Abstract
The stability and activity of tumor suppressor p53 are tightly regulated and partially depend on the p53 proline-rich domain (PRD). We recently analyzed mice expressing p53 with a deletion of the PRD (p53ΔP). p53ΔP, a weak transactivator hypersensitive to Mdm2-mediated degradation, is unable to suppress oncogene-induced tumors. This phenotype could result from the loss of two motifs: Pin1 sites proposed to influence p53 stabilization and PXXP motifs proposed to mediate protein interactions. We investigated the importance of these motifs by generating mice encoding point mutations in the PRD. p53TTAA contains mutations suppressing all putative Pin1 sites in the PRD, while p53AXXA lacks PXXP motifs but retains one intact Pin1 site. Both mutant proteins accumulated in response to DNA damage, although the accumulation of p53TTAA was partially impaired. Importantly, p53TTAA and p53AXXA are efficient transactivators and potent suppressors of oncogene-induced tumors. Thus, Pin1 sites in the PRD may modulate p53 stability but do not significantly affect function. In addition, PXXP motifs are not essential, but structure dictated by the presence of prolines, PXXXXP motifs that may mediate protein interactions, and/or the length of this region appears to be functionally significant. These results may explain why the sequence of the p53 PRD is so variable in evolution.
p53 is a transcription factor that is very unstable and present at low levels in the absence of stress. Various types of DNA damage, overexpressed activated oncogenes, and other stresses stabilize and activate p53 to induce the transcription of numerous target genes involved in cell cycle arrest, apoptosis, DNA repair, and other programs (14). p53 has been reported to induce apoptosis through transcription-independent mechanisms (12) by processes that appear to be modulated by-products generated by p53 transactivation (6). Together, p53-mediated stress responses ensure cell cycle entry and progression of only those cells with intact genomes and normally functioning growth control programs.
The p53 proline-rich domain (PRD) contributes to the regulation of p53 stability, transactivation ability, and induction of transcription-independent apoptosis. This domain was first identified by Walker and Levine as a region enriched in prolines and containing several repeats of the amino acid motif PXXP (where P indicates proline and X indicates any amino acid) (31) (Fig. 1A). This region could be significant for p53 regulation since PXXP motifs create binding sites for Src homology 3 (SH3) domain-containing proteins and can modulate signal transduction (16). For example, the p53 PXXP motifs may contribute to interactions with the transcription coactivator p300 (9). Furthermore, the prolyl isomerase Pin1 was proposed to affect proline conformational changes that may reduce Mdm2 binding and enable p53 accumulation (34, 36, 37). Pin1 specifically binds sites consisting of a phosphorylated serine or threonine residue preceding a proline (pS/T-P) (35); several such sites exist in p53, but the Thr81-Pro82 site in the PRD was recently found to exert a predominant role in regulating p53 stability (2). This is consistent with previous evidence that the PRD modulates Mdm2 binding (3, 10). The functional importance of the PRD is also suggested by transfection studies using a p53 PRD deletion (p53ΔP) showing that the PRD is dispensable for the cell cycle arrest response (25, 30, 38) but is essential for apoptosis (1, 7, 24, 25, 30, 38; for an exception, see reference 11).
FIG. 1.
Targeting of the p53AXXA and p53TTAA mutations at the mouse p53 locus. (A) Murine p53 is a protein of 390 amino acids with five proposed domains: the transactivation domain (TAD), proline-rich domain (PRD), specific DNA binding domain (DBD), tetramerization domain (4D), and C-terminal regulatory domain (CT). Below are comparisons of amino acid residues 67 to 95 of WT murine p53 and of the p53ΔP, p53AXXA, and p53TTAA mutants. Prolines (P) are shown in boldface type, PXXP motifs are underlined in red, and putative Pin1 sites are in green. (B) Strategy for targeting the mutations in ES cells. The 11 exons of WT p53 are contained in a 17-kb-long EcoRI fragment. The targeting constructs (below) were in four parts: the 5′ homology region (exons 1 to 6) (4* designates the mutated exon), the floxed Neo gene (LNeoL), the 3′ homology region (exons 7 to 9), and the thymidine kinase gene (TK). Targeted recombinants are G418 and ganciclovir resistant. They result from a double crossover as described in the text. Recombinants are detected by Southern blotting with the indicated probe containing a shorter 9-kb band. Cre expression in the male germ line subsequently allows the excision of Neo. (C) Screening of recombinant ES cells and germ line transmission of the p53AXXA mutation. (Left) G418- and ganciclovir-resistant clones analyzed as described in B. Clone e contains both a WT and a recombinant 9-kb-long band. (Right) Genotyping of pups from the breeding of p53+/AXXA mice. Primers flanking exon 4 were used for PCR, and amplified products were then digested by BstBI, as the AXXA mutation introduces a BstBI site absent from the WT sequence. (D) Screening of recombinant ES cells and germ line transmission of the p53TTAA mutation. (Left) Only clone d contains the recombinant 9-kb band (the smears above 17 kb result from incomplete digestions). (Right) Genotyping of pups from the breeding of p53+/TTAA mice. Primers flanking exon 4 were used for PCR, and amplified products were then digested by HinP1I, as the TTAA mutation introduces a HinP1I site that is absent from the WT sequence.
