD33N/R47Q, ModD33N/H46D, and Mod(mdg4)ΔDD, immunolocalization was similar to mod(mdg4)+ flies (Fig. 5), confirming that these proteins interact with Su(Hw). Proteins lacking insulator function did not associate with chromosomes, including Mod(mdg4)ΔBTB, ModH46D, ModD33N, Mod(mdg4)Gaf, and ModD33N/H46DΔDD.
FIG. 5.
Localization of mutant Mod(mdg4) proteins on polytene chromosomes of the y2scD1; mod(mdg4)u1/mod(mdg4)u1 line. Salivary glands were dissected from third-instar larvae. The upper panel shows 4′,6′-diamidino-2-phenylindole (DAPI)-stained chromosomes. The lower panel represents the localization of Mod(mdg4) on polytene chromosomes; the white arrows indicate the Mod(mdg4) bands. The mutant Mod(mdg4) proteins were detected using a polyclonal antibody against the Mod(mdg4)-67.2 isoform and Cy3-conjugated secondary antibodies.
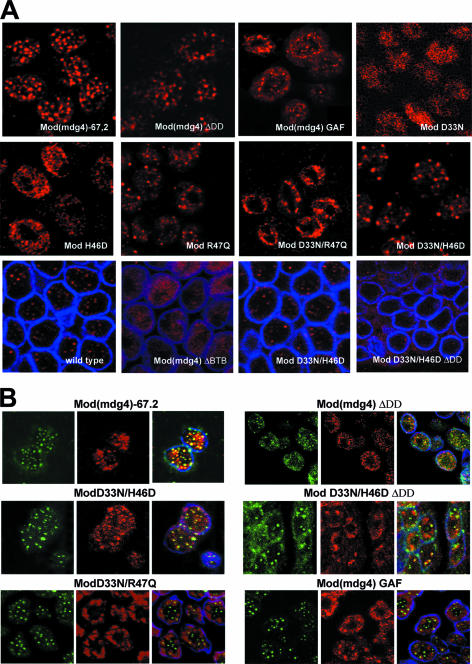
In a second assay, we determined whether mutant Mod(mdg4) proteins reconstituted nuclear speckles that represent a coalescence of the Su(Hw)-Mod(mdg4) protein complexes in diploid nuclei (Fig. 6). Previous studies suggest that formation of these speckles correlates with insulator function (16, 38). In the transgenic lines, Mod(mdg4)-67.2, ModR47Q, ModD33N/H46D, and Mod(mdg4)DDD proteins displayed a wild-type punctate pattern (Fig. 6A). We also found that the Mod(mdg4)-67.2, ModD33N/H46D, and Mod(mdg4)ΔDD proteins colocalize with the Su(Hw) protein (Fig. 6B), confirming that the interaction between these proteins is nuclear. The Mod(mdg4)Gaf protein produced a more diffuse pattern with many fuzzy speckles. The Su(Hw) and Mod(mdg4)Gaf proteins displayed only partial colocalization on the nuclear periphery. Other mutant proteins, Mod(mdg4)ΔBTB, ModD33N/R47Q, ModH46D, and ModD33N, showed diffuse nuclear localization predominantly at the nuclear periphery (Fig. 6A). Interestingly, ModD33N/H46DΔDD displayed unusual patterns: it formed spackles at the nuclear periphery and cytoplasm. The Su(Hw) protein only occasionally colocalizes with ModD33N/R47Q or ModD33N/H46DΔDD aggregates, suggesting that these Mod(mdg4) mutants and Su(Hw) failed to interact in nuclei.
FIG. 6.
Immunolocalization of insulator proteins in diploid cells of larval imaginal discs. (A) Localization of Mod(mdg4)-67.2 and the mutant Mod(mdg4) proteins. (B) Colocalization of Su(Hw) and Mod(mdg4) proteins. Interphase diploid cells were obtained from imaginal discs of y2scD1; mod(mdg4)u1/mod(mdg4)u1 lines and y2scD1; mod(mdg4)u1/mod(mdg4)u1 lines expressing Mod(mdg4)-67.2 or one of the mutant Mod(mdg4) proteins. The distribution of proteins was detected with polyclonal antibody against the Mod(mdg4)-67.2 isoform (red), anti-lamin (blue), and anti-Su(Hw) (green) antibodies.
