Abstract
The coactivator TIF2 was predicted to interact with an unknown factor to modify both the relative inhibition in glucocorticoid receptor (GR)-mediated gene repression and several parameters of agonists and antisteroids in GR-regulated induction. Here, we describe the isolation and characterization of the predicted factor as a new 1,277-amino-acid endogenous protein (STAMP). STAMP associates with coactivators (TIF2 and SRC-1) and is selective for a subset of the steroid/nuclear receptors including GRs. Transfected STAMP increases the effects of TIF2 in GR-mediated repression and induction. Conversely, the levels of both induction and repression of endogenous genes are reduced when STAMP small interfering RNAs are used to lower the level of endogenous STAMP. Endogenous STAMP colocalizes with GR in intact cells and is recruited to the promoters of endogenous GR-induced and -repressed genes. We suggest that STAMP is an important new, downstream component of GR action in both gene activation and gene repression.
Glucocorticoid receptors (GRs) mediate both gene induction and gene repression by increasing or decreasing, respectively, the rates of gene transcription, thereby altering the levels of gene expression. For gene induction, GRs typically bind directly to a glucocorticoid response element (GRE) consisting of the consensus palindromic sequence of AGAACANNNTGTTCT, where N is any nucleotide. No such common DNA sequence exists among genes that are down-regulated by GRs due to the diversity of mechanisms involved in GR-mediated repression (3). While gene induction by GRs has been more extensively studied and has, in combination with induction by other steroid/nuclear receptors, provided most of the data for constructing the current model of GR action (54, 55), gene repression by GRs is of particular clinical importance due to the widespread use of glucocorticoids to suppress inflammatory responses (12, 24, 45). One common mode of suppression is for GRs to be “tethered” to DNA-bound jun-fos dimers of AP-1, thereby reducing the transactivation activity of AP-1 (24, 45). The responses in GR-regulated induction, and often in GR-directed repression, are augmented by cofactors such as the coactivators TIF2 and hBRM (20, 21, 41, 44, 49, 57), which are recruited to DNA-associated GRs (34, 40, 61).
An important property of GR induction is the position of the dose-response curve. The 50% effective concentration (EC50) of a dose-response curve specifies the steroid concentration required for 50% of maximal induction and numerically indicates differences in induction potency or sensitivity among steroid-responsive genes. A lower EC50 value affords more gene activation at the subsaturating, physiological concentrations of steroid. Thus, changes in the EC50 are a powerful method for effecting differential gene expression during development and homeostasis (15, 18).
It has recently become clear that the EC50 of all genes regulated by a given receptor-steroid complex is not constant. Even for a specific gene, the EC50 is not fixed but varies with the levels of numerous factors that include coactivators (46, 47). Changes in the concentrations of these factors also modify the amount of residual partial agonist activity of antisteroids, which has widespread clinical potential for reducing the number of unwanted side effects that often result during endocrine therapies with antisteroids due to their indiscriminant blockage of agonist actions for many responsive genes (46, 47).
The regions of the coactivators TIF2 and SRC-1 that suffice to modulate the EC50 and partial agonist activity of GR-inducible genes are separable from activation domain 1 (AD1) and AD2 (20) that are critical for increasing the total levels of gene activation (4, 23, 31, 37, 56). The modulatory domain of TIF2 (amino acids 624 to 1010, or TIF2.4 [56]) interacts with receptors and other proteins via three LXXLL motifs, or receptor interaction domains (RIDs) (1, 11, 27, 28, 33). However, an additional binding site was implicated by the ability of overexpressed TIF2.4 fragments lacking the LXXLL motifs to competitively inhibit several biological activities of TIF2.4 with GRs (20). These studies also supported previous studies suggesting that two mechanistic pathways, each with possibly different cofactors, are responsible for the coactivator-mediated increase in total level of gene expression versus modulation of EC50 and partial agonist activity for GRs (8, 10, 20, 25, 39, 52, 53, 58, 62): progesterone receptors (PRs) (16, 50) and mineralocorticoid receptors (59).
For GR-mediated repression of AP-1 induced responses, amino acids 648 to 1007 of GRIP1/TIF2 lack AD1 and AD2 but suffice for TIF2 augmentation of GR-mediated repression (41). This repressive domain is only slightly smaller than the above region of TIF2 that is required for the modulation of both the EC50 and partial agonist activity in GR induction (20). Thus, it is reasonable to propose that the molecular mechanisms of several of the properties of GR-mediated induction and repression involve an unidentified transcription factor(s).
We now identify and characterize the predicted cofactor that participates both in the modulation of GR induction properties and in GR repression. The accumulated properties of this novel protein suggest that it is a new coregulator in GR-mediated induction and repression.
MATERIALS AND METHODS
Unless otherwise indicated, all operations were performed at 0°C.
Chemicals.
Dexamethasone (Dex) and phorbol 12-myristate 13-acetate (PMA) were purchased from Sigma. Dex-21-mesylate (Dex-Mes) was synthesized as previously described (48). R1881 (Liang-Nian Song, Georgetown University, Washington, DC) and roziglitazone and 9-cis-retinoic acid (Kai Ge, National Institute of Diabetes and Digestive and Kidney Diseases [NIDDK], NIH) were obtained as gifts. [35S]methionine and [32P]dCTP are from Amersham. Restriction enzymes and DNA polymerase were obtained from New England Biolabs, Amersham Biosciences, or Promega.
Antibodies.
Antihemagglutinin (anti-HA) mouse monoclonal antibody (Roche), anti-FLAG mouse monoclonal antibody (Sigma), anti-GR mouse and rabbit monoclonal antibodies (BUGR-2 and PA1-511A; Affinity BioReagents), anti-TIF2 mouse monoclonal antibody (item 610984; BD Biosciences), and mouse monoclonal anti-GAL DNA binding domain (DBD) and anti-VP16 antibodies (Santa Cruz Biotechnology) are commercially available. STAMP antibodies 1 and 2 were raised in New Zealand White rabbits by Covance (Denver, PA), using the KLH-conjugated STAMP C-terminal CRRATSQKASKGSSAEGQ (amino acids [aa] 1234 to 1250) and N-terminal CESLNSKAKLLIAALYERKLLS (aa 525 to 544) peptides (Princeton Biomolecules, Langhorne, PA) as antigens. α-STAMP 1 and 2 antibodies were purified by antigenic peptide affinity column chromatography.
Plasmids.
GR, GREtkLUC, TIF2, TIF2.4, SRC1, SRC1-1139C, VP16-GR, pBAL-GR, glutathione S-transferase (GST)-TIF2.4, GAL-TIF2.4, and GRIP (HA-GRIP) have been previously described (20). VP16 chimeras of full-length androgen, estrogen (α and β), thyroid (β), and retinoid (α) receptors were generously provided by Donald McDonnell (Duke University, Durham, NC) (5). KIAA0998 plasmid was donated by Takahiro Nagase (Kazusa DNA Research Institute, Japan). AP-1-Luc (Inez Rogatsky, University of California-San Francisco Medical School, San Francisco, CA), TREtkLUC(F2) (Paul Yen, Johns Hopkins Bayview Medical Center, Baltimore, MD) and AR, AREtkLUC, VP16/peroxisome proliferator-activated receptor γ, and VP16/retinoid X receptor α (Kai Ge, NIDDK, NIH) were gifts. pSos-TIF2.4 was constructed by inserting the NcoI/NotI fragment of Gal-TIF2.4 into pSos (Stratagene). Full-length STAMP was constructed by assembling the BamHI/XbaI fragment of KIAA0998 clone (Japanese Kazusa DNA Institute) with the EcoRV/BamHI fragment of IMAGE clone 3632160 (Open Biosystems) and inserting the product into the EcoRV/XbaI sites of pCMV-Sport6 (Invitrogen).
The following primers were used to construct different fragments of STAMP or other constructs: STAMP 3′ Primer 2, 5′-GCT CTA GAT GCA CCC AGG AGT GGT GAA CAG G-3′; STAMP 5′ Primer 2, 5′-AAG ATG AGA CAG GAA TCT GTG CC-3′; 220F, 5′-TAC GAT ATC TGA TGG CCC GGG ACC TGG AGG AAA C-3′; 525R, 5′-AGT AAG AAG GGC TTC AGG-3′; Xb2092R, 5′-CGT CTA GAC TAC CAA AGC TGT CAA TGA GGG TG-3′; ER2093F, 5′-GGA ATT CGA AAA TAC ACC CAA AGA AAA TTC C-3′; Xb2729R, 5′-GCT CTA GAC ACC TTT TGC CCC ACT ATC AGA A-3′; ER2728F, 5′-GGA ATT CGA TCA CCC TGA GAC TAT AAT GG-3′; Xb3091R, 5′-CGT CTA GAG GCC AGT AGG GCT TGG GAT GTT C-3′; ER3091F, 5′-GGA ATT CCT GCC ACG CTG TCG ATC AGG AAG-3′; ER3605F, 5′-GGA ATT CAC AGG GGT GGT CCC CCA GCA C-3′; Xb3605R, 5′-CGT CTA GAT AGC CTG GCT GTA CAC GTT GTT TTC-3′; TIF2-1140F, 5′-GGA ATT CGC ACA AAT GGC CCA GGG TAG C-3′; and TIF2-1464R, 5′-CGG GAT CCT CAG CAA TAT TTC CGT GTT GTG TC-3′.
HA-pSG5-STAMP (HA/STAMP) was constructed by joining the EcoRV/BamHI fragment of the 5′ STAMP PCR product of 220F and 525R primers with the BamHI/NotI fragment of KIAA0998, followed by insertion into the EcoRV/NotI site of a modified HA-pSG5. The modified HA-pSG5 was prepared by inserting the annealed double-strand oligonucleotide (GAA TTC CCG GGA TAT CGT CGA CCC ACG CGT CCG GGG CGG CCG CTC TAG AGT ATC CCT CGA GGA TCC) into the EcoRI/BamHI Sites of HA-pSG5 (Mike Stallcup, University of Southern California, Los Angeles, CA), thus introducing new EcoRV and NotI Sites. pFlag/STAMP (Flag/STAMP) was constructed by inserting the EcoRV/XbaI fragment of HA/STAMP into pFlag-CMV2 (Sigma). GAL4 DBD with a fragment of STAMP containing amino acids 623 to 834 [GAL/STAMP(623-834)] (Gal/6-6CL1) and VP16/6-6CL1 were constructed by inserting the 1.2-kb EcoRI fragment of the original 6-6CL1 clone from the yeast library screening into the PM and VP16 plasmids (Clontech), respectively. The following constructs were also made as follows: for GAL/STAMP(N623) (GAL fused to the N terminus of amino acids 1 to 623 of STAMP), the PCR-amplified fragment prepared with 220F and Xb2092R primers was then cut and inserted into the EcoRV/XbaI sites of PM plasmid; for GAL/STAMP(N834), the PCR-amplified fragment prepared with 220F and Xb2729R primers was then cut and inserted into the EcoRV/XbaI sites of PM plasmid; for GAL/STAMP(834C) (GAL fused to amino acid 834 to the C terminus of STAMP), the PCR-amplified fragment prepared with ER2728F and STAMP 3′ Primer 2 was then cut and inserted into the EcoRI/XbaI sites of PM plasmid; for GAL/STAMP(1127C), the PCR-amplified fragment prepared with ER3605F and STAMP 3′ Primer 2 was then cut and inserted into the EcoRI/XbaI sites of PM plasmid; for GAL/STAMP(834-956), the PCR-amplified fragment prepared with ER2728F and Xb3091R primers was then cut and inserted into the EcoRI/XbaI sites of PM plasmid; for GAL/STAMP(956-1127), the PCR-amplified fragment prepared with ER3091F and Xb3605R primers were then cut and inserted into the EcoR1/XbaI sites of PM plasmid; for GST/STAMP(956C), the PCR-amplified fragment prepared with ER3091F and STAMP 3′ Primer 2 was then cut and inserted into the EcoR1/XhoI sites of pGEX6p1 (Amersham Pharmacia).
