Abstract
RDH12 codes for a member of the family of short-chain alcohol dehydrogenases/reductases proposed to function in the visual cycle that supplies the chromophore 11-cis retinal to photoreceptor cells. Mutations in RDH12 cause severe and progressive childhood onset autosomal-recessive retinal dystrophy, including Leber congenital amaurosis. We generated Rdh12 knockout mice, which exhibited grossly normal retinal histology at 10 months of age. Levels of all-trans and 11-cis retinoids in dark- and light-adapted animals and scotopic and photopic electroretinogram (ERG) responses were similar to those for the wild type, as was recovery of the ERG response following bleaching, for animals matched for an Rpe65 polymorphism (p.L450M). Lipid peroxidation products and other measures of oxidative stress did not appear to be elevated in Rdh12−/− animals. RDH12 was localized to photoreceptor inner segments and the outer nuclear layer in both mouse and human retinas by immunohistochemistry. The present findings, together with those of earlier studies showing only minor functional deficits in mice deficient for Rdh5, Rdh8, or Rdh11, suggest that the activity of any one isoform is not rate limiting in the visual response.
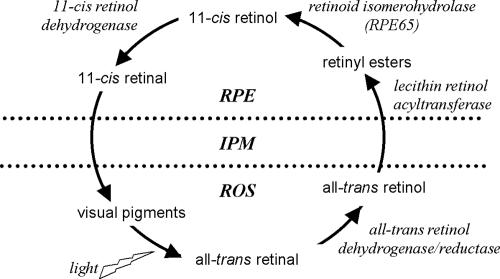
Retinoid dehydrogenases/reductases (RDHs) perform critical oxidation-reduction reactions in the visual cycle mechanism that converts vitamin A (all-trans retinol) to 11-cis retinal, the light-absorbing chromophore of the visual pigments in photoreceptor cells (Fig. 1) (14). One of these reactions involves the oxidation of 11-cis retinol produced by retinoid isomerohydrolase activity in the retinal pigment epithelium (RPE), resulting in formation of 11-cis retinal that is delivered to the photoreceptor cell outer segments. The second reaction involves reduction of all-trans retinal released from rhodopsin and cone opsins upon bleaching, resulting in formation of all-trans retinol that is returned to the RPE for trans-to-cis isomerization. A number of RDH isoforms are expressed in the RPE and/or the neuroretina that differ in terms of substrate specificity and sites of expression.
FIG. 1.
Schematic of the visual cycle mechanism showing the interconversion of vitamin A analogs and necessary enzyme activities. RPE, retinal pigment epithelium; IPM, interphotoreceptor matrix; ROS, rod outer segments.
Within the RPE, RDH5 is thought to be the key enzyme involved in converting 11-cis retinol to 11-cis retinal (25). Mutations in RDH5 in humans result in fundus albipunctatus, a form of congenital stationary night blindness (31). Since Rdh5 knockout mice exhibit very mild visual disturbances (7), it has been suggested that Rdh5 activity may be redundant with that of other isoforms. Consistent with this notion, mice deficient for Rdh11 and for prRdh, two isoforms expressed in the photoreceptors, exhibit a mild phenotype without signs of retinal degeneration (15, 16). Functional interaction of RDH5 with RDH11 has been proposed (16), but studies showing that RDH11 is expressed in the photoreceptor cell inner segment complicate this interpretation (15). RDH10 is expressed in both the RPE and Müller cells and is specific for the oxidation of all-trans retinol (30). Its activity may be important for an auxiliary light-dependent visual cycle (5). At least five RDH isoforms, including RDH8 (prRDH), RDH11, RDH12, RDH14, and DHRS3 (retSDR1), have been reported to be expressed in the photoreceptor cells (10, 11, 20). In vitro, each exhibits substrate specificity compatible with a role in converting all-trans retinal to all-trans retinol in the recovery phase of the visual cycle.
Mutations in RDH12 cause a severe form of autosomal-recessive retinal dystrophy (arRD) with childhood onset that is often diagnosed as Leber congenital amaurosis (13, 18, 27). The severe phenotype associated with RDH12 mutations is consistent with a nonredundant role of RDH12 in photoreceptor physiology, leading to the notion that it may play a unique role in the visual cycle mechanism. RDH12 has also been proposed to detoxify medium-chain aldehydes potentially present in the photoreceptors as a result of lipid peroxidation (2).
We disrupted the Rdh12 gene in mice, characterized the associated phenotype in functional assays, and localized its expression in the retina. Our findings suggest that the murine phenotype associated with Rdh12 deficiency does not result from disruption of visual cycle function and leave open the possibility that multiple RDH isoforms may contribute to the regulation of the oxidation state of retinoids in the outer retina.
MATERIALS AND METHODS
All experimental procedures complied with the regulations of the Ministry of Science and Public Health of the City State of Hamburg, Germany, the University of Michigan Committee on Use and Care of Animals, and the regulations of Augenklinik Campus Virchow-Klinikum Charité, Berlin, Germany. Mice were reared in a 12-h-12-h light-dark cycle (∼800-lux room light, as indicated below) and euthanized by CO2 inhalation.
Generation of Rdh12 knockout mice.
An Rdh12-carrying clone isolated from a 129/SvJ mouse genomic λ library (Stratagene) was used to construct the targeting vector. A 12.9-kb XmaI/NotI fragment including exons 1 to 5 of the Rdh12 gene was cloned into the pKO-V901 plasmid (Lexicon Genetics) with a phosphoglycerate kinase gene (pgk) promoter-driven diphtheria toxin A cassette. A pgk promoter-driven neomycin resistance cassette flanked by loxP sites was ligated into the ApaI site in intron 3. A third loxP site and an additional EcoRV site were inserted into the BstBI site in the 5′ region of the Rdh12 gene. The construct was electroporated into R1 mouse embryonic stem (ES) cells. Neomycin-resistant clones were analyzed by Southern analysis, using an external, ∼300-bp probe to screen for an additional EcoRV site incorporated into the targeted allele. Correctly targeted ES cells were transfected with a plasmid expressing Cre recombinase to remove the neomycin cassette and exons 1 to 3. Two independent embryonic stem cell clones were injected into C57BL/6 blastocysts to generate chimeras that were backcrossed with C57BL/6 mice. Genomic DNAs from tail biopsies were prepared by proteinase K (Roche) digestion and phenol-chloroform extraction. Mice were genotyped by Southern analysis (as described above) or by PCR to assay the presence or absence of wild-type and knockout alleles. For amplification of the knockout allele, the sense primer F1 (5′-GGAATGCACAGGCTTCAGCATG-3′) and the antisense primer R1 (5′-CTGCTATCATCCTGGACACC-3′) were used. For amplification of the wild-type allele, primers F2 (5′-GGGGTCTGCAACCCTATAGG-3′) and R2 (5′-GGACTCTCCATTGCCACTAGG-3′) were used. Both primer pairs F1/R1 and F2/R2 amplify ∼500-bp fragments.
Mice were also genotyped for an Rpe65 polymorphism, p.L450M, which affects the efficiency of 11-cis retinal synthesis (28). PCR products (∼300 bp) were obtained (sense primer, 5′-GCATACGGACTTGGGTTGAATCAC-3′; antisense primer, 5′-GGTTGAGAAACAAAGATGGGTTCAG-3′) and digested with the restriction enzyme MwoI, which recognizes the L450 allele, and the pattern of cutting was evaluated on agarose gels. Studies were performed in a mixed 129SV;C57BL/6 background, using the F3 and F4 generations. Littermates matched for Rpe65 genotype, as indicated, served as controls.
