Abstract
LINE-1 (L1) retrotransposons comprise a large fraction of genomic DNAs of many organisms. Many L1 elements are active and may generate potentially deleterious mutations by inserting into genes, yet little is known about the control of retrotransposition by the host. Here we examined whether retrotransposition depends on the cell cycle by using a retrotransposition assay with cultured human cells. We show that in both cancer cells and primary human fibroblasts, retrotransposition was strongly inhibited in the cells arrested in the G1, S, G2, or M stage of the cell cycle. Retrotransposition was also inhibited during cellular senescence in primary human fibroblasts. The levels of L1 transcripts were strongly reduced in arrested cells, suggesting that the reduction in L1 transcript abundance limits retrotransposition in nondividing cells. We hypothesize that inhibition of retrotransposition in nondividing cells protects somatic tissues from accumulation of deleterious mutations caused by L1 elements.
Long interspersed nuclear elements 1 (L1 elements) are active and abundant retrotransposons. There are approximately 500,000 L1 elements in the human genome, comprising ∼17% of the genome mass (18, 32). Given their abundance, L1 elements could potentially wreak havoc in the genome by creating mutations and genomic instability (8, 35). Indeed, there are multiple examples in which an L1 insertion was associated with human disease (reviewed in reference 27). Examples of such insertions are insertions into the APC tumor suppressor gene, which caused colon cancer (21), and into factor VIII, which resulted in hemophilia (16). Yet in most cases, retrotransposons do not destroy their hosts, as multiple factors restrict L1 activity.
Furthermore, L1 retrotransposition seems to be limited to specific cell types. Full-length sense-stranded L1 transcripts have been detected in male germ cells and are rare in somatic tissues (3). In transgenic mice, retrotransposition of enhanced green fluorescent protein (EGFP)-tagged L1 was detected in germ cells (24, 26, 29), in early embryogenesis (29), and recently in neuronal precursor cells (24). Retrotransposition has not been observed in other somatic tissues. In cultured cells, retrotransposition of L1 under control of its native promoter was detected in a large variety of transformed cell lines but not in primary cells. It is not known what limits retrotransposition in primary cells and in somatic tissues. A common characteristic of male germ cells, early embryonic tissues, and transformed cell lines is active cell division; in contrast, cells within most somatic tissues divide rarely, suggesting that cell division may be required for L1 retrotransposition.
Here, we examined whether cell divisions are required for L1 retrotransposition. In order to study retrotransposition in arrested cells, we established a retrotransposition assay with primary human fibroblasts. In contrast to cancer cells, primary cells have intact cell cycle checkpoints and can be arrested at any stage of the cell cycle, including G1, or can enter G0 arrest upon serial passaging. The majority of cells in somatic tissues are arrested in G1/G0, which represents a normal physiological state for animal cells. To detect retrotransposition, we used a cassette containing EGFP-tagged L1 expressed under control of its endogenous promoter (28). We observed L1 retrotransposition in actively dividing IMR-90 cells. However, retrotransposition was strongly inhibited in primary fibroblasts arrested in the G1, G0, S, G2, or M stage of the cells cycle. We also show that retrotransposition is inhibited in HeLa cells arrested in S or G2/M. These results indicate that L1 retrotransposition requires host cell divisions. The requirement for cell divisions may represent a mechanism that limits L1 retrotransposition and prevent genomic instability in somatic tissues.
MATERIALS AND METHODS
Plasmids.
The construction of episomal vectors pL1RP-EGFP and pL1RP(JM111)-EGFP containing EGFP-based retrotransposition cassettes was described previously (28). In this study L1RP was replaced with a more active L1 element, LRE3 (4). These constructs were kindly provided by John V. Moran (University of Michigan).
Cell culture.
All the cells were cultured in a humidified incubator with 5% CO2 and 5% O2 at 37°C. HeLa cells were grown in high-glucose Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum and penicillin-streptomycin. IMR-90 cells were grown in Eagle's minimum essential medium with Earle's salts supplemented with 15% fetal calf serum, nonessential amino acids, 1 mM sodium pyruvate, and penicillin-streptomycin. In all the experiments except the study of retrotransposition during cellular senescence, IMR-90 cells at population doubling (PD) 28 were used.
Transfection of cells.
HeLa cells were split 1day before the transfection, plated at a density of 1 × 106 cells/10-cm plate, and then transfected with FuGENE6 transfection reagent (Roche). Transfection was performed with 18 μl of FuGENE6 and 11 μg of DNA in a total volume of 600 μl. IMR-90 cells were split 2 days before the transfection, plated at a density of 5 × 105 cells/10-cm plate for young cells or 1 × 106 cells/10-cm plate for senescent cells, and then transfected with an Amaxa Nucleofector using 5 μg of DNA per transfection. Transfection programs were U20 for young cells and presenescent cells and P22 for senescent cells. Transfection efficiency was determined by transfecting the cells with pEGFP-N1 (Clontech).
FACS analysis of L1 retrotransposition.
Cells were harvested, washed in 1× phosphate-buffered saline (PBS), and kept on ice prior to fluorescence-activated cell sorter (FACS) analysis. FACS analysis was performed on a FACSCalibur instrument (BD Biosciences), using red-versus-green fluorescence plots. The gating for EGFP-positive cells was determined by analyzing cells transfected with pEGFP-N1 and pUC19 (EGFP negative). A minimum of 20,000 cells per sample were analyzed. For the samples with low retrotransposition frequencies, up to 50,000 cells were scored. Data were analyzed using CellQuest software.
Cell cycle arrest.