VP16/SRC(1139C) (the VP16 activation domain fused to amino acid 1139 to the C terminus of SRC-1) was constructed by inserting the EcoRI/XbaI fragment of SRC1-1139C into VP16. VP16/TIF2(1140C) was constructed by inserting into VP16 the EcoR1/BamHI fragment from the PCR product of TIF2 with the TIF2-1140F and TIF2-1464R primers.
Yeast Sos-Ras two-hybrid library screen.
The yeast Sos-recruitment two-hybrid system (Cyto Trap Vector Kit) and the human fetal brain Cyto Trap Plasmid cDNA Library were purchased from Stratagene and employed according to the manufacturer's instructions. Briefly, about 50 μg of pSos bait construct (pSos-TIF2.4) and 50 μg of pMyr cDNA plasmid library (Stratagene) were cotransformed into 10 ml of cdc25H yeast-competent cells, spread onto 40, 100-mm plates of synthetic drop-out glucose agar lacking uracil and leucine [SD/Glu(-UL)], and grown for 2 to 4 days at 25°C. Colonies were replica plated onto SD galactose agar lacking uracil and leucine [SD/Gal(-UL)] plates and grown for 6 days at 37°C. Candidate “interactor” colonies were selected and “patched” onto SD/Glu(-UL) plates, grown for 2 days at 25°C for Gal repression. These colonies were “repatched” onto two serial interaction plates for 4 days at 37°C: one with SD/Glu(-UL) and the other with SD/Gal(-UL). The colonies that grew on SD/Gal(-UL) but not on SD/Glu(-UL) plates were selected for further study. The cDNA plasmid from these putative colonies was isolated and cotransformed with bait or empty pSos plasmid into cdc25H yeast cells to see whether the candidate cDNA clone specifically interacted with the bait in yeast.
Cell culture, transient transfection, and reporter analysis.
Triplicate samples of cells were transiently transfected in 24-well plates with luciferase reporter plasmids as described for CV-1 (20). For U2OS.rGR cells (41), triplicate samples were seeded into 24-well plates in Dulbecco's modified Eagle's medium (DMEM)-10% fetal bovine serum (FBS) at 20,000 cells per well and transfected the following day in FBS-free DMEM by using 0.8 μl of Lipofectamine and 1.6 μl of PLUS reagent (Invitrogen) per well according to the manufacturer's instructions. The total transfected DNA was adjusted to 150 ng/well of a 24-well plate with pBluescriptII SK+ (Stratagene). The molar amount of plasmids expressing different protein constructs was kept constant with added empty plasmid or plasmid expressing human serum albumin (58). phRG-TK Renilla (Promega) (10 ng/well of a 24-well plate) was included as an internal control. After transfection (3 h), cells were refed with DMEM-10% FBS, allowed to recover for 3 h, and refed with DMEM-10% FBS containing appropriate hormone dilutions. Twelve hours later, the cells were lysed and assayed for reporter gene activity using dual luciferase assay reagents according to the manufacturer's instructions (Promega, Madison, WI). Luciferase activity was measured by an EG&G Berthold's luminometer (Microlumat LB 96P). In both cases, the data were normalized either for total protein or Renilla null luciferase activity and expressed as a percentage of the maximal response with Dex before being plotted ± standard error of the mean, unless otherwise noted.
mRNA, total RNA extraction, and reverse transcription-PCR (RT-PCR).
Human testis poly(A)+ RNA was purchased from BD Clontech. CV-1 cell total RNA was prepared by growing CV-1 cells to confluence in 60-mm dishes for 2 days, lysing the cells with TRIzol (Invitrogen) reagent, and extracting the total RNA according to the manufacturer's instructions. First-strand cDNA was synthesized by SuperScript II RNase H reverse transcriptase (Invitrogen). PCR was performed with STAMP 5′ Primer 2 and 3′ Primer 2 and Pfu Ultra High-Fidelity DNA Polymerase (Stratagene) according to the manufacturer's instructions to amplify the full-length STAMP. For quantitative real-time PCR (qRT-PCR), total RNA was extracted by TRIzol reagent (Invitrogen) according to the manufacturer's instructions. First-strand cDNA was synthesized by SuperScript II reverse transcriptase. The relative levels of target mRNAs were quantitated using SyberGreen and the ABI 7900HT real-time PCR system for insulin-like growth factor binding protein 1 (IGFBP1), glucocorticoid-induced leucine zipper (GILZ), LAD1, and interferon regulatory factor 8 (IRF8) (primers were those specified by Chen et al. [9]). The other genes (collagenase 3, glyceraldehyde-3-phosphate dehydrogenase, and STAMP; primers from ABI are Hs00233992-m1, 4310884E, and Hs00209404-m1, respectively) were quantitated by Taqman.
Northern blotting.
STAMP(2093-2426) probe was prepared by cutting the original 6-6CL1 clone with EcoRI/SalI and purifying the 300-bp fragment from an agarose (2%) gel. The probe (along with a control β-actin probe from BD Clontech) was labeled with [32P]dCTP using Prime-a-Gene Labeling System (Promega) according to the manufacturer's instructions. 32P-labeled probes were hybridized with Multiple Tissue Northern blotting (BD Clontech) membranes according to the manufacturer's instructions.
Mammalian two-hybrid and pull-down assays.
Mammalian two-hybrid and pull-down assays were conducted as previously described (20).
Immunoprecipitation assays.
Immunoprecipitations were conducted as described previously (58) with the following modifications. The day before transfection, Cos-7 cells were seeded into 150-mm dishes at 2,000,000 cells per dish containing 20 ml of medium. On the next day, 30 μg of DNA/dish was transfected with 60 μl of FuGene reagent. After 2 days of growth, cells were treated with ethanol (EtOH) with or without Dex, washed once with 20 ml of PBS 2 h later, lysed for 5 min at room temperature with 1.6 ml of CytoBuster protein extraction reagent (Novagen) containing protease inhibitor cocktail (Roche) per 150-mm dish, collected with a cell scraper, and centrifuged for 5 min at 16,000 × g (4°C). Aliquots (700 μl) of supernatant were incubated on a roller drum (3 rpm) with either 50 μl of HA-matrix (Roche) (overnight at 4°C) or 80 μl or 50% slurry protein G (Amersham Pharmacia) (4°C for 1 h) and then centrifuged for 1 min at 13,000 rpm (4°C). The supernatant was incubated with 10 μl of antibody for 1 h (at 4°C on a roller drum) and then overnight at 4°C with 70 μl of protein G. On the next day, the antibody complexes were centrifuged (1 min at 16,000 × g at 4°C), washed three times with 1 ml of lysis buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 0.1% NP-40), extracted with 20 μl of 2× sodium dodecyl sulfate (SDS) loading buffer (95°C for 3 min), and separated by 8 to 10% SDS-polyacrylamide gel electrophoresis (PAGE).
Immunocytochemistry.
Cells were cultured in phenol red-free medium with 10% charcoal-dextran-stripped fetal bovine serum in 2 to 4 chambers per slide (Nalge Nunc) for 2 to 4 days. Cells were then left untreated or treated with steroid, washed with 1 ml of cold phosphate-buffered saline (PBS), and fixed with 1 ml of freshly prepared 4% paraformaldehyde (20 min at room temperature). Cells were washed with PBS (5 min), treated with 1 ml of 100% methanol (10 min), washed with PBS (10 min), and treated with 1 ml of blocking buffer (1× PBS, 0.1% Tween 20, 10% goat immunoglobulin G [IgG], 1% bovine serum albumin) for 30 min, all at room temperature. After incubation with 250 to 500 μl of the first antibody (1:250 dilution of purified α-STAMP 1 or α-GR) in blocking buffer (2 h at room temperature), cells were washed twice with PBS-0.05% Tween 20 (each for 10 min) and treated with 250 to 500 μl of the fluorescence-coupled second antibody (1:1,000 dilution of anti-mouse or -rabbit antibody; Jackson ImmunoResearch) in blocking buffer (at room temperature for 1 h in the dark) before being washed (in 1 ml of PBS-0.05% Tween 20 for 10 min) and treated with 1 μg/ml of 4,6-diamidino-2-phenyindole (Sigma) in PBS-0.05% Tween 20 (5 min in the dark). Cells were washed (in PBS-0.05% Tween 20 for 10 min) before being examined with a fluorescent microscope.
ChIP and reChIP assays.
The basic method of Ma et al. (32) was modified as follows. Two days before harvest, U2OS.rGR cells were seeded into 150-mm dishes at 3,000,000 cells per dish in phenol red-free medium with 10% charcoal-dextran-stripped FBS. Each dish was transfected on the next day with 15 μg of HA-STAMP and 60 μl of Lipofectamine (no transfection for endogenous proteins). On the third day, after brief ligand treatment, formaldehyde (37%) was added directly into the medium to a final concentration of 1% (at 37°C for 10 min), which was followed by glycine to a final concentration of 0.125 M (room temperature for 5 min with shaking). Cells were washed with (4°C) cold PBS twice and harvested by scraping into 5 ml of cold (4°C) PBS with a protease inhibitor cocktail (Roche). After centrifugation (at 1200 × g for 10 min), the cell pellet was resuspended in 1 ml of hypotonic buffer (at 0°C for 10 min) and lysed by passing through a 25-gauge needle five times. The nuclei were collected by centrifugation (at 15,000 × g for 10 min) and resuspended in 250 μl of nuclear lysis buffer per dish (at 0°C for 10 min). The nuclear pellet is sonicated with a Fisher Scientific Ultrasonic Dismembrator (model 500) (10 to 12 pulses of a cycle of 20 s on and 40 s off at 22% power, thus generating fragments of purified DNA of 200 to 600 bp). After centrifugation (15,000 × g for 10 min), 100 μl of supernatant was diluted into 1 ml of immunoprecipitation dilution buffer and treated with 20 μl of preblocked protein G beads (Amersham Pharmacia) with gentle mixing (at 4°C for 1 h on a rotating drum at 4 rpm). After centrifugation (15,000 × g for 10 min), the supernatant was treated with 2 μg of anti-GR (PA1-516; Affinity BioReagents), anti-HA (sc7392; Santa Cruz), or anti-STAMP (Covance) antibody (at 4°C overnight). The next morning, 40 μl of preblocked protein G beads was added (at 4°C for 2 h). The pellet was centrifuged (5,500 × g for 1 min), washed sequentially by 1 ml each of wash buffers I, II, and III, three times with 1 ml of Tris-EDTA buffer, and then eluted twice with 150 μl of elution buffer. In the chromatin reimmunoprecipitation (reChIP) assays, complexes were eluted by incubation for 30 min at 37°C in 25 μl of 10 mM dithiothreitol. The supernatant was diluted 1:50 into immunoprecipitation dilution buffer followed by reimmunoprecipitation with the second antibody and processed as above. The eluents were combined, adjusted to 300 mM NaCl (at 65°C overnight; total volume, 300 μl) to reverse the cross-linking, and then treated with 10 μl of EDTA (0.5 M), 40 μl of 1 M Tris-HCl (pH 6.5), and 2 μl of proteinase K (10 μg/μl; Sigma) at 45°C for 1 h. The DNA was purified by a QIAGEN PCR purification kit according to the manufacturer's instructions. The immunoprecipitated DNA was amplified by PCR using the published primers for the collagenase 3 and hsp70 genes (44). Primers for the human IGFBP promoter (+31 to −204), which are 5′-CCC CCT AAC AAC GGG ACA AAC AGT A-3′ (forward) and 5′-ATG CTC GCT GGA TGG GAT GG-3′ (reverse), were selected from the known structure of the human IGFBP gene (17, 51).
siRNA assays.
