Abstract
Understanding the relationship between microbial community dynamics and functional instability is an important step towards designing reliable biological water treatment systems. In this study, the community dynamics of two dispersed-growth denitrifying reactors were examined during periods of functional stability and instability. In both reactors during the period of functional instability, the effluent chemistry changed over time, with periods of high nitrate concentrations followed by periods of fluctuating nitrite concentrations. Community structure was examined by clone library analysis of the 16S rRNA gene. Community dynamics were investigated with terminal restriction fragment (T-RF) length polymorphism, and the functional diversity represented by T-RFs was assessed through nitrate reduction assays of representative isolates. During the period of functional instability, the community structure changed considerably, and the dynamics correlated significantly with effluent chemistry. The nitrite concentration was significantly correlated with the relative abundances of the nitrate-reducing Delftia- and Achromobacter-like T-RFs. The isolate representing the Acidovorax-like T-RF reduced nitrate directly to nitrogen in batch assays without the accumulation of any intermediates. The Acidovorax-like T-RF relative abundance was significantly negatively correlated with nitrite concentration, indicating that it was associated with good functional performance. The results of this study reveal a clear relationship between community dynamics and functional instability and the importance of diversity among nitrate-reducing populations within a denitrifying community.
Biological water treatment systems must be functionally stable and able to continuously treat contaminants over long times while relying upon the activity of complex microbial communities. However, the ecological principles underlying microbial community structure, dynamics, and functional stability and how they are related are poorly understood. The elucidation of such principles and the application of them to engineering design, inoculation, and operation could potentially improve system stability (4, 10, 28).
The development of molecular techniques has allowed the investigation of diversity and community dynamics in many functionally stable and unstable biological water treatment systems (11, 16, 17, 25, 26, 29-31, 34). However, findings have not been consistent across studies, hindering the formulation of general principles. For instance, in one comparative study of two nitrifying units, the more stable system was also more diverse (29), but the opposite was found in a comparison of two activated sludge communities treating coking effluent (25). In studies of community dynamics, functional stability was not necessarily accompanied by community stability (11, 16, 17, 30), and functional instability occurred in systems with both stable and dynamic communities (26, 31, 34). When community dynamics have been observed during periods of functional instability, there has not been an obvious connection between changes in community structure and the changes in functional performance (26, 31).
Denitrification is a critical process in many engineering applications including nitrogen removal from wastewater and bioremediation of polluted groundwater, and it is also an important process in the global nitrogen cycle. In engineering applications, the complete reduction of nitrate to nitrogen gas without the accumulation of intermediates is required. Nitrite is a particularly problematic intermediate, because it is more toxic than nitrate, has a lower maximum contaminant level than nitrate (33a), inhibits bacterial activity (1, 19, 20), and poses a eutrophication threat to nutrient-limited environments. Therefore, functional stability for denitrifying systems is defined here as the maintenance of the complete reduction of nitrate to nitrogen over time without the accumulation of nitrite.
From an ecological perspective, denitrifying communities are particularly interesting for studies of functional stability due to the diversity among denitrifying species in their tendencies to accumulate the denitrification intermediates, especially nitrite and nitrous oxide (2, 5, 27). Additionally, not all nitrate-reducing bacteria are true denitrifiers, because they cannot all completely reduce nitrate to nitrogen. Some, referred to as nitrate respirers, are capable of reduction to nitrite only (33). The exploration of functional diversity in engineered denitrifying systems through isolations has revealed the presence of nitrate respirers as well as denitrifiers with varied tendencies to accumulate intermediates (12, 17, 27).
The functional diversity of nitrate-reducing bacteria suggests that system function may depend upon the functional composition of the denitrifying community and that functional stability could, in turn, be related to community dynamics. The link between functional composition and system function in denitrifying environments has been examined with culture-based methods and mathematical modeling in soils, laboratory bioreactors, and full-scale treatment systems (6, 7, 26, 27, 35). In studies of soil-denitrifying communities and constructed communities in chemostats, the system function (i.e., accumulation or absence of nitrite or nitrous oxide) was reflected in the functional composition of isolates from the community (i.e., the tendency of isolates to produce nitrite or nitrous oxide) (6, 7, 27). Examinations of community dynamics have been restricted to functional characterization of isolates over time (26), limiting the establishment of a link between the function of individual populations, their dynamics, and functional performance.
In this investigation, model dispersed-growth reactors were established from a previously described pilot-scale denitrifying fluidized bed reactor (FBR) (17). After functionally stable operation for over 160 days, one of the reactors experienced a complete loss of denitrification activity followed by over 150 days of functional instability. Frequent molecular analysis of community dynamics during this period was coupled with the occasional functional characterization of representative isolates. From this data set, strong, significant correlations between the functional composition of the community, its dynamics, and functional instability were established.
MATERIALS AND METHODS
Reactor specifications and operation.
Reactor vessels consisted of sterile 2-liter Erlenmeyer flasks fitted with sterile rubber stoppers and glass tubing ports for feed, waste, sampling, and gas venting. Reactors were fed constantly. The feed solution was provided at a rate of 1 ml min−1 with a peristaltic pump consisting of a Masterflex 7520-50 console drive (Cole-Parmer, Barrington, IL) and a dual-channel Masterflex 77202-50 pump head (Cole-Parmer), resulting in a delivered volume of 180 ml every 3 h. Waste pumps consisting of a Masterflex 7553-10 drive and a Masterflex 7016 pump head (Cole-Parmer) were controlled by timers. Every 3 h, the pumps were turned on and run for a time period sufficient for the reactor contents to be pumped to the level of waste tubing (tube used for removal of culture). The position of this tube was set such that the volume of reactor contents after pumping was 1.8 liters, resulting in an average volume of 1.9 liters. To allow for the pressure changes that accompanied the volume changes of each cycle and to prevent air leaks into the system, the gas vent line was connected to a balloon with a sterile syringe filter. Constant mixing was provided by a Nova 2300 platform shaker (New Brunswick Scientific Co., Inc., Edison, NJ) operated at 100 rpm.