The cells were harvested 24 h after transfection and plated at a density of 1 × 106 cells/plate, and drugs were added at the following concentrations: 5 μg/ml aphidicolin, 76 μg/ml hydroxyurea, 0.4 μg/ml nocodazole, and 0.1 μg/ml colchicine. For the low-serum treatment, transfected cells were placed in medium with 0.1% fetal calf serum.
Cell cycle analysis.
The proportion of cells arrested at each stage of cell cycle was determined using propidium iodide (PI) staining. Cells were fixed in 70% ethanol (1 × 106 cells/3 ml ethanol/tube) overnight at 4°C. Fixed cells were washed twice with 1× PBS and then incubated in 500 μl of 1× PBS containing 20 μg/ml PI in the presence of RNase A (1-mg/ml final concentration) for at least 30 min at room temperature. PI fluorescence was determined by flow cytometry on a FACSCalibur (BD Biosciences). A minimum of 20,000 cells per sample were analyzed. Data were collected and analyzed using CellQuest software.
The proportion of cells arrested in the M stage of the cell cycle was determined by PI staining combined with staining with M-stage-specific TG-3 antibodies (20). Cells were fixed in 70% ethanol (1 × 106 cells/3 ml ethanol/tube) overnight at 4°C. Fixed cells were washed once with 1× PBS, followed by a wash with 1× PBS-0.5% bovine serum albumin (BSA)-0.5% Tween 20, and then cells were treated with 1× PBS-2% fetal bovine serum at room temperature for 20 min. Cells were incubated with 7 μl of TG-3 hybridoma culture supernatant/10 μl (kindly provided by Peter Davies, Albert Einstein College of Medicine) in the dark at 4°C overnight. The cells were then washed twice in 1× PBS-0.5% BSA-0.5% Tween 20 and were incubated in 0.7 μg of fluorescein isothiocyanate (FITC)-conjugated rat anti-mouse IgM antibody/10 μl (BD PharMingen) in the dark at 4°C for 2 h. The cells were then washed twice in 1× PBS-0.5% BSA/0.5% Tween 20 and incubated in 500 μl of 1× PBS containing 20 μg/ml PI in the presence of RNase A (1 mg/ml final) for at least 30 min at room temperature. PI fluorescence (DNA content) and FITC fluorescence (TG-3 labeling) were determined by flow cytometry on a FACSCalibur (BD Biosciences). A minimum of 20,000 cells/sample were analyzed. Data were collected and analyzed using CellQuest software.
Analysis of L1 RNA.
IMR-90 cells were transfected with LRE3 and treated to induce cell cycle arrest as described above. Untransfected cells were used as a negative control. Total RNA was isolated on day 4 posttransfection, using an RNeasy kit (QIAGEN). RNA (0.5 μg) was reverse transcribed and PCR amplified using the Titan one-tube reverse transcription-PCR (RT-PCR) kit (Roche) with primers to LRE3-EGFP and actin. The primers to LRE3-EGFP anneal to open reading frame 2 (ORF2) and EGFP sequences flanking β-globin intron. The spliced L1 RNA yields a 1.8-kb PCR product. The sequences of the L1 primers were 5′-GACGGCGACGTAAACGGCCACAAGTTCAGC-3′ and 5-AGAAACTACCATCAGAGTGAACAGGCAACC-3′. Primers to β-actin were obtained from Ambion. RT-PCR products were quantified using ImageQuant software version 1.2. L1 RNA levels were normalized by expression of actin. The experiments were repeated three times.
RESULTS
Detection of L1 retrotransposition in IMR-90 normal human fibroblasts and in HeLa cells.
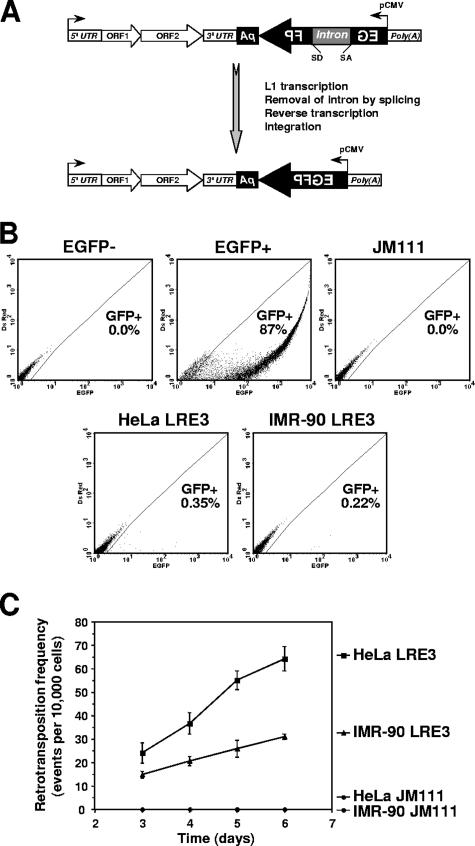
To examine L1 retrotransposition in nondividing cells, we established a retrotransposition assay with normal human fibroblasts because normal fibroblasts can be induced to enter G1/G0 arrest, which mimics the physiological state of cells in somatic tissues. We also analyzed HeLa cells, since this cell line has traditionally been used in L1 studies. To detect retrotransposition, we chose an EGFP retrotransposition cassette (28), as the fluorescent marker can be used in nondividing cells. The cassette contains an L1 element harboring an engineered EGFP gene in its 3′ untranslated region (3′UTR). The EGFP gene is controlled by a cytomegalovirus promoter and is inserted in the opposite orientation to the L1 transcript. The EGFP gene is interrupted by a γ-globin intron in the same orientation as the L1 transcript. In this arrangement, EGFP-positive cells arise only when the L1 transcript is spliced, reverse transcribed, and integrated (Fig. 1A). We used retrotransposition cassettes containing either an active L1 element (LRE3) or a retrotransposition-incompetent L1 element (JM111) (4, 22, 28).