Four different small interfering RNA (siRNA) oligonucleotides for STAMP were designed and synthesized by QIAGEN: (i) GCCAGCTGAGTACGCGGAATT, (ii) CACCCTCTCTGAAGCACAAAA, (iii) CAGCACTGACTATAACCTAAT, and (iv) GAGGGCAATGAGGCCAAAATA. Control β-actin and lamin siRNAs were purchased from QIAGEN. U2OS.rGR cells were seeded at 50,000 cells per well in 24-well plates the day before transfection. DNA (STAMP plasmid and AP-1/Luc reporter; total, 150 ng) together with 500 ng of siRNA were mixed with 1.6 ml of Lipofectamine 2000 (Invitrogen), allowed to stand at room temperature for 15 min, and then added to FBS-free DMEM prewashed cells. Four hours later, the cells were refed with DMEM-10% FBS (containing G418), allowed to recover for 2 h, and refed with DMEM-10% FBS (containing G418) containing appropriate hormone dilutions with or without PMA. The cells were harvested, and luciferase activity was determined as described above in the standard transient transfection studies, or mRNA levels were quantitated by RT-PCR as described above.
RESULTS
Isolation of predicted TIF2.4-binding protein.
The activity of TIF2 and its derivatives (Fig. 1A) in modulating GR transactivation properties is quantitated by the ability of ectopic coactivator to shift the EC50 value of the dose-response curve to lower concentrations of agonist steroid and to increase the partial agonist activity of antiglucocorticoids (8, 20, 52, 53). Using this assay in CV-1 cells, we previously established that TIF2.4 retains most of the modulatory activity of TIF2. Using TIF2.4 as the bait with a human fetal brain library in the Sos-Ras yeast two-hybrid screen that regulates cell growth (Fig. 1B) (2), we isolated 6-6CL1 (Fig. 1C) with an open reading frame for 214 amino acids. In mammalian two-hybrid assays, 6-6CL1 fused to the VP16 activation domain interacts with TIF2.4 fused to the GAL4 DBD (Fig. 1D). The same two-hybrid assay reveals negligible association of 6-6CL1 with p300, CBP, and p/CAF (not shown), which are cofactors that bind to GR/TIF2 complexes (6, 7, 31, 56). 6-6CL1 also interacts with TIF2.4 when oppositely oriented fusion constructs are used (Fig. 1E).
FIG. 1.
Isolation and characterization of partial clone of TIF2.4 binding protein. (A) Schematic diagram of constructs of the coactivators TIF2 and SRC-1. Abbreviations: CBP, CBP interaction domain; Q rich, glutamine rich; Neg, negative suppressor domain (38). (B) Cartoon of the Sos-Ras two-hybrid screen. The bait, present as a chimera with Sos, will activate the membrane-bound Ras protein in a GDP-dependent coupled reaction to cause colony growth only if it binds to a target protein that is fused to a membrane localization sequence. (C) Growth of yeast clone 6-6CL1 from Sos-Ras two-hybrid screen for TIF2.4-interacting proteins. (D and E) Selectivity of 6-6CL1 interactions with TIF2.4 in mammalian two-hybrid assays. The ability of the indicated GAL-DBD and VP16-AD plasmids to activate the GAL4-regulated reporter, pFRLuc, in transiently transfected CV-1 cells is plotted. Similar results were obtained in three additional experiments. (F and G) Comparison of binding to 6-6CL1 by similarly positioned domains of SRC-1 and TIF2. The interaction of GAL with and without 6-6CL1 constructs with VP16 chimeras of SRC-1 (F) or TIF2 (G) segments was determined as in described for panels D and E. Similar results for SRC-1 were obtained in two additional experiments. ND, not determined. All error bars indicate the standard deviations of triplicate experiments.
SRC-1, like TIF2, modulates the dose-response curve and partial agonist activity of GR complexes (52). The C-terminal 303-amino-acid fragment, SRC-1(1139C) (Fig. 1A), possesses the most modulatory activity (20) and displays a strong interaction with 6-6CL1 in the mammalian two-hybrid assay (Fig. 1F). In contrast, while the TIF2 and SRC-1 domains are similarly organized (35, 60), the spatially analogous segment of TIF2, TIF2(1140C) (Fig. 1A), does not interact with 6-6CL1 (Fig. 1G). Western blots (not shown) demonstrate that TIF2(1140C) inactivity is not due to inadequate protein expression. This ability of different domains of TIF2 and SRC-1 to both alter GR induction properties and interact with 6-6CL1 is strong evidence that 6-6CL1 is part of a protein that is involved in the documented modulatory activity of each coactivator (20, 52). For this reason, the predicted full-length protein is called STAMP (SRC-1 and TIF2-associated modulatory protein).
Examination of GenBank sequences revealed that 6-6CL1 corresponds to part of a human protein segment of unknown function (36) (GenBank accession no. AB023215, NM_015072, and BC002766). The assembly of these sequences yields a 4.6-kb cDNA (GenBank accession no. AY237126) that encodes a 1,277-amino-acid protein (Fig. 2A), with a predicted molecular mass of 143 kDa, and resides on chromosome 14q24.3 with 32 exons (GenBank accession no. NM_015072). There is a weaker Kozak sequence (purine at −3 but no G at +4) (29) for an alternative start site four amino acids (italicized residues in Fig. 2A) upstream of the predicted major start site. Therefore, heterogeneity in the amino terminal sequence of STAMP may exist among tissues. The presence of multiple stop codons in all reading frames upstream of the start of translation and at the end of the open reading frame argue that this clone encodes the full-length STAMP protein.
FIG. 2.
Full-length cDNA and predicted protein sequence for STAMP. (A) Human STAMP. Stop codons flanking the coding sequence are given in bold type. Double daggers indicate the positions of two sets of consensus Kozak sequence nucleotides for the start of translation. The first methionine may not be the major start site as only the nucleotide at −3 to the ATG fits the consensus Kozak sequence (29). The asterisk marks the end of the predicted protein sequence. A poly(A) signal sequence is marked by double underlining. (B) Identification of endogenous STAMP mRNA in human testis library by RT-PCR. Human STAMP primers (single underlined sequences in panel A) were used to amplify STAMP mRNA sequences present in a human testis mRNA library (predicted 4.1-kb band is indicated by arrow). (C) Northern blots of mRNAs from different tissues. 32P-labeled STAMP oligonucleotide (bp 2093 to 2426) was used to probe a membrane containing the mRNAs of human tissues (BD Clonetech). A common species at about 4.6 kb was detected in all samples by radioautography. β-Actin mRNA levels (control) were determined with a 32P-labeled actin fragment (BD Clontech).
RT-PCR of mRNA from human testis using internal primers (Fig. 2A, underlined sequences) produces the predicted 4.1-kb product (Fig. 2B). DNA sequencing of this RT-PCR product confirms the above assembled full-length cDNA sequence. This argues that an mRNA encoding the full-length STAMP protein exists in intact cells. Northern blots reveal the predicted 4.6-kb mRNA in 12 tissues (Fig. 2C). Except in heart and skeletal muscle, the STAMP mRNA levels are low, in which case the STAMP protein levels may also be low. However, these data suggest that STAMP is a ubiquitously expressed protein in humans. Among the cell lines used in this study, qRT-PCR revealed that the most frequently used CV-1 cells contain the least STAMP mRNA (relative amounts in CV-1, Cos-7, U2OS.rGR, and U2OS cells are 1, 1.2, 1.7, and 3.6, respectively), which is 800 times less than that of β-actin mRNA (data not shown).
RT-PCR of CV-1 cell mRNA yields a monkey cDNA (GenBank accession no. AY383558) encoding a protein that is 96.7% identical to human STAMP protein (not shown). A mouse cDNA encodes a protein of unknown function (GenBank accession no. XM_126935) that is 83.3% identical to the C-terminal 705 amino acids of human STAMP (not shown). Thus, the STAMP gene and protein are highly conserved among these different species.
Whole-cell localization of STAMP.
Two anti-STAMP antibodies have been prepared. The concentration of STAMP in human osteosarcoma cell (U2OS.rGR) (42) cytosols is too low for definitive identification with purified α-STAMP antibody 1 (Fig. 3A, lane 1), as expected from the low levels of STAMP mRNA in most tissues (Fig. 2C). However, immunoprecipitation of cell lysates with or without Dex by STAMP antibody 1 (lanes 2 and 3), but not with added immunogenic peptide (lane 4) or by preimmune serum (lane 5), concentrated a protein that was visualized by Western blotting with STAMP antibody 1. The second antibody, STAMP antibody 2, gave similar results (not shown). Both the specifically precipitated and crude endogenous proteins comigrate with authentic STAMP (Fig. 3A, lanes 2 and 3 versus 6). We conclude that STAMP is an endogenous protein of U2OS.rGR cells.
FIG. 3.
Endogenous STAMP of U2OS.rGR cells. (A) Detection by Western blotting. Lysates from cells treated with Dex (100 nM; 40 min) or untreated were separated on SDS-PAGE gels directly (lane 1) or first immunoprecipitated either with anti-STAMP antibody with or without competing antigenic peptide (lanes 2 to 4) or with preimmune serum (lane 5), followed by Western blotting with STAMP antibody 1 (lanes 1 to 6). (B) Immunocytochemical localization. U2OS.rGR cells were left untreated or treated with 100 nM Dex (35 min) as described in Materials and Methods to locate STAMP (green), GRs (red), or colocalized GR and STAMP (yellow). IP, immunoprecipitation.