Transcript analysis.
For Northern analysis, total RNAs were isolated from mouse eyes by using an RNeasy kit (QIAGEN) according to the manufacturer's instructions. Total RNA (5 μg) was electrophoresed and blotted onto membranes by standard protocols, hybridized with a random-primed [α-32P]dATP-labeled full-length Rdh12 cDNA probe (accession number gi:58037513), and exposed to X-ray film.
For quantitative PCR, total RNAs were isolated from separately dissected retinas and RPE (n = 8 per genotype), and first-strand cDNAs were generated using SuperScript II (Invitrogen) and random hexamers (Invitrogen). Visual cycle genes were amplified with the primers shown below, using real-time PCR with SYBR green PCR master mix (Applied Biosystems) and a Rotor-Gene real-time cycler (Corbett Research). The cycling protocol was 95°C for 5 min, 95°C for 10 s, 60°C for 15 s, and 72°C for 20 s, with steps 2 to 4 repeated 40 times. Each sample was amplified in duplicate and gave consistent results, with the amplification efficiency normalized to that of Hprt.
The following primers (sense and antisense) were used: for Rdh12, GATACTGCAGTGCTTTTGCCTATGG and GAGCCGCCATAGCAAACACAGCAGG; for Rdh11, GGTGCTAACACAGGCATTGGGAAG and AGGTCCAGTTTCCGTACGAAGACC; for Rdh5, GTGTCTCCAAGTTTGGCCTGGAGG and CCAACAAGCCTTCAGGGTGCTCTCC; for Rpe65, TGTGCAGTTCCCCTGCAGTGATCG and CCATGCTTTCATTGGACTCGAAACA; for Cralbp, AGTCGGGACAAGTATGGTCGAGTG and CAGAAGCCGTTGATTTGGGTTTCC; for Lrat, CAGTTGGGACTGACTCCATACAGC and AGACTGCTTCTCTGATCACGAATG; for Abcr, GTCGCAGTTCATGGACACCCTGAG and GAACTTGGGAGTTGACCAGAAGATG; for Irbp, CACACTGATGCCCTGATCATCGAC and TGGGTCCAAAGTTCGCTGACCGAG; for Crbp, GGAACTATATCATGGACTTCCAAG and CTCCTTCTCTCCCTTCTGCACACA; for Rho, CACTTCACCATTCCTATGATCGTC and AGCCAGCAGATCAGGAAGAAGATG; for Rdh13, CATTCGAGGAGAGACCTTGAATCCC and CATCTCAAAGCCATCCTCAGTGGTCC; for Rdh14, CAGCTGGTCGTTAAGGAGCTGGACC and CAAACTGCATCTCAAACCCATCTTC; for Rdh8, GGAGCTCTTCCGCTCTGTGGGACAG and CTTAAGCGCTGTCAGTGGGAGGTAG; for retSdr1, CACCACCGTTCTGCCCTTTCACACC and GGGCCTGGTTTTGTTGCACAGCATC; and for Hprt, GTTCTTTGCTGACCTGCTGGA and TCCCCCGTTGACTGATCATT.
Morphological analysis.
For high-power micrographs and electron microscopy, mice were perfused with 4% paraformaldehyde and 1% glutaraldehyde in phosphate-buffered saline (PBS). Tissues were postfixed in 1% OsO4, dehydrated, and embedded in Epon. Sections (0.5 μm) were stained with methylene blue. Ultrathin sections (60 nm) were stained with uranyl acetate and lead citrate and viewed and photographed using a Zeiss EM 902 microscope.
Antibody development.
Antibodies specific for human or mouse RDH12 were raised against synthetic peptide sequences chosen on the basis of predicted antigenicity and uniqueness within the family of RDH proteins. Peptides were coupled via an N-terminal cysteine to the keyhole limpet hemocyanin carrier protein. For mouse Rdh12, rabbits were immunized against the peptide CKRMWVSSRARNKKT (amino acids 285 to 299; Protein Data Bank accession number gi:58037513) or C-SPFFKSTSQGAQ (amino acids 252 to 263, with an additional N-terminal cysteine), and each resulting antiserum (termed CT or SQ, respectively) was purified on an affinity matrix made by conjugation with the same peptide. For the generation of an antibody against human RDH12, mice were immunized with the peptide C-DCKRTWVSPRARNNKT (amino acids 284 to 299, with an additional N-terminal cysteine; Protein Data Bank accession number gi:19343614), and a hybridoma cell line secreting monoclonal antibody (MAb) 2C9 was generated and used to elicit ascites fluid in mice, from which immunoglobulin G was purified on protein A-Sepharose by using standard protocols (12).
Western analysis.
Mouse eyes were enucleated and homogenized in sodium dodecyl sulfate sample buffer, insoluble material was removed by low-speed centrifugation, and proteins were separated by polyacrylamide gel electrophoresis and blotted onto membranes by using standard protocols. Blots were probed by incubation with the affinity-purified SQ rabbit antiserum diluted 1:500, and reactivity was detected using a horseradish peroxidase-coupled secondary antibody (1:4,000; GE Healthcare) and a chemiluminescence kit (Renaissance; DuPont). Human retinas were dissected from postmortem donor eyes (Midwest Eye Bank and Transplantation Center), homogenized in sodium dodecyl sulfate sample buffer, electrophoresed, and blotted as described for mouse eyes. Blots were probed by incubation with MAb 2C9 immunoglobulin G (∼0.5 μg/ml), and reactivity was detected using an alkaline phosphatase-conjugated secondary antibody (1:500; Molecular Probes). Western analysis of COS-7 cells transfected with cDNA constructs encoding either mouse or human protein with an amino-terminal His tag (pcDNA3.1/HIS) and probed with anti-Xpress antibody served as a positive control.
Immunohistochemistry.
Mouse eyes were enucleated, and corneas were removed. The eye cups were fixed in 4% paraformaldehyde in PBS for 30 min at 4°C, washed three times for 5 min each with PBS, placed in 20% sucrose in PBS at 4°C overnight, and then flash frozen in Tissue-Tek O.C.T. Sections (8 μm) were blocked with 3% normal goat serum and 0.3% Triton X-100 in PBS for 30 min, incubated with primary antibody (SQ [1:500] or anti-rhodopsin) or fluorescein isothiocyanate-conjugated peanut agglutinin (PNA)-lectin (0.05 mg/ml; Molecular Probes) in blocking solution overnight, washed three times for 5 min each with PBS, incubated with Alexa Fluor 488- or 555-conjugated secondary antibody (1:3,000; Molecular Probes) and Toto-3 for counterstaining of nuclei (1:10,000; Molecular Probes) in blocking solution for 2 h, washed, and covered with coverslips. Human eyes were dissected to obtain pieces of retina/RPE/choroid/sclera that were fixed in 4% paraformaldehyde for 15 min, placed in 20% sucrose for 5 min, and then flash frozen in Tissue-Tek O.C.T. Sections (10 μm) were blocked with 20% sheep serum-0.2% Triton X-100 in PBS, incubated with primary antibody (2C9, ∼1 μg/ml; anti-rhodopsin, 1:1,000; or anti-prRDH, 1:25) or FITC-conjugated PNA-lectin (0.05 mg/ml; Molecular Probes) in 2% sheep serum-0.2% Triton X-100 in PBS, washed, and incubated in the same buffer with Alexa Fluor 488- or 555-conjugated secondary antibody. Specimens were viewed and photographed either with a confocal microscope (Leica TCS) or on a Nikon Eclipse E800 microscope with a Nikon DMX1200 digital camera, using the manufacturer's data acquisition software.