We used homologous recombination to generate mice that encode p53 lacking the PRD (28). Consistent with transfection studies, endogenous mouse p53ΔP was more sensitive to Mdm2-mediated degradation than was wild-type (WT) p53 (p53WT). However, in contrast to transfection analyses, p53ΔP/ΔP cells did not undergo DNA damage-induced cell cycle arrest but did elicit apoptotic responses that were generally compromised relative to wild-type cells. We suggested that the differences between in vivo and transfection analyses of this and other mouse mutants (4, 5, 15, 17, 26) are likely derived from the inability of transfection approaches to enable the expression of mutant proteins at physiologically relevant levels (28, 29). Our study left open the important question of whether deleting the PRD generated the observed phenotype due to the loss of important functional motifs, such as PXXP motifs or Pin1 binding sites, or by generating a substantial structural alteration. This report answers this question through the generation of two additional mutant mouse strains with point mutations in the p53 PRD. One mouse strain encodes p53TTAA, in which threonines 76 and 86 were mutated to alanines to remove putative Pin1 binding sites. The second mouse strain encodes p53AXXA, a p53 in which four prolines were mutated into alanines, to remove all PXXP motifs from the PRD.
MATERIALS AND METHODS
Targeting constructs.
To make the AXXA mutation, a 0.7-kb XhoI-XbaI fragment containing part of exon 4 was subcloned into pBluescript KSII (Stratagene) for PCR mutagenesis (QuickChange site-directed mutagenesis kit). Two rounds of mutagenesis were required to introduce the mutation of four prolines into alanines. The first round, with primers 5′-ACCGAGACCCCTGGGGCAGTGGCCGCTGCCCCAGCCACTC-3′ and 5′-GAGTGGCTGGGGCAGCGGCCACTGCCCCAGGGGTCTCGGT-3′, mutated Pro79 and Pro82. The second round, with primers 5′-GGCCGCTGCCGCAGCCACTGCATGGCCCCTG-3′ and 5′-CAGGGGCCATGCAGTGGCTGCGGCAGCGGCC-3′, mutated Pro84 and Pro87 and introduced a BstBI site. To make the TTAA mutation, the 0.7-kb XhoI-XbaI fragment was mutated first with primers 5′-GACCCTGTCACCGAGGCGCCTGGGCCAGTGGCC-3′ and 5′-GGCCACTGGCCCAGGCGCCTCGGTGACAGGGTC-3′, which mutated Thr76 into Ala and added a HinP1I restriction site, and then with primers 5′-CCTGCCCCAGCCGCTCCATGGCCCC-3′ and 5′-GGGGCCATGGAGCGGCTGGGGCAGG-3′, which mutated Thr86 into Ala. Targeting constructs were assembled in pSP72N (Promega), and the sequences of exons and intron-exon junctions were verified by prior targeting.
Targeting/genotyping.
PrmCre 129/SvJae embryonic stem (ES) cells were electroporated with targeting constructs linearized with NotI. We screened G418- and ganciclovir-resistant colonies for the presence of the EcoRI restriction site within the Neo cassette using Southern blot hybridization of EcoRI-digested DNA with a 600-bp fragment from intron 9 (external to the targeting construct) as a probe. The presence of the mutation was confirmed independently by PCR amplification using primers flanking exon 4 and digestion with BstBI or HinP1I, depending on the mutation. ES cells with the targeted mutations were injected into blastocysts, which were then implanted into pseudopregnant females. Germ line transmission was verified by genotyping mouse embryonic fibroblasts (MEFs) from breedings of chimeras with p53+/− mice. RNAs isolated from p53AXXA/− and p53TTAA/− MEFs were sequenced to verify that their sequences (available upon request) were identical to that of p53WT except for the desired mutations.
Cells and reagents.
Primary MEFs were isolated from 13.5-day-old embryos, genotyped, and cultured, unless noted otherwise, for a maximum of four passages in Dulbecco's modified Eagle's medium with 15% fetal bovine serum, 2-mercaptoethanol (100 μM), l-glutamine (2 mM), and antibiotics. Cells were irradiated at room temperature with a 60Co γ-irradiator or treated overnight with adriamycin (ADR) (Sigma) at 0.25, 0.5, or 1 μg/ml.
Glutathione S-transferase pull-down and Western blots.
Glutathione S-transferase pull-down experiments were performed essentially as described previously (36, 37); cells were treated for 14 h with 10 μM nutlin or UV irradiated with 20 J·m−2 14 h before cell lysates were prepared. In the case of phosphatase treatment, samples were treated for 30 min at 30°C with 7 U μl−1 of Lambda protein phosphatase (New England Biolabs). Protein lysates were prepared and analyzed on sodium dodecyl sulfate-polyacrylamide gels as described previously (28). Blots were probed with primary antibodies against p53 (CM-5), p53 phospho-Ser18 (Cell Signaling Technologies), MDM2 (2A10), p21 (C-19; Santa Cruz), and α-actin (Sigma). Peroxidase-conjugated secondary antibodies were detected using Pierce Supersignal West Pico chemiluminescent substrate.
Quantitative PCR.
Total RNA was extracted using an RNeasy mini kit (QIAGEN) and reverse transcribed using Superscript III RT (Invitrogen). Real-time quantitative PCR was performed using an ABI PRISM 7700 sequence detection system with Platinum SYBR Green Mastermix as described previously (28). Primer sequences for quantification are available upon request.
Retrovirus preparation and infection of MEFs.
Cells were infected with the pWZL-E1A12S virus or were sequentially infected with the pWZL-E1A12S- and the pBABE-HrasV12-containing viruses as previously described (28).
Flow cytometry.
