Abstract
Chromosomal translocations are frequently associated with soft-tissue sarcomas. Fusion proteins generated by such translocations often play critical roles in tumorigenesis. Therefore, it is important to understand the function of the fusion protein to develop therapeutic interventions. The t(X;18)(p11.2;q11.2) translocation found in synovial sarcomas results in a fusion between the SYT gene on chromosome 18 and an SSX gene on the X chromosome. Although SYT-SSX fusion proteins appear to trigger synovial sarcoma development, little is known about the downstream targets of SYT-SSX. We found that the SYT-SSX fusion protein produces a dominant-negative function for SYT, which is a transcriptional coactivator. We then analyzed the gene expression profiles of SYT-SSX1-expressing HeLa cells using oligonucleotide microarrays and found that the SYT-SSX1 fusion protein directly down-regulated the expression of COM1, a regulator of cell proliferation. COM1 was found to be expressed at relatively low levels in synovial sarcoma tissues and cell lines. We then investigated the impact of conditional COM1 expression in the synovial sarcoma cell line. Increased COM1 expression resulted in induced apoptosis and in reduced cell growth and colony formation activity. Our results suggested that restoration of COM1 expression may be of therapeutic benefit in synovial sarcoma.
Synovial sarcomas are aggressive tumors of adolescent and young adults that account for about 10% of all soft-tissue sarcomas. Synovial sarcomas are subdivided into biphasic and monophasic forms by their histomorphologic appearance. Biphasic synovial sarcomas contain epithelial cells arranged in glandular structures and spindle cells, whereas monophasic types are entirely composed of spindle cells. Cytogenetic analysis indicates that the chromosomal translocation t(X;18)(p11.2;q11.2) is present in the majority of these tumors, with molecular analysis of translocation breakpoints showing a disruption of the SYT (for “synovial sarcoma translocated”) gene on chromosome 18q11.2 and juxtaposition to one of the SSX (for “synovial sarcoma X breakpoint”) genes on Xp11.2 in a mutually exclusive fashion, with the result being a chimeric SYT-SSX protein (5, 9). The SSX gene family consists of nine contiguous members, SSX1 to SSX9, encoded on the X chromosome (12). Although the chromosomal arrangement of the SSX genes could in principle allow for heterogeneity at the breakpoint, fusions of SYT with SSX1, SSX2, and SSX4 have been reported in synovial sarcomas (6, 8, 9, 23). The identity of the rearranged SSX gene has significant clinical impact (16).
The normal SYT gene is ubiquitously expressed in a wide range of human tissues and cell lines (6). In contrast, SSX transcripts show a very restricted distribution in adult human tissues, and the expression is confined to the testis and at very low levels in the thyroid (6, 11, 26). SYT contains a transcriptional activating domain, rich in glycine, proline, glutamine, and tyrosine (QPGY domain). On the other hand, SSX possesses two transcriptional repressor domains, a Krüppel-associated box (KRAB) and an SSX repressor domain (SSXRD) (6, 17). Both gene products, together with the fusion proteins, are localized in the nucleus but lack obvious DNA binding motifs. Recently, it was shown that SYT interacts with a putative transcriptional factor, AF10, an acetyltransferase p300, a component of histone deacetylase complex mSin3A, and a component of SWI/SNF chromatin remodeling complexes BRM and Brg1 (7, 10, 13, 14, 19, 25). We have already shown that the transcriptional activity of SYT was regulated by SWI/SNF chromatin remodeling complexes (13). These suggested that presumably their transcriptional regulatory functions are exerted through interactions with other nuclear proteins.
COM1 (candidate of metastasis 1) protein, also known as p8, is a regulator of cell proliferation and was initially identified from metastatic breast cancer (22). In contrast, it has been reported that COM1 inhibits the growth of pancreatic cancer cells (18) and the breast cancer cell line MCF7 (2) and that COM1−/− mouse embryonic fibroblasts (MEFs) show more rapid growth than COM1+/+ MEFs (28). These findings suggest that COM1 mediates both growth inhibition and stimulation in a cell- or tissue-specific manner.
Although these SYT-SSX fusion proteins appear to trigger synovial sarcoma development, the biological functions of the SYT-SSX fusion proteins remain unclear. In this report, we provide evidence that the SYT-SSX1 fusion protein directly down-regulates the expression of the COM1 gene by the dominant-negative function for the SYT. The down-regulation of COM1 plays an important role in synovial sarcoma growth, and restoration of COM1 expression may be of therapeutic benefit in synovial sarcoma.
MATERIALS AND METHODS
Tissue samples and cell lines.
Tumor tissues were obtained with informed consent from patients who received surgical treatment at the National Cancer Center Hospital, Japan. Synovial sarcoma cell lines SYO-1, 1273/99, and HS-SY-II were obtained from A. Kawai (National Cancer Center, Tokyo, Japan), O. Larsson (Karolinska Hospital, Stockholm, Sweden), and H. Sonobe (Kouchi Medical School, Kouchi, Japan), respectively. Other tumor cell lines were obtained from the American Type Culture Collection (ATCC). All cell lines were maintained in Dulbecco's modified Eagle's medium (DMEM) with 10% calf serum, 10% fetal calf serum, or 20% fetal calf serum containing F-12 nutrient mixture.
Plasmid construction.
Gal4-SYT, Gal4-SYT-SSX1, Flag-tagged SYT, and Flag-tagged SYT-SSX1 were described previously (13). COM1 cDNA was isolated from mRNA of HeLa cells by PCR and sequenced. Flag-tagged COM1 cDNA was inserted in pCMV-tag2 (Stratagene) and pTRE2-hyg (Clontech). Hemagglutinin (HA)-tagged SYT-SSX1 and SYT were inserted in pCMV-tag2. The promoter DNAs of COM1 (−4000 to +22), PLAB (−1600 to +20), and CHOP (−2000 to +17) that contain transcriptional start sites and upstream regions were isolated from human genomic DNA by PCR. These DNAs were sequenced and inserted in pGL3-basic (Promega) that contains a firefly luciferase gene.
Reporter assay.
Transfections of expression vectors, pG5luc, pGL3-basic vectors containing several target promoters, and pRL-TK were carried out by using Lipofectamine Plus reagent (Invitrogen). After 36 h of transfection, the cells in 24-well plates were washed by phosphate-buffered saline (PBS) and analyzed for luciferase activities in triplicate in each transfection experiment by using the dual-luciferase reporter assay system (Promega). All experiments were repeated at least three times.
Immunoprecipitation.
Immunoprecipitation assays were performed essentially as described previously (13) with the following modifications. The expression vectors were transfected in HEK293T cells by Lipofectamine Plus reagent. After 36 h, the cells were washed and suspended in lysis buffer (10 mM Tris-HCl [pH 7.8], 1 mM EDTA, 10% glycerol, 0.05% NP-40, 0.3 M NaCl, 5 mM 2-mercaptoethanol, and 1 mM Pefablock [protease inhibitors; Roche]). After lysates were centrifuged for 30 min at 15,000 rpm at 4°C, supernatants were incubated with 30 μl of anti-HA-agarose (Sigma) at 4°C overnight and washed with lysis buffer four times. The bound proteins were separated by sodium dodecyl sulfate-12% polyacrylamide gel electrophoresis and analyzed with anti-HA antibody (Santa Cruz) and anti-Flag antibody M2 (Sigma), respectively. The ECL System (Amersham Pharmacia Biotech) was used for detection of signal on RX-V X-ray films (Fuji).