The reactors discussed in the present study, designated DGR1 and DGR2, were the last two in a series of five identical dispersed-growth reactors originally inoculated with the effluent from a previously described pilot-scale denitrifying FBR (17). The FBR had been inoculated with a denitrifying enrichment culture developed from groundwater from well TPB-16 at the Field Research Center in Oak Ridge, Tennessee (http://www.esd.ornl.gov/nabirfrc/), and was operated under nonsterile conditions for 2 years prior to inoculation of the first dispersed-growth reactor. This study examined a 335-day period of operation of DGR1, starting on day 0, when the reactor was inoculated. DGR2 was inoculated from DGR1 after 6 months of operation. Nitrate and nitrite concentrations were frequently monitored throughout the course of operation, and nitrous oxide was occasionally measured. Liquid samples (1.5 ml) for nitrate and nitrite analyses were centrifuged at 16,000 × g for 10 min, decanted, and stored at −20°C. Nitrous oxide samples were removed from the reactor headspace with a Hamilton syringe and analyzed immediately. Liquid concentrations of nitrous oxide were then calculated using a Henry's constant of 2.4 · 10−2 mole liter−1 atm−1 (36). Both reactors were operated for an additional year following the period that is presented in this study.
The sterile feed solutions consisted of nitrate, lactic acid and ethanol as electron donors, nutrient salts, and trace elements. Nitrate concentrations varied, and nutrient salts were provided in proportion to the nitrate concentration. For example, the feed solution initially contained (per liter) 1.9 g NaNO3, 2.3 g KNO3, and nutrient salts in the following amounts: 42 mg of Na3P3O9, 21 mg of NaSO4, 32 mg of MgCl2 · 6H2O, and 46 mg of CaCl2 · 2H2O. Trace elements were provided in the following amounts (per liter), regardless of nitrate concentration: 0.3 mg of FeCl2 · 4H2O, 0.1 mg of ZnSO4 · 7H2O, 85 μg of MnSO4 · H2O, 60 μg of H3BO3, 19 μg of CoCl2 · 6H2O, 4 μg of CuSO4 · 5H2O, 28 μg of NiSO4 · 6H2O, and 40 μg of Na2MoO4 · 2H2O. The pH of the feed solution was adjusted to a range of 2.4 to 4.3, depending upon influent nitrate and lactic acid concentrations, with 2 N NaOH. The remaining acidity was neutralized to circumneutral pH by alkalinity produced during denitrification.
The electron donors ethanol and lactic acid were provided in equal concentrations on a chemical oxygen demand (COD) basis. The COD/N ratio was adjusted to minimize the COD in the effluent and ensure the complete reduction of nitrate and nitrite. Upon start-up of the first reactor, a COD/N ratio of 5.6 resulted in residual nitrogen oxides in the reactor effluent. Ethanol and lactate concentrations were therefore increased until no residual nitrogen oxides remained, resulting in a COD/N ratio of 8.2. Subsequent changes to the COD/N ratio were undertaken in order to minimize COD in the effluent while maintaining the complete removal of nitrate and nitrite, as will be discussed in Results.
Isolations.
As previously described, isolate G1 was obtained from the pilot-scale FBR used as an inoculum for the reactors in this study (17). One month after the period of DGR1 operation examined in this study, additional isolations were made from DGR1 and from a culture of DGR1 diluted 1/10 in a phosphate buffer (2.6 g liter−1 K2HPO4, 2.0 g liter−1 KH2PO4). This diluted culture was regrown by spiking four times with a feed solution containing (per liter) 310 mg NaNO3, 110 mg C2H6O, 270 mg NaC3H5O3, and nutrients and trace elements in the same proportions as the reactor feed solution. For isolation, a dilution series of each sample was prepared in a phosphate buffer (2.6 g liter−1 K2HPO4, 2.0 g liter−1 KH2PO4, 2.9 g liter−1 NaCl), and aliquots of this dilution series were plated onto Luria-Bertani medium and incubated aerobically at 30°C. Forty-nine isolates representing 13 morphologically distinct colony types were picked, restreaked to purity, grown aerobically in liquid LB cultures, and stored in 20% glycerol at −80°C. Partial 16S rRNA gene sequences were obtained for 32 of the isolates, representing all of the morphological groups (see below for sequencing procedures).
Isolates closely related to the Acidovorax, Delftia, and Achromobacter species were assayed for the reduction of nitrate and the production of nitrite, nitrous oxide, and nitrogen gas. Batch assays were carried out in sealed anaerobic serum bottles with a helium headspace and the following medium composition (per liter): 210 mg NaNO3, 139 mg NaC3H5O3 or 57 mg C2H6O, 2.6 g NaHCO3, and other nutrients in the same proportions as in the feed solutions for the reactors. Isolates were grown aerobically in LB medium, centrifuged at 5,000 × g for 20 min, and washed three times before inoculation. Each wash step consisted of the removal of the supernatant, suspension of the cell pellet in bicarbonate buffer (2.6 g liter−1 NaHCO3), and centrifugation at 5,000 × g for 15 min. Liquid samples from batch assays were removed, centrifuged at 16,000 × g for 10 min, decanted, and stored at −20°C for nitrate and nitrite analysis. Gas samples for nitrogen and nitrous oxide analysis were removed with Hamilton syringes and analyzed immediately.
Microbial community analysis. (i) DNA extraction and purification.
For DGR1, samples for microbial community analysis were archived occasionally during the first 235 days and every 2 to 3 days thereafter. Samples (4 ml) from DGR2 were archived every 2 to 3 days for the 160 days of operation reported on in this study. For archiving, the culture was removed from reactor sampling ports with a sterile syringe and centrifuged for 2 min at 16,000 × g, the supernatant was decanted, and the cell pellet was stored at −20°C until analysis. Samples for 16S rRNA gene phylogenetic analysis of isolates were prepared from cultures aerobically grown on LB medium, centrifuged at 16,000 × g, decanted, and stored at −20°C until analysis. DNA from all samples was extracted using an UltraClean Microbial DNA isolation kit according to the manufacturer's instructions (Mo Bio Laboratories, Carlsbad, CA).
(ii) PCR amplification and purification.