FIG. 1.
L1 retrotransposition in IMR-90 normal human fibroblasts and in HeLa cells. (A) Diagram of the reporter cassette for detection of L1 retrotransposition. The cassette consists of a full-length L1 element containing a 5′UTR harboring an internal promoter, two open reading frames (ORF1 and ORF2), and a 3′UTR with a poly(A) tail at the end. The 3′UTR contains an inverted copy of the EGFP gene interrupted by an intron that is in the same orientation as L1. Retrotransposition removes the intron and activates the EGFP gene. (B) Detection of retrotransposition events by flow cytometry. Cells were analyzed on red-versus-green fluorescence plots. Gating was determined using cells transfected with a negative control plasmid (EGFP−) and the pEGFP vector (EGFP+). Representative FACS traces of HeLa and IMR-90 cells transfected with an active L1 element (LRE3) and of IMR-90 cells transfected with an inactive L1 element (JM111) are shown. (C) Retrotransposition frequencies on days 3 to 6 after transfection. Cells were transfected with active L1 (LRE3) or retrotransposition-deficient L1 (JM111), and the number of green cells was analyzed by flow cytometry. Twenty thousand cells were analyzed in each sample. The retrotranspostion frequency was calculated as a ratio of GFP-positive cells obtained in transfection with L1 to GFP-positive cells obtained in transfection with pEGFP vector × 10,000 cells. The experiments were repeated at least three times, and standard deviations are shown.
We transfected IMR-90 fetal lung fibroblasts and HeLa cells with episomal vectors containing an EGFP retrotransposition cassette with either LRE3 or JM111, or with pEGFP vector as a control of transfection efficiency. Fibroblasts were transfected using Amaxa Nucleofector, which typically transfects these cells at ∼90% efficiency. The appearance of EGFP-positive cells was analyzed by flow cytometry on days 3 to 6 posttransfection. Cells were analyzed on red-versus-green fluorescence plots (Fig. 1B), which eliminates the background of autofluorescent cells and increases the sensitivity of detection (31). Upon transfection with LRE3, retrotransposition was observed in both IMR-90 and HeLa cells, although the retrotransposition frequency was consistently higher in HeLa cells (Fig. 1B and C). No green cells were detected after transfection with retrotransposition-defective L1, suggesting that GFP-positive cells observed after transfection with LRE3 resulted from authentic retrotransposition events. We confirmed that the green fluorescent cells contained retrotransposed L1 by testing for correct splicing of the EGFP gene, which is expected to occur during retrotransposition. Genomic DNA was extracted from the cells on day 4 after transfection with LRE3 and PCR amplified with primers flanking the γ-globin intron within the EGFP gene. A product corresponding in size to the correctly spliced EGFP gene was obtained from both IMR-90 and HeLa cells (data not shown). This result demonstrates that L1 retrotransposition can occur in primary human fibroblasts.
L1 retrotransposition is inhibited in arrested cells.
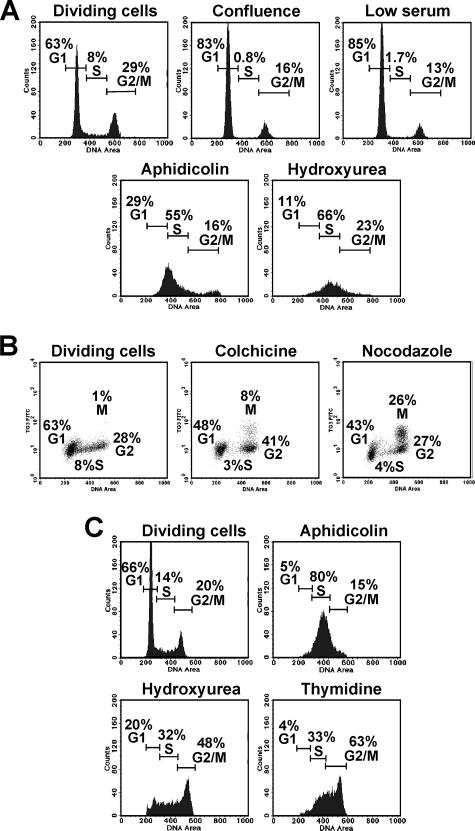
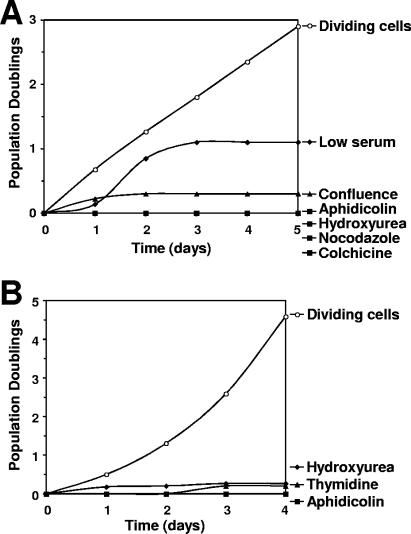
To examine whether L1 retrotransposition is controlled by the cell cycle, we first determined the treatments that arrest IMR-90 and HeLa cells at various cell cycle stages. Conditions of confluence or low serum caused IMR-90 cells to arrest in the G1 stage (Fig. 2A). Treatment with aphidicolin and hydroxyurea arrested IMR-90 in the S stage (Fig. 2A). These drugs affect DNA replication by inhibiting DNA polymerase α (aphidicolin) (14) or by disrupting nucleotide pools (hydroxyurea) (30). Colchicine arrested IMR-90 cells in the G2 stage, and nocodazole arrested IMR-90 cells in the M stage (Fig. 2B). Nocodazole and colchicine are microtubule poisons: colchicine prevents microtubule polymerization (19), and nocodazole inhibits microtubule dynamics (15). All these treatments caused prolonged growth arrest in IMR-90 cells (Fig. 3A), with less than 10% cell death during the first 5 days.
