Abstract
CUG-BP1/CELF1 is a multifunctional RNA-binding protein involved in the regulation of alternative splicing and translation. To elucidate its role in mammalian development, we produced mice in which the Cugbp1 gene was inactivated by homologous recombination. These Cugbp1−/− mice were viable, although a significant portion of them did not survive after the first few days of life. They displayed growth retardation, and most Cugbp1−/− males and females exhibited impaired fertility. Male infertility was more thoroughly investigated. Histological examination of testes from Cugbp1−/− males showed an arrest of spermatogenesis that occurred at step 7 of spermiogenesis, before spermatid elongation begins, and an increased apoptosis. A quantitative reverse transcriptase PCR analysis showed a decrease of all the germ cell markers tested but not of Sertoli and Leydig markers, suggesting a general decrease in germ cell number. In wild-type testes, CUG-BP1 is expressed in germ cells from spermatogonia to round spermatids and also in Sertoli and Leydig cells. These findings demonstrate that CUG-BP1 is required for completion of spermatogenesis.
Following transcription, a eukaryotic pre-mRNA undergoes nuclear maturation. The mature mRNA is then exported to the cytoplasm, where its stability and translation are finely regulated. It is increasingly clear that a strong link exists between the nuclear and the cytoplasmic events that affect a pre-mRNA and the resulting mRNA. This is illustrated by the observation that a number of RNA-binding proteins, such as those belonging to the CELF or BRUNOL family, control both the splicing of pre-mRNA and the cytoplasmic fate of mature mRNA.
The vertebrate CELF family consists of six members, and the name (CUG-BP1 and ETR-3 Like Factors) was derived from the two founding members, CUG-BP1/CELF1 and ETR-3/CUG-BP2/CELF2 (19). The same family was also named BRUNOL (Bruno-like) due to the sequence similarity that its members share with Drosophila melanogaster Bruno (15). The biochemical functions of these proteins are numerous. In mammals, CELF1 to CELF6 were shown to regulate the alternative splicing of some pre-mRNA by stimulating either inclusion or skipping of nonconstitutive exons (for a review, see reference 1). Furthermore, in the cytoplasm, by binding to the 5′ regions of the cognate mRNAs, CUG-BP1/CELF1 stimulates the translation of the cdk inhibitor p21 and determines the translation initiation codon of the transcription factor C/EBPβ (41, 42). CUG-BP2/CELF2 controls the stability and the translation of cyclooxygenase-2 mRNA by binding to the 3′ untranslated region (3′UTR) (26). In Xenopus embryos, EDEN-BP (embryo deadenylation element-binding protein, the name of Xenopus CUG-BP1, which is 88% identical to CUG-BP1) is required for the rapid deadenylation [poly(A) tail shortening] of certain mRNAs that contain binding sites in the 3′UTR (32). This deadenylation is associated with translational repression (12). Human CUG-BP1 can rescue the deadenylation activity when added to an EDEN-BP-depleted Xenopus embryos extract, indicating that EDEN-BP and CUG-BP1 are functionally equivalent (31). The capacity of CUG-BP1/CELF1 to act as an deadenylation factor was recently confirmed with human cells extracts (25). Finally, in a nonvertebrate, Bruno controls the translation of oskar, gurken, and mitotic cyclin mRNAs during Drosophila oogenesis, again by binding to the 3′UTR (13, 38, 48).
Downregulation or mutant analyses have been undertaken to decipher the developmental roles of CELF proteins in some model species. Etr-1 was inactivated by RNA interference in Caenorhabditis elegans. This resulted in an embryonic lethal phenotype, with defects in muscle formation and paralysis (24), but the links between Etr-1 and muscle formation were not investigated. In Drosophila, strong alleles of Bruno result in oogenesis arrest, possibly due to an overexpression of mitotic cyclins in meiotic cells (38, 48). In Xenopus, EDEN-BP/CUG-BP1 was inactivated in embryos by injection of morpholino antisense oligonucleotides or neutralizing antibodies. This demonstrated the requirement of EDEN-BP for somitic segmentation, the process that leads to the appearance in the developing embryo of repeated, metameric mesodermal units termed somites (14).
In contrast to these results, virtually no data are available concerning the developmental functions of CELF proteins in mammals. Here, we report the production of mice in which the Cugbp1 gene was inactivated by homologous recombination in embryonic stem (ES) cells. The Cugbp1−/− mice were viable, but a significant portion of them did not survive after the first few days of life. They displayed growth retardation, and above all, both male and female Cugbp1−/− mice exhibited impaired fertility. We have focused on a description of male hypofertility. Analysis of wild-type testes showed a broad expression of CUG-BP1 in somatic and germ cells, and a combination of approaches demonstrated an increased apoptosis, a reduction of germ cell markers, and an arrest of spermiogenesis in cugbp1−/− testes.
MATERIALS AND METHODS
Targeted disruption of Cugbp1 gene.
Two contiguous 6.5- and 2.2-kb HindIII fragments, covering 3 kb of the 5′ untranslated region, exon 1 to exon 4, and part of intron 4, were isolated from bacterial artificial chromosomes purchased from Research Genetics, Inc. These fragments were used to generate the 5′ and 3′ targeting arms. The 2.2-kb 5′ short KpnI-BglII arm, containing a portion of the 5′ untranslated region and the beginning of exon 1, was fused in frame with nls-lacZ (nuclear localization signal-β-galactosidase-simian virus 40 pA cassette isolated from the plasmid pSKTNLSLACZ, a gift from S. Tajbakhsh), leaving the first 10 amino acids of the Cugbp1 reading frame intact (in exon 1) (see Fig. 1A). Correct in-frame fusion was confirmed by sequencing. Several consecutive cloning steps were required to generate the 3′ homology arm, using the plasmid pPGKDTAlox2pPGKNEO (kindly provided by P. Soriano) as a backbone for the targeting vector. This plasmid contains a cloning site surrounded with a loxP-flanked neomycin resistance cassette (pgk-NEO) and a pgk-DTA cassette encoding the A subunit of the diphtheria toxin gene. The 3′ long arm encompassing the genomic fragment starting in exon 1 at BglII downstream of the nls-lacZ fusion point and extending for 5 kb was cloned in the polylinker of the backbone plasmid. Insertion of this arm was such that the pgk-DTA cassette was positioned at the 3′ end of the final construct. Finally, the 5′ arm fused with nls-lacZ was introduced in the above vector upstream of the pgk-NEO cassette in the right orientation relative to the 3′ arm. The integrity of the targeting vector (17 kb) was verified by restriction digestion.
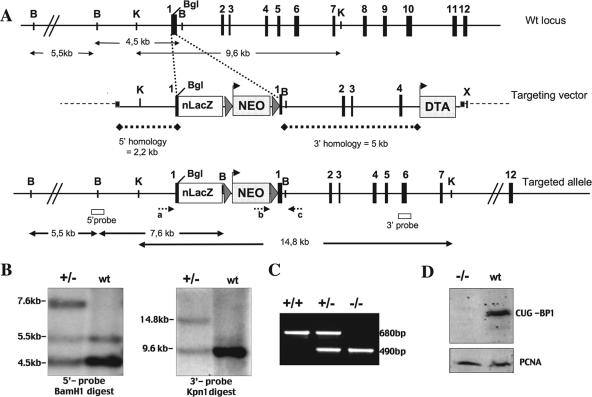
FIG. 1.
Generation of Cugbp1-deficient mice. (A) Schematic representation of the genomic structure of the Cugbp1 locus (top), the targeting vector, and the targeted allele. Exons (solid boxes), LoxP sequences (triangles), and positions of primers for genotype analysis (small arrows a, b, and c) and probes used for Southern blots (open boxes) are indicated. The NEO box flanked with LoxP sequences and the DTA box represent positive and negative selection cassettes. Both are under a pgk promoter. The nls LacZ (nLacZ) cassette in frame with the Cugbp1 coding sequence downstream of the translation start site of Cugbp1 in exon 1 is also shown. Restriction sites relevant to the targeting construct and to the screening strategies are BamHI (B), KpnI (K), XhoI (X), and BglII (BgI). (B) Southern blot analysis of the recombinant and wild-type (wt) ES cell clones: genomic DNA digested with BamH1 or Kpn1 was subjected to Southern blotting and probed with the 5′ and 3′ probes, respectively, depicted in panel A. (C) Results of PCR genotyping using the primers (a, b, and c) shown in panel A. Amplification with primers a and c (wild-type allele) yielded a 680-bp PCR product, and amplification with primers a and c (recombinant allele) yielded a 490-bp PCR product. (D) Western blot of fibroblast whole-cell extract from mutant and wild-type mice, blotted with anti-CUG-BP1 monoclonal antibody (upper panel) or anti-PCNA antibody (lower panel).