For clone library construction and isolate sequencing, 16S rRNA genes were amplified from community DNA with a GeneAmp PCR System 9700 thermocycler (Applied Biosystems, Pleasanton, CA) using primer pair 8F (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1392R (5′-ACGGGCGGTGTGTRC-3′) (23) (QIAGEN Operon, Alameda, CA). For terminal restriction fragment (T-RF) length polymorphism (T-RFLP), the same primer sequences were used, and primer 8F was fluorescently labeled with hexachlorofluorescein (QIAGEN Operon, Alameda, CA). All PCR amplification mixtures (50 μl) contained 1× PCR buffer (100 mM KCl, 20 mM Tris HCl [pH 8.0], 0.1 mM EDTA, 1 mM dithiothreitol, 50% glycerol, 0.5% Tween, and 0.5% Nonidet P-40), 2.5 mM MgCl2, 5% dimethyl sulfoxide, 0.4 mM of each deoxynucleoside triphosphate (Invitrogen, Carlsbad, CA), 12.5 pmol of each primer, 1.5 U of Taq DNA polymerase in storage buffer B (Promega, Madison, WI), and 40 ng of template DNA. Samples were placed into a thermocycler that was preheated to 94°C. The PCR thermal cycling parameters were as follows: 94°C for 45 s, 55°C for 30 s, and 72°C for 1.5 min for 30 cycles and 72°C for 7 min. PCR products were analyzed on 1.8% (wt/vol) Tris-borate-EDTA agarose gels, and the product size was confirmed. Products for three replicate PCR amplifications were combined for each sample and purified with a Montage PCR centrifugal device (Millipore, Billerica, MA).
(iii) Cloning, sequence determination, and phylogenetic analysis.
PCR products from DGR1 on day 257 and from DGR2 on day 65 were cloned into the pCR2.1-TOPO cloning vector according to the manufacturer's specifications (TOPO TA Cloning Version Q; Invitrogen). Clones were checked for inserts and orientation by PCR amplification with primer 1392R and either M13f (5′-CCCAGTCACGACGTTGTAAAACG-3′) (QIAGEN) or M13r (5′-CAGGAAACAGCTATGAC-3′) (Invitrogen). PCR conditions were the same as detailed above for genomic DNA amplification. Positive clones were grown up in LB medium with 15% (vol/vol) glycerol, frozen at −20°C, and sent to MCLab (South San Francisco, CA) for sequencing with either primer M13f or M13r, depending upon the insert orientation.
The same DNA and PCR protocols used for T-RFLP were also used to prepare 16S rRNA gene PCR products from isolates for sequencing except that primer 8F was unlabeled. Products from isolates were prepared (150 ng PCR product and 9 pmol of primer 8F in a total volume 15 μl) and sent to MCLab for sequencing.
Clone sequences, isolate sequences, and sequences of the closest BLAST matches were aligned with 16S rRNA gene sequences from the Ribosomal Database Project (RDP) (9) in ARB (24). The sequence alignment was exported from ARB to PAUP version 4.0, and a similarity matrix was constructed. Clone sequences were grouped based on a 97% similarity criterion. Phylogenetic analysis of sequences of representative clones from each group, isolates, and database sequences was performed using the neighbor-joining method within PAUP. A bootstrap analysis of 100 replicates was also performed using PAUP.
(iv) Restriction digests and T-RFLP data analysis.
The PCR product purified for T-RFLP analysis (10 ng μl−1) was cleaved with 1 U μl−1 of HaeIII (Invitrogen) in buffer provided by the manufacturer for 3 h in a 37°C water bath. The enzyme was deactivated in a 65°C water bath for 10 min, and digests were desalted by purification with the Montage PCR centrifugal device (Millipore). Restriction digest products were analyzed on an ABI PRISM 3100 genetic analyzer at the Genomics Technology Support Facility, Michigan State University.
T-RFLP profiles were standardized based on methods described previously by Dunbar et al. and Kaplan et al. (13, 22). The total signal was calculated as the sum of peak heights. Profiles were standardized to the sample with the lowest total signal, and peaks with adjusted heights lower than the threshold (50 units) were removed. The relative abundance of each T-RF was calculated as the ratio of the peak height for that T-RF to the sum of peak heights for all T-RFs in the profile and was expressed as a percentage. Histograms were prepared from relative abundance data.
In silico digestions of clone and isolate sequences were determined in the sequence alignment window of ARB. Isolate sequences were not determined for the first 50 bp downstream of primer 8F, because this primer was used for sequence determinations. Isolate sequences were well aligned. It was assumed that if no restriction site was present in the first 50 bp of the closely related clones, it was also not present in the isolates. When a restriction site was present in the first 50 bp of the clones, an in silico analysis of the isolate sequences could not be obtained and was recorded as being unknown. Principal-component analysis (PCA) of T-RFLP profiles was performed with the ADE4 software package (32) in R 1.9.0. ADE4 and R1.9.0 were downloaded from the Comprehensive R Archive Network (http://cran.r-project.org).
(v) Calculations of Pearson's correlation coefficients.
To measure the strength and direction of linear relationships between effluent concentrations and T-RF relative abundance, Pearson correlation coefficients were calculated from the two data sets (3). The significance of the calculated correlation coefficients was determined with a t test, and P values at which the null hypothesis was rejected for given t values were then calculated in R.
Analytical methods.
Nitrate and nitrite concentrations were measured using an ion chromatograph (Dionex, Sunnyvale, CA). Nitrous oxide was measured on an HP5890 gas chromatograph equipped with a CARBONPLT column and an electron capture detector (Agilent Technologies, Palo Alto, CA). Nitrogen was quantified by a gas chromatograph with a thermal conductivity detector (Gow-Mac Instrument Company, Lehigh, PA). Biomass concentrations were measured as volatile suspended solids (VSS) according to standard method 2540E (14). COD was measured using high-range COD reagent vials from Hach Company (Loveland, CO).
RESULTS
Functional stability. (i) Reactor DGR1.
Reactor DGR1 was inoculated on day zero. After a 2-day start-up period during which nitrite accumulated to 2.5 mM, the reactor community was functionally stable for 160 days. During this period, the community reduced a feed concentration of 45 mM NO3− without detectable nitrate or nitrite in the effluent (data not shown) at a COD/N ratio of 8.2. The COD/N ratio was decreased from 8.2 to 6.0 between days 52 and 86, resulting in a decrease in effluent soluble COD concentrations from 2.1 g COD liter−1 to 655 ± 172 mg COD liter−1 on average. Nitrous oxide levels between days 145 and 153 ranged from 6 to 12 mM N2O-N, indicating incomplete nitrate reduction at that time. Feed nitrate concentrations and dilution rates were constant throughout this period of operation.