FIG. 2.
Analysis of cell cycle arrest. (A) Cell cycle distribution of IMR-90 normal human fibroblasts on day 3 after the indicated treatments. The number of cells is plotted against the DNA content determined by PI staining. (B) Analysis of M-stage arrest in IMR-90 cells treated as indicated. FITC fluorescence corresponding to cells stained with M-stage-specific antibody TG-3 is plotted against DNA content. (C) Cell cycle distribution of HeLa cells on day 2 after treatments. Twenty thousand cells were analyzed in each sample.
FIG. 3.
Growth arrest of IMR-90 normal human fibroblasts (A) and HeLa cells (B). Cells were treated as described in Materials and Methods.
HeLa cells have a G1 checkpoint defect, and therefore confluence and low serum concentrations do not arrest these cells. Aphidicolin and hydroxyurea arrested HeLa cells in the S and S/G2/M stages, respectively (Fig. 2C). Thymidine arrested HeLa cells in S/G2/M (Fig. 2C). Nocodazole and colchicine were not used for HeLa cells, since they caused massive cell death. Treatments with aphidicoline, hydroxyurea, and thymidine arrested growth of HeLa cells for at least 4 days (Fig. 3B), with less than 20% cell death.
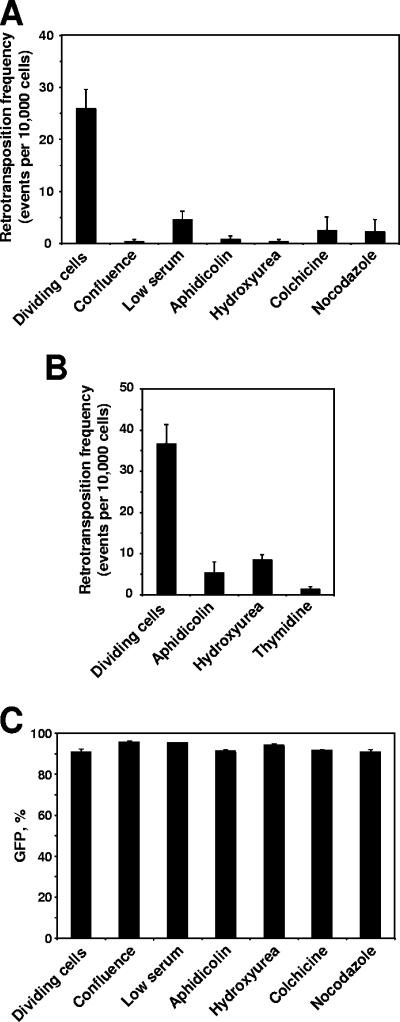
We then tested L1 retrotransposition in arrested cells. Cells were transfected with LRE3 or JM111, and 24 h later the cells were harvested and products of multiple transfections were pooled together to equalize any possible differences in transfection efficiency. Cells were then counted, plated, and treated with drugs or were placed in medium with 1% serum for the low-serum treatment. To obtain confluent cells, the cells were plated at high density. Retrotransposition was analyzed by flow cytometry on day 5 after transfection for IMR-90 cells and day 4 after transfection for HeLa cells. Retrotransposition was strongly inhibited in all the arrested cells regardless of the cell cycle stage (Fig. 4A and B). The inhibition was less efficient in the cells arrested by low serum concentration (Fig. 4A), which can be explained by delayed induction of the growth arrest (Fig. 3A). To rule out a possibility that the treatments affect the expression of the EGFP reporter, we transfected IMR-90 cells with a pEGFP-N1 plasmid carrying the EGFP gene and subjected the cells to growth-arresting treatments. The percentage of GFP-positive cells was determined on day 5 after transfection by using FACS analysis. The treatments did not affect EGFP expression (Fig. 4C). These results indicate that cell divisions are required for L1 retrotransposition.
FIG. 4.
Inhibition of L1 retrotransposition by growth arrest. (A) Retrotransposition frequency in IMR-90 normal human fibroblasts at 4 days after induction of cell cycle arrest. Cells were transfected with an L1 retrotransposition cassette, and 24 h after transfection the cells were treated to induce cell cycle arrest. Retrotransposition frequency was determined by flow cytometry. At least 20,000 cells were scored for each sample. For the samples with low retrotransposition frequencies, up to 50,000 cells were scored. (B) Retrotransposition frequency in HeLa cells 3 days after induction of cell cycle arrest. (C) IMR-90 cells transfected with EGFP expression vector pEGFP-N1. Growth arrest does not affect EGFP expression. All the experiments were repeated at least three times, and error bars represent standard deviations.
The levels of L1 transcripts are reduced in nondividing cells.
