Abstract
We report the characterization of the yeast Npa2p (Urb2p) protein, which is essential for 60S ribosomal subunit biogenesis. We identified this protein in a synthetic lethal screening with the rsa3 null allele. Rsa3p is a genetic partner of the putative RNA helicase Dbp6p. Mutation or depletion of Npa2p leads to a net deficit in 60S subunits and a decrease in the levels all 27S pre-rRNAs and mature 25S and 5.8S rRNAs. This is likely due to instability of early pre-60S particles. Consistent with a role of Npa2p in 60S subunit biogenesis, green fluorescent protein-tagged Npa2p localizes predominantly to the nucleolus and TAP-tagged Npa2p sediments with large complexes in sucrose gradients and is associated mainly with 27SA2 pre-rRNA-containing preribosomal particles. In addition, we reveal a genetic synthetic interaction between Npa2p, several factors required for early steps of 60S subunit biogenesis (Dbp6p, Dbp7p, Dbp9p, Npa1p, Nop8p, and Rsa3p), and the 60S protein Rpl3p. Furthermore, coimmunoprecipitation and gel filtration analyses demonstrated that at least Npa2p, Dbp6p, Npa1p, Nop8p, and Rsa3p are present together in a subcomplex of low molecular mass whose integrity is independent of RNA. Our results support the idea that these five factors work in concert during the early steps of 60S subunit biogenesis.
The synthesis of eukaryotic ribosomes is a complex and highly energy-consuming process (53, 103). Ribosome biogenesis takes place primarily in the nucleolus, but some events occur in the nucleoplasm, where the preribosomal subunits gain export competence, and in the cytoplasm, where the last steps in the maturation of the ribosomal subunits (r-subunits) occur (94, 96). Although ribosome biogenesis is conserved throughout eukaryotes (39, 90), it has been best characterized in the yeast Saccharomyces cerevisiae (for reviews, see references 33, 58, and 100). In yeast, three of the four rRNAs (18S, 5.8S, and 25S rRNAs) are transcribed as a single precursor by RNA polymerase I, whereas RNA polymerase III separately transcribes the pre-5S rRNA (for a review, see reference 73). Concomitantly with transcription, the pre-rRNA intermediates are extensively modified (for a review, see reference 13). These precursors are then processed by a complex series of endo- and exonucleolytic reactions (see Fig. 1), which requires small nucleolar RNAs and nonribosomal proteins (r-proteins) (for reviews, see references 58 and 101). While some of these protein factors have clear functions in pre-rRNA processing and modification (e.g., nucleases and base methylases), the precise functions of most of them remain unclear.
FIG. 1.
Pre-rRNA processing in S. cerevisiae. (A) Structure and processing sites of the 35S pre-rRNA. This precursor contains the sequences for the mature 18S, 5.8S, and 25S rRNAs that are separated by two internal transcribed spacer sequences, ITS1 and ITS2, and flanked by two external transcribed spacer sequences (ETS), 5′ ETS and 3′ ETS. The mature rRNA species are shown as bars and the transcribed spacer sequences as lines. The processing sites and the various probes used are indicated. (B) Schematic representation of the pre-rRNA processing pathway of the 35S pre-rRNA and pre-5S rRNA. Cleavage and trimming reactions are indicated. The data presented in this study suggest that Npa2p is required for efficient processing of the 27S pre-rRNAs. For reviews of pre-rRNA processing and the known processing enzymes, see references 75 and 101.
Pre-rRNA processing does not occur on naked RNA molecules. Instead, pre-rRNA molecules at all stages of maturation associate with most r-proteins and nonribosomal proteins to form preribosomal particles (r-particles) (27, 37, 94, 104). Recent advances in the proteomic field have facilitated the identification of the protein components of preribosomal particles (for reviews, see references 14, 31, 33, and 95). These analyses have also redefined the model of the r-subunit assembly pathway. In this model, the 90S preribosomal particles contain the complete machinery responsible for cleavages of the 35S pre-rRNA at sites A0 to A2 as well as several large and small r-proteins but lack most of the factors involved in 60S r-subunit formation (5, 23, 41, 49; for reviews, see references 14, 21, 31, and 33). Following pre-rRNA cleavage at site A2, the last 90S particle gives rise to early 43S and 66S preribosomal particles, which contain 20S and 27SA2 pre-rRNAs, respectively. It appears that most of the factors associated with 90S particles, with few exceptions, are released after this cleavage step (23, 41, 72, 82). The early 43S preribosomal particle is rapidly exported to the cytoplasm, where 20S pre-rRNA is processed to mature 18S rRNA and the last modification and assembly reactions take place (8, 96). Only a few factors and 40S ribosomal proteins have been described so far as being needed to finish the maturation of 40S r-subunits from early 43S preribosomal particles (22, 29, 32, 43, 52, 65, 82, 89, 98). The study of the different purified pre-60S complexes is consistent with the presence of distinct pre-60S intermediates. These intermediates are termed, according to their position in the pathway, early, medium, late, and cytoplasmic pre-60S r-particles (4, 19, 28, 44, 72, 80, 81). Much less is known about the role of 60S r-proteins in 60S r-subunit maturation (17, 18, 25, 35, 60, 71, 97). The earliest 66S preribosomal particle is likely the result of the association of about 50 nonribosomal proteins and several large r-proteins with the 27SA2 pre-rRNA (19; see Discussion). The complexity of the pre-60S r-particles decreases during their maturation (36, 72, 80), and the export-competent pre-60S particle has completed the pre-rRNA processing reactions (4, 72). As for the pre-40S particles, last assembly reactions occur in the cytoplasm (46, 55, 86).
Despite the success of the proteomic approach in defining preribosomal particles and identifying their components, many challenges clearly remain for a complete understanding of ribosome assembly. Many of the nonribosomal factors that have been identified in preribosomal particles or in systematic analyses remain uncharacterized so far. For those characterized, we generally lack an understanding of their precise function in ribosome assembly and we do not know their substrates and their interacting partners. Except in a few instances, we understand neither the order of recruitment of the different factors to the preribosomal particles (81) nor their precise time of action. We are interested in the identification of partners of the putative RNA helicase Dbp6p, which is required for 60S r-subunit assembly and has been proposed to act at an early step during this process (56). Our initial genetic and functional analyses have revealed specific interactions between Dbp6p, different 60S r-subunit assembly factors (Dbp7p, Dbp9p, Nop8p, Npa1p/Urb1p, and Rsa3p), and the 60S r-protein Rpl3p (16, 78). Here, we provide evidence that the predominantly nucleolar Npa2p protein is an additional member of the aforementioned network of proteins. Our results indicate that Npa2p is required for 27S pre-rRNA processing and suggest that Npa2p has an early role during 60S r-subunit assembly similar to that described previously for Dbp6p (56), Dbp7p (12), Dbp9p (11), Nop8p (108), Npa1p (19, 70, 78), or Rsa3p (16, 78). Recently, some of us have reported the composition of very early pre-60S r-particles purified using Npa1p-TAP as bait (19). These particles also contain Dbp6p, Dbp7p, Dbp9p, Npa2p, and Nop8p but seem to lack Rsa3p (19). Npa2p (alternatively named Urb2p) has also been independently identified in a systematic functional analysis of essential yeast genes (70). Therein, Npa2p was described as a protein required for ribosome biogenesis that copurified with Npa1p (70). In agreement with these reports, in this study, we found that Npa2p is present in a very early pre-60S r-particle(s) containing 27SA2 pre-rRNA and that it physically interacts with Npa1p. Moreover, we provide evidence that Npa2p also intimately interacts with Dbp6p, Nop8p, and Rsa3p even in the absence of RNA. We conclude that Npa2p forms a RNA-independent heteromeric subcomplex required during early maturation of nascent 60S r-subunits.
MATERIALS AND METHODS
Strains, media, and genetic manipulations.
Most yeast strains used in this study (see Table S1 in the supplemental material) are derivatives of strain W303 (MATa/MATα ade2-1/ade2-1 his3-11,15/his3-11,15 leu2-3,112/leu2-3,112 trp1-1/trp1-1 ura3-1/ura3-1 can1-100/can1-100). The isolation of synthetic lethal (sl) mutants has been previously described (78). Strain IVY317 [YCplac33-NPA2] was obtained after four consecutive backcrosses of W303-1A with a meiotic npa2::KanMX4 segregant of Y26839 (Euroscarf collection) containing the YCplac33-NPA2 plasmid. Strain YO795 (NAP2-TAP) was obtained as follows: a gene cassette flanked on the 5′ side by the last 52 nucleotides of the NPA2 open reading frame (ORF) and on the 3′ side by a segment of the NPA2 terminator and containing the TAP tag sequence followed by a URA3 marker gene from Kluyveromyces lactis was PCR amplified using plasmid pBS1539 (76) and oligonucleotides YJR041C-TAP1 (5′-TTTCAAAGCACTTTACCTCCAATACAAAAAGGTTGGTAAATGGCGCGAAGATTCCATGGAAAAGAGAAG-3′) and YJR041C-TAP2 (5′-ACTTGTTTAAGCTCCGTCACCCTGTTATTAAACGTGAGCAGAGAAATGCCTTTACGACTCACTATAGGG-3′). This cassette were integrated into strain YO341, creating strain YO795.
Growth and handling of yeast were performed by established procedures (3, 54). Tetrad dissections were performed using a Singer MSM manual micromanipulator. Escherichia coli strain DH5α was used for cloning and propagation of plasmids (79).
Cloning of NPA2.
Strain sl3-4, which has a slow growth (sg) phenotype at any temperature tested, was transformed with a YCplac111-based yeast genomic library (57), and about 10,000 transformants were screened for wild-type growth on plates with synthetic dextrose without leucine (SD-Leu) at 30 and 37°C. One plasmid, pIV223, containing a ca. 8.7-kb insert, complemented both the sg and the sl phenotypes of the sl3-4 mutant. The sl phenotype of strain sl2-3, which grows as the wild-type strain at any temperature, was also complemented by pIV223. The sequence of the terminal regions shows that the library insert contained YJR040W (GEF1) and YJR041C (NPA2/URB2) as sole complete ORFs. Further subcloning of YJR041C as a 5.2-kb SphI-BbeI fragment, which was blunt ended, into SmaI-restricted YCplac111 (40) (hereafter named YCplac111-NPA2) and YCplac33 (40) (hereafter named YCplac33-NPA2) confirmed that use of YJR041C was sufficient to complement the sg and sl phenotypes of the sl3-4 mutant and the sl phenotype of the sl2-3 mutant.
Plasmids.
YCplac33-NPA2eGFP was constructed as follows: a 5.1-kb NruI-NarI fragment from pIV223 was blunt ended and cloned into SmaI-restricted YCplac33-yeGFP/TCYC1 (a gift from M. Hall). One candidate in the appropriate orientation, pIV1-eGFP, was selected. Then, a PCR was performed using pIV223 as a template and the oligonucleotides 5′-1813-NPA2 (5′-GAGGAGACAAATATCACG-3′, placed 60 bp upstream from the sole XbaI site present in the YJR041C ORF) and 3′-XBA1-NPA2 (5′-GCTCTAGAATCTTCGCGCCATTTAC-3′, complementary to the end of the YJR041C ORF but lacking the stop codon; an XbaI site is underlined). The PCR product was digested with XbaI and cloned into pIV1-eGFP, which was also restricted with XbaI. YCplac33-NPA2-eGFP is one candidate in the appropriate orientation.