Analysis of retinoid content.
Mice were allowed to dark adapt or were exposed to various light regimens and then were euthanized, and their eyes were enucleated and frozen in liquid N2 in the dark. Retinoids were extracted using a modification of a previously described method (3). Briefly, under dim red light and on ice, two eyes were homogenized in 1 ml chloroform:methanol:hydroxylamine (2 M) (3:6:1) and incubated at room temperature for 2 min. Next, 200 μl chloroform and 240 μl water were added, and each sample was vortexed and centrifuged at 14,000 rpm for 5 min. The lower phase was collected, the solvent was evaporated under nitrogen, and the sample was dissolved in hexane. Retinoids in the extracts were identified and quantified by high-performance liquid chromatography (HPLC) analysis, using a Waters Alliance separation module and photodiode array detector with a Supelcosil LC-31 column (25 cm by 4.6 mm by 3 μm) developed with 5% 1,4-dioxane in hexane. For quantification of retinyl esters, the column effluent at 3 to 6 min was collected and saponified by incubation in ethanolic KOH (40 mM) at 55°C for 30 min, and the products were then extracted into hexane and subjected to HPLC analysis as described above. Peak identification was done by comparison to retention times of standard compounds and evaluation of wavelength maxima. Quantitative analysis was done by comparison of peak areas at 325 nm for all-trans retinol, 318 nm for 11-cis retinol, 357 and 361 nm for syn- and anti-all-trans retinal oxime, respectively, 347 and 351 nm for syn- and anti-11-cis retinal oxime, respectively, and 325 nm for all-trans retinal esters, using published extinction coefficients (24). Data were analyzed using analysis of variance (α = 0.05) (Excel software), and statistical differences were designated for P values of <0.05.
Assays of oxidative stress indicators.
Mice were subjected to bleaching light (5,000 lux for 30 min) before being sacrificed. Lipid peroxidation products potentially resulting from photooxidation (2) were assayed as thiobarbituric reactive substances (TBARs) in retinal homogenates in RIPA buffer (4). The transcript abundance of heme oxygenase 1 (HO-1), a marker for oxidative stress, was assayed in retinal total RNA by using quantitative PCR (sense primer, 5′-GCATGCCCCAGGATTTGTC-3′; antisense primer, 5′-CTGGCCCTTCTGAAAGTTCCTCATG-3′) and was normalized with respect to Hprt expression (sense primer, 5′-GCAAGCTTGCTGGTGAAAGGAC-3′; antisense primer, 5′-CCTGAAGTACTCATTATAGTCAAGGGC-3′).
ERG recordings.
Electroretinograms (ERGs) were recorded from 8-month-old anesthetized mydriatic Rdh12+/+ and Rdh12−/− mice (n = 5 per genotype) as described previously (23). For scotopic ERGs, mice were allowed to adapt to the dark for at least 3 h. The flash energies used ranged from 10−4 to 3 cd m−2, divided into eight steps of 0.4 or 0.6 log cd m−2. Flash stimuli were delivered in a Ganzfeld globe (Toennies, Hoechberg, Germany) and had a duration of 50 μs. Subsequently, scotopic a-wave recording using relatively high stimulus energies was performed. The high-energy stimuli were generated by a photoflash. Four flash energies were applied, ranging from 1.5 to 2.5 log cd m−2. Above these energies, the scotopic a-wave amplitude saturated. To prevent rod adaptation, no averaging was performed. The interstimulus interval was at least 2 minutes in duration. Immediately after scotopic ERG recordings, mice were exposed to a background light of 60 cd m−2 for 10 min. Photopic ERGs were then recorded by averaging the responses to 10 single flashes of 15 and 25 cd m−2. In a second experiment to compare dark adaptation capabilities, mice were allowed to adapt to the dark for at least 2 h, and recording was performed using the 10−2 mcd m−2 (rod dominated) and 3 mcd m−2 (combined scotopic rod-cone response) stimuli. Light adaptation was then obtained using 125 cd m−2 of white light delivered in the Ganzfeld globe for 5 min. In the dark again, the ERG response was recorded every 2 min, using a flash intensity of 10−2 cd m−2 for 20 min. In an additional experiment, higher levels of bleaching were applied to 5-month-old mice (n = 4 per genotype). The animals were allowed to adapt to the dark for 12 h and were subsequently exposed to 5,000 lux of white light for 10 min (delivered by a halogen lamp). After 90 min of dark adaptation, dark-adapted ERGs were recorded. For statistical analysis, analysis of variance for repeated measurements and the t test (with Bonferroni correction, if necessary) were applied. In addition, the Mann-Whitney U test was applied to cross-check the results, as the distribution of the data is unknown.
RESULTS
Disruption of Rdh12.
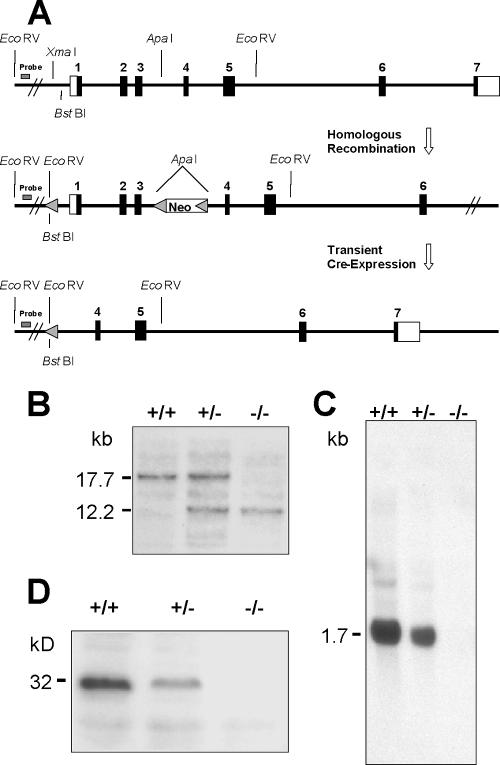
The gene targeting strategy used resulted in deletion of Rdh12 exons 1 to 3, encoding the N-terminal one-third of the protein (Fig. 2A). To exclude side effects from the neomycin selection cassette, the neo gene was removed from the targeted allele by Cre-mediated excision at the cell culture level. Matings of heterozygous null (Rdh12+/−) animals resulted in homozygous null (Rdh12−/−) offspring at the expected Mendelian ratio (∼25%). Genotypes were determined by Southern analysis showing the presence of an additional EcoRV site in the targeted allele (Fig. 2B) or by PCR analysis showing amplification of the wild-type or knockout allele (data not shown). Rdh12−/− mice were viable, appeared to develop normally, and could not be distinguished from heterozygous or wild-type littermates by mere observation. No Rdh12 transcripts were detected in the eyes of Rdh12−/− mice by Northern analysis using a full-length murine cDNA probe (Fig. 2C). Western blot analysis for Rdh12 in whole-eye lysates from Rdh+/+, Rdh+/−, and Rdh−/− mice showed that the protein was reduced in heterozygous animals and absent in mice that were homozygous null for Rdh12 (Fig. 2D).
FIG. 2.
Generation of Rdh12 knockout mice. (A) Construct design for deletion of mouse Rdh12 exons 1 to 3, using loxP sites and Cre-mediated deletion in ES cells. (B) Southern analysis of Rdh12 expression in wild-type and heterozygous and homozygous null mice. (C) Northern analysis of Rdh12 transcript abundance in all three genotypes. (D) Western analysis of Rdh12 protein abundance, using an antipeptide antibody specific for the mouse protein.