Given the low background with rabbit IgG (Fig. 3B top), endogenous STAMP could be visualized by indirect immunofluorescence in U2OS.rGR cells with purified STAMP antibody 1 (Fig. 3B, middle and bottom). In the absence of steroid, STAMP is both cytoplasmic and nuclear and occasionally colocalizes with the endogenous GR, as indicated by the yellow in the merged pictures. Thirty-five minutes after the addition of Dex, the abundance of yellow in the merged pictures indicates that both STAMP and GR are extensively colocalized in the nucleus, although some cytoplasmic STAMP remains in some cells (Fig. 3B, bottom, and data not shown).
Activity of STAMP in GR-mediated induction.
We next asked whether exogenous STAMP can modulate the EC50 of glucocorticoids and the partial agonist activity of antiglucocorticoids in transient transfection assays where GR induces a GREtkLUC reporter (8, 20, 52, 53). The effects of Flag/STAMP or TIF2 alone on the dose-response curve are small (Fig. 4A). More TIF2 has no additional effect (8; also data not shown). However, a significant further left shift in the dose-response curve occurs with both STAMP and TIF2. When the relative change in EC50 from four experiments is normalized to that of GR alone, STAMP plus TIF2 produces a greater change in EC50 to lower steroid concentrations than expected from the responses of STAMP and TIF2 alone (Fig. 4B). Similarly, the effect of STAMP plus TIF2 on the partial agonist activity of Dex-Mes is greater than the sum of the individual factors (Fig. 4B). This is consistent with STAMP's being a limiting factor in the presence of exogenous GR and TIF2. At the same time, STAMP increases the relative induction (above basal activity) with 1 μM Dex both with and without added TIF2 (Fig. 4B).
FIG. 4.
Modulation by STAMP of GR-mediated gene induction and repression. (A) Effect on the dose-response curve and partial agonist activity for GR-regulated induction. CV-1 cells were transiently transfected with GR (6 ng) with or without TIF2 (20 ng) and with or without STAMP (160 ng) plasmids and GREtkLUC reporter. Normalized luciferase activities, expressed as a percentage of the maximal response with 1 μM Dex, are plotted as described in Materials and Methods. (B) Relative changes in GR induction parameters by STAMP and TIF2. The average relative increase in each parameter from four experiments, as shown in panel A, was determined as follows: for EC50s, relative increase = EC50(GR)/EC50(GR+factor); for partial agonist activity and relative Dex induction, relative increase = activityGR+factor/activityGR. (C) Ectopic STAMP augments GR repression of an AP-1-induced gene. U2OS.rGR cells were transiently transfected with or without TIF2 (20 ng) and with or without STAMP (100 ng) plasmids plus AP-1/Luc reporter (20 ng) and induced with EtOH or 25 ng/ml PMA with or without 0.1 μM Dex. The average relative increase in luciferase activity over the basal level response with EtOH (n = 4) is plotted. The numbers in parentheses above the data for “PMA+Dex” indicate the relative repression caused by Dex with or without the various factors. (D) Requirement of functional TIF2 for STAMP activity. U2OS.rGR cells were transiently transfected and treated with PMA with or without Dex and processed as described in panel C. The average relative repression by Dex (n = 4 to 5) is plotted.  , P ≤ 0.031 between samples containing equal amounts of TIF2 and TIF2m123. (E) ChIP assays. The Dex-induced (0.1 μM plus 25 ng/ml PMA; 40 min) binding of endogenous STAMP and GR to the promoters of endogenous GR-repressed (coll3) and -induced (IGFBP1) genes in U2OS.rGR cells was determined as described in Materials and Methods with purified anti-STAMP 1 and anti-GR. ChiP/reChIP assays (first antibody, anti-STAMP 1; second antibody, anti-GR or -IgG) were similarly processed. As a control for specificity, the same treatment does not coprecipitate hsp70 DNA sequences. Similar results were obtained in one or two additional experiments.
, P ≤ 0.031 between samples containing equal amounts of TIF2 and TIF2m123. (E) ChIP assays. The Dex-induced (0.1 μM plus 25 ng/ml PMA; 40 min) binding of endogenous STAMP and GR to the promoters of endogenous GR-repressed (coll3) and -induced (IGFBP1) genes in U2OS.rGR cells was determined as described in Materials and Methods with purified anti-STAMP 1 and anti-GR. ChiP/reChIP assays (first antibody, anti-STAMP 1; second antibody, anti-GR or -IgG) were similarly processed. As a control for specificity, the same treatment does not coprecipitate hsp70 DNA sequences. Similar results were obtained in one or two additional experiments.
Activity of STAMP in GR-mediated repression.
TIF2 also augments GR repression of an AP-1-induced gene (41, 44). Furthermore, the TIF2 domain required to increase GR repression is within the fragment (TIF2.4) that binds STAMP. We therefore asked if STAMP increases the repressive activity of GR in the AP-1 responsive bioassay system with U2OS.rGR cells. The relative induction by PMA (25 ng/ml) is unchanged by added TIF2, STAMP, or TIF2 plus STAMP (Fig. 4C). In the presence of Dex-bound GRs, induction by AP-1 is further reduced by TIF2 as previously reported (41). Interestingly, added STAMP also augments GR repression. The effect of STAMP plus TIF2 is greater than that of either component alone (P = 0.029; Mann-Whitney test), consistent with a cooperative action of these cofactors.
The requirement of TIF2 for the expression of STAMP activity was investigated by comparing the relative repression seen with the wild-type TIF2 to the mutant TIF2m123 (Fig. 1A), which does not interact with GR (14, 20, 56). We added less STAMP here (50 versus 100 ng) to reduce the relative repression with just STAMP added. The data of Fig. 4D show that TIF2m123 either by itself or in combination with STAMP is unable to augment the repression by GR-Dex complexes of PMA-stimulated gene expression. These results suggest that STAMP requires TIF2 binding to GR for full activity and has little ability to directly affect GR-mediated repression.
The endogenous collagenase 3 (coll3) gene in U2OS.rGR cells is repressed by GR. TIF2 potentiates GR repression of coll3 and is recruited to the coll3 promoter in concert with GR-agonist complexes (44). Because STAMP augments the ability of TIF2 to increase GR repression of a transiently transfected gene (Fig. 4C), we used ChIP assays to ask if endogenous STAMP is also recruited by endogenous GR/TIF2 complexes to the endogenous coll3 gene. We also examined GR and STAMP binding to an endogenous GR-induced gene, IGFBP-1 (43). Preliminary experiments revealed that the recruitment of HA-STAMP and endogenous GR to the coll3 promoter increased from 20 to 40 min of steroid treatment, which was about when nuclear colocalization of GR and STAMP was visualized (Fig. 3B). Figure 4E shows that endogenous STAMP and GR are specifically recruited to the promoters of both the GR-repressed coll3 gene and the GR-induced IGFBP gene. ChiP/reChIP assays give a weak but reproducible signal (Fig. 4E), indicating that endogenous STAMP and GR are in a common complex bound to the coll3 promoter.
Inhibition of STAMP actions by STAMP siRNA.
We next asked if STAMP siRNAs can reduce STAMP protein levels and reverse the effects of STAMP. We first looked at the effects on exogenous STAMP because the levels of endogenous STAMP are so low (Fig. 3A). siRNA actions are not completely specific (22). Therefore, we prepared four STAMP siRNAs. The probability that each siRNA would have at least 11 nucleotides in common with any other gene, and thus silence the expression of a gene other than STAMP, is exceedingly small. Each STAMP siRNA significantly reduces the level of overexpressed STAMP protein (Fig. 5A, open arrows), with little or no effect on the levels of ectopic GR (thin arrows) or nonspecific protein (asterisks), while a nonspecific control (β-actin siRNA; right-hand panel) has negligible effects on any protein (Fig. 5A). Thus, the STAMP siRNAs are relatively specific in reducing the levels of expressed STAMP protein. Figure 5B shows that each STAMP siRNA is effective in reversing the ability of transfected STAMP to increase the relative repression by exogenous GR of an AP-1 reporter relative to the β-actin siRNA control. Finally, the ability of added STAMP siRNA 3 or 4 alone to reduce the repressive activity of GR-agonist complexes with the coll3 gene, as determined by quantitative RT-PCR in Fig. 5C, argues that endogenous STAMP assists endogenous GR in inhibiting an endogenous AP-1-induced gene.
FIG. 5.
Modulation by STAMP siRNA of GR-mediated gene expression. (A) Selective repression of STAMP protein levels by STAMP siRNAs. Cos-7 cell extracts after transient transfection with HA/STAMP (2 μg) and GR (1 μg) plasmids with or without 5 μg of one of the four STAMP siRNAs or β-actin siRNA (right-hand panel) per 60-mm dish were Western blotted with anti-HA (for STAMP) or anti-GR to reveal STAMP (open arrow), GR (thin arrows), and nonspecifically detected proteins ( ). (B and C) STAMP siRNAs inhibit STAMP activity for exogenous (B) and endogenous (C) components of gene repression. U2OS.rGR cells were either transiently transfected with HA/STAMP (100 ng) plasmid and AP-1/Luc reporter (20 ng) with or without 500 ng of STAMP or β-actin siRNAs (B) or were transfected with or without 500 ng of STAMP or lamin siRNAs (C) and treated with PMA with or without Dex. Cells were processed and the relative repression by Dex of exogenous AP-1/Luc (B) or endogenous collagenase 3 mRNA (C) was calculated and plotted as described in the legend of Fig. 4D (n = 4 to 5 [B] and 6 [C])
). (B and C) STAMP siRNAs inhibit STAMP activity for exogenous (B) and endogenous (C) components of gene repression. U2OS.rGR cells were either transiently transfected with HA/STAMP (100 ng) plasmid and AP-1/Luc reporter (20 ng) with or without 500 ng of STAMP or β-actin siRNAs (B) or were transfected with or without 500 ng of STAMP or lamin siRNAs (C) and treated with PMA with or without Dex. Cells were processed and the relative repression by Dex of exogenous AP-1/Luc (B) or endogenous collagenase 3 mRNA (C) was calculated and plotted as described in the legend of Fig. 4D (n = 4 to 5 [B] and 6 [C])  , P ≤ 0.032;
, P ≤ 0.032; 
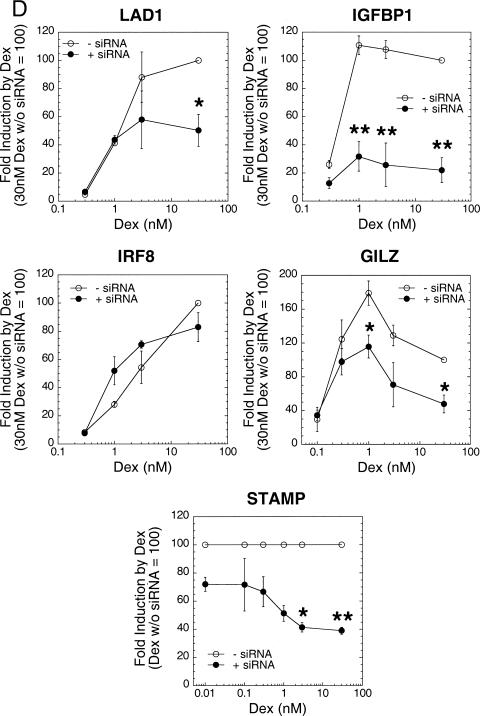
 , P = 0.0022. (D) Effect of STAMP siRNAs on induction of endogenous GR-regulated genes. The amount of mRNA of the indicated genes present in U2OS.rGR cells after transient transfection with 500 ng of STAMP (filled circles; STAMP SMARTpool from Dharmacon) or nonspecific siRNAs (open circles; nonspecific control siRNA no. 2 from Dharmacon) and treated with the indicated concentrations of Dex were determined by qRT-PCR and plotted as a percentage of the response to 30 nM Dex with nonspecific siRNA for each gene except STAMP, where the data are expressed as a percentage of STAMP mRNA with nonspecific siRNA at each concentration of Dex.