For cell cycle analyses, log-phase cells were irradiated with 6 or 12 Gy γ-irradiation and incubated for 24 h. Cells were then pulse-labeled for 1 h with bromodeoxyuridine (10 μM); fixed in 70% ethanol; double stained with fluorescein isothiocyanate (FITC), antibromodeoxyuridine, and propidium iodide; and then analyzed using a FACScan apparatus. Apoptosis assays were performed either on E1A-expressing MEFs that were treated for 24 h with 0, 0.25, 0.5, and 1 μg/ml adriamycin or on thymocytes from mice that were treated 6 h earlier with 0 or 10 Gy whole-body γ-irradiation. In both assays, harvested cells were stained with annexin V-FITC and analyzed using FACScan.
Tumorigenicity.
We injected 5 × 106 E1A- and Ras-expressing MEFs (E1A+Ras MEFs) subcutaneously into the rear flanks of 6-week-old female athymic nude mice. After 15 days, mice were sacrificed, and tumors were isolated and weighed.
Protein sequence analysis.
The GenBank accession numbers of analyzed sequences are as follows: NM_000546, XP_511957, U48956, X16384, X90592, CAI52016, NP_112251, U48616, U50395, U07182, U43902, AJ001022, AJ009673, NM_001009294, NP_001003210, NP_999310, CAA57348, U74486, D49825, and NM_001009403.
RESULTS
Targeting point mutations in the PRD using homologous recombination.
The p53ΔP mutation that we generated deleted residues 75 to 91, thereby removing six prolines and all PXXP motifs and putative Pin1 sites of the mouse PRD (28) (Fig. 1A). We next generated an AXXA mutation, which changed prolines 79, 82, 84, and 87 into alanines, thus mutating both PXXP motifs but leaving one putative Pin1 site intact. We also generated a TTAA mutation, which consisted of the mutation of threonines 76 and 86 into alanines, thereby preserving both PXXP motifs while eliminating both putative Pin1 sites (Fig. 1A). Importantly, Thr76 and Thr86 can be phosphorylated by mitogen-activated protein kinases (22), which could convert Thr76-Pro77 and Thr86-Pro87 into bona fide putative Pin1 sites.
We used homologous recombination to introduce the desired mutations into the endogenous Trp53 locus as described previously (28). The targeting constructs had similar structures, with a mutated exon 4 and a neo gene flanked by loxP sites (Fig. 1B). The ES cells used for targeting contain PrmCre, a Cre recombinase transgene controlled by the protamine promoter to enable Cre expression specifically in spermatocytes (23). Targeted recombinants were identified by Southern blotting (Fig. 1C and D), confirmed by PCR (not shown), and injected into blastocysts to generate chimeric mice. Due to the expression of PrmCre, the loxP-flanked selectable marker was excised when passed through the male germ line (Fig. 1B). PCR using primers flanking exon 4 and specific restriction enzymes were used to demonstrate germ line transmission of the mutations (Fig. 1C and D). We isolated RNAs from p53AXXA/− and p53TTAA/− MEFs and sequenced their entire coding regions. The sequences of p53AXXA and p53TTAA were identical to that of wild-type p53 except for the intended introduced mutations.
Intercrosses of p53+/− mice lead to a reduced yield of p53−/− animals resulting from a frequent defect in neural tube closure, most likely caused by defective apoptosis (see reference 21 for a review). In striking contrast, we previously showed that the distribution of animals from p53+/ΔP intercrosses conforms to Mendelian ratios (28). Here, we determined the distribution of 196 animals from p53+/AXXA intercrosses and 199 animals from p53+/TTAA intercrosses. Similar to the p53ΔP mutation, we found that both distributions conformed to Mendelian ratios (according to χ2 assays, with υ = 2, χ2 = 2.69 [<9.21], and υ = 2, χ2 = 1.22 [<9.21], respectively), and no developmental abnormalities were noticed.
Analysis of the accumulation and activation of mutant p53 proteins.
p53ΔP accumulates after DNA damage but is much more sensitive to Mdm2-dependent degradation than p53WT (28). p53ΔP also transactivates p21 and Mdm2 weakly in response to γ-irradiation (28). To test whether these properties result from a loss of PXXP motifs or Pin1 sites, we directly compared the accumulation and transactivation abilities of p53WT, p53ΔP, p53AXXA, and p53TTAA in response to 6 Gy irradiation. As previously described, p53ΔP accumulates about as well as p53WT after irradiation but only because p53ΔP/ΔP cells contain very little Mdm2 under these conditions. In contrast, irradiation of p53AXXA/AXXA and p53TTAA/TTAA MEFs led to much higher Mdm2 levels than in p53ΔP/ΔP cells, to more closely resemble those observed in WT cells (Fig. 2A). Note that p53AXXA migrates faster than p53WT, presumably due to proline substitutions. Importantly, p53AXXA accumulation after stress appeared to be close to normal, but that of p53TTAA was compromised, especially soon after irradiation (Fig. 2A). Since p53AXXA retains one putative Pin1 site, but p53TTAA has lost both, the data suggest that Pin1 sites in the PRD may affect the regulation of p53 stabilization after stress. Consistent with this, we observed a stabilization of p53WT and p53AXXA, but not p53TTAA, in several independent half-life measurements within 2 h of irradiation (data not shown). Of note, however, p53TTAA levels still increased 24 h after irradiation (Fig. 2A), suggesting that Pin1 sites in the PRD are not the only determinants of p53 stress-induced accumulation. Consistent with this, we observed the phosphorylation-dependent binding of Pin1 to p53TTAA in UV-irradiated MEFs. These data imply that Pin1 sites outside the PRD can mediate Pin1 binding, even in the absence of the Pin1 sites normally present in the PRD. One inference from these data is that Pin1 sites within the PRD are not essential for Pin1-p53 interactions (see Fig. S1 in the supplemental material).