The Rdh12−/− mice were derived from a cross between the 129/Sv and C57BL/6 mouse strains, which differ with respect to a p.L450M polymorphism in Rpe65 that confers resistance to light damage due to decreased synthesis of 11-cis retinal (6, 28). When the Rdh12 knockout animals were typed for this single-nucleotide polymorphism, all three genotypes were detected. Since the regulation of 11-cis retinal synthesis by Rpe65 impacts rates of pigment regeneration (28), the Rpe65 genotype was considered in phenotypic comparisons of Rdh12−/− and wild-type animals, as indicated below.
RDH12 immunolocalization in mice and humans.
For localization of Rdh12 expression in mouse eyes, polyclonal antibodies CT and SQ were elicited by immunization with synthetic peptides corresponding to p.C285-p.T299 and p.S252-p.Q263 of the mouse Rdh12 sequence, respectively. Immunohistochemical analysis with each antibody showed similar immunoreactivities in wild-type retina/RPE/choroid cryosections that were absent in Rdh12−/− mice (shown for SQ antibody in Fig. 3D). The signal in both cases was sensitive to paraformaldehyde fixation, with optimal staining achieved with sections fixed for short times (30 min) or without paraformaldehyde (e.g., with cold methanol).
FIG. 3.
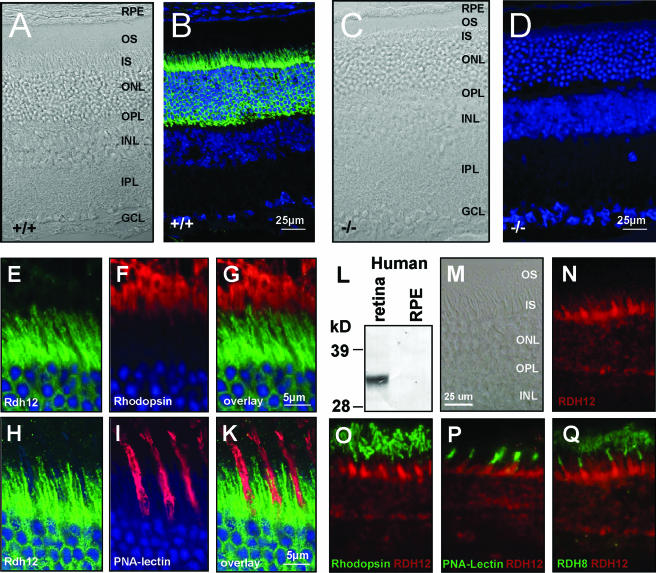
RDH12 localizes to photoreceptor inner segments. Cryosections of wild-type (A, B, and E to K) and Rdh12−/− (C and D) eyes are shown. (B, D, and E to K) Murine sections were counterstained with TOTO-3 to label cell nuclei, shown in blue. (M to Q) Cryosections of human eyes. (A, C, and M) Phase-contrast microscopy images. (D) Absence of Rdh12 staining in tissue from Rdh12−/− mice confirms the specificity of the antibody. Rdh12 labeling (green) did not colocalize with rhodopsin (red) (E to G) or PNA-lectin (red) (H to K). (L) A monoclonal antibody directed against human RDH12 detects a corresponding band in retina lysates but not in RPE. (N) In the human retina, RDH12 localizes to photoreceptor inner segments as well and does not overlap with (O) rhodopsin, (P) PNA-lectin, or (Q) prRDH. OS, outer segments; ONL, outer nuclear layer; IS, inner segments; INL, inner nuclear layer; IPL, inner plexiform layer; GCL, ganglion cell layer; RPE, retinal pigment epithelium.
The pattern of Rdh12 immunoreactivity present in mouse retina/RPE/choroid sections was consistent with expression in the photoreceptor inner segments and outer nuclear layer, with no apparent expression in the RPE or photoreceptor outer segments (Fig. 3A to D). Double-labeling experiments using rhodopsin antibodies specific for the rod outer segment (Fig. 3E to G) and PNA-lectin staining of cone sheaths (Fig. 3H to K) showed no overlap with Rdh12 immunoreactivity.
For localization of RDH12 expression in human eyes, the mouse MAb 2C9, generated by immunization with a synthetic peptide corresponding to p.D284-p.T299 of the human RDH12 sequence, was used. Specificity was established by Western analysis of COS-7 cells transfected with a cDNA encoding Xpress-tagged human RDH12, in which MAb 2C9 was shown to recognize the same bands as those recognized by anti-Xpress antibody that correspond to glycosylated and unglycosylated forms of RDH12 (13; data not shown). These immunoreactive bands were not seen in Western blots of untransfected cells or COS-7 cells transfected with RDH11- or RDH5-encoding cDNAs (data not shown). MAb 2C9 also recognized RDH12 as an ∼33-kDa protein on immunoblots of lysates of human retinas but not RPE/choroid (Fig. 3L). As in the mouse, the pattern of MAb 2C9 immunoreactivity in human eyes was consistent with RDH12 expression in photoreceptor inner segments and the outer nuclear layer (Fig. 3N), with no evidence of overlap with rhodopsin expression in the rod outer segments (Fig. 3O), with PNA-lectin labeling of cone sheaths (Fig. 3P), or with prRDH expressed in rod and cone outer segments (Fig. 3Q) (20).
The similar localization of Rdh12 and RDH12 proteins in mouse and human photoreceptors, respectively, may indicate an analogous physiological role of the enzymes in both species and suggests that the Rdh12−/− mouse is likely to recapitulate, at least in part, deficits resulting from RDH12 loss-of-function mutations in humans.
Retinal histology of Rdh12−/− mice.
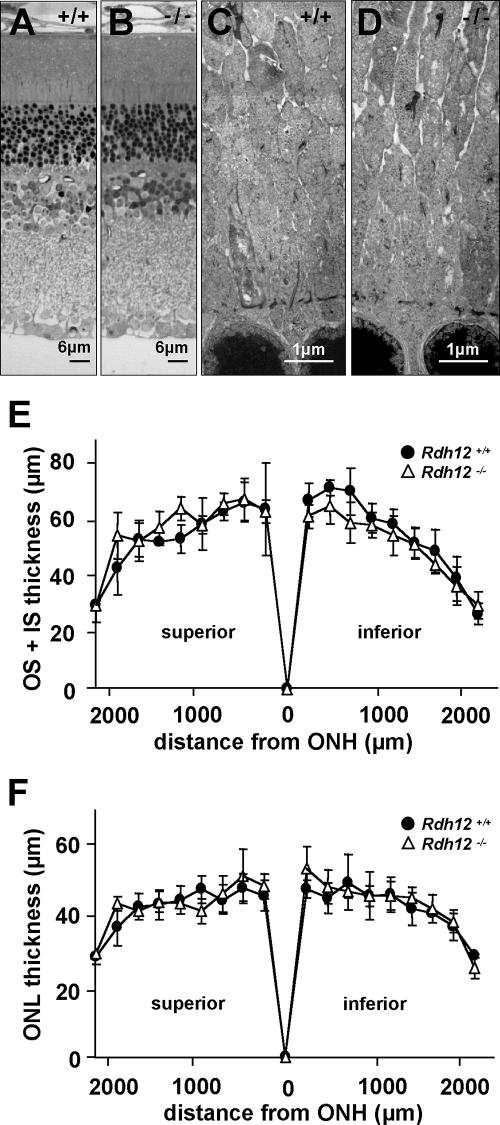
Light micrographs of retina/RPE/choroid sections from Rdh12−/− and wild-type mice showed comparable retinal histologies at 7 months of age (Fig. 4A and B), with normal lamination, numbers of cells, and apposition with the RPE. Similarly, transmission electron micrographs at 7 months of age revealed no major differences in retinal architecture or in the ultrastructure of photoreceptor outer and inner segments and of the outer nuclear layer (Fig. 4C and D). There was no evidence of accumulated bone spicule pigment, photoreceptor debris, retinyl ester droplets, retinal detachment, or altered ratios of rods to cones. Although the thickness of the outer nuclear layer did not appear to be decreased at 10 months of age, morphometric analysis showed slightly decreased outer-plus-inner-segment lengths in the inferior retinas of Rdh12−/− animals (Fig. 4E and F).