, P = 0.0022. (D) Effect of STAMP siRNAs on induction of endogenous GR-regulated genes. The amount of mRNA of the indicated genes present in U2OS.rGR cells after transient transfection with 500 ng of STAMP (filled circles; STAMP SMARTpool from Dharmacon) or nonspecific siRNAs (open circles; nonspecific control siRNA no. 2 from Dharmacon) and treated with the indicated concentrations of Dex were determined by qRT-PCR and plotted as a percentage of the response to 30 nM Dex with nonspecific siRNA for each gene except STAMP, where the data are expressed as a percentage of STAMP mRNA with nonspecific siRNA at each concentration of Dex.
Four endogenous GR-inducible genes (LAD1, IGFBP1, IRF8, and GILZ) in U2OS.rGR cells (43) were selected to examine the role of endogenous STAMP on the induction properties of GR-regulated genes. A commercially available mixture of STAMP siRNAs, which was slightly more active than our individual siRNAs used above, lowered the mRNA levels of both STAMP and the GR-induced genes (Fig. 5D and data not shown). STAMP siRNAs significantly reduced the relative induction by high concentrations of Dex for all genes but IRF8. Thus, STAMP does not affect equally all GR-inducible genes, and lowering the levels of endogenous STAMP does not have nonspecific effects on gene transcription in general. STAMP siRNAs do lower the level of endogenous STAMP protein but not GR, but the very low levels of STAMP (Fig. 3A and data not shown) preclude a quantification of the reduction. We therefore determined the levels of STAMP mRNA. Unexpectedly, the ability of STAMP siRNAs to reduce the levels of STAMP mRNA increased at higher Dex concentrations (Fig. 5D). This nonlinear response makes it impossible to determine whether the position of the dose-response curve is altered at lower endogenous STAMP concentrations. Nevertheless, the ability of reduced levels of endogenous STAMP mRNA and protein to modify the properties of both endogenous GR-induced and -repressed genes strongly argues for several physiological roles of STAMP.
Interactions of STAMP with TIF2 and GR.
The data of Fig. 4 and 5 support the hypothesis that the biological effects of STAMP are mediated by STAMP binding to TIF2 and possibly GR. Coimmunoprecipitation assays with cytosols from Cos-7 cells that were cotransfected with HA/GRIP1(TIF2), GR, and Flag/STAMP or Flag itself (Fig. 6A) show that GRIP1 (panel B) and GR (panel C) are both associated with STAMP (panel A) in a STAMP-dependent manner (lanes 2 and 3 with Flag-STAMP versus lane 4 with Flag). The agonist Dex does not alter the ability of GR to be coimmunoprecipitated by STAMP (lane 2 versus 3). Conversely, GR and Flag-STAMP are coimmunoprecipitated with HA/GRIP by anti-HA antibody (not shown). Similar experiments show coprecipitation of HA/STAMP with GR and Flag/STAMP with HA/GRIP (not shown). Finally, endogenous STAMP and GR occur as a complex in cells, as shown by their coimmunoprecipitation with anti-STAMP antibody (Fig. 6B). The control for this experiment, showing the presence of STAMP in the expected lanes, is presented above in lanes 1 to 6 of Fig. 3A.
FIG. 6.
Binding of STAMP to TIF2 and GR. (A) Whole-cell coimmunoprecipitation of STAMP, GR, and TIF2/GRIP1. Cos-7 cells that had been cotransfected with Flag/STAMP (or Flag itself), HA/GRIP1, and GR plasmids were treated for 2 h with EtOH with or without 1 μM Dex before being lysed. The pellets obtained after treating the lysate with anti-Flag antibody to immunoprecipitate Flag/STAMP were separated on SDS-PAGE gels and Western blotted with anti-Flag (a), anti-HA (b), or anti-GR antibodies (c) (lane 1, 5.7% of input lysate). Similar results were obtained in a second experiment. (B) Coimmunoprecipitation of endogenous STAMP and GR. Lysates from U2OS.rGR cells (with or without 100 nM Dex; 40 min) were separated on SDS-PAGE gels directly (lane 1, 2.5% of input in other lanes) or first immunoprecipitated with anti-STAMP antibody with or without competing antigenic peptide or with preimmune serum and then Western blotted with anti-GR antibody. A Western blot showing levels of STAMP in each lane is given in Fig. 3A. Similar results were obtained in a second experiment. (C and D) Regions of STAMP that associate with TIF2.4 and GR in two-hybrid assays. CV-1 cells were transiently transfected with VP16 or VP16/TIF2.4 (C) or with VP16/GR with or without 1 μM Dex (D) plus GAL fusions of STAMP segments and FRLuc reporter. The average relative luciferase activities (n = 2 to 5) are plotted as for Fig. 1D and E. The values in parentheses in panel D indicate the relative increased interaction with or without 1 μM Dex. (E and F) Domains of STAMP that bind to TIF2.4 and GR in cell-free pull-down assays. [35S]methionine-labeled, in vitro translated STAMP fragments (E) or wild-type GR (F) that were retained by GST-chimeras of TIF2.4 (E) and STAMP segments (F) immobilized on glutathione-Sepharose beads were separated on SDS-PAGE gels and visualized by autoradiography. Specific binding is seen as the binding to the GST-chimera sample that is in excess of that to the GST control. “Input” was 10% of the [35S]methionine label that was initially loaded onto the matrix beads. Similar results were obtained in a second experiment. (G) Synergistic interactions of GR, STAMP, and TIF2 in three-hybrid assays. CV-1 cells were transiently transfected with either GAL/STAMP(623-834) or -/STAMP(623C) plus TIF2.4 (or TIF2.4m123) and VP16 or VP16/GR with or without 1 μM Dex in addition to FRLuc reporter. The average relative luciferase activities (n = 2 to 4) are plotted as for Fig. 1D and E. (H and I) STAMP domains involved in the modulation of GR-mediated gene induction. CV-1 cells were transiently transfected with GR (6 ng) with or without TIF2 (20 ng) and with or without STAMP (160 ng) plasmids, GREtkLUC reporter, and Renilla control plasmid and induced with EtOH with or without the indicated concentrations of Dex or 1 μM Dex-Mes. Luciferase activities, normalized to the internal Renilla control values, were then expressed as a percentage of the maximal response with 1 μM Dex as described in Materials and Methods and plotted as described in the legend of Fig. 4 for STAMP(623C) (H) and STAMP(N834) (I). Similar results were obtained in one or two additional experiments.
Two-hybrid assays were preformed to identify the regions of STAMP that bind to TIF2 and GR. Figure 6C shows that the majority of STAMP interactions with TIF2.4 are mediated by amino acids 623 to 834 of STAMP. We call this region the coactivator interaction domain, or CID. Western blots (not shown) and activity in the assay of Fig. 6D indicate that the weak response of the amino and carboxyl fragments (N623 and 834C) is not due to poor protein expression. It should be noted that neither any segment of STAMP (Fig. 6C) nor full-length STAMP (not shown) displays any intrinsic transactivation activity, as indicated by the inability of various constructs to induce the FRLuc reporter in the absence of TIF2.4.
A region of STAMP C-terminal of the CID interacts with GR in a steroid-responsive manner (Fig. 6D). Again, Western blots (not shown) and the activity in Fig. 6C argue that the relative inactivity of GAL/N834 and GAL/623-834 is not due to low protein expression. The numbers in parentheses above the bars for VP16/GR (Dex) represent the relative induction with and without Dex. The C-terminal half of STAMP (aa 834 to 1277, or 834C) interacts most strongly with agonist-bound GRs. There is substantial ligand-independent association of GR with STAMP(834-956) while the largest agonist-induced changes are restricted to the two STAMP domains of amino acids 956 to 1127 and 1127 to 1277 (1127C) (Fig. 6D). Neither half of GR (N-terminal [N523] or C-terminal [407C] half) interacted with STAMP(834C) in this assay under conditions that give a strong response with GR(407C) and TIF2.4 (not shown). Thus, a single domain appears to be sufficient for STAMP-TIF2 (and TIF2-GR) binding while both N- and C-terminal domains of GR are required for its interaction with STAMP.
The binding of [35S]methionine-labeled, in vitro translated STAMP constructs to GST/TIF2.4 in pull-down assays supports the conclusions of the above two-hybrid assay. There is only a weak interaction with the full-length STAMP, but the fragment encompassing amino acids 623 to 834 binds strongly to TIF2.4 (Fig. 6E). This binding is selective as neither the amino terminal (N623) nor the C-terminal (834C) fragment exhibits significantly more binding to GST/TIF2.4 than to GST (Fig. 6E). Similarly, the two-hybrid assay interactions of GR with STAMP are supported by pull-down assays that show [35S]methionine-labeled full-length GRs binding to GST/STAMP(956C) (Fig. 6F). This binding is steroid independent, as observed in coimmunoprecipitation assays (Fig. 6A).
The data of Fig. 6C to E indicate that TIF2 and GR bind to separate but adjacent domains of STAMP. Thus, a complex in which each molecule is attached to the other two is possible. Such a ternary complex is consistent with the coimmunoprecipitation results of Fig. 6A and B and the additive effects of TIF2 and STAMP in GR-mediated induction and repression (Fig. 4A to C). This hypothesis was tested directly by asking if TIF2.4 would inhibit or augment the association of GR with STAMP in a three-hybrid assay. As seen in Fig. 6G, TIF2.4 (but not the mutant TIF2.4m123) synergistically increases the interaction of GR in the presence of Dex with the C-terminal half of STAMP [STAMP(623C)], which contains both the TIF2 and GR binding domains. This result is incompatible with the idea that TIF2.4 competes with GR for binding to STAMP and strongly supports the existence of a stabilized ternary STAMP/GR/TIF2 complex. The loss of this synergistic interaction in reactions with the smaller STAMP construct, GAL/STAMP(623-834) (Fig. 6G), is expected because only TIF2, and not GR, binds efficiently to this central domain of STAMP (Fig. 6C to E).
The N-terminal domain of STAMP contains the only previously characterized, functional domain, which is a tubulin-tyrosine ligase (TTL) domain at amino acids 113 to 391 (Fig. 6E). STAMP lacking the TTL domain [STAMP(623C)] retains 85% of the capacity of full-length STAMP to increase the relative induction with Dex and 40% of the ability to increase the partial agonist activity of Dex-Mes and decrease the EC50 for Dex induction (Fig. 6H). In contrast, STAMP with just the TTL and CID domains [STAMP(N834)] has negligible activity and shows signs of being a dominant negative construct (Fig. 6I). We conclude that the TTL domain is not essential for any of the identified activities of STAMP.