To generate pTAPC111-NPA2, pTAPC111-NOP8 (pDK961; a generous gift from D. Kressler) was digested with EcoRI and BamHI and blunt ended. This double digestion releases the NOP8 ORF but retains the TAP tag with the vector. Then, a 5.1-kb NruI-NarI fragment from pIV223 was blunt ended and cloned in the appropriate orientation to generate pIV230. Finally, the aforementioned XbaI-restricted PCR product was cloned into XbaI-restricted pIV230. pTAPC111-NPA2 is one candidate in the appropriate orientation.
pRS414-NPA2 was obtained by cloning a blunt-ended 5.1-kb SphI-BbeI fragment from pIV223 into the SmaI site of pRS414 (87).
The plasmid YCplac22-NOP8-HA was constructed by cloning a ca. 2.3-kb EcoRI-HindIII fragment from pHAC111-NOP8 (pDK646; a generous gift from D. Kressler) into the EcoRI-HindIII-restricted YCplac22 plasmid (87).
pHAC111-NPA1 was constructed by cloning of a 5.9-kb ApaI-NsiI blunt-ended fragment from pIV222 (78) into SmaI-restricted pHAC33 (a gift from M. Hall). One candidate in the appropriate orientation, pIVN1-HA, was selected. Then a PCR was performed using YCplac111-NPA1 as a template and the oligonucleotides NPA1StuIUP and NPA1StopLO (78). The PCR product was digested with StuI and BamHI and ligated into pIVN1-HA restricted with the same enzymes. pHAC33-NPA1 is a correct candidate of this cloning. PHAC111-NPA1 was obtained after subcloning of a 6.6-kb PvuII fragment of pHAC111-NPA1 into SmaI-restricted YCplac33 plasmid.
YCplac33-RSA3-eGFP and YCplac22-HA-DBP7 were generous gifts from D. Kressler. YCplac22-HA-DBP9 and pRS414-HA-DBP6 have been previously described (11). pHAC33-RSA3 has also been previously reported (16).
Construction of a GAL::ZZ-NPA2 allele and in vivo depletion of Npa2p.
Strain YH378 (GAL::ZZ-NPA2) was obtained as follows. A gene cassette flanked on the 5′ side by a segment of the NPA2 promoter and on the 3′ side by the 5′ end of the NPA2 ORF and containing the HIS3 gene marker and the GAL10 promoter followed by the ZZ tag sequence was PCR amplified using plasmid pTL27 (61) and oligonucleotides pGAL-YJR041C1 (5′-AGAGGGCACTTGGTCACAACTACAGAATTGTTTACTAGCATAGGAACATCCTCTTGGCCTCCTCTAGT-3′) and pGAL-YJR041C3bis (5′-TTTCGACAAATCTTGGGCATTGTCTGGGATAGATAGTTCTTCTGTAAGATCACCCATATTCGCGTCTACTTTCGG-3′). This cassette was integrated into strain YDL402 (61), creating strain GAL::ZZ-NPA2.
For in vivo depletion, the GAL::ZZ-NPA2 strain was grown in YPGal+Suc (1% yeast extract, 2% peptone, 2% galactose, 4% sucrose) medium at 30°C until reaching the midexponential phase (optical density at 600 nm [OD600], 0.8). Cells were harvested, washed, and used to inoculate cultures in YPD (1% yeast extract, 2% peptone, 2% glucose) medium. Cell growth was monitored over a period of 30 h, during which the cultures were regularly diluted into fresh YPD medium to maintain exponential growth. As a control, the wild-type YDL402 strain was used. At different time points, samples were collected to perform protein and RNA extractions and polysome analysis.
Fluorescence microscopy.
Strain IVY317 [YCplac111-NPA2] was transformed with YCplac33-NPA2eGFP followed by plasmid segregation on SD-Ura plates. This strain grows at the same rate as a wild-type strain.
For localization, IVY317 [YCplac33-NPA2eGFP] was first transformed with pUN100-DsRedNOP1 (kindly provided by J. Bassler). Then, several transformants were grown to mid-log phase in SD-Leu-Ura liquid medium at 30°C, washed, and resuspended in water. Acquisition was performed using a Leica DMR microscope equipped with a DC camera following the instructions of the manufacturer. Digital images were processed with Adobe Photoshop 7.0 (Adobe Systems).
Sucrose gradient centrifugation.
Polysome and r-subunit preparation and analyses were performed as previously described (56). Gradient analysis was performed with an ISCO UA-6 system with continuous monitoring at A254. Analyses of proteins and pre-rRNAs from gradient fractions were carried out exactly as previously described (15).
Synthetic interaction crosses.
To determine synthetic lethal or enhancement interactions, crosses involving the npa2-1 or the npa1-1 mutant and selected mutants affecting assembly of 60S r-subunits were performed. Information on the crosses can be found in the supplemental material.
Antibodies and Western blotting.
Rabbit anti-protein A antibodies were purchased from Sigma and HA.11 monoclonal antibodies from Covance. Rabbit polyclonal anti-Has1p antibodies have been previously described (26). Anti-Nog2p antibodies were a gift from M. Fromont-Racine (81). Anti-Nop1p antibodies (MCA-28F2) were obtained from EnCor Biotechnology. Anti-Nop7p antibodies were a gift from B. Stillman (24). Anti-Rpl3p monoclonal antibodies were a gift from J. Warner (102). Anti-Rpl12p and anti-Rpp0p were a gift from J. P. G. Ballesta (9). Anti-Rsp8p was a gift from G. Dieci (67). Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blotting were performed following standard procedures (79). Antibody binding was detected with horseradish peroxidase-conjugated secondary antibodies (Bio-Rad) and a chemiluminescence system (Pierce).
RNA extractions, Northern hybridization, and primer extension.
RNA extraction, Northern hybridization, and primer extension analyses were carried out according to standard procedures (99). In all experiments, RNA was extracted from samples corresponding to 10 OD600 units of exponentially grown cells and 5 or 2.5 μg was loaded in gels or used in primer extension reactions. Sequences of oligonucleotides used for RNA hybridization and primer extension analyses have been described previously (20, 78).
Immunoprecipitations.
Two liters of the appropriate cells were grown in YPD or specific SD medium to an OD600 of 0.8 and then harvested. The cell pellets were washed twice with cold water and frozen with liquid nitrogen. Frozen cell pellets were broken with dry ice in a RM100 motorized mortar grinder (Retsch) as previously described (84). Broken cells were resuspended in 2 ml of cold IPP150 buffer (10 mM Tris-HCl [pH 8.0], 150 mM NaCl, 0.1% Triton X-100) plus 0.5 mM dithiothreitol, 0.2 mM EDTA (pH 8.0), 20% glycerol, and protease inhibitors (Complete; Roche). Total cell extracts were obtained by centrifugation at 20,000 × g in a 75VTi rotor (Beckman) for 1 h at 4°C. The supernatants were used for immunoprecipitation or subjected to a second centrifugation at 180,000 × g in the same rotor for 45 min at 4°C. The latter supernatants were named ultracentrifuged extracts. Aliquots of 2 ml of total or ultracentrifuged extracts were mixed with 200 μl of immunoglobulin G-Sepharose 6 Fast Flow (IgG-Sepharose) beads (Amersham) and incubated for 2 h at 4°C with end-over-end tube rotation. When indicated, the ultracentrifuged extracts were treated with 100 ng/ml RNase A before mixing with the IgG-Sepharose beads. After incubation, the beads were washed eight times with 1.5 ml of the same buffer at 4°C, the IgG-Sepharose beads were collected, and the bound proteins were eluted with 900 μl 0.5 M acetic acid and concentrated by lyophilization. Total, nonbound and bound proteins were separated on 6 and 12% SDS-PAGE gels and subjected to Western blot analysis using the appropriate antibodies.
For the immunoprecipitation analysis (see Fig. 7), the protocols used were those previously described in reference 63.
FIG. 7.
Genetic partners of Npa2p. Solid lines represent synthetic lethality, and dashed lines represent synthetic enhancement. This paper shows the genetic interaction of npa2 and npa1 alleles, with mutations in the genes coding the indicated proteins. Note that the genetic network formed by Dpb6p, Dbp7p, Dbp9p, Nop8p, Rpl3p, and Rsa3p has been previously described (78). See Materials and Methods, Results, and the supplemental material for more details.
RESULTS
Isolation of npa2 mutants that exhibit a synthetic lethal phenotype with the rsa3 null allele.
We have previously identified a genetic network of interaction between Dpb6p, Dbp7p, Dbp9p, Rpl3p, Nop8p, and Rsa3p (16). More recently, we have extended the genetic network by performing a synthetic lethal (sl) screening with the rsa3 null allele (78). This screening confirmed the interactions between RSA3 and DBP6, DBP9, RPL3, and NOP8 and revealed additional interactions with alleles of NPA1 (78). Two sl mutants, the sl3-4 and sl2-3 strains, remained uncharacterized (see Materials and Methods). To identify the corresponding mutated genes, we first transformed the sl3-4 strain with a yeast genomic library and recovered the library plasmid of the sole colony that grew as the wild-type strain at 30°C and lost the sl phenotype (restored red-white sectoring and growth on 5-fluoroorotic acid [5-FOA]). Sequence and subcloning analysis of the insert showed that ORF YJR041C, which we named NPA2, corresponds to the gene defined by the sl3-4 mutation. NPA2 also complemented the sl phenotype of the sl2-3 strain.
To isolate the sl mutation from the sl3-4 mutant, hereafter npa2-1, we crossed this strain to an isogenic wild-type strain, W303-1A; the resulting diploid was sporulated and tetrads were dissected. In all complete tetrads, two sg spore clones were obtained. For further analyses, the meiotic segregants IVY255 and IVY251 were selected (see Table S1 in the supplemental material). Further genetic analysis confirmed that the npa2-1 mutation is linked to the NPA2 gene (data not shown). Figure 2 shows that the npa2-1 mutation confers a sg phenotype and causes lethality in combination with the rsa3 null allele. The sg phenotype of npa2-1 is enhanced at 37°C (data not shown).
FIG. 2.
The npa2-1 mutation confers a slow growth phenotype and is synthetically lethal with the rsa3 null allele. Strains IVY251 (npa2-1), carrying the plasmid YCplac33-NPA2, and YMD3-2D (Δrsa3) were crossed, the resulting diploid was sporulated, and tetrads were dissected. Complete tetrads were streaked on YPD plates and restreaked on 5-FOA-containing plates to counterselect YCplac33-NPA2. A representative tetratype tetrad is shown on a YPD plate (top half) or on a 5-FOA containing plate (bottom half). Plates were incubated at 30°C for 4 days.
Npa2p is an essential protein that localizes in the nucleolus.