Thus, functional inactivation of the BTB domain or deletion of the DD domain is directly correlated with the inability of the mutant protein to interact with Su(Hw) on polytene chromosomes and to form nuclear speckles.
The functionality of the mutant Mod(mdg4) proteins does not correlate with their ability to interact with CP190.
A second insulator protein that interacts with Mod(mdg4)-67.2 is CP190 (38). If this interaction is critical for the Mod(mdg4)-67.2 activity, then the function of the mutant Mod(mdg4) proteins might be directly correlated with an ability to interact with CP190. To test this assumption, we determined whether the mutant Mod(mdg4) proteins interacted with CP190 in the yeast two-hybrid system.
Pai et al. (38) reported that CP190 fails to interact with itself but associates with Mod(mdg4)-67.2 in the yeast two-hybrid assay. This unexpected finding might be explained if CP190 showed the same orientation effects described for Mod(mdg4)67.2. For this reason, we tested interactions using C-terminal fusions of the GALAD. When GAL4AD was fused to the C terminus of CP190 (CP190-GAL4AD), a weak interaction was observed (data not shown). Further tests indicate that the C-terminal part of CP190 negatively influenced the activity of the fused GAL4AD, complicating the interpretation of these results. Tests of a C-terminally truncated CP1901-765 coexpressed with the full-length GAL4BD-CP190 supported strong interaction (Table 4). As deletion of the C-terminal acidic domain of CP190 does not significantly influence function (38), we used the CP1901-765-GALAD protein in further experiments.
TABLE 4.
Summary of interactions with CP190a
| Interacting proteins | Strength of interaction |
|---|---|
| CP190, CP190 | +++ |
| CP190, Mod(mdg4) | +++ |
| CP190, ModR47Q | +++ |
| CP190, Mod(mdg4)ΔDD | +++ |
| CP190, Mod(mdg4)ΔBTB | − |
| CP190, ModH46D | − |
| CP190, ModD33N | − |
| CP190, ModD33N/R47Q | + |
| CP190, ModD33N/H46D | + |
| CP190, ModD33N/H46DΔDD | + |
| CP190, Mod(mdg4)GAF | +++ |
| CP190, GAF | − |
The GAL4AD was on the C terminus of the fused proteins. The GAL4BD domain was fused to full-length CP190, while GAL4AD was fused to the C terminus of the truncated CP1901-765 protein. The relative strength of the two-hybrid interaction was similar in both directions. No growth occurred after transformation with single plasmids, indicating that interactions between the proteins are required for expression of the reporter genes (data not shown). Equivalent expression of the chimeric proteins in yeast was confirmed by immunoblotting with GAL4 BD or AD monoclonal antibodies (Fig. 2A and data not shown). Designations are as defined for Table 1.
Each of the Mod(mdg4) mutants was tested in the yeast two-hybrid assay for its ability to interact with CP190 (Table 4). CP190 was unable to interact with Mod(mdg4)ΔBTB, ModH46D, and ModD33N and displayed weak interactions with ModD33N/R47Q, ModD33N/H46D, and ModD33N/H46DΔDD and strong interactions with Mod(mdg4), ModR47Q, Mod(mdg4)ΔDD, and Mod(mdg4)Gaf. Interestingly, Mod(mdg4)Gaf and Mod(mdg4)-67.2 interact with CP190 with similar efficiency (Table 4). Pai et al. (38) showed that GAF did not interact with CP190 in the yeast two-hybrid assay, a finding supported in our studies (Table 4). Thus, these data suggest that Mod(mdg4)-67.2 interacts with CP190 in a BTB-independent manner. Further, the observation that Mod(mdg4)Gaf lacks insulator properties suggests that the functionality of Mod(mdg4) mutants is not directly correlated with their ability to interact with CP190.
DISCUSSION
The BTB domain is a versatile protein-protein interaction motif that participates in a wide range of cellular functions, including transcriptional regulation, cytoskeleton dynamics, ion channel assembly and targeting proteins for ubiquitination (43). Here we demonstrated that the integrity of the BTB domain is critical for the Mod(mdg4)-67.2 activity in insulation.