FIG. 4.
Retinal histology and thickness measurements for Rdh12−/− versus wild-type mice. (A and B) Semithin sections of Rdh12−/− and wild-type retinas/RPE/choroid stained with methylene blue. (C and D) Transmission electron micrographs of photoreceptor inner segments in Rdh12−/− and wild-type mice at 7 months of age. (E and F) Plots of photoreceptor layer and total retinal thicknesses for Rdh12−/− and wild-type mice, with measurements made on sections parallel to the vertical meridian of the eye and with standard deviations shown. •, wild type; ▵, Rdh12−/−. OS, outer segment; IS, inner segment; ONL, outer nuclear layer; ONH, optic nerve head.
Expression of visual cycle genes in Rdh12−/− mice.
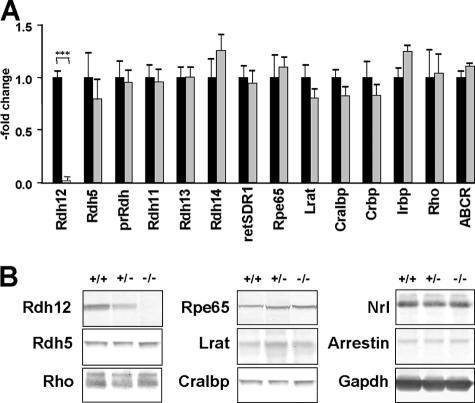
Analysis of transcript abundance in whole eyes from Rdh12−/− and wild-type mice showed approximately equivalent expression of the visual cycle genes Rdh5, Rpe65, Lrat, Cralbp, Crbp, Irbp, Rho, and Abcr in assays using real-time PCR (Fig. 5A). In addition, there was no apparent change in the expression of other Rdh isoforms expressed in photoreceptors, including Rdh8 (prRdh), Rdh11, Rdh13, Rdh14, and retSDR1, that might be suggestive of a compensatory response. Similarly, Western analysis of eyes from Rdh12−/− and wild-type animals showed no differences in protein levels of Rdh5, Rpe65, Lrat, Cralbp, Rho, Nrl, arrestin, and Gapdh (Fig. 5B).
FIG. 5.
Visual cycle gene expression in Rdh12−/− versus wild-type mice. (A) Transcript abundance assayed using quantitative real-time PCR, normalized to Hprt expression and shown as an x-fold increase compared to the wild-type abundance. (B) Western analysis of protein expression in whole-eye protein extracts, using the following antibodies and dilutions: anti-Rdh5, 1:1,000; anti-Rpe65, 1:5,000; anti-Lrat, 1:1,000; anti-Cralbp, 1:1,000; anti-rhodopsin, 1:1,000; anti-Nrl, 1:1,000; anti-arrestin, 1:1,000; and anti-Gapdh, 1:4,000.
Analysis of oxidative stress in Rdh12−/− mice.
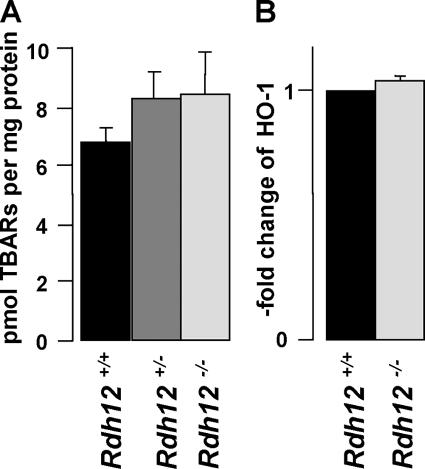
Indicators of oxidative stress were assayed in retinas from Rdh12−/−, Rdh12+/−, and wild-type mice subjected to bleaching light (5,000 lux for 30 min) before being sacrificed. Levels of lipid peroxidation products present in retinal homogenates, assayed as TBARs, were not significantly increased due to Rdh12 loss of function (Fig. 6A). In addition, transcript levels for HO-1, an enzyme that is upregulated in response to oxidative stress (19), were not markedly increased (Fig. 6B). Thus, despite the ability of RDH12 to recognize C9 aldehydes resulting from lipid photooxidation (2), our data do not indicate that Rdh12 plays a major role in detoxifying toxic aldehydes in the retina, at least in the short term.
FIG. 6.
Analysis of oxidative stress in Rdh12−/− mice. (A) Comparison of TBARs in retinal homogenates as an index of lipid peroxidation and oxidative stress. (B) Assay of HO-1 expression using quantitative real-time PCR with retinal cDNAs, with transcript levels being normalized to Hprt and shown as x-fold increases compared to the wild-type level.
Retinoid analysis with Rdh12−/− mice.
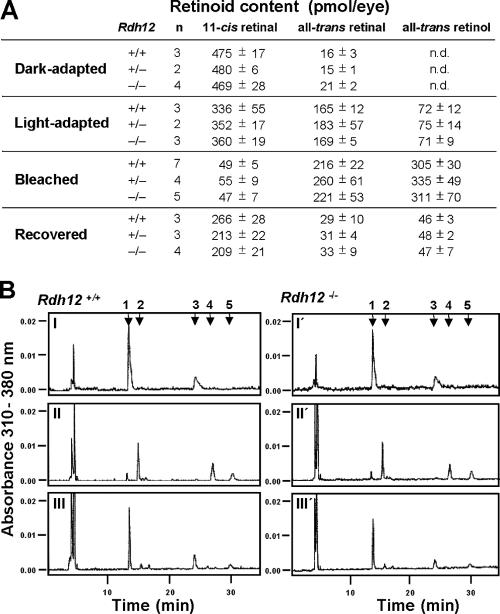
To assay the steady-state function of the visual cycle, HPLC analysis was used to compare the retinoid contents of whole eyes from Rdh12−/−, Rdh12+/−, and wild-type mice that were dark adapted or light adapted and subjected to intense bleaching light (5,000 lux for 20 min), or been subjected to bleaching and allowed to recover in the dark (30 min). Under dark-adapted and light-adapted conditions, the retinoid contents of eyes from knockout and wild-type animals were equivalent and unaffected by the p.L450M polymorphism of Rpe65. In all cases, 11-cis retinal predominated in the dark-adapted state, and all-trans retinal and all-trans retinol predominated in the light-adapted state, with no significant differences between genotypes (Fig. 7A and B). In addition, there were no significant differences in retinyl ester content between knockout and wild-type animals, with low levels of all-trans retinyl esters present in the dark- and light-adapted states that increased upon bleaching (data not shown). However, the Rpe65 genotype was a major factor in determining rates of 11-cis retinal regeneration during recovery in the dark after bleaching for both Rdh12−/− and wild-type mice. In both cases, recovery in p.L450/p.L450 and p.L450/p.M450 animals was significantly faster than that in p.M450/p.M450 animals (data not shown). We therefore excluded the last genotype in such comparisons and found that 11-cis retinal, all-trans retinal, and all-trans retinol contents were equivalent in eyes from Rdh12−/− and wild-type animals that were subjected to bleaching and allowed to recover in the dark for 30 min (Fig. 7A and B).