NPA2 is an essential gene that encodes a relatively large protein of 1,147 amino acids (135.2 kDa). A detailed sequence analysis revealed no obvious protein motifs for Npa2p, but its first 748 amino acids have a predicted structure related to proteins of the HEAT-repeat family (Yeast Resource Center Informatics Platform; http://www.yeastrc.org) (45). We also found an IMP dehydrogenase-GMP reductase domain between amino acids 520 and 855 (InterProScan; http://www.ebi.ac.uk/InterProScan), although the biological relevance of this finding is unclear. Searches in the preribosomal network (http://pre-ribosome.de) (68) found Npa2p associated with early pre-60S particles. Moreover, Npa2p has been described to be required for pre-rRNA processing (70, 74) and it has been found by some of us as a component of a very early pre-60S particle(s) containing Npa1p-TAP (19).
For a first hint of the function of Npa2p, we determined its subcellular localization. To do this, we constructed a C-terminal green fluorescent protein-tagged NPA2 allele expressed under the control of its cognate promoter (see Materials and Methods). The resulting strain was further transformed with pUN100-DSRed-NOP1, which expressed DsRed-tagged Nop1p (35). We detected the green fluorescence of Npa2p-green fluorescent protein in the nucleolus, since it mostly colocalized with the red signal of DsRed-Nop1p (see Fig. S1 in the supplemental material). Our results are in full agreement with those of Huh et al. (50) and consistent with a role of this protein in ribosome biogenesis.
Mutation or depletion of Npa2p leads to a deficiency in 60S r-subunits.
To investigate the role of Npa2p in ribosome biogenesis, we first analyzed polysome profiles of npa2-1 and NPA2 isogenic strains. For the NPA2 strain, we detected a typical wild-type profile (Fig. 3A). For the npa2-1 strain, we detected a deficit of free 60S versus 40S r-subunits, an overall decrease in the level of 80S ribosomes, and an accumulation of half-mer polysomes (Fig. 3B). We also performed polysome profile analysis upon depletion of Npa2p. For this, we first constructed a conditional GAL::ZZ-NPA2 strain (YH378), which contains the NPA2 gene under the control of the GAL10 promoter (see Materials and Methods). In galactose-sucrose containing medium, the GAL::ZZ-NPA2 strain had the same growth behavior as the otherwise isogenic strain, indicating that the GAL::ZZ-NPA2 allele is fully functional. As expected, only residual growth of the GAL::ZZ-NPA2 strain was observed on glucose-containing plates (see Fig. S2 in the supplemental material). After a shift from galactose-sucrose to glucose-containing liquid medium, the growth rate of the GAL::ZZ-NPA2 strain remained similar to that of the wild-type strain during at least the first 6 h but then progressively decreased to a doubling time of >8 h after 30 h, as compared with the 2 h doubling time for YDL402. A concomitant depletion of Npa2p was observed by Western blotting (see Fig. S2 in the supplemental material). The GAL::ZZ-NPA2 strain and its isogenic wild-type counterpart (YDL402) were grown at 30°C in medium containing galactose and sucrose as a carbon source and then shifted to glucose-containing medium for 6, 12, and 24 h. Extracts were prepared at the different time points, and polysome profiles were analyzed. Wild-type profiles were obtained for both strains in galactose-sucrose medium and for YDL402 at any time after the shift (Fig. 3C and data not shown). In similarity to the npa2-1 mutant results, the Npa2p-depleted strain displayed a deficit in 60S r-subunits. This deficit was observed after 6 h and became more pronounced after 12 and 24 h in glucose-containing medium (Fig. 3D to F). These results show clearly that Npa2p is required for 60S r-subunit accumulation.
FIG. 3.
The npa2-1 mutation and the depletion of Npa2p result in a deficit in free 60S r-subunits and in the accumulation of half-mer polysomes. W303-1B (NPA2) (A) and IVY251 (npa2-1) (B) strains were grown in YPD at 30°C. Strain YH378 (GAL::ZZ-NPA2) was grown at 30°C in YPGal+Suc (C) or shifted to YPD for 6 h (D), 12 h (E), and 24 h (F). Cells were harvested at an OD600 of 0.8, and cell extracts were resolved in 7% to 50% polysome sucrose gradients. The A254 was continuously measured. Sedimentation is shown from left to right. The peaks of free 40S and 60S r-subunits, 80S free couples-monosomes, and polysomes are indicated. Half-mers are labeled by arrows.
Npa2p is required for normal pre-rRNA processing.
To determine whether the deficit in 60S r-subunits in the npa2 mutant strains is associated with defects in pre-rRNA processing, we analyzed the steady-state levels of pre- and mature rRNA species by Northern blotting and primer extension. First, total RNA was isolated from the GAL::ZZ-NPA2 and the isogenic NPA2 strain at various time points after transfer from galactose-sucrose to glucose-containing medium and analyzed by Northern hybridization. As shown in Fig. 4, a significant decrease in levels of mature rRNAs was observed after depletion of Npa2p. The 35S pre-rRNA and the aberrant 23S (that extends from +1 to site A3) and 21S (that extends from site A1 to site A3) pre-rRNA species slightly accumulated, with ongoing depletion compared to result seen with the wild-type strain in glucose medium. These accumulations are likely due to delayed processing at site A2 and to a lesser extent at sites A0 and A1. Probably in part as a consequence of the A2 cleavage delay, the levels of the 20S and the 27SA2 precursors were significantly affected. Importantly, there was a drastic reduction in the levels of the 27SB pre-rRNA species but a less pronounced reduction of 7S pre-rRNAs (Fig. 4A to B). Finally, we detected mild accumulation of an aberrant A2-C2 fragment at early time points of depletion, which suggests that depletion of Npa2p allows premature cleavage of 27SA2 pre-rRNA at site C2.
FIG. 4.
Effects of Npa2p depletion on steady-state levels of pre-rRNAs and mature rRNAs. Strains YDL402 (NPA2) and YH378 (GAL::ZZ-NPA2) were grown in YPGal+Suc medium and then shifted to YPD medium. Cells were harvested at the indicated times, and total RNA was extracted. (A) Equal amounts of total RNA (5 μg) were resolved on a 1.2% agarose-formaldehyde gel, transferred onto a nylon membrane, and hybridized consecutively with different probes. (B) Equal amounts of total RNA (2.5 μg) were resolved on a 7% polyacrylamide-urea gel, transferred onto a nylon membrane, and hybridized consecutively with different probes. (C) Equal amounts of total RNA (5 μg) were used for primer extension analysis. Probe g was labeled and used for the reactions. Note that this probe allows detection of 27SA2 (as the stop at site A2), 27SA3 (as the stop at site A3), both 27SBs (as stops at sites B1L and B1S), and 25.5S (as the stop at site C2). Probe names are indicated between parentheses (see Fig. 1 for their locations in the 35S pre-rRNA). Signal intensities were measured by scanning, and values obtained (indicated below each lane) were normalized to those obtained for the wild-type strain grown in galactose-sucrose medium, arbitrarily set at 100.
In order to discriminate between the 27SA2 and 27SA3 pre-rRNAs, distinguish between the 27SBL and 27SBS pre-rRNAs, and detect the 25.5S pre-rRNA species, we performed primer extension analyses. As shown in Fig. 4C, the quantities of the cDNAs terminating at sites A3, B1L, and B1S and to a lesser extent those terminating at site A2 were found to be significantly decreased upon depletion of Npa2p. Moreover, primer extension analysis through site C2 showed a clear reduction in the level of the 25.5S pre-rRNA upon depletion of Npa2p.
Pre-rRNA processing was similarly affected in the npa2-1 mutant after a shift for 9 h to 37°C. The 35S and 23S pre-rRNAs accumulated, the levels of the 20S and 27SA precursors were reduced markedly, and the levels of the 7S pre-rRNA, 18S, and 25S rRNA were reduced mildly; however, no clear alteration in the levels of 5.8S and 5S was detected (data not shown).
Altogether, our results suggest that the deficit in 60S r-subunit levels upon depletion or mutation of Npa2p is a consequence of impaired formation of the 27S precursors, thus leading to decreased levels of the mature 25S and 5.8S rRNAs. The phenotypes discussed above are most likely due to improper early assembly and instability of early pre-60S r-particles.
Npa2p is a component of preribosomal particles containing 27SA2 pre-rRNA.
We previously showed that Npa2p is associated with Npa1p, itself predominantly present within very early pre-60S r-particle(s) (19). This finding suggested that Npa2p is also a component of such particles. To confirm this inference, we first analyzed the sedimentation behavior of the Npa2p-TAP fusion protein in low Mg2+ sucrose density gradients. This kind of gradient has proved very useful to study the sedimentation of 60S preribosomal particles (15, 78, 93), which are labile and partially dissociated when subjected to standard polysome sucrose gradients (92) (see below). Strains expressing Npa2p-TAP grow normally, indicating that the tagged protein is functional (data not shown). As shown in Fig. 5A, most Npa2p-TAP was found associated with high-molecular-mass particles. We detected association with high-molecular-mass particles for other proteins, including Dbp6p, Has1p, Npa1p, Nop8p, and Rsa3p (Fig. 5A). All these factors, except Rsa3p, have also been described as components of the Npa1p-TAP purified particle(s) (19). Northern hybridization showed that the maximum of the peak of Npa2p-TAP coincides with the peak of the 27SA2 pre-rRNA (Fig. 5B).
FIG. 5.
Npa2p is present in large complexes, most likely pre-60S r-particles. Total cell extracts were obtained from IVY317 (Npa2p-TAP, Has1p, Rpl3p), IVY325 (Npa1p-HA), IVY347 (HA-Dbp6p), IVY411 (Nop8p-HA), and IVY409 (Rsa3p-HA) cells following growth at 30°C. About 15 A254 units of cell extract were resolved in 7% to 50% sucrose gradients containing a low concentration of Mg2+ to dissociate ribosomes into subunits. Sedimentation is shown from left to right. The sedimentation positions of 40S and 60S ribosomal subunits are indicated. (A) Fractions were collected from the gradients, and proteins were extracted from the same volume of each fraction, resolved on 7% SDS-PAGE gels, and subjected to Western blotting. The blots were decorated with specific antibodies detecting the proteins indicated. (B) RNA was extracted from equal volumes of each fraction, resolved on a 1.2% agarose-formaldehyde gel, and transferred onto a nylon membrane. The same membrane was hybridized with different probes. Probe names are indicated between parentheses. T stands for total extract, and numbers correspond to fraction numbers.
We also performed coimmunoprecipitation experiments with Npa2p-TAP and nontagged control strains and analyzed coprecipitated RNAs by Northern hybridization and primer extension. As is consistent with the previous published data for Npa1p-TAP (19), by far the most efficiently coprecipitated pre-rRNA with Npa2p-TAP was the 27SA2 pre-rRNA (Fig. 6A and B). A small fraction of 35S pre-rRNA could also be coprecipitated (Fig. 6B). The 27SB and 7S precursors were precipitated only at background levels, and no significant precipitation was seen for the snRNA U4, which was used as a negative control (Fig. 6).
FIG. 6.