Stable and conditional gene expression cell lines.
Flag-tagged SYT-SSX1 and Flag-tagged COM1 cDNAs were transfected in HeLa cells. Cells were selected by G418 sulfate (0.5 mg/ml), and a single colony was isolated. SYO-1 cells containing Tet-Off vector (Clontech) were isolated with G418 sulfate (1 mg/ml). Flag-tagged COM1 cDNA in pTRE2-hyg were transfected in the SYO-1 cells containing Tet-Off vector. Cells were selected by hygromycin (0.2 mg/ml) with G418 sulfate (1 mg/ml) and doxycycline (1 μg/ml), and a single colony was isolated.
RT-PCR.
Total RNAs were isolated from cell lines and tissues by Trizol reagent (Invitrogen). First-strand cDNAs were obtained from total RNA (5 μg) by a superscript first-strand synthesis system (Invitrogen). Reverse transcriptase PCR (RT-PCR) was performed using the following primer sets: COM1 forward primer, GCA GAG ACA GAC AAA GCG TTAG; COM1 reverse primer, AGA CTC AGT CAG CGG GAA TAAG; PLAB forward primer, GAG TTG CAC TCC GAA GAC TCC; PLAB reverse primer, GAG AGA TAC GCA GGT GCA GG; CHOP forward primer, AAA ATC AGA GCT GGA ACC TGAG; CHOP reverse primer, TCT TCC TCT TCA TTT CCA GGAG.
Oligonucleotide microarray.
The protocol used for the sample preparation and microarray processing is available from Affymetrix (Santa Clara, CA). Briefly, 5 μg purified total RNA was used. After the cRNA was linearly amplified with T7 polymerase, the biotinylated cRNA was cleaned with an RNeasy Mini Column (QIAGEN), fragmented to 50 to 200 nucleotides, and then hybridized to Human Genome U95A ver.2 arrays (Affymetrix). The stained microarray was scanned with a GeneArray Scanner (Affymetrix), and the signal was calculated with the Affymetrix software Microarray Suite 5.0. All of the data were scaled with the global scaling method to adjust the target intensity to 1,000.
siRNA and chromatin immunoprecipitation (ChIP) analysis.
For the small interfering RNA (siRNA) experiments, 20 nM of control siRNA (QIAGEN) and SYT-SSX-specific siRNA (UGA CCA GAU CAU GCC CAA GTT; Nippon Gene) were transfected into SYO-1 cells (1 × 106) using Cell-Line Nucleofector kit V (Amaxa Biosystems). For the chromatin immunoprecipitation (ChIP) analysis, HEK293T cells were transfected with the vector alone (5 μg) or HA-tagged SYT-SSX1 (5 μg). After 3 days of incubation, the cells were treated with 1% formaldehyde to cross-link proteins to DNA and processed for ChIP assay as described previously (33) with anti-HA antibody.
Cell growth and colony formation analysis.
In cell growth analysis, cells (5 × 104) were placed in 100-mm tissue culture dishes for withdrawal of doxycycline. After incubation at 37°C for 2 to 11 days, cell numbers were counted in several periods. Colony formation analysis was then performed. Cells (5 × 102 to approximately 5 × 104) were placed in DMEM (10% fetal calf serum) containing 0.33% agarose (Wako) and overlaid on a layer of DMEM in 0.5% agarose. After incubation at 37°C for 24 days, colonies were stained with 0.05% crystal violet and colony numbers were counted.
Immunofluorescence microscopy.
At 3 days after the cessation of doxycycline treatment, SYO-1 cells conditionally expressing COM1 were removed to chamber slides, incubated for 24 h more, and fixed with 3% formaldehyde in PBS for 10 min. After washing with PBS, the cells were permeabilized with 0.1% polyoxyehylene octylphenyl ether in PBS for 10 min and washed by PBS. The slide was incubated in blocking buffer (2% normal swine serum in PBS) for 30 min. Cells were incubated with primary antibody for 1 h at room temperature, washed by PBS three times, and incubated with secondary antibody for 1 h at room temperature. We used anti-Flag antibody M2 as a primary antibody. The slide was mounted with 4′,6′-diamidino-2-phenylindole (DAPI) and visualized under a laser-scanning microscope (LSM510; Carl Zeiss).
Detection of apoptosis by caspase assay.
We used a carboxyfluorescein fluorochrome-labeled inhibitors of caspases (FLICA) apoptosis detection kit for apoptosis analysis. The protocol used for sample preparation and caspase assay processing is available from Immunochemistry Technologies, LLC (Bloomington, MN). Cells (1 × 105) were seeded onto a sterile glass coverslip in a 35-mm culture dish. After incubation at 37°C for about 24 h, cells were treated in medium with or without doxycycline (1 μg/ml). Cells were incubated at 37°C for 72 h, and the 30× FLICA solution was added to the medium at a 1:30 ratio. After incubation at 37°C for about 1 h, the medium was removed and cells were incubated with Hoechst stain solution (1 μg/ml) at 37°C for 5 min. Cells were then washed twice with 2 ml 1× wash buffer and fixed with 1% formaldehyde at 4°C for 24 h. Cells were protected from light throughout the procedure. Cells were then visualized under a laser-scanning microscope (LSM510; Carl Zeiss) using a band pass filter (excitation, 490 nm; emission, >520 nm) to view the green fluorescence of caspase-positive cells. DNA was visualized using Hoechst.
RESULTS AND DISCUSSION
SYT-SSX1 down-regulates the expression of SYT target genes.
Translation of the SYT-SSX1 fusion gene yields a chimeric protein in which the C-terminal 8 amino acids of the SYT protein are replaced by 78 amino acids (residues 111 to 188) from the C terminus of the SSX1 protein (5, 9). The fusion protein contains both a potential transcription-activating domain (QPGY domain) from the SYT protein and a repressing domain (SSXRD domain) from the SSX1 protein (5, 9). However, it lacks obvious DNA binding motifs, which suggests that SSX1 may exert its transcriptional regulatory effects via interaction with DNA binding proteins. While SYT reportedly shows transcriptional activity, the transcriptional activity of SYT-SSX1 is virtually nonexistent (3, 13, 25), suggesting that the fused SSXRD domain of SSX1 inhibits SYT-mediated transcriptional activation.