FIG. 2.
Accumulation and transactivation ability of p53AXXA and p53TTAA in response to DNA damage. (A) Irradiated MEFs with p53AXXA or p53TTAA mutations display high levels of Mdm2 and p21. MEFs prepared from WT, p53AXXA/AXXA (AXXA), p53TTAA/TTAA (TTAA), and p53ΔP/ΔP (ΔP) embryos were left untreated (−) or submitted to 6 Gy irradiation, and protein extracts were then prepared 2 or 24 h after irradiation and immunoblotted with antibodies to p53, Mdm2, p21, and actin. Low and high exposure times (lo x/hi x) are shown for p21. (B) p53TTAA accumulation is partially impaired after adriamycin treatment. WT, p53AXXA/AXXA, and p53TTAA/TTAA MEFs were left untreated (−) or cultured for 24 h in a medium with 0.5 μg/ml ADR, and protein extracts were immunoblotted as described above. (C) Unlike p53ΔP, p53AXXA and p53TTAA transactivate p21 and Mdm2 efficiently after irradiation. RNAs were prepared from MEFs treated as in A and quantified using real-time PCR. Results are from two independent experiments, each resulting from quantifications in triplicates. For each gene, results are expressed relative to mRNA levels in WT unstressed cells.
To further investigate the accumulation defect in of the p53TTAA mutant, we next analyzed cells incubated for 24 h with ADR, which, unlike irradiation, represents a continuous stress. Consistent with a partial defect in p53TTAA stress-induced accumulation, we observed that p53TTAA accumulated after ADR treatment but less than p53WT or p53AXXA (Fig. 2B). Importantly, however, the transcriptional capacity of p53WT, p53TTAA, and p53AXXA again appeared to be similar, since all ADR-treated MEFs expressed high levels of Mdm2 and p21 (Fig. 2B).
The results described above suggest that unlike p53ΔP, p53AXXA and p53TTAA are efficient transactivators of the Mdm2 and p21 genes. We tested this directly by quantifying Mdm2 and p21 mRNA levels in WT, p53ΔP/ΔP, p53AXXA/AXXA, and p53TTAA/TTAA MEFs before and after irradiation (Fig. 2C). While p53ΔP is a poor transactivator under these conditions, p53AXXA and p53TTAA appear to be similar to p53WT. Consistent with this, p53ΔP/ΔP MEFs proliferate at a faster rate than WT MEFs, most likely due to their reduced p21 levels (28). By contrast, at all passages, the growth kinetics of p53AXXA/AXXA and p53TTAA/TTAA MEFs were more similar to that of WT MEFs and were considerably slower than that of p53ΔP/ΔP MEFs (Fig. 3A). These data show that removing both PXXP motifs or eliminating both putative Pin1 sites has little if any effect on proliferation rate.
FIG. 3.
The p53AXXA and p53TTAA mutations have little effect on cell proliferation or DNA damage responses. (A) The proliferation rates of p53AXXA/AXXA and p53TTAA/TTAA MEFs differ from those of p53ΔP/ΔP cells. The proliferation of p53−/− (KO), WT, p53ΔP/ΔP (ΔP), p53AXXA/AXXA (AXXA), and p53TTAA/TTAA (TTAA) MEFs was determined using a 3T3 protocol. Each point represents the mean value for two independent MEFs, the value for each MEF resulting from counts of duplicate plates. (B) Unlike p53ΔP/ΔP MEFs, p53AXXA/AXXA and p53TTAA/TTAA MEFs arrest cycling upon DNA damage. Cell cycle control in p53−/−, WT, p53ΔP/ΔP, p53AXXA/AXXA, and p53TTAA/TTAA MEFs was analyzed by fluorescence-activated cell sorter (FACS) analysis. Asynchronous cell populations were left untreated or treated with 6 or 12 Gy γ-irradiation and then processed for fluorimetric analysis 24 h later. The graph shows the mean G1/S ratios and standard deviations (SD) calculated from at least six independent experiments carried out, for each genotype, from at least two independent MEFs. Asterisks indicate significant differences after stress. (C) Efficient apoptosis in p53AXXA/AXXA and p53TTAA/TTAA E1A-expressing MEFs. Apoptosis was analyzed in p53−/−, WT, p53ΔP/ΔP, p53AXXA/AXXA, and p53TTAA/TTAA MEFs expressing the E1A oncogene. Cells were left untreated or treated with indicated doses of adriamycin for 24 h and then harvested and stained with annexin-FITC before FACS analysis. The bar graph shows the mean percentage of apoptotic cells and SD calculated from at least four independent experiments carried out, for each genotype, from at least two independent E1A-MEFs. (D) Efficient apoptosis in irradiated p53AXXA/AXXA and p53TTAA/TTAA thymocytes. WT, p53−/−, p53ΔP/ΔP, p53AXXA/AXXA, and p53TTAA/TTAA mice were left untreated or submitted to 10 Gy of whole-body γ-irradiation. Thymocytes were stained with annexin-FITC and analyzed by FACS analysis 6 h after irradiation. For each genotype, results are from four to eight mice analyzed.
p53TTAA and p53AXXA exhibit normal cell cycle arrest and apoptotic responses.