CK35 embryonic stem (ES) cells (18) were maintained as previously described (34). The cells were cultured on mitomicyn C-treated Neor primary fibroblasts in the presence of 103 U/ml of leukemia inhibitory factor (ESGRO). A total of 1.6 × 107 ES cells were electroporated with 20 μg of XhoI-linearized targeting vector. After 24 h, G418 (0.3 mg/ml; Invitrogen) was added to the culture medium, and resistant ES clones were recovered 10 to 12 days later in duplicate cultures for 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) staining and DNA preparation. Homologous recombination events were identified by Southern blot analysis with genomic DNA corresponding to X-Gal-positive clones.
DNA samples were either digested with BamHI and probed with a 5′ external probe (380-bp PCR product of a genomic fragment) or digested with KpnI and probed with a 3′ external probe (a 560-bp PCR product of a genomic fragment; see Fig. 1 for digest and probe localization). Using an internal probe in the neomycin gene, ES clones were also checked for a single integration at the CUG-BP1 locus (data not shown).
Two ES cell clones exhibiting the correct targeting event were injected into C57BL/6N blastocysts and transferred into (C57BL/6N × CBA)F1 pseudopregnant females. The resulting chimeric males were mated with C57BL/6N or 129/Sv females. Germ line transmission of the targeted CUG-BP1 locus was confirmed by PCR genotyping of mouse tail tips with a three-primer strategy: a common antisense primer (c) (5′-GGACCACCAGAGCTACAGACA-3′) can pair with a wild-type allele-specific sense primer (a) (5′-ACCACCCAGACCAACCAGAT-3′) or with a mutant allele-specific sense primer (b) (5′-GGGAGGATTGGGAAGACAAT-3′). Amplification of the wild-type allele results in a 490-bp band, while amplification of the mutant allele yields a product of 683 bp (see Fig. 1 for primer localization). The PCR amplification protocol was as follows: 94°C for 4 min, followed by 35 cycles at 94°C for 60s, 58°C for 60 s, and 72°C for 60 s.
CUG-BP1 protein levels were analyzed by Western blotting in fibroblasts from ear biopsies obtained from adult mice. Wild-type or Cugbp1−/− protein extracts corresponding to 3 × 105 cells were resolved by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Western analysis was performed using anti-CUG-BP1 antibodies (monoclonal 3B1; Abcam). Immunoreactive proteins were visualized by using the ECF detection system (Amersham Pharmacia) on a STORM 860 molecular imager.
The phenotypic data of this study were obtained by using mice with a mixed (C57BL/6 × 129sv) genetic background in the F2 generation. The mutant phenotype was recapitulated in mice derived from the two independent ES clones.
Embryo collection, whole mount staining.
Pregnant mice from heterozygous CUG-BP1 intercrosses were scored for fertilization by the appearance of a vaginal plug, and deciduae were isolated from euthanized females. The day of vaginal plug appearance is defined as 0.5. Mouse oocytes and preimplantation embryos were obtained from superovulated female mice, and embryos at different cleavage stages were collected from ampullae or by oviduct flushing at an appropriate time after human chorionic gonadotrophin injection according to standard procedures (27).
The preimplantation embryos were rinsed with phosphate-buffered saline (PBS) and fixed in 2% paraformaldehyde (PFA) and 0.02% glutaraldehyde in phosphate-buffered saline for 10 to 20 min. Postimplantation embryos were fixed in 4% PFA for 15 min to 1 h, depending on the stage of the embryo. lacZ expression was assessed by X-Gal staining, as previously described (27, 47). Stained embryos were stored at 4°C in phosphate-buffered saline-10% glycerol.
For whole-mount immunochemistry, mouse fetuses were fixed at 4°C in MEMFA, rinsed in PBS, dehydrated in H20-methanol, and stored at −20°C in methanol. After rehydratation in PBT (PBS plus Triton [0.5%]), embryos were incubated with polyclonal anti-Xenopus EDEN-BP (which is 88% identical to CUG-BP1) antibody (32) at a dilution of 1/1,000 in PBT with 20% goat serum (Sigma) during 48 h at 4°C. Preimmune serum was used at the same dilution as a negative control. Washes were then performed during 12 h in PBT, and embryos were incubated with a secondary antibody coupled to horseradish peroxidase (Jackson) during 36 h at 4°C. The peroxidase activity was revealed with 3,3′-diaminobenzidine tetrachloride (peroxidase substrate kit; Vector, Biovalley, CA) as a substrate, and the reaction was stopped by rinsing in PBT.
Histology and section staining.
CUG-BP1-deficient and wild-type adult mature mice were killed by cervical dislocation, and gonads were recovered and fixed overnight in Bouin solution at room temperature. Paraffin sections (7 μm) were made and stained with hematoxylin-eosin (Shandon) or with the periodic acid Schiff reagent (to visualize the spermatid acrosome) counterstained with hematoxylin.
To reveal β-galactosidase activity, freshly dissected adult testes were fixed in 4% PFA for 1 h 30 min at 4°C and then rinsed in PBS (20 min, 4°C). The tissues were then included in Tissue Tek compound and immersed in liquid nitrogen. Sections (12 μm) were made with a cryostat and recovered on polylysine-coated slides. The slides were then incubated with freshly prepared X-Gal for up to 2 h at 37°C in a humid atmosphere. As a control, sections from wild-type testes, which do not express the lacZ gene, were treated in parallel.
Embryos were fixed in 4% PFA-PBS for 15 min, soaked in 20% (wt/vol) sucrose, embedded in O.C.T., frozen, and stored at −80°C. Cryostat sections (15 to 25 μm) were briefly fixed in 2% PFA and X-Gal stained as described above.
For detection of the CUG-BP1 protein, testes were included in Tissue Teck compound before being frozen in liquid nitrogen. Cryostat sections, after drying, were fixed in 4% PFA at 4°C for 40 min and then rinsed in PBS (10 min, three times). Alternatively, testes were fixed in 4% PFA for 2 h 30 min at 4°C, rinsed in PBS, and carried through a series of sucrose solutions (12%, 15%, and 18%) in PBS at 4°C. Finally the tissues were included in Tissue Tek compound and immersed in isopentane cooled to −80°C.
Immunostaining was performed with the Vectastain Elite ABC kit (Vector Laboratories) according to the manufacturer's instructions. To reveal CUG-BP1, the sections were incubated overnight (4°C) with either the anti-CUG-BP1 monoclonal antibody (3B1; AbCam) used at a dilution of 1/200 in PBS or the polyclonal anti-Xenopus EDEN-BP antibody (32) used at a dilution of 1/100 in PBS. To avoid the problem of detecting endogenous mouse immunoglobulin Gs with the secondary antibody when using monoclonal antibodies, the Ultravision mouse tissue detection kit (antimouse) (Lab Vision Corp., Fremont, CA) was used. Terminal deoxynucleotidyl transferase-mediated dUTP nick end labeling (TUNEL) staining was performed as described previously (8) using an in situ cell death detection kit (Roche).
Real-time RT-PCR.
Testes were harvested in TRI Reagent (1 ml TRI Reagent/testis; Euromedex). RNA was recovered according to the manufacturer's instructions and quantified by measurement of the optical density at 260 nm. Reverse transcription was carried out following standard procedures using random primers and Superscript II reverse transcriptase (RT) (Invitrogen), except for “RT−” controls, where the enzyme was omitted. Real-time PCR was done with an ABI Prism 7000 device (Applied) using SybrGreen master mix and the primers given below. For each mRNA sample from individual testes, quantifications were made in triplicate. We checked that the RT− controls gave no amplification or gave amplification at a CT value far above that obtained with the corresponding RT+ conditions. Relative mRNA quantities were given by determining the difference between the CT value and the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) CT value according to the following formula: relative quantity = 2exp(CT GAPDH − CT).
Primer sequences for protamine 1 (accession number Xl4003), protamine 2 (NM_008933), proacrosin (D00754), Ldh3 (X04752), GAPDH (XM_354654), LH receptor (M81310), and prostaglandin D (PGD) synthetase (AB006361) have been published (30, 50). The other primers were designed using Primerexpress software (ABI Prism). With the aid of the FastDB databank (7), we selected only pairs of primers that hybridize to different but constitutive exons. The sequences are as follows: hsp70-2 (BC052350), ACTCCGACCAGTCAGGATGTCT and TTGGCGATGATCTCCACCTT; androgen binding protein (MMU85644), AAGTACCTCAGCAATGGCCCA and TGGATCCCAGGTTCGAAACTC; miwi (NM_021311), CCAGCATGAAGACCTCATTGG and TTCCCCATTCCGAGTCTGACT.