Melnick et al. (33, 34) showed that the alignment of charged residues within the BTB pocket of the PLZF and Bcl-6 proteins was required for autonomous transcriptional repression by the homodimers. When two conserved charged residues in the pocket, D35 and R49, were switched to polar amino acids, the full-length PLZF or Bcl-6 proteins harboring the double mutant BTB domain were severely impaired for transcriptional repression yet could still oligomerize and localize to characteristic nuclear speckles. Here we demonstrate that the Mod(mdg4)-67.2 protein carrying mutations in analogous residues in the BTB domain loses much of its function as a part of the gypsy and 1A2 insulators. Elimination of a negative charge in the pocket by the single alteration D33N almost completely compromised the integrity of the BTB domain of Mod(mdg4)-67.2 and inactivated its function, suggesting that the net charge may be important for proper folding of the BTB domain. The opposite charge alteration, replacing a positive charge with a neutral amino acid (R47Q) had a minor effect, suggesting that the overall charge may be less important than the identity of the residue changed. The substitution of the most conserved H46 with a negatively charged aspartic acid (D) completely inactivated Mod(mdg4)-67.2. While H46 is not involved in formation of the pocket structure, the additional D33N substitution that reestablishes the overall negative charge in the pocket of the ModD33N/H46D double mutant restores Mod(mdg4)-67.2 insulator function, even though the D33N/H46D BTB domain partially loses the ability to dimerize. Taken together, we predict that the net charge is essential for proper folding of the BTB domain.
Here we found that Mod(mdg4)-67.2 contains a second domain involved in homodimerization of the protein. When the BTB domain partially lost the ability to dimerize, as in the mutant ModD33N/H46D protein, the second domain directed proper nuclear localization and formation of the insulator complex on the DNA. Thus, the DD domain contributes to dimerization of Mod(mdg4)-67.2 that is essential for insulator function.
Interestingly, we found that interaction between the Mod(mdg4)-67.2 and GAF proteins requires both the BTB domain and a second protein interaction domain (35). We show that the specificity of the interaction between Mod(mdg4)-67.2 and CP190 is determined by an unidentified domain beside BTB. Based on these findings, we suggest that interactions between BTB-containing proteins may be supported by additional protein-protein interaction domains.
Mod(mdg4)-67.2 facilitates binding of Su(Hw) to insulator sequences in vivo (16). Here we found that inactivation of the BTB domain by point mutations like H46D, D33N, or BTB deletion render the mutant Mod(mdg4) protein unable to associate with polytene chromosomes. These proteins also only partially colocalize with the Su(Hw) protein. As these mutant proteins still interact with the Su(Hw) protein in the yeast two-hybrid assay, we postulate that specific interactions mediated by the BTB domain of Mod(mdg4) with unidentified protein(s) are required for efficient recruitment of the Su(Hw)/Mod(mdg4) complex to the insulator sequences and nuclear bodies formed by the insulator proteins. This model is supported by the inability of the chimeric Mod(mdg4)Gaf protein, containing the GAF BTB domain, to interact with Su(Hw) binding sites in vivo and extensively form the nuclear speckles with Su(Hw), while in two-hybrid assays Mod(mdg4)Gaf interacts with Su(Hw) with the same efficiency as Mod(mdg4)-67.2. These data imply that the interaction with Su(Hw) is not sufficient to recruit Mod(mdg4) to Su(Hw) binding sites. Further, the interaction of Mod(mdg4)-67.2 with CP190 does not appear to be critical for the recruitment of Mod(mdg4)-67.2 to insulators. Based on these results, we speculate that an unidentified protein is required for recruiting the Su(Hw)-Mod(mdg4)-67.2 complex to the Su(Hw) insulator and propose that the BTB domain of the Mod(mdg4)-67.2 facilitates or directly interacts with this hypothetical protein.
Supplementary Material
Acknowledgments
We thank Paul Fisher for the anti-lamin antibody, Dale Dorsett for providing the mod(mdg4)-67.2, full-length su(Hw) cDNA clone, and plasmid with the Su(Hw) promoter, and Mark Brennan for providing the mod(mdg4)T6 line.