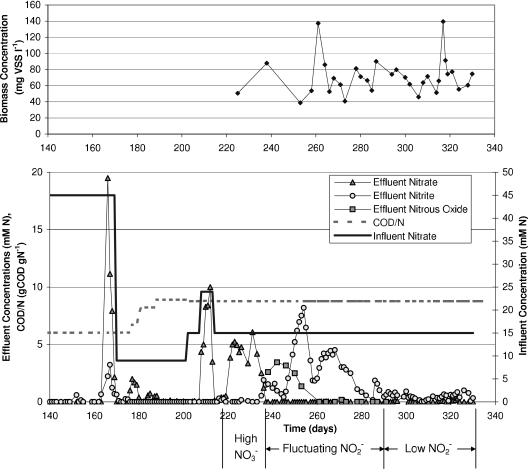
Effluent and operational data for DGR1 beginning on day 140 are presented in Fig. 1. On day 163, a slight decrease in performance occurred as 0.65 mM NO2− was detected in the DGR1 effluent. By the time the next sample was taken on day 166, the reactor effluent contained 19.5 mM NO3− and 2.2 mM NO2− (Fig. 1). Because less denitrification was occurring, less bicarbonate was produced to neutralize the acidic feed, resulting in a pH of 4.5 in the reactor. In an effort to recover denitrification activity, the feed and waste pumps were turned off, and sterile sodium hydroxide was added to increase the pH to 7.5. By day 170, all of the nitrate and nitrite had been consumed (Fig. 1), and the feed was resumed at a nitrate concentration of 9 mM NO3−. The COD/N ratio was adjusted thereafter by following a typical operational control strategy of increasing COD when incomplete denitrification products were observed without residual electron donors in the effluent.
FIG. 1.
Functional performance and operating conditions in reactor DGR1 for days 140 to 335. (A) Biomass concentrations in mg VSS liter−1. (B) Effluent concentrations of nitrate, nitrite, and nitrous oxide, influent concentration of nitrate, and influent ratio of electron donor to electron acceptor in units of chemical oxidation demand of donor to nitrate nitrogen (COD/N).
Once DGR1 was functionally stable at the 9 mM NO3− level, a series of step changes in the feed nitrate concentration was initiated by using the same magnitude of feed concentration increases as that during the start-up of the first reactor. The reactor community was able to completely remove nitrate and nitrite at a feed concentration of 9 mM and 15 mM NO3−, but nitrate accumulated at a feed concentration of 24 mM NO3− (COD/N ratio of 8.8) (Fig. 1), and lactate and ethanol concentrations increased (data not shown), indicating that the electron donor was not limited. The feed nitrate concentration was then restored to 15 mM, but functional stability did not recover.
The functional instability from day 214 to day 335 in DGR1 was characterized by three distinct behaviors: a period of high nitrate accumulation, a period of fluctuating nitrite, and a period of much lower nitrite concentrations with less fluctuation. Nitrate accumulated to 6.6 ± 2.6 mM between day 221 and day 233. As nitrate concentrations declined between days 236 and 291, nitrite concentrations increased and fluctuated, reaching a maximum of 8.2 mM NO2− on day 254. Nitrous oxide concentrations reached 3.4 mM N2O-N and then declined to below the limit of detection by day 261. From day 293 to day 334, nitrite concentrations were much lower, below 0.9 mM, gradually declining, and then gradually increasing. Biomass concentrations fluctuated between 39 and 139 mg VSS liter−1 throughout this period.
(ii) Reactor DGR2.
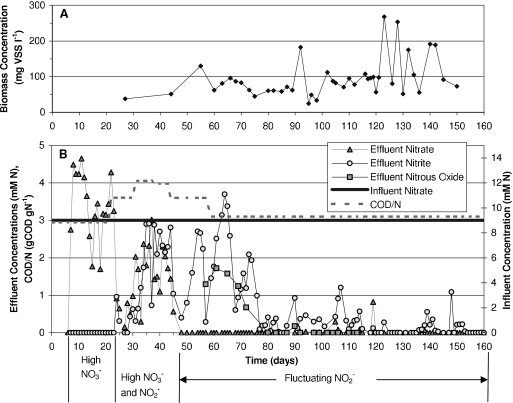
On day 174 of DGR1 operation, after the initial recovery of nitrate and nitrite removal at the 9 mM level, a new reactor was inoculated and designated DGR2 (Fig. 2). This reactor never attained functional stability, and its performance was characterized by three distinct periods of functional performance: a period of high nitrate accumulation, a period of high nitrate and nitrite, and a period of fluctuating nitrite. Nitrous oxide was monitored from days 57 to 113 of DGR2 operation, with elevated values until about day 73, after which it declined. Biomass concentrations in DGR2 varied between 25 and 270 mg VSS liter−1.
FIG. 2.
Functional performance and operating conditions in reactor DGR2 for days 0 to 160. (A) Biomass concentrations in mg VSS liter−1. (B) Effluent concentrations of nitrate, nitrite, and nitrous oxide, influent concentration of nitrate, and influent ratio of electron donor to electron acceptor in units of chemical oxidation demand of donor to nitrate nitrogen (COD/N).
Community dynamics.
To determine if the functional instability was related to microbial community structure, samples taken from DGR1 and DGR2 throughout operation were analyzed by T-RFLP of the 16S rRNA gene. Microbial community dynamics, or changes in microbial community structure over time, were assessed by PCA.
(i) Reactor DGR1.
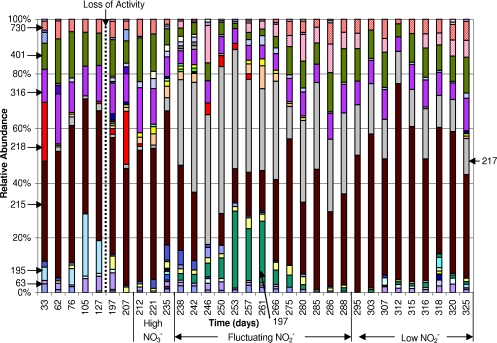
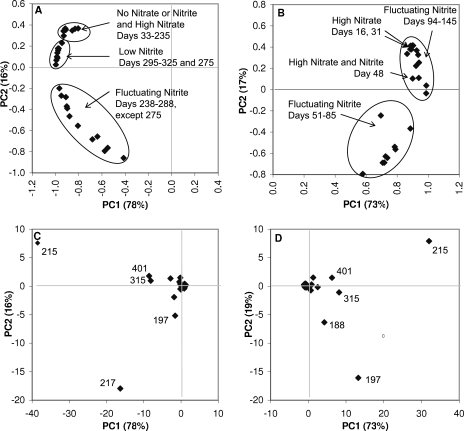
Five samples were analyzed from the period before the loss of activity, when no nitrate or nitrite was detected, on days 33, 62, 76, 105, and 127 (Fig. 3). The results of PCA clustered these samples together (Fig. 4A), suggesting that the community structure was stable during this period. All profiles were dominated by the 215-bp T-RF, which ranged in relative abundance from 37 to 50%. Other significant T-RFs with relative abundances greater than 5% were the 62-, 195-, 316-, 401-, and 730-bp fragments. Samples taken during recovery from the disturbance (days 197 and 207) and during the period of functional instability characterized by high nitrate concentrations (days 212, 221, and 235) had community structures resembling those of the predisturbance samples, as supported by the clustering of these samples with the predisturbance samples in the PCA. They were also dominated by the 215-bp T-RF, but the 195-bp T-RF was no longer present.
