Abstract
Lactococcus lactis subsp. cremoris Ropy352 produces two distinct heteropolysaccharides, phenotypically described as ropy and mucoid, when cultured in nonfat milk. One exopolysaccharide precipitated with 50% ethanol as a series of elongated threads and was composed of glucose and galactose in a molar ratio of 3:2. The second exopolysaccharide precipitated with 75% ethanol as a fine flocculant and consisted of galactose, glucose, and mannose with a molar ratio of 67:21:12. A mutant strain, L. lactis subsp. cremoris EK240, lacking the ropy phenotype did not produce the exopolysaccharide that precipitated with 50% ethanol; however, it produced the exopolysaccharide that precipitated with 75% ethanol, indicating that the former exopolysaccharide is essential for the ropy phenotype. Cultures of L. lactis subsp. cremoris Ropy352 in 10% nonfat milk reached a viscosity of 25 Pa-s after 24 h, while those of the nonropy L. lactis subsp. cremoris EK240 mutant did not change. A mutation abolishing ropy exopolysaccharide expression mapped to a region on a plasmid containing two open reading frames, epsM and epsN, encoding novel glycosyltransferases bordered by ISS1 elements oriented in the same direction. Sequencing of this plasmid revealed two other regions involved in exopolysaccharide expression, an operon located between partial IS981 and IS982 elements, and an independent gene, epsU. Two and possibly three of these regions are involved in L. lactis subsp. cremoris Ropy352 exopolysaccharide expression and are arranged in a novel fashion different from that of typical lactococcal exopolysaccharide loci, and this provides genetic evidence for exopolysaccharide gene reorganization and evolution in Lactococcus.
The use of lactic acid bacteria (LAB) for the manufacture of fermented milk products has been ongoing for centuries, with exopolysaccharides (EPS) produced by this group becoming ever more important in food production. Consumers are increasingly demanding fresh and natural foods free of overprocessing and additives such as stabilizers and preservatives (56). As a group, LAB are generally recognized as safe, and EPS from LAB are considered natural products (31). The EPS from LAB provide unique characteristics that are proving extremely useful in stabilizing dairy products and improving their rheological properties (45). The compositions and structures of these EPS are highly diverse, yet all are reported to have similar thickening effects (7, 11, 43). Ropy strains of LAB yield fermented milk products having smoother body, higher viscosity, and less syneresis than products made with nonropy strains (6, 55) and are of particular interest in Scandinavian milk products such as viili and langmjolk (27). Ropy starter cultures also provide benefits for yogurt production. In Mexico, where the milk supply routinely faces shortages, yogurt can be made with less total milk solids if a ropy starter culture is used (55). However, the usefulness and desirability of EPS-producing strains are not without problems. Depending on the culture medium and conditions, some strains can produce excessive ropiness and undesirable sensory characteristics (48, 55). Although much research focuses on monoculturing of yogurts for testing, mixed cultures of ropy and nonropy strains are recommended to achieve a proper balance (7, 27, 30). Since LAB are perceived as natural and have historically been used in foods, they are a good source of more and potentially superior polymers for use in healthy foods (31).
Several structures of LAB EPS have been recently reviewed (25). Nakajima et al. (33, 34) have described a high-molecular-weight EPS produced by Lactococcus lactis subsp. cremoris STB 0495 cultured on whey permeate medium. This EPS was composed of d-glucose, d-galactose, l-rhamnose, and phosphate, while β linkages preponderated in its structure. Marshall et al. (29) described two EPS from L. lactis subsp. cremoris LC330 grown on a defined medium that are quite different from one another; one is a neutral, high-molecular-weight polysaccharide consisting of galactose, glucose, and glucosamine, whereas the second polysaccharide is much lower in molecular weight and contains glucose, galactose, rhamnose, glucosamine, and phosphate.
In the lactococcal strains examined so far, the genetic loci for EPS expression have been reported to be associated with plasmids of various sizes, i.e., 4.5 MDa (53), 17 MDa (35), 18.5 MDa (52), 19.8 MDa (24), 26.5 MDa (50), 30 MDa (35, 54), and 38.3 MDa (15). The presence of the EPS genes on plasmids has been implicated as the cause of EPS expression instability with higher temperatures and frequent transfers (11, 52). More recently, the presence of mobile IS elements bordering EPS operons has also been thought to provide a means for instability of EPS expression (5, 16). Indeed, many strains of Lactococcus have been shown to carry multiple copies of the insertion sequences ISS1 and IS981 (37). Mobile elements are well known for enabling genetic exchange among different genera (9), and these events are suspected of contributing to the evolution of several cellular behaviors including the organization of EPS loci (5, 26), degradation of complex xenobiotics (46, 49), spread of antibiotic resistance (10), and bacterial pathogenesis (2, 32).
EPS-expressing LAB have been studied on the genetic level to determine the organization and characterization of the genes necessary for EPS expression. Members of the Lactobacillus (25), Lactococcus (50), and Streptococcus (43) genera have been studied, and the genes necessary for EPS expression were found to be grouped into a single locus. A general organization of genes involved in EPS expression has emerged beginning with regulation, chain length determination, synthesis of the repeating units, polymerization, and finally export (22, 25, 44); however, there are exceptions (5, 51).
In our previous studies of L. lactis subsp. cremoris Ropy352, we found that this strain produces two distinct EPS, phenotypically described as ropy and mucoid, and that mutations could be isolated impacting one EPS but not the other EPS (23). This report examines the EPS operon and associated gene cluster responsible for ropy EPS expression in the L. lactis subsp. cremoris Ropy352 strain. The chemical composition and physical properties of two EPS produced by L. lactis subsp. cremoris Ropy352 and a nonropy mutant when cultured in nonfat milk medium are also described.
MATERIALS AND METHODS
Bacterial strains and media.
Frozen stock lactococcal cultures were maintained in 11% reconstituted nonfat dry milk containing 20% glycerol at −70°C. The strains used included L. lactis subsp. cremoris Ropy352 (13); L. lactis subsp. cremoris Ropy352 nonropy mutant EK240 (L. lactis subsp. cremoris Ropy352 EK238 [23] cured of pGh9:ISS1); spontaneous L. lactis subsp. cremoris Ropy352 nonropy strains EK316, EK317, and EK318 (this study); and strain EK396 (L. lactis MG1363 [17] harboring pEK396). The plasmids used included pEPS352 (native ropy EPS plasmid), pGh9:ISS1 (28), pEK238 (pGh9:ISS1 with a fragment of the insertion locus [see Fig. 4] isolated with HindIII), and pEK396 (pGh9:ISS1 with a fragment of the insertion locus [see Fig. 4] isolated with EcoRI). Strains were cultured on whey agar as previously described (23). Isolated colonies were transferred to M17-glucose broth (47), grown at 30°C without shaking for 24 h, and used for further manipulations or inoculations. The ropy EPS phenotype was assayed on whey-glucose agar as previously described (23). Colonies were scored as ropy if visible strings greater than 5 mm in length were observed and nonropy if no strings were observed (53).
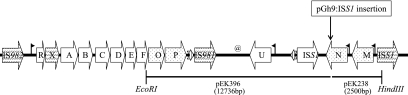
FIG. 4.
Genetic organization of the ropy eps gene cluster of plasmid pEPS352 of L. lactis subsp. cremoris Ropy352. Restriction sites relevant for pGh9:ISS1-mediated subcloning are shown. The symbol @ indicates the replication origin used for recovery of the EcoRI fragment. Large block arrows with diagonal dashes indicate partial ORFs. The large block arrows filled with dots correspond to novel genes described in the text. The pGh9:ISS1 insertion site is indicated. Putative RBS-promoter sequences for the epsN, epsM, and epsU genes and the EPS352 gene operon are indicated with black flags. The corresponding plasmid of the pGh9:ISS1-mediated subcloned fragment is indicated below the arrows. The small block arrows with vertical lines are the two fragments of epsH as described in the text.
Electroporation, DNA isolation, PCR, and sequencing.
Electroporation of Lactococcus was carried out as previously described (23). Plasmid DNA from Lactococcus was isolated as previously described (36). Plasmid DNA was electrophoresed in 0.7% agarose gels to generate plasmid profiles. DNA sequencing was done on an Applied Biosystems 3730 Genetic Analyzer with Big Dye Terminator chemistry at the Center for Gene Research and Biotechnology's Central Services Laboratory (Oregon State University, Corvallis). PCR-based walking was performed as previously described (21).
Production and isolation of EPS.
All EPS production medium consisted of 10% nonfat milk, which was prepared by dissolving dry nonfat milk powder in deionized water at room temperature for 1 h and then sterilizing the mixture for 12 min at 120°C. Typically, 1 liter of milk medium was placed in a 2.8-liter Fernbach flask for culture. When culturing L. lactis subsp. cremoris Ropy352, the milk medium was inoculated from single colonies that tested positive for the ropy phenotype on whey agar plates. After a 16-h static culture at 30°C, the milk solution had changed to a more gel-like consistency. When culturing L. lactis subsp. cremoris EK240, individual colonies that were not ropy were picked from whey agar plates and cultured as described above. In each case, the culture broths were transferred to 500-ml centrifuge bottles and the insoluble fraction was pelleted at 10,000 × g for 20 min. The clarified supernatant was then transferred to dialysis tubing with a 6- to 8-kDa MWCO (Spectra/Por 1; Spectrum Laboratories, Inc., Laguna Hills, CA) and dialyzed against water containing 0.02% sodium azide for at least 24 h.
The volume of the contents of the dialysis tubing was then measured, and an equal volume of absolute ethanol was added while the solution was stirred in an ice bath. L. lactis subsp. cremoris Ropy352 cultures yielded a precipitate consisting of elongated threads that formed in the mixture and were collected by centrifugation as described above. To the remaining supernatant, 2 additional volumes of absolute ethanol were added and the mixture was allowed to stand at 4°C for 24 h. After this time, a fine flocculent precipitate formed and was collected by centrifugation. When treating the supernatant obtained from 16-h cultures of L. lactis subsp. cremoris EK240, no precipitates formed when 1 volume of ethanol was added, so 2 additional volumes of ethanol were added and the mixture was treated as just described. The ropy fraction from L. lactis subsp. cremoris Ropy352 was resuspended in a small volume of deionized water, and solid trichloroacetic acid was added to yield a 15% (wt/vol) solution in order to precipitate residual protein. The solution was allowed to stand for 10 min in an ice bath and then centrifuged at 30,000 × g for 30 min at 4°C. The supernatant containing the EPS was again dialyzed against deionized water. The same procedure to remove residual protein was applied to the flocculant (F) fraction from both L. lactis subsp. cremoris Ropy352 and EK240. Total polysaccharide recovery was determined from the dry weight of the final lyophilized material. For both EPS (ropy and F fractions), the final protein concentration was determined by the bicinchoninic acid method (BCA assay; Pierce Chemical Co., Rockford, IL) and phosphorus content was measured as described by Ames (1).
Characterization of EPS by size exclusion chromatography (SEC) with multiangle laser light scattering (MALLS) detection.
Purified EPS (ropy fraction from L. lactis subsp. cremoris Ropy352 and F fractions from L. lactis subsp. cremoris Ropy352 and EK240) were filtered through a 0.45-μm syringe filter and injected onto a high-performance liquid chromatography (HPLC) apparatus equipped with a Showdex OH-pak KB-806 M column (0.8 by 30 cm; Waters Corp., Milford, MA) equilibrated in water pumped at a flow rate of 1 ml/min at room temperature. A Wyatt Technology DAWN EOS detector with a 690-nm laser coupled with a Wyatt Optilab DSP interferometric refractometer was used to analyze the column effluent. The signal from the detectors was analyzed with the ASTRA software (version 4.73.04) from Wyatt Technology (Santa Barbara, CA).
Characterization of purified EPS by gas chromatography-mass spectrometry (GC-MS).
The sugar compositions of these two EPS fractions have been previously described (23). To characterize the linkage sites in the EPS, a sample of each EPS, obtained as described above, was permethylated with sodium methylsulfinylmethanide and methyl iodide in dimethyl sulfoxide (40). The methylated EPS was then hydrolyzed at 120°C for 1 h with 2 M trifluoroacetic acid. The trifluoroacetic acid was removed by passing the sample over a minicolumn (0.5 by 5 cm) containing AG1-X8 anion-exchange resin (Bio-Rad Laboratories, Hercules, CA) and then drying the effluent under reduced pressure (SpeedVac; Savant Instruments, Inc., Farmingdale, NY). The sugars were then converted to the peracetylated aldononitrile (PAAN) derivatives (40) and analyzed by GC-MS on a cross-linked methyl silicone column (25 mm by 0.22 mm [inside diameter] by 0.1 μm [thickness]; Hewlett Packard, Wilmington, DE). The column temperature was held for 3 min at 130°C, increased by 5°C/min to 165°C, and then held at 165°C for 10 min; helium was used as the carrier gas.
Viscometry.
The viscosity of the intact fermented milk culture produced by L. lactis subsp. cremoris Ropy352 and the nonropy mutant L. lactis subsp. cremoris EK240 was characterized in a rotary viscometer (LV2000; Cannon Instrument Co., State College, PA) at room temperature with the L1 spindle. The viscosity of a supernatant obtained by centrifugation of the intact culture was obtained in the same fashion. The initial viscosity at the lowest spindle speed, 0.3 rpm, was first recorded, and subsequent measurements were made at increasing spindle speeds to estimate the shear sensitivity of the intact culture and the clarified supernatant.
Acid titration of phosphate groups in the EPS.
The degree of protonation of the phosphate groups in the ropy and F fraction EPS was determined by titration with base. For the ropy polysaccharide, 0.5 mg in 5 ml water was first decationized by treatment with AG 50W-X1 cation-exchange resin (hydrogen form; Bio-Rad Laboratories). The solution was then titrated by adding 20-μl aliquots of 0.1 M NaOH and recording the pH after each addition. The same procedure was used for the F fraction polysaccharide.
Nucleotide sequence accession number.
The complete sequence of the ropy eps gene cluster of plasmid pEPS352 is available in the GenBank database under accession number EF192213.
RESULTS
Properties of fermented milk products.
After 24 h, the milk medium inoculated with L. lactis subsp. cremoris strain Ropy352 became quite firm, although there was a small amount of liquid permeate on the surface of the culture. The viscosity of the intact L. lactis subsp. cremoris Ropy352 culture was 25.6 Pa-s when measured at the slowest spindle speed setting on the viscometer, which was 0.3 rpm. At higher spindle speeds, the viscosity decreased substantially, reaching a value of only 2 Pa-s at 60 rpm, indicating a large degree of shear sensitivity in the culture. Additionally, after centrifugation of the intact L. lactis subsp. cremoris Ropy352 culture to sediment cells and insoluble matter, the viscosity of the remaining supernatant reached a value of only 10.0 Pa-s at a spindle speed setting of 0.3 rpm. This is a significantly lower value than for the intact culture, which indicates that it is not the polysaccharide alone that is responsible for the elevated viscosity in intact cultures of L. lactis subsp. cremoris Ropy352. Despite the significant reduction in viscosity in the clarified supernatant, the solution could still be described as being ropy in nature, although not to the same extent as seen in the intact L. lactis subsp. cremoris Ropy352 culture. In contrast to the results derived from L. lactis subsp. cremoris Ropy352, the viscosity of cultures of the nonropy mutant, L. lactis subsp. cremoris EK240, did not change from that of the starting milk medium during the same time period.
Production of EPS in milk.
The EPS responsible for the ropy characteristic of L. lactis subsp. cremoris strain Ropy352 was purified by precipitation with 50% ethanol, followed by removal of residual protein by precipitation with trichloroacetic acid. The ropy fraction contained 204 mg/liter polysaccharide and was essentially protein free (<20 μg/mg polysaccharide). The ropy fraction also contained 2.3 μg phosphorus/mg polysaccharide. Further addition of ethanol (to 75% [vol/vol], final concentration) to the 50% ethanol supernatant yielded a second EPS, the F fraction. The F fraction contained 7 mg/liter polysaccharide after it was deproteinized. It contained 1.7 μg phosphorus/mg polysaccharide.
In contrast to L. lactis subsp. cremoris Ropy352, the nonropy mutant, L. lactis subsp. cremoris EK240, did not produce an EPS that precipitated with 50% ethanol. However, an EPS did precipitate with 75% ethanol (F fraction). The residual protein concentration was negligible (<20 μg/mg polysaccharide), and phosphorus was present at 1.5 μg/mg polysaccharide.
Molecular mass analysis.
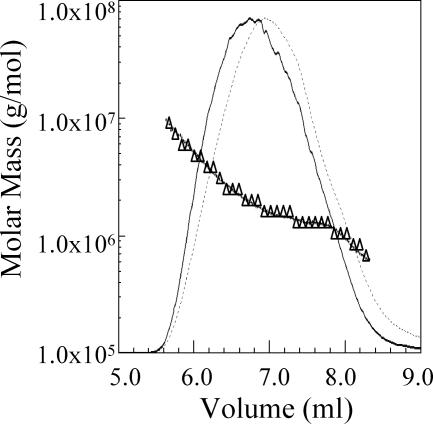
The purified ropy fraction EPS was analyzed by HPLC SEC-MALLS. The ropy EPS eluted from the SEC column as a single, moderately broad peak (Fig. 1). Analysis of the molar mass along the peak indicated significant heterogeneity in mass in the ropy fraction, which is seen as a downward-sloping line on a plot of molar mass versus time. The polysaccharide emerging at the front of the peak had a mass close to 107 Da, and at the top of the peak the average mass was approximately 2 × 106 Da. The EPS that emerged at the end of the peak had a mass of approximately 8 × 105 Da. An analysis of the root mean square radius of the EPS showed that it too was moderately heterogeneous, ranging from a high of approximately 250 nm down to around 70 nm, with a mean value for the root mean square radius of 100 nm.
FIG. 1.
HPLC SEC-MALLS molar mass distribution plot of purified ropy fraction EPS from L. lactis subsp. cremoris Ropy352. The solid line is the 90° light-scattering signal; the dashed line is the refractive-index signal. Δ corresponds to molar mass on the y axis.
Sugar linkages.
The predominant sugar found in the ropy EPS, at 36 mol%, is (1,4)-linked glucose (Table 1). The only sugar found as a terminal nonreducing end group was galactose at 27 mol%; this quantity is indicative of a highly branched structure. A (1,4,6)-linked glucose residue was found at a concentration of 21 mol%; the three linkage sites indicate that it is a branch point in this structure. The least represented sugar was the (1,4)-linked galactose, which occurred at a concentration of 15 mol%.
TABLE 1.
Identification of permethylated PAAN derivatives from L. lactis subsp. cremoris Ropy352 and EK240 polysaccharides
| PAAN methyl sugar | Linkage site(s) | Ropy352 ropy fraction (mol%) | Ropy352 F fraction (mol%) | EK240 F fraction (mol%) |
|---|---|---|---|---|
| 2,3,4,6-Tetra-O-methyl galactose | 1 | 27 | 42 | 36 |
| 2,3,6-Tri-O-methyl galactose | 1,4 | 15 | NDa | ND |
| 2,4,6-Tri-O-methyl galactose | 1,3 | ND | 21 | 22 |
| 2,3,4-Tri-O-methyl galactose | 1,6 | ND | 4 | 10 |
| 2,3,6-Tri-O-methyl glucose | 1,4 | 36 | 10 | 9 |
| 2,3,4-Tri-O-methyl glucose | 1,6 | ND | ND | 6 |
| 3,4,6-Tri-O-methyl mannose | 1,2 | ND | 5 | 5 |
| 2,3-Di-O-methyl glucose | 1,4,6 | 21 | 5 | ND |
| 3,4-Di-O-methyl glucose | 1,2,6 | ND | 6 | 2 |
| 2,4-Di-O-methyl mannose | 1,3,6 | ND | 7 | 10 |
ND, not detected.
In the F fraction EPS from L. lactis subsp. cremoris Ropy352, terminal nonreducing galactose is the most predominant residue at 42 mol%. Linear (1,3)-linked galactose (21 mol%) and (1,4)-linked glucose (10 mol%) residues were also determined. Mannose, as the (1,2)- and (1,3,6)-linked branches, was also present in the F fraction EPS obtained from both sources. While the overall ratios of galactose-glucose-mannose were similar between the F fractions from L. lactis subsp. cremoris Ropy352 (67:21:12) and the L. lactis subsp. cremoris EK240 mutant (68:17:15), there were some differences in the sugar linkages found in the two sources. The F fraction from L. lactis subsp. cremoris Ropy352 did not contain any (1,6)-linked glucose, which was present in the F fraction from L. lactis subsp. cremoris EK240 at 6 mol%. Conversely, the F fraction EPS from L. lactis subsp. cremoris Ropy352 had 5 mol% (1,4,6)-linked glucose, while this branched sugar was not detected in the F fraction EPS from L. lactis subsp. cremoris EK240.
Degree of phosphate protonation.
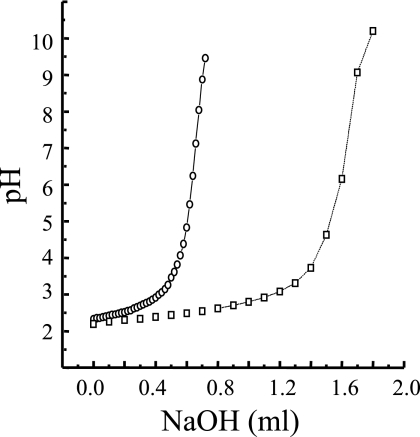
As sodium hydroxide was added to the polysaccharide solution, there is only one inflection in the titration profiles, indicating that the phosphate group in the ropy and F fraction EPS is in the form of a phosphodiester linkage rather than as the monoester, which would have shown two inflection points (Fig. 2). Incubation of the decationized polymer at 100°C for 30 min did not change the titration pattern for the ropy EPS, which suggests that the phosphate does not occur in a simple sugar phosphodiester bridge in the side chain as is found in phosphomannans (42).
FIG. 2.
Titration of the purified ropy and F fraction polysaccharides from L. lactis subsp. cremoris Ropy352. Symbols: □, ropy polysaccharide; ○, F fraction polysaccharide.
Plasmid localization of the ropy EPS phenotype.
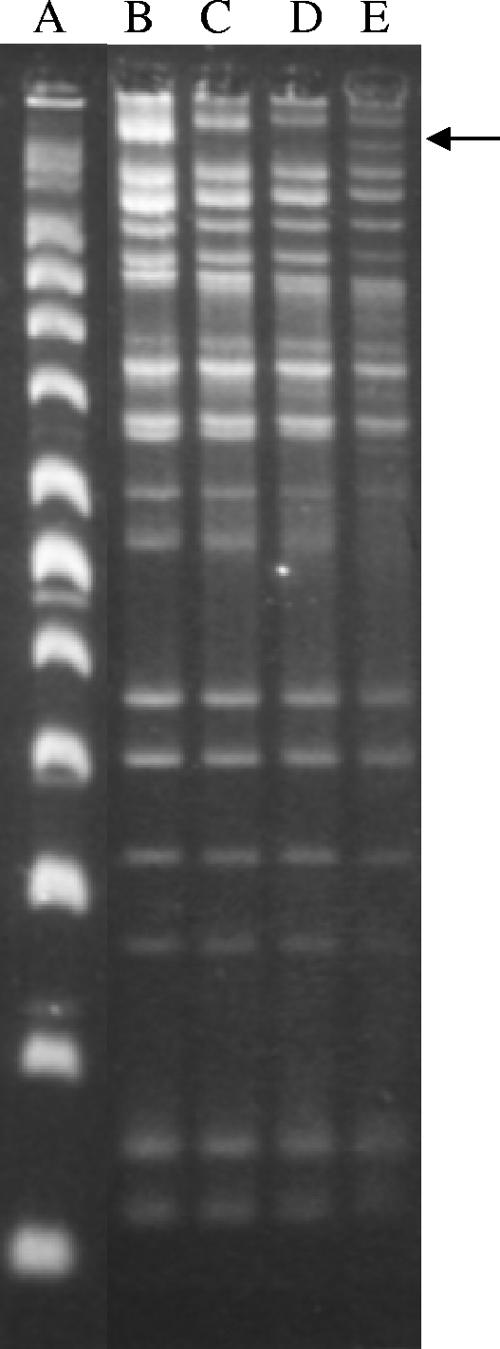
L. lactis subsp. cremoris Ropy352 contains at least 18 and possibly as many as 20 native plasmids that range in size from an estimated 2.5 kb to 34 kb (Fig. 3). During routine maintenance of the L. lactis subsp. cremoris Ropy352 strain on whey agar plates, three nonropy colonies spontaneously arose, L. lactis subsp. cremoris Ropy352 nonropy strains EK316, EK317, and EK318 (this study). These three nonropy colonies, however, still expressed the mucoid phenotype. The plasmid profiles of the three spontaneous nonropy colonies of L. lactis subsp. cremoris Ropy352 were examined and found to be devoid of a large plasmid further referred to as pEPS352 (Fig. 3). The total plasmid DNA of the spontaneous nonropy strains was tested by PCR with primer sequences internal to the ropy EPS loci described in the following sections. Plasmid pEPS352 was absent, and no products were observed (data not shown), indicating that these strains had lost the genes necessary for ropy EPS expression. These results suggested that the ropy EPS locus was located on a large plasmid, pEPS352.
FIG. 3.
Plasmid profiles of L. lactis subsp. cremoris Ropy352 and its spontaneous nonropy mutants. Lane A, supercoiled plasmid ladder (16,210, 14,174, 12,138, 10,102, 8,066, 7,045, 6,030, 5,012, 3,990, 2,972, and 2,067 bp). Lanes B, C, and D, spontaneous nonropy mutants L. lactis subsp. cremoris EK316, EK317, and EK318, respectively. Lane E, wild-type L. lactis subsp. cremoris Ropy352. The arrow indicates the plasmid absent from the nonropy mutants.
Isolation of DNA fragments surrounding the Gh9:ISS1 insertion site.
The transposon-based mutagenic tool pGh9:ISS1 allows the isolation of DNA flanking either side of the Gh9:ISS1 insertion site by HindIII and EcoRI digestion (28). A HindIII DNA fragment of 2,016 bp (pEK238) from L. lactis subsp. cremoris EK240 was isolated adjacent to the Gh9:ISS1 insertion. However, isolating flanking DNA on the other side of the Gh9:ISS1 insertion with EcoRI proved to be more difficult. Hypothesizing that the ropy EPS genes might reside on a large plasmid, plasmid DNA was isolated from the nonropy mutant, L. lactis subsp. cremoris EK240 containing the pGh9:ISS1 insertion, restricted with EcoRI, diluted, and ligated to form covalently closed plasmids. The plasmids were electroporated into L. lactis MG1363 and cultured at 37°C to block replication driven by the replication origin (ori) of pGh9:ISS1. This led to erythromycin-resistant transformants carrying a large plasmid consisting of pGh9:ISS1 and an EcoRI DNA fragment of 12,736 bp (pEK396). Presumably, the EcoRI DNA fragment came from pEPS352 (Fig. 4) and contained this native plasmid's replication origin (ori) because replication would not be able to be sustained by the pGh9:ISS1 replication origin (ori) because of the culturing temperature.
ORF identification and organization of the ropy EPS loci.
The genetic organization of a 22.4-kb region of the plasmid, pEPS352, identified as containing the Gh9:ISS1-interrupted EPS locus is shown in Fig. 4. Two fragments, one on either side of the Gh9:ISS1 insertion, were recovered by the strategy outlined above. The sequences bordering these fragments were isolated from the wild-type L. lactis subsp. cremoris Ropy352 strain by PCR-based walking. The EPS locus was organized into three separate yet sequential regions containing putative components of the ropy EPS expression system, namely, the epsMN region, the epsU region, and the EPS operon (epsR, -X, -A, -B, -C, -D, -E, -F, -O, and -P) region. Sequence data of these regions revealed 19 complete open reading frames (ORFs) having ATG as a start codon and putative ribosomal binding sites (RBS), as well as many incomplete or fragmented ORFs.
EpsMN region.
Sequence analysis of this region showed three complete ORFs (epsM, epsN, and ISS1) having putative RBS (12) just upstream from their start codons and one partial ORF (ISS1) missing a start codon and putative RBS (Fig. 4). The predicted gene products of two of the complete ORFs (epsM and epsN) showed similarity to glycosyltransferases involved in EPS expression in a variety of gram-positive and gram-negative organisms. Interestingly, the top matches for gram-positive dairy organisms are all Streptococcus thermophilus (Table 2). The ORF epsM shows only limited sequence similarity to glycosyltransferases present in the database, while epsN exhibits a very high sequence similarity (94%) to S. thermophilus eps7F whereas the rest of the significant sequence similarities are much lower (32 to 34%). The G+C contents of these two ORFs are 30.2 and 27%, respectively, which are lower than the reported 37.2 to 39.8% G+C content of S. thermophilus (14) and the 35.3% G+C content of Lactococcus as calculated from the chromosomal nucleotide analysis of L. lactis subsp. lactis IL-1403 (AE005176) in the codon usage database (http://www.kazusa.or.jp/codon) or the 34 to 36% G+C content previously reported (8). The Gh9:ISS1 insertion site was localized to the terminal end of epsN, a putative glycosyltransferase (Fig. 4). Glycosyltransferases are responsible for the transfer of sugar residues to a growing EPS backbone. A mutation blocking the ability of epsN to add the proper sugar residue to a growing EPS chain would presumably affect the ropy phenotype of L. lactis subsp. cremoris Ropy352. Also present in this region are two ISS1 ORFs facing in the opposite direction relative to epsM and epsN yet facing in the same direction relative to each other. This is a typical arrangement for native ISS1 units after having been transposed (28). The ISS1 ORF located 3′ to epsN is complete, while the ISS1 ORF located 5′ to epsM is missing the first 82 amino acids (aa) and is probably not functional. In the nonropy mutant L. lactis subsp. cremoris EK240, the PCR was used to verify that no other insertions had occurred in any of the other putative EPS genes, epsR-P, epsM, or epsU, including the upstream promoter regions. PCR products were exactly the same as in the wild type, except for the Gh9:ISS1 insertion localized to epsN (data not shown).
TABLE 2.
Properties and similarities of novel enzymes encoded by pEPS352 eps
| pEPS352 gene | Predicted protein sizea | pI of protein | G+C content (%) | Putative translation start sequenceb | Similar polypeptide(s) | % Identity/similarity | Organismc | Putative function of gene product |
|---|---|---|---|---|---|---|---|---|
| epsM | 331/38.6 | 9.61 | 27 | GGAGtataaaattgATG | EpsG | 35/54 | S. thermophilus (1) | Glycosyltransferase |
| CpsI | 35/54 | S. thermophilus (2) | Glycosyltransferase | |||||
| EpsH | 35/52 | S. thermophilus (1) | Glycosyltransferase | |||||
| EpsI | 33/55 | S. thermophilus (3) | Glycosyltransferase | |||||
| epsN | 329/38.3 | 8.69 | 30.2 | AGGAGtagaataaagagATG | Eps7F | 94/96 | S. thermophilus (4) | Glycosyltransferase |
| EpsH | 34/52 | S. thermophilus (1) | Glycosyltransferase | |||||
| EpsV | 32/55 | S. thermophilus (5) | Glycosyltransferase | |||||
| Eps9I | 32/53 | S. thermophilus (6) | Glycosyltransferase | |||||
| epsO | 235/27.8 | 7.74 | 30.2 | GAGAAgatATG | Eps7I | 86/91 | S. thermophilus (4) | Putative glycosyltransferase |
| EpsQ | 41/61 | L. lactis subsp. cremoris (7) | Putative glycosyltransferase | |||||
| Eps10 | 41/59 | S. thermophilus (8) | Putative glycosyltransferase | |||||
| WciT | 38/56 | S. pneumoniae (9) | Putative glycosyltransferase | |||||
| epsP | 364/42.2 | 9.89 | 26.9 | GGAAaataattactATG | Eps7H | 78/82 | S. thermophilus (4) | Putative repeating-unit polymerase |
| Orf7B | 78/79 | S. thermophilus (4) | Putative repeating-unit polymerase | |||||
| CpsIVH | 22/41 | S. agalactiae (10) | Putative repeating-unit polymerase | |||||
| Eps4N | 19/40 | S. thermophilus (11) | Putative repeating-unit polymerase | |||||
| epsU | 472/53.5 | 9.61 | 30.2 | GGAGGgttattaATG | CpsU | 97/98 | S. thermophilus (12) | Glycosyltransferase |
| EpsU | 97/98 | S. thermophilus (5) | Glycosyltransferase | |||||
| EpsI | 97/98 | S. thermophilus (1) | Glycosyltransferase | |||||
| Eps7M | 96/97 | S. thermophilus (4) | Glycosyltransferase |
Number of amino acids/molecular mass in kilodaltons.
The lactococcal 16S rRNA sequence of the 3′ end is 3′-UCUUUCCUCC-5′ (12). Uppercase letters indicate putative RBS, and boldface indicates the start codon.
The epsU and plasmid replication region.
The epsU and plasmid replication region was sequenced and found to encode seven complete ORFs (epsU, repA, repC, resolvase, and three hypothetical ORFs [Fig. 5 ]) and four incomplete ORFs. This region contains a single gene that is possibly involved in ropy EPS expression and several other genes involved in plasmid replication. epsU shows 97% sequence similarity to epsU of S. thermophilus CNRZ368 (5) and limited sequence similarity to repeat unit polymerases involved in polymer biosynthesis. Contained in the intergenic region between the complete ISS1 sequence downstream of epsN and epsU is an incomplete ORF encoding the last 214 aa of epsH of S. thermophilus Sfi39 (AF373595). Contained in the intergenic regions between epsU, resolvase, RepA, RepC, and IS981 are three complete hypothetical proteins and several incomplete genes related to plasmid replication including a resolvase, RepB, and a mobilization protein fragment. An origin of replication composed of four direct repeats of 19 bp each was found directly upstream of RepA, a typical organization for a bacterial replication origin (18). The discovery of genes involved in replication and an associated binding site in this region demonstrates that our hypothesis was correct that the L. lactis subsp. cremoris Ropy352 EPS locus resides on a large plasmid and that a native origin of replication resides on the 12,736-bp EcoRI fragment of pEK396 and is able to replicate pEK396 independently of the replication origin (ori) associated with pGh9:ISS1.
FIG. 5.
Genetic organization of the pEPS352 replication-epsU region of L. lactis subsp. cremoris Ropy352. Large block arrows with diagonal dashes indicate partial ORFs. Putative RBS-promoter sequences are indicated with black flags. The small block arrow with vertical lines is the fragment of epsH as described in the text. The symbol @ indicates the replication origin.
The eps operon region.
Plasmid pEPS352 contains an operon of 10 genes (epsR, -X, -A, -B, -C, -D, -E, -F, -O, and -P) and is flanked on both ends by partial IS elements (Fig. 4). The IS element upstream of the operon shows 62% sequence similarity to other IS elements found in dairy lactococci. The downstream IS element shows 91% sequence similarity to IS981 but is missing the first 38 aa, start codon, and RBS and is presumably not functional. Between the downstream IS981 element and epsP is a small fragment of the same epsH ORF seen in the epsU region. The 5′ 39-aa end of epsH is located between epsP and IS981, while the 3′ 214-aa end is located between ISS1 and epsU. These two fragments are facing in opposite directions and encode different parts of the epsH gene, suggesting that an insertional event occurred at some point to interrupt this gene. epsR shows 98 and 97% identity to epsR of L. lactis subsp. cremoris HO2 (AF142639) and L. lactis NIZO B40 (AF036485), respectively. epsX showed 100% identity over 149 aa to epsX of L. lactis NIZO B40; however, the amino-terminal 106 aa are missing and no start codon was found, suggesting that epsX of L. lactis subsp. cremoris Ropy352 may not be expressed. epsA shows 91% identity to epsA of L. lactis subsp. cremoris HO2 and L. lactis NIZO B40. The epsB-epsF region shows a high degree of sequence similarity to the same region of L. lactis NIZO B891 (AF100298). The DNA sequence of this region showed a 98% sequence similarity with just 5 bp changes over 2,884 bp verified by three independent PCR events. The ORF epsO shows 86% identity with eps7I of S. thermophilus (AF454498) and lower sequence similarities with putative glycosyltransferases. The ORF epsP shows only limited sequence similarities with putative polysaccharide polymerases from Streptococcus species (Table 2).
DISCUSSION
Industrial interest in ropy strains of LAB such as L. lactis subsp. cremoris is on the rise for the purpose of developing and improving the production of fermented milk products. EPS involved in the development of the ropy property of fermented milk products have been chemically characterized to various extents (23, 27, 29, 33, 34, 41, 51). The compositions and some of the physical properties of the EPS produced by L. lactis subsp. cremoris Ropy352 and L. lactis subsp. cremoris EK240, a mutant lacking the ropy phenotype, were elucidated in this study, and our results show that they are different from those that have been described to date. We isolated and characterized two distinct EPS and have shown that one of them is responsible for the ropy characteristic that is found in the intact cultured milk product. The sugar composition of the ropy EPS is 57% glucose and 42% galactose, and it also contains phosphorus. The ropy EPS from strain L. lactis subsp. cremoris Ropy352 is most like that from L. lactis NIZO B891 (51). The GC-MS data for linkage analysis were similar; however, the L. lactis NIZO B891 EPS has acetyl groups to the internal glucose of the disaccharide side chain, which were not detected in L. lactis subsp. cremoris Ropy352. Additionally, L. lactis subsp. cremoris Ropy352 contains phosphate in a diphosphate linkage that was not released by autohydrolysis. The EPS from L. lactis subsp. cremoris LC330 (29) was also phosphorylated but differed from the ropy polysaccharide of L. lactis subsp. cremoris Ropy352 in that it contained a large proportion of rhamnose and a small amount of glucosamine. The authors reported that this low-molecular-weight phosphopolysaccharide was a minor component of cultures relative to a high-molecular-weight neutral polysaccharide. However, this low-molecular-weight phosphopolysaccharide was consistently produced and was not affected by variations in growth conditions and it was not reported whether the polysaccharide retained its ropy property after purification. Another EPS that produces highly viscous solutions is made by Lactobacillus sake 0-1; it contains glucose and rhamnose and also phosphorus (39). The phosphate in the polysaccharide produced by L. sake 0-1 occurred as glycerol 3-phosphate attached to the 4 position of a side chain rhamnose. Other structural analyses of polysaccharides made by LAB have shown that some of them are homopolysaccharides of galactose, such as that made by L. lactis subsp. cremoris H414 (20), and can also impart a ropy character to a solution. Thus, it would appear that no one particular structural feature must exist in order to impart the ropy property to these polymers.
Removal of the cells and other insoluble material from the 24-h L. lactis subsp. cremoris Ropy352 cultured milk caused a considerable drop in viscosity with retention of ropiness. Apparently, it is the interaction of the ropy polysaccharide with other cellular components, such as the intrinsic protein or capsular polysaccharides (CPS) that remain attached to the cell walls, that is responsible for the dramatic change in viscosity during growth while also providing the desirable ropiness evident when the fluid is manipulated. The exact nature of this interaction remains to be determined, but it warrants further investigation if these compounds are to be effectively exploited in foods or other industrial applications.
The organization of EPS genes in L. lactis subsp. cremoris Ropy352 is different from that of other lactococcal operons encoding EPS biosynthesis. Three ORFs, epsM, epsN, and epsU, complete with putative promoter regions, RBS, and ATG start codons exist separately and are oriented in the opposite direction of an operon containing 10 genes predicted to be involved in EPS expression. Previously, we showed that the ropy EPS locus of L. lactis subsp. cremoris Ropy352 had the potential to be plasmid borne, as demonstrated by the ability to transfer the Gh9:ISS1-interrupted EPS locus, marked by erythromycin resistance, to L. lactis MG1363 (23). In this study, we demonstrated that the spontaneous loss of the ropy phenotype along with the loss of a large plasmid supports the conclusion that the ropy EPS locus is plasmid borne. This is in agreement with other described lactococcal plasmid-borne EPS loci (15, 24, 35, 50, 52-54).
Sequence data show that the ISS1 insertion conferred by pGh9:ISS1 mapped to epsN. Analysis of the three genetic loci (the epsMN region, epsU, and the EPS operon) on pEPS352 revealed no other insertions other than in epsN. The ORFs epsM, epsN, and epsU show strong sequence similarity to glycosyltransferases; however, epsU also shows limited sequence similarity to repeat unit transporters. Isolation and characterization of EPS produced by the nonropy L. lactis subsp. cremoris EK240 mutant strain support the claim made by the genetic analysis, that impairing the expression of epsN, an enzyme responsible for the transfer of sugar moieties to a growing EPS chain, eliminated production of the ropy EPS, while production of the mucoid EPS was unaffected. Blocking expression of one glycosyltransferase, epsN, could be expected to lead to the observed nonropy phenotype by creating problems with recognition of the incomplete heteropolysaccharide by the polymerizing and exporting enzymes. One study observed that EPS expressed by an exoV mutant lacked only a pyruvyl modification to the polymer, but this difference was enough to eliminate production of the EPS, presumably because of the inability of the polymerization or export machinery to recognize the unmodified EPS (19).
The structure of the L. lactis subsp. cremoris Ropy352 EPS operon (epsR, -X, -A, -B, -C, -D, -E, -F, -O, and -P) is similar to that of EPS operons described in L. lactis strains NIZO B40 and NIZO B891 (51). The operons share the epsRXABCDEF genes, but in L. lactis subsp. cremoris Ropy352 these genes are followed by two more genes, epsO and epsP, that are presumably involved in polymerization and export. It would be interesting to determine if L. lactis NIZO B891 also has these two genes. The ability to interrupt one of the isolated glycosyltransferase genes, epsN, and disrupt ropy EPS biosynthesis suggests that the expression of the ropy phenotype of L. lactis subsp. cremoris Ropy352 is the result of at least two independent genetic loci, a glycosyltransferase cluster consisting of epsM and epsN and an operon of 10 genes (epsR, -X, -A, -B, -C, -D, -E, -F, -O, and -P). What role epsU plays, if any, in expression of the ropy or mucoid EPS phenotype has yet to be determined.
Typical of EPS operon organization in Lactococcus and Streptococcus is the presence of IS elements flanking the operon. The CPS type 1 operon in Streptococcus pneumoniae is flanked by a pair of IS1167 elements, both facing in the same direction (32). In a lactococcal strain, an EPS region involved in preventing phage adsorption is flanked by IS elements (15). In L. lactis subsp. cremoris Ropy352, the EPS operon (epsR, -X, -A, -B, -C, -D, -E, F, -O, and -P) is flanked by a pair of partial IS elements, IS981 and IS982, and the epsMN cassette is flanked by complete and incomplete ISS1 elements. This organization is similar to the IS element organization reported for S. thermophilus CRNZ368, and the authors noted that the epsL-IS981SC region was likely transferred to S. thermophilus CRNZ368 from L. lactis via horizontal transfer (5). L. lactis subsp. cremoris Ropy352 and S. thermophilus CRNZ368 share another organizational similarity. Both strains contain IS element-flanked intact ORFs for genes potentially involved in EPS biosynthesis that are separated from the EPS operon, the epsU and epsMN genes of L. lactis subsp. cremoris Ropy352 and the epsUV genes of S. thermophilus CRNZ368. Interestingly, no EPS expression from S. thermophilus CRNZ368 under the conditions tested was noted (5). One study found that genes necessary for the transport of a CPS across the inner membrane in Escherichia coli are separated from the genes necessary for production and export of the polymer (38). This type of mosaic structure is also seen in the alkane degradation genes of Pseudomonas putida (49). What is striking in the alkane degradation example is that two genes involved in degradation are separated from the main operon by IS elements and are facing in the opposite direction, very similar to the structure and orientation observed for epsMN in the ropy EPS loci of L. lactis subsp. cremoris Ropy352. These mosaic areas of IS elements found to be flanking specific genes are formed in regions of DNA not necessarily needed for survival. Through the activity of mobile genetic units such as IS sequences, competitive advantages may be realized by gaining or losing genes or rearranging genes which lead to the expression of functions needed under unpredictable or hostile growth conditions.
Analysis of the EPS operons in lactococcal and streptococcal strains provides a clear example of IS-mediated evolution. Glycosyltransferases involved in polymer biosynthesis have been previously reported to be localized to the central region of a single EPS operon (22). This is the first report that two additional glycosyltransferases, bounded by ISS1 elements, are located separately and in the opposite transcriptional direction from the EPS operon in a lactococcal strain. The low G+C content of the ISS1-epsMN-ISS1 region and the relationship of the structure of these two genes with the ISS1 elements suggest that it was transposed from a source other than lactococcal or streptococcal origins. On the basis of the evidence obtained, we hypothesize that through horizontal transfers and IS-mediated rearrangements, transcriptionally distant yet functionally coupled glycosyltransferases operate in concert with an EPS operon to produce a unique ropy EPS in L. lactis subsp. cremoris Ropy352. This type of genetic organization has not previously been reported for a ropy EPS polymer and is probably a manifestation of the activity of the mobile genetic elements present in LAB. Indeed, as more EPS and CPS operons are sequenced, it is likely that IS elements will be found in close association with these operons and affiliated eps genes, underscoring the natural evolutionary origin of the vast diversity of polymers found in the LAB family.
There are obvious benefits for the food and beverage industry in designing and constructing custom-made LAB EPS with specific functional characteristics. Previous work shows that heterologous complementation of priming glycosyltransferases by different genera in Lactococcus can be achieved (51). Similarly, the exoA gene product, a glucosyltransferase from Rhizobium meliloti, was shown to complement an amsE mutant of Erwinia amylovora to mucoidy; however, no mention was made of how the polymer structurally or compositionally compares to the wild-type polymer (3). The evidence obtained in this study demonstrates heterologous expression and function of additional glycosyltransferase activity. The organization of the genetic regions necessary for expression of the L. lactis subsp. cremoris Ropy352 EPS illustrates that it may be possible to provide a glycosyltransferase activity in trans, and not transcriptionally coupled with the EPS operon, to create a novel polymer. One potential problem would be the failure of the polymerization and export machinery to recognize the novel polymer, yet there are conflicting reports in this area. Transport of an altered S. thermophilus polymer in L. lactis was shown to occur as the altered polymer was present in the supernatant, but at only 0.001% compared to wild-type S. thermophilus levels (4, 44). Additionally, expression of an altered polymer was observed in L. lactis at 5% of wild-type levels (4, 44). This work shows that glycosyltransferases may have multiple specificities for donor and acceptor molecules and that the polymerase and exporters are active on repeating units that differ in backbone, as well as side chain, composition. Our results suggest that the specificity of the polymerization and export enzymes in L. lactis subsp. cremoris Ropy352 is high enough that blocking the function of just one glycosyltransferase abolishes ropy EPS production. However, we do not know whether that glycosyltransferase plays a role in construction of the backbone or side chain modification, as this could determine whether the polymer is recognized by the polymerization and export enzymes. As more details emerge about the organization, function, and specificity of the genes and enzymes involved in EPS expression in LAB, we are nearing the threshold of being able to use a rational design strategy to mimic the natural activity of IS elements in EPS construction by providing additional glycosyltransferase activities via plasmids to create novel polymers.
Acknowledgments
We thank M. Skinner for help with determining the origin of replication of pEPS352, James J. Nicholson for assistance in the GC-MS analysis of the polysaccharides, and Todd A. Lanser for help with bacterial culturing.
E. P. Knoshaug was supported by a Tartar Award. This work was supported by grants from the U.S. Department of Agriculture, the National Dairy Promotion and Research Board (Dairy Management Inc.), and the Harry B. and Ralph H. Levey Philanthropic Fund.
Footnotes
Published ahead of print on 22 November 2006.
REFERENCES
- 1.Ames, B. 1966. Assay of inorganic phosphate, total phosphate and phosphatases, vol. 8. Academic Press, Inc., New York, NY.
- 2.Bennett, P. 2004. Genome plasticity: insertion sequence elements, transposons and integrons, and DNA rearrangement. Methods Mol. Biol. 266:71-113. [DOI] [PubMed] [Google Scholar]
- 3.Bernhard, F., D. Coplin, and K. Geider. 1993. A gene cluster for amylovoran synthesis in Erwinia amylovora: characterization and relationship to cps genes in Erwinia stewartii. Mol. Gen. Genet. 239:158-168. [DOI] [PubMed] [Google Scholar]
- 4.Boels, I., M. Beerthuyzen, M. Kosters, M. V. Kaauwen, M. Kleerebezem, and W. M. de Vos. 2004. Identification and functional characterization of the Lactococcus lactis rfb operon, required for dTDP-rhamnose biosynthesis. J. Bacteriol. 186:1239-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bourgoin, F., A. Pluvinet, B. Gintz, B. Decaris, and G. Guedon. 1999. Are horizontal transfers involved in the evolution of the Streptococcus thermophilus exopolysaccharide synthesis loci? Gene 233:151-161. [DOI] [PubMed] [Google Scholar]
- 6.Bouzar, F., J. Cerning, and M. Desmazeaud. 1997. Exopolysaccharide production and texture-promoting abilities of mixed-strain starter cultures in yogurt production. J. Dairy Sci. 80:2310-2317. [Google Scholar]
- 7.Bouzar, F., J. Cerning, and M. Desmazeaud. 1996. Exopolysaccharide production in milk by Lactobacillus delbrueckii subsp. bulgaricus CNRZ 1187 and by two colonial variants. J. Dairy Sci. 79:205-211. [Google Scholar]
- 8.Bridge, P., and P. Sneath. 1983. Numerical taxonomy of Streptococcus. J. Gen. Microbiol. 129:565-596. [DOI] [PubMed] [Google Scholar]
- 9.Burrus, V., G. Pavlovic, B. Decaris, and G. Guedon. 2002. Conjugative transposons: the tip of the iceberg. Mol. Microbiol. 46:601-610. [DOI] [PubMed] [Google Scholar]
- 10.Burrus, V., and M. Waldor. 2004. Shaping bacterial genomes with integrative and conjugative elements. Res. Microbiol. 155:376-386. [DOI] [PubMed] [Google Scholar]
- 11.Cerning, J., C. Bouillanne, M. Landon, and M. Desmazeaud. 1992. Isolation and characterization of exopolysaccharides from slime-forming mesophilic lactic acid bacteria. J. Dairy Sci. 75:692-699. [Google Scholar]
- 12.Chiaruttini, C., and M. Milet. 1993. Gene organization, primary structure and RNA processing analysis of a ribosomal RNA operon in Lactococcus lactis. J. Mol. Biol. 230:57-76. [DOI] [PubMed] [Google Scholar]
- 13.Dierksen, K., W. Sandine, and J. Trempy. 1997. Expression of ropy and mucoid phenotypes in Lactococcus lactis. J. Dairy Sci. 80:1528-1536. [DOI] [PubMed] [Google Scholar]
- 14.Farrow, J., and M. Collins. 1984. DNA base composition, DNA-DNA homology and long chain fatty acid studies on Streptococcus thermophilus and Streptococcus salivarius. J. Gen. Microbiol. 130:357-362. [DOI] [PubMed] [Google Scholar]
- 15.Forde, A., and G. Fitzgerald. 2003. Molecular organization of exopolysaccharide (EPS) encoding genes on the lactococcal bacteriophage adsorption blocking plasmid, pCI658. Plasmid 49:130-142. [DOI] [PubMed] [Google Scholar]
- 16.Gancel, F., and G. Novel. 1994. Exopolysaccharide production by Streptococcus salivarius subsp. thermophilus cultures. 2. Distinct modes of polymer production and degradation among clonal variants. J. Dairy Sci. 77:689-695. [Google Scholar]
- 17.Gasson, M. J. 1983. Plasmid complements of Streptococcus lactis NCDO 712 and other lactic streptococci after protoplast-induced curing. J. Bacteriol. 154:1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Giraldo, R., J. Andreu, and R. Diaz-Orejas. 1998. Protein domains and conformational changes in the activation of RepA, a DNA replication initiator. EMBO J. 17:4511-4526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Glucksmann, M., T. Reuber, and G. Walker. 1993. Genes needed for the modification, polymerization, export, and processing of succinoglycan by Rhizobium meliloti: a model for succinoglycan biosynthesis. J. Bacteriol. 175:7045-7055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gruter, M., B. Leeflang, J. Kuiper, J. Kamerling, and J. Vliegenthart. 1992. Structure of the exopolysaccharide produced by Lactococcus lactis subsp. cremoris H414 grown in a defined medium or skimmed milk. Carbohydr. Res. 231:273-291. [DOI] [PubMed] [Google Scholar]
- 21.Harrison, R., J. Miller, M. D'Souza, and G. Kampo. 1997. Easy gene walking. BioTechniques 22:650-653. [DOI] [PubMed] [Google Scholar]
- 22.Jolly, L., and F. Stingele. 2001. Molecular organization and functionality of exopolysaccharide gene clusters in lactic acid bacteria. Int. Dairy J. 11:733-745. [Google Scholar]
- 23.Knoshaug, E., J. Ahlgren, and J. Trempy. 2000. Growth associated exopolysaccharide expression in Lactococcus lactis subspecies cremoris Ropy352. J. Dairy Sci. 83:633-640. [DOI] [PubMed] [Google Scholar]
- 24.Kojic, M., M. Vujcic, A. Banina, P. Cocconcelli, J. Cerning, and L. Topisirovic. 1992. Analysis of exopolysaccharide production by Lactobacillus casei CG11, isolated from cheese. Appl. Environ. Microbiol. 58:4086-4088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lamothe, G., L. Jolly, B. Mollet, and F. Stingele. 2002. Genetic and biochemical characterization of exopolysaccharide biosynthesis by Lactobacillus delbrueckii subsp. bulgaricus. Arch. Microbiol. 178:218-228. [DOI] [PubMed] [Google Scholar]
- 26.Laws, A., and V. Marshall. 2001. The relevance of exopolysaccharides to the rheological properties in milk fermented with ropy strains of lactic acid bacteria. Int. Dairy J. 11:709-721. [Google Scholar]
- 27.Macura, D., and T. Townsley. 1984. Scandinavian ropy milk—identification and characterization of endogenous streptococci and their extracellular excretion. J. Dairy Sci. 67:735-744. [Google Scholar]
- 28.Maguin, E., H. Prevost, S. D. Ehrlich, and A. Gruss. 1996. Efficient insertional mutagenesis in lactococci and other gram-positive bacteria. J. Bacteriol. 178:931-935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marshall, V., E. Cowie, and R. Moreton. 1995. Analysis and production of two exopolysaccharides from Lactococcus lactis subsp. cremoris LC330. J. Dairy Res. 62:621-628. [Google Scholar]
- 30.Marshall, V., and H. Rawson. 1999. Effects of exopolysaccharide-producing strains of thermophilic lactic acid bacteria on the texture of stirred yoghurt. Int. J. Food Sci. Technol. 34:137-143. [Google Scholar]
- 31.Morris, V. 1993. Fermentation-derived polysaccharides for use in foods. Food Biotechnol. 58:199-201. [Google Scholar]
- 32.Muñoz, R., M. Mollerach, R. López, and E. García. 1999. Characterization of the type 8 capsular gene cluster of Streptococcus pneumoniae. J. Bacteriol. 181:6214-6219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nakajima, H., T. Hirota, T. Toba, T. Itoh, and S. Adachi. 1992. Structure of the extracellular polysaccharide from slime-forming Lactococcus lactis subspecies cremoris SBT 0495. Carbohydr. Res. 224:245-253. [DOI] [PubMed] [Google Scholar]
- 34.Nakajima, H., S. Toyoda, T. Toba, T. Itoh, T. Mukai, H. Kitazawa, and S. Adachi. 1990. A novel phosphopolysaccharide from slime-forming Lactococcus lactis subspecies cremoris SBT 0495. J. Dairy Sci. 73:1472-1477. [Google Scholar]
- 35.Neve, H., A. Geis, and M. Teuber. 1988. Plasmid-encoded functions of ropy lactic acid streptococcal strains from Scandinavian fermented milk. Biochimie 70:437-442. [DOI] [PubMed] [Google Scholar]
- 36.O'Sullivan, D. J., and T. Klaenhammer. 1993. Rapid mini-prep isolation of high-quality plasmid DNA from Lactococcus and Lactobacillus spp. Appl. Environ. Microbiol. 59:2730-2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Polzin, K., D. Romero, M. Shimizu-Kadota, T. Klaenhammer, and L. McKay. 1993. Copy number and location of insertion sequences ISS1 and IS981 in lactococci and several other lactic acid bacteria. J. Dairy Sci. 76:1243-1252. [DOI] [PubMed] [Google Scholar]
- 38.Rigg, G., B. Barrett, and I. Roberts. 1998. The localization of KpsC, S and T, and KfiA, C and D proteins involved in the biosynthesis of the Escherichia coli K5 capsular polysaccharide: evidence for a membrane-bound complex. Microbiology 144:2905-2914. [DOI] [PubMed] [Google Scholar]
- 39.Robijn, G. W., D. J. van den Berg, H. Haas, J. P. Kamerling, and J. F. Vliegenthart. 1995. Determination of the structure of the exopolysaccharide produced by Lactobacillus sake 0-1. Carbohydr. Res. 276:117-136. [DOI] [PubMed] [Google Scholar]
- 40.Seymour, F., R. Plattner, M. Slodki, and R. Stodola. 1976. Methylation and acetolysis of extracellular d-mannans from yeast. Carbohydr. Res. 48:225-237. [DOI] [PubMed] [Google Scholar]
- 41.Shellhaass, S. 1983. Ph.D. thesis. Characterization of the exocellular slime produced by bacterial starter cultures used in the manufacture of fermented dairy products. University of Minnesota, Minneapolis.
- 42.Slodki, M. 1966. The structure of extracellular phosphorylated galactans from Sporobolomyces yeasts. J. Biol. Chem. 241:2700-2706. [PubMed] [Google Scholar]
- 43.Stingele, F., J. Neeser, and B. Mollet. 1996. Identification and characterization of the eps (exopolysaccharide) gene cluster from Streptococcus thermophilus Sfi6. J. Bacteriol. 178:1680-1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stingele, F., S. Vincent, E. Faber, J. Newell, J. Kamerling, and J. Neeser. 1999. Introduction of the exopolysaccharide gene cluster from Streptococcus thermophilus Sfi6 into Lactococcus lactis MG1363: production and characterization of an altered polysaccharide. Mol. Microbiol. 32:1287-1295. [DOI] [PubMed] [Google Scholar]
- 45.Sutherland, I. 1998. Novel and established applications of microbial polysaccharides. Trends Biotechnol. 16:41-46. [DOI] [PubMed] [Google Scholar]
- 46.Tan, H. 1999. Bacterial catabolic transposons. Appl. Microbiol. Biotechnol. 51:1-12. [DOI] [PubMed] [Google Scholar]
- 47.Terzaghi, B. E., and W. Sandine. 1975. Improved medium for lactic streptococci and their bacteriophages. Appl. Microbiol. 29:807-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Toba, T., H. Uemuar, T. Mukai, T. Fujii, T. Itoh, and S. Adachi. 1991. A new fermented milk using capsular polysaccharide-producing Lactobacillus kefiranofaciens isolated from kefir grains. J. Dairy Res. 58:497-502. [Google Scholar]
- 49.van Beilen, J., S. Panke, S. Lucchini, A. Franchini, M. Rothlisberger, and B. Witholt. 2001. Analysis of Pseudomonas putida alkane-degradation gene clusters and flanking insertion sequences: evolution and regulation of the alk genes. Microbiology 147:1621-1630. [DOI] [PubMed] [Google Scholar]
- 50.van Kranenburg, R., J. D. Marugg, I. I. van Swam, N. J. Willem, and W. M. de Vos. 1997. Molecular characterization of the plasmid-encoded eps gene cluster essential for exopolysaccharide biosynthesis in Lactococcus lactis. Mol. Microbiol. 24:387-397. [DOI] [PubMed] [Google Scholar]
- 51.van Kranenburg, R., H. R. Vos, I. I. van Swam, M. Kleerebezem, and W. M. de Vos. 1999. Functional analysis of glycosyltransferase genes from Lactococcus lactis and other gram-positive cocci: complementation, expression, and diversity. J. Bacteriol. 181:6347-6353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vedamuthu, E., and J. Neville. 1986. Involvement of a plasmid in production of ropiness (mucoidness) in milk cultures by Streptococcus cremoris MS. Appl. Microbiol. 51:677-682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vescovo, M., G. Scolari, and V. Bottazzi. 1989. Plasmid-encoded ropiness production in Lactobacillus casei subsp. casei. Biotechnol. Lett. II:709-712. [Google Scholar]
- 54.von Wright, A., and S. Tynkkynen. 1987. Construction of Streptococcus lactis subsp. lactis strains with a single plasmid associated with mucoid phenotype. Appl. Environ. Microbiol. 53:1385-1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wacher-Rodarte, C., M. Galvan, A. Farres, F. Gallardo, V. Marshall, and M. Garcia-Garibay. 1993. Yogurt production from reconstituted skim milk powders using different polymer and non-polymer forming starter cultures. J. Dairy Res. 60:247-254. [Google Scholar]
- 56.Zink, D. 1997. The impact of consumer demands and trends on food processing. Emerg. Infect. Dis. 3:467-469. [DOI] [PMC free article] [PubMed] [Google Scholar]