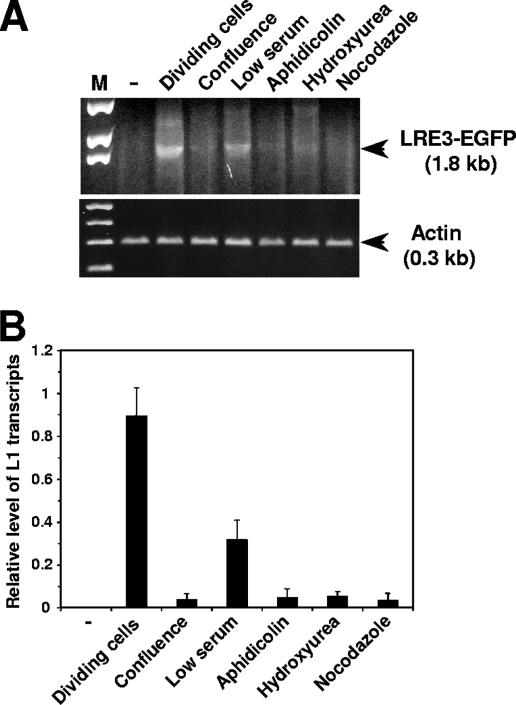
To determine whether L1 transcription is affected in nondividing cells, we tested the level of L1 transcripts. IMR-90 cells were transfected with LRE3, and growth arrest was induced as described above. Cells were harvested on day 4 after transfection, and L1 transcripts were analyzed by RT-PCR. Actin transcripts were amplified as an input standard. L1 RNA levels were strongly reduced in arrested cells (Fig. 5). Only trace amounts of L1 transcripts were observed in the cells arrested by confluence, aphidicolin, hydroxyurea, and nocodazole (Fig. 5). The reduction was less severe in the cells treated with low serum concentrations, which is in accordance with the slow onset of cell cycle arrest following serum withdrawal (Fig. 3A). These results suggest that low L1 transcript levels limit retrotransposition in nondividing cells.
FIG. 5.
L1 RNA levels in arrested cells. (A) IMR-90 cells were transfected with an L1 retrotransposition cassette (LRE3-EGFP) and treated to induce cell cycle arrest. Cells were harvested on day 4 after transfection, and total RNA was isolated and analyzed by RT-PCR. Actin was used as an input control. Spliced L1 RNA yields a 1.8-kb PCR product. The negative control is untransfected IMR-90 cells. (B) Quantification of RT-PCR results. L1 expression was normalized by expression of actin. The experiments were repeated three times, and error bars show standard deviations.
L1 retrotransposition is inhibited in replicatively senescent cells.
Normal human fibroblasts do not divide indefinitely and after 60 to 70 PDs enter a growth arrest called replicative senescence (11, 12). Replicative senescence is believed to be a tumor suppressor mechanism that limits cell proliferation (5). Since senescent cells accumulate in aging tissues (6, 13) and aging is associated with genomic instability (9), we were interested in testing whether L1 retrotransposition is activated in senescent cells.
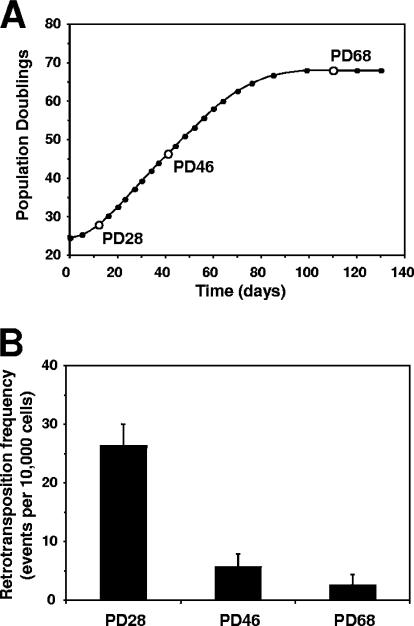
IMR-90 cells were passaged until they reached senescence at PD 68 (Fig. 6A), and aliquots of cells were frozen at every passage. Young (PD 28), middle-aged (PD46), and senescent (PD 68) cells were transfected with LRE3 or pEGFP-N1 plasmid to determine transfection efficiency. L1 retrotransposition was analyzed 5 days after transfection. The retrotransposition frequency declined with increasing replicative age and became extremely low in senescent cells (Fig. 6B). Thus, replicative senescence inhibited L1 retrotranspostion, which is in line with a tumor suppressor function of replicative senescence.
FIG. 6.
Inhibition of L1 retrotransposition during replicative senescence. (A) Growth curve for IMR-90 normal human fibroblasts. Open circles represent passages at which L1 retrotransposition was analyzed. (B) Retrotransposition frequency in IMR-90 cells of different replicative ages. The cells were transfected with an L1 retrotransposition cassette and analyzed by flow cytometry on day 5 after transfection. Retrotranspostion frequency was calculated as the ratio of GFP-positive cells obtained in transfection with L1 to GFP-positive cells obtained in transfection with pEGFP vector × 10,000 cells. All the experiments were repeated at least three times, and error bars represent standard deviations.
DISCUSSION
L1 retrotransposition in primary cells.
It has long been believed that L1 retrotransposition can occur only in germ cells and transformed or immortalized cells. This belief was challenged when L1 retrotransposition was observed in mouse neural precursor cells (24) and, most recently, in human fibroblasts and hepatocytes (17). In the latter study, however, the L1 element was controlled by a ubiquitous phosphoglycerate kinase 1 (PGK) promoter, and it was concluded that augmenting L1 transcription allows retrotransposition to occur in cell types that do not support expression of endogenous L1 elements (17). Here we have demonstrated that L1 driven by its native promoter is capable of retrotransposition in actively dividing primary human fibroblasts, suggesting that endogenous L1 elements may be active in dividing primary somatic human cells.
There are two possible explanations why earlier studies failed to detect L1 retrotransposition in these cells. First, the frequency of retrotransposition in these cells is low, possibly due to a lower mitotic index in normal cells than in cancer cells. Therefore, a sensitive detection method is needed to observe L1 retrotransposition in these cells. Here we used FACS analysis on two channels (red versus green), which eliminates the background of autofluorescent cells and increases sensitivity. With our FACS settings, no green cells were detected among the control cells transfected with inactive retrotransposon JM111, indicating that the background is virtually zero. We estimate that with this detection method, retrotransposition frequencies of as low as 1 per 10,000 cells can be detected. The second possible explanation is that there is a variation between different lines of normal human fibroblasts with respect to L1 retrotransposition. Each cell line is derived from a different donor, and individuals in the human population may differ in retrotransposon activity.