This work was supported by the Molecular and Cellular Biology program of Russian Academy of Science and by a stipend from the Center for Medical Studies, University of Oslo to A.G., by a Joint Research Project SCOPES grant from the Swiss National Science Foundation to V.P and P.G., and by an International Research Scholar award from the Howard Hughes Medical Institute to P.G. The work was also supported by the National Institute of Health grant GM42539 to P.G.
Footnotes
Published ahead of print on 13 November 2006.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Ahmad, K. F., C. K. Engel, and G. G. Prive. 1998. Crystal structure of the BTB domain from PLZF. Proc. Natl. Acad. Sci. USA 95:12123-12128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahmad, K. F., A. Melnick, S. Lax, D. Bouchard, J. Liu, C. L. Kiang, S. Mayer, S. Takahashi, J. D. Licht, and G. G. Prive. 2003. Mechanism of SMRT corepressor recruitment by the BCL6 BTB domain. Mol. Cell 12:1551-1564. [DOI] [PubMed] [Google Scholar]
- 3.Buchner, K., P. Roth, G. Schotta, V. Krauss, H. Saumweber, G. Reuter, and R. Dorn. 2000. Genetic and molecular complexity of the position effect variegation modifier mod(mdg4) in Drosophila. Genetics 155:141-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cai, H. N., and M. Levine. 1997. The gypsy insulator can function as a promoter-specific silencer in the Drosophila embryo. EMBO J. 16:1732-1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Campuzano, S., L. Carramolino, C. V. Cabrera, M. Ruiz-Gomez, R. Villares, A. Boronat, and J. Modolell. 1985. Molecular genetics of the achaete-scute gene complex of D. melanogaster. Cell 40:327-338. [DOI] [PubMed] [Google Scholar]
- 6.Capelson, M., and V. G. Corces. 2004. Boundary elements and nuclear organization. Biol. Cell 96:617-629. [DOI] [PubMed] [Google Scholar]
- 7.Dorn, R., H. Morawietz, G. Reuter, and H. Saumweber. 1993. Identification of an essential Drosophila gene that is homologous to the translation initiation factor eIF-4A of yeast and mouse. Mol. Gen. Genet. 237:233-240. [DOI] [PubMed] [Google Scholar]
- 8.Dorn, R., and V. Krauss. 2003. The modifier of mdg4 locus in Drosophila: functional complexity is resolved by trans splicing. Genetica 117:165-177. [DOI] [PubMed] [Google Scholar]
- 9.Dorsett, D. 1990. Potentiation of a polyadenylylation site by a downstream protein-DNA interaction. Proc. Natl. Acad. Sci. USA 87:4373-4377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dorsett, D. 1999. Distant liaisons: long-range enhancer-promoter interactions in Drosophila. Curr. Opin. Genet. Dev. 9:505-514. [DOI] [PubMed] [Google Scholar]
- 11.Espinas, M. L., E. Jimenez-Garcia, A. Vaquero, S. Canudas, J. Bernues, and F. Azorin. 1999. The N-terminal POZ domain of GAGA mediates the formation of oligomers that bind DNA with high affinity and specificity. J. Biol. Chem. 274:16461-16469. [DOI] [PubMed] [Google Scholar]
- 12.Gause, M., P. Morcillo, and D. Dorsett. 2001. Insulation of enhancer-promoter communication by a gypsy transposon insert in the Drosophila cut gene: cooperation between suppressor of hairy-wing and modifier of mdg4 proteins. Mol. Cell. Biol. 21:4807-4817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gdula, D. A., and V. G. Corces. 1997. Characterization of functional domains of the su(Hw) protein that mediate the silencing effect of mod(mdg4) mutations. Genetics 145:153-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Georgiev, P., and M. Kozycina. 1996. Interaction between mutations in the suppressor of Hairy wing and modifier of mdg4 genes of Drosophila melanogaster affecting the phenotype of gypsy-induced mutations. Genetics 142:425-436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gerasimova, T. I., D. A. Gdula, D. V. Gerasimov, O. Simonova, and V. G. Corces. 1995. A Drosophila protein that imparts directionality on a chromatin insulator is an enhancer of position-effect variegation. Cell 82:587-597. [DOI] [PubMed] [Google Scholar]