FIG. 3.
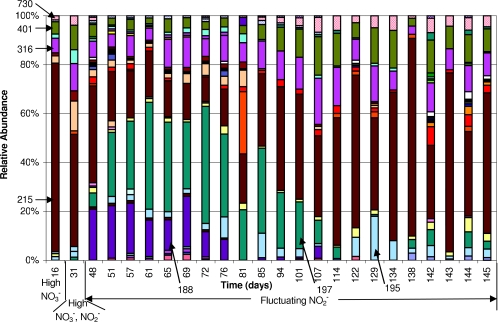
Histograms of T-RF relative abundances in reactor DGR1 for HaeIII T-RFLP profiles. The relative abundance is the ratio of the peak height of a given T-RF in a given sample to the sum of all T-RFs in that sample expressed as a percentage. Arrows indicate the sizes of the restriction fragments for most T-RFs in base pairs.
FIG. 4.
PCA of the T-RFLP profiles. (A) Score plot of samples from DGR1. Of the total variability, 78% and 16% were accounted for by the first and second principal components (PC1 and PC2), respectively. (B) Score plot of samples from DGR2. Of the total variability, 73% and 19% were accounted for by the first and second principal components, respectively. (C) Loading plot of DGR1. Labels indicate T-RF lengths of T-RFs with high loadings. (D) Loading plot of DGR2. Labels indicate T-RF lengths of T-RFs with high loadings.
The community structure changed drastically from days 235 to 238 as demonstrated by the departure of the day 238 structure from the original cluster in the PCA. The rest of the samples from the period of fluctuating nitrite concentrations, days 242 to 288, with the exception of day 275, were also separate from the original cluster of samples, indicating dissimilarity with the original community structure. Additionally, the community was more dynamic during the period of fluctuating nitrite concentrations, as reflected in the larger spread of samples in this cluster in the PCA. In particular, the relative abundances of the 197-, 215-, and 217-bp T-RFs varied greatly and contributed significantly to the overall variability of the data set (Fig. 4C). Prior to day 238, the 217-bp T-RF was present in relative abundances of less than 5%, but during the period of high nitrite fluctuation, the relative abundance of this T-RF became more variable, reaching a maximum relative abundance of 54% on day 250. Similarly, the 197-bp T-RF was not present before day 238 and varied significantly, while the nitrite concentrations fluctuated. A maximum relative abundance of 28% was measured for the 197-bp T-RF on day 253. In contrast, the previously dominant 215-bp T-RF fluctuated to lower relative abundances and was not even detected on day 246. When the nitrite concentrations were lower (from days 293 to 334), the T-RFLP profile returned to being dominated by the 215-bp T-RF, with low relative abundances of the 197- and 217-bp T-RFs.
(ii) Reactor DGR2.
Immediately after the start-up of DGR2, when effluent nitrate concentrations were high (days 16 and 31), the T-RFLP profile was dominated by the 215-bp T-RF (Fig. 5). As found in DGR1, samples from the period of nitrite fluctuation were scattered in the PCA (Fig. 4B), demonstrating that the community was dynamic. In this reactor, samples from days 51 to 85, when nitrite fluctuated to high levels, were more similar to each other than to samples from days 94 to 145, when nitrite fluctuated to low levels. Similarly to DGR1, the 215- and 197-bp T-RFs had high loadings on the principal components (Fig. 4D). The relative abundance of the 215-bp T-RF varied to lower values (10 to 15%) during the period of nitrite fluctuation but eventually returned to high values, reaching a maximum of 80% on day 138. The 197-bp T-RF appeared and varied significantly as nitrite concentrations were varying as in DGR1 but at higher relative abundances than in DGR1, reaching a maximum of 47% on day 72. In contrast to DGR1, a new T-RF that was 188 bp in length appeared in DGR2, but the 217-bp T-RF did not. Additionally, the 195-bp T-RF reappeared in DGR2, although it was detected only in predisturbance samples in DGR1.
FIG. 5.
Histograms of T-RF relative abundances in reactor DGR2 for HaeIII T-RFLP profiles. The relative abundance is the ratio of the peak height of a given T-RF in a given sample to the sum of all T-RFs in that sample expressed as a percentage. Arrows indicate the sizes of the restriction fragments for most T-RFs in base pairs.
Community structure.
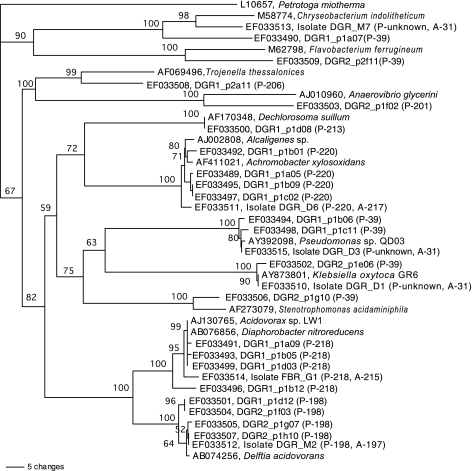
Clone libraries were constructed from samples collected on day 257 from DGR1 and on day 65 from DGR2. The selection of these specific samples allowed for the correlation of clone sequences with T-RFs that were particularly interesting in the T-RFLP analysis. Eighty-four clones were sequenced and grouped based on a 97% similarity criterion. Representative clones of each group were included in the phylogenetic analysis presented in Fig. 6. The majority of clone sequences belonged to the Proteobacteria. The majority of clone sequences from DGR1 were closely related to Achromobacter xylosoxidans (99 to 100% similarity), Acidovorax sp. strain LW1 (98 to 100% similarity), Delftia acidovorans (98 to 100% similarity), and Pseudomonas sp. strain QD03 (99 to 100% similarity), accounting for 41, 27, 10, and 10% of the clones from DGR1, respectively. When examined with a 97% similarity criterion, the sequences closely related to Acidovorax formed four distinct clusters. Clone sequences closely related to Delftia acidovorans (98 to 100% similarity) accounted for 79% of the clones in the library of DGR2. Several additional sequences closely related to species in the γ-Proteobacteria and Flavobacteria were also detected.