Npa2p interacts predominantly with the 27SA2 pre-rRNA. (A) Northern analysis of pre-rRNAs and mature rRNAs precipitated with Npa2p-TAP or from extracts lacking a tagged protein. Precipitations were performed using IgG-Sepharose beads and total cellular extracts. RNA was extracted from the pellets obtained after precipitation (lanes IP) or from an amount of total extract corresponding to 1/30 of that used for the precipitation (lanes T). (B) Primer extension analysis of pre-rRNAs precipitated with Npa2p-TAP. Precipitation was performed as described for panel A. (C) Northern analysis of snRNA U4 precipitated with Npa2p-TAP. Precipitation was performed as described for panel A. Signal intensity was measured by phosphorimager scanning to derive the percentage of input RNA precipitated together with Npa2p-TAP (indicated below each IP lane). bg, background value. Probe names are indicated between parentheses.
We conclude that Npa2p is a specific component of a very early pre-60S ribosomal particle(s).
Synthetic lethality with npa1 and npa2 alleles (genetic partners of Npa2p).
To obtain more details about the function of Npa2p, we have tested the synthetic interaction relationships between the npa2-1 mutant and the set of mutants that show genetic interaction with the rsa3 null allele (78). These mutants map in genes for the 60S r-subunit assembly factors Dbp6p, Dbp7p, Dbp9p, Nop8p, and Npa1p and the 60S r-subunit protein Rpl3p (16, 78). As a specificity control, we selected Spb4p, since we had previously shown that there is no synthetic interaction between the spb4-1 mutant and mutant alleles of DBP6, DBP7, or RSA3 (16, 78). As additional controls, we selected different has1 alleles, two nop4 mutants, and the dbp3 and nop6 null alleles (see Table S1 in the supplemental material). Has1p, Nop4p, and Dbp3p are 60S r-subunit biogenesis factors that have been described as components of the Npa1p-TAP purified particle(s) (19). Nop6p is a predicted 60S r-subunit biogenesis factor which was found associated with Npa2p in a large-scale affinity purification study (47).
As a result, we detected synthetic interaction only between npa2-1 and mutant alleles of DBP6, DBP7, DBP9, NOP8, RSA3, and RPL3 but not with npa1-1 (Fig. 7). In addition, none of the combinations of npa2-1 with the tested has1 and nop4 alleles, the spb4-1 mutant, or the dbp3 and nop6 null alleles showed synthetic interaction (Fig. 7). Interestingly, when similar sl analyses were performed with the npa1-1 mutant, we found identical results: the npa1-1 showed synthetic interaction only with mutant alleles of DBP6, DBP7, DBP9, NOP8, RSA3, and RPL3 (Fig. 7). The specific genetic interactions between Npa2p or Npa1p and Dbp6p, Dbp7p, Dbp9p, Nop8p, Rsa3p, and Rpl3p strongly suggest that all these proteins functionally interact during early assembly of 60S r-subunits.
Npa2p forms a complex with other trans-acting factors involved in early steps of 60S r-subunit biogenesis (protein partners of Npa2p).
We next asked whether the set of proteins that genetically interact with Npa2p are also able to interact physically. For this purpose, we first analyzed the sedimentation profile of Npa2p and other 60S r-subunit assembly factors on polysome sucrose gradients. This kind of gradient has been reported to allow dissociation of a number of pre-60S r-particle components; thus, 27S precursors sediment more slowly than 25S rRNA (92) (see Fig. 8B).
FIG. 8.
Npa2p, Npa1p, Dbp6p, Nop8p, and Rsa3p dissociate from preribosomes upon polysome gradient fractionation. Total cell extracts were obtained from IVY317 (Npa2p-TAP, Has1p, and Rpl3p), IVY325 (Npa1p-HA), IVY347 (HA-Dbp6p), IVY411 (Nop8p-HA), and IVY409 (Rsa3p-HA) cells following growth at 30°C. About 15 A254 units of cell extract were resolved in 7% to 50% standard sucrose gradients for polysome analysis. Sedimentation is shown from left to right. The peaks of 40S and 60S ribosomal subunits and 80S couples-monosomes are indicated. The analysis of proteins (A) and RNA (B) from the gradients was performed as described in the legend of Fig. 5.
As expected for a component of a pre-60S ribosomal particle, a fraction of Npa2p-TAP was present in large complexes and cosedimented with 27SA pre-rRNA (Fig. 8A, lanes 12 to 15). However, most Npa2p was reproducibly detected in fractions 6 and 7, where no pre-rRNAs are detected (Fig. 8A, lanes 6 and 7). Interestingly, we found a similar sedimentation pattern for Dbp6p, Npa1p, Nop8p, and Rsa3p (Fig. 8A, lanes 6 and 7) and Dbp7p and Dbp9p (data not shown). A different behavior was observed for Has1p, which is enriched in fractions 11 to 13 containing 27SA2 pre-rRNA, and for Rpl3p, which sedimented with the bulk of the 60S r-subunits and ribosomes (Fig. 8). These results indicate that Npa2p, Dbp6p, Dbp7p, Dbp9p, Nop8p, Npa1p, and Rsa3p are present in pre-60S r-particles but that they can be released from them as a complex(es) of low molecular mass.
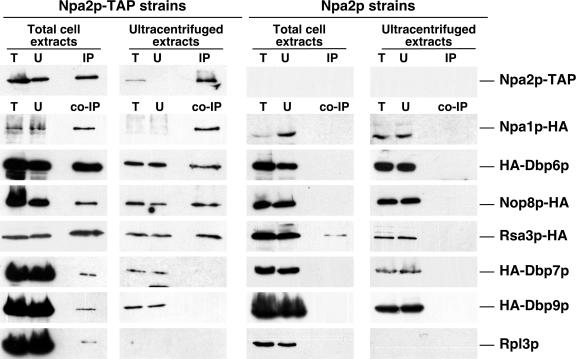
To further analyze this small complex(es), we decided to adopt the procedure described by Hughes and coworkers (59), which allowed them to report the copurification of Npa2p with Npa1p (70). Briefly, large complexes such as preribosomal particles and r-subunits present in total extracts are pelleted by a high-speed centrifugation step (180,000 × g for 45 min) before precipitation of Npa2p-TAP and analysis of coimmunoprecipitating proteins (Fig. 9). When the high-speed centrifugation step was omitted, Dbp6p, Dbp7p, Dbp9p, Nop8p, Npa1p, and Rsa3p were found associated with Npa2p-TAP (Fig. 9). Neither Nog2p, Nop1p, nor Nop7p was found associated with Npa2p-TAP (data not shown). Rpl3p, Rpl12p, and Rpp0p but not Rps8p were found associated with Npa2p-TAP (Fig. 9 and data not shown). Interestingly, when the high-speed centrifugation step was performed, only Dbp6p, Nop8p, Npa1p, and Rsa3p were found associated with Npa2p-TAP. Neither Dbp7p nor Dbp9p and the tested r-proteins coprecipitated with Npa2p-TAP (Fig. 9 and data not shown). When the nontagged Npa2p strains were used as a control, no coimmunoprecipitation was detected except for Rsa3p from total cell extracts (Fig. 9). We suspect that Rsa3p has some unspecific affinity for Sepharose supports. We conclude that Npa2p interacts intimately with Dbp6p, Nop8p, Npa1p, and Rsa3p.
FIG. 9.
Coimmunoprecipitation (co-IP) of Dbp6p, Npa1p, Nop8p, and Rsa3p with Npa2p. Strains IVY317 (Npa2p-TAP and Rpl3p), IVY325 (Npa1p-HA), IVY347 (HA-Dbp6p), IVY411 (Nop8p-HA), IVY409 (Rsa3p-HA), IVY422 (HA-Dbp7p), and IVY423 (HA-Dbp9p) harboring an NPA2-TAP construct (Npa2p-TAP strains) or the untagged NPA2 gene (Npa2p strains) on a plasmid were grown at 30°C to mid-log phase. Cells were broken in a mortar grinder and processed as total or ultracentrifuged extracts by centrifugation as described in Materials and Methods. Extracts were then subjected to immunoprecipitation (IP) with IgG-Sepharose beads. Totals of 0.25% of extract (T), 0.25% of unbound proteins (U), and 1/8 of the immunoprecipitated fraction (IP and co-IP) were subjected to SDS-PAGE and then analyzed by Western blotting using specific antibodies to detect the different proteins.
In order to test whether or not RNA mediates the interaction between the factors discussed above, an RNase treatment of the supernatant obtained following high-speed centrifugation of extracts was performed before immunoprecipitation. As seen in Fig. 10, the interaction between Npa2p and Dbp6p, Nop8p, Npa1p, and Rsa3p is kept even after RNase treatment. We conclude that Npa2p is present in a stable heteromeric subcomplex(es) with Dbp6p, Npa1p, Nop8p, and Rsa3p which is independent of the presence of pre-rRNA.
FIG. 10.
Coimmunoprecipitation (co-IP) of Dbp6p, Npa1p, Nop8p, and Rsa3p with Npa2p is independent of RNA. Strains IVY317 (Npa2p-TAP), IVY325 (Npa1p-HA), IVY347 (HA-Dbp6p), IVY411 (Nop8p-HA), and IVY409 (Rsa3p-HA) harboring a NPA2-TAP construct were grown at 30°C to mid-log phase. Cells were broken in a mortar grinder and processed as ultracentrifuged extracts as described in Materials and Methods. Half of each extract was treated with 100 ng/ml RNase A for 1 h at 4°C, and both treated and untreated extracts were then subjected to immunoprecipitation (IP) with IgG-Sepharose beads. Precipitated proteins were analyzed as described in the legend of Fig. 9.
To define better this subcomplex(es), gel filtration fractionation of RNase-treated high-speed centrifuged extracts was performed by chromatography through a Superose 6HR column and fractions were analyzed by Western blotting. As shown in Fig. 11A, Npa2p-TAP reproducibly peaked in fractions 24 to 28, with a maximum at fraction 26. This indicates that Npa2p is present in a complex of about 550 to 600 kDa. We also studied the elution patterns of Dbp6p, Nop8p, Npa1p, and Rsa3p and found that a substantial proportion of all these proteins coeluted with Npa2p-TAP (Fig. 11A). This was more evident for Dpb6p and Npa1p, while Nop8p and Rsa3p were eluted together in intermediate fractions. The combined molecular masses of Npa2p-TAP and its partner proteins yield a mass of about 500 kDa, which is consistent with a heteromeric nature of the Npa2p-containing complex. Some Dbp6p was reproducibly eluted in fractions 30 to 32, indicating that Dpb6p forms also a smaller complex of about 150 kDa which clearly does not represent monomeric Dbp6p. Some Npa2p-TAP eluted at a position close to the void volume (fraction 16), thus corresponding to a complex of about 1 MDa. This presumably represents the population of Npa2p that it is complexed with the above-mentioned protein partners and fragments of pre-rRNA that are broken during the high-speed ultracentrifugation. Indeed, when the treatment with RNase is avoided after high-speed ultracentrifugation, most Npa2p is detected in fraction 16. Under these experimental conditions, most Dbp6p, Nop8p, Npa1p, and Rsa3p coeluted with Npa2p (Fig. 11B). We suggest that this 1 MDa subcomplex corresponds to the light peak containing Npa2p that is detected upon polysome sucrose gradient fractionation (Fig. 8A, fractions 6 and 7). Interestingly, part of Dbp6p was still eluted in fractions 30 to 32, indicating that the existence of this Dbp6p subcomplex of 150 kDa is independent of the presence of RNA (Fig. 11B).
