Abstract
The antibiotic resistances of 45 lactic acid bacteria strains belonging to the genera Lactobacillus, Streptococcus, Lactococcus, Pediococcus, and Leuconostoc were investigated. The objective was to determine antibiotic resistances and to verify these at the genetic level, as is currently suggested by the European “qualified presumption of safety” safety evaluation system for industrial starter strains. In addition, we sought to pinpoint possible problems in resistance determinations. Primers were used to PCR amplify genes involved in β-lactam antibiotic, chloramphenicol, tetracycline, and erythromycin resistance. The presence of ribosomal protection protein genes and the ermB gene was also determined by using a gene probe. Generally, the incidences of erythromycin, chloramphenicol, tetracycline, or β-lactam resistances in this study were low (<7%). In contrast, aminoglycoside (gentamicin and streptomycin) and ciprofloxacin resistances were higher than 70%, indicating that these may constitute intrinsic resistances. The genetic basis for ciprofloxacin resistance could not be verified, since no mutations typical of quinolone resistances were detected in the quinolone determining regions of the parC and gyrA genes. Some starter strains showed low-level ampicillin, penicillin, chloramphenicol, and tetracycline resistances, but no known resistance genes could be detected. Although some strains possessed the cat gene, none of these were phenotypically resistant to chloramphenicol. Using reverse transcription-PCR, these cat genes were shown to be silent under both inducing and noninducing conditions. Only Lactobacillus salivarius BFE 7441 possessed an ermB gene, which was encoded on the chromosome and which could not be transferred in filter-mating experiments. This study clearly demonstrates problems encountered with resistance testing, in that the breakpoint values are often inadequately identified, resistance genes may be present but silent, and the genetic basis and associated resistance mechanisms toward some antibiotics are still unknown.
Lactic acid bacteria (LAB) have a long and safe history of use in the production and consumption of fermented foods and beverages (5, 29, 50). LAB are consumed in enormous quantities, primarily in fermented foods. According to the International Dairy Federation, the average annual consumption of fermented milk products is 22 kg per capita in Europe (30). In total, this amounts to about 8.5 billion kg of fermented milk per year. However, this figure does not take into account the LAB used in other food fermentations (e.g., vegetable and meat) or probiotic strains, and so the actual amount can thus be expected to be far greater.
Bacteria used as starter cultures for the production of foods could possibly contain antibiotic resistance genes (8, 46). In recent years, studies on the selection for and dissemination of antibiotic resistances have focused mainly on clinically relevant bacterial species. More recently, it was speculated that food bacteria may act as reservoirs of antibiotic resistance genes (13, 26). Fermented foods, therefore, may be important vehicles of enormous amounts of living bacteria, with biotechnical use as starter cultures, into the human body. These may carry transferable antibiotic resistances, which might be transferred to commensal or pathogenic bacteria. Recently, the European Food Safety Authority (EFSA) has taken responsibility to launch the European initiative toward a “qualified presumption of safety” (QPS) concept which, similar to the GRAS system in the United States, is aimed to allow strains with an established history and safety status to enter the market without extensive testing requirements (11). The presence of transmissible antibiotic resistance markers in the evaluation of strains is thus an important safety criterion.
LAB often harbor plasmids of different sizes, and some antibiotic resistance determinants located on plasmids have been reported to occur in Lactococcus lactis and various Lactobacillus and Enterococcus species (14). Among the LAB, antibiotic resistance of the enterococci has been subject to intense study (19, 27, 28), particularly because strains of these bacteria cause numerous and serious infections in humans (32, 34). In contrast, fewer physiological and molecular data are available on the antibiotic resistances of lactobacilli present in fermented foods. Determination of antibiotic resistances among LAB is confounded by problems regarding the use of media and MIC breakpoints for the genera or species. Generally, the choice of medium has been shown to have a profound impact on the MICs of LAB (13, 20). Furthermore, MIC breakpoint values have been shown to be species specific and thus vary between species of the same genera (8). The objective of the present study was not only to determine the spectrum and incidences of antibiotic resistance of LAB starter strains but also to verify these resistances with the underlying genetic mechanism. Furthermore, we sought to elucidate mechanisms of LAB resistance to antibiotics such as ciprofloxacin, which thus far have not been intensively investigated. Therefore, the antibiotic resistances of 40 commercial LAB starter strains and 5 probiotic strains were determined by using the E-test, and we attempted to identify the mechanisms for the antibiotic resistance by using PCR amplification of and/or gene probe hybridization with antibiotic resistance genes. The observed physiological resistances were correlated with the genetic data and the reported MIC breakpoint values to pinpoint potential problems in safety evaluations as suggested by the European QPS system.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
A total of 45 LAB strains belonging to the genera Lactobacillus, Leuconostoc, Lactococcus, Pediococcus, and Streptococcus, including 40 industrial starter cultures used for the manufacture of various fermented foods, and 5 strains that were either being developed as probiotic strains (3 strains) or were from a commercial probiotic product (2 strains), were investigated (Table 1) . A total of 43 strains were obtained from Danisco, while two strains were isolated from a probiotic product from SymbioPharm. The investigations were done in full cooperation and agreement with the producers of the strains. Strains were routinely grown aerobically at 30°C or, in the case of S. thermophilus strains, at 40°C in MRS broth pH 6.0 (Merck, Darmstadt, Germany).
TABLE 1.