RESULTS
Generation of Cugbp1 null mice.
To generate a Cugbp1 null allele, we constructed a targeting vector in which the coding sequence of nls (nuclear localizing sequences)-lacZ was inserted in frame in exon 1 of Cugbp1. This insertion is predicted to generate a null allele and allow the synthesis of β-galactosidase in place of CUG-BP1 in order to easily monitor Cugbp1 expression in situ. The targeting vector contained, in addition, 2.2 and 5 kb of 5′ and 3′ homology, respectively, a floxed Neo cassette for positive selection, and a DT-A cassette for negative selection. After electroporation of the targeting vector into 129Sv CK35 ES cells (18), Neor-selected clones were monitored for LacZ expression: 17% (49 out of 288) were X-Gal positive, and among these, 24 out of 31 tested using the appropriate probes were shown to have the correct insertion in both the 3′ and 5′ regions (Fig. 1; Materials and Methods). Two targeted ES clones were injected into C57BL/6N blastocysts, and both produced chimeras which transmitted the disrupted allele to the progeny. Heterozygotes obtained from germ line transmitting chimeras were then intercrossed and the progeny genotyped. RT-PCR was performed on RNA of fibroblasts isolated from Cugbp1−/− (−/−) and Cugbp1+/+ (+/+) mice. Using primers flanking the region of insertion of nls-LacZ, the expected amplified fragments were obtained from +/+ fibroblast RNA; in contrast, no amplification could be obtained in the case of the mutant fibroblasts (data not shown). In addition, CUG-BP1 protein synthesis was monitored in −/− or +/+ fibroblasts. As can be seen in Fig. 1D, CUG-BP1 protein synthesis is completely abolished in Cugbp1−/− fibroblasts. Taken together, these results show that the targeting vector allowed us to generate a null allele.
Genotyping was done both with embryos from cugbp1+/− (+/−) intercrosses collected by cesarean section just before birth (E18.5) and with born animals. In embryos, the proportions of the three genotypes (cugbp1+/+, cugbp1+/−, and cugbp1−/− [+/+, +/−, and −/−]) were not statistically different from the expected Mendelian distribution (P = 0.17, χ2 test; Table 1). Furthermore, no resorption sites were found among a total of 14 pregnant mice analyzed, indicating that there was no intrauterine mortality. In contrast, when the same analysis was performed around 8 to 10 days after birth, we found that the proportion of −/− progeny was significantly different from the expected Mendelian frequency, with homozygous mutants being underrepresented (P = 2.8 E−06, χ2 test; Table 1). This was most probably due to the fact that the −/− progeny has difficulty thriving due to their runt condition (see the “growth retardation” section below). In fact, we occasionally observed the death, 1 or 2 days after birth, of some overtly smaller and presumably −/− progeny. In each instance where genotyping was possible, we could verify that they were indeed −/− mutants. Furthermore, a proportion of the dead progeny may have been eaten by the mother and therefore escaped our analysis. Collectively, these results suggest that while there is some perinatal mortality of the −/− progeny, the latter are nevertheless viable and therefore may reach the adult state.
TABLE 1.
Genotype analysis of Cugbp1+/− intercross progenya
| Mouse group (nb) | No. of mice observed per genotype
|
P value (χ2 test) | ||
|---|---|---|---|---|
| +/+ | +/− | −/− | ||
| E18.5 (102) | 31 | 53 | 18 | 0.17 |
| P8-10 (299) | 90 | 172 | 37 | 2.8 E-06 |
Late embryos (E18.5) or young mice (8 to 10 days after birth [P8-10]) resulting from intercrosses between heterozygous (+/−) mice were genotyped. A χ2 test was used to evaluate the fitting of the observed distribution with the expected Mendelian ratio of 1:2:1 (+/+, +/−, −/−).
n, no. of mice analyzed.
CUG-BP1-deficient mice display growth retardation.
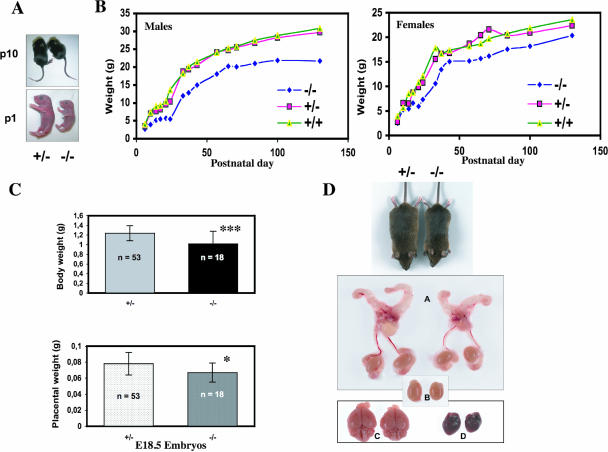
Among littermates generated by interbreeding heterozygous +/− mice, we noticed that homozygous mutant progeny were somewhat smaller, as illustrated in Fig. 2A for individuals at 1 or 10 days after birth (P1 or P10). We therefore analyzed in more detail the weight and growth characteristics of the −/− progeny compared to those of the wild-type and heterozygous mice. As can be seen in Fig. 2B, the rate of weight gain of both male and female homozygous mutant mice is lower than that of both the heterozygous or wild-type littermates. This was reflected, for example, by the observation that 14-week-old male mutant mice (n = 6) weighed on average 23% less than heterozygous mice. At the same age, mutant females (n = 6) weighed 16% less. This difference remained stable throughout their life (about 2 years), and Cugbp1−/− mice never reached the size of their littermates. In addition, the growth deficiency was already apparent just before birth, when the weights of Cugbp1+/− (n = 53) and Cugbp1−/− (n = 18) E18.5 embryos collected by cesarean section were compared (Fig. 2C). Cugbp1−/− E18.5 embryos were 18% smaller than the heterozygous ones. This could be due to an impaired placental function, since the placentas of E18.5 Cugbp1−/− conceptuses were also found to display a significantly lower weight than the Cugbp1+/− ones (Fig. 2C). We also compared the weights of total body and organs of adult −/− and +/− males (n = 4 for each genotype; age range, 7 to 12 months). As expected from the growth curves, the −/− mouse weights were about 65% of those of the +/− mice. Furthermore, except for the spleen, the other organs, including the heart, lung, liver, brain, kidney, and testes, were smaller (Fig. 2D).
FIG. 2.
Analysis of growth phenotype. (A) Representative pictures of heterozygous and Cugbp1−/− littermates of newborn (P1) or 10-day-old (P10) mice. (B) Postnatal growth curve of male or female littermates. Mice were weighed over a period of 130 days. Results are plotted as the mean body weight of female −/− (n = 3 to 6), male −/− (n = 3 to 6), female +/− (n = 3 to 18), male +/− (n = 4 to 23), female +/+ (n = 3 to 6), or male +/+ (n = 3 to 10) mice. (C) Body or placenta weight of E18.5 +/− (n = 53) or −/− (n = 18) conceptuses. Fetal body weights (means ± standard deviation) were as follows: Cugbp1+/−, 1.238 ± 0.153 g; Cugbp1−/−, 1.018 ± 0.259 g; ***, P < 0.001 (Fisher test). Placental weights (means ± standard deviations) were as follows: Cugbp1+/−, 0,078 ± 0,014; Cugbp1−/−, 0,067 ± 0,012. *, P = 0.016 (Fisher test). (D) Macroscopic appearance of selected organs from a 4-month-old Cugbp1-deficient male (right) and a heterozygous male littermate (left). A, reproductive tract; B, testes; C, brain; D, heart.
Expression of CUG-BP1 in whole embryos.
To analyze the normal expression of CUG-BP1 during development of the mouse embryo, two approaches were used: measurement of CUG-BP1 promoter activity revealed by detecting β-galactosidase activity and study of the protein expression by immunohistochemistry.