FIG. 11.
Npa2p forms a discrete complex of about 600 kDa. Strains IVY317 (Npa2p-TAP), IVY325 (Npa1p-HA), IVY347 (HA-Dbp6p), IVY411 (Nop8p-HA), and IVY409 (Rsa3p-HA) harboring a NPA2-TAP construct were grown at 30°C to mid-log phase. (A) Cells were broken in a mortar grinder, processed as ultracentrifuged extracts as described in Materials and Methods, and treated with 100 ng/ml RNase A for 1 h at 4°C. Extracts were then subjected to gel filtration chromatography through a Superose 6 HR column. Fractions were collected and subjected to SDS-PAGE and Western blotting using specific antibodies to detect the different proteins analyzed. T, total loaded extract; numbers correspond to fraction numbers. Arrows indicate elution positions of molecular mass standards. (B) Gel filtration analysis of ultracentrifuged extracts not treated with RNase.
We conclude that Npa2p can be released from early pre-60S r-particles as a >1 MDa complex that contains RNA. An RNase treatment allows the identification of a smaller stable complex of 550 to 600 kDa that contains at least Npa2p, Dbp6p, Nop8p, Npa1p, and Rsa3p.
DISCUSSION
To identify proteins that function together with the putative RNA helicase Dbp6p in the synthesis of 60S r-subunits, we screened for mutations that are sl with the rsa3 null allele, which is in turn sl with selected dbp6 alleles (16). We have characterized in detail the proteins corresponding to most sl mutations that are the 60S r-subunit biogenesis factors, Dbp7p, Dbp9p, Nop8p, Npa1p, Rsa3p, and the 60S r-protein Rpl3p (78). The data presented here establish that alleles of the NPA2/URB2 gene are also sl with rsa3.
We demonstrate that Npa2p is an early-acting nucleolar component of the 60S r-subunit synthesis machinery. Inactivation of Npa2p in the npa2-1 mutant or depletion of Npa2p results in a net deficit in 60S r-subunits. This is due to a strong deficit in the steady-state levels of 27SA2 pre-rRNA and its pre-rRNA derivatives 27SB, 7S, and 25.5S, resulting in greatly diminished levels of mature 25S and 5.8S rRNAs. It seems likely that in the absence of Npa2p assembly of early pre-60S r-particles does not occur properly and therefore that 27SA2 pre-rRNA is efficiently degraded. An aberrant A2-C2 fragment accumulated, suggesting that improper assembly of early pre-60S r-particles also allows premature processing of the 27SA2 pre-rRNA fragment at site C2. Accumulation of the A2-C2 fragment has been previously observed in Ssf1p- and Npa1p-depleted cells and in the cic1-2 and npa1-1 mutants at 37°C (19, 28, 30, 78). In addition, mutation or depletion of Npa2p leads to mild accumulation of 35S pre-rRNA and appearance of aberrant 23S pre-rRNA. This is due to delayed processing at sites A0 to A2, which is frequently observed in most strains deficient for 60S r-subunit accumulation and is most likely the consequence of the lack of proper recycling of trans-acting factors involved in pre-rRNA processing at these sites (34; for further discussion, see reference 101). Mutations or depletion of Dbp6p (56), Dbp7p (12), Dbp9p (11), Nop8p (108), Rsa3p (16, 78), Npa1p (78), and Rpl3p (I. V. Rosado, unpublished results) leads to pre-rRNA processing defects very similar to those of the npa2-defective strains. We believe that Npa2p is a member of a subset of 60S r-subunit assembly factors that operate together, mediating similar structural changes in early 66S preribosomal particles. In agreement with this hypothesis, we found that an Npa2p-TAP construct associates primarily with early 66S preribosomal particles containing 27SA2 pre-rRNAs. A lesser amount of 35S pre-rRNA copurifies with Npa2p-TAP, which is consistent with a role of Npa2p in an earlier assembly step. The lack of association with 27SB pre-rRNAs may reflect the fact that Npa2p dissociates from early 66S preribosomal particles upon 27SA pre-rRNA processing. This idea is further supported by the fact that Nog2p, which associates with pre-60S r-particles after 27SB pre-rRNA is generated (81), does not coimmunoprecipitate with Npa2p (data not shown). Attempts to explore the pre-rRNAs associated with Dbp6p have so far failed (J. de la Cruz, unpublished data; see also reference 16), and analysis of the RNAs associated with Dbp7p, Dbp9p, Nop8p, or Rsa3p has not yet been performed. Npa1p has a pattern of association to pre-60S particles similar to that of Npa2p (19). Moreover, Dbp6p, Dbp7p, Dbp9p, and Nop8p are also stably associated with the Npa1p-containing particle (19), and comparisons with selected purified pre-60S r-particles strongly suggest that at least Dbp7p, Dbp9p, and Nop8p may dissociate from these particles following 27SA pre-rRNA processing (68, 72, 81).
The hypothesis of functional associations between Npa2p and Dbp6p, Dbp7p, Dbp9p, Nop8p, Npa1p, Rsa3p, and Rpl3p is further supported by our genetic data that show synthetic lethality or enhancement in interactions between Npa2p or Npa1p and the rest of the above-named proteins. This clearly reflects specific functional interactions rather than mere cumulative effects on diminishing 60S r-subunit levels. Indeed, previous studies indicate that most combinations of double mutants with 60S r-subunit deficiency do not result in lethality (for examples, see references 1, 16, and 57). Moreover, the npa2 and npa1 mutations are not sl with mutants in other 60S r-subunit biogenesis factors such as Dbp3p (105), Has1p (26), Nop4p (88), and Spb4p (15).
Our data show that Dbp6p, Dbp7p, Dbp9p, Nop8p, Npa1p, and Rsa3p and r-protein Rpl3p, Rpl12p, and Rpp0p are efficiently immunoprecipitated with tagged Npa2p from total cell extracts while Nog2p, Nop1p, Nop7p, and Rps8p are not. This result, together with that of the similar sedimentation patterns of Dbp6p, Dbp7p, Dbp9p, Npa1p, Nop8p, and Rsa3p factors in r-subunit sucrose gradients, strongly suggests that all these proteins exist within early 66S preribosomal particles. Interestingly, Dbp6p, Nop8p, Npa1p, Npa2p, and Rsa3p were found in fractions near the top of polysome sucrose gradients, suggesting that these factors may dissociate from early 66S preribosomal particles during polysome fractionation. Consistent with this idea, we found that Dbp6p, Nop8p, Npa1p, and Rsa3p were still efficiently immunoprecipitated with tagged Npa2p from high-speed supernatants. This result confirms the data by Mnaimneh et al. (70), who reported an Npa1p-Npa2p interaction, and extends the list of partners to Dbp6p, Nop8p, and Rsa3p. In addition, it suggests that all these proteins could be organized in a discrete subcomplex. One important observation is that this subcomplex contains RNA, most likely in the form of fragments of 27SA pre-rRNA. However, the integrity of the subcomplex is RNA independent, since the interaction is not disrupted even after an exhaustive RNase treatment. Gel filtration chromatography of RNase-treated high-speed supernatants further revealed that the Npa2p-containing subcomplex has an apparent molecular mass of about 550 to 600 kDa and that substantial amounts of Dbp6p, Nop8p, Npa1p, and Rsa3p were eluted together with Npa2p, which strengthens the hypothesis that these four proteins are components of the complex. We also found Dbp6p in a smaller complex of about 150 kDa. Further studies are required to define the nature of this smaller complex. We conclude that Dbp6p, Nop8p, Npa1p, Npa2p, and Rsa3p define both a structural and functional subcomplex involved in 60S r-subunit biogenesis. Our results are consistent with the recent data from a genome-wide characterization of yeast protein complexes (38). Thus, when we make use of the informatic tool described in that reference to analyze core components of protein macrocomplexes (see http://yeast-complexes.embl.de), we find Npa2p in the same network as Dbp6p, Nop8p, Npa1p, and Rsa3p among other most likely unrelated proteins such as Arg1p, Leu4p, Leu9p, and Pcr1p. Neither Gavin et al. nor we have found evidence for a genetic or physical interaction of the Npa2p-containing subcomplex with Nip7p (reference 38 and data not shown). Nip7p is a nucleolar 60S r-subunit biogenesis factor (109) which has been described as interacting with Nop8p, as determined by two-hybrid analysis and coimmunoprecipitation experiments using total cell extracts (108). Consistent with a lack of functional relationship between the Npa2p-containing subcomplex and Nip7p, the deficiency of Nip7p leads to a delayed processing of 27SB pre-rRNA species instead of instability of 27SA2 pre-rRNA (109).
The specific sl interactions between rpl3 alleles and mutants in all components of the Npa2p-containing subcomplex, Dbp7p, and Dbp9p may result from the fact that these factors are required for the efficient assembly of Rpl3p into early 66S preribosomal particles. The Npa2p-containing complex, Dbp7p, and Dbp9p could allow dissociation of Rpl3p from Rrb1p, which has been proposed to function as the chaperone for free Rpl3p prior to or during its loading onto pre-60S r-subunits (51, 83). In agreement with this model, the rrb1-TAP allele, which displays a sg and a ts phenotype (83), is sl with dbp6, dbp9, npa1, npa2, and nop8 alleles and enhances the growth defect of dbp7 and rsa3 null alleles (I. V. Rosado and J. de la Cruz, unpublished results). Both genetic and molecular approaches are required to study properly how Rpl3p assembles onto nascent 60S r-subunits.
Few additional low-molecular-mass complexes involved in yeast ribosome biogenesis have been reported so far. In addition to snoRNPs, RNase P, and RNase MRP (for reviews, see references 6 and 107), only a few other complexes have been analyzed in depth: the exosome (for a review, see reference 69), the UTP A-C subcomplexes (59), the t-UTP complex (37), the Erb1p-Nop7p-Ytm1p subcomplex (59, 66), the Arx1p-Alb1p complex (64), and the Rix1p complex (36, 59). However, subcomplexes are likely a common feature in ribosome biogenesis. This hypothesis is supported by the fact that many r-subunit biogenesis factors fractionate near the top of polysome sucrose gradients. Throughout the literature, we have found many examples of such factors, including Noc1p (67), Nop53p (91), Rix7p (34), Rrp1p (49), Rcl1p (7), and Ssf1p (28). Moreover, some of these complexes are conserved in higher eukaryotes, such as the exosome (10) or the PeBoW complex consisting of the proteins WDR12, Bop1, and Pes1, which are the mammalian homologues of Ytm1p, Erb1p, and Nop7p, respectively (48, 62). Regarding the Npa2p-containing subcomplex, so far, recognizable mammalian homologues have been identified for Nop8p and Npa1p (19, 85). Further work is needed to determine whether mammalian Nop8p and Npa1p interact with each other and putative mammalian homologues of Dpb6p, Npa2p, and Rsa3p. Moreover, the issue has to be addressed of whether the Npa2p-containing subcomplex and the complexes mentioned above exist independently of preribosomes or only in the context of preribosomal particles.