Strains and plasmids used in this study
| Strain category and species (no. of strains) or plasmid | Strain no., antibiotic resistance phenotype, or antibiotic resistance gene (source or reference) |
|---|---|
| Starter strainsa and their use for | |
| production of: | |
| Cheese | |
| Lactococcus lactis subsp. lactis (11) | BFE 7400 to BFE 7410 |
| Lactococcus lactis subsp. diacetylis (5) | BFE 7411 to BFE 7415 |
| Leuconostoc mesenteroides | BFE 7416 |
| Leuconostoc pseudomesenteroides | BFE 7417 |
| Yogurt | |
| Streptococcus thermophilus (11) | BFE 7418 to BFE 7428 |
| Lactobacillus acidophilus (1) | BFE 7429 |
| Lactobacillus delbrueckii subsp. | |
| bulgaricus (2) | BFE 7430, BFE 7431 |
| Lactobacillus helveticus (1) | BFE 7432 |
| Sausage | |
| Lactobacillus plantarum (2) | BFE 7433, BFE 7440 |
| Pediococcus acidilactici (1) | BFE 7434 |
| Lactobacillus curvatus (1) | BFE 7435 |
| Pediococcus pentosaceus (1) | BFE 7436 |
| Lactobacillus pentosus (1) | BFE 7437 |
| Lactococcus lactis subsp. lactis (1) | BFE 7439 |
| Probiotic strains | |
| Lactobacillus farciminis (1) | BFE 7438 |
| Lactobacillus salivarius (1) | BFE 7441 |
| Lactobacillus rhamnosus (1) | BFE 7442 |
| Lactobacillus acidophilus (1) | BFE 7444 |
| Lactobacillus casei (1) | BFE 7445 |
| Control strains for antibiotic | |
| resistance study | |
| E. faecium FAIR-E 25b | Penr Cmr Tetr Eryr Cir (19)c |
| E. faecalis FAIR-E 85 | Penr Cmr Tetr Eryr Cir Smr Gmr |
| E. faecalis FAIR-E 260 | Penr Cmr Tetr Eryr Cir Gmr |
| E. faecalis FAIR-E 265 | Cmr Tetr Eryr Cir Smr Gmr |
| L. lactis MG1363 | lmrA+ |
| Plasmids | |
| pMG36e | ermB (48) |
| pUC19 | bla (New England Biolabs) |
Starters were obtained from a major commercial starter strain producer and deposited in our BFE collection.
The enterococci were derived from the EU study Enterococci in Food Fermentations (FAIR-CT97-3078). The FAIR-E culture collection is administered by the BCCM/LMG (Bacteria Collection of the Laboratory of Microbiology, University of Ghent).
Penr, penicillin resistant; Cmr, chloramphenicol resistant; Tetr, tetracycline resistant; Eryr, erythromycin resistant; Cir, ciprofloxacin resistant; Smr, streptomycin resistant; Gmr, gentamicin resistant.
Antibiotic susceptibility testing and MIC determination.
E-test strips (Viva Diagnostika, Cologne, Germany) for the determination of either ampicillin, penicillin G, erythromycin, chloramphenicol, gentamicin, streptomycin, tetracycline, or ciprofloxacin resistance were used according to the manufacturer's instructions. Briefly, a bacterial suspension was made by picking colonies from MRS or M17 (Difco, Heidelberg, Germany) agar plates using a sterile loop and then adding these to quarter-strength Ringer's solution (Merck, Darmstadt, Germany) to reach a density corresponding to a McFarland value of 0.5. In a pilot study, we found that in order to ensure the good growth of streptococci, it was necessary to swab the strains onto MRS and M17-agar plates with an inoculum density of McFarland 1 to 2. Using a sterile swab, the suspension was swabbed in three directions onto 4-mm-thick agar plates. Care was taken to use only agar plates with a layer thickness of 4 ± 0.5 mm in order to standardize the diffusion of the antibiotic. The plates were incubated in an anaerobic chamber (95% CO2, 5% N2; Don Whitley Scientific, Shipley, United Kingdom) at either 37°C (streptococci) or 30°C (all other LAB strains) for 48 h before reading.
DNA preparation and manipulations.
Total genomic DNA from each isolate was extracted and purified by using the method of Pitcher et al. (37) as modified for gram-positive bacteria by Björkroth and Korkeala (3). Small-scale isolation of plasmid-DNA was done as described by van Belkum and Stiles (48). Large-scale plasmid isolation was done by equilibrium centrifugation using the cesium chloride-ethidium bromide gradient centrifugation method as described by Sambrook et al. (38). Agarose gel electrophoresis and Southern blotting were carried out by standard procedures (38). Labeling of DNA probes using a DIG dUTP DNA labeling and detection kit (catalog no. 1093657; Roche, Mannheim, Germany) was performed according to the manufacturer's instructions.
PCR detection of resistance genes.
PCR amplification of genes associated with resistance to chloramphenicol (cat, the chloramphenicol acetyltransferase gene), β-lactam antibiotics (bla, the β-lactamase gene), macrolides (the ermA, ermB, ermC, msrA/B, ereA, ereB, mphA, and mefA/E genes), and tetracycline [the ribosomal protection proteins tet(M), tet(O), tet(S), and tet(W) or the efflux proteins tet(K) and tet(L)] was done in 50-μl volumes that contained 30 pmol of each specific primer, 1× Taq DNA polymerase buffer (Amersham Biosciences, Freiburg, Germany), each deoxynucleoside triphosphate at a concentration of 200 