Abstract
Vibrio splendidus is a dominant culturable Vibrio in seawater, and strains related to this species are also associated with mortality in a variety of marine animals. The determinants encoding the pathogenic properties of these strains are still poorly understood; however, the recent sequencing of the genome of V. splendidus LGP32, an oyster pathogen, provides an opportunity to decipher the basis of the virulence properties by disruption of candidate genes. We developed a novel suicide vector based on the pir-dependent R6K replicative origin, which potentially can be transferred by RP4-based conjugation to any Vibrio strain and which also carries the plasmid F toxin ccdB gene under control of the PBAD promoter. We demonstrated that this genetic system allows efficient counterselection of integrated plasmids in the presence of arabinose in both V. splendidus and Vibrio cholerae and thus permits efficient markerless allelic replacement in these species. We used this technique to construct several mutants of V. splendidus LGP32, including a derivative with a secreted metalloprotease gene, vsm, deleted. We found that this gene is essential for LGP32 extracellular product toxicity when the extracellular products are injected into oysters but is not necessary for virulence of bacteria in the oyster infection model when bacteria are injected.
Vibrio splendidus is a dominant culturable Vibrio in coastal marine sediments, seawater, and bivalves, including oysters (23). This organism has long been considered to be an environmental organism without any pathogenic significance. However, over the last few years, different strains phenotypically related to this species have been associated with mortality mainly in mollusks, shrimps, gorgonians, and fish (for a review, see reference 35). Compared to human pathogen species, little is known about Vibrio pathogenesis in marine animals, and despite descriptions of invasiveness and extracellular product (ECP) toxicity, no data are available for a group related to V. splendidus (26, 37, 48, 56).
The different types of enzymatic activities that have been shown to play a role in the virulence of a variety of pathogenic bacteria include extracellular proteases; for example, such proteases have been described for Vibrio cholerae (7), Vibrio vulnificus (33), and Vibrio anguillarum (42), although a direct role of these proteases in virulence has not been demonstrated. For example, it has been shown that the V. cholerae metalloprotease cleavage activity is essential for activating the A subunit of the cholera enterotoxin (12), as well as for degrading intestinal mucin and facilitating the action of cholera toxin (7). In the case of V. vulnificus infection, a metalloprotease has been shown to cause a hemorrhagic reaction by degrading type IV collagen in basement membranes (44). Finally, the empA-encoded metalloprotease of V. anguillarum has been shown to be involved in the invasive mechanism of this fish pathogen (49).
We recently completed sequencing of the genome of V. splendidus strain LGP32 in order to obtain access to its full gene repertoire (F. Le Roux, M. Zouine, N. Chakroun, J. Binesse, D. Saulnier, L. Ma, C. Rusniok, C. Buchriser, and D. Mazel, unpublished data). The strain that we used is an oyster (Crassostrea gigas) pathogen (23, 24). We identified a gene, vsm, potentially encoding a zinc-containing metalloprotease, which could play a role in pathogenesis (45). Interestingly, we found that the vsm predicted product exhibits 95% identity with the product of the V. anguillarum vam gene, which has been shown to be involved in the virulence properties of this fish pathogen (42).
Gene knockout is often essential for formal demonstration of the predicted or supposed role of a gene candidate. However, this strategy is limited to species in which the available genetic tools can be used. There can be limitations at several levels, from DNA delivery inside the cells to the allelic exchange efficiency. DNA transformation, whether it is natural or artificial, is either inoperative or inefficient in numerous species. In V. splendidus, attempts to transfer plasmids using electroporation were ineffective (unpublished results), which prevented use of the Wanner red-swap recombination strategy (14) to perform allelic exchange.
In many cases, exogenous DNA delivery can be achieved by using conjugation with broad-host-range plasmids, and several systems based on the IncPα plasmid RP4 (RK2) transfer functions have been described (54). In most cases the subsequent step, allelic replacement or integration of the incoming DNA, is achieved through use of a nonreplicative DNA molecule. The most popular system for gram-negative species is the system using conditionally replicative R6K plasmid derivatives, such as pGP704 (41). R6K replication is dependent on binding of the pir-encoded Π protein, and transcomplementation of a pir-dependent plasmid derivative by Π proteins expressed from another replicon can be performed (32). Based on this seminal observation, several plasmids carrying the R6Kγ origin of replication that can be replicated only in strains expressing pir have been constructed. When these plasmids also carry an RP4 transfer origin, they can be transferred to various bacterial cells through the broad-host-range conjugation system of RP4. Since these plasmids behave as suicide vectors in pir recipients, they have been successfully used to create mutants through gene disruption by insertion (41) or transposon mutagenesis (29). A wide range of gram-negative bacteria can be engineered with such tools, and most proteobacteria can be used as recipients for conjugation (reference 14 and references therein). Several counterselectable markers have been described (for a review, see reference 48), and some of them have been successfully used in R6K-oriTRP4 derivatives for positive selection of replaced alleles (19, 51, 55).
One of the methods consists of using a wild-type (WT) rpsL gene in a streptomycin-resistant rpsL mutant background (the streptomycin-sensitive wild type is transdominant) (16). However, this strategy requires that cognate rpsL genes be cloned and Smr mutant strains be used.
Due to its general efficiency in gram-negative bacteria and to the simplicity of the counterselection protocol, the Bacillus subtilis levansucrase gene sacB has gained considerable notoriety since 1985, when it was first introduced (25), and it is now certainly the most commonly used of the different counterselectable markers. sacB-based suicide vectors, such as pCVD442 (19), have been successfully used for allelic replacement in V. cholerae (18). However, even though sacB has occasionally been used successfully, the use of this gene for allelic replacement in many Vibrio species or in other marine bacterial species is seriously impeded by the necessary absence of NaCl in the counterselection medium (6). This is the case, for example, with V. splendidus, which cannot grow when NaCl is not present in the medium.
In order to overcome this limitation, we developed a novel suicide vector, based on the pSW family (17), which can be mobilized by the RP4 transfer machinery and which carries the ccdB gene of the Escherichia coli F plasmid under control of the arabinose PBAD promoter (52) as a counterselection marker. We demonstrated that this system allowed positive selection for the loss of vector sequences after homologous recombination in V. splendidus and V. cholerae, with high efficiency. This strategy was used to construct a V. splendidus strain with the vsm gene deleted in order to establish the contribution of the Vsm metalloprotease to the virulence properties of this strain during infection of oysters.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
The bacterial strains and plasmids used in this study are described in Tables 1 and 2. V. splendidus strain LGP32 was isolated from the hemolymph of oysters suffering from outbreaks of summer mortality and was demonstrated to be pathogenic for oysters and clams (23). V. cholerae and E. coli strains were grown in Luria-Bertani (ML) broth or, in case of Π3813, Mueller-Hinton broth at 37°C. V. splendidus strains were grown in Luria-Bertani broth containing 0.5 M NaCl, in marine broth (MB), or on marine agar (MA) at 20°C. All media were obtained from Difco. Antibiotics were used at the following concentrations: ampicillin, 100 μg/ml; chloramphenicol, 12.5 μg/ml; erythromycin, 200 μg/ml; kanamycin, 25 μg/ml; nalidixic acid, 30 μg/ml; spectinomycin, 50 μg/ml; and tetracycline, 15 μg/ml. Thymidine and diaminopimelate were added when necessary at a final concentration of 0.3 mM. Induction of ccdB expression under control of the PBAD promoter was achieved by addition of 0.2% l-arabinose to the growth media, and the expression was repressed by addition of 1% d-glucose.