FIG. 7.
Retinoid content of eyes from Rdh12−/− mice. (A) Retinoid content given in absolute amounts. (B) HPLC analysis of retinoids extracted from eyes from (I to III) Rdh12+/+ and (I′ to III′) Rdh12−/− mice that were (I) dark adapted, (II) bleached (5,000 lux for 10 min), or (III) bleached and allowed to recover (5,000 lux for 10 min, dark for 30 min). Retinaldehydes were extracted as syn- and anti-retinal oximes and summed. 1, syn-11-cis retinal oxime; 2, syn-all-trans retinal oxime; 3, anti-11-cis retinal oxime; 4, all-trans retinol; 5, anti-all-trans retinal oxime. n.d., not detected.
Analysis of visual function in Rdh12−/− mice.
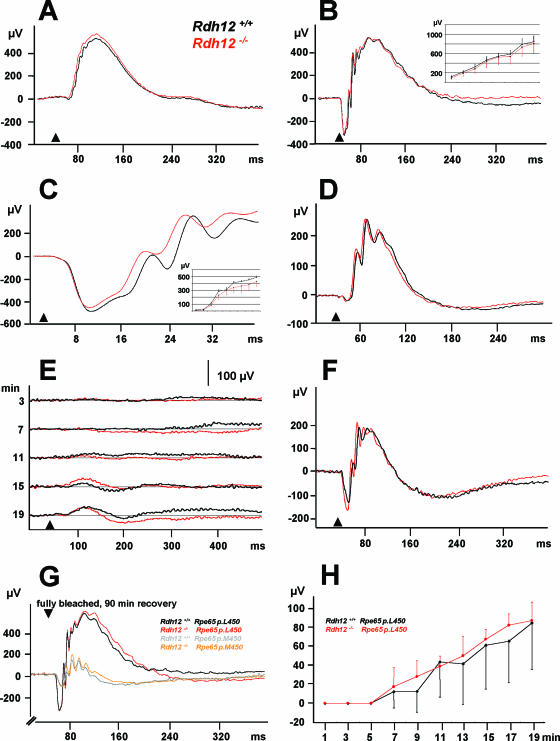
To test whether visual function was impaired in Rdh12−/− mice, ERGs were recorded from 8-month-old anesthetized Rdh12+/+ and Rdh12−/− animals. Neither the scotopic, rod-dominated ERG response (stimulus of 10−2 cd m−2) (Fig. 8A), the combined rod-cone response (stimulus of 3 cd m−2) (Fig. 8B), nor the cone response to a stimulus of 25 cd m−2 on a 60 cd m−2 white background (photopic ERG) (Fig. 8D) revealed any abnormalities in Rdh12−/− mice after complete dark adaptation. The comparison of a-waves after a stimulus of 300 cd m−2 demonstrated a normal phototransduction gain (Fig. 8C). The time course of the recovery of the rod-dominated ERG response was unaltered as well (Fig. 8E), suggesting a comparable regeneration of 11-cis retinal in Rdh12−/− mice. After 20 min of dark adaptation, the combined scotopic rod-cone response to a 3-cd m−2 stimulus reached about 50% (Fig. 8F) of the amplitude of the complete dark-adapted response (Fig. 8B).
FIG. 8.
ERG recordings for 8-month-old animals. (A to F) Panels A to D represent the averages for five Rdh12+/+ and five Rdh12−/− mice. Arrowheads, stimulus (flash). (A) Scotopic ERG, rod-dominated response (stimulus, 10−2 cd m−2). (B) Scotopic ERG, combined rod-cone response (stimulus, 3 cd m−2). (Inset) Amplitude versus log intensity as a function of scotopic b-wave amplitude (stimuli, 10−4 to 3 cd m−2 [x axis]). (C) Scotopic ERG, comparison of “a” waves (for details, see the text) (stimulus, 300 cd m−2). (Inset) Amplitude versus log intensity as a function of scotopic a-wave amplitude (stimuli, 4 × 10−3 to 300 cd m−2 [x axis]). (D) Cone response (stimulus of 25 cd m−2 on a 60-cd m−2 white background [photopic ERG]). (E) Recovery of the ERG response after delivery of an intense white light (125 cd m−2 for 10 min). Each set of lines represents the averages for three Rdh12+/+ and four Rdh12−/− mice. Arrowheads, stimulus (flash). (F) After 20 min of dark adaptation, the combined scotopic rod-cone response (3 cd m−2) reached about 50% of the amplitude of the complete dark-adapted response (B). (G) The combined rod-cone response 90 min after full bleaching depends on the Rpe65 polymorphism but not on the Rdh12 genotype (n = 4 for each genotype). (H) The recovery after high-energy bleaching also does not depend on the Rdh12 genotype.
Additionally, we performed adaptation experiments with both Rdh12−/− and wild-type mice controlled for the p.L450M polymorphism of Rpe65. As expected, p.M450/p.M450 animals recovered at a lower rate than p.L450/p.L450 animals (22). However, scotopic and photopic recordings were independent of the Rdh12 genotype, both with the protocol used for the data set displayed in Fig. 8G and after adaptation to a higher light energy (5,000 lux) (Fig. 8H).
DISCUSSION
Mutations in human RDH12 cause a severe form of child hood onset arRD associated with massive accumulation of bone spicule pigment, RPE atrophy, and macular scarring (13, 18, 27). Young children have useful visual function, whereas the condition progresses to legal blindness in young adulthood. RDH12 activity has been proposed to be necessary for the recovery phase of the visual cycle on the basis of in vitro studies showing that recombinant RDH12 efficiently converts all-trans retinal to all-trans retinol and that RDH12 transcripts are present in the outer nuclear layer in monkey and mouse retinas (2, 11). It follows that RDH12 loss-of-function mutations should decrease visual cycle efficiency, potentially resulting in reduced 11-cis retinal synthesis and the buildup of potentially toxic aldehyde intermediates in the retina. Mutations in the visual cycle gene RPE65 also cause severe and early-onset arRD (9, 17), with loss of function resulting in loss of 11-cis retinal synthesis, constitutive opsin activity, and destabilization and mislocalization of pigment proteins (8, 22, 32).
Rdh12−/− mice kept in a regular light-dark cycle did not develop any histological signs of retinal degeneration within the first 10 months of life, as assessed at the light microscope or ultrastructural level. Analysis of ocular retinoids showed no significant differences in 11-cis retinal contents and rates of recovery following exposure to full bleaching light, with no accumulation of all-trans retinal or other aldehydes potentially resulting from lipid peroxidation (2). In addition, an enzyme marker of oxidative stress, heme oxygenase 1 (19), was not upregulated. ERG analysis showed only minor alterations in responses in the dark-adapted state or for recovery after bleaching in both young and old animals. In addition, we found that RDH12 expression localizes to the photoreceptor inner segment and the outer nuclear layer in both mouse and human retinas and is not present in outer segments, where phototransduction takes place. This expression pattern is similar to that reported for RDH11 and contrasts with those for DHRS3 (retSDR1) and RDH8 (prRDH), reported to be present in cone and rod outer segments, respectively (10, 20).