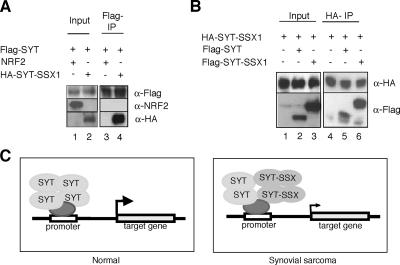
Recently, it was shown that SYT forms homooligomers through its C-terminal QPGY domain (21). To determine whether SYT-SSX1 interacts with normal SYT molecules via QPGY domains, we performed immunoprecipitation studies. When Flag-tagged SYT was cotransfected with HA-tagged SYT-SSX1 or NRF2 (negative control) into HEK293T cells, SYT-SSX1 was coimmunoprecipitated by SYT using the anti-Flag antibody (Fig. 1A, lanes 2 and 4) but NRF2 was not (Fig. 1A, lanes 1 and 3). We also found that SYT was coimmunoprecipitated by SYT-SSX1 using the anti-HA antibody (Fig. 1B, lanes 2 and 5). These results suggested that SYT-SSX1 specifically bound SYT, and the fused domain of SSX1 showed no effect for the binding in this cell line. Next, to determine whether SYT-SSX1 forms homooligomers, HA-tagged SYT-SSX1 was cotransfected with Flag-tagged SYT-SSX1 into HEK293T cells. We found that HA-SYT-SSX1 bound Flag-SYT-SSX1 (Fig. 1B, lanes 3 and 6), suggesting that SYT-SSX1 formed homooligomers, as does SYT (21). Based on our results, we developed a molecular model of synovial sarcoma in which the SYT-SSX1 fusion protein formed heterooligomers with SYT at a target gene's promoters and inhibited the transcriptional activity of SYT through the dominant-negative function of SYT-SSX1 (Fig. 1C). In that way, down-regulation by the dominant-negative SYT-SSX1 might lead to synovial sarcoma.
FIG. 1.
Dominant-negative function of SYT-SSX1 for forming heterooligomers with SYT. (A) SYT bound SYT-SSX1. FLAG-tagged SYT was transfected with NRF2 (lanes 1 and 3) or HA-tagged SYT-SSX1 (lanes 2 and 4) into HEK293T cells. FLAG-tagged SYT was immunoprecipitated from whole-cell lysates (Inp; left panel) using anti-FLAG antibody-conjugated agarose (FLAG-IP; right panel). Bound proteins were washed and detected with anti-FLAG antibody (SYT), anti-NRF2 antibody (NRF2), or anti-HA antibody (SYT-SSX1). SYT bound SYT-SSX1 but not NRF2. (B) SYT-SSX1 bound SYT-SSX1 itself. HA-tagged SYT-SSX1 was transfected with empty vector (lanes 1 and 4), Flag-tagged SYT (lanes 2 and 5), or Flag-tagged SYT-SSX1 (lanes 3 and 6) into HEK293T cells. HA-tagged SYT-SSX1 was immunoprecipitated from whole-cell lysates (Inp; left panel) using anti-HA antibody-conjugated agarose (HA-IP; right panel). Bound proteins were washed and detected with anti-HA antibody (SYT-SSX1) or anti-Flag antibody (SYT and SYT-SSX1). (C) A model of SYT target gene repression by SYT-SSX. In normal cells, SYT with its homooligomers activates target gene expression. In synovial sarcoma cells, SYT-SSX inhibits the transcriptional activity of SYT by a dominant-negative function by forming heterooligomers with SYT. The gene down-regulation by the dominant-negative function of SYT-SSX1 may lead to synovial sarcoma.
The SYT-SSX fusion protein directly down-regulates gene expression of COM1.
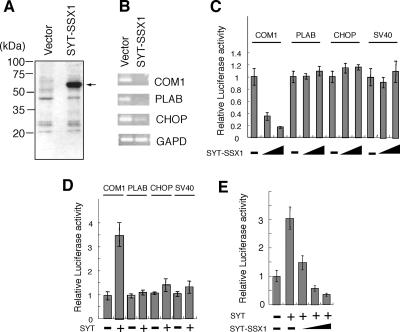
Although SYT-SSX fusion protein reportedly alters expression of several genes (19, 29-31), the direct target genes of SYT-SSX are unknown. To identify the direct gene targets of SYT-SSX, we established HeLa cell lines that stably expressed Flag-tagged SYT-SSX1. Expression of SYT-SSX1 fusion protein was first confirmed using the anti-Flag antibody M2 (Fig. 2A). SYT-SSX1 mRNA expression levels were about fivefold higher than those of endogenous SYT (data not shown). RNA was extracted from cells transfected with either the SYT-SSX1 construct or with an empty vector, and the expression levels of about 12,000 genes were compared using an oligonucleotide microarray (HG-U95A ver2 Array; Affymetrix; see the microarray data in the supplemental material). As listed in Table 1, analysis of mRNA expression profiles revealed a number of candidate SYT-SSX1 targets, i.e., those which exhibited at least a threefold decrease in expression. Of these, we focused on three cell proliferation regulators, COM1 (22, 24), PLAB (32), and CHOP (20). To confirm the microarray data, we performed reverse transcriptase PCR (RT-PCR) to analyze the expression levels of these three genes using the same RNA as that used for the array analysis. By comparing SYT-SSX1- and empty vector-transfected cell lines, we found that expression of the three candidate genes was indeed down-regulated in SYT-SSX1-expressing cells (Fig. 2B). When we analyzed gene expression levels in three other SYT-SSX1-transfected HeLa cell lines, results similar to those shown in Fig. 2B were obtained (data not shown; also see Fig. S1A in the supplemental material).
FIG. 2.
Down-regulation of COM1 by SYT-SSX1 fusion protein. (A) Western blot analysis of SYT-SSX1 cDNA-expressing HeLa cells and empty vector-transfected HeLa cells. Whole-cell lysates (40 μg) were analyzed by anti-Flag antibody. (B) Expression levels of COM1, PLAB, and CHOP in SYT-SSX1 cDNA-expressing HeLa cells and empty vector-transfected HeLa cells. RT-PCR was performed for 30 cycles. Amplification of the glyceraldehyde-3-phosphate dehydrogenase (GAPD) gene was used as a positive control. (C) Luciferase activity of COM1, PLAB, and CHOP promoters in the presence of the SYT-SSX1 fusion protein. pGL3-basic reporter plasmid (100 ng) containing COM1, PLAB, or CHOP promoter sequence was cotransfected with increasing amounts (0, 100, or 500 ng) of SYT-SSX1 into NIH 3T3 cells. pGL3-basic plasmid containing an SV40 promoter sequence (pGL3 promoter vector) was used for a negative control. Luciferase activity in the cell lysate was normalized with Renilla luciferase activity of pRL-TK as an internal control. The activity for each promoter in the absence of SYT-SSX1 was set at 1. (D) Luciferase activity of COM1, PLAB, and CHOP promoters in the presence of the SYT protein. pGL3-basic reporter plasmid (100 ng) containing COM1, PLAB, or CHOP promoter sequence was cotransfected with (300 ng) or without SYT into NIH 3T3 cells. pGL3-basic plasmid containing an SV40 promoter sequence (pGL3-promoter vector) was used for a negative control. The activity of each promoter in the absence of SYT was set at 1. SYT activated the COM1 promoter but not the PLAB and CHOP promoters. (E) Luciferase activity of SYT-SSX1 in the presence of SYT. pGL3-basic reporter plasmid (100 ng) containing a COM1 promoter sequence was cotransfected with SYT (200 ng) and SYT-SSX1 (0, 200, 400, or 600 ng) into NIH 3T3 cells. The activity in the absence of SYT and SYT-SSX1 was set at 1.
TABLE 1.