L1 retrotransposition requires cells divisions.
Here we present a systematic analysis of L1 retrotransposition in cultured cells arrested at various stages of the cell cycle. We show that cell divisions are required for L1 retrotransposition. Retrotransposition was strongly inhibited in the cells arrested in the G1, S, G2, and M stages and in replicatively senescent cells. Residual retrotransposition events detected in arrested cells could have occurred during the first 2 days after transfection before the onset of cell cycle arrest, or alternatively, inhibition of retrotransposition in arrested cells may not be absolute, allowing for some residual activity.
Cell cycle arrest was induced both by drug treatments and by physiological conditions such as serum deprivation and confluence. The drugs that inhibit DNA replication could potentially affect the retrotransposition process directly rather than through inhibition of the cell cycle. In contrast, confluence and serum deprivation induce physiological G1 arrest, which does not affect the transcription and repair capacities of the cell (31). All of the different types of cell cycle arrest inhibited L1 retrotransposition, suggesting that the requirement for cell divisions is universal and is not an artifact of the treatment.
We have shown that the level of L1 transcripts is strongly reduced in arrested cells, suggesting that L1 transcription may be the limiting step for L1 retrotransposition in nondividing cells. Multiple lines of evidence suggest that L1 transcription is regulated by the host. Sense-strand full-length L1 transcripts have been detected in spermatocytes but are rare or undetectable in somatic tissues (3, 26). The L1 promoter contains binding sites for transcription factors of the SRY family. The Sox11 transcription factor was shown to transactivate the L1 promoter (36), the Sox2 transcription factor has been implicated in repression of L1 retrotransposition (24), and the RUNX3 transcription factor was shown to regulate L1 promoter activity. Thus, a specific transcription factor may repress L1 transcription in nondividing cells. Recent reports showing regulation of L1 by RNA interference (33, 38) suggest another intriguing possibility, i.e., that the amount of L1 RNA in nondividing cells is reduced by a posttranscriptional RNA interference mechanism.
Recent report by Kubo et al. concluded that L1 retrotransposition can occur in nondividing cells (17). The fundamental difference between this study and our work is that Kubo et al. used an L1 element driven by the ubiquitous PGK promoter. Since we find L1 mRNA levels to be the limiting factor in retrotransposition in nondividing cells, the use of the PGK promoter is likely to account for the differences in our conclusions. Furthermore, Kubo et al. reported that L1 retrotransposition frequency was reduced threefold in G1/S-arrested glioma cells, and no retrotransposition occurred in G0-arrested cells (17). Therefore, even when L1 is transcribed from a strong ubiquitous promoter, some factors still restrict retrotransposition in nondividing cells. In summary, considering the differences in our experimental systems, our results are consistent with the results of Kubo et al. Both studies show a significant reduction of retrotransposition in arrested cells (a 3- to 100-fold reduction in the study by Kubo et al. and a 10- to 70-fold reduction in our study).
The requirement for cell divisions may contribute to cell and tissue specificity of L1 retrotransposition.
Our results may explain previous observations related to the lack of L1 retrotransposition in various experimental systems. Preferential L1 retrotransposition in early embryogenesis (29) and in male germ cells (26) can in part be explained by active cell divisions in these tissues, whereas other somatic tissues contain relatively low numbers of dividing cells. Our results may also explain a delayed appearance of the first retrotransposition events in cultured cell assay. It was noted that the first retrotransposition events can be detected only at 48 h after transfection with L1 element, and this delay cannot be entirely accounted for by the time required for plasmid transfection and expression (28). It is likely that the delay is caused in part by transient arrest of cell divisions due to the stress of transfection.
Inhibition of L1 retrotransposition in nondividing cells promotes genome stability.
L1, if unleashed, can be a powerful mutagen, and multiple factors that limit L1 retrotransposition in the cell have been identified. These include methylation of CpG dinucleotides in the L1 5′UTR (2, 10, 25, 37, 39), RNA interference (33, 38), and APOBEC3 proteins (1, 23, 34). Complex genomic rearrangements, which are likely to involve host DNA repair activities, have also been associated with L1 retrotransposition in HeLa cells (7), suggesting that host processes may act to restrict L1 integration and limit the number of active L1 elements (7).
Furthermore, L1 is most active in the male germ line. This tissue specificity is explained by L1 being a genetic parasite, which ensures its spread to future generations by replicating in the germ line. However, active retrotransposition in differentiated somatic tissues may also be detrimental to the host, since it would cause genomic instability and interfere with tissue function. We show that L1 retrotransposition is inhibited in nondividing cells, which represents a novel relationship between the host cell and L1. A block of retrotransposition in nondividing cells ensures genome stability in somatic tissues and possibly evolved to maintain the fine balance between a genomic parasite and its host.
Acknowledgments
We thank John V. Moran for critical reading of the manuscript and for providing L1 retrotransposition constructs, and we thank Peter C. Keng for advice on FACS analysis.
V.G. is supported by grants from the National Institutes of Health, the Ellison Medical Foundation, the American Federation for Aging Research, and the Komen Foundation.
Footnotes
Published ahead of print on 4 December 2006.