STAMP interactions with other steroid/nuclear receptors.
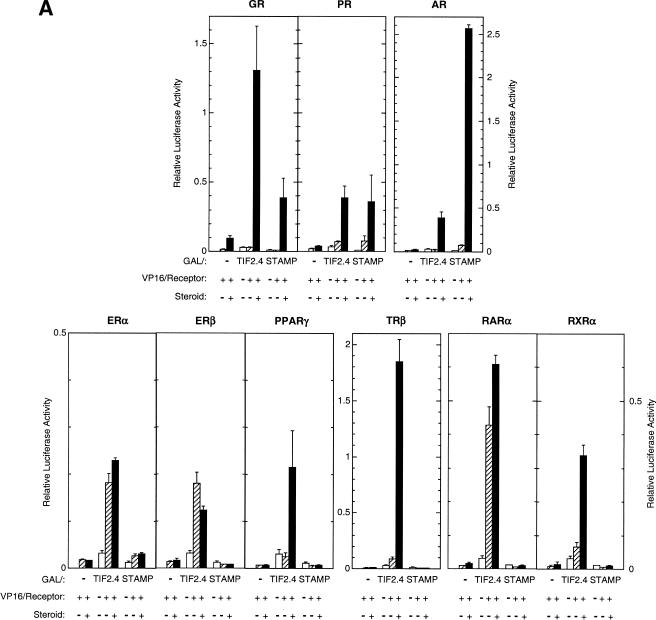
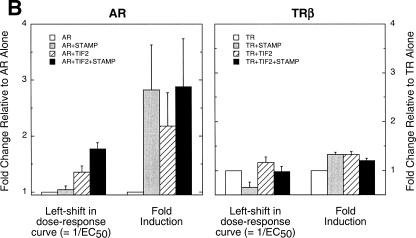
STAMP(834C) also interacts with agonist-bound, full-length PRs (B-form) and androgen receptors (ARs) but not estrogen α or β, peroxisome proliferator-activated receptor γ2, thyroid hormone receptor β(TRβ), retinoid X receptor α, or retinoid α receptors in two-hybrid assays (Fig. 7A). The lack of interaction with all but GR, PR, and AR is not due to low expression or function of the other receptors as seen by their robust activity with GAL/TIF2.4 (Fig. 7A). The ability of STAMP to supplement the whole-cell actions of TIF2 with full-length AR and TRβ was therefore examined. STAMP augments the left-shift of the dose-response curve of ARs in the presence of TIF2 but not when added by itself (Fig. 7B). STAMP increases the relative induction by AR, but exogenous TIF2 produces no additional effect. In contrast to AR, added STAMP has little ability with TRβ either to cause a left shift in the dose-response curve or to increase the relative induction, regardless of whether TIF2 is present (Fig. 7B). Thus, the effects of STAMP on AR and GR transactivation are not due to nonspecific effects on general transcription. Collectively, these data suggest that STAMP association and activity with receptors are more discriminatory than those of the coactivator TIF2 and may be selective for the subclass of receptors that bind to a GRE (3).
FIG. 7.
Interaction of STAMP with or without TIF2 with steroid/nuclear receptors. (A) Association of STAMP with assorted steroid or nuclear receptors. CV-1 cells were transfected as described in the legend of Fig. 6D with GAL, or GAL fused to TIF2.4 or STAMP(834C), plus VP16 fused to the indicated full-length receptors and incubated with the appropriate steroid (1 μM Dex, 20 nM R5020, 1 nM R1881, 0.1 μM triiodothyronine, or 1 μM estradiol, roziglitazone, or 9-cis-retinoic acid). The average relative luciferase activities are plotted (n = 2 to 3). (B) Specificity of STAMP biological activity. CV-1 cells were transiently transfected with AR or TRβ with or without TIF2 and with or without STAMP plasmids plus GREtkLUC or TREtkLUC reporter and induced for 20 h with EtOH with or without 10 nM R1881 or 5 μM T3 (thyroid hormone). The average (n = 3 to 4) relative changes in EC50 and induction levels were plotted as described in the legend of Fig. 4B.
DISCUSSION
We previously reported that the ability of the coactivator TIF2 to modulate the position of the dose-response curve of GR-agonist complexes and the partial agonist activity of GR-antagonist complexes is mediated by a protein that interacts with amino acids 624 to 1010 of TIF2 (Fig. 1A) (20). Here, we isolate a novel protein with comodulatory activity that binds to this region (TIF2.4) in both mammalian two-hybrid and pull-down assays. The full-length protein has been cloned; it resides on human chromosome 14q24.3 and is predicted to contain 1,277 amino acids with a calculated molecular mass of 143 kDa. The predicted ∼4.6-kb mRNA is detected in all human tissues examined (Fig. 2C) while low levels of STAMP protein are found in U2OS.rGR cells (Fig. 3A). This new protein also interacts with the same region of the coactivator SRC-1(Fig. 1F and G) that modulates GR transactivation properties (20). For this reason, we call this protein STAMP.
Unexpectedly, STAMP also binds to GRs in both mammalian two-hybrid and pull-down assays (Fig. 6D and F). The whole-cell colocalization of STAMP and GR is more prevalent with Dex and is predominantly nuclear (Fig. 3C) at times when STAMP and GR coassociate with the promoter region of an endogenous regulated gene (Fig. 4E and data not shown). Coimmunoprecipitation assays suggest the formation of ternary cellular complexes of TIF2 and GR with endogenous and exogenous STAMP (Fig. 6A and B). This conclusion is supported by the additive effects of STAMP and TIF2 in GR-regulated gene expression (Fig. 4A to D) and further confirmed by the observation that TIF2.4 synergistically increases the interactions of STAMP and GR in a three-hybrid assay (Fig. 6G). Such a complex appears topologically feasible because the binding sites that have been identified on STAMP for TIF2 and GR (Fig. 6C to G) and on TIF2 for GR and STAMP (20) do not overlap.
The biological and physiological relevance of STAMP is indicated by its presence as an endogenous protein (Fig. 3) and the fact that STAMP enhances TIF2 actions both in GR-mediated induction and repression (Fig. 4A to C) and in AR-regulated induction (Fig. 7B). Furthermore, the reduction by STAMP siRNAs of exogenous and endogenous STAMP reduces the activity of GRs in both gene repression (Fig. 5B and C) and gene induction (Fig. 5D). We propose that STAMP acts at the molecular level by binding to both TIF2 and GR to augment the modulatory activity of TIF2 in altering the EC50 and partial agonist activity in gene induction by GRs in a manner that requires both the central CID and the C-terminal RID of STAMP (Fig. 6G to I). STAMP appears to be inactive with those receptors that do not bind STAMP (Fig. 7A and B). Collectively, these data suggest that both the TIF2 and receptor binding domains of STAMP (i.e., CID and RID) are necessary, while neither is sufficient, for STAMP activity in our assays. The importance of the one previously characterized domain in STAMP, a TTL domain, is currently obscure because it is not required for STAMP modulatory activities (Fig. 6H).
Our attempts to demonstrate the effects of STAMP on the dose-response curves of endogenous genes have been frustrated by the generation of non-Michaelis-Menten curves (26) with four different genes. This behavior has been seen both when reducing endogenous STAMP with siRNAs (Fig. 5D) and when increasing either STAMP in U2OS.rGR cells or STAMP plus GR in U2OS cells (data not shown). The different shapes of the dose-response curves with different endogenous genes (Fig. 5D) suggest that promoter architecture may be an important determinant here. Thus, while STAMP clearly modifies the total expression of endogenous, GR-regulated genes, much remains to determine STAMP's effect on the dose-response curve of any endogenous gene.
These mechanistic proposals are supported by the observations that endogenous STAMP is localized to the promoters of endogenous GR-induced and -repressed genes (Fig. 4E), binds to TIF2 (Fig. 1D, G, and E and 6A, C, and E), associates with GR in a complex both in solution (Fig. 6A, B, D, and F) and on the promoter of a endogenous GR-regulated gene (Fig. 4E), is required for the full responses of both induced and repressed endogenous GR-regulated genes (Fig. 5B to D), and is inactive in the presence of a mutant TIF2 that does not bind to GR (Fig. 4D). However, the insensitivity of IRF8 induction by GR to lower STAMP levels (Fig. 5D) reveals that not all GR-induced endogenous genes require STAMP, thus indicating an additional level of mechanistic specificity. Interestingly, a similar differential requirement of the mediator subunits Med1 and Med14 has been observed for Dex induction of these same genes (9). A GenBank search failed to uncover any proteins related to STAMP. Thus, STAMP appears to be a unique protein that is a physiologically relevant new component of GR transcriptional actions in intact cells. Mice lacking STAMP will be useful in confirming these hypotheses.
Despite the fact that GR induces and suppresses gene expression by different mechanisms, STAMP increases both the inductive (Fig. 4B) and the repressive activity of GR with and without TIF2 (Fig. 4C), as expected from the colocalization of GR and STAMP in ChIP assays (Fig. 4E). The original definition of a coactivator was a factor that augments the activity of an agonist steroid (38). By this definition, the actions of STAMP are mechanistically consistent because STAMP potentiates the actions of Dex, a glucocorticoid agonist, both in GR-mediated gene induction and gene repression. The response to exogenous STAMP in the absence of added TIF2 (Fig. 4C) presumably reflects limiting endogenous STAMP concentrations cooperating with endogenous TIF2. Importantly, added STAMP augments TIF2 activities by further shifting the dose-response curve of a GR-inducible reporter gene to lower concentrations of agonist steroid and by increasing the partial agonist activity of antiglucocorticoids (Fig. 4). Preliminary results indicate a similar shift in the dose-response curve for GR-regulated gene repression (Y. Sun and S. S. Simons, unpublished). Therefore, we propose that STAMP affects multiple aspects of GR-regulated gene expression in whole cells for both GR-mediated induction and repression in a manner that requires the presence of STAMP/GR/TIF2 complexes.
STAMP is required for both the biological activity (Fig. 4 and 5) and whole-cell interactions (Fig. 6D and 7A) of steroid-bound but not steroid-free GRs. In contrast, cell-free interactions of STAMP and GR are steroid independent (Fig. 6A, B, and F). This binding of steroid-free GRs appears to be due to a lower affinity of steroid-free versus steroid-bound GRs for STAMP, as seen by the less intense colocalization of STAMP with ligand-free GRs in the whole-cell immunofluorescence studies of Fig. 3B, and would not be observed in two-hybrid assays (Fig. 6D and 7A), which cannot detect low-affinity (≥100 nM) interactions (13).
STAMP contains several GR interaction domains between amino acids 834 and 1277 (Fig. 6). While these domains do not contain any LXXLL motifs, which are critical for coactivator binding to steroid/nuclear receptors (5, 11, 33), numerous ΦXXΦΦ motifs (where Φ is a hydrophobic residue) are present. The fact that sequences of TIF2 and SRC-1 with functional modulatory activity (20) are located in regions of each protein with very different amino acid sequences and only 38% homology suggests that features other than primary sequence are also important. Thus, the interactions of STAMP with TIF2, SRC-1, and GR may involve protein-protein-induced conformational changes that will be most easily observed in X-ray and/or nuclear magnetic resonance studies.