Gene expression profiles: genes down-regulated by SYT-SSX1
| Gene | Accession no. | Signal intensity
|
Ratioa | |
|---|---|---|---|---|
| Vector | SYT-SSX1 | |||
| BAX | NM_004324 | 7,587 | 1,459 | 0.192 |
| B4GALT1 | NM_001497 | 18,152 | 4,746 | 0.261 |
| CHOP | NM_004083 | 4,183 | 510 | 0.122 |
| COM1 | NM_012385 | 2,968 | 237 | 0.080 |
| ORP150 | NM_006389 | 17,324 | 5,326 | 0.307 |
| PLAB | NM_004864 | 11,376 | 1,718 | 0.151 |
Signal intensity of SYT-SSX1/signal intensity of vector.
Next, to determine whether these genes were directly down-regulated by SYT-SSX1, we used PCR and genomic DNA to amplify the promoter DNA sequence from each of the three genes, inserted each promoter sequence upstream of a luciferase gene in a reporter plasmid, and analyzed the impact of SYT-SSX1 expression. While SYT-SSX1 down-regulated COM1 promoter activity, promoter activities of PLAB, CHOP, and simian virus 40 (SV40) (control) were not affected in NIH 3T3 cells (Fig. 2C). In contrast to SYT-SSX1, we found that SYT up-regulated COM1 promoter activity but not PLAB and CHOP promoter activities in NIH 3T3 cells (Fig. 2D). Next, to examine whether SYT-SSX1 repressed the transcriptional activation of SYT on the COM1 promoter, we analyzed COM1 promoter activity in NIH 3T3 cells cotransfected with SYT and with different doses of SYT-SSX1. The results shown in Fig. 2E showed that SYT up-regulated COM1 promoter activity, whereas that activity was decreased by SYT-SSX1. Those results strongly suggested that SYT-SSX1 inhibited the transcriptional activity of SYT by its dominant-negative function on the COM1 promoter, as shown in Fig. 1C.
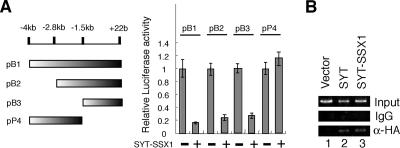
To identify the promoter region in which SYT-SSX1 down-regulates activity, we analyzed COM1 promoter deletion mutants which were 4 kb, 2.8 kb, and 1.5 kb upstream from the transcriptional start site (Fig. 3A, left panel). We found that SYT-SSX1 affected the region 1.5 kb upstream of the COM1 start site but not the region kb −4 kb to −1.5 (Fig. 3A, right panel). To examine whether SYT-SSX1 or SYT functions 1.5 kb upstream from the COM1 promoter, we analyzed the binding activities of SYT-SSX1 and SYT using chromatin immunoprecipitation (ChIP) analysis. For this purpose, HEK293T cells were transfected with the vector alone, HA-tagged SYT-SSX1, or HA-tagged SYT. After 3 days of incubation, the cells were treated with 1% formaldehyde to cross-link proteins to DNA and processed for ChIP assay with anti-HA antibody. The results in Fig. 3B showed that SYT-SSX1 or SYT bound to the COM1 promoter region. These data suggest that COM1 could be a direct target of SYT, a target which is down-regulated by the SYT-SSX1 fusion gene product in synovial sarcoma. Because SYT-SSX1 and SYT lack obvious DNA binding motifs, we speculate that both SYT-SSX1 and SYT bind to the COM1 promoter region through DNA binding proteins which recognize the 1.5-kb upstream region of the COM1 promoter. In future work, we will search the critical promoter region affected by SYT-SSX1 repression and attempt to identify the DNA binding proteins that recognize the region. These studies will provide improved understanding of the molecular mechanisms by which SYT-SSX1 contributes to synovial sarcomas.
FIG. 3.
SYT located at the promoter region of COM1. (A) Luciferase activity of deletion mutants of the COM1 promoter in the presence of SYT-SSX1 protein. pB1 (kb −4 ∼ +22), pB2(kb −2.8 ∼ +22), or pB3(kb −1.5 ∼ +22) was the pGL3-basic reporter plasmid containing deletion mutants (the 4-kbp, 2.8-kbp, or 1.5-kbp upstream region, respectively, from the transcriptional start site) of the COM1 promoter. pP4 (kb −4 ∼ −1.5) was the pGL3 promoter vector with an SV40 promoter containing the region from kb 1.5 to 4 of the COM1 promoter. These constructs (100 ng) were cotransfected with (500 ng) or without SYT-SSX1 into NIH 3T3 cells. The activity for each promoter in the absence of SYT-SSX1 was set at 1. (B) Binding activity of SYT-SSX1 or SYT to the 1.5-kbp upstream region from the transcriptional start site of the COM1 promoter. HEK293T cells were transfected with the vector alone (5 μg), HA-tagged SYT-SSX1 (5 μg), or HA-tagged SYT (5 μg). After 3 days of incubation, the cells were treated with 1% formaldehyde to cross-link proteins to DNA and processed by ChIP assay with anti-HA antibody or immunoglobulin G (IgG) (negative control). DNA was amplified by PCR performed for 35 cycles.
COM1 is expressed at low levels in synovial sarcoma tissues and cell lines.
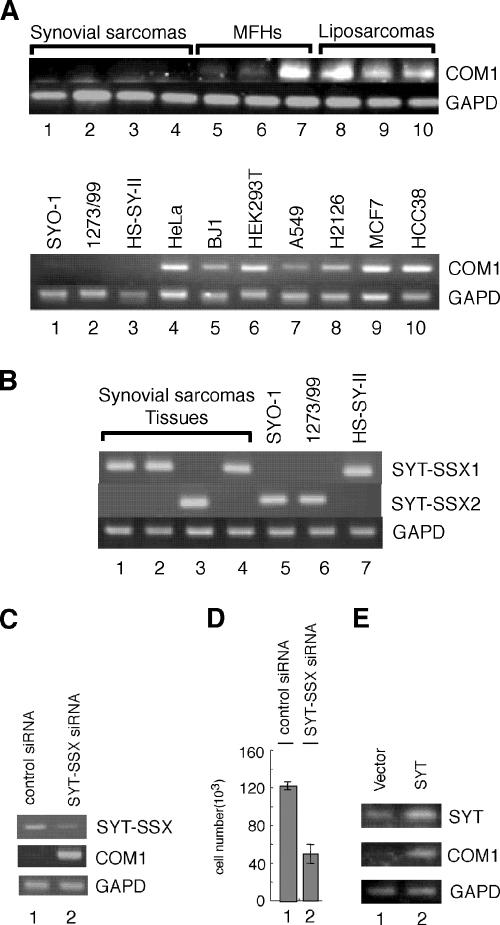
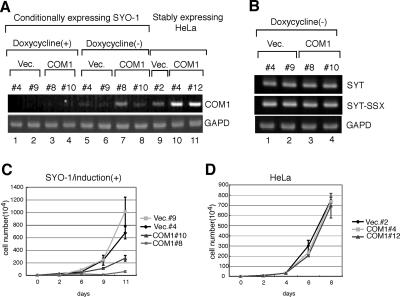
To examine the expression of COM1 in synovial sarcoma, we performed RT-PCR analysis. COM1 expression was not detected in either synovial sarcoma tissues (Fig. 4A, upper panel, lanes 1 to 4) or in cell lines (Fig. 4A, lower panel, lanes 1 to 3; SYO-1, 1273/99, and HS-SY-II cell lines) after 30 cycles of PCR. However, COM1 expression was detected in other types of sarcoma, including malignant fibrous histiocytomas and liposarcomas, under the same RT-PCR conditions (Fig. 4A, upper panel, lanes 5 to 10). Expression was also observed in other cell lines (Fig. 4A, lower panel, lanes 4 to 10; the fibroblast cell line BJ1 and other tumor-derived cell lines). These results suggested that COM1 expression was reduced in synovial sarcoma tissues and cell lines. Next, we analyzed expression levels of SYT-SSXs (SYT-SSX1 and SYT-SSX2) in synovial sarcoma tissues and cell lines. We determined that SYT-SSX1 was expressed in three synovial sarcoma tissues and the HS-SY-II cell line (Fig. 4B, upper panel, lanes 1, 2, 4, and 7) and that SYT-SSX2 was expressed in one synovial sarcoma tissue and in the SYO-1 and 1273/99 cell lines (Fig. 4B, middle panel, lanes 3, 5, and 6). Thus, the lower level of expression of COM1 in both synovial sarcoma tissues and cell lines correlated with the expression of the SYT-SSX fusion gene.