FIG. 6.
Phylogenetic analysis of representative partial 16S rRNA gene sequences. The tree was constructed with the neighbor-joining method using sequences of 480 nucleotides. Petrotoga miotherma was the outgroup species. Bootstrap values below 50 (out of 100) are not shown. Sequences from the RDP are listed with genus and species names and accession numbers. Clone sequences are named beginning with DGR1 or DGR2, indicating the reactor of origin, and isolates sequence names begin with isolate. T-RF sizes are given in parentheses, with P- indicating predicted sizes from in silico digestion and A- indicating actual sizes from T-RFLP.
Correlation of functional instability and community dynamics.
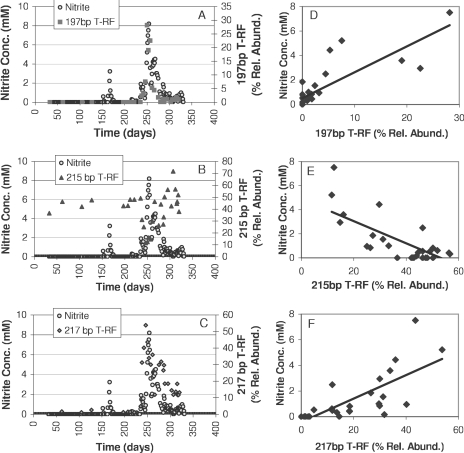
The relationships between the relative abundances of the 188-, 197-, 215-, and 217-bp T-RFs and nitrite concentrations were carefully examined, because the relative abundances of these T-RFs underwent large variations and had high loadings on the principal components while nitrite concentrations fluctuated. When nitrite concentrations were plotted along with relative abundances, the 197-, 215-, and 217-bp T-RF relative abundances varied in relation to the nitrite concentrations in DGR1 and DGR2. The relationships between nitrite concentrations and relative abundances for DGR1 are shown in Fig. 7, and similar relationships were observed in DGR2 (plots not shown). Pearson's correlation coefficients were calculated to quantify the strength and significance of the relationship of nitrite and relative abundance (Table 1). In both of the reactors, the 197-bp T-RF had a highly significant positive correlation with nitrite concentration. In contrast, the 215-bp T-RF had a significant and strong negative correlation with nitrite concentration in both reactors. The 217-bp T-RF, found only in DGR1, also had a significant and strong positive correlation to nitrite concentration.
FIG. 7.
Relationships between effluent chemistry and T-RF dynamics in DGR1. (A to C) Effluent nitrite concentrations (Conc.) and T-RF relative abundances (Rel. Abund.) over time. (D to F) Effluent nitrite concentrations versus T-RF relative abundance.
TABLE 1.
Pearson correlation coefficients for nitrite concentration versus relative abundance of different T-RFs
| T-RF (bp) | Genus represented in clone library | Pearson correlation coefficient (P value)a
|
|
|---|---|---|---|
| DGR1 | DGR2 | ||
| 197 | Delftia | 0.818 (1.9E−8) | 0.636 (2.0E−3) |
| 215 | Acidovorax | −0.778 (2.0E−5) | −0.653 (1.0E−3) |
| 217 | Achromobacter | 0.692 (2.1E−5) | Not present |
P values at which the null hypothesis is rejected are given in parentheses.
Biomass concentrations also varied greatly during the period of functional instability in both reactors (Fig. 1 and 2). However, there was no consistent pattern or relationship between this variation and fluctuations in nitrite concentrations.
Functional diversity of isolates.
In an effort to determine if there was a physiological explanation for the observed correlations, representative bacteria of several T-RFs were isolated from the community. The isolates obtained represented most of the genera found in the clone library, including Acidovorax, Delftia, Achromobacter, Pseudomonas, Chryseobacterium, and Klebsiella (Fig. 6). T-RFLP of these isolates and in silico digestion of isolate and clone sequences allowed the correspondence of representative isolates to particular T-RFs. Isolate G1, closely related to Acidovorax sp. strain LW1 (99% similarity), yielded a strong signal at 215 bp in T-RFLP electropherograms as well as weaker signals at 316, 401, and 730 bp. These sizes correspond to the second, third, and fourth occurrence of the HaeIII restriction sites in the clone sequences. The presence of these additional fragments in the T-RFLP profiles may be due to either incomplete digestion or the formation of pseudo-T-RFs (15). A 197-bp T-RF was obtained for isolate M2, closely related to Delftia acidovorans (100% similarity), with additional T-RFs at 215, 316, and 401 bp. The T-RF for isolate D6, closely related to Achromobacter species (99% similarity), was 217 bp in length, with additional T-RFs of 315 and 401 bp. In silico prediction of restriction fragment lengths was performed to verify that the isolates were representative of sequences contributing to the T-RFs. The analysis was done in the sequence alignment window of ARB, and the results are presented in Fig. 6. The restriction sites of isolates were perfectly aligned with those of clones, and predicted fragments were the same size as those measured in T-RFLP of the isolates, within several base pairs. A discrepancy of several base pairs is within the range that was observed in a detailed comparison of actual and predicted T-RF lengths (21).
Isolates were assayed for the reduction of nitrate and the production of nitrite, nitrous oxide, and nitrogen gas (Table 2). The electron donors lactate and ethanol were tested in separate assays. The representative Acidovorax-like isolate, G1, reduced nitrate directly to nitrogen without the accumulation of any denitrification intermediates. In isolate M2 and isolate D6, representing Delftia and Achromobacter, respectively, nitrate was stoichiometrically reduced to nitrite without the detectable production of nitrous oxide or nitrogen. Results were the same with growth on lactate or ethanol.
TABLE 2.