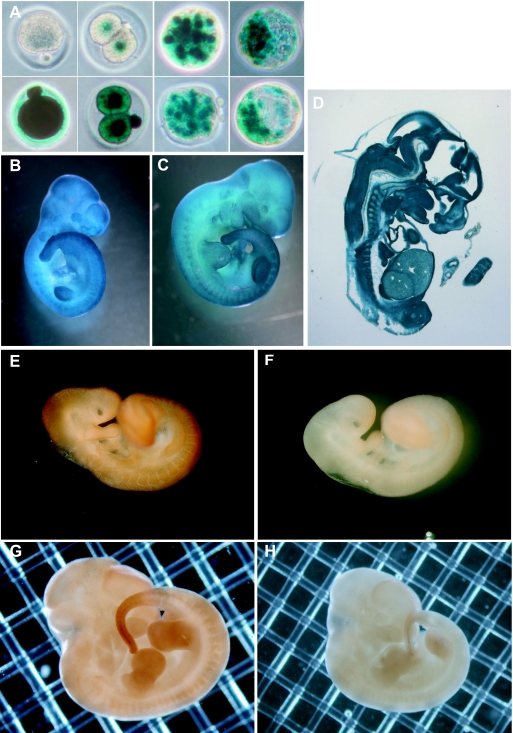
We first examined CUG-BP1 expression in preimplantation development. To that end, embryos were collected at different times after mating, and β-galactosidase expression was monitored. A cross between a heterozygous male and a wild-type female showed that Cugbp1 promoter activity initiates at the two-cell stage, i.e., at the time of the first main zygotic activation, and then continues at least until the blastocyst stage (Fig. 3A, upper panels). The reciprocal cross showed that Cugbp1 promoter activity is very strong in the oocyte and is sustained during the preimplantation period (Fig. 3A, lower panels).
FIG. 3.
CUG-BP1 expression during embryo development. (A to D) Expression of CUG-BP1 analyzed by X-Gal staining. (A) Upper panels, left, an oocyte from a +/+ female; then (left to right), two-cell, morulae, and blastocyst embryos derived from crosses between +/+ superovulated females and +− males. Lower panels, left, an oocyte from a +/− female; then (left to right), two-cell, morulae, and blastocyst embryos derived from reciprocal crosses between +/− superovulated females and +/+ males. (B) Ten-day-postcoitum embryo. (C) Eleven-day-postcoitum embryo. (D) Section of a 12-dpc embryo. (E to H) Expression of CUG-BP1 analyzed by immunohistochemistry of wild-type embryos. (E) Nine-day-postcoitum embryo stained with anti-EDEN-BP antibody. (F) Nine-day-postcoitum embryo stained with preimmune serum. (G) Twelve-day-postcoitum embryo stained with anti-EDEN-BP antibody. (H) Twelve-day-postcoitum embryo stained with preimmune serum.
To evaluate the expression pattern of CUG-BP1 later in development, embryos were obtained at different times postcoitum from crosses between heterozygous males and females and stained for β-galactosidase. This showed that CUG-BP1 was widely expressed at 10 and 11 days postcoitum (dpc), with the highest levels of expression observed in the tail region, somites, cephalic structures, and limb buds (Fig. 3B and C). This was confirmed by immunohistochemical staining of wild-type embryos with an anti-EDEN-BP antibody that yielded similar results for 9- and 12-dpc embryos (Fig. 3E and G). Preimmune serum used as a negative control assessed signal specificity (Fig. 3F and H). That the endogenous CUG-BP1 protein had the same temporal and spatial pattern of expression as β-galactosidase was also observed for a number of other developmental stages (data not shown).
To determine Cugbp1 expression in internal tissues, β-galactosidase staining was performed on sections of +/− 12-dpc embryos. Results shown in Fig. 3D indicated that expression was extensive although variable in intensity.
Impaired fertility in mice lacking CUG-BP1.
When attempting to cross −/− mice, we rapidly realized that they were highly hypofertile or even sterile. The fertility of homozygous mutant mice of both sexes was more accurately monitored using the following protocol. Adult mutant males were mated singly with two heterozygous or wild-type females, and two mutant females were mated with one heterozygous or wild-type male. The matings were performed for 3 months, during which the number of litters and of pups born was systematically monitored. Control crosses were performed by mating one heterozygous male with two heterozygous females.
Fifteen homozygous mutant males were tested. The vast majority (11 [73%]) had dramatically impaired fertility. Five of them did not produce a single litter during the 3-month period, while among the remaining 6, 3 gave only 1 litter (average litter size, 3.6), 2 gave 2 litters (average litter size, 4.0), and the last 1, 3 litters (average litter size, 3.0). Comparison with the control cross (average number of litters, 4.6; average litter size, 7.6) reveals a highly significant difference (P = 0.017 and 0.019 for the number of litters and the litter size, respectively; Mann-Whitney test). Finally, a minority of mutant males (n = 4 [26%]) appeared to be normally fertile, since the average number of litters (3.75) and the average litter size observed (7.9) were not significantly different from those observed with the control (P = 0.076 and 0.7, respectively; Mann-Whitney test). This indicates that the sterility phenotype observed for mutant males is not fully penetrant, presumably due to the mixed genetic background.
Thirteen females were mated to either wild-type or heterozygous males during the 3-month period. Each of them gave only 1 litter, with an average litter size of 2.38. During the same time period, 6 heterozygous females of the control crosses produced a total of 14 litters (2.3 per female), with a mean litter size of 7.57. We conclude from these results that the fertility of homozygous mutant females is severely impaired.
There did not seem to be a problem of sexual behavior with either mutant males or females, as demonstrated by the frequent presence of vaginal plugs. Taken together, these results indicate that disruption of Cugbp1 results in severely impaired fertility for both male and female mice without obvious anomalies in sexual behavior.
Disruption of cugbp1 causes spermatogenesis defects.
To search for a cause of the decreased fertility, ovaries and testes were removed from the −/− and wild-type mature animals, and histological sections were performed. The general histology of the ovaries was normal, and no obvious defect in the population ratios of the different follicular stages was observed (data not shown). The reasons for decreased fertility of −/− females remain therefore to be investigated. In contrast, a clear difference was observed between the wild-type and most −/− testes. Figure 4 shows testis and epididymis sections of one +/+ and two −/− mature males. The seminiferous tubules from the wild-type animal showed a normal pattern of spermatogenesis, with the presence of all spermatogenic stages from spermatogonia to spermatozoa. As expected, almost all seminiferous tubules contained elongated spermatids (Fig. 4A and B). The epididymis from the wild-type animal was filled with spermatozoa (Fig. 4C).
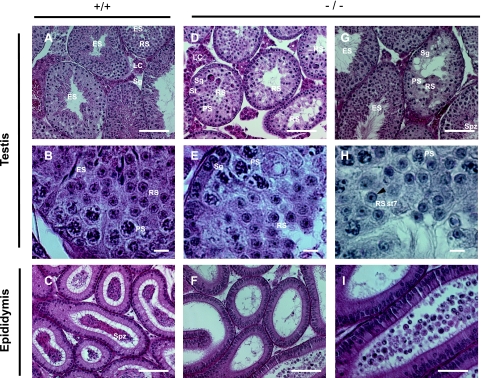
FIG. 4.
Histological comparison of testes and epididymes from wild-type and Cugbp1−/− mice. (A) Low-magnification section of a testis from a mature, wild-type male. (B) Higher magnification of the same section shown in panel A. (C) Low-magnification section of the epididymis from the same male. (D) Low-magnification section of a testis from a mature, Cugbp1−/− male with a strong spermiogenesis arrest. (E) Higher magnification of the same section shown in panel D. (F) Low-magnification section of the epididymis from the same male. (G) Low-magnification section of a testis from a mature, Cugbp1−/− male with a mild spermiogenesis arrest. (H) High-magnification section of a testis from a mature, Cugbp1−/− male with a strong spermiogenesis arrest, stained with periodic acid Schiff and counterstained with hematoxylin. The arrowhead indicates the cap-shaped acrosome of the round spermatid (RS), and St7 is step 7 of spermiogenesis. (I) Higher magnification of F. Except for H, all the sections were stained with hematoxylin and eosin. LC, Leydig cells; Sg, spermatogonia; PS, pachytene spermatocyte; RS, round spermatid; ES, elongated spermatid; Spz, spermatozoa. For panels A, C, D, and F, bar = 100 μm; for panels B, E, and H, bar = 10 μm; for panels G and I, bar = 50 μm.
The testes from 17 −/− males were similarly analyzed. For a minority of them (6/17 [35%]), we could find no difference with the sections from a +/+ testis. In contrast, 11 (65%) of the −/− testes showed a different picture. In 7 of the −/− testes, spermatogonia, pachytene spermatocytes, and round spermatids were present but all the seminiferous tubules were devoid of elongated spermatid and spermatozoa. This corresponds to the “strong” phenotype shown in Fig. 4D and E. Furthermore, some seminiferous tubules of the −/− testes showed vacuoles and seemed to contain fewer germ cells. Hence, in these mice, spermiogenesis (differentiation of round spermatids into spermatozoa) is blocked before spermatid elongation. Of these seven animals, three had been tested for their fertility by mating before sections were performed, and all of them were found to be sterile (no litter). In the four other affected −/− testes, spermiogenesis was blocked in most tubules, but some tubules still harbored elongated spermatids or spermatozoa. This corresponds to the “mild” phenotype shown in Fig. 4G (compare the central tubule where spermatid elongation did not occur and the tubules with elongated spermatids or spermatozoa). The fertility of two of these four animals (including the one shown in Fig. 4G) had been tested before sections were performed, and these two animals were found to be sterile. This suggests that even if some spermatozoa are produced by these animals, this is not sufficient to confer fertility.