FIG. 4.
Lower-level expression of COM1 in synovial sarcoma tissues and cell lines and conditional expression of COM1 in synovial sarcoma cell lines. (A) COM1 expression in sarcoma tissues (four synovial sarcoma, three malignant fibrous histiocytoma [MFHs], and three liposarcoma tissues) (upper panel) and in synovial sarcoma cell lines (SYO-1, 1273/99, and HS-SY-II), fibroblast cell line BJ-1, and tumor-derived cell lines (lower panel) was analyzed by RT-PCR performed for 30 cycles using COM1-specific primers. The glyceraldehyde-3-phosphate dehydrogenase (GAPD) gene was used as a positive control as described in the legend to Fig. 2B. (B) Expression levels of SYT-SSXs (SYT-SSX1 and SYT-SSX2) in synovial sarcoma tissues (lanes 1 to 4) and cell lines (lanes 5 to 7). SYT-SSX expression was analyzed by RT-PCR performed for 30 cycles using SYT-SSX1- or SYT-SSX2-specific primers. The GAPD gene was used as a positive control as described in the legend to Fig. 2B. (C) COM1 expression was increased by the knock-down of SYT-SSX2 in synovial sarcoma cell line SYO-1. The control siRNA (lane 1) or the siRNA specific for the SYT-SSX fused junction region (lane 2) was transfected into SYO-1 cells. Three days after transfection, cells were collected. The levels of SYT-SSX and COM1 expression were analyzed by RT-PCR, which was performed for 30 cycles. (D) The cell growth of SYO-1 cells in the knock-down of SYT-SSX. The control siRNA (lane 1) or the siRNA specific for the SYT-SSX fused junction region (lane 2) was transfected into SYO-1 cells. Seven days after transfection, cells were collected and the numbers were counted. The knock-down of SYT-SSX by the siRNA specific for the SYT-SSX fused junction region induced cell growth inhibition in SYO-1 cells. (E) COM1 expression levels after the addition of SYT in SYO-1 cells. SYT expression vector (5 μg) or empty vector (5 μg) was transfected into SYO-1 cells. Three days after transfection, cells were collected. The levels of SYT and COM1 expression were analyzed by RT-PCR, which was performed for 30 cycles.
To confirm that expression of SYT-SSX reduced expression of COM1 in synovial sarcoma cell lines, we employed a small interference RNA (siRNA) to suppress SYT-SSX fusion. For this purpose, SYO-1 cells were transfected with control siRNA or SYT-SSX-specific siRNA (which targeted the fused region of SYT-SSX). After 3 days of incubation, expression of SYT-SSX2 and COM1 was analyzed by RT-PCR. We found that the SYT-SSX-specific siRNA reduced SYT-SSX2 expression in SYO-1 cells (Fig. 4C, upper panel, lane 1 versus lane 2). Furthermore, SYT-SSX-specific siRNA strongly increased COM1 expression (Fig. 4C, middle panel, lane 1 versus lane 2).
To determine whether knockdown of SYT-SSX2 expression impacted growth of SYO-1 cells, we analyzed growth for 7 days after transfection of the cells with SYT-SSX-specific siRNA. The knockdown of SYT-SSX expression reduced growth of SYO-1 cells 3 days after the transfection of SYT-SSX-specific siRNA (data not shown). As displayed in Fig. 4D, after 7 days, the number of cells in cultures transfected with SYT-SSX-specific siRNA was approximately one third of that in cultures transfected with control siRNA. These results suggested that the SYT-SSX fusion protein led to down-regulation of COM1 expression and stimulated growth in synovial sarcoma cell line SYO-1. Next, we determined how the level of endogenous COM1 expression changed in response to overexpression of SYT in SYO-1 cells. SYT cDNA was transiently transfected into SYO-1 cells. Results demonstrated that COM1 expression increased in SYO-1 cells (Fig. 4E, middle panel). These experiments with SYO-1 cells strongly suggested that SYT-SSX inhibited the transcriptional activity of SYT by its dominant-negative function (Fig. 1C).
Increased COM1 expression in the synovial sarcoma cell line reduced in vitro cell growth and colony formation.
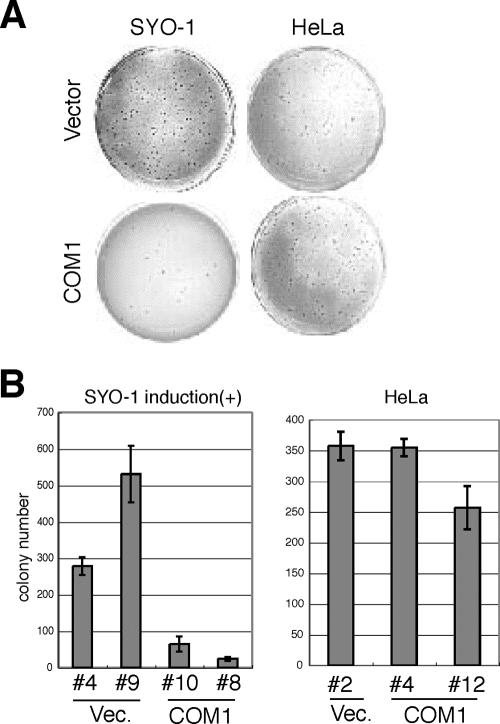
Since COM1 is highly expressed in metastatic breast cancer and T-cell leukemia (22, 24), it was believed to possess mitogenic activity (27). However, it has also been reported that COM1 inhibits the growth of pancreatic cancer cells and the breast cancer cell line MCF7 (2, 18). These findings suggest that COM1 mediates either growth inhibition or stimulation in a cell- or tissue-specific manner. To determine how reduced expression of COM1 affected synovial sarcoma, we attempted to produce SYO-1 and HeLa cells that stably expressed COM1. Although we readily isolated HeLa cells that stably expressed COM1, analogous SYO-1 cells could not be isolated (data not shown). Therefore, we attempted to produce SYO-1 cells that conditionally expressed COM1. SYO-1 cells containing a Tet-Off vector were transfected with pTRE2 vector containing COM1 and treated with doxycycline. After the withdrawal of doxycycline, SYO-1 cells conditionally expressing COM1 were analyzed for COM1 expression levels by RT-PCR analysis. Two days after doxycycline withdrawal, we observed a severalfold increase in COM1 mRNA levels (Fig. 5A, lanes 3 and 4 versus lanes 7 and 8). Induction of COM1 in SYO-1 cells conditionally expressing COM1 did not affect the expression levels of either SYT or SYT-SSX2 (Fig. 5B). These cell lines were then used to examine the effects of COM1 expression on cell growth. Growth of SYO-1 cells containing empty vector or COM1 expression vector with doxycycline was normal (see Fig. S1B in the supplemental material). In contrast, COM1 expression reduced growth of SYO-1 cells 3 days after the cessation of doxycycline treatment (Fig. 5C). On the other hand, COM1 expression showed no effects on the growth of HeLa cells (Fig. 5D). We then analyzed the effects of COM1 expression on colony formation in soft agar and found that increased COM1 expression following doxycycline withdrawal reduced colony formation in SYO-1 cells but not in HeLa cells (Fig. 6). Cell growth and colony formation activity of SYO-1 cells transfected with empty vector were almost the same as those seen with nontransfected SYO-1 cells (data not shown). These results suggested that reduced expression of COM1 may play a role in synovial sarcoma growth.