- 16.Gerasimova, T. I., and V. G. Corces. 1998. Polycomb and trithorax group proteins mediate the function of a chromatin insulator. Cell 92:511-521. [DOI] [PubMed] [Google Scholar]
- 17.Geyer, P. K., C. Spana, and V. G. Corces. 1986. On the molecular mechanism of gypsy-induced mutations at the yellow locus of Drosophila melanogaster. EMBO J. 5:2657-2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Geyer, P. K., and V. G. Corces. 1987. Separate regulatory elements are responsible for the complex pattern of tissue-specific and developmental transcription of the yellow locus in Drosophila melanogaster. Genes Dev. 1:996-1004. [DOI] [PubMed] [Google Scholar]
- 19.Geyer, P. K., and V. G. Corces. 1992. DNA position-specific repression of transcription by a Drosophila zinc finger protein. Genes Dev. 6:1865-1873. [DOI] [PubMed] [Google Scholar]
- 20.Ghosh, D., T. I. Gerasimova, and V. G. Corces. 2001. Interactions between the Su(Hw) and Mod(mdg4) proteins required for gypsy insulator function. EMBO J. 20:2518-2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Golovnin, A., I. Birukova, O. Romanova, M. Silicheva, A. Parshikov, E. Savitskaya, V. Pirrotta, and P. Georgiev. 2003. An endogenous Su(Hw) insulator separates the yellow gene from the Achaete-scute gene complex in Drosophila. Development 130:3249-3258. [DOI] [PubMed] [Google Scholar]
- 22.Hagstrom, K., M. Muller, and P. Schedl. 1996. Fab-7 functions as a chromatin domain boundary to ensure proper segment specification by the Drosophila bithorax complex. Genes Dev. 10:3202-3215. [DOI] [PubMed] [Google Scholar]
- 23.Harrison, D. A., D. A. Gdula, R. S. Coyne, and V. G. Corces. 1993. A leucine zipper domain of the suppressor of Hairy-wing protein mediates its repressive effect on enhancer function. Genes Dev. 7:1966-1978. [DOI] [PubMed] [Google Scholar]
- 24.Holdridge, C., and D. Dorsett. 1991. Repression of hsp70 heat shock gene transcription by the suppressor of hairy-wing protein of Drosophila melanogaster. Mol. Cell. Biol. 11:1894-1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hoover, K. K., T. I. Gerasimova, A. J. Chien, and V. G. Corces. 1992. Dominant effects of suppressor of Hairy-wing mutations on gypsy-induced alleles of forked and cut in Drosophila melanogaster. Genetics 132:691-697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jack, J., D. Dorsett, Y. Delotto, and S. Liu. 1991. Expression of the cut locus in the Drosophila wing margin is required for cell type specification and is regulated by a distant enhancer. Development 113:735-747. [DOI] [PubMed] [Google Scholar]
- 27.Karess, R. E., and G. M. Rubin. 1984. Analysis of P transposable element functions in Drosophila. Cell 38:135-146. [DOI] [PubMed] [Google Scholar]
- 28.Katsani, K. R., M. A. Hajibagheri, and C. P. Verrijzer. 1999. Co-operative DNA binding by GAGA transcription factor requires the conserved BTB/POZ domain and reorganizes promoter topology. EMBO J. 18:698-708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim, J., B. Shen, C. Rosen, and D. Dorsett. 1996. The DNA-binding and enhancer-blocking domains of the Drosophila suppressor of Hairy-wing protein. Mol. Cell. Biol. 16:3381-3392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kuhn, E. J., and P. K. Geyer. 2003. Genomic insulators: connecting properties to mechanism. Curr. Opin. Cell Biol. 15:259-265. [DOI] [PubMed] [Google Scholar]
- 31.LaJeunesse, D., and A. Shearn. 1996. E (z): a polycomb group gene or a trithorax group gene? Development 122:2189-2197. [DOI] [PubMed] [Google Scholar]