Mouse spermiogenesis can be divided into 16 steps according to modifications of the acrosome shape (36). To determine the step at which spermiogenesis is arrested in −/− males, sections were stained with periodic acid Schiff to reveal glycoproteins that are present within the acrosome or the proacrosomal vesicles (Fig. 4H). Round spermatids with an extended cap-shaped acrosome could be detected, showing that the fusion of proacrosomal vesicles was complete in these spermatids. This corresponds to step 7 of round spermatids. In addition, spermatogenesis in −/− mice was also distinguished from that in wild-type mice by the presence of multinucleated giant cells (Fig. 4D). Such giant cells were previously shown to correspond to apoptotic germ cells (28). Finally, in agreement with the absence of elongated spermatid and spermatozoa in the seminiferous tubules from −/− mice with the strongest phenotype, the epididymis from the testes of these animals was devoid of spermatozoa (Fig. 4F). In some cases, degenerated round spermatids were found in the epididymis (Fig. 4I). Together, these results show that CUG-BP1 plays a key role in spermatid elongation and, therefore, production of spermatozoa.
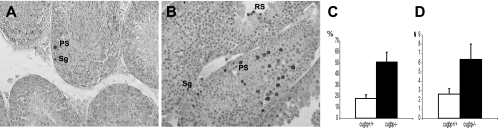
Next, apoptosis was compared for +/+ and −/− testes. Sections of +/+ and −/− testes were TUNEL stained (Fig. 5). Some germ cells (spermatogonia and spermatocytes) from a minority of tubules in +/+ testes were found to be TUNEL positive (Fig. 5A), indicating that apoptosis-induced DNA fragmentation had occurred in these cells. However, more TUNEL-positive cells were found in the −/− testis. Furthermore, round spermatids, in addition to spermatogonia and spermatocytes, were found to be positive (Fig. 5B). The percentage of seminiferous tubules containing at least one positive cell was significantly higher in −/− testes than in +/+ testes (Fig. 5C), and in the tubules containing at least one positive cell, the average number of positive cells was significantly higher in −/− testes than in +/+ testes (Fig. 5D). Therefore, germ cells from −/− testes are more likely to enter into apoptosis than germ cells from +/+ testes.
FIG. 5.
Apoptosis in testes of Cugbp1+/+ and Cugbp1−/− males. Sections of testes from +/+ (A) or −/− (B) males were subjected to TUNEL staining. Spermatogonia (Sg) and pachytene spermatocyte (PS) and a round spermatid (RS) in B that are TUNEL positive are depicted. (C) Percentages of seminiferous tubules containing at least one TUNEL-positive cell in +/+ or −/− testes. P = 1.7 E−06, Student test. (D) Average number of TUNEL-positive cells per tubule containing at least one positive cell. P = 5 E−04, Student test.
Expression of germ cell markers is altered in −/− testes.
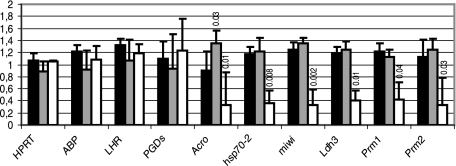
As a first step toward understanding the molecular basis of the spermatogenesis defects illustrated in Fig. 4 and 5, we analyzed the expression of a number of markers by real-time RT-PCR. ABP (androgen-binding protein) is a Sertoli marker (40). We found that the expression of this mRNA was very similar in +/+, +/−, and −/− testes, suggesting that the function of Sertoli cells is not altered in −/− mice (Fig. 6). Similarly, the expression of LH receptor and of PGD synthetase, two Leydig markers (30), was the same in all three genotypes, suggesting again that the primary cause for the observed spermatogenesis defects is not an alteration of Leydig cells (Fig. 6). We next examined the expression of six germ cell markers. Four of them, proacrosin/acrosomal serine protease (acro), hsp70-2, miwi, and Ldh3/LdhC4, are transcribed at the beginning of meiosis (early spermatocytes I) and thereafter (10, 16, 22, 37). The other two, protamines 1 and 2, are transcribed only in postmeiotic cells (spermatids) (21). We found that the amount of all these markers was significantly less in −/− testes than in +/+ testes. In contrast, except for proacrosin, the amounts of the markers were not different in +/+ and in +/− testes (Student test; P > 0.1). Finally, and as expected, the expression level of the ubiquitously expressed hypoxanthine phosphoribosyltransferase mRNA was the same in the testes of all three genotypes.
FIG. 6.
Expression of Sertoli, Leydig, and germ cell markers. Relative quantities of the ubiquitously expressed GAPDH mRNA and of the indicated mRNAs were quantified by real-time RT-PCR. The amounts of the indicated mRNA relative to that of the GAPDH mRNA were calculated for +/+ (black bars; n = 3), +/− (gray bars; n = 4), and −/− (white bars; n = 3 [2 −/− mice with a histological “strong phenotype” and 1 mouse with a histological “mild phenotype”]; see Fig. 4) mice. Results are expressed as the mean (± standard deviation) of the ratios for each genotype, normalized to 1 for the average value of the three phenotypes. Statistical comparison between the +/+ genotype and the other genotypes was done with a Student test. When the P values were less than 0.1, they were indicated above the corresponding bars. ABP, androgen binding protein; LHR, LH receptor; PGDs, PGD synthetase; Acro, proacrosin; Prm1 and Prm2, protamines 1 and 2.
These real-time RT-PCR results suggest a decrease in the number of germ cells in −/− mice. Since even early markers are downregulated, this decrease probably concerns all the germ cells and not only elongating spermatids and spermatozoa (see Discussion).
CUG-BP1 expression in testes.
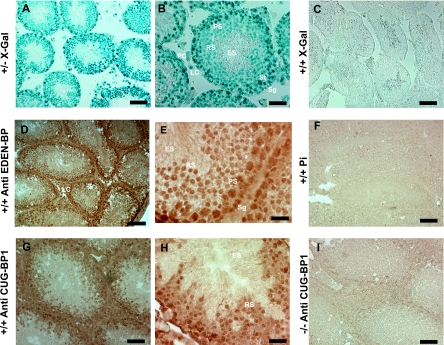
The above results show that the inactivation of Cugbp1 results in a blockage of spermatid elongation and a general decrease of germ cell markers. These two alterations may be independent consequences of the absence of CUG-BP1, or one of the alterations may be a consequence of the other. Each of these hypotheses requires that CUG-BP1 is expressed in particular cells in the testes. We examined Cugbp1 expression in testicular cells. To that end, histological sections were made from Cugbp1+/− testis and the promoter activity was revealed by β-galactosidase labeling (Fig. 7A and B). The lacZ gene inserted into the Cugbp1 gene contains a nuclear localization signal, so that the β-galactosidase staining is nuclear. In Fig. 7A is shown a transverse section of a testis harboring several seminiferous tubules at different stages of the cycle of the seminiferous epithelium. With higher magnification (Fig. 7B), Leydig cells, peritubular cells, Sertoli cells, spermatogonia, pachytene spermatocytes, and round spermatids could be observed. In this seminiferous tubule, the elongated spermatids are at step 14/15 of the spermiogenesis (36). All of these cells except the elongated spermatids are β-galactosidase positive, showing that the Cugbp1 promoter is active from spermatogonia to round spermatids. It was expected that the more advanced elongated spermatids in this seminiferous tubule would not be stained for β-galactosidase, since transcription stops around step 10 of spermiogenesis (17). That the observed β-galactosidase activity is due to Cugbp1 gene activity was shown by the absence of any X-Gal staining in the testes for wild-type animals (Fig. 7C).
FIG. 7.
Expression of CUG-BP1 in the testis. (A and B) Testes from heterozygous males were used to analyze promoter activity of Cugbp1 by detection of β-galactosidase activity. B is a higher magnification of A. Localization of the staining is nuclear because of the presence of nls in the construction. (C) Testis from a wild-type (+/+) male was used as a negative control for X-Gal staining. (D to H) Testes from wild-type (+/+) mice were used for immunohistochemical localization of CUG-BP1. Immunohistochemistry was performed with an anti-EDEN-BP antibody (D and E), the corresponding preimmune serum (F), or anti-CUG-BP1 antibody (G and H). (I) Immunohistochemical staining performed on a testis from a −/− mouse with the anti-CUG-BP1 antibody. In panels D to G and I, testes were fixed in paraformaldehyde after congelation, whereas in panel H, the testis was fixed before congelation, which allowed preservation of the cytoplasmic compartment. LC, Leydig cell; St, Sertoli cell; PC, peritubular cell; Sg, spermatogonia; PS, pachytene spermatocyte; RS, round spermatid; ES, elongated spermatid. For panels A, D, C, and F, bar = 100 μm; for panels B, G, and I, bar = 50 μm; for panels E and H, bar = 25 μm.