FIG. 5.
Conditional expression of COM1 induces slow growth rates in SYO-1 cells. (A) Expression of COM1 in SYO-1 cells that conditionally express COM1 using the Tet-Off system. Clones #8 and #10 are shown with doxycycline (+) and at 48 h after doxycycline withdrawal (−). Empty vector-transfected (Vec.) SYO-1 cells (clones #4 and #9) are also shown as controls. Expression of COM1 in HeLa cells that stably express COM1 (clones #4 and #12) and empty vector-transfected HeLa cells (clone #2) are also shown. COM1 expression was analyzed by RT-PCR, which was performed for 30 cycles. (B) Expression of SYT or SYT-SSX in SYO-1 cells that conditionally express COM1 using the Tet-Off system. RNAs were extracted from SYO-1 cells as described in the legend to panel A. Expression levels of SYT and SYT-SSX were analyzed by RT-PCR. The overexpression of COM1 showed no effects for the expression levels of SYT or SYT-SSX in SYO-1 cells. (C) Analysis of cell growth of SYO-1 cells that conditionally express COM1 or empty vector after the withdrawal of doxycycline [induction (+)] for 11 days. (D) Analysis of cell growth of HeLa cells that stably express COM1 or empty vector for 8 days.
FIG. 6.
Conditional expression of COM1 reduces colony formation activity in SYO-1 cells. (A) Analysis of colony formation activity in soft agar of SYO-1 or HeLa cells that express COM1 or empty vector after 24 days. (B) Average numbers of formed colonies depicted in Fig. 4B are shown. Vec., empty vector.
The restoration of COM1 expression induced apoptosis in SYO-1 cells.
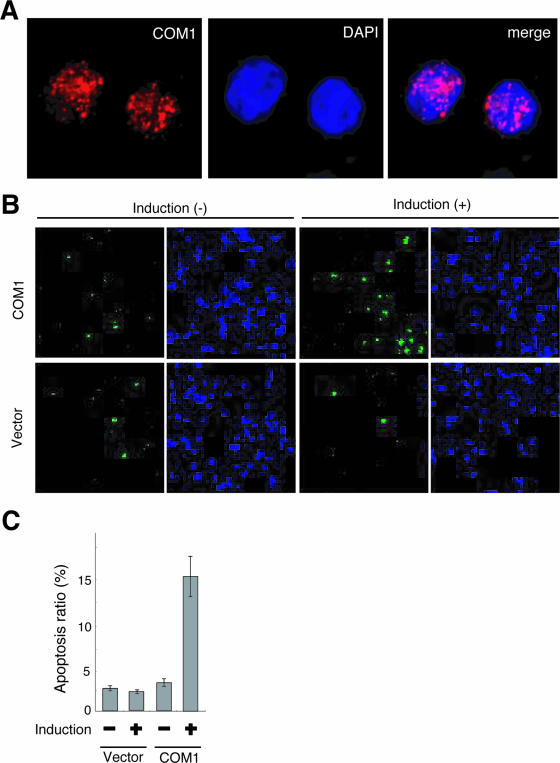
In preliminary results using COS-7 cells transiently transfected with COM1 cDNA, nuclear localization of COM1 was observed (27). In human thyroid cancer cells, COM1 was detected in either the nucleus or cytoplasm depending on the stage of the disease (15). Based on those studies, we analyzed subcellular location of Flag-tagged COM1 protein in SYO-1 cells using confocal laser-scanning microscopy. Red COM1 signals were observed within the nucleus with a punctate pattern and in regions with little blue DAPI signal (Fig. 7A), which suggested that COM1 resided mainly in areas of euchromatin within SYO-1 cell nuclei. However, the mechanism of growth inhibition mediated by COM1 expression in SYO-1 cell nuclei is still unclear. It was reported that COM1+/+ MEFs were more sensitive than COM1−/− MEFs to apoptosis (28). Therefore, to examine whether restoration of COM1 expression induces apoptosis in SYO-1 cells, we analyzed apoptosis of SYO-1 cells conditionally expressing COM1 using the covalent binding of a fluorochrome inhibitor to the active caspases (1). The apoptotic cells produced green signals (Fig. 7B). SYO-1 cells conditionally expressing COM1 showed a fourfold increase in the number of apoptotic cells 3 days after doxycycline withdrawal (Fig. 7B and C). In contrast, we did not detect any differences in the number of apoptotic cells in control vector-expressing SYO-1 cells after doxycycline withdrawal (Fig. 7B and C). This result suggested that the restoration of COM1 expression induced apoptosis in SYO-1 cells. Recently, it was reported that COM1 mediated apoptotic effects via up-regulation of the endoplasmic reticulum stress-related genes ATF4, CHOP, and TRB3 (4). We have found that the restoration of COM1 induced CHOP expression in SYO-1 cells (data not shown). These results may indicate that endoplasmic reticulum stress-related genes are involved in SYO-1 cell apoptosis mediated by COM1.
FIG. 7.
Restored COM1 protein localized in euchromatin and induced apoptosis in SYO-1 cells. (A) Localization of COM1 protein. Expression of COM1 in SYO-1 cells that conditionally express COM1 using the Tet-Off system. Clone #8 is shown at 72 h after doxycycline withdrawal. The signals were detected by indirect immunofluorescence with anti-FLAG antibody (COM1, left panel) and DAPI (middle panel). Merged signals (merge, right panel) showed that COM1 signals (red) were observed within the nucleus with a punctate pattern and in regions with little blue DAPI signal. (B) Apoptosis induction in SYO-1 cells by expression of COM1. Apoptosis was analyzed by caspase activity using a carboxyfluorescein FLICA apoptosis detection kit. The COM1-transfected SYO-1 cell (clone #8) is shown with doxycycline [induction(−)] and at 72 h after doxycycline withdrawal [induction (+)]. Empty vector-transfected SYO-1 cells (clone #4) are also shown as a control. The green signals are fluorescence of caspase-positive cells (left panel), and the blue signals are DNA visualized by Hoechst (right panel). (C) Average ratio of apoptosis (caspase-positive) cells depicted in Fig. 6B are shown. SYO-1 cells conditionally expressing COM1 showed a fourfold increase in apoptotic cell number 3 days after doxycycline withdrawal.