We next investigated the consequences of the p53AXXA and p53TTAA mutations on the cell cycle arrest response. While p53−/− and p53ΔP/ΔP MEFs failed to arrest after either 6 Gy or 12 Gy irradiation, both irradiation doses led to similar significant arrest responses in WT, p53AXXA/AXXA, and p53TTAA/TTAA MEFs (Fig. 3B). However, it remains possible that the arrest response is slightly less efficient in p53AXXA/AXXA MEFs. Nevertheless, the ability of both p53AXXA/AXXA and p53TTAA/TTAA MEFs to arrest following irradiation indicates that neither the PXXP nor the Pin1 motifs of the PRD are required for cell cycle arrest.
We then determined whether the AXXA and TTAA mutations affect the apoptotic response. MEFs prepared from WT, p53−/−, p53ΔP/ΔP, p53AXXA/AXXA, and p53TTAA/TTAA mice were infected with a retrovirus expressing the E1A gene (18), and their apoptotic responses to ADR were compared. Consistent with our previous analysis (28), a low dose of adriamycin (0.25 μg/ml) induced apoptosis in 60% of WT cells, while most (ca. 85%) p53ΔP/ΔP cells exhibited no apoptosis. However, at higher doses (0.5 and 1 μg/ml), the fraction of apoptotic p53ΔP/ΔP cells increased to close to WT levels, while few p53−/− cells underwent apoptosis (Fig. 3C). At all ADR doses, WT, p53AXXA/AXXA, and p53TTAA/TTAA genotypes did not differ significantly in apoptosis induction (Fig. 3C). This suggests that the AXXA and TTAA mutations have very little, if any, effect on the apoptotic response. We analyzed the apoptotic response in thymocytes of irradiated mice to further investigate apoptosis in a more relevant in vivo setting. Ten grays of whole-body γ-irradiation of WT mice induced apoptosis in about one-third of thymocytes (Fig. 3D). Under these conditions, the apoptotic response in p53ΔP/ΔP thymocytes was barely detectable. In contrast, a clear apoptotic response was observed in the thymocytes from p53AXXA/AXXA or p53TTAA/TTAA mice (Fig. 3D). These results are consistent with those obtained in E1A-MEFs and indicate that p53AXXA/AXXA and p53TTAA/TTAA cells retain an efficient apoptotic response.
p53TTAA and p53AXXA efficiently suppress oncogene-induced tumors.
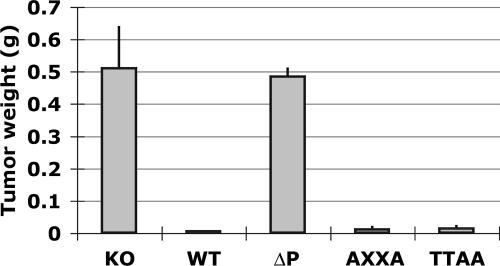
Our results indicate that unlike p53ΔP, p53AXXA and p53TTAA have retained the ability to induce cell cycle arrest and an efficient apoptotic response. Because these damage responses are essential to p53 function as a tumor suppressor, we next compared the abilities of p53ΔP, p53AXXA, and p53TTAA to suppress tumors. We previously showed that p53ΔP is a rather efficient suppressor of spontaneous tumors but a poor suppressor of oncogene-induced tumors (28). We thus compared the abilities of p53ΔP, p53AXXA, and p53TTAA to suppress oncogene-induced xenograft tumors. MEFs from WT, p53−/−, p53ΔP/ΔP, p53AXXA/AXXA, and p53TTAA/TTAA mice were infected with retroviruses expressing the E1A and Ras oncogenes, and a constant number of E1A+Ras MEFs from the different genotypes was injected into the flanks of nude mice. Tumors were allowed to grow for 15 days, mice were then sacrificed, and tumor weights were determined (Fig. 4). Under these conditions, no tumors were observed from injected E1A+Ras WT MEFs, whereas tumors of equivalent sizes were observed from injected E1A+Ras p53−/− MEFs or E1A+Ras p53ΔP/ΔP MEFs. However, p53AXXA and p53TTAA were both efficient suppressors of oncogene-induced tumors (Fig. 4).
FIG. 4.
Unlike p53ΔP, p53AXXA and p53TTAA are efficient suppressors of oncogene-induced tumorigenesis. MEFs prepared from p53−/− (KO), WT, p53ΔP/ΔP (ΔP), p53AXXA/AXXA (AXXA), and p53TTAA/TTAA (TTAA) mice were infected with retroviruses conferring expression of E1A and Ras oncogenes. E1A- and Ras-expressing MEFs were then injected into the flanks of nude mice. After 15 days, mice were sacrificed, and the weights of xenograft tumors were determined. Means and SD are from three mice for each genotype.
DISCUSSION
These studies used point mutations in the p53 PRD to investigate its role in p53 regulation. The TTAA mutation, which eliminates both putative Pin1 sites of the PRD, as well as two potential phosphorylation sites, may compromise p53 accumulation, but this did not measurably affect transactivation, cell proliferation rate, DNA damage responses, or tumor suppression. These results suggest a lack of concordance between the regulation of p53 stability and its activity. Importantly, a similar lack of correlation was suggested by the p53S23A mutant, which transactivates Mdm2 and p21 as efficiently as p53WT after γ-irradiation, despite its reduced accumulation (19). The mechanisms underlying such a disconnect are presently unknown but further support the notion that p53 stability and activity are regulated in distinct ways, as recently proposed by two independent studies (13, 28). In any case, the present results suggest that, similarly to what we recently found for C-terminal lysines (17), the putative Pin1 sites in the PRD may fine-tune the p53 pathway but are dispensable for p53 functional outputs that have an impact on stress-activated growth control. To test this point further, we aligned the sequences from the 20 mammalian p53 PRDs that are currently available. Walker and Levine initially defined the PRD as the region homologous to residues 61 to 94 of human p53 by comparing the sequences of human, monkey, mouse, rat, and chicken p53 (31). We used the same criterion to compare the 20 mammalian PRDs. Consistent with our conclusion that Pin1 sites in the PRD are not essential for p53 function, we found that the p53 PRD from rabbit, guinea pig, dog, pig, and sheep are devoid of putative Pin1 sites (Fig. 5). Together, the data indicate that Pin1 sites in the PRD may modulate p53 stabilization but are not essential for p53 activation and tumor suppression. Thus, we suggest that if Pin1 plays an important role in tumor suppression, it may do so by interacting with p53 at Pin1 sites outside of the PRD, such as S37 and S312 (a possibility supported by our observation that Pin1 still binds to the p53TTAA mutant) (see Fig. S1 in the supplemental material) and/or by regulating other important target proteins such as c-Myc (33).