In conclusion, we show that Npa2p is necessary for early steps in the biogenesis of 60S r-subunits and present evidence for a direct functional and physical interaction between 60S r-subunit biogenesis factors Nop8p, Npa1p, Npa2p, and Rsa3p and the putative RNA helicase Dbp6p. Recently, it has been reported that the nucleolar protein Esf2p interacts directly with the putative RNA helicase Dbp8p and that this interaction substantially increases Dbp8p ATPase activity (42). Similarly to Dbp8p, Dbp5p requires activation by Gle1p and inositol hexakisphosphate (2, 106) and eIF4A by eIF4B, eIF4H, and eIF4F (77). Further studies are required to determine whether or not Nop8p, Npa2p, Npa1p, and Rsa3p modulate the in vivo and in vitro activities of Dbp6p.
Supplementary Material
Acknowledgments
We are indebted to all colleagues who contributed plasmids and reagents to this study. We are grateful to members of the Chávez laboratory for discussions, S. Diaz-Troya and J. L. Crespo for their valued help with high-performance liquid chromatography, and M. J. Quintero for technical assistance.
This work was supported by grants from the Spanish Ministry of Science and Technology and FEDER (BFU2004-00252/BMC) to J.D.L.C. and from l'Agence Nationale de la Recherche and from La Ligue Nationale contre le Cancer (Equipe Labelisée) to Y.H. We also thank the Andalusian Government (CVI271), the CNRS, and the Université Paul Sabatier for support.
Footnotes
Published ahead of print on 4 December 2006.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Adams, C. C., J. Jakovljevic, J. Roman, P. Harnpicharnchai, and J. L. Woolford, Jr. 2002. Saccharomyces cerevisiae nucleolar protein Nop7p is necessary for biogenesis of 60S ribosomal subunits. RNA 8:150-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alcázar-Román, A. R., E. J. Tran, S. Guo, and S. R. Wente. 2006. Inositol hexakisphosphate and Gle1 activate the DEAD-box protein Dbp5 for nuclear mRNA export. Nat. Cell Biol. 8:711-716. [DOI] [PubMed] [Google Scholar]
- 3.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1994. Saccharomyces cerevisiae, p. 13.0.1-13.14.17. In Current protocols in molecular biology, vol. 2. John Wiley & Sons, Inc., New York, NY. [Google Scholar]
- 4.Bassler, J., P. Grandi, O. Gadal, T. Lessmann, E. Petfalski, D. Tollervey, J. Lechner, and E. Hurt. 2001. Identification of a 60S preribosomal particle that is closely linked to nuclear export. Mol. Cell 8:517-529. [DOI] [PubMed] [Google Scholar]
- 5.Bernstein, K. A., J. E. Gallagher, B. M. Mitchell, S. Granneman, and S. J. Baserga. 2004. The small-subunit processome is a ribosome assembly intermediate. Eukaryot. Cell 3:1619-1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bertrand, E., and M. J. Fournier. 2004. The snoRNPs and related machines: ancient devices that mediate maturation of rRNA and other RNAs, p. 222-256. In M. O. J. Olson (ed.), Nucleolus. Kluwer Academic Publishers, New York, NY.
- 7.Billy, E., T. Wegierski, F. Nasr, and W. Filipowicz. 2000. Rcl1p, the yeast protein similar to the RNA 3′-phosphate cyclase, associates with U3 snoRNP and is required for 18S rRNA biogenesis. EMBO J. 19:2115-2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brand, R. C., J. Klootwijk, T. J. M. van Steenbergen, A. J. de Kok, and R. J. Planta. 1977. Secondary methylation of yeast ribosomal precursor RNA. Eur. J. Biochem. 75:311-318. [DOI] [PubMed] [Google Scholar]
- 9.Briones, E., C. Briones, M. Remacha, and J. P. G. Ballesta. 1998. The GTPase center protein L12 is required for correct ribosomal stalk assembly but not for Saccharomyces cerevisiae viability. J. Biol. Chem. 273:31956-31961. [DOI] [PubMed] [Google Scholar]
- 10.Brouwer, R., G. J. Pruijn, and W. J. van Venrooij. 2001. The human exosome: an autoantigenic complex of exoribonucleases in myositis and scleroderma. Arthritis Res. 3:102-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Daugeron, M.-C., D. Kressler, and P. Linder. 2001. Dbp9p, a putative ATP-dependent RNA helicase involved in 60S-ribosomal-subunit biogenesis, functionally interacts with Dbp6p. RNA 7:1317-1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Daugeron, M.-C., and P. Linder. 1998. Dbp7p, a putative ATP-dependent RNA helicase from Saccharomyces cerevisiae, is required for 60S ribosomal subunit assembly. RNA 4:566-581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Decatur, W. A., and M. J. Fournier. 2002. rRNA modifications and ribosome function. Trends Biochem. Sci. 27:344-351. [DOI] [PubMed] [Google Scholar]
- 14.de la Cruz, J., D. Kressler, and P. Linder. 2004. Ribosomal subunit assembly, p. 258-285. In M. O. J. Olson (ed.), Nucleolus. Kluwer Academic Publishers, New York, NY.
- 15.de la Cruz, J., D. Kressler, M. Rojo, D. Tollervey, and P. Linder. 1998. Spb4p, an essential putative RNA helicase, is required for a late step in the assembly of 60S ribosomal subunits in Saccharomyces cerevisiae. RNA 4:1268-1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de la Cruz, J., T. Lacombe, O. Deloche, P. Linder, and D. Kressler. 2004. The putative RNA helicase Dbp6p functionally interacts with Rpl3p, Nop8p and the novel trans-acting factor Rsa3p during biogenesis of 60S ribosomal subunits in Saccharomyces cerevisiae. Genetics 166:1687-1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deshmukh, M., J. Stark, L. C. Yeh, J. C. Lee, and J. L. Woolford, Jr. 1995. Multiple regions of yeast ribosomal protein L1 are important for its interaction with 5S rRNA and assembly into ribosomes. J. Biol. Chem. 270:30148-30156. [DOI] [PubMed] [Google Scholar]
- 18.Deshmukh, M., Y.-F. Tsay, A. G. Paulovich, and J. L. Woolford, Jr. 1993. Yeast ribosomal protein L1 is required for the stability of newly synthesized 5S rRNA and the assembly of 60S ribosomal subunits. Mol. Cell. Biol. 13:2835-2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dez, C., C. Froment, J. Noaillac-Depeyre, B. Monsarrat, M. Caizergues-Ferrer, and Y. Henry. 2004. Npa1p, a component of very early pre-60S ribosomal particles, associates with a subset of small nucleolar RNPs required for peptidyl transferase center modification. Mol. Cell. Biol. 24:6324-6337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dez, C., J. Noaillac-Depeyre, M. Caizergues-Ferrer, and Y. Henry. 2002. Naf1p, an essential nucleoplasmic factor specifically required for accumulation of box H/ACA small nucleolar RNPs. Mol. Cell. Biol. 22:7053-7065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dez, C., and D. Tollervey. 2004. Ribosome synthesis meets the cell cycle. Curr. Opin. Microbiol. 7:631-637. [DOI] [PubMed] [Google Scholar]
- 22.Dong, J., R. Lai, J. L. Jennings, A. J. Link, and A. G. Hinnebusch. 2005. The novel ATP-binding cassette protein ARB1 is a shuttling factor that stimulates 40S and 60S ribosome biogenesis. Mol. Cell. Biol. 25:9859-9873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dragon, F., J. E. Gallagher, P. A. Compagnone-Post, B. M. Mitchell, K. A. Porwancher, K. A. Wehner, S. Wormsley, R. E. Settlage, J. Shabanowitz, Y. Osheim, A. L. Beyer, D. F. Hunt, and S. J. Baserga. 2002. A large nucleolar U3 ribonucleoprotein required for 18S ribosomal RNA biogenesis. Nature 417:967-970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Du, Y. C., and B. Stillman. 2002. Yph1p, an ORC-interacting protein: potential links between cell proliferation control, DNA replication, and ribosome biogenesis. Cell 109:835-848. [DOI] [PubMed] [Google Scholar]
- 25.Eisinger, D. P., F. A. Dick, and B. L. Trumpower. 1997. Qsr1p, a 60S ribosomal subunit protein, is required for joining of 40S and 60S subunits. Mol. Cell. Biol. 17:5136-5145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Emery, B., J. de la Cruz, S. Rocak, O. Deloche, and P. Linder. 2004. Has1p, a member of the DEAD-box family, is required for 40S ribosomal subunit biogenesis in Saccharomyces cerevisiae. Mol. Microbiol. 52:141-158. [DOI] [PubMed] [Google Scholar]
- 27.Fath, S., P. Milkereit, A. V. Podtelejnikov, N. Bischler, P. Schultz, M. Bier, M. Mann, and H. Tschochner. 2000. Association of yeast RNA polymerase I with a nucleolar substructure active in rRNA synthesis and processing. J. Cell Biol. 149:575-590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fatica, A., A. D. Cronshaw, M. Dlakic, and D. Tollervey. 2002. Ssf1p prevents premature processing of an early pre-60S ribosomal particle. Mol. Cell 9:341-351. [DOI] [PubMed] [Google Scholar]
- 29.Fatica, A., M. Oeffinger, M. Dlakic, and D. Tollervey. 2003. Nob1p is required for cleavage of the 3′ end of 18S rRNA. Mol. Cell. Biol. 23:1798-1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fatica, A., M. Oeffinger, D. Tollervey, and I. Bozzoni. 2003. Cic1p/Nsa3p is required for synthesis and nuclear export of 60S ribosomal subunits. RNA 9:1431-1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fatica, A., and D. Tollervey. 2002. Making ribosomes. Curr. Opin. Cell Biol. 14:313-318. [DOI] [PubMed] [Google Scholar]
- 32.Ferreira-Cerca, S., G. Poll, P. E. Gleizes, H. Tschochner, and P. Milkereit. 2005. Roles of eukaryotic ribosomal proteins in maturation and transport of pre-18S rRNA and ribosome function. Mol. Cell 20:263-275. [DOI] [PubMed] [Google Scholar]
- 33.Fromont-Racine, M., B. Senger, C. Saveanu, and F. Fasiolo. 2003. Ribosome assembly in eukaryotes. Gene 313:17-42. [DOI] [PubMed] [Google Scholar]
- 34.Gadal, O., D. Strauss, J. Braspenning, D. Hoepfner, E. Petfalski, P. Philippsen, D. Tollervey, and E. Hurt. 2001. A nuclear AAA-type ATPase (Rix7p) is required for biogenesis and nuclear export of 60S ribosomal subunits. EMBO J. 20:3695-3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gadal, O., D. Strauβ, J. Kessl, B. Trumpower, D. Tollervey, and E. Hurt. 2001. Nuclear export of 60S ribosomal subunit depends on Xpo1p and requires a nuclear export sequence-containing factor, Nmd3p, that associates with the large subunit protein Rpl10p. Mol. Cell. Biol. 21:3405-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Galani, K., T. A. Nissan, E. Petfalski, D. Tollervey, and E. Hurt. 2004. Rea1, a dynein-related nuclear AAA-ATPase, is involved in late rRNA processing and nuclear export of 60 S subunits. J. Biol. Chem. 279:55411-55418. [DOI] [PubMed] [Google Scholar]
- 37.Gallagher, J. E., D. A. Dunbar, S. Granneman, B. M. Mitchell, Y. Osheim, A. L. Beyer, and S. J. Baserga. 2004. RNA polymerase I transcription and pre-rRNA processing are linked by specific SSU processome components. Genes Dev. 18:2506-2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gavin, A. C., P. Aloy, P. Grandi, R. Krause, M. Boesche, M. Marzioch, C. Rau, L. J. Jensen, S. Bastuck, B. Dumpelfeld, A. Edelmann, M. A. Heurtier, V. Hoffman, C. Hoefert, K. Klein, M. Hudak, A. M. Michon, M. Schelder, M. Schirle, M. Remor, T. Rudi, S. Hooper, A. Bauer, T. Bouwmeester, G. Casari, G. Drewes, G. Neubauer, J. M. Rick, B. Kuster, P. Bork, R. B. Russell, and G. Superti-Furga. 2006. Proteome survey reveals modularity of the yeast cell machinery. Nature 440:631-636. [DOI] [PubMed] [Google Scholar]
- 39.Gerbi, S. A., and A. V. Borovjagin. 2004. Pre-ribosomal RNA processing in multicellular organisms. In M. O. J. Olson (ed.), Nucleolus. Kluwer Academic Publishers, New York, NY.