In summary, we found that the COM1 gene could be a direct target of SYT and that COM1 down-regulation by the dominant-negative activity of the SYT-SSX1 fusion gene product with translocation appeared to be involved in synovial sarcoma development. Our results indicate that the restoration of COM1 expression might be of therapeutic benefit in synovial sarcoma. Our study also helped to elucidate signaling pathways of the SYT-SSX fusion protein. We suggest that further explorations of the molecular mechanisms of growth inhibition mediated by COM1 could improve our understanding of synovial sarcoma progression and lead to new therapeutic approaches.
Supplementary Material
Acknowledgments
We thank Teruhiko Yoshida (NCCRI, Japan), Shinya Tanaka (Hokkaido Univ, Japan), Akira Kawai (NCCH, Japan), Susumu Hirose (NIG, Japan), and Toyomasa Katagiri (Tokyo Univ, Japan) for valuable suggestions. We also thank Rie Ito, Yoko Kitajima, and Reiko Odagawa (NCCRI, Japan) for technical support.
This work was supported by a Grant-in-Aid for Cancer Research from the Ministry of Health, Labor, and Welfare of Japan; a grant from the program for promotion of Fundamental Studies in Health Sciences of the National Institute of Biomedical Innovation (NiBio); and a grant from the program for promotion of Fundamental Studies in Health Sciences of the Organization for Pharmaceutical Safety and Research, Ministry of Education, Culture, Sports, Science, and Technology, Japan.
Footnotes
Published ahead of print on 13 November 2006.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Bedner, E., P. Smolewski, P. Amstad, and Z. Darzynkiewicz. 2000. Activation of caspases measured in situ by binding of fluorochrome-labeled inhibitors of caspases (FLICA): correlation with DNA fragmentation. Exp. Cell Res. 259:308-313. [DOI] [PubMed] [Google Scholar]
- 2.Bratland, A., K. Risberg, G. M. Maelandsmo, K. B. Gutzkow, O. E. Olsen, A. Moghaddam, M. Y. Wang, C. M. Hansen, H. K. Blomhoff, J. P. Berg, O. Fodstad, and A. H. Ree. 2000. Expression of a novel factor, com1, is regulated by 1,25-dihydroxyvitamin D3 in breast cancer cells. Cancer Res. 60:5578-5583. [PubMed] [Google Scholar]
- 3.Brett, D., S. Whitehouse, P. Antonson, J. Shipley, C. Cooper, and G. Goodwin. 1997. The SYT protein involved in the t(X;18) synovail sarcoma translocation is a transcriptional activator localised in nuclear bodies. Hum. Mol. Genet. 6:1559-1564. [DOI] [PubMed] [Google Scholar]
- 4.Carracedo, A., M. Lorente, A. Egia, C. Blazquez, S. Garcia, V. Giroux, C. Malicet, R. Villuendas, M. Gironella, L. Gonzalez-Feria, M. A. Piris, J. L. Iovanna, M. Guzman, and G. Velasco. 2006. The stress-regulated protein p8 mediates cannabinoid-induced apoptosis of tumor cells. Cancer Cell 4:301-312. [DOI] [PubMed] [Google Scholar]
- 5.Clark, J., P. J. Rocques, A. J. Crew, S. Gill, J. Shipley, A. M. Chan, B. A. Gusterson, and C. S. Cooper. 1994. Identification of novel genes, SYT and SSX, in the t(X;19)(p11.2;q11.2) translocation found in human synovial sarcoma. Nat. Genet. 7:502-508. [DOI] [PubMed] [Google Scholar]
- 6.Crew, A. J., J. Clark, C. Fisher, S. Gill, R. Grimer, A. Chand, J. Shipley, B. A. Gusterson, and C. S. Cooper. 1995. Fusion of SYT to two genes, SSX1 and SSX2, encoding proteins with homology to the Kuppel-associated box in human synovial sarcoma. EMBO J. 14:2333-2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Bruijn, D. R., N. R. dos Santos, J. Thijssen, M. Balemans, S. Debernardi, B. Linder, B. D. Young, and A. G. van Kessel. 2001. The synovial sarcoma associated protein SYT interacts with the acute leukemia associated protein AF10. Oncogene 20:3281-3289. [DOI] [PubMed] [Google Scholar]
- 8.de Leeuw, B., M. Balemans, D. Olde Weghuis, and A. G. van Kessel. 1995. Identification of two alternative fusion genes, SYT-SSX1 and SYT-SSX2, in t(X;18)(p11.2;q11.2)-positive synovial sarcomas. Hum. Mol. Genet. 4:1097-1099. [DOI] [PubMed] [Google Scholar]
- 9.dos Santos, N. R., D. R. de Bruijn, and A. G. van Kessel. 2001. Molecular mechanisms underlying human synovial sarcoma development. Genes Chromosomes Cancer 30:1-14. [DOI] [PubMed] [Google Scholar]
- 10.Eid, J. E., A. L. Kung, R. Scully, and D. M. Livingston. 2000. p300 Interacts with the nuclear proto-oncoprotein SYT as part of the active control of cell adhesion. Cell 102:839-848. [DOI] [PubMed] [Google Scholar]
- 11.Gure, A. O., O. Tureci, U. Sahin, S. Tsang, M. J. Scanlan, E. Jager, A. Knuth, M. Pfreundschuh, L. J. Old, and Y. T. Chen. 1997. SSX: a multigene family with several members transcribed in normal testis and human cancer. Int. J. Cancer 72:965-971. [DOI] [PubMed] [Google Scholar]
- 12.Gure, A. O., I. J. Wei, L. J. Old, and Y. T. Chen. 2002. The SSX gene family: characterization of 9 complete genes. Int. J. Cancer 101:448-453. [DOI] [PubMed] [Google Scholar]
- 13.Ishida, M., S. Tanaka, M. Ohki, and T. Ohta. 2004. Transcriptional co-activator activity of SYT is negatively regulated by BRM and Brg1. Genes Cells 9:419-428. [DOI] [PubMed] [Google Scholar]
- 14.Ito, T., M. Ouchida, S. Ito, Y. Jitsumori, Y. Morimoto, T. Ozaki, A. Kawai, H. Inoue, and K. Shimizu. 2004. SYT, a partner of SYT-SSX oncoprotein in synovial sarcomas, interacts with mSin3A, a component of histone deacetylase complex. Lab. Investig. 84:1484-1490. [DOI] [PubMed] [Google Scholar]
- 15.Ito, Y., H. Yoshida, Y. Motoo, E. Miyoshi, J. L. Iovanna, C. Tomoda, T. Uruno, Y. Takamura, A. Miya, K. Kobayashi, F. Matsuzuka, N. Matsuura, K. Kuma, and A. Miyauchi. 2003. Expression and cellular localization of p8 protein in thyroid neoplasms. Cancer Lett. 201:237-244. [DOI] [PubMed] [Google Scholar]