FIG. 5.
Conservation of an enrichment in prolines, but not of PXXP or Pin1 motifs, in mammalian p53 PRDs. The sequences for the 20 known mammalian p53 PRDs were aligned. The PRD is defined here as proposed previously by Walker and Levine (31), i.e., the region homologous to residues 61 to 94 of human p53 (or, as labeled here to facilitate comparison with Fig. 1A, residues 58 to 91 of murine p53). PXXP motifs are underlined in red, and putative Pin1 sites (pS/T-P) are in green.
We also show that the AXXA mutation, which removed both PXXP sites and one Pin1 site of the PRD, did not significantly affect p53 accumulation, transactivation, cell proliferation, or the suppression of xenograft tumors and exhibited little if any effect on cell cycle control or apoptosis. The lower G1/S ratios (Fig. 3B) or a lower proportion of apoptotic thymocytes (Fig. 3D) may suggest a subtle effect of the AXXA mutation, but differences were barely significant in the former case and not statistically significant in the latter case. Therefore, any potential differences are very modest, and the AXXA mutation clearly does not recapitulate the phenotype observed with p53ΔP. We conclude that although PXXP motifs of the PRD may play some role in the regulation of p53, they are not essential for p53 tumor suppressor functions, like Pin1 sites. Again, the sequence comparison of 20 mammalian PRDs supports this conclusion, since we found that all tested regions are enriched in prolines (18 to 35% of all residues), but the number and positions of PXXP are extremely variable, and the p53 PRD from guinea pig is devoid of any PXXP motif (Fig. 5). Transfection studies suggested that PXXP motifs are required for efficient interactions with p300 (9), but our present data do not support this conclusion. Rather, our results and the sequence comparison of mammalian PRDs suggest two nonexclusive possibilities that may explain the differences between the p53AXXA and p53ΔP mutants. First, although PXXP was first described as the core peptide ligand motif for SH3 domains, evidence from several proteins later suggested that ligands to SH3 are enriched in prolines but may rely on variable peptide sequences (16). This suggests that protein-protein interactions at the p53 PRD in guinea pig, which cannot occur through PXXP motifs, could actually occur through multiple PXXXXP motifs (Fig. 5). In support of a biological role of PXXXXP motifs, when a phage-displayed library encoding peptides of the form X6PXXPX6 was used to identify ligands to SH3 domains, 72 of the 99 ligands contained a PXXXXP motif (8, 27). Interestingly, our mutant mouse p53AXXA has lost both its PXXP motifs but retains two PXXXXP motifs (at residues 67 to 72 and 72 to 77) (Fig. 1A), which could be sufficient to ensure protein-protein interactions. According to this hypothesis, p53ΔP would be enfeebled because it retains only one PXXXXP motif (at residues 67 to 72), which would not be enough to ensure efficient protein interactions. Another possibility is that the proline-rich domain has a structural role. Although the structure of the p53 PRD has not been determined, it has been proposed that repeats of proline-rich sequences may be structured as stiff “elbow-hinged” peptide chains and that such a structure may be important to enable protein interactions (32). In addition, the PRD could act as a spacer between the transactivation domain (which interacts with the transcription machinery, cofactors like p300, and inhibitors like Mdm2 and Mdm4) and the DNA binding domain. In any case, the sequence of the PRD is not apparently constrained in evolution so that even multiple point mutations such as those in p53AXXA would have little effect, particularly if the remaining prolines are sufficient to maintain a stiff structure. On the contrary, the p53ΔP mutant would have a dramatic effect because in this case, the spacer/stiff chain is shortened, which could change the p53 structure sufficiently to alter its ability to activate transcription and/or interact with other proteins.
Our results clearly show that PXXP motifs and Pin1 sites in the proline-rich domain are largely dispensable for p53 tumor suppressor functions and do not recapitulate the phenotype observed with their deletion. Our point mutations in PXXP motifs indicate that the proline-rich domain does not strictly rely on these peptide sequences, so alternate motifs like PXXXXP, or the maintenance of a proper structure, are more important. Furthermore, our mutation of putative Pin1 sites combined with other studies of mouse mutants that alter posttranslational modification sites in p53 suggest that such sites are not the on-off switches that enable diverse stresses to activate p53. Rather, consistent with other analyses, we postulate that the more critical regulatory targets in the p53 system are its negative regulators Mdm2 and Mdm4 (MdmX) and the various factors that control their abundance, stability, interactions with p53, and subcellular locations (13, 20, 28, 29).
Supplementary Material
Acknowledgments
We thank G. Campbell and B. Jaroszynski for technical assistance.
This work was supported by NIH grant 100845 to G.M.W. F.T. was supported in part by a fellowship from the Association pour la Recherche sur le Cancer.