- 32.Mazur, A. M., P. G. Georgiev, and A. K. Golovnin. 2005. The acid domain located at the C terminus of the Su(Hw) protein represses transcription in the yeast two-hybrid system. Dokl. Biochem. Biophys. 400:1-3. [DOI] [PubMed] [Google Scholar]
- 33.Melnick, A., K. F. Ahmad, S. Arai, A. Polinger, H. Ball, K. L. Borden, G. W. Carlile, G. G. Prive, and J. D. Licht. 2000. In-depth mutational analysis of the promyelocytic leukemia zinc finger BTB/POZ domain reveals motifs and residues required for biological and transcriptional functions. Mol. Cell. Biol. 20:6550-6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Melnick, A., G. Carlile, K. F. Ahmad, C. L. Kiang, C. Corcoran, V. Bardwell, G. G. Prive, and J. D. Licht. 2002. Critical residues within the BTB domain of PLZF and Bcl-6 modulate interaction with corepressors. Mol. Cell. Biol. 22:1804-1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Melnikova, L., F. Juge, N. Gruzdeva, A. Mazur, G. Cavalli, and P. Georgiev. 2004. Interaction between the GAGA factor and Mod(mdg4) proteins promotes insulator bypass in Drosophila. Proc. Natl. Acad. Sci. USA 101:14806-14811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Modolell, J., and S. Campuzano. 1998. The achaete-scute complex as an integrating device. Int. J. Dev. Biol. 42:275-282. [PubMed] [Google Scholar]
- 37.Pagans, S., M. Ortiz-Lombardia, M. L. Espinas, J. Bernues, and F. Azorin. 2002. The Drosophila transcription factor tramtrack (TTK) interacts with Trithorax-like (GAGA) and represses GAGA-mediated activation. Nucleic Acids Res. 30:4406-4413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pai, C.-Y., E. P. Lei, D. Ghosh, and V. G. Corces. 2004. The centrosomal protein CP190 is a component of the gypsy chromatin insulator. Mol. Cell 16:737-748. [DOI] [PubMed] [Google Scholar]
- 39.Parnell, T. J., M. M. Viering, A. Skjesol, C. Helou, E. J. Kuhn, and P. K. Geyer. 2003. An endogenous Suppressor of hairy-wing insulator separates regulatory domains in Drosophila. Proc. Natl. Acad. Sci. USA 100:13436-13441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Platero, J. S., E. J. Sharp, P. N. Adler, and J. C. Eissenberg. 1996. In vivo assay for protein-protein interactions using Drosophila chromosomes. Chromosoma 104:393-404. [DOI] [PubMed] [Google Scholar]
- 41.Read, D., M. J. Butte, A. F. Dernburg, M. Frasch, and T. B. Kornberg. 2000. Functional studies of the BTB domain in the Drosophila GAGA and Mod(mdg4) proteins. Nucleic Acids Res. 28:3864-3870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Spana, C., and V. G. Corces. 1990. DNA bending is a determinant of binding specificity for a Drosophila zinc finger protein. Genes Dev. 4:1505-1515. [DOI] [PubMed] [Google Scholar]
- 43.Stogios, P. G., G. S. Downs, J. S. Jauhal, S. K. Nandra, and G. G. Prive. 2005. Sequence and structural analysis of BTB domain proteins. Genome Biol. 6:R82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.West, A. G., M. Gaszner, and G. Felsenfeld. 2002. Insulators: many functions, many mechanisms. Genes Dev. 16:271-288. [DOI] [PubMed] [Google Scholar]
- 45.West, A. G., and P. Fraser. 2005. Remote control of gene transcription. Hum. Mol. Genet. 14:101-111. [DOI] [PubMed] [Google Scholar]
- 46.Wilkins, R. S., and J. T. Lis. 1999. DNA distortion and multimerization: novel functions of the glutamine-rich domain of GAGA factor. J. Mol. Biol. 285:515-525. [DOI] [PubMed] [Google Scholar]
- 47.Zollman, S., D. Godt, G. G. Prive, J. L. Couderc, and F. A. Laski. 1994. The BTB domain, found primarily in zinc finger proteins, defines an evolutionarily conserved family that includes several developmentally regulated genes in Drosophila. Proc. Natl. Acad. Sci. USA 91:10717-10721. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.