- 40.Gietz, R. D., and A. Sugino. 1988. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene 74:527-534. [DOI] [PubMed] [Google Scholar]
- 41.Grandi, P., V. Rybin, J. Bassler, E. Petfalski, D. Strauss, M. Marzioch, T. Schafer, B. Kuster, H. Tschochner, D. Tollervey, A. C. Gavin, and E. Hurt. 2002. 90S pre-ribosomes include the 35S pre-rRNA, the U3 snoRNP, and 40S subunit processing factors but predominantly lack 60S synthesis factors. Mol. Cell 10:105-115. [DOI] [PubMed] [Google Scholar]
- 42.Granneman, S., C. Lin, E. A. Champion, M. R. Nandineni, C. Zorca, and S. J. Baserga. 2006. The nucleolar protein Esf2 interacts directly with the DExD/H box RNA helicase, Dbp8, to stimulate ATP hydrolysis. Nucleic Acids Res. 34:3189-3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Granneman, S., M. R. Nandineni, and S. J. Baserga. 2005. The putative NTPase Fap7 mediates cytoplasmic 20S pre-rRNA processing through a direct interaction with Rps14. Mol. Cell. Biol. 25:10352-10364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Harnpicharnchai, P., J. Jakovljevic, E. Horsey, T. Miles, J. Roman, M. Rout, D. Meagher, B. Imai, Y. Guo, C. J. Brame, J. Shabanowitz, D. F. Hunt, and J. L. Woolford, Jr. 2001. Composition and functional characterization of yeast 66S ribosome assembly intermediates. Mol. Cell 8:505-515. [DOI] [PubMed] [Google Scholar]
- 45.Hazbun, T. R., L. Malmstrom, S. Anderson, B. J. Graczyk, B. Fox, M. Riffle, B. A. Sundin, J. D. Aranda, W. H. McDonald, C. H. Chiu, B. E. Snydsman, P. Bradley, E. G. Muller, S. Fields, D. Baker, J. R. Yates III, and T. N. Davis. 2003. Assigning function to yeast proteins by integration of technologies. Mol. Cell 12:1353-1365. [DOI] [PubMed] [Google Scholar]
- 46.Hedges, J., M. West, and A. W. Johnson. 2005. Release of the export adapter, Nmd3p, from the 60S ribosomal subunit requires Rpl10p and the cytoplasmic GTPase Lsg1p. EMBO J. 24:567-579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ho, Y., A. Gruhler, A. Heilbut, G. D. Bader, L. Moore, S. L. Adams, A. Millar, P. Taylor, K. Bennett, K. Boutilier, L. Yang, C. Wolting, I. Donaldson, S. Schandorff, J. Shewnarane, M. Vo, J. Taggart, M. Goudreault, B. Muskat, C. Alfarano, D. Dewar, Z. Lin, K. Michalickova, A. R. Willems, H. Sassi, P. A. Nielsen, K. J. Rasmussen, J. R. Andersen, L. E. Johansen, L. H. Hansen, H. Jespersen, A. Podtelejnikov, E. Nielsen, J. Crawford, V. Poulsen, B. D. Sorensen, J. Matthiesen, R. C. Hendrickson, F. Gleeson, T. Pawson, M. F. Moran, D. Durocher, M. Mann, C. W. Hogue, D. Figeys, and M. Tyers. 2002. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415:180-183. [DOI] [PubMed] [Google Scholar]
- 48.Hölzel, M., M. Rohrmoser, M. Schlee, T. Grimm, T. Harasim, A. Malamoussi, A. Gruber-Eber, E. Kremmer, W. Hiddemann, G. W. Bornkamm, and D. Eick. 2005. Mammalian WDR12 is a novel member of the Pes1-Bop1 complex and is required for ribosome biogenesis and cell proliferation. J. Cell Biol. 170:367-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Horsey, E. W., J. Jakovljevic, T. D. Miles, P. Harnpicharnchai, and J. L. Woolford, Jr. 2004. Role of the yeast Rrp1 protein in the dynamics of pre-ribosome maturation. RNA 10:813-827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huh, W. K., J. V. Falvo, L. C. Gerke, A. S. Carroll, R. W. Howson, J. S. Weissman, and E. K. O'Shea. 2003. Global analysis of protein localization in budding yeast. Nature 425:686-691. [DOI] [PubMed] [Google Scholar]
- 51.Iouk, T. L., J. D. Aitchison, S. Maguire, and R. W. Wozniak. 2001. Rrb1p, a yeast nuclear WD-repeat protein involved in the regulation of ribosome synthesis. Mol. Cell. Biol. 21:1260-1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jakovljevic, J., P. A. de Mayolo, T. D. Miles, T. M. Nguyen, I. Leger-Silvestre, N. Gas, and J. L. Woolford, Jr. 2004. The carboxy-terminal extension of yeast ribosomal protein S14 is necessary for maturation of 43S preribosomes. Mol. Cell 14:331-342. [DOI] [PubMed] [Google Scholar]
- 53.Jorgensen, P., J. L. Nishikawa, B. J. Breitkreutz, and M. Tyers. 2002. Systematic identification of pathways that couple cell growth and division in yeast. Science 297:395-400. [DOI] [PubMed] [Google Scholar]
- 54.Kaiser, C., S. Michaelis, and A. Mitchell. 1994. Methods in yeast genetics: a Cold Spring Harbor Laboratory course manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 55.Kallstrom, G., J. Hedges, and A. Johnson. 2003. The putative GTPases Nog1p and Lsg1p are required for 60S ribosomal subunit biogenesis and are localized to the nucleus and cytoplasm, respectively. Mol. Cell. Biol. 23:4344-4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kressler, D., J. de la Cruz, M. Rojo, and P. Linder. 1998. Dbp6p is an essential putative ATP-dependent RNA helicase required for 60S-ribosomal-subunit assembly in Saccharomyces cerevisiae. Mol. Cell. Biol. 18:1855-1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kressler, D., M. Doère, M. Rojo, and P. Linder. 1999. Synthetic lethality with conditional dbp6 alleles identifies the previously uncharacterized RSA1 gene, encoding a protein involved in a late nucleoplasmic step of 60S-ribosomal-subunit assembly. Mol. Cell. Biol. 19:8633-8645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kressler, D., P. Linder, and J. de la Cruz. 1999. Protein trans-acting factors involved in ribosome biogenesis in Saccharomyces cerevisiae. Mol. Cell. Biol. 19:7897-7912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Krogan, N. J., W. T. Peng, G. Cagney, M. D. Robinson, R. Haw, G. Zhong, X. Guo, X. Zhang, V. Canadien, D. P. Richards, B. K. Beattie, A. Lalev, W. Zhang, A. P. Davierwala, S. Mnaimneh, A. Starostine, A. P. Tikuisis, J. Grigull, N. Datta, J. E. Bray, T. R. Hughes, A. Emili, and J. F. Greenblatt. 2004. High-definition macromolecular composition of yeast RNA-processing complexes. Mol. Cell 13:225-239. [DOI] [PubMed] [Google Scholar]
- 60.Kruiswijk, T., R. J. Planta, and J. M. Krop. 1978. The course of the assembly of ribosomal subunits in yeast. Biochim. Biophys. Acta 517:378-389. [DOI] [PubMed] [Google Scholar]
- 61.Lafontaine, D., and D. Tollervey. 1996. One-step PCR mediated strategy for the construction of conditionally expressed and epitope tagged yeast proteins. Nucleic Acids Res. 24:3469-3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lapik, Y. R., C. J. Fernandes, L. F. Lau, and D. G. Pestov. 2004. Physical and functional interaction between Pes1 and Bop1 in mammalian ribosome biogenesis. Mol. Cell 15:17-29. [DOI] [PubMed] [Google Scholar]
- 63.Lebaron, S., C. Froment, M. Fromont-Racine, J. C. Rain, B. Monsarrat, M. Caizergues-Ferrer, and Y. Henry. 2005. The splicing ATPase Prp43p is a component of multiple preribosomal particles. Mol. Cell. Biol. 25:9269-9282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lebreton, A., C. Saveanu, L. Decourty, J. C. Rain, A. Jacquier, and M. Fromont-Racine. 2006. A functional network involved in the recycling of nucleocytoplasmic pre-60S factors. J. Cell Biol. 173:349-360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Léger-Silvestre, I., P. Milkereit, S. Ferreira-Cerca, C. Saveanu, J. C. Rousselle, V. Choesmel, C. Guinefoleau, N. Gas, and P. E. Gleizes. 2004. The ribosomal protein Rps15p is required for nuclear exit of the 40S subunit precursors in yeast. EMBO J. 23:2336-2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Miles, T. D., J. Jakovljevic, E. W. Horsey, P. Harnpicharnchai, L. Tang, and J. L. Woolford, Jr. 2005. Ytm1, Nop7, and Erb1 form a complex necessary for maturation of yeast 66S preribosomes. Mol. Cell. Biol. 25:10419-10432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Milkereit, P., O. Gadal, A. Podtelejnikov, S. Trumtel, N. Gas, E. Petfalski, D. Tollervey, M. Mann, E. Hurt, and H. Tschochner. 2001. Maturation and intranuclear transport of pre-ribosomes requires Noc proteins. Cell 105:499-509. [DOI] [PubMed] [Google Scholar]
- 68.Milkereit, P., H. Kuhn, N. Gas, and H. Tschochner. 2003. The pre-ribosomal network. Nucleic Acids Res. 31:799-804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mitchell, P., and D. Tollervey. 2000. Musing on the structural organization of the exosome complex. Nat. Struct. Biol. 7:843-846. [DOI] [PubMed] [Google Scholar]
- 70.Mnaimneh, S., A. P. Davierwala, J. Haynes, J. Moffat, W. T. Peng, W. Zhang, X. Yang, J. Pootoolal, G. Chua, A. Lopez, M. Trochesset, D. Morse, N. J. Krogan, S. L. Hiley, Z. Li, Q. Morris, J. Grigull, N. Mitsakakis, C. J. Roberts, J. F. Greenblatt, C. Boone, C. A. Kaiser, B. J. Andrews, and T. R. Hughes. 2004. Exploration of essential gene functions via titratable promoter alleles. Cell 118:31-44. [DOI] [PubMed] [Google Scholar]
- 71.Moritz, M., B. A. Pulaski, and J. L. Woolford, Jr. 1991. Assembly of 60S ribosomal subunits is perturbed in temperature-sensitive yeast mutants defective in ribosomal protein L16. Mol. Cell. Biol. 11:5681-5692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nissan, T. A., J. Bassler, E. Petfalski, D. Tollervey, and E. Hurt. 2002. 60S pre-ribosome formation viewed from assembly in the nucleolus until export to the cytoplasm. EMBO J. 21:5539-5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nomura, M., Y. Nogi, and M. Oakes. 2004. Transcription of rDNA in the yeast Saccharomyces cerevisiae, p. 128-153. In M. O. J. Olson (ed.), Nucleolus. Kluwer Academic Publishers, New York, NY.