Footnotes
Published ahead of print on 11 December 2006.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Baptiste, N., P. Friedlander, X. Chen, and C. Prives. 2002. The proline-rich domain of p53 is required for cooperation with anti-neoplastic agents to promote apoptosis of tumor cells. Oncogene 21:9-21. [DOI] [PubMed] [Google Scholar]
- 2.Berger, M., N. Stahl, G. Del Sal, and Y. Haupt. 2005. Mutations in proline 82 of p53 impair its activation by Pin1 and Chk2 in response to DNA damage. Mol. Cell. Biol. 25:5380-5388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berger, M., R. V. Sionov, A. J. Levine, and Y. Haupt. 2001. A role for the polyproline domain of p53 in its regulation by Mdm2. J. Biol. Chem. 276:3785-3790. [DOI] [PubMed] [Google Scholar]
- 4.Bruins, W., E. Zwart, L. D. Attardi, T. Iwakuma, E. M. Hoogervorst, R. B. Beems, B. Miranda, C. T. van Oostrom, J. van den Berg, G. J. van den Aardweg, G. Lozano, H. van Steeg, T. Jacks, and A. de Vries. 2004. Increased sensitivity to UV radiation in mice with a p53 point mutation at Ser389. Mol. Cell. Biol. 24:8884-8894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chao, C., M. Hergenhahn, M. D. Kaeser, Z. Wu, S. Saito, R. Iggo, M. Hollstein, E. Appella, and Y. Xu. 2003. Cell type- and promoter-specific roles of Ser18 phosphorylation in regulating p53 responses. J. Biol. Chem. 278:41028-41033. [DOI] [PubMed] [Google Scholar]
- 6.Chipuk, J. E., L. Bouchier-Hayes, T. Kuwana, D. D. Newmeyer, and D. R. Green. 2005. PUMA couples the nuclear and cytoplasmic proapoptotic function of p53. Science 309:1732-1735. [DOI] [PubMed] [Google Scholar]
- 7.Chipuk, J. E., T. Kuwana, L. Bouchier-Hayes, N. M. Droin, D. D. Newmeyer, M. Schuler, and D. R. Green. 2004. Direct activation of Bax by p53 mediates mitochondrial membrane permeabilization and apoptosis. Science 303:1010-1014. [DOI] [PubMed] [Google Scholar]
- 8.D'Erchia, A. M., G. Pesole, A. Tullo, C. Saccone, and E. Sbisa. 1999. Guinea pig p53 mRNA: identification of new elements in coding and untranslated regions and their functional and evolutionary implications. Genomics 58:50-64. [DOI] [PubMed] [Google Scholar]
- 9.Dornan, D., H. Shimizu, L. Burch, A. J. Smith, and T. R. Hupp. 2003. The proline repeat domain of p53 binds directly to the transcriptional coactivator p300 and allosterically controls DNA-dependent acetylation of p53. Mol. Cell. Biol. 23:8846-8861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dumaz, N., D. M. Milne, L. J. Jardine, and D. W. Meek. 2001. Critical roles for the serine 20, but not the serine 15, phosphorylation site and for the polyproline domain in regulating p53 turnover. Biochem. J. 359:459-464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Edwards, S. J., L. Hananeia, M. R. Eccles, Y. F. Zhang, and A. W. Braithwaite. 2003. The proline-rich region of mouse p53 influences transactivation and apoptosis but is largely dispensable for these functions. Oncogene 22:4517-4523. [DOI] [PubMed] [Google Scholar]
- 12.Erster, S., and U. M. Moll. 2004. Stress-induced p53 runs a direct mitochondrial death program: its role in physiologic and pathophysiologic stress responses in vivo. Cell Cycle 3:1492-1495. [DOI] [PubMed] [Google Scholar]
- 13.Francoz, S., P. Froment, S. Bogaerts, S. De Clercq, M. Maetens, G. Doumont, E. Bellefroid, and J. C. Marine. 2006. Mdm4 and Mdm2 cooperate to inhibit p53 activity in proliferating and quiescent cells in vivo. Proc. Natl. Acad. Sci. USA 103:3232-3237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harms, K., S. Nozell, and X. Chen. 2004. The common and distinct target genes of the p53 family transcription factors. Cell. Mol. Life Sci. 61:822-842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jhappan, C., T. M. Yusufzai, S. Anderson, M. R. Anver, and G. Merlino. 2000. The p53 response to DNA damage in vivo is independent of DNA-dependent protein kinase. Mol. Cell. Biol. 20:4075-4083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kay, B. K., M. P. Williamson, and M. Sudol. 2000. The importance of being proline: the interaction of proline-rich motifs in signaling proteins with their cognate domains. FASEB J. 14:231-241. [PubMed] [Google Scholar]
- 17.Krummel, K. A., C. J. Lee, F. Toledo, and G. M. Wahl. 2005. The C-terminal lysines fine-tune p53 stress responses in a mouse model, but are not required for stability control or transactivation. Proc. Natl. Acad. Sci. USA 102:10188-10193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lowe, S. W., H. E. Ruley, T. Jacks, and D. E. Housman. 1993. p53-dependent apoptosis modulates the cytotoxicity of anticancer agents. Cell 74:957-967. [DOI] [PubMed] [Google Scholar]