TABLE 1.
Bacterial strains used and constructed in this study
| Bacterial strain | Description | Reference or source |
|---|---|---|
| LGP32 | Vibrio splendidus | 24 |
| N16961 | Vibrio cholerae | 28 |
| DH5α | F−supE44 ΔlacU169 (φ80lacZΔM15) ΔargF hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | Laboratory collection |
| B462 | lacIqthi-1 supE44 endA1 recA1 hsdR17 gyrA462 zei-298::Tn10 (Tcr) | L. Van Melderen |
| Π3813 | B462 ΔthyA::(erm-pir-116) (Ermr) | This study |
| S17-1 | F− RP4-2-Tc::Mu aph::Tn7 recA (Smr) | 54 |
| β2163 | F− RP4-2-Tc::Mu ΔdapA::(erm-pir) (Kmr Emr) | 17 |
| β3914 | β2163 gyrA462 zei-298::Tn10 (Kmr Emr Tcr) | This study |
| GG784 | TG1 F′ (ΔccdA::speC rexBAD ccdB) (Spr) | 52 |
| δ2989 | LGP32 Δvsm | This study |
| δ4175 | LGP32 ΔluxU | This study |
| δ3453 | LGP32 ΔluxM | This study |
| δ6720−vsm | δ2989 ΔISVs1 orfB::araC-PBADvsm | This study |
| δintIA | N16961 ΔintIA | This study |
TABLE 2.
Plasmids used and constructed in this study
| Plasmid | Description | Source or reference |
|---|---|---|
| pUC18 | ori ColE1 (Apr) | Pharmacia |
| pKOBEGA | pSC101ts::PBADredγδβα (Apr) | 10 |
| pSU18 | ori p15A (Cmr) | 1 |
| PBADgfp | gfp cassette (S65T, F64L) (Apr) | J. M. Ghigo |
| PSU18T-PBADgfp | pSU18::oriTRP4-araC-PBADgfp (Cmr) | This study |
| PSU18T-PBADgfp2 | pSU18::oriTRP4-araC-PBADgfp (Cmr) | This study |
| PSU18T-PBADvsm | pSU18::oriTRP4-araC-PBADvsm (Cmr) | This study |
| pSW23T | oriVR6KγoriTRP4 (Cmr) | 17 |
| pSW29T | oriVR6KγoriTRP4 (Kmr) | 17 |
| pSW4426T | pSW23T::aadA7-araC-PBADccdB (Spr Cmr) | This study |
| pSW4427T | pSW29T::aadA7-araC-PBADccdB (Spr Kmr) | This study |
| pSWδ2989T | pSW4426T, Δvsm (Spr Cmr) | This study |
| pSWδ4175T | pSW4426T, ΔluxU (Spr Cmr) | This study |
| pSWδ3453T | pSW4426T, ΔluxM (Spr Cmr) | This study |
| pSWδ5679T | pSW4426T, ΔgyrA (Spr Cmr) | This study |
| pSWδintIAT | pSW4427T, ΔintIA (Spr Kmr) | This study |
| pSWδ6720T-vsm | pSW4426T, ΔISVs1 orfB::araC-PBADvsm (Spr Cmr) | This study |
PCR.
PCRs performed for plasmid assembly were done in 50-μl mixtures by using the Pfu DNA polymerase (Promega) and following the manufacturer's instructions. Other PCRs were performed in 50-μl mixtures using the Bioline Taq polymerase according to the manufacturer's instructions. The primers used are listed in Table S1 in the supplemental material. The conditions used for amplification were as follows: 94°C for 3 min, followed by 30 cycles of 94°C for 30 s, 10°C less than the melting temperature for 30 s, and 72°C for 60 s per kb.
Construction of CcdB-resistant E. coli strains.
As the gyrA462 mutation has been shown to prevent toxic interaction of the gyrase with CcdB (13), we used this genetic background to prevent selection of a ccdB-inactivating mutation. We previously developed a set of strains carrying pir alleles in different genetic contexts (17). One of these strains carries a pir-116 allele inserted into the thymidylate synthase gene (thyA) of E. coli.
The ΔthyA::(erm-pir-116) E. coli Π3813 strain was constructed by allelic replacement of the chromosomal thyA allele of B462 with ΔthyA::(erm-pir-116), as described previously (17). This thymidine auxotroph strain is CcdB resistant and permits R6K vector replication.
The chromosomal gyrA462 allele was moved from S17-1 to ΔdapA::(erm-pir) E. coli strain β2163 by P1 transduction and Tetr selection of cotransduced zei298::Tn10 to construct strain β3914. To easily discriminate the transductant carrying the gyrA462 allele from transductants still carrying gyrA, we used Nalr/Nals screening. Indeed, Nal resistance is linked to mutations in gyrA, but gyrA462 does not confer this phenotype. Then we selected a spontaneous Nalr derivative of β2163 and used it as the recipient for gyrA462-zei298::Tn10 cotransduction. After transduction, the colonies which had acquired the gyrA462 allele together with zei298::Tn10 were expected to be Nals. In agreement with this, Tcr Nals colonies were found to be insensitive to CcdB toxicity. One clone was designated β3914 and used in this study.
Plasmid construction.
(i) pSU18T-PBADgfp. A 270-bp fragment harboring the RP4 origin of transfer (oriTRP4) was amplified from pSW23T by PCR using primers OriT-Xba and OriT-Pst. After XbaI-PstI digestion, the fragment generated was cloned in pSU18, a p15A derivative compatible with ColE1 and R6K derivatives, yielding pSU18T. A 1,978-bp fragment harboring the araC gene, the PBAD promoter, and the gfp gene was amplified from the PBADgfp plasmid by using primers PBADGFP-Kpn and PBADGFP-Xba. After KpnI-XbaI digestion, the fragment generated was cloned in pSU18T. Green fluorescent protein (GFP) expression was confirmed by epifluorescence microscopy.
(ii) Suicide vectors.
A 2,645-bp fragment harboring a transcriptional terminator, the aadA7 gene, the araC gene, the PBAD promoter, and the ccdB gene was PCR amplified using GG784 DNA (52) as the template and primers ccdB1 and ccdB2. After XbaI-SacI digestion, this fragment was ligated to the suicide vectors pSW23T and pSW29T and digested with the same restriction enzymes, yielding pSW4426T and pSW4427T, respectively.