Our analysis of the ocular phenotype of Rdh12−/− mice showed much less severe consequences than those associated with human RDH12 mutations. The findings that Rdh12−/− mice retain nearly normal visual cycle throughput and that expression of the Rdh12 protein is confined to photoreceptor inner segments are consistent with at least two different interpretations. The first is that RDH12 activity is not essential for regeneration of the 11-cis retinal chromophore in mice or humans and, instead, functions in an unrelated aspect of photoreceptor physiology not addressed by our studies. In this case, the absence of a rapid retinal degeneration phenotype in Rdh12−/− mice could be due to species differences related to retinal physiology, compensation of Rdh12 loss of function by another RDH isoform, or disease onset linked to time in years rather than relative age, a notion consistent with the mild pathology observed in very young children that progresses to devastating retinal degeneration over the course of ∼20 years.
A second possibility is that RDH12 activity is normally required for visual cycle function but that another RDH isoform can compensate for Rdh12 loss of function in mice but not in humans. Such a scenario has been proposed for RDH5, as human mutations result in fundus albipunctatus but Rdh5−/− mice do not exhibit the flecked retinas or ERG responses characteristic of the disease (31). Expression of RDH12 in photoreceptor inner, but not outer, segments would require that reduction of all-trans retinal occur at a site distant from its release from bleached photopigments, potentially involving a novel trafficking mechanism. Although it is somewhat counterintuitive, perhaps the dual-substrate specificity of RDH12 necessitates this compartmentalization in order to protect 11-cis retinal from reduction in the outer segment. So far, our analysis of Rdh12−/− mice has not shown upregulation of any other isoforms that might compensate for the loss of function.
Although analysis of the retinal phenotype of the Rdh12−/− mouse does not yet permit us to distinguish between these two alternatives, it is clear that the outcomes of RDH12 and RPE65 losses of function are fundamentally different, with mutations in the latter resulting in uniformly severe functional deficits in canines, mice, and humans (21, 26, 29). In addition, the genetic background of Rdh12−/− mice with respect to an Rpe65 polymorphism (p.L450M) that affects rates of 11-cis retinal synthesis is a major factor that needs to be controlled for in comparisons of visual performance and visual cycle activity in Rdh12−/− versus wild-type mice. In fact, consideration of the Rpe65 genotype is also likely to be important for studies of other visual cycle genes, especially other RDH isoforms, but was not always done in previous studies of the effects of Rdh5, Rdh11, and Rdh8 (prRDH) loss of function on specific in vivo functional roles.
Our understanding of RDH12 in vivo function is likely to be improved by developing appropriate challenges of the relevant physiological pathway(s) in knockout animals. This approach has proven successful in eliciting profound phenotypes in a number of strains of knockout mice in which no or mild differences from the wild type were initially apparent (1). Mice deficient in multiple RDHs may also be useful for identifying a minimal set of isoforms essential for visual cycle function. Our targeting strategy will also allow the tissue-specific and time-controlled disruption of Rdh12 for future studies. Identifying the functional correlates between the Rdh12−/− mouse phenotype and human disease will be critical for establishing an animal model useful for evaluating potential modes of therapeutic intervention.
Acknowledgments
Grant support for this study was given by the Deutsche Forschungsgemeinschaft (I.K. and C.A.H. [SFB444 and FG604]), EV-GENORET (LSHG-CT-2005-512036) (A.G.), the National Institutes of Health (R01-EY12298, P30-EY07003, M01-RR00042, and P60DK-20572), the Foundation for Fighting Blindness, and Research to Prevent Blindness (D.A.T.).
Gifts of antibodies were given by the following people: anti-rhodopsin, Barry E. Knox at SUNY Upstate; anti-prRDH antibodies, Amir Rattner and Jeremy Nathans at Johns Hopkins University; anti-RDH5, Kryzsztof Palczewski at Case Western Reserve University; anti-LRAT, Dean Bok at University of California-Los Angeles; anti-CRALBP, John Crabb at Cleveland Clinic; and anti-NRL, Anand Swaroop at University of Michigan. Technical support was given by Irm Hermans-Borgmeyer and Antje K. Hübner (Rdh12−/− mice); by Mitchell Gillett (morphology); by Elizabeth Smith (hybridomas); by Ali Derin, Susanne Conrad, and Austra Liepa (animal care); and by Naheed Khan (electrophysiology).
Footnotes
Published ahead of print on 27 November 2006.
REFERENCES
- 1.Argmann, C. A., P. Chambon, and J. Auwerx. 2005. Mouse phenogenomics: the fast track to “systems metabolism.” Cell Metab. 2:349-360. [DOI] [PubMed] [Google Scholar]
- 2.Belyaeva, O. V., O. V. Korkina, A. V. Stetsenko, T. Kim, P. S. Nelson, and N. Y. Kedishvili. 2005. Biochemical properties of purified human retinol dehydrogenase 12 (RDH12): catalytic efficiency toward retinoids and C9 aldehydes and effects of cellular retinol-binding protein type I (CRBPI) and cellular retinaldehyde-binding protein (CRALBP) on the oxidation and reduction of retinoids. Biochemistry 44:7035-7047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bligh, E. G., and W. J. Dyer. 1959. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37:911-917. [DOI] [PubMed] [Google Scholar]
- 4.Callaway, J. K., P. M. Beart, and B. Jarrott. 1998. A reliable procedure for comparison of antioxidants in rat brain homogenates. J. Pharmacol. Toxicol. Methods 39:155-162. [DOI] [PubMed] [Google Scholar]
- 5.Chen, P., W. Hao, L. Rife, X. P. Wang, D. Shen, J. Chen, T. Ogden, G. B. Van Boemel, L. Wu, M. Yang, and H. K. Fong. 2001. A photic visual cycle of rhodopsin regeneration is dependent on Rgr. Nat. Genet. 28:256-260. [DOI] [PubMed] [Google Scholar]
- 6.Danciger, M., M. T. Matthes, D. Yasamura, N. B. Akhmedov, T. Rickabaugh, S. Gentleman, T. M. Redmond, M. M. La Vail, and D. B. Farber. 2000. A QTL on distal chromosome 3 that influences the severity of light-induced damage to mouse photoreceptors. Mamm. Genome 11:422-427. [DOI] [PubMed] [Google Scholar]
- 7.Driessen, C. A., H. J. Winkens, K. Hoffmann, L. D. Kuhlmann, B. P. Janssen, A. H. Van Vugt, J. P. Van Hooser, B. E. Wieringa, A. F. Deutman, K. Palczewski, K. Ruether, and J. J. Janssen. 2000. Disruption of the 11-cis-retinol dehydrogenase gene leads to accumulation of cis-retinols and cis-retinyl esters. Mol. Cell. Biol. 20:4275-4287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fan, J., M. L. Woodruff, M. C. Cilluffo, R. K. Crouch, and G. L. Fain. 2005. Opsin activation of transduction in the rods of dark-reared Rpe65 knockout mice. J. Physiol. 568:83-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gu, S. M., D. A. Thompson, C. R. Srikumari, B. Lorenz, U. Finckh, A. Nicoletti, K. R. Murthy, M. Rathmann, G. Kumaramanickavel, M. J. Denton, and A. Gal. 1997. Mutations in RPE65 cause autosomal recessive childhood-onset severe retinal dystrophy. Nat. Genet. 17:194-197. [DOI] [PubMed] [Google Scholar]