In order to evaluate the expression of CUG-BP1 in elongated spermatids, a study of the endogenous CUG-BP1 protein was performed by immunohistochemistry. This expression was revealed by staining sections from wild-type testis with anti-EDEN-BP polyclonal antibody or anti-CUG-BP1 monoclonal antibody. Similar staining patterns were obtained with both antibodies (Fig. 7D to I). As observed for β-galactosidase staining, the endogenous CUG-BP1 protein was detected in all the germ cells of the seminiferous tubules except in the central region that contains the elongated spermatids and spermatozoa (Fig. 7D and G). It was also detected in the Leydig cells of the interstitial tissue (Fig. 7D). The specificity of this staining for CUG-BP1 is attested by its absence from testes obtained from −/− animals (Fig. 7I; also data not shown). Also, no staining was observed with the preimmune serum for the anti-EDEN-BP antibody (Fig. 7F). Analysis of higher-magnification pictures again indicated that CUG-BP1 is expressed in the somatic cells of the seminiferous tubule, such as peritubular and Sertoli cells (Fig. 7E; also data not shown) but also in spermatogonia, Pachytene spermatocytes, and round spermatids. It was absent from elongated spermatids (Fig. 7E). Numerous fixation conditions were tested to obtain CUG-BP1 antigenicity preservation associated with the best histological images (allowing identification of cell type). Direct freezing, followed by paraformaldehyde postfixation of the sections, gave the best results and corresponds to the pictures shown (except Fig. 7H). However, under these conditions, cellular membranes are not well preserved and the limits of the cytoplasms are not clearly defined. An alternative protocol (paraformaldehyde fixation prior to freezing) allowed us to conclude that CUG-BP1 is expressed in both the nuclear and cytoplasmic compartments of the stained cells, notably of the round spermatids (Fig. 7H).
In conclusion, analysis of CUG-BP1 expression at the gene and protein levels showed that this factor is broadly expressed in the mouse testes. Indeed, somatic cells (Leydig, Sertoli, and peritubular cells) but also germ cells from spermatogonia to round spermatids expressed CUG-BP1.
DISCUSSION
To understand the biological functions of CUG-BP1 in mammals, we have generated mice in which this gene was inactivated by homologous recombination. Mice devoid of CUG-BP1 display a number of phenotypic traits, including perinatal mortality and a growth retardation that is already observed in embryos and is not compensated in postnatal life. In addition, a remarkable feature of these mice is their marked hypofertility, both for males and females, despite apparently normal sexual behavior. Gene inactivation of RNA-binding proteins that led to both male and female infertility has been described previously. For instance, inactivating CPEB blocks germ cells at the pachytene stage of meiosis (39), and inactivating TIAR leads to a depletion of primordial germ cells (3). In these cases, both male and female germ cells are affected. The situation is different for CUG-BP1, since we could find no evidence of a difference between the germ cells of Cugbp1−/− and wild-type females. Histological examination of ovaries did not show any obvious alteration in the number or nature of oocytes or follicles. Hence, CUG-BP1−/− females may be infertile due to an impaired gestation for reasons that remain to be explored.
In contrast, we found that spermatogenesis showed alterations in about two-thirds of the −/− males, while it was apparently normal in the remaining one-third. The difference between these individuals is probably due to the mixed genetic background. This altered spermatogenesis had three characteristics, a blockage of spermiogenesis at stage 7 in all or most of the seminiferous tubules, an increased apoptosis, and a down-regulation of six germ cell markers. In wild-type mice, proacrosin and hsp70-2 mRNA persist until the round spermatid stage, but their abundance strongly decreases in elongating spermatids (16, 37). Accordingly, we think that the downregulation of these markers observed in −/− testes cannot be explained by the depletion of elongated spermatids. Rather, we favor the hypothesis that the downregulation of all the germ cell markers tested in −/− testes is due to a general decrease of the number of germ cells, including spermatogonia, spermatocytes, and round spermatids. One cause for this decrease may be the increased apoptosis, but we cannot rule out that, in addition, spermatogonia divide to a lesser extent. Nevertheless, the decrease in germ cell number would be evocative of the phenotype of mice inactivated for TIAR, an RNA-binding protein involved in the control of alternative splicing and mRNA stability, like CUG-BP1. TIAR−/− mice show a complete depletion of primordial germ cells and consequently of more advanced germ cells (3).
What are the relationships between the spermiogenesis blockage and the decrease in germ cell abundance? Spermatogenesis is a highly controlled process. There seem to be a number of cross talks in seminiferous tubules in order to maintain a critical cell ratio between the different germ cell stages and between these cells and Sertoli cells (35). Accordingly, the arrest of spermiogenesis may cause spermatogonia to reduce proliferation or favor the entry of spermatogonia or spermatocytes into apoptosis. Alternatively, these two events may be independent consequences of cugbp1 inactivation. CUG-BP1 is expressed both in spermatogonia and in round spermatids, so it could be independently required for both spermatogonia proliferation and spermatid differentiation.
A number of genes whose inactivation alters spermatogenesis have been identified. However, only a few gene inactivations alter spermiogenesis. Interestingly, five of the genes whose inactivation leads to spermiogenesis defects encode proteins implicated in posttranscriptional regulations of gene expression, like CUG-BP1. These are the following: (i) MSY2, which participates in storing dormant mRNAs (51); (ii) MIWI, a cytoplasmic protein found associated with, and possibly regulating, the stability, of certain mRNAs (10); (iii) tPAP, a cytoplasmic, testis-specific poly(A) polymerase, the enzyme that elongates the poly(A) tail of mRNAs (16); (iv) PRBP, a double-stranded RNA-binding protein (53); and (v) GRTH/Ddx25, an RNA helicase (43). In addition, some genes that do not encode proteins involved in posttranscriptional processes but whose disruption leads to a blockage of spermiogenesis have also been described. This is the case for CREM, TLF/TRF2, RNF17, and TSLC1/IGFS4 (5, 23, 28, 33, 45, 49, 52).
What is the link between the absence of CUG-BP1 and the blockage of spermiogenesis at stage 7? It is highly probable that in wild-type mice CUG-BP1 regulates a number of RNAs and that it is the deregulation of these mRNAs, following the inactivation of CUG-BP1, that is responsible for the altered spermiogenesis. This raises the question of the identity of these RNA targets of CUG-BP1 but also of the type of regulation that CUG-BP1 exerts on them. CUG-BP1 is a multifunctional protein that can regulate both alternative splicing and translation (see the introduction). Importantly, we have shown that CUG-BP1 is expressed in both the nucleus and the cytoplasm of germ cells from spermatogonia to spermatids, so it could play a role in both splicing and cytoplasmic regulation in any of these cells.
The testis is an organ where several events of alternative splicing take place (for reviews, see references 11 and 46). For instance, during meiosis, the splicing pattern of Crem changes, causing a switch from an inactive to an active form of the protein (for a review, see reference 4). Disruption of the Crem gene impairs spermiogenesis at stage 4, and it is possible that preventing the change in its splicing pattern would have the same consequences (5, 28). CUG-BP1 may regulate the alternative splicing of one or several pre-mRNAs (as yet unidentified) in spermatogonia, spermatocytes, or early spermatids, leading to the appearance of isoforms that are required at stage 7 of spermiogenesis to allow elongation of round spermatids.
Proteins that stimulate translation and are required for spermatogenesis have also been described. For instance, PRBP is required to activate the translation of protamine mRNAs (53). Generally, since polyadenylation is associated with translational stimulation (for a review, see reference 9), the testis-specific poly(A) polymerase tPAP (16) or the polyadenylation-promoting factor CPEB (39) stimulates the translation of its target mRNA. Disruption of either of these genes abrogates spermatogenesis. CUG-BP1 was shown to stimulate translation of the mRNA encoding the cdk inhibitor p21 (41) and of a reporter RNA when tethered to the 3′UTR (2). CUG-BP1 may therefore be required to stimulate the translation of one or several mRNAs during spermatogenesis, and the lack or decreased concentration of the corresponding proteins may be responsible for impaired spermiogenesis in mice devoid of CUG-BP1. As for splicing, the hypothetical stage of spermatogenesis where the translation of some mRNAs has to be stimulated in a CUG-BP1-dependent manner may be during or even before spermiogenesis.