Our data indicate that STAMP is intimately involved in the modulation of several parameters of GR-mediated induction and repression in both CV-1 and U2OS.rGR cells (Fig. 4 and 5). AR properties in CV-1 cells are also affected (Fig. 7B). Therefore, the actions of STAMP are not limited to one receptor or one cell line. The theoretical ability to modify the endogenous levels of STAMP in cells and tissues thus offers interesting new possibilities for modifying the amount of response to endogenous levels of glucocorticoid hormones and to pharmacological doses of antiglucocorticoids.
The chromosomal localization of STAMP at 14q24.3 places it near two other modulators of steroid receptor action (SKIP and SLIRP) and the orphan receptor ERRβ. Both SKIP and SLIRP also modify the transactivation properties of glucocorticoid receptors and other steroid receptors (19, 63), thus raising the interesting possibility that this locus of chromosome 14 may coordinately express several factors involved in steroid receptor transcription. It is of further note that SKIP binds to the coactivator SRC-1 and can act as either a coactivator or a corepressor, depending upon the cell line, in a manner that appears to depend upon the ratio of corepressor NCoR and comodulator p300 (30). This is similar to the ability of the ratio of coactivators to corepressors to determine the transactivation properties of glucocorticoid receptors (reviewed in references 46 and 47).
The specificity of STAMP interactions with steroid or nuclear receptors in mammalian two-hybrid assays is of note. STAMP associates with, and significantly modifies the induction parameters of, only a subset of the receptors that bind to the coactivator TIF2 (Fig. 7). Whether this specificity is related to the singular ability of GR, PR, and AR to bind to the DNA sequences of functional GREs remains to be determined. However, STAMP appears to be unique in selectively associating with and modifying the induction properties of GR, PR, and AR but not estrogen receptors (α or β) or nuclear receptors. The physiological consequences of these discriminatory cell-free and whole-cell properties await exploration.
Acknowledgments
We thank Kai Ge, Donald McDonnell, Takahiro Nagase, Inez Rogatsky, Liang-Nian Song, and Keith Yamamoto for generously donating reagents; Ami Aronheim (TIIT, Israel) for help on the yeast Sos-recruitment system; Tao Tao (NCBI, NIH) for assistance with the bioinformatic analysis; Dongqing Wang (NIDDK, NIH) for technical assistance; and Inez Rogatsky, Gordon Hager (NCI, NIH), and members of the Steroid Hormones Section for helpful comments about the manuscript.
This research was supported by the Intramural Research Program of the NIH, NIDDK.
Footnotes
Published ahead of print on 20 November 2006.
REFERENCES
- 1.Anafi, M., Y. F. Yang, N. A. Barlev, M. V. Govindan, S. L. Berger, T. R. Butt, and P. G. Walfish. 2000. GCN5 and ADA adaptor proteins regulate triiodothyronine/GRIP1 and SRC-1 coactivator-dependent gene activation by the human thyroid hormone receptor. Mol. Endocrinol. 14:718-732. [DOI] [PubMed] [Google Scholar]
- 2.Aronheim, A. 1997. Improved efficiency Sos recruitment system: expression of the mammalian GAP reduces isolation of Ras GTPase false positives. Nucleic Acids Res. 25:3373-3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beato, M., G. Chalepakis, M. Schauer, and E. P. Slater. 1989. DNA regulatory elements for steroid hormones. J. Steroid Biochem. 32:737-748. [DOI] [PubMed] [Google Scholar]
- 4.Brown, K., Y. Chen, T. M. Underhill, J. S. Mymryk, and J. Torchia. 2003. The coactivator p/CIP/SRC-3 facilitates retinoic acid receptor signaling via recruitment of GCN5. J. Biol. Chem. 278:39402-39412. [DOI] [PubMed] [Google Scholar]
- 5.Chang, C.-Y., J. D. Norris, H. Gron, L. A. Paige, P. T. Hamilton, D. J. Kenan, D. Fowlkes, and D. P. McDonnell. 1999. Dissection of the LXXLL nuclear receptor-coactivator interaction motif using combinatorial peptide libraries: discovery of peptide antagonists of estrogen receptors α andβ. Mol. Cell. Biol. 19:8226-8239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen, D., S. M. Huang, and M. R. Stallcup. 2000. Synergistic, p160 coactivator-dependent enhancement of estrogen receptor function by CARM1 and p300. J. Biol. Chem. 275:40810-40816. [DOI] [PubMed] [Google Scholar]
- 7.Chen, H., R. J. Lin, R. L. Schiltz, D. Chakravarti, A. Nash, L. Nagy, M. L. Privalsky, Y. Nakatani, and R. M. Evans. 1997. Nuclear receptor coactivator ACTR is a novel histone acetyltransferase and forms a multimeric activation complex with P/CAF and CBP/p300. Cell 90:569-580. [DOI] [PubMed] [Google Scholar]
- 8.Chen, S., N. J. Sarlis, and S. S. Simons, Jr. 2000. Evidence for a common step in three different processes for modulating the kinetic properties of glucocorticoid receptor-induced gene transcription. J. Biol. Chem. 275:30106-30117. [DOI] [PubMed] [Google Scholar]
- 9.Chen, W., I. Rogatsky, and M. J. Garabedian. 2006. MED14 and MED1 differentially regulate target-specific gene activation by the glucocorticoid receptor. Mol. Endocrinol. 20:560-572. [DOI] [PubMed] [Google Scholar]
- 10.Cho, S., B. L. Kagan, J. A. Blackford, Jr., D. Szapary, and S. S. Simons, Jr. 2005. Glucocorticoid receptor ligand binding domain is sufficient for the modulation of both the dose-response curve of receptor-agonist complexes and the partial agonist activity of receptor-antisteroid complexes by glucocorticoid receptors, coactivator TIF2, and Ubc9. Mol. Endocrinol. 19:290-311. [DOI] [PubMed] [Google Scholar]
- 11.Darimont, B. D., R. L. Wagner, J. W. Apriletti, M. R. Stallcup, P. J. Kushner, J. D. Baxter, R. J. Fletterick, and K. R. Yamamoto. 1998. Structure and specificity of nuclear receptor-coactivator interactions. Genes Dev. 12:3343-3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Bosscher, K., B. W. Vanden, and G. Haegeman. 2003. The interplay between the glucocorticoid receptor and nuclear factor-κB or activator protein-1: molecular mechanisms for gene repression. Endocrinol. Rev. 24:488-522. [DOI] [PubMed] [Google Scholar]
- 13.de Felipe, K. S., B. T. Carter, E. A. Althoff, and V. W. Cornish. 2004. Correlation between ligand-receptor affinity and the transcription readout in a yeast three-hybrid system. Biochemistry 43:10353-10363. [DOI] [PubMed] [Google Scholar]
- 14.Ding, X. F., C. M. Anderson, H. Ma, H. Hong, R. M. Uht, P. J. Kushner, and M. R. Stallcup. 1998. Nuclear receptor-binding sites of coactivators glucocorticoid receptor interacting protein 1 (GRIP1) and steroid receptor coactivator 1 (SRC-1): Multiple motifs with different binding specificities. Mol. Endocrinol. 12:302-313. [DOI] [PubMed] [Google Scholar]
- 15.Dubrulle, J., and O. Pourquie. 2004. fgf8 mRNA decay establishes a gradient that couples axial elongation to patterning in the vertebrate embryo. Nature 427:419-422. [DOI] [PubMed] [Google Scholar]
- 16.Giannoukos, G., D. Szapary, C. L. Smith, J. E. W. Meeker, and S. S. Simons, Jr. 2001. New antiprogestins with partial agonist activity: potential selective progesterone receptor modulators (SPRMs) and probes for receptor- and coregulator-induced changes in progesterone receptor induction properties. Mol. Endocrinol. 15:255-270. [DOI] [PubMed] [Google Scholar]
- 17.Goswami, R., R. Lacson, E. Yang, R. Sam, and T. Unterman. 1994. Functional analysis of glucocorticoid and insulin response sequences in the rat insulin-like growth factor-binding protein-1 promoter. Endocrinology 134:736-743. [DOI] [PubMed] [Google Scholar]
- 18.Gurdon, J. B., and P. Y. Bourillot. 2001. Morphogen gradient interpretation. Nature 413:797-803. [DOI] [PubMed] [Google Scholar]
- 19.Hatchell, E. C., S. M. Colley, D. J. Beveridge, M. R. Epis, L. M. Stuart, K. M. Giles, A. D. Redfern, L. E. Miles, A. Barker, L. M. MacDonald, P. G. Arthur, J. C. Lui, J. L. Golding, R. K. McCulloch, C. B. Metcalf, J. A. Wilce, M. C. Wilce, R. B. Lanz, B. W. O'Malley, and P. J. Leedman. 2006. SLIRP, a small SRA binding protein, is a nuclear receptor corepressor. Mol. Cell 22:657-668. [DOI] [PubMed] [Google Scholar]
- 20.He, Y., D. Szapary, and S. S. Simons, Jr. 2002. Modulation of induction properties of glucocorticoid receptor-agonist and -antagonist complexes by coactivators involves binding to receptors but is independent of ability of coactivators to augment transactivation. J. Biol. Chem. 277:49256-49266. [DOI] [PubMed] [Google Scholar]
- 21.Hong, H., K. Kohli, A. Trivedi, D. L. Johnson, and M. R. Stallcup. 1996. GRIP1, a novel mouse protein that serves as a transcriptional coactivator in yeast for the hormone binding domains of steroid receptors. Proc. Natl. Acad. Sci. USA 93:4948-4952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jackson, A. L., S. R. Bartz, J. Schelter, S. V. Kobayashi, J. Burchard, M. Mao, B. Li, G. Cavet, and P. S. Linsley. 2003. Expression profiling reveals off-target gene regulation by RNAi. Nat. Biotechnol. 21:635-637. [DOI] [PubMed] [Google Scholar]
- 23.Kalkhoven, E., J. E. Valentine, D. M. Heery, and M. G. Parker. 1998. Isoforms of steroid receptor co-activator 1 differ in their ability to potentiate transcription by the oestrogen receptor. EMBO J. 17:232-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karin, M., and L. Chang. 2001. AP-1-glucocorticoid receptor crosstalk taken to a higher level. J. Endocrinol. 169:447-451. [DOI] [PubMed] [Google Scholar]
- 25.Kaul, S., J. A. Blackford, Jr., S. Cho, and S. S. Simons, Jr. 2002. Ubc9 is a novel modulator of the induction properties of glucocorticoid receptors. J. Biol. Chem. 277:12541-12549. [DOI] [PubMed] [Google Scholar]