REFERENCES
- 1.Bogerd, H. P., H. L. Wiegand, A. E. Hulme, J. L. Garcia-Perez, K. S. O'Shea, J. V. Moran, and B. R. Cullen. 2006. Cellular inhibitors of long interspersed element 1 and Alu retrotransposition. Proc. Natl. Acad. Sci. USA 103:8780-8785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bourc'his, D., and T. H. Bestor. 2004. Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature 431:96-99. [DOI] [PubMed] [Google Scholar]
- 3.Branciforte, D., and S. L. Martin. 1994. Developmental and cell type specificity of LINE-1 expression in mouse testis: implications for transposition. Mol. Cell. Biol. 14:2584-2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brouha, B., C. Meischl, E. Ostertag, M. de Boer, Y. Zhang, H. Neijens, D. Roos, and H. H. Kazazian, Jr. 2002. Evidence consistent with human L1 retrotransposition in maternal meiosis I. Am. J. Hum Genet. 71:327-336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Campisi, J. 2001. Cellular senescence as a tumor-suppressor mechanism. Trends Cell Biol. 11:S27-31. [DOI] [PubMed] [Google Scholar]
- 6.Dimri, G. P., X. Lee, G. Basile, M. Acosta, G. Scott, C. Roskelley, E. E. Medrano, M. Linskens, I. Rubelj, O. Pereira-Smith, M. Peacocke, and J. Campisi. 1995. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc. Natl. Acad. Sci. USA 92:9363-9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gilbert, N., S. Lutz, T. A. Morrish, and J. V. Moran. 2005. Multiple fates of L1 retrotransposition intermediates in cultured human cells. Mol. Cell. Biol. 25:7780-7795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gilbert, N., S. Lutz-Prigge, and J. V. Moran. 2002. Genomic deletions created upon LINE-1 retrotransposition. Cell 110:315-325. [DOI] [PubMed] [Google Scholar]
- 9.Gorbunova, V., and A. Seluanov. 2005. Making ends meet in old age: DSB repair and aging. Mech. Ageing Dev. 126:621-628. [DOI] [PubMed] [Google Scholar]
- 10.Hata, K., and Y. Sakaki. 1997. Identification of critical CpG sites for repression of L1 transcription by DNA methylation. Gene 189:227-234. [DOI] [PubMed] [Google Scholar]
- 11.Hayflick, L. 1965. The limited in vitro lifetime of human diploid cell strains. Exp. Cell Res. 37:614-636. [DOI] [PubMed] [Google Scholar]
- 12.Hayflick, L., and P. S. Moorhead. 1961. The serial cultivation of human diploid strains. Exp. Cell Res. 25:585-621. [DOI] [PubMed] [Google Scholar]
- 13.Herbig, U., M. Ferreira, L. Condel, D. Carey, and J. M. Sedivy. 2006. Cellular senescence in aging primates. Science 311:1257. [DOI] [PubMed] [Google Scholar]
- 14.Ikegami, S., T. Taguchi, M. Ohashi, M. Oguro, H. Nagano, and Y. Mano. 1978. Aphidicolin prevents mitotic cell division by interfering with the activity of DNA polymerase-alpha. Nature 275:458-460. [DOI] [PubMed] [Google Scholar]
- 15.Jordan, M. A., D. Thrower, and L. Wilson. 1992. Effects of vinblastine, podophyllotoxin and nocodazole on mitotic spindles. Implications for the role of microtubule dynamics in mitosis. J. Cell Sci. 102:401-416. [DOI] [PubMed] [Google Scholar]
- 16.Kazazian, H. H., Jr., C. Wong, H. Youssoufian, A. F. Scott, D. G. Phillips, and S. E. Antonarakis. 1988. Haemophilia A resulting from de novo insertion of L1 sequences represents a novel mechanism for mutation in man. Nature 332:164-166. [DOI] [PubMed] [Google Scholar]
- 17.Kubo, S., C. Seleme Mdel, H. S. Soifer, J. L. Perez, J. V. Moran, H. H. Kazazian, Jr., and N. Kasahara. 2006. L1 retrotransposition in nondividing and primary human somatic cells. Proc. Natl. Acad. Sci. USA 103:8036-8041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lander, E. S., L. M. Linton, B. Birren, C. Nusbaum, M. C. Zody, J. Baldwin, K. Devon, K. Dewar, M. Doyle, W. FitzHugh, R. Funke, D. Gage, K. Harris, A. Heaford, J. Howland, L. Kann, J. Lehoczky, R. LeVine, P. McEwan, K. McKernan, J. Meldrim, J. P. Mesirov, C. Miranda, W. Morris, J. Naylor, C. Raymond, M. Rosetti, R. Santos, A. Sheridan, C. Sougnez, N. Stange-Thomann, N. Stojanovic, A. Subramanian, D. Wyman, J. Rogers, J. Sulston, R. Ainscough, S. Beck, D. Bentley, J. Burton, C. Clee, N. Carter, A. Coulson, R. Deadman, P. Deloukas, A. Dunham, I. Dunham, R. Durbin, L. French, D. Grafham, S. Gregory, T. Hubbard, S. Humphray, A. Hunt, M. Jones, C. Lloyd, A. McMurray, L. Matthews, S. Mercer, S. Milne, J. C. Mullikin, A. Mungall, R. Plumb, M. Ross, R. Shownkeen, S. Sims, R. H. Waterston, R. K. Wilson, L. W. Hillier, J. D. McPherson, M. A. Marra, E. R. Mardis, L. A. Fulton, A. T. Chinwalla, K. H. Pepin, W. R. Gish, S. L. Chissoe, M. C. Wendl, K. D. Delehaunty, T. L. Miner, A. Delehaunty, J. B. Kramer, L. L. Cook, R. S. Fulton, D. L. Johnson, P. J. Minx, S. W. Clifton, T. Hawkins, E. Branscomb, P. Predki, P. Richardson, S. Wenning, T. Slezak, N. Doggett, J. F. Cheng, A. Olsen, S. Lucas, C. Elkin, E. Uberbacher, M. Frazier, et al. 2001. Initial sequencing and analysis of the human genome. Nature 409:860-921. [DOI] [PubMed] [Google Scholar]