Results of isolate nitrate reduction assaysa
| Isolate | T-RF (bp) | Closest relative | Compound detected
|
||
|---|---|---|---|---|---|
| NO2− | N2O | N2 | |||
| G1 | 215 | Acidovorax | − | − | + |
| D6 | 217 | Achromobacter | + | − | − |
| M2 | 197 | Delftia | + | − | − |
Compounds detected during nitrate reduction are indicated with a +. The size of T-RF generated from T-RFLP of the isolate and the genus of the closest relative from the RDP are given.
DISCUSSION
Ensuring functional stability is a major challenge for the design and operation of biological water treatment systems. However, very little is known about the microbial community dynamics within functionally unstable systems or how those dynamics might relate to functional instability. In this study, the opportunity to examine this relationship arose after a denitrification failure in reactor DGR1 and during the subsequent period of functional instability in reactors DGR1 and DGR2. In both reactors, the microbial communities were dynamic, fluctuating during the periods of functional instability. Strong, statistically significant correlations between the relative abundances of several T-RFs and nitrite concentrations in reactor effluents were found, revealing a definite relationship between dynamics in the community structure and functional instability.
Previous work established a correlation between the function of denitrification, as measured by denitrification rates, and denitrifier abundance (18). The correlations observed in this work underscore the importance of functional diversity within the nitrate-reducing community. The 197-bp and 217-bp T-RFs had strong positive correlations with effluent nitrite concentrations. They were represented by isolates closely related to the genera Delftia and Achromobacter, respectively, both of which were nitrate respirers, capable of reducing nitrate only to nitrite. The strong correlation of relative abundance of nitrate respirers and effluent nitrite concentrations demonstrates that their presence and dynamics in denitrifying reactors can negatively influence the overall functional performance. The isolate representative of the 215-bp T-RF was closely related to a species of Acidovorax and reduced nitrate to nitrogen without the accumulation of any intermediate products. This T-RF was negatively correlated with effluent nitrite concentrations and was associated with good functional performance, suggesting the potential importance of maintaining populations with complete denitrifying capabilities for functional stability.
The pattern of dampening oscillations observed in both community dynamics and effluent chemistry in both reactors suggests two possible phenomena: (i) complex interactions between populations competing for nitrate and nitrite and (ii) trophic interactions between phage and bacteria. During the period of functional instability, concentrations of nitrate and nitrite fluctuated, changing environmental conditions and altering the competitive advantages of different populations. In both reactors, the T-RFs corresponding to nitrate-respiring Delftia and Achromobacter species increased in relative abundance as effluent composition shifted from nitrate to nitrite, perhaps because growth of these organisms was favored under high nitrate concentrations. Subsequently, the T-RF corresponding to the completely denitrifying Acidovorax species returned to high relative abundance, possibly due to consumption by these organisms of the residual nitrite left by the nitrate respirers. Some features of the functional instability may also be explained by predator-prey interactions between phage and bacteria. Using fluorescence microscopy and transmission electron microscopy, bacteriophage were shown to be present in the reactors (data not shown). Plaque assays testing for phage were carried out on the Acidovorax-like isolate, and plaques were detected (data not shown). The other isolates were not tested. Although we do not have the data on phage dynamics over time that would be necessary to evaluate this hypothesis, the observation of oscillations in the reactors in spite of constant influent conditions suggests the possibility that such ecological interactions drove the observed functional instability. The periodic, rather than continuous, removal of effluent could have contributed to the competition or predation phenomena by allowing the accumulation of particular bacterial or phage populations that would have been completely washed out of a continuous system.
The identification of populations that appear to promote or disrupt functional stability is an important step in linking microbial ecology to functional stability. A subsequent challenge is to use such information to design, inoculate, and maintain systems, ensuring the presence and maintenance of populations that promote functional stability. A solid theoretical understanding of microbial community assembly and invasion in these systems is required before the goal of rational design to promote functionally stable microbial communities can be realized (4, 10, 28).
Acknowledgments
This research was supported in part by the Natural and Accelerated Bioremediation Research Program, Biological and Environmental Research, U.S. Department of Energy (grant DOEAC05-00OR22725), and in part by a National Science Foundation Doctoral Dissertation Improvement grant (grant DEB-0408108) awarded to C. M. Jessup. M. E. Gentile was supported by a fellowship from the U.S. Environmental Protection Agency Science To Achieve Results Program.
Footnotes
Published ahead of print on 1 December 2006.
REFERENCES
- 1.Almeida, J., S. Julio, M. Reis, and M. Carrondo. 1995. Nitrite inhibition of denitrification by Pseudomonas fluorescens. Biotechnol. Bioeng. 46:194-201. [DOI] [PubMed] [Google Scholar]
- 2.Betlach, M., and J. Tiedje. 1981. Kinetic explanation for accumulation of nitrite, nitric oxide, and nitrous oxide during bacterial denitrification. Appl. Environ. Microbiol. 42:1074-1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bluman, A. 2001. Elementary statistics: a step by step approach, 4th ed. McGraw-Hill, New York, NY.
- 4.Briones, A., and L. Raskin. 2003. Diversity and dynamics of microbial communities in engineered environments and their implications for process stability. Curr. Opin. Biotechnol. 14:270-276. [DOI] [PubMed] [Google Scholar]
- 5.Carlson, C., and J. Ingraham. 1983. Comparison of denitrification by Pseudomonas stutzeri, Pseudomonas aeruginosa, and Paracoccus denitrificans. Appl. Environ. Microbiol. 45:1247-1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cavigelli, M., and G. Robertson. 2000. The functional significance of denitrifier community composition in a terrestrial ecosystem. Ecology 81:1402-1414. [Google Scholar]
- 7.Cavigelli, M., and G. Robertson. 2001. Role of denitrifier diversity in rates of nitrous oxide consumption in a terrestrial ecosystem. Soil Biol. Biochem. 33:297-310. [Google Scholar]
- 8.Reference deleted.