- 74.Peng, W. T., M. D. Robinson, S. Mnaimneh, N. J. Krogan, G. Cagney, Q. Morris, A. P. Davierwala, J. Grigull, X. Yang, W. Zhang, N. Mitsakakis, O. W. Ryan, N. Datta, V. Jojic, C. Pal, V. Canadien, D. Richards, B. Beattie, L. F. Wu, S. J. Altschuler, S. Roweis, B. J. Frey, A. Emili, J. F. Greenblatt, and T. R. Hughes. 2003. A panoramic view of yeast noncoding RNA processing. Cell 113:919-933. [DOI] [PubMed] [Google Scholar]
- 75.Raué, H. A. 2004. Pre-ribosomal RNA processing and assembly in Saccharomyces cerevisiae: the machine that makes the machine, p. 199-222. In M. O. J. Olson (ed.), Nucleolus. Kluwer Academic Publishers, New York, NY.
- 76.Rigaut, G., A. Shevchenko, B. Rutz, M. Wilm, M. Mann, and B. Séraphin. 1999. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17:1030-1032. [DOI] [PubMed] [Google Scholar]
- 77.Rogers, G. W., Jr., N. J. Richter, W. F. Lima, and W. C. Merrick. 2001. Modulation of the helicase activity of eIF4A by eIF4B, eIF4H, and eIF4F. J. Biol. Chem. 276:30914-30922. [DOI] [PubMed] [Google Scholar]
- 78.Rosado, I. V., and J. de la Cruz. 2004. Npa1p is an essential trans-acting factor required for an early step in the assembly of 60S ribosomal subunits in Saccharomyces cerevisiae. RNA 10:1073-1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 80.Saveanu, C., D. Bienvenu, A. Namane, P. E. Gleizes, N. Gas, A. Jacquier, and M. Fromont-Racine. 2001. Nog2p, a putative GTPase associated with pre-60S subunits and required for late 60S maturation steps. EMBO J. 20:6475-6484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Saveanu, C., A. Namane, P. E. Gleizes, A. Lebreton, J. C. Rousselle, J. Noaillac-Depeyre, N. Gas, A. Jacquier, and M. Fromont-Racine. 2003. Sequential protein association with nascent 60S ribosomal particles. Mol. Cell. Biol. 23:4449-4460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Schäfer, T., D. Strauss, E. Petfalski, D. Tollervey, and E. Hurt. 2003. The path from nucleolar 90S to cytoplasmic 40S pre-ribosomes. EMBO J. 22:1370-1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Schaper, S., M. Fromont-Racine, P. Linder, J. de la Cruz, A. Namade, and M. Yaniv. 2001. A yeast homolog of chromatin assembly factor 1 is involved in early ribosome assembly. Curr. Biol. 11:1885-1890. [DOI] [PubMed] [Google Scholar]
- 84.Schultz, M. C. 1999. Chromatin assembly in yeast cell-free extracts. Methods 17:161-172. [DOI] [PubMed] [Google Scholar]
- 85.Sekiguchi, T., Y. Todaka, Y. Wang, E. Hirose, N. Nakashima, and T. Nishimoto. 2004. A novel human nucleolar protein, Nop132, binds to the G proteins, RRAG A/C/D. J. Biol. Chem. 279:8343-8350. [DOI] [PubMed] [Google Scholar]
- 86.Senger, B., D. L. Lafontaine, J. S. Graindorge, O. Gadal, A. Camasses, A. Sanni, J. M. Garnier, M. Breitenbach, E. Hurt, and F. Fasiolo. 2001. The nucle(ol)ar Tif6p and Efl1p are required for a late cytoplasmic step of ribosome synthesis. Mol. Cell 8:1363-1373. [DOI] [PubMed] [Google Scholar]
- 87.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sun, C., and J. L. Woolford, Jr. 1994. The yeast NOP4 gene product is an essential nucleolar protein required for pre-rRNA processing and accumulation of 60S ribosomal subunits. EMBO J. 13:3127-3135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tabb, A. L., T. Utsugi, C. R. Wooten-Kee, T. Sasaki, S. A. Edling, W. Gump, Y. Kikuchi, and S. R. Ellis. 2001. Genes encoding ribosomal proteins Rps0A/B of Saccharomyces cerevisiae interact with TOM1 mutants defective in ribosome synthesis. Genetics 157:1107-1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Takahashi, N., M. Yanagida, S. Fujiyama, T. Hayano, and T. Isobe. 2003. Proteomic snapshot analyses of preribosomal ribonucleoprotein complexes formed at various stages of ribosome biogenesis in yeast and mammalian cells. Mass Spectrom. Rev. 22:287-317. [DOI] [PubMed] [Google Scholar]
- 91.Thomson, E., and D. Tollervey. 2005. Nop53p is required for late 60S ribosome subunit maturation and nuclear export in yeast. RNA 11:1215-1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Tollervey, D., H. Lehtonen, R. Jansen, H. Kern, and E. C. Hurt. 1993. Temperature-sensitive mutations demonstrate roles for yeast fibrillarin in pre-rRNA processing, pre-rRNA methylation, and ribosome assembly. Cell 72:443-457. [DOI] [PubMed] [Google Scholar]
- 93.Torchet, C., and S. Hermann-Le Denmat. 2000. Bypassing the rRNA processing endonucleolytic cleavage at site A2 in Saccharomyces cerevisiae. RNA 6:1498-1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Trapman, J., J. Retèl, and R. J. Planta. 1975. Ribosomal precursor particles from yeast. Exp. Cell Res. 90:95-104. [DOI] [PubMed] [Google Scholar]
- 95.Tschochner, H., and E. Hurt. 2003. Pre-ribosomes on the road from the nucleolus to the cytoplasm. Trends Cell Biol. 13:255-263. [DOI] [PubMed] [Google Scholar]
- 96.Udem, S. A., and J. R. Warner. 1973. The cytoplasmic maturation of a ribosomal precursor ribonucleic acid in yeast. J. Biol. Chem. 248:1412-1416. [PubMed] [Google Scholar]
- 97.van Beekvelt, C. A., M. de Graaff-Vincent, A. W. Faber, J. van't Riet, J. Venema, and H. A. Raué. 2001. All three functional domains of the large ribosomal subunit protein L25 are required for both early and late pre-rRNA processing steps in Saccharomyces cerevisiae. Nucleic Acids Res. 29:5001-5008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Vanrobays, E., J. P. Gelugne, P. E. Gleizes, and M. Caizergues-Ferrer. 2003. Late cytoplasmic maturation of the small ribosomal subunit requires RIO proteins in Saccharomyces cerevisiae. Mol. Cell. Biol. 23:2083-2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Venema, J., R. J. Planta, and H. A. Raué. 1998. In vivo mutational analysis of ribosomal RNA in Saccharomyces cerevisiae, p. 257-270. In R. Martin (ed.), Protein synthesis: methods and protocols, vol. 77. Humana Press, Totowa, NJ. [DOI] [PubMed] [Google Scholar]
- 100.Venema, J., and D. Tollervey. 1995. Processing of pre-ribosomal RNA in Saccharomyces cerevisiae. Yeast 11:1629-1650. [DOI] [PubMed] [Google Scholar]
- 101.Venema, J., and D. Tollervey. 1999. Ribosome synthesis in Saccharomyces cerevisiae. Annu. Rev. Genet. 33:261-311. [DOI] [PubMed] [Google Scholar]
- 102.Vilardell, J., and J. R. Warner. 1997. Ribosomal protein L32 of Saccharomyces cerevisiae influences both the splicing of its own transcript and the processing of rRNA. Mol. Cell. Biol. 17:1959-1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Warner, J. R. 1999. The economics of ribosome biosynthesis in yeast. Trends Biochem. Sci. 24:437-440. [DOI] [PubMed] [Google Scholar]
- 104.Warner, J. R., and R. Soeiro. 1967. Nascent ribosomes from HeLa cells. Proc. Natl. Acad. Sci. USA 58:1984-1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Weaver, P. L., C. Sun, and T.-H. Chang. 1997. Dbp3p, a putative RNA helicase in Saccharomyces cerevisiae, is required for efficient pre-rRNA processing predominantly at site A3. Mol. Cell. Biol. 17:1354-1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Weirich, C. S., J. P. Erzberger, J. S. Flick, J. M. Berger, J. Thorner, and K. Weis. 2006. Activation of the DExD/H-box protein Dbp5 by the nuclear-pore protein Gle1 and its coactivator InsP6 is required for mRNA export. Nat. Cell Biol. 8:668-676. [DOI] [PubMed] [Google Scholar]
- 107.Xiao, S., F. Scott, C. A. Fierke, and D. R. Engelke. 2002. Eukaryotic ribonuclease P: a plurality of ribonucleoprotein enzymes. Annu. Rev. Biochem. 71:165-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Zanchin, N. I. T., and D. S. Goldfarb. 1999. Nip7p interacts with Nop8p, an essential nucleolar protein required for 60S ribosome biogenesis, and the exosome subunit Rrp43p. Mol. Cell. Biol. 19:1518-1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zanchin, N. I. T., P. Roberts, A. DeSilva, F. Sherman, and D. S. Goldfarb. 1997. Saccharomyces cerevisiae Nip7p is required for efficient 60S ribosome subunit biogenesis. Mol. Cell. Biol. 17:5001-5015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.