- 26.Kim, Y., Y. Sun, C. Chow, Y. G. Pommier, and S. S. Simons, Jr. 2006. Effects of acetylation, polymerase phosphorylation, and DNA unwinding in glucocorticoid receptor transactivation. J. Steroid Biochem. Mol. Biol. 100:3-17. [DOI] [PubMed] [Google Scholar]
- 27.Kino, T., and G. P. Chrousos. 2003. Tumor necrosis factor alpha receptor- and Fas-associated FLASH inhibit transcriptional activity of the glucocorticoid receptor by binding to and interfering with its interaction with p160 type nuclear receptor coactivators. J. Biol. Chem. 278:3023-3029. [DOI] [PubMed] [Google Scholar]
- 28.Kino, T., O. Slobodskaya, G. N. Pavlakis, and G. P. Chrousos. 2002. Nuclear receptor coactivator p160 proteins enhance the HIV-1 long terminal repeat promoter by bridging promoter-bound factors and the Tat-P-TEFb complex. J. Biol. Chem. 277:2396-2405. [DOI] [PubMed] [Google Scholar]
- 29.Kozak, M. 1989. The scanning model of translation: an update. J. Cell Biol. 108:229-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leong, G. M., N. Subramaniam, L. L. Issa, J. B. Barry, T. Kino, P. H. Driggers, M. J. Hayman, J. A. Eisman, and E. M. Gardiner. 2004. Ski-interacting protein, a bifunctional nuclear receptor coregulator that interacts with N-CoR/SMRT and p300. Biochem. Biophys. Res. Commun. 315:1070-1076. [DOI] [PubMed] [Google Scholar]
- 31.Ma, H., H. Hong, S.-M. Huang, R. A. Irvine, P. Webb, P. J. Kushner, G. A. Coetzee, and M. R. Stallcup. 1999. Multiple signal input and output domains of the 160-kilodalton nuclear receptor coactivator proteins. Mol. Cell. Biol. 19:6164-6173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ma, H., Y. Shang, D. Y. Lee, and M. R. Stallcup. 2003. Study of nuclear receptor-induced transcription complex assembly and histone modification by chromatin immunoprecipitation assays. Methods Enzymol. 364:284-296. [DOI] [PubMed] [Google Scholar]
- 33.McInerney, E. M., D. W. Rose, S. E. Flynn, S. Westin, T.-M. Mullen, A. Krones, J. Inostroza, J. Torchia, R. T. Nolte, N. Assa-Munt, M. V. Milburn, C. K. Glass, and M. G. Rosenfeld. 1998. Determinants of coactivator LXXLL motif specificity in nuclear receptor transcriptional activation. Genes Dev. 12:3357-3368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McKenna, N. J., R. B. Lanz, and B. W. O'Malley. 1999. Nuclear receptor coregulators: cellular and molecular biology. Endocrine Rev. 20:321-344. [DOI] [PubMed] [Google Scholar]
- 35.McKenna, N. J., and B. W. O'Malley. 2002. Combinatorial control of gene expression by nuclear receptors and coregulators. Cell 108:465-474. [DOI] [PubMed] [Google Scholar]
- 36.Nagase, T., K. Ishikawa, M. Suyama, R. Kikuno, M. Hirosawa, N. Miyajima, A. Tanaka, H. Kotani, N. Nomura, and O. Ohara. 1999. Prediction of the coding sequences of unidentified human genes. XIII. The complete sequences of 100 new cDNA clones from brain which code for large proteins in vitro. DNA Res. 6:63-70. [DOI] [PubMed] [Google Scholar]
- 37.Onate, S. A., V. Boonyaratanakornkit, T. E. Spencer, S. Y. Tsai, M.-J. Tsai, D. P. Edwards, and B. W. O'Malley. 1998. The steroid receptor coactivator-1 contains multiple receptor interacting and activation domains that cooperatively enhance the activation function 1 (AF1) and AF2 domains of steroid receptors. J. Biol. Chem. 273:12101-12108. [DOI] [PubMed] [Google Scholar]
- 38.Onate, S. A., S. Y. Tsai, M.-J. Tsai, and B. W. O'Malley. 1995. Sequence and characterization of a coactivator for the steroid hormone receptor superfamily. Science 270:1354-1357. [DOI] [PubMed] [Google Scholar]
- 39.Oshima, H., and S. S. Simons, Jr. 1993. Sequence-selective interactions of transcription factor elements with tandem glucocorticoid-responsive elements at physiological steroid concentrations. J. Biol. Chem. 268:26858-26865. [PubMed] [Google Scholar]
- 40.Robyr, D., A. P. Wolffe, and W. Wahli. 2000. Nuclear hormone receptor coregulators in action: diversity for shared tasks. Mol. Endocrinol. 14:329-347. [DOI] [PubMed] [Google Scholar]
- 41.Rogatsky, I., H. F. Luecke, D. C. Leitman, and K. R. Yamamoto. 2002. Alternate surfaces of transcriptional coregulator GRIP1 function in different glucocorticoid receptor activation and repression contexts. Proc. Natl. Acad. Sci. USA 99:16701-16706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rogatsky, I., J. M. Trowbridge, and M. J. Garabedian. 1997. Glucocorticoid receptor-mediated cell cycle arrest is achieved through distinct cell-specific transcriptional regulatory mechanisms. Mol. Cell. Biol. 17:3181-3193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rogatsky, I., J. C. Wang, M. K. Derynck, D. F. Nonaka, D. B. Khodabakhsh, C. M. Haqq, B. D. Darimont, M. J. Garabedian, and K. R. Yamamoto. 2003. Target-specific utilization of transcriptional regulatory surfaces by the glucocorticoid receptor. Proc. Natl. Acad. Sci. USA 100:13845-13850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rogatsky, I., K. A. Zarember, and K. R. Yamamoto. 2001. Factor recruitment and TIF2/GRIP1 corepressor activity at a collagenase-3 response element that mediates regulation by phorbol esters and hormones. EMBO. J. 20:6071-6083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schaaf, M. J., and J. A. Cidlowski. 2002. Molecular mechanisms of glucocorticoid action and resistance. J Steroid Biochem. Mol. Biol. 83:37-48. [DOI] [PubMed] [Google Scholar]
- 46.Simons, Jr., S. S. 2003. The importance of being varied in steroid receptor transactivation. TIPS 24:253-259. [DOI] [PubMed] [Google Scholar]
- 47.Simons, Jr., S. S. 2006. How much is enough? Modulation of dose-response curve for steroid receptor-regulated gene expression by changing concentrations of transcription factor. Curr. Topics Med. Chem. 6:271-285. [DOI] [PubMed] [Google Scholar]
- 48.Simons, Jr., S. S., M. Pons, and D. F. Johnson. 1980. α-Keto mesylate: a reactive thiol-specific functional group. J. Org. Chem. 45:3084-3088. [Google Scholar]
- 49.Singh, P., S. W. Chan, and W. Hong. 2001. Retinoblastoma protein is functionally distinct from its homologues in affecting glucocorticoid receptor-mediated transcription and apoptosis. J. Biol. Chem. 276:13762-13770. [DOI] [PubMed] [Google Scholar]
- 50.Song, L.-N., B. Huse, S. Rusconi, and S. S. Simons, Jr. 2001. Transactivation specificity of glucocorticoid vs. progesterone receptors: role of functionally different interactions of transcription factors with amino- and carboxyl-terminal receptor domains. J. Biol. Chem. 276:24806-24816. [DOI] [PubMed] [Google Scholar]
- 51.Suwanichkul, A., M. L. Cubbage, and D. R. Powell. 1990. The promoter of the human gene for insulin-like growth factor binding protein-1. Basal promoter activity in HEP G2 cells depends upon liver factor B1. J. Biol. Chem. 265:21185-21193. [PubMed] [Google Scholar]
- 52.Szapary, D., Y. Huang, and S. S. Simons, Jr. 1999. Opposing effects of corepressor and coactivators in determining the dose-response curve of agonists, and residual agonist activity of antagonists, for glucocorticoid receptor regulated gene expression. Mol. Endocrinol. 13:2108-2121. [DOI] [PubMed] [Google Scholar]
- 53.Szapary, D., M. Xu, and S. S. Simons, Jr. 1996. Induction properties of a transiently transfected glucocorticoid-responsive gene vary with glucocorticoid receptor concentration. J. Biol. Chem. 271:30576-30582. [DOI] [PubMed] [Google Scholar]
- 54.Truss, M., and M. Beato. 1993. Steroid hormone receptors: interaction with deoxyribonucleic acid and transcription factors. Endocrine Rev. 14:459-479. [DOI] [PubMed] [Google Scholar]
- 55.Tsai, M.-J., and B. W. O'Malley. 1994. Molecular mechanisms of action of steroid/thyroid receptor superfamily members. Annu. Rev. Biochem. 63:451-486. [DOI] [PubMed] [Google Scholar]
- 56.Voegel, J. J., M. J. S. Heine, M. Tini, V. Vivat, P. Chambon, and H. Gronemeyer. 1998. The coactivator TIF2 contains three nuclear receptor-binding motifs and mediates transactivation through CBP binding-dependent and -independent pathways. EMBO J. 17:507-519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Voegel, J. J., M. J. S. Heine, C. Zechel, P. Chambon, and H. Gronemeyer. 1996. TIF2, a 160-kDa transcriptional mediator for the ligand-dependent activation function AF-2 of nuclear receptors. EMBO J. 15:3667-3675. [PMC free article] [PubMed] [Google Scholar]
- 58.Wang, Q., J. A. Blackford, Jr., L.-N. Song, Y. Huang, and S. S. Simons, Jr. 2004. Equilibrium interactions of corepressors and coactivators modulate the properties of agonist and antagonist complexes of glucocorticoid receptors. Mol. Endocrinol. 18:1376-1395. [DOI] [PubMed] [Google Scholar]
- 59.Wang, Q., W. F. Richter, S. L. Anzick, P. S. Meltzer, and S. S. Simons, Jr. 2004. Modulation of transcriptional sensitivity of mineralocorticoid and estrogen receptors. J. Steroid Biochem. Mol. Biol. 91:197-210. [DOI] [PubMed] [Google Scholar]
- 60.Xu, J., and Q. Li. 2003. Review of the in vivo functions of the p160 steroid receptor coactivator family. Mol. Endocrinol. 17:1681-1692. [DOI] [PubMed] [Google Scholar]
- 61.Xu, L., C. K. Glass, and M. G. Rosenfeld. 1999. Coactivator and corepressor complexes in nuclear receptor function. Curr. Opin. Cell Biol. 9:140-147. [DOI] [PubMed] [Google Scholar]
- 62.Zeng, H., S. Y. Plisov, and S. S. Simons, Jr. 2000. Ability of the glucocorticoid modulatory element (GME) to modify glucocorticoid receptor transactivation indicates parallel pathways for the expression of GME and glucocorticoid response element activities. Mol. Cell Endocrinol. 162:221-234. [DOI] [PubMed] [Google Scholar]
- 63.Zhang, C., D. R. Dowd, A. Staal, C. Gu, J. B. Lian, W. A. J. van, G. S. Stein, and P. N. MacDonald. 2003. Nuclear coactivator-62 kDa/Ski-interacting protein is a nuclear matrix-associated coactivator that may couple vitamin D receptor-mediated transcription and RNA splicing. J. Biol. Chem. 278:35325-35336. [DOI] [PubMed] [Google Scholar]