- 9.Cole, J. R., B. Chai, T. L. Marsh, R. J. Farris, Q. Wang, S. A. Kulam, S. Chandra, D. M. McGarrell, T. M. Schmidt, G. M. Garrity, and J. M. Tiedje. 2003. The Ribosomal Database Project (RDP-II): previewing a new autoaligner that allows regular updates and the new prokaryotic taxonomy. Nucleic Acids Res. 31:442-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Curtis, T., I. Head, and D. Graham. 2003. Theoretical ecology for engineering biology. Environ. Sci. Technol. 37:64A-70A. [DOI] [PubMed] [Google Scholar]
- 11.Cytryn, E., D. Minz, I. Gelfand, A. Neori, A. Gieseke, D. De Beer, and J. Van Rijn. 2005. Sulfide-oxidizing activity and bacterial community structure in a fluidized bed reactor from a zero-discharge mariculture system. Environ. Sci. Technol. 39:1802-1810. [DOI] [PubMed] [Google Scholar]
- 12.Drysdale, G., H. Kasan, and F. Bux. 2001. Assessment of denitrification by the ordinary heterotrophic organisms in an NDBEPR activated sludge system. Water Sci. Technol. 43(1):147-154. [PubMed] [Google Scholar]
- 13.Dunbar, J., L. Ticknor, and C. Kuske. 2000. Assessment of microbial diversity in four southwestern United States soils by 16S rRNA gene terminal restriction fragment analysis. Appl. Environ. Microbiol. 66:2943-2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eaton, A., L. Clesceri, and A. Greenberg. 1995. Standard methods for the examination of water and wastewater. American Public Health Association, Washington, DC.
- 15.Egert, M., and M. Friedrich. 2003. Formation of pseudo-terminal restriction fragments, a PCR-related bias affecting terminal restriction fragment length polymorphism analysis of microbial community structure. Appl. Environ. Microbiol. 69:2555-2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fernandez, A., S. Y. Huang, S. Seston, J. Xing, R. Hickey, C. Criddle, and J. Tiedje. 1999. How stable is stable? Function versus community composition. Appl. Environ. Microbiol. 65:3697-3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gentile, M., T. Yan, S. M. Tiquia, M. Fields, J. Nyman, J. Zhou, and C. Criddle. 2006. Stability in a denitrifying fluidized bed reactor. Microb. Ecol. 52:311-321. [DOI] [PubMed] [Google Scholar]
- 18.Ginige, M. P., J. Keller, and L. L. Blackall. 2005. Investigation of an acetate-fed denitrifying microbial community by stable isotope probing, full-cycle rRNA analysis, and fluorescent in situ hybridization-microautoradiography. Appl. Environ. Microbiol. 71:8683-8691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Glass, C., and J. Silverstein. 1998. Denitrification kinetics of high nitrate concentration water: pH effect on inhibition and nitrite accumulation. Water Res. 32:831-839. [Google Scholar]
- 20.Glass, C., J. Silverstein, and J. Oh. 1997. Inhibition of denitrification in activated sludge by nitrite. Water Environ. Res. 69:1086-1093. [Google Scholar]
- 21.Kaplan, C., and C. Kitts. 2003. Variation between observed and true terminal restriction fragment length is dependent on true TRF length and purine content. J. Microbiol. Methods 54:121-125. [DOI] [PubMed] [Google Scholar]
- 22.Kaplan, C. W., J. C. Astaire, M. E. Sanders, B. S. Reddy, and C. L. Kitts. 2001. 16S ribosomal DNA terminal restriction fragment pattern analysis of bacterial communities in feces of rats fed Lactobacillus acidophilus NCFM. Appl. Environ. Microbiol. 67:1935-1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lane, D. 1991. 16S/23S rRNA sequencing, p. 115-175. In E. Stackebrant and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley & Sons, New York, NY.
- 24.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Forster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. Konig, T. Liss, R. Lussmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K. H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Manefield, M., A. S. Whiteley, R. I. Griffiths, and M. J. Bailey. 2002. RNA stable isotope probing, a novel means of linking microbial community function to phylogeny. Appl. Environ. Microbiol. 68:5367-5373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martienssen, M., and R. Schops. 1997. Biological treatment of leachate from solid waste landfill sites—alterations in the bacterial community during the denitrification process. Water Res. 31:1164-1170. [Google Scholar]
- 27.Martienssen, M., and R. Schops. 1999. Population dynamics of denitrifying bacteria in a model biocommunity. Water Res. 33:639-646. [Google Scholar]
- 28.Rittmann, B. E., M. Hausner, F. Loffler, N. G. Love, G. Muyzer, S. Okabe, D. B. Oerther, J. Peccia, L. Raskin, and M. Wagner. 2006. A vista for microbial ecology and environmental biotechnology. Environ. Sci. Technol. 40:1096-1103. [DOI] [PubMed] [Google Scholar]
- 29.Rowan, A. K., J. R. Snape, D. Fearnside, M. R. Barer, T. P. Curtis, and I. M. Head. 2003. Composition and diversity of ammonia-oxidising bacterial communities in wastewater treatment reactors of different design treating identical wastewater. FEMS Microbiol. Ecol. 43:195-206. [DOI] [PubMed] [Google Scholar]
- 30.Smith, N., Z. Yu, and W. Mohn. 2003. Stability of the bacterial community in a pulp mill effluent treatment system during normal operation and a system shutdown. Water Res. 37:4873-4884. [DOI] [PubMed] [Google Scholar]
- 31.Stamper, D., M. Walch, and R. Jacobs. 2003. Bacterial population changes in a membrane bioreactor for graywater treatment monitored by denaturing gradient gel electrophoretic analysis of 16S rRNA gene fragments. Appl. Environ. Microbiol. 69:852-860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thioulouse, J., D. Chessel, S. Doledec, and J.-M. Olivier. 1997. ADE-4: a multivariate analysis and graphical display software. Stat. Comput. 7:75-83. [Google Scholar]
- 33.Tiedje, J. 1994. Denitrifiers, p. 245-267. In R. Weaver, S. Angle, P. Bottomley, et al. (ed.), Methods of soil analysis, part 2. Microbiological and biochemical properties. Soil Science Society of America, Madison, WI.
- 33a.U.S. Government Printing Office. 2002. National primary drinking water regulations. CFR title 40. U.S. Government Printing Office, Washington, DC.
- 34.von Canstein, H., Y. Li, A. Felske, and I. Wagner-Dobler. 2001. Long-term stability of mercury-reducing microbial biofilm communities analyzed by 16S-23S rDNA interspacer region polymorphism. Microb. Ecol. 42:624-634. [DOI] [PubMed] [Google Scholar]
- 35.Wilderer, P., W. Jones, and U. Dau. 1987. Competition in denitrification systems affecting reduction rate and accumulation of nitrite. Water Res. 21:239-245. [Google Scholar]
- 36.Wilhelm, E., R. Battino, and R. J. Wilcock. 1977. Low-pressure solubility of gases in liquid water. Chem. Rev. 77:219-262. [Google Scholar]