Finally, in Xenopus embryos, EDEN-BP causes mRNA deadenylation, which is associated with translational repression (12, 32). Human CUG-BP1 can replace EDEN-BP in a Xenopus egg extract after immunodepletion of the endogenous protein (31). Recently, the capacity of CUG-BP1 to act as a deadenylation factor was confirmed with a mammalian cell extract (25). Therefore, in some instances, CUG-BP1 can act as a translational repressor of target mRNAs, and inactivating CUG-BP1 may cause the inappropriate expression of one or several target mRNAs, potentially in any germ cell between spermatogonia and round spermatid, which could cause the observed arrest in spermiogenesis. That translational repression is important for spermiogenesis is illustrated by two observations. MSY2 is one of the Y-box proteins, a family of DNA and RNA binding proteins that are involved in silencing a number of mRNAs. Inactivation of MSY2 impairs spermiogenesis (51). Another example of translational repression in testis is given by the protamine mRNAs. These mRNAs accumulate during early spermiogenesis but are translated only days later, in elongated spermatids. Causing the premature translation of protamine 1 blocks spermiogenesis (20).
These data highlight the need for the identification, on a systematic basis, of the pre-mRNA or mRNA targets of CUG-BP1. As for several RNA-binding proteins, there is no strict RNA consensus sequence for CUG-BP1 binding. Indeed, published RNA sequences that interact with this protein include CUG repeats, GCN repeats, and UG- or UG/UA-rich regions (reviewed in reference 1). This diversity in the sequences recognized by CUG-BP1 precludes the use of a purely bioinformatic approach to identify its RNA targets. Coimmunoprecipitation approaches (29, 44), which were recently adapted to mouse testis (6), are more likely to allow the identification of the targets of CUG-BP1 and hence the elucidation of the molecular link between CUG-BP1 and spermatogenesis.
Acknowledgments
We thank S. Tajbakhsh for helpful discussion and suggestions about the targeting vector. We are extremely grateful to M. Kress for excellent advice on molecular biology and for his invaluable help concerning the design of the sequential steps of the targeting construct as well as his constant support and fruitful discussion during this work. We are also grateful to X. Montagutelli for his help with statistical analysis. We acknowledge T. Rouxel for excellent photographic assistance. We are also grateful to the technical staff of the Laboratoire d'Histologie et de Biologie Cellulaire of the Medical Faculty of Rennes for access to their histological equipment. Microscope observations of histological sections were made on the Platefome de Microscopie of the IFR 140 (University of Rennes). We sincerely thank X. Jaglin for technical assistance and R. Habert and V. Rouiller-Fabre for discussions on immunohistochemical analyses.
This work was supported by grants from the Centre National de la Recherche Scientifique, the Institut Pasteur, the Association de la Recherche sur le Cancer (contract no. 4791), and the Ministère Délégué à la Recherche ACI BCMS314 (to L.P.).
Footnotes
Published ahead of print on 27 November 2006.
REFERENCES
- 1.Barreau, C., L. Paillard, A. Mereau, and H. B. Osborne. 2006. Mammalian CELF/Bruno-like RNA-binding proteins: molecular characteristics and biological functions. Biochimie 88:515-525. [DOI] [PubMed] [Google Scholar]
- 2.Barreau, C., T. Watrin, H. Beverley Osborne, and L. Paillard. 2006. Protein expression is increased by a class III AU-rich element and tethered CUG-BP1. Biochem. Biophys. Res. Commun. 347:723-730. [DOI] [PubMed] [Google Scholar]
- 3.Beck, A. R., I. J. Miller, P. Anderson, and M. Streuli. 1998. RNA-binding protein TIAR is essential for primordial germ cell development. Proc. Natl. Acad. Sci. USA 95:2331-2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Behr, R., and G. F. Weinbauer. 2001. cAMP response element modulator (CREM): an essential factor for spermatogenesis in primates? Int. J. Androl. 24:126-135. [DOI] [PubMed] [Google Scholar]
- 5.Blendy, J. A., K. H. Kaestner, G. F. Weinbauer, E. Nieschlag, and G. Schutz. 1996. Severe impairment of spermatogenesis in mice lacking the CREM gene. Nature 380:162-165. [DOI] [PubMed] [Google Scholar]
- 6.Cho, Y. S., N. Iguchi, J. Yang, M. A. Handel, and N. B. Hecht. 2005. Meiotic messenger RNA and noncoding RNA targets of the RNA-binding protein Translin (TSN) in mouse testis. Biol. Reprod. 73:840-847. [DOI] [PubMed] [Google Scholar]
- 7.de la Grange, P., M. Dutertre, N. Martin, and D. Auboeuf. 2005. FAST DB: a website resource for the study of the expression regulation of human gene products. Nucleic Acids Res. 33:4276-4284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Delbes, G., C. Levacher, C. Pairault, C. Racine, C. Duquenne, A. Krust, and R. Habert. 2004. Estrogen receptor beta-mediated inhibition of male germ cell line development in mice by endogenous estrogens during perinatal life. Endocrinology 145:3395-3403. [DOI] [PubMed] [Google Scholar]
- 9.de Moor, C. H., H. Meijer, and S. Lissenden. 2005. Mechanisms of translational control by the 3′ UTR in development and differentiation. Semin. Cell Dev. Biol. 16:49-58. [DOI] [PubMed] [Google Scholar]
- 10.Deng, W., and H. Lin. 2002. miwi, a murine homolog of piwi, encodes a cytoplasmic protein essential for spermatogenesis. Dev. Cell 2:819-830. [DOI] [PubMed] [Google Scholar]
- 11.Elliott, D. J. 2004. The role of potential splicing factors including RBMY, RBMX, hnRNPG-T and STAR proteins in spermatogenesis. Int. J. Androl. 27:328-334. [DOI] [PubMed] [Google Scholar]
- 12.Ezzeddine, N., L. Paillard, M. Capri, D. Maniey, T. Bassez, O. Aït-Ahmed, and B. Osborne. 2002. EDEN dependent translational repression of maternal mRNAs is conserved between Xenopus and Drosophila. Proc. Natl. Acad. Sci. USA 99:257-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Filardo, P., and A. Ephrussi. 2003. Bruno regulates gurken during Drosophila oogenesis. Mech. Dev. 120:289-297. [DOI] [PubMed] [Google Scholar]
- 14.Gautier-Courteille, C., C. Le Clainche, C. Barreau, Y. Audic, A. Graindorge, D. Maniey, H. B. Osborne, and L. Paillard. 2004. EDEN-BP-dependent post-transcriptional regulation of gene expression in Xenopus somitic segmentation. Development 131:6107-6117. [DOI] [PubMed] [Google Scholar]
- 15.Good, P., O. Chen, S. Warner, and D. Herring. 2000. A family of human RNA-binding proteins related to the Drosophila Bruno translational regulator. J. Biol. Chem. 275:28583-28592. [DOI] [PubMed] [Google Scholar]
- 16.Kashiwabara, S., J. Noguchi, T. Zhuang, K. Ohmura, A. Honda, S. Sugiura, K. Miyamoto, S. Takahashi, K. Inoue, A. Ogura, and T. Baba. 2002. Regulation of spermatogenesis by testis-specific, cytoplasmic poly(A) polymerase TPAP. Science 298:1999-2002. [DOI] [PubMed] [Google Scholar]
- 17.Kierszenbaum, A. L., and L. L. Tres. 1975. Structural and transcriptional features of the mouse spermatid genome. J. Cell Biol. 65:258-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kress, C., S. Vandormael-Pournin, P. Baldacci, M. Cohen-Tannoudji, and C. Babinet. 1998. Nonpermissiveness for mouse embryonic stem (ES) cell derivation circumvented by a single backcross to 129/Sv strain: establishment of ES cell lines bearing the Omd conditional lethal mutation. Mammal. Genome 9:998-1001. [DOI] [PubMed] [Google Scholar]
- 19.Ladd, A. N., N. Charlet-B, and T. Cooper. 2001. The CELF family of RNA binding proteins is implicated in cell-specific and developmentally regulated alternative splicing. Mol. Cell. Biol. 21:1285-1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee, K., H. S. Haugen, C. H. Clegg, and R. E. Braun. 1995. Premature translation of protamine 1 mRNA causes precocious nuclear condensation and arrests spermatid differentiation in mice. Proc. Natl. Acad. Sci. USA 92:12451-12455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mali, P., A. Kaipia, M. Kangasniemi, J. Toppari, M. Sandberg, N. B. Hecht, and M. Parvinen. 1989. Stage-specific expression of nucleoprotein mRNAs during rat and mouse spermiogenesis. Reprod. Fertil. Dev. 1:369-382. [DOI] [PubMed] [Google Scholar]