- 16.Ladanyi, M., C. R. Antonescu, D. H. Leung, J. M. Woodruff, A. Kawai, J. H. Healey, M. F. Brennan, J. A. Bridge, J. R. Neff, F. G. Barr, J. D. Goldsmith, J. S. Brooks, J. R. Goldblum, S. Z. Ali, J. Shipley, C. S. Cooper, C. Fisher, B. Skytting, and O. Larsson. 2002. Impact of SYT-SSX fusion type on the clinical behavior of synovial sarcoma: a multi-institutional retrospective study of 243 patients. Cancer Res. 62:135-140. [PubMed] [Google Scholar]
- 17.Lim, F. L., M. Soulez, D. Koczan, H. J. Thiesen, and J. C. Knight. 1998. A KRAB-related domain and a novel transcription repression domain in protein encoded by SSX genes that are disrupted in human sarcomas. Oncogene 17:2013-2018. [DOI] [PubMed] [Google Scholar]
- 18.Malicet, C., N. Lesavre, S. Vasseur, and J. L. Iovanna. 2003. p8 inhibits the growth of human pancreatic cancer cells and its expression is induced through pathways involved in growth inhibition and repressed by factors promoting cell growth. Mol. Cancer 2:37-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nagai, M., S. Tanaka, M. Tsuda, S. Endo, H. Kato, H. Sonobe, A. Minami, H. Hiraga, H. Nishihara, H. Sawa, and K. Nagashima. 2001. Analysis of transforming activity of human synovial sarcoma-associated chimeric protein SYT-SSX1 bound to chromatin remodeling factor hBRM/hSNF1αlpha. Proc. Natl. Acad. Sci. USA 98:3843-3848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Papathanasiou, M. A., N. C. Kerr, J. H. Robbins, O. W. McBride, I. Alamo, Jr., S. F. Barrett, I. D. Hickson, and A. J. Fornace, Jr. 1991. Induction by ionizing radiation of the gadd45 gene in cultured human cells: lack of mediation by protein kinase C. Mol. Cell. Biol. 11:1009-1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Perani, M., C. J. Ingram, C. S. Cooper, M. D. Garrett, and G. H. Goodwin. 2003. Conserved SNH domain of the proto-oncoprotein SYT interacts with components of the human chromatin remodelling complexes, while the QPGY repeat domain forms homo-oligomers. Oncogene 22:8156-8167. [DOI] [PubMed] [Google Scholar]
- 22.Ree, A. H., M. Tvermyr, O. Engebraaten, M. Rooman, O. Rosok, E. Hovig, L. A. Meza-Zepeda, O. S. Bruland, and O. Fodstad. 1999. Expression of a novel factor in human breast cancer cells with metastatic potential. Cancer Res. 59:4675-4680. [PubMed] [Google Scholar]
- 23.Skytting, B., G. Nilsson, B. Brodin, Y. Xie, J. Lundeberg, M. Uhlen, and O. Larsson. 1999. A novel fusion gene, SYT-SSX4, in synovial sarcoma. J. Natl. Cancer Inst. 91:974-975. [DOI] [PubMed] [Google Scholar]
- 24.Soulier, J., A. Madani, V. Cacheux, M. Rosenzwajg, F. Sigaux, and M. H. Stern. 1994. The MTCP-1/c6.1B gene encodes for a cytoplasmic 8 kD protein overexpressed in T cell leukemia bearing a t(X;14) translocation. Oncogene 9:3565-3570. [PubMed] [Google Scholar]
- 25.Thaete, C., D. Brett, P. Monaghan, S. Whitehouse, G. Rennie, E. Rayner, C. S. Cooper, and G. H. Goodwin. 1999. Functional domains of SYT and SYT-SSX synovial sarcoma translocation proteins and co-localization with the SNF protein BRM in the nucleus. Hum. Mol. Genet. 8:585-591. [DOI] [PubMed] [Google Scholar]
- 26.Tureci, O., U. Sahin, I. Schobert, M. Koslowski, H. Scmitt, H. J. Schild, F. Stenner, G. Seitz, H. G. Rammensee, and M. Pfreundschuh. 1996. The SSX-2 gene, which is involved in the t(X;18) translocation of synovial sarcomas, codes for the human tumor antigen HOM-MEL-40. Cancer Res. 56:4766-4772. [PubMed] [Google Scholar]
- 27.Vasseur, S., G. Vidal Mallo, F. Fiedler, H. Bodeker, E. Canepa, S. Moreno, and J. L. Iovanna. 1999. Cloning and expression of the human p8, a nuclear protein with mitogenic activity. Eur. J. Biochem. 59:670-675. [DOI] [PubMed] [Google Scholar]
- 28.Vasseur, S., A. Hoffmeister, A. Garcia-Montero, G. V. Mallo, R. Feil, S. Kuhbandner, J. C. Dagorn, and J. L. Iovanna. 2002. p8-deficient fibroblasts grow more rapidly and are more resistant to adriamycin-induced apoptosis. Oncogene 21:1685-1694. [DOI] [PubMed] [Google Scholar]
- 29.Xie, Y., B. Skytting, G. Nilsson, A. Gasbarri, K. Haslam, A. Bartolazzi, B. Brodin, N. Mandahl, and O. Larsson. 2002. SYT-SSX is critical for cyclin D1 expression in synovial sarcoma cells: a gain of function of the t(X;18)(p11.2;q11.2) translocation. Cancer Res. 62:3861-3867. [PubMed] [Google Scholar]
- 30.Xie, Y., B. Skytting, G. Nilsson, R. J. Grimer, C. D. Mangham, C. Fisher, J. Shipley, B. Bjerkehagen, O. Myklebost, and O. Larsson. 2002. The SYT-SSX1 fusion type of synovial sarcoma is associated with increased expression of cyclin A and D1. A link between t(X;18)(p11.2; q11.2) and the cell cycle machinery. Oncogene 21:5791-5796. [DOI] [PubMed] [Google Scholar]
- 31.Xie, Y., M. Tornkvist, Y. Aalto, G. Nilsson, L. Girnita, B. Nagy, S. Knuutila, and O. Larsson. 2003. Gene expression profile by blocking the SYT-SSX fusion gene in synovial sarcoma cells. Identification of XRCC4 as a putative SYT-SSX target gene. Oncogene 22:7628-7631. [DOI] [PubMed] [Google Scholar]
- 32.Yokoyama-Kobayashi, M., M. Saeki, S. Sekine, and K. Kato. 1997. Human cDNA encoding a novel TGF-beta superfamily protein highly expressed in placenta. J. Biochem. (Tokyo) 122:622-626. [DOI] [PubMed] [Google Scholar]
- 33.Zhang, D., H. G. Yoon, and J. Wong. 2005. JMJD2A is a novel N-CoR-interacting protein and is involved in repression of the human transcription factor achaete scute-like homologue 2 (ASCL2/Hash2). Mol. Cell. Biol. 25:6404-6414. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.