- 19.Luduena, R. F., A. Banerjee, and I. A. Khan. 1992. Tubulin structure and biochemistry. Curr. Opin. Cell Biol. 4:53-57. [DOI] [PubMed] [Google Scholar]
- 20.McCollum, G., P. C. Keng, J. C. States, and M. J. McCabe, Jr. 2005. Arsenite delays progression through each cell cycle phase and induces apoptosis following G2/M arrest in U937 myeloid leukemia cells. J. Pharmacol. Exp. Ther. 313:877-887. [DOI] [PubMed] [Google Scholar]
- 21.Miki, Y., I. Nishisho, A. Horii, Y. Miyoshi, J. Utsunomiya, K. W. Kinzler, B. Vogelstein, and Y. Nakamura. 1992. Disruption of the APC gene by a retrotransposal insertion of L1 sequence in a colon cancer. Cancer Res. 52:643-645. [PubMed] [Google Scholar]
- 22.Moran, J. V., S. E. Holmes, T. P. Naas, R. J. DeBerardinis, J. D. Boeke, and H. H. Kazazian, Jr. 1996. High frequency retrotransposition in cultured mammalian cells. Cell 87:917-927. [DOI] [PubMed] [Google Scholar]
- 23.Muckenfuss, H., M. Hamdorf, U. Held, M. Perkovic, J. Lower, K. Cichutek, E. Flory, G. G. Schumann, and C. Munk. 2006. APOBEC3 proteins inhibit human LINE-1 retrotransposition. J. Biol. Chem. 281:22161-22172. [DOI] [PubMed] [Google Scholar]
- 24.Muotri, A. R., V. T. Chu, M. C. Marchetto, W. Deng, J. V. Moran, and F. H. Gage. 2005. Somatic mosaicism in neuronal precursor cells mediated by L1 retrotransposition. Nature 435:903-910. [DOI] [PubMed] [Google Scholar]
- 25.Nur, I., E. Pascale, and A. V. Furano. 1988. The left end of rat L1 (L1Rn, long interspersed repeated) DNA which is a CpG island can function as a promoter. Nucleic Acids Res. 16:9233-9251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ostertag, E. M., R. J. DeBerardinis, J. L. Goodier, Y. Zhang, N. Yang, G. L. Gerton, and H. H. Kazazian, Jr. 2002. A mouse model of human L1 retrotransposition. Nat. Genet. 32:655-660. [DOI] [PubMed] [Google Scholar]
- 27.Ostertag, E. M., and H. H. Kazazian, Jr. 2001. Biology of mammalian L1 retrotransposons. Annu. Rev. Genet. 35:501-538. [DOI] [PubMed] [Google Scholar]
- 28.Ostertag, E. M., E. T. Prak, R. J. DeBerardinis, J. V. Moran, and H. H. Kazazian, Jr. 2000. Determination of L1 retrotransposition kinetics in cultured cells. Nucleic Acids Res. 28:1418-1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Prak, E. T., A. W. Dodson, E. A. Farkash, and H. H. Kazazian, Jr. 2003. Tracking an embryonic L1 retrotransposition event. Proc. Natl. Acad. Sci. USA 100:1832-1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pratt, W. M., R. W. Ruddon, W. D. Ensminger, and J. Maybaum. 1994. The anticancer drugs, p. 97-98. Oxford University Press, New York, NY.
- 31.Seluanov, A., D. Mittelman, O. M. Pereira-Smith, J. H. Wilson, and V. Gorbunova. 2004. DNA end joining becomes less efficient and more error-prone during cellular senescence. Proc. Natl. Acad. Sci. USA 101:7624-7629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smit, A. F. 1996. The origin of interspersed repeats in the human genome. Curr. Opin. Genet. Dev. 6:743-748. [DOI] [PubMed] [Google Scholar]
- 33.Soifer, H. S., A. Zaragoza, M. Peyvan, M. A. Behlke, and J. J. Rossi. 2005. A potential role for RNA interference in controlling the activity of the human LINE-1 retrotransposon. Nucleic Acids Res. 33:846-856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stenglein, M. D., and R. S. Harris. 2006. APOBEC3B and APOBEC3F inhibit L1 retrotransposition by a DNA deamination-independent mechanism. J. Biol. Chem. 281:16837-16841. [DOI] [PubMed] [Google Scholar]
- 35.Symer, D. E., C. Connelly, S. T. Szak, E. M. Caputo, G. J. Cost, G. Parmigiani, and J. D. Boeke. 2002. Human L1 retrotransposition is associated with genetic instability in vivo. Cell 110:327-338. [DOI] [PubMed] [Google Scholar]
- 36.Tchenio, T., J. F. Casella, and T. Heidmann. 2000. Members of the SRY family regulate the human LINE retrotransposons. Nucleic Acids Res. 28:411-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thayer, R. E., M. F. Singer, and T. G. Fanning. 1993. Undermethylation of specific LINE-1 sequences in human cells producing a LINE-1-encoded protein. Gene 133:273-277. [DOI] [PubMed] [Google Scholar]
- 38.Yang, N., and H. H. Kazazian, Jr. 2006. L1 retrotransposition is suppressed by endogenously encoded small interfering RNAs in human cultured cells. Nat. Struct. Mol. Biol. 13:763-771. [DOI] [PubMed] [Google Scholar]
- 39.Yoder, J. A., C. P. Walsh, and T. H. Bestor. 1997. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet. 13:335-340. [DOI] [PubMed] [Google Scholar]