- 22.Markert, C. L., T. M. Amet, and E. Goldberg. 1998. Human testis-specific lactate dehydrogenase-C promoter drives overexpression of mouse lactate dehydrogenase-1 cDNA in testes of transgenic mice. J. Exp. Zool. 282:171-178. [PubMed] [Google Scholar]
- 23.Martianov, I., G. M. Fimia, A. Dierich, M. Parvinen, P. Sassone-Corsi, and I. Davidson. 2001. Late arrest of spermiogenesis and germ cell apoptosis in mice lacking the TBP-like TLF/TRF2 gene. Mol. Cell 7:509-515. [DOI] [PubMed] [Google Scholar]
- 24.Milne, C. A., and J. Hodgkin. 1999. ETR-1, a homologue of a protein linked to myotonic dystrophy, is essential for muscle development in Caenorhabditis elegans. Curr. Biol. 9:1243-1246. [DOI] [PubMed] [Google Scholar]
- 25.Moraes, K. C., C. J. Wilusz, and J. Wilusz. 2006. CUG-BP binds to RNA substrates and recruits PARN deadenylase. RNA 12:1084-1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mukhopadhyay, D., C. W. Houchen, S. Kennedy, B. K. Dieckgraefe, and S. Anant. 2003. Coupled mRNA stabilization and translational silencing of cyclooxygenase-2 by a novel RNA binding protein, CUGBP2. Mol. Cell 11:113-126. [DOI] [PubMed] [Google Scholar]
- 27.Nagy, A., M. Gertsenstein, K. Vintersten, and R. Behringer. 2003. Manipulating the mouse embryo: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 28.Nantel, F., L. Monaco, N. S. Foulkes, D. Masquilier, M. LeMeur, K. Henriksen, A. Dierich, M. Parvinen, and P. Sassone-Corsi. 1996. Spermiogenesis deficiency and germ-cell apoptosis in CREM-mutant mice. Nature 380:159-162. [DOI] [PubMed] [Google Scholar]
- 29.Niranjanakumari, S., E. Lasda, R. Brazas, and M. A. Garcia-Blanco. 2002. Reversible cross-linking combined with immunoprecipitation to study RNA-protein interactions in vivo. Methods 26:182-190. [DOI] [PubMed] [Google Scholar]
- 30.O'Shaughnessy, P. J., L. Willerton, and P. J. Baker. 2002. Changes in Leydig cell gene expression during development in the mouse. Biol. Reprod. 66:966-975. [DOI] [PubMed] [Google Scholar]
- 31.Paillard, L., V. Legagneux, and H. B. Osborne. 2003. A functional deadenylation assay identifies human CUG-BP as a deadenylation factor. Biol. Cell 95:107-113. [DOI] [PubMed] [Google Scholar]
- 32.Paillard, L., F. Omilli, V. Legagneux, T. Bassez, D. Maniey, and H. B. Osborne. 1998. EDEN and EDEN-BP, a cis element and an associated factor that mediates sequence-specific mRNA deadenylation in Xenopus embryos. EMBO J. 17:278-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pan, J., M. Goodheart, S. Chuma, N. Nakatsuji, D. C. Page, and P. J. Wang. 2005. RNF17, a component of the mammalian germ cell nuage, is essential for spermiogenesis. Development 132:4029-4039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Robertson, E., and A. Bradley. 1986. Production of permanent cell lines from early embryos and their use in studying developmental problems. In J. Pedersen (ed.), Experimental approaches to mammalian embryonic development. IRL Press, Oxford, United Kingdom.
- 35.Rodriguez, I., C. Ody, K. Araki, I. Garcia, and P. Vassalli. 1997. An early and massive wave of germinal cell apoptosis is required for the development of functional spermatogenesis. EMBO J. 16:2262-2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Russell, L., R. Ettlin, A. SinhaHikim, and E. Clegg. 1990. Histological and histopathological evaluation of the testis. Cache River Press, St. Louis, MO.
- 37.Sarge, K. D., O. K. Park-Sarge, J. D. Kirby, K. E. Mayo, and R. I. Morimoto. 1994. Expression of heat shock factor 2 in mouse testis: potential role as a regulator of heat-shock protein gene expression during spermatogenesis. Biol. Reprod. 50:1334-1343. [DOI] [PubMed] [Google Scholar]
- 38.Sugimura, I., and M. A. Lilly. 2006. Bruno inhibits the expression of mitotic cyclins during the prophase I meiotic arrest of Drosophila oocytes. Dev. Cell. 10:127-135. [DOI] [PubMed] [Google Scholar]
- 39.Tay, J., and J. D. Richter. 2001. Germ cell differentiation and synaptonemal complex formation are disrupted in CPEB knockout mice. Dev. Cell 1:201-213. [DOI] [PubMed] [Google Scholar]
- 40.Terada, K., K. Yomogida, T. Imai, H. Kiyonari, N. Takeda, T. Kadomatsu, M. Yano, S. Aizawa, and M. Mori. 2005. A type I DnaJ homolog, DjA1, regulates androgen receptor signaling and spermatogenesis. EMBO J. 24:611-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Timchenko, N. A., P. Iakova, Z. J. Cai, J. R. Smith, and L. T. Timchenko. 2001. Molecular basis for impaired muscle differentiation in myotonic dystrophy. Mol. Cell. Biol. 21:6927-6938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Timchenko, N. A., A. L. Welm, X. Lu, and L. T. Timchenko. 1999. CUG repeat binding protein (CUGBP1) interacts with the 5′ region of C/EBPβ mRNA and regulates translation of C/EBPβ isoforms. Nucleic Acids Res. 27:4517-4525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tsai-Morris, C. H., Y. Sheng, E. Lee, K. J. Lei, and M. L. Dufau. 2004. Gonadotropin-regulated testicular RNA helicase (GRTH/Ddx25) is essential for spermatid development and completion of spermatogenesis. Proc. Natl. Acad. Sci. USA 101:6373-6378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ule, J., K. Jensen, A. Mele, and R. B. Darnell. 2005. CLIP: a method for identifying protein-RNA interaction sites in living cells. Methods 37:376-386. [DOI] [PubMed] [Google Scholar]
- 45.van der Weyden, L., M. J. Arends, O. E. Chausiaux, P. J. Ellis, U. C. Lange, M. A. Surani, N. Affara, Y. Murakami, D. J. Adams, and A. Bradley. 2006. Loss of TSLC1 causes male infertility due to a defect at the spermatid stage of spermatogenesis. Mol. Cell. Biol. 26:3595-3609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Venables, J. P. 2002. Alternative splicing in the testes. Curr. Opin. Genet. Dev. 12:615-619. [DOI] [PubMed] [Google Scholar]
- 47.Vernet, M., C. Bonnerot, P. Briand, and J. F. Nicolas. 1993. Application of LacZ gene fusions to preimplantation development. Methods Enzymol. 225:434-451. [DOI] [PubMed] [Google Scholar]
- 48.Webster, P. J., L. Liang, C. A. Berg, P. Lasko, and P. M. Macdonald. 1997. Translational repressor bruno plays multiple roles in development and is widely conserved. Genes Dev. 11:2510-2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yamada, D., M. Yoshida, Y. N. Williams, T. Fukami, S. Kikuchi, M. Masuda, T. Maruyama, T. Ohta, D. Nakae, A. Maekawa, T. Kitamura, and Y. Murakami. 2006. Disruption of spermatogenic cell adhesion and male infertility in mice lacking TSLC1/IGSF4, an immunoglobulin superfamily cell adhesion molecule. Mol. Cell. Biol. 26:3610-3624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang, J., S. Medvedev, P. P. Reddi, R. M. Schultz, and N. B. Hecht. 2005. The DNA/RNA-binding protein MSY2 marks specific transcripts for cytoplasmic storage in mouse male germ cells. Proc. Natl. Acad. Sci. USA 102:1513-1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang, J., S. Medvedev, J. Yu, L. C. Tang, J. E. Agno, M. M. Matzuk, R. M. Schultz, and N. B. Hecht. 2005. Absence of the DNA-/RNA-binding protein MSY2 results in male and female infertility. Proc. Natl. Acad. Sci. USA 102:5755-5760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang, D., T. L. Penttila, P. L. Morris, M. Teichmann, and R. G. Roeder. 2001. Spermiogenesis deficiency in mice lacking the Trf2 gene. Science 292:1153-1155. [DOI] [PubMed] [Google Scholar]
- 53.Zhong, J., A. H. Peters, K. Lee, and R. E. Braun. 1999. A double-stranded RNA binding protein required for activation of repressed messages in mammalian germ cells. Nat. Genet. 22:171-174. [DOI] [PubMed] [Google Scholar]