- 10.Haeseleer, F., J. Huang, L. Lebioda, J. C. Saari, and K. Palczewski. 1998. Molecular characterization of a novel short-chain dehydrogenase/reductase that reduces all-trans-retinal. J. Biol. Chem. 273:21790-21799. [DOI] [PubMed] [Google Scholar]
- 11.Haeseleer, F., G. F. Jang, Y. Imanishi, C. A. Driessen, M. Matsumura, P. S. Nelson, and K. Palczewski. 2002. Dual-substrate specificity short chain retinol dehydrogenases from the vertebrate retina. J. Biol. Chem. 277:45537-45546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harlow, E., and D. Lane. 1988. Antibodies: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 13.Janecke, A. R., D. A. Thompson, G. Utermann, C. Becker, C. A. Hübner, E. Schmid, C. L. McHenry, A. R. Nair, F. Ruschendorf, J. Heckenlively, B. Wissinger, P. Nurnberg, and A. Gal. 2004. Mutations in RDH12 encoding a photoreceptor cell retinol dehydrogenase cause childhood-onset severe retinal dystrophy. Nat. Genet. 36:850-854. [DOI] [PubMed] [Google Scholar]
- 14.Jang, G. F., J. K. McBee, A. M. Alekseev, F. Haeseleer, and K. Palczewski. 2000. Stereoisomeric specificity of the retinoid cycle in the vertebrate retina. J. Biol. Chem. 275:28128-28138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kasus-Jacobi, A., J. Ou, D. G. Birch, K. G. Locke, J. M. Shelton, J. A. Richardson, A. J. Murphy, D. M. Valenzuela, G. D. Yancopoulos, and A. O. Edwards. 2005. Functional characterization of mouse RDH11 as a retinol dehydrogenase involved in dark adaptation in vivo. J. Biol. Chem. 280:20413-20420. [DOI] [PubMed] [Google Scholar]
- 16.Kim, T. S., A. Maeda, T. Maeda, C. Heinlein, N. Kedishvili, K. Palczewski, and P. S. Nelson. 2005. Delayed dark adaptation in 11-cis-retinol dehydrogenase-deficient mice: a role of RDH11 in visual processes in vivo. J. Biol. Chem. 280:8694-8704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marlhens, F., C. Bareil, J. M. Griffoin, E. Zrenner, P. Amalric, C. Eliaou, S. Y. Liu, E. Harris, T. M. Redmond, B. Arnaud, M. Claustres, and C. P. Hamel. 1997. Mutations in RPE65 cause Leber's congenital amaurosis. Nat. Genet. 17:139-141. [DOI] [PubMed] [Google Scholar]
- 18.Perrault, I., S. Hanein, S. Gerber, F. Barbet, D. Ducroq, H. Dollfus, C. Hamel, J. L. Dufier, A. Munnich, J. Kaplan, and J. M. Rozet. 2004. Retinal dehydrogenase 12 (RDH12) mutations in Leber congenital amaurosis. Am. J. Hum. Genet. 75:639-646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Poss, K. D., and S. Tonegawa. 1997. Reduced stress defense in heme oxygenase 1-deficient cells. Proc. Natl. Acad. Sci. USA 94:10925-10930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rattner, A., P. M. Smallwood, and J. Nathans. 2000. Identification and characterization of all-trans-retinol dehydrogenase from photoreceptor outer segments, the visual cycle enzyme that reduces all-trans-retinal to all-trans-retinol. J. Biol. Chem. 275:11034-11043. [DOI] [PubMed] [Google Scholar]
- 21.Redmond, T. M., S. Yu, E. Lee, D. Bok, D. Hamasaki, N. Chen, P. Goletz, J. X. Ma, R. K. Crouch, and K. Pfeifer. 1998. Rpe65 is necessary for production of 11-cis-vitamin A in the retinal visual cycle. Nat. Genet. 20:344-351. [DOI] [PubMed] [Google Scholar]
- 22.Rohrer, B., H. R. Lohr, P. Humphries, T. M. Redmond, M. W. Seeliger, and R. K. Crouch. 2005. Cone opsin mislocalization in Rpe65−/− mice: a defect that can be corrected by 11-cis retinal. Investig. Ophthalmol. Vis. Sci. 46:3876-3882. [DOI] [PubMed] [Google Scholar]
- 23.Ruether, K., D. van de Pol, G. Jaissle, W. Berger, R. P. Tornow, and E. Zrenner. 1997. Retinoschisislike alterations in the mouse eye caused by gene targeting of the Norrie disease gene. Investig. Ophthalmol. Vis. Sci. 38:710-718. [PubMed] [Google Scholar]
- 24.Saari, J. C., G. G. Garwin, F. Haeseleer, G. F. Jang, and K. Palczewski. 2000. Phase partition and high-performance liquid chromatography assays of retinoid dehydrogenases. Methods Enzymol. 316:359-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Simon, A., U. Hellman, C. Wernstedt, and U. Eriksson. 1995. The retinal pigment epithelial-specific 11-cis retinol dehydrogenase belongs to the family of short chain alcohol dehydrogenases. J. Biol. Chem. 270:1107-1112. [PubMed] [Google Scholar]
- 26.Thompson, D. A., P. Gyurus, L. L. Fleischer, E. L. Bingham, C. L. McHenry, E. Apfelstedt-Sylla, E. Zrenner, B. Lorenz, J. E. Richards, S. G. Jacobson, P. A. Sieving, and A. Gal. 2000. Genetics and phenotypes of RPE65 mutations in inherited retinal degeneration. Investig. Ophthalmol. Vis. Sci. 41:4293-4299. [PubMed] [Google Scholar]
- 27.Thompson, D. A., A. R. Janecke, J. Lange, K. L. Feathers, C. A. Hübner, C. L. McHenry, D. W. Stockton, G. Rammesmayer, J. R. Lupski, G. Antinolo, C. Ayuso, M. Baiget, P. Gouras, J. R. Heckenlively, A. den Hollander, S. G. Jacobson, R. A. Lewis, P. A. Sieving, B. Wissinger, S. Yzer, E. Zrenner, G. Utermann, and A. Gal. 2005. Retinal degeneration associated with RDH12 mutations results from decreased 11-cis retinal synthesis due to disruption of the visual cycle. Hum. Mol. Genet. 14:3865-3875. [DOI] [PubMed] [Google Scholar]
- 28.Wenzel, A., C. E. Reme, T. P. Williams, F. Hafezi, and C. Grimm. 2001. The Rpe65 Leu450Met variation increases retinal resistance against light-induced degeneration by slowing rhodopsin regeneration. J. Neurosci. 21:53-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wrigstad, A., K. Narfstrom, and S. E. Nilsson. 1994. Slowly progressive changes of the retina and retinal pigment epithelium in Briard dogs with hereditary retinal dystrophy. A morphological study. Doc. Ophthalmol. 87:337-354. [DOI] [PubMed] [Google Scholar]
- 30.Wu, B. X., Y. Chen, J. Fan, B. Rohrer, R. K. Crouch, and J. X. Ma. 2002. Cloning and characterization of a novel all-trans retinol short-chain dehydrogenase/reductase from the RPE. Investig. Ophthalmol. Vis. Sci. 43:3365-3372. [PubMed] [Google Scholar]
- 31.Yamamoto, H., A. Simon, U. Eriksson, E. Harris, E. L. Berson, and T. P. Dryja. 1999. Mutations in the gene encoding 11-cis retinol dehydrogenase cause delayed dark adaptation and fundus albipunctatus. Nat. Genet. 22:188-191. [DOI] [PubMed] [Google Scholar]
- 32.Znoiko, S. L., B. Rohrer, K. Lu, H. R. Lohr, R. K. Crouch, and J. X. Ma. 2005. Downregulation of cone-specific gene expression and degeneration of cone photoreceptors in the Rpe65−/− mouse at early ages. Investig. Ophthalmol. Vis. Sci. 46:1473-1479. [DOI] [PubMed] [Google Scholar]