- 19.MacPherson, D., J. Kim, T. Kim, B. K. Rhee, C. T. Van Oostrom, R. A. DiTullio, M. Venere, T. D. Halazonetis, R. Bronson, A. De Vries, M. Fleming, and T. Jacks. 2004. Defective apoptosis and B-cell lymphomas in mice with p53 point mutation at Ser 23. EMBO J. 23:3689-3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marine, J. C., S. Francoz, M. Maetens, G. Wahl, F. Toledo, and G. Lozano. 2006. Keeping p53 in check: essential and synergistic functions of Mdm2 and Mdm4. Cell Death Differ. 13:927-934. [DOI] [PubMed] [Google Scholar]
- 21.Miller, F. D., C. D. Pozniak, and G. S. Walsh. 2000. Neuronal life and death: an essential role for the p53 family. Cell Death Differ. 7:880-888. [DOI] [PubMed] [Google Scholar]
- 22.Milne, D. M., D. G. Campbell, F. B. Caudwell, and D. W. Meek. 1994. Phosphorylation of the tumor suppressor protein p53 by mitogen-activated protein kinases. J. Biol. Chem. 269:9253-9260. [PubMed] [Google Scholar]
- 23.O'Gorman, S., N. A. Dagenais, M. Qian, and Y. Marchuk. 1997. Protamine-Cre recombinase transgenes efficiently recombine target sequences in the male germ line of mice, but not in embryonic stem cells. Proc. Natl. Acad. Sci. USA 94:14602-14607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roth, J., P. Koch, A. Contente, and M. Dobbelstein. 2000. Tumor-derived mutations within the DNA-binding domain of p53 that phenotypically resemble the deletion of the proline-rich domain. Oncogene 19:1834-1842. [DOI] [PubMed] [Google Scholar]
- 25.Sakamuro, D., P. Sabbatini, E. White, and G. C. Prendergast. 1997. The polyproline region of p53 is required to activate apoptosis but not growth arrest. Oncogene 15:887-898. [DOI] [PubMed] [Google Scholar]
- 26.Sluss, H. K., H. Armata, J. Gallant, and S. N. Jones. 2004. Phosphorylation of serine 18 regulates distinct p53 functions in mice. Mol. Cell. Biol. 24:976-984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sparks, A. B., J. E. Rider, N. G. Hoffman, D. M. Fowlkes, L. A. Quillam, and B. K. Kay. 1996. Distinct ligand preferences of Src homology 3 domains from Src, Yes, Abl, Cortactin, p53bp2, PLCgamma, Crk, and Grb2. Proc. Natl. Acad. Sci. USA 93:1540-1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Toledo, F., K. A. Krummel, C. J. Lee, C. W. Liu, L. W. Rodewald, M. Tang, and G. M. Wahl. 2006. A mouse p53 mutant lacking the proline rich domain rescues Mdm4 deficiency and provides insight into the Mdm2-Mdm4-p53 regulatory network. Cancer Cell 9:273-285. [DOI] [PubMed] [Google Scholar]
- 29.Toledo, F., and G. M. Wahl. 2006. Regulating the p53 pathway: in vitro hypotheses, in vivo veritas. Nat. Rev. Cancer 6:909-923. [DOI] [PubMed] [Google Scholar]
- 30.Venot, C., M. Maratrat, C. Dureuil, E. Conseiller, L. Bracco, and L. Debussche. 1998. The requirement for the p53 proline-rich functional domain for mediation of apoptosis is correlated with specific PIG3 gene transactivation and with transcriptional repression. EMBO J. 17:4668-4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Walker, K., and A. Levine. 1996. Identification of a novel p53 functional domain that is necessary for efficient growth suppression. Proc. Natl. Acad. Sci. USA 93:15335-15340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Williamson, M. P. 1994. The structure and function of proline-rich regions in proteins. Biochem. J. 297:249-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wulf, G., G. Finn, F. Suizu, and K. P. Lu. 2005. Phosphorylation-specific prolyl isomerization: is there an underlying theme? Nat. Cell Biol. 7:435-441. [DOI] [PubMed] [Google Scholar]
- 34.Wulf, G. M., Y. C. Liou, A. Ryo, S. W. Lee, and K. P. Lu. 2002. Role of Pin1 in the regulation of p53 stability and p21 transactivation, and cell cycle checkpoints in response to DNA damage. J. Biol. Chem. 277:47976-47979. [DOI] [PubMed] [Google Scholar]
- 35.Yaffe, M. B., M. Schutkowski, M. Shen, X. Z. Zhou, P. T. Stukenberg, J. U. Rahfeld, J. Xu, J. Kuang, M. W. Kirschner, G. Fischer, L. C. Cantley, and K. P. Lu. 1997. Sequence-specific and phosphorylation-dependent proline isomerization: a potential mitotic regulatory mechanism. Science 278:1957-1960. [DOI] [PubMed] [Google Scholar]
- 36.Zacchi, P., M. Gostissa, T. Uchida, C. Salvagno, F. Avolio, S. Volinia, Z. Ronai, G. Blandino, C. Schneider, and G. Del Sal. 2002. The prolyl isomerase Pin1 reveals a mechanism to control p53 functions after genotoxic insults. Nature 419:853-857. [DOI] [PubMed] [Google Scholar]
- 37.Zheng, H., H. You, X. Z. Zhou, S. A. Murray, T. Uchida, G. Wulf, L. Gu, X. Tang, K. P. Lu, and Z. X. Xiao. 2002. The prolyl isomerase Pin1 is a regulator of p53 in genotoxic response. Nature 419:849-853. [DOI] [PubMed] [Google Scholar]
- 38.Zhu, J., J. Jiang, W. Zhou, K. Zhu, and X. Chen. 1999. Differential regulation of cellular target genes by p53 devoid of the PXXP motifs with impaired apoptotic activity. Oncogene 18:2149-2155. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.