Several V. splendidus genes, including luxU, luxM, ISVisp1 orfB, vsm, and gyrA, as well as the V. cholerae superintegron integrase gene intIA, were PCR amplified from genomic DNA using primers luxU-1 and luxU-2, primers luxM-1 and luxM-2, primers ISvisp1-1 and ISvisp1-2, primers vsm-1 and vsm-2, primers gyrA-1 and gyrA-2, and primers intIA1 and intIA2, respectively. Amplicons were digested with EcoRI and cloned in the EcoRI site of pUC18 (Pharmacia). The corresponding alleles carrying an internal deletion were constructed by inverse PCR using primers luxU-3 and luxU-4 (ΔluxU allele), primers luxM-3 and luxM-4 (ΔluxM allele), primers ISvisp1-3 and ISvisp1-4 (ISVisp1 ΔorfB allele), primers vsm-3 and vsm-4 (Δvsm allele), primers gyrA-3 and gyrA-4 (ΔgyrA allele), and primers intIA3 and intIA4 (ΔintIA allele), XhoI digestion, and self-ligation. The different alleles were then recovered after EcoRI digestion and gel extraction and introduced by ligation into pSW4426T and/or pSW4427T previously linearized with EcoRI.
(iii) Vector construction for ectopic complementation.
Complementation experiments were performed by introducing vsm under PBAD promoter control into the nonessential gene coding for ISVisp1 transposase present in the chromosome, using the strategy describe above. An inverse PCR was performed with primers GFP3 and GFP4 using pSU18T-PBADgfp as the template. The resulting amplicon was digested by EcoRI and self-ligated, yielding pSU18T-PBADgfp2. The vsm gene was PCR amplified from V. splendidus genomic DNA with primers vsm-1 and vsm-7, digested by EcoRI-XbaI, and cloned in pSU18T-PBADgfp2 with the gfp gene deleted after EcoRI-XbaI digestion. This yielded pSU18T-PBADvsm.
In order to perform ectopic complementation from the chromosome, the araC-PBADvsm amplicon was amplified from pSU18T-PBADvsm using primers araC-S-Xho and vsm-7, XhoI digested, and cloned in the XhoI site of the ISVisp1 ΔorfB allele carried by pSWδ6720T. This yielded pSWδ6720T-vsm.
Conjugation.
Overnight cultures of a donor and a recipient were diluted 1:100 in culture media without antibiotic and grown at 30°C to an optical density at 600 nm (OD600) of 0.3. The different conjugation experiments were performed by a filter mating procedure as described previously (5) with a donor/recipient ratio of 1/10. Conjugation was performed overnight on filters incubated on plates containing ML supplemented with diaminopimelic acid (and containing NaCl in the case of V. splendidus) at 30°C. Counterselection of a ΔdapA donor was performed by plating on a medium lacking diaminopimelic acid but supplemented with 1% glucose and either chloramphenicol or kanamycin. The first recombination frequency was calculated by using the number of transconjugants and the total number of recipients. Antibiotic-resistant colonies were isolated, grown in ML (containing NaCl in the case of V. splendidus) to the late logarithmic phase, and spread on plates containing 0.2% arabinose. Mutants were screened by PCR using primers 5 and 6 flanking the different genes targeted.
Immunoblotting.
Cells (2 ml of an overnight culture) were centrifuged for 5 min at 5,000 rpm, resuspended in 50 μl of bacterial protein extraction reagent (Pierce) supplemented with 1× protease inhibitor (Complete; Roche), vortexed vigorously for 1 min, and centrifuged for 5 min at 13,000 rpm. As measured by the Bradford assay, equal amounts of protein were loaded and separated on a 10% polyacrylamide-sodium dodecyl sulfate (SDS) gel and transferred to a nitrocellulose membrane (Bio-Rad) by electroblotting. The membrane was incubated with the primary antibody (rabbit polyclonal anti-GFP [Sigma]) and then with horseradish peroxidase-conjugated goat anti-rabbit immunoglobulins (Amersham). The proteins were visualized with an enhanced chemiluminescence kit (Amersham) as instructed by the supplier.
Preparation of V. splendidus extracellular products.
Bacterial ECPs were produced by the cellophane overlay method described by Liu (36). Tubes containing 5 ml MB were inoculated with one colony of the LGP32 wild type or mutant and incubated at 20°C for 24 h. Then 500 μl of each culture was transferred onto a sterile cellophane film placed on the surface of an MA plate. After incubation at 20°C for 24 h, the cellophane overlay was transferred to an empty petri dish. Cells were washed off the cellophane film using 2 ml of phosphate-buffered saline and were removed by centrifugation at 13,000 rpm at 20°C for 30 min. The supernatant containing the ECPs was sterilized by filtration (0.22 μm) and stored at −80°C until it was used. The protein concentration of the ECPs was determined by the method of Bradford (8) with bovine serum albumin (Sigma) as the standard and normalized.
Detection of protease activity.
Protease activity was determined using azocasein (Sigma) as the substrate. Briefly, crude ECPs (250 μl) were added to 250 μl of azocasein (5 mg·ml−1 in 50 mM Tris-HCl buffer, pH 8.0) and 245 μl of distilled water. The mixture was incubated at 20°C for 10 min. The undigested substrate was precipitated by adding 500 μl of 10% trichloroacetic acid to the reaction mixture, followed by centrifugation at 13,000 rpm and 4°C for 5 min. The supernatant (500 μl) was neutralized by addition of an equal volume of 1 N NaOH. After mixing, the absorbance at 440 nm was determined for triplicate samples.
In addition, the protease activity of separated proteins in an SDS-polyacrylamide gel was detected by copolymerizing 0.2% gelatin in the polyacrylamide matrix (30). After electrophoresis, the gel was soaked in 2.5% Triton X-100 for 2 h at room temperature, incubated overnight at 37°C in 50 mM Tris-HCl (pH 7.5)-200 mM NaCl-5 mM CaCl2, and then fixed and stained with 0.4% Coomassie brilliant blue in 30% methanol-10% acetic acid and destained in 30% methanol-10% acetic acid.
Virulence studies using oysters.
Bacteria were grown with constant agitation at 20°C for 36 h in MB, harvested, and resuspended in sterile seawater (121°C for 15 min) at an OD600 of 1. This OD600 corresponded to a bacterial concentration of 109 to 2 × 109 CFU ml−1 as determined by conventional dilution plating on marine agar (data not shown). Oysters were inoculated intramuscularly with bacterial strains or ECPs (5 μg/g of oyster) as described previously (23, 34). After injection, the oysters were transferred to aquaria (15 to 20 oysters per 2.5-liter aquarium) containing aerated 5-μm-filtered seawater at 20°C, kept under static conditions, and fed daily with a mixture of planktonic algae (Isochrisis galbana and Chaetoceros calcitrans). Each bacterial treatment was performed in duplicate, and mortality was recorded daily.
Nucleotide sequence accession numbers.
The nucleotide sequences of the luxU, luxM, vsm, gyrA, and ISVisp1 genes and the pSW4426T and pSW4427T cloning vectors have been deposited in the GenBank database under accession numbers DQ987705, DQ987706, DQ987707, DQ987708, DQ987704, DQ995482, and DQ995483.
RESULTS
Construction of a plasmid vector allowing controlled expression in V. splendidus.
We constructed a p15A derivative plasmid, pSU18T-PBADgfp, carrying the green fluorescent protein gene under control of the positively regulated arabinose-inducible PBAD promoter and an origin of transfer from RP4. Conjugative transfer of this plasmid from E. coli to V. splendidus was observed at a frequency of 10−3 transconjugant per recipient. Western blot analysis using commercial polyclonal antibodies resulted in detection of GFP with a molecular mass of about 30 kDa in extracts of LGP32 transconjugants when arabinose was added to the culture media, while no signal was obtained when transconjugants were grown in the presence of glucose or, in the case of LGP32, WT extracts (Fig. 1). Thus, the PBAD promoter appeared to be tightly and properly regulated in LGP32 by the araC gene product, which activated transcription in response to its natural inducer, arabinose.
FIG. 1.
Detection of GFP by immunoblotting in protein extract of WT LGP32 (lane 1) or transconjugants containing plasmid pSU18T-PBADgfp (lanes 2 to 4). GFP expression was repressed in the absence (lane 2) or in the presence (lane 3) of 1% glucose and was induced in the presence of 0.2% arabinose (lane 4). Cells were grown overnight before protein extraction and GFP detection. The expression of GFP in strains was confirmed by epifluorescence microscopy.
Development of a two-step allelic replacement method using CcdB as a positive selection marker.
CcdB is a very strong gyrase inhibitor and was originally discovered in the postsegregational killing operon carried on plasmid F, ccdBA (2). The ccdB gene has been employed as a potent counterselection marker in a number of commonly used applications (3, 4, 52).
Starting with pSW23T (Cmr) and pSW29T (Kmr), both carrying oriVR6Kγ and oriTRP4, we constructed the derivatives pSW4426T and pSW4427T carrying the ccdB gene under control of PBAD-araC. In order to avoid natural selection of inactive ccdB mutants due to leaking of PBAD, we obtained CcdB-resistant E. coli (pir+) strains Π3813 and β3914 by replacement of the gyrA gene with the gyrA462 allele, which is not sensitive to CcdB poisoning (2).
Using pSW4426T, we constructed three suicide vector derivatives with the luxU, luxM, and gyrA alleles deleted in order to create LGP32 mutants with mutations at these loci by allelic exchange. These Cmr plasmids were designed to allow selection of strains in which the plasmid was integrated into the target gene by a first homologous recombination event (with ccdB repression) and to allow positive selection of a second recombination event resulting in suicide vector loss (ccdB expression) (Fig. 2).
FIG. 2.
Schematic diagram of the two-step allelic exchange procedure. Integration can occur by recombination on either side of the plasmid allele of interest (a or b). For excision to lead to successful allelic exchange, recombination must occur in the second region of homology. Only strains in which the PBADccdB-containing plasmid has been excised can survive on arabinose-containing medium. When the 5′ and 3′ regions, which flank the deletion in the chosen gene, are similar sizes, selection for plasmid loss should result in a 50:50 ratio of wild type to mutant when nonessential genes are targeted.
The conjugation and integration frequencies of these plasmids in the LGP32 genome were found to be 10−8 to 10−7 recombinant per recipient, depending on the size of the region flanking the deletion in the fragment (Table 3). Several Cmr colonies were reisolated and grown overnight, and 107 CFU was spread on a plate containing 0.2% arabinose. One hundred colonies were obtained and found to be Cms. Screening for mutants among the surviving colonies was performed by PCR using primers flanking the target gene in the genome; 40 to 50% of the colonies were mutants in the case of ΔluxU and ΔluxM allele replacement, whereas only wild-type genotypes were detected in the case of the essential gyrA gene.
TABLE 3.
Insert size dependence for integration efficiency and proportion of mutants obtained using the two-step allelic replacement procedure
| Open reading frame | Size (bp) | Integration efficiency | Proportion of mutants |
|---|---|---|---|
| luxU | 140 | 10−8 | 3/8 |
| luxM | 570 | 2 × 10−7 | 4/8 |
| vsm | 600 | 5 × 10−7 | 18/18 |
| gyrA | 600 | 10−8 | 0/8 |
To validate the use of our strategy with another Vibrio species, V. cholerae, we constructed an integron integrase allele, ΔintIA, and cloned it into pSW4427T, a suicide vector containing a Kmr gene instead of a Cmr gene. In V. cholerae N16961, the frequency of conjugation and integration was found to be 10−6 recombinant/recipient. Several isolates were grown without selection, and colonies were selected on arabinose-supplemented medium. Thirty Kms colonies were then tested by PCR; nine isolates were found to carry the deleted allele, while the rest carried the WT intIA gene (data not shown).
Construction and characterization of V. splendidus Δvsm mutant.
We constructed pSWδ2989T, a suicide vector having an internal deletion in the vsm gene, to create an LGP32 mutant with a mutation at this locus by allelic exchange. After conjugative transfer and selection with arabinose, screening for mutants among the surviving colonies was performed by PCR using primers flanking the vsm gene in the genome and amplifying 1,800 bp from the wild type and 1,300 bp from mutants (Table 3). Surprisingly, all 18 colonies tested were found to carry the mutated allele, instead of the 50% theoretically expected, when mutant constructs having 5′ and 3′ flanking regions that were similar sizes were used as recombination targets. One of these isolates was designated δ2989 and used in this study.
The δ2989 mutant was tested for the loss of metalloprotease activity. The proteolytic activity of δ2989 ECPs (Fig. 3A, lanes 3 and 4) was determined by an azocasein assay and was shown to be eightfold lower than that of the wild type (Fig. 3A, lanes 1 and 2). In addition, a gelatin-SDS-polyacrylamide gel was used to detect extracellular proteins with protease activity. In the case of LGP32 ECPs, two predominant protease bands (at estimated molecular masses of 30 to 40 kDa) and one minor band (at an estimated molecular mass of 70 kDa) were observed as strong zones of clearing (Fig. 3B, lanes 1 and 2). These bands were missing from the δ2989 gel (Fig. 3B, lanes 3 and 4).
FIG. 3.
Determination of the proteolytic activity of ECPs by the azocasein assay (absorbance at 440 nm) (A) and with a gelatin-SDS-polyacrylamide gel (proteases which degraded gelatin were detected by zones of clearing) (B). ECPs of LGP32 (lanes 1 and 2), δ2989 (lanes 3 and 4), and δ6720-vsm (lanes 5 and 6) were prepared using MA (lanes 1, 3, and 5) or MA containing arabinose (lanes 2, 4, and 6) as described in Materials and Methods. Bacterial ECPs were produced by the cellophane overlay method as described by Liu (36). (A) The error bars indicate standard deviations. (B) The molecular masses on the right indicate the positions of molecular mass markers migrating in the nondenaturing conditions used.
Ectopic complementation of the vsm mutation.
ISVisp1 is an insertion sequence specific for V. splendidus (deposited in the IS database at http://www-is.biotoul.fr/is.html), and one copy is present in the LGP32 genome (Mazel et al., unpublished data). As this locus is probably not essential, we used it as a platform for ectopic expression in a transcomplementation experiment with the Δvsm mutant. The vsm gene under control of araC-PBAD was cloned in the ISVisp1 ΔorfB allele carried by pSWδ6720T (Cmr). The resulting plasmid was transferred by conjugation to strain δ2989, in which it could integrate into the genome through homologous recombination with the ISVisp1 orfB gene. Cmr clones were obtained, and four of these clones were grown without selection and then subjected to selection on arabinose-supplemented medium. Ten colonies having the Cms phenotype expected after a second recombination event were tested by PCR, and one-half of them were found to carry the ΔorfB::araC-PBAD-vsm allele. One of these isolates was designated δ6720-vsm and used in this study.
As shown in Fig. 3A, the metalloprotease activity was restored by ectopic complementation, as the activity detected for the δ6720-vsm ECPs when the organism was grown in the presence of arabinose was similar to the activity detected for WT LGP32 ECPs and clearly greater than the activity detected for the δ2989 ECPs. Furthermore, gelatin-SDS-polyacrylamide gel analysis of δ6720-vsm ECPs revealed the same two predominant protease bands that were observed for WT LGP32 ECPs, but only when arabinose was added to the media (Fig. 3B, lane 6).
Oyster experiments.
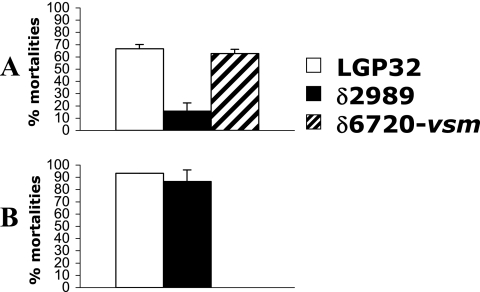
In order to evaluate their toxicities, the ECPs (5 μg/g of oysters) were injected into oyster adductor muscle (Fig. 4A). Two days postinjection, 68% mortality was observed in the case of LGP32 ECPs and 15% mortality was observed in the case of δ2989 ECPs. Ectopic expression of vsm driven by arabinose was found to restore the toxicity of ECPs as mortality rates similar to those obtained with WT LGP32 ECPs were obtained with δ6720-vsm ECPs. These data demonstrate the role of the Vsm metalloprotease in ECP toxicity. However, when the levels of virulence of the LGP32 and δ2989 strains were compared after injection of living bacteria into oysters, similar mortality rates were obtained for the two strains (Fig. 4B). These results suggest that vsm expression is not essential for full bacterial virulence.
FIG. 4.
Comparison of levels of oyster mortality after intramuscular injection of ECPs (5 μg/g of oysters) from wild-type strain LGP32 and the δ2989 and δ6720-vsm mutants (A) or of living organisms (109 to 2 × 109 CFU ml−1). Experiments were performed in triplicate (15 to 20 oysters per aquarium). The mean mortality rates were determined 2 days after injection of ECPs (A) or strains (B). The error bars indicate standard deviations. Data from each experiment were statistically analyzed using the chi-square test and StatView software. The differences were significant only for comparisons of ECPs of LGP32 and the δ2989 mutants (χ2 = 27.62, P < 0.0001) and for comparisons of ECPs of the δ6720-vsm and δ2989 mutants (χ2 = 23.69, P < 0.0001). Sterile seawater was injected as a negative control, and no mortality was observed (data not shown).
DISCUSSION
Since the first report on the complete genome sequence of Haemophilus influenzae in 1995, more than 300 other prokaryotic genome sequences have been completed and another 750 projects are under way (22). Genomics-based approaches have significantly increased our understanding of the physiology and pathogenicity of many microbes and have provided insights into the mechanisms and history of genome evolution. Paradoxically, only a limited number of bacterial species are amenable to genetic manipulation, which is often essential for demonstration of the proposed or suspected function of a gene candidate.
Parallel to the complete sequencing of the V. splendidus genome, we developed a gene knockout strategy which was used here to investigate the role of a secreted metalloprotease in the toxicity of extracellular products and in virulence in vivo.
In the first step, we established conditions for transferring a mobilizable plasmid in V. splendidus through the RP4 conjugative machinery and the functionality of the PBAD promoter in this strain using a replicative vector expressing the GFP.
The second step was to establish a genetic system for allelic replacement in V. splendidus. In 1988, Miller and Mekalanos set up a system allowing single-gene disruption through a homologous recombination event between a targeted gene and a truncated version of it carried by a suicide vector introduced into a strain by conjugation (41). Since then, workers have had considerable success with this strategy. However, it has several important limitations. First, the chromosomes of mutants constructed by this technique carry a partial duplication of the targeted gene, which can be the source of reversion events at a high frequency. Second, the presence of a copy of the vector backbone prevents further mutant construction by the same method in these strains, as recombination between the incoming vector and the chromosomal copy cannot be counterselected. Finally, the mutations obtained through vector integration can also have polar or other uncontrolled effects on the genes located in the neighborhood of the targeted gene.
To overcome these limitations, Donnenberg and Kaper developed a two-step strategy that allowed positive selection of clones in which a second recombination event leading to the loss of the vector and either the mutated or the WT allele occurred after the first integration event (19). The positive selection was linked to the presence in the vector backbone of a copy of the B. subtilis sacB gene. sacB encodes an enzyme, levansucrase, and its activity has been shown to be toxic for gram-negative organisms when they are grown in the presence of sucrose (25). When a mutant which contains an integrated copy of a suicide vector also carries sacB due to recombination into a target locus, upon exposure to sucrose, the daughter cells that underwent a second recombination event resulting in loss of the suicide vector are the only cells that thrive. If the targeted gene is not vital, the second recombination is expected to lead either to restoration of the WT allele or to allelic substitution at a 1:1 ratio. If the gene is essential, the isolates that survive in the presence of sucrose carry the WT allele. However, it is known that SacB toxicity is susceptible to the presence of sodium chloride in the selective medium (6). As addition of sodium chloride to media is absolutely necessary for the growth of many Vibrio species, such as V. splendidus, we had to develop and use a different counterselective marker in order to be able to use a similar strategy for mutant construction in such species.
We developed a novel suicide vector, based on the pSW family (17), which can be mobilized by the RP4 transfer machinery and which carries the ccdB gene of the E. coli F plasmid under control of the arabinose PBAD promoter (52) as a counterselective marker. Indeed, ccdB encodes a very efficient gyrase inhibitor, which has been observed to work on a broad spectrum of bacteria and has been used as a counterselective marker for the development of several cloning vectors ( 3, 4). We constructed two vectors, pSW4426T, which carries a Cmr marker and the araC-PBAD-ccdB cassette, and pSW4427T, which is identical to pSW4426T except that it carries a Kmr marker. When these plasmids carry the mutated allele, allelic replacement can be assayed in a two-step procedure, first through selection via the plasmid resistance markers of clones of the recipient strain that have integrated the suicide plasmid through homologous recombination with the WT allele, and second through transfer of the selected clones in a medium supplemented with arabinose to induce the expression of the lethal CcdB protein. As described above, if the targeted gene is not vital, the second recombination leads to either WT allele restoration or to allelic substitution with the same proportion (50%); if the gene is essential, the surviving isolates carry only the WT allele.
In order to validate our technique, V. splendidus ΔluxU, ΔluxM, and ΔgyrA alleles were constructed and cloned in pSW4426T, and allelic replacement was tested for each of them using the protocol described above. As expected, the ΔluxU and ΔluxM alleles were successfully substituted for the WT alleles at a frequency of about 50% in the cells that survived in the presence of arabinose, while in the case of the essential gyrA gene, the cells that survived in the presence of arabinose were found to carry only the WT allele. We also observed that the frequency of conjugation plus insertion correlated with the size of the DNA allowing the first recombination event.
We tested this two-step knockout strategy with another Vibrio species, V. cholerae, using to the superintegron integrase gene intIA (formerly called intI4 [40]) as the target. The ΔintIA allele was cloned into the Kmr vector pSW4427T, and allelic replacement was successfully assayed in strain N16961. As observed for luxU and luxM gene replacement in V. splendidus, clones that survived in the presence of arabinose were found to carry the WT and ΔintIA alleles at nearly identical proportions, showing that IntIA is not essential in V. cholerae in laboratory culture conditions.
Construction and characterization of a V. splendidus metalloprotease mutant.
As mentioned above, we have completed sequencing of the V. splendidus LGP32 genome (Mazel et al., unpublished data). Among the genes which could play a role in pathogenesis, we identified a zinc-containing metalloprotease gene, vsm, the predicted product of which exhibits 95% identity with the product of the V. anguillarum vam gene, which has been associated the virulence properties of this fish pathogen (42).
Vibrio zinc-containing metalloproteases are classified into three distinct categories according to their amino acid sequences (for a review, see reference 45). The class I Vibrio metalloproteases contain a large signal peptide region and a zinc-binding motif that includes an extra glutamic acid located 19 bases downstream from the second histidine residue (HEXXH-19 amino acids-E), whereas metalloproteases belonging to classes II and III have only a HEXXH motif. The V. splendidus vsm gene product contains the characteristic class I metalloprotease zinc-binding motif. In addition to this HEXXH-19 amino acid-E motif, analysis of the primary structure of the predicted Vsm protein through sequence alignment predicted the existence of a second consensus sequence, GXXNEXXSD, which, when associated with the HEXXH motif, constitutes the features that define the thermolysin family.
The metalloproteases belonging to the thermolysin family are synthesized as inactive precursors, which mature through successive processing stages. According to previous reports, an N-terminal peptide is cleaved during passage through the inner membrane in a signal peptide-dependent manner. In the periplasm the N-terminal propeptide is then cleaved by an autoproteolytic mechanism, and the mature protein is generated. A second processing at the carboxy terminus by autocatalytic cleavage has been described in other Vibrio species (15, 27, 33, 42). In these species, the mature protease has been proposed to consist of two domains, an N-terminal domain mediating the proteolytic action and a C-terminal domain that may be implicated in attachment to protein substrates (46).
In order to characterize the role of the thermolysin-related Vsm protease, we constructed a Δvsm mutant strain using the technique described above with plasmid pSWδ2989T. Cms arabinose-resistant isolates were obtained at a frequency similar to the frequencies of the lux alleles used in the validation process. However, PCR analysis and DNA sequencing revealed that surviving clones were Δvsm mutants in all cases and that none of them exhibited WT vsm allele restoration. The probability of this happening by chance is extremely low (1 in 262,144). This could be an indication that vsm expression is somehow deleterious in laboratory culture conditions and that the deleted allele has a strong selective advantage in these conditions compared to the wild-type allele. Interestingly, we did not observe any recombinational bias when we constructed the ectopically complemented vsm strain (see Results). In this case, we reintroduced the vsm gene under PBAD promoter control; thus, in the absence of arabinose, the ectopic vsm gene is silent, likely relieving any selective pressure against the recombinant.
The complete open reading frame of vsm encodes a 610-amino-acid polypeptide corresponding to a putative preproprotein with a theoretical molecular mass of 67 kDa. Cleavage of the preproprotein at the N-terminal amino acid side should lead to a mature protein that is 412 amino acids long and has a calculated molecular mass of 41.3 kDa. A second processing at the carboxy terminus could led to a shorter, approximately 30-kDa protein.
Analysis of proteins with protease activity in LGP32 ECPs performed using a gelatin-SDS-polyacrylamide gel revealed three strong zones of clearing, two predominant bands corresponding to proteases having molecular masses of approximately 30 to 40 kDa and a minor band corresponding to a protease having a molecular mass of approximately 70 kDa. These bands could correspond to the different processing stages of Vsm, as they were not detected in mutant strain δ2989. However, an alternative hypothesis, that these bands could correspond to other proteases whose processing is controlled by Vsm, cannot be excluded.
In order to demonstrate that this phenotype was due to the vsm deletion and not to an indirect effect of the deletion, we performed ectopic complementation through expression of the vsm gene under control of the PBAD promoter from another locus of the V. splendidus genome, the single-copy ISVisp1 gene. We observed that when grown in presence of arabinose, this strain displayed ECP protease activity identical to the WT ECP protease activity, demonstrating the direct relationship between vsm deletion and the ECP activity.
In the past, the pathogenesis of bacterial infections has frequently been associated with the production of extracellular proteases (38, 39, 57). The most generally accepted belief is that these proteases facilitate the spread of the pathogen into the host by causing extensive tissue damage and up-regulate bacterial growth by degrading numerous host proteins to provide readily available nutrients. Among the bacteria in the genus Vibrio, different proteases have been characterized and reported to play important roles in the pathogenicity of V. cholerae (20) V. anguillarum (49), V. vulnificus (43), and V. mimicus (11), to name a few.
As previous work demonstrated that ECPs from V. splendidus were implicated in the virulence process (26), we hypothesized that Vsm might directly contribute to oyster toxicity, by analogy with extracellular proteases produced by other shellfish-pathogenic vibrios (9, 50).
When injected into oysters, the LGP32 ECPs exhibited lethality, suggesting that they contained one or more toxic factors responsible either directly or indirectly for some of the pathological processes observed during infection. The lethal effect was dramatically reduced in mutant δ2989 and was restored by ectopic complementation, suggesting that the Vsm metalloprotease has a role in ECP toxicity. However, similar mortality rates were obtained when strain LGP32 or strain δ2989 was injected into oysters, showing that vsm expression is not necessary for bacterial virulence in this infection model.
In previous studies workers have examined the contribution to virulence of various Vibrio metalloproteases in animal experimental models by using mutants with deletions at the protease gene (20, 31, 42, 53). No conclusive evidence concerning the role of the protease in virulence was found, since mutants deficient in protease exhibited levels of virulence comparable to those of their parental strains. Shao and Hor suggested that in V. vulnificus other factors may be overactive in the absence of the metalloprotease and proposed a possible multifactor interaction in bacterial virulence, involving the protease to an undefined extent. So far, it is not known whether the Vsm metalloprotease is predominant during oyster infection or whether additional and/or coregulated virulence factors are involved in the pathogenesis. There are only a few examples of toxins (such as diphtheria or tetanus toxin) which act as single determinants to produce disease. Microbial pathogenesis is often multifactorial, and pathogens use several biochemical mechanisms operating in concert to produce infection and disease (21). For instance, the HA/P metalloprotease from V. cholerae was reported to activate proteolytically both the El Tor cytolysin/hemolysin (47) and cholera toxin, an ADP-ribosylating enterotoxin inducing a highly secretory diarrhea (7). Thus, it could be hypothesized that Vsm metalloprotease may similarly interact with other virulence factors in V. splendidus ECPs to potentiate their expression and/or effects on the host. Research is now under way to identify the protein targets that are processed by Vsm in the ECP fraction.
Supplementary Material
Acknowledgments
We acknowledge J. M. Ghigo for providing strains and for plasmid construction, L. Van Melderen for providing gyrA462 strains and for help with strain construction, Y. Labreuche and J. L. Nicolas for participating in metalloprotease analyses, and D. Rowe-Magnus for critical reading of the manuscript.
This study was carried out with financial assistance from the Institut Pasteur, the Centre National de la Recherche Scientifique (CNRS-URA 2171), the Institut Français de Recherche pour l'Exploitation de la Mer (IFREMER), and the Institut de Génomique Marine (contrat Ministère de la Recherche no. 0425).
Footnotes
Published ahead of print on 22 November 2006.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Bartolome, B., Y. Jubete, E. Martinez, and F. de la Cruz. 1991. Construction and properties of a family of pACYC184-derived cloning vectors compatible with pBR322 and its derivatives. Gene 102:75-78. [DOI] [PubMed] [Google Scholar]
- 2.Bernard, P., and M. Couturier. 1992. Cell killing by the F plasmid CcdB protein involves poisoning of DNA-topoisomerase II complexes. J. Mol. Biol. 226:735-745. [DOI] [PubMed] [Google Scholar]
- 3.Bernard, P., P. Gabant, E. M. Bahassi, and M. Couturier. 1994. Positive-selection vectors using the F plasmid ccdB killer gene. Gene 148:71-74. [DOI] [PubMed] [Google Scholar]
- 4.Betton, J. M. 2004. Cloning vectors for expression-PCR products. BioTechniques 37:346-347. [DOI] [PubMed] [Google Scholar]
- 5.Biskri, L., M. Bouvier, A. M. Guerout, S. Boisnard, and D. Mazel. 2005. Comparative study of class 1 integron and Vibrio cholerae superintegron integrase activities. J. Bacteriol. 187:1740-1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blomfield, I. C., V. Vaughn, R. F. Rest, and B. I. Eisenstein. 1991. Allelic exchange in Escherichia coli using the Bacillus subtilis sacB gene and a temperature-sensitive pSC101 replicon. Mol. Microbiol. 5:1447-1457. [DOI] [PubMed] [Google Scholar]
- 7.Booth, B. A., M. Boesman-Finkelstein, and R. A. Finkelstein. 1984. Vibrio cholerae hemagglutinin/protease nicks cholera enterotoxin. Infect. Immun. 45:558-560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 9.Brown, C., and G. Roland. 1984. Characterization of exotoxin produced by a shellfish-pathogenic Vibrio sp. J. Fish Dis. 7:117-126. [Google Scholar]
- 10.Chaveroche, M. K., J. M. Ghigo, and C. d'Enfert. 2000. A rapid method for efficient gene replacement in the filamentous fungus Aspergillus nidulans. Nucleic Acids Res. 28:E97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chowdhury, M. A., S. Miyoshi, and S. Shinoda. 1991. Role of Vibrio mimicus protease in enterotoxigenicity. J Diarrhoeal Dis. Res. 9:332-334. [PubMed] [Google Scholar]
- 12.Crowther, R. S., N. W. Roomi, R. E. Fahim, and J. F. Forstner. 1987. Vibrio cholerae metalloproteinase degrades intestinal mucin and facilitates enterotoxin-induced secretion from rat intestine. Biochim. Biophys. Acta 924:393-402. [DOI] [PubMed] [Google Scholar]
- 13.Dao-Thi, M. H., L. Van Melderen, E. De Genst, H. Afif, L. Buts, L. Wyns, and R. Loris. 2005. Molecular basis of gyrase poisoning by the addiction toxin CcdB. J. Mol. Biol. 348:1091-1102. [DOI] [PubMed] [Google Scholar]
- 14.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.David, V. A., A. H. Deutch, A. Sloma, D. Pawlyk, A. Ally, and D. R. Durham. 1992. Cloning, sequencing and expression of the gene encoding the extracellular neutral protease, vibriolysin, of Vibrio proteolyticus. Gene 112:107-112. [DOI] [PubMed] [Google Scholar]
- 16.Dean, D. 1981. A plasmid cloning vector for the direct selection of strains carrying recombinant plasmids. Gene 15:99-102. [DOI] [PubMed] [Google Scholar]
- 17.Demarre, G., A. M. Guerout, C. Matsumoto-Mashimo, D. A. Rowe-Magnus, P. Marlière, and D. Mazel. 2005. A new family of mobilizable suicide plasmids based on the broad host range R388 plasmid (IncW) or RP4 plasmid (IncPα) conjugative machineries and their cognate E.coli host strains. Res. Microbiol. 156:245-255. [DOI] [PubMed] [Google Scholar]
- 18.Ding, Y., B. M. Davis, and M. K. Waldor. 2004. Hfq is essential for Vibrio cholerae virulence and downregulates sigma expression. Mol. Microbiol. 53:345-354. [DOI] [PubMed] [Google Scholar]
- 19.Donnenberg, M. S., and J. B. Kaper. 1991. Construction of an eae deletion mutant of enteropathogenic Escherichia coli by using a positive-selection suicide vector. Infect. Immun. 59:4310-4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Finkelstein, R. A., M. Boesman-Finkelstein, Y. Chang, and C. C. Hase. 1992. Vibrio cholerae hemagglutinin/protease, colonial variation, virulence, and detachment. Infect. Immun. 60:472-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Finlay, B. B., and S. Falkow. 1997. Common themes in microbial pathogenicity revisited. Microbiol. Mol. Biol. Rev. 61:136-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fraser-Liggett, C. M. 2005. Insights on biology and evolution from microbial genome sequencing. Genome Res. 15:1603-1610. [DOI] [PubMed] [Google Scholar]
- 23.Gay, M., F. C. Berthe, and F. Le Roux. 2004. Screening of Vibrio isolates to develop an experimental infection model in the Pacific oyster Crassostrea gigas. Dis. Aquat. Organ. 59:49-56. [DOI] [PubMed] [Google Scholar]
- 24.Gay, M., T. Renault, A. M. Pons, and F. Le Roux. 2004. Two vibrio splendidus related strains collaborate to kill Crassostrea gigas: taxonomy and host alterations. Dis. Aquat. Org. 62:65-74. [DOI] [PubMed] [Google Scholar]
- 25.Gay, P., D. Le Coq, M. Steinmetz, T. Berkelman, and C. I. Kado. 1985. Positive selection procedure for entrapment of insertion sequence elements in gram-negative bacteria. J. Bacteriol. 164:918-921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gomez-Leon, J., L. Villamil, M. L. Lemos, B. Novoa, and A. Figueras. 2005. Isolation of Vibrio alginolyticus and Vibrio splendidus from aquacultured carpet shell clam (Ruditapes decussatus) larvae associated with mass mortalities. Appl. Environ Microbiol. 71:98-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hase, C. C., and R. A. Finkelstein. 1991. Cloning and nucleotide sequence of the Vibrio cholerae hemagglutinin/protease (HA/protease) gene and construction of an HA/protease-negative strain. J. Bacteriol. 173:3311-3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heidelberg, J. F., J. A. Eisen, W. C. Nelson, R. A. Clayton, M. L. Gwinn, R. J. Dodson, D. H. Haft, E. K. Hickey, J. D. Peterson, L. Umayam, S. R. Gill, K. E. Nelson, T. D. Read, H. Tettelin, D. Richardson, M. D. Ermolaeva, J. Vamathevan, S. Bass, H. Qin, I. Dragoi, P. Sellers, L. McDonald, T. Utterback, R. D. Fleishmann, W. C. Nierman, O. White, S. L. Salzberg, H. O. Smith, R. R. Colwell, J. J. Mekalanos, J. C. Venter, and C. M. Fraser. 2000. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature 406:477-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Herrero, M., V. de Lorenzo, and K. N. Timmis. 1990. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J. Bacteriol. 172:6557-6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heussen, C., and E. B. Dowdle. 1980. Electrophoretic analysis of plasminogen activators in polyacrylamide gels containing sodium dodecyl sulphate and copolymerized substrates. Anal. Biochem. 102:196-202. [DOI] [PubMed] [Google Scholar]
- 31.Jeong, K. C., H. S. Jeong, J. H. Rhee, S. E. Lee, S. S. Chung, A. M. Starks, G. M. Escudero, P. A. Gulig, and S. H. Choi. 2000. Construction and phenotypic evaluation of a Vibrio vulnificus vvpE mutant for elastolytic protease. Infect. Immun. 68:5096-5106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kolter, R., M. Inuzuka, and D. R. Helinski. 1978. Trans-complementation-dependent replication of a low molecular weight origin fragment from plasmid R6K. Cell 15:1199-1208. [DOI] [PubMed] [Google Scholar]
- 33.Kothary, M. H., and A. S. Kreger. 1987. Purification and characterization of an elastolytic protease of Vibrio vulnificus. J. Gen. Microbiol. 133:1783-1791. [DOI] [PubMed] [Google Scholar]
- 34.Labreuche, Y., P. Soudant, M. Gonçalves, C. Lambert, and J. L. Nicolas. 2006. Effects of extracellular products from the pathogenic Vibrio aestuarianus strain 01/32 on lethality and cellular immune responses of the oyster Crassostrea gigas. Dev. Comp. Immunol. 30:367-379. [DOI] [PubMed] [Google Scholar]
- 35.Le Roux, F., and B. Austin. 2006. Vibrio splendidus, p. 285-296. In F. L. Thompson, B. Austin, and J. Swings (ed.), The vibrios. ASM Press, Washington, DC.
- 36.Liu, P. V. 1957. Survey of hemolysin production among species of pseudomonads. J. Bacteriol. 74:718-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lodeiros, C., J. Bolinches, C. P. Dopazo, and A. E. Toranzo. 1987. Bacillary necrosis in hatcheries of Ostrea edulis in Spain. Aquaculture 65:15-29. [Google Scholar]
- 38.Maeda, H. 1996. Role of microbial proteases in pathogenesis. Microbiol. Immunol. 40:685-699. [DOI] [PubMed] [Google Scholar]
- 39.Maeda, H., and T. Yamamoto. 1996. Pathogenic mechanisms induced by microbial proteases in microbial infections. Biol. Chem. Hoppe-Seyler 377:217-226. [DOI] [PubMed] [Google Scholar]
- 40.Mazel, D., B. Dychinco, V. A. Webb, and J. Davies. 1998. A distinctive class of integron in the Vibrio cholerae genome. Science 280:605-608. [DOI] [PubMed] [Google Scholar]
- 41.Miller, V. L., and J. J. Mekalanos. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 170:2575-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Milton, D. L., A. Norqvist, and H. Wolf-Watz. 1992. Cloning of a metalloprotease gene involved in the virulence mechanism of Vibrio anguillarum. J. Bacteriol. 174:7235-7244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miyoshi, N., S. Miyoshi, K. Sugiyama, Y. Suzuki, H. Furuta, and S. Shinoda. 1987. Activation of the plasma kallikrein-kinin system by Vibrio vulnificus protease. Infect. Immun. 55:1936-1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miyoshi, S., H. Nakazawa, K. Kawata, K. Tomochika, K. Tobe, and S. Shinoda. 1998. Characterization of the hemorrhagic reaction caused by Vibrio vulnificus metalloprotease, a member of the thermolysin family. Infect. Immun. 66:4851-4855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Miyoshi, S., and S. Shinoda. 2000. Microbial metalloproteases and pathogenesis. Microbes Infect. 2:91-98. [DOI] [PubMed] [Google Scholar]
- 46.Miyoshi, S., Y. Sonoda, H. Wakiyama, M. M. Rahman, K. Tomochika, S. Shinoda, S. Yamamoto, and K. Tobe. 2002. An exocellular thermolysin-like metalloprotease produced by Vibrio fluvialis: purification, characterization, and gene cloning. Microb. Pathog. 33:127-134. [DOI] [PubMed] [Google Scholar]
- 47.Nagamune, K., K. Yamamoto, A. Naka, J. Matsuyama, T. Miwatani, and T. Honda. 1996. In vitro proteolytic processing and activation of the recombinant precursor of El Tor cytolysin/hemolysin (pro-HlyA) of Vibrio cholerae by soluble hemagglutinin/protease of V. cholerae, trypsin, and other proteases. Infect. Immun. 64:4655-4658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nicolas, J. L., S. Corre, G. Gauthier, R. Robert, and D. Ansquer. 1996. Bacterial problems associated with scallop Pecten maximus larval culture. Dis. Aquat. Org. 27:67-76. [Google Scholar]
- 49.Norqvist, A., B. Norrman, and H. Wolf-Watz. 1990. Identification and characterization of a zinc metalloprotease associated with invasion by the fish pathogen Vibrio anguillarum. Infect. Immun. 58:3731-3736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nottage, A. S., and T. H. Birkbeck. 1987. Purification of a proteinase produced by the bivalve pathogen Vibrio alginolyticus NCMB 1339. J. Fish Dis. 10:211-220. [Google Scholar]
- 51.Philippe, N., J. P. Alcaraz, E. Coursange, J. Geiselmann, and D. Schneider. 2004. Improvement of pCVD442, a suicide plasmid for gene allele exchange in bacteria. Plasmid 51:246-255. [DOI] [PubMed] [Google Scholar]
- 52.Roux, A., C. Beloin, and J. M. Ghigo. 2005. Combined inactivation and expression strategy to study gene function under physiological conditions: application to identification of new Escherichia coli adhesins. J. Bacteriol. 187:1001-1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shao, C. P., and L. I. Hor. 2000. Metalloprotease is not essential for Vibrio vulnificus virulence in mice. Infect. Immun. 68:3569-3573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Simon, R., U. B. Priefer, and A. Puhler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram negative bacteria. Bio/Technology 1:784-791. [Google Scholar]
- 55.Skrzypek, E., P. L. Haddix, G. V. Plano, and S. C. Straley. 1993. New suicide vector for gene replacement in Yersiniae and other gram-negative bacteria. Plasmid 29:160-163. [DOI] [PubMed] [Google Scholar]
- 56.Sugumar, G., T. Nakai, Y. Hirata, D. Matsubara, and K. Muroga. 1998. Vibrio splendidus biovar II as the causative agent of bacillary necrosis of Japanese oyster Crassostrea gigas larvae. Dis. Aquat. Org. 33:111-118. [DOI] [PubMed] [Google Scholar]
- 57.Travis, J., J. Potempa, and H. Maeda. 1995. Are bacterial proteinases pathogenic factors? Trends Microbiol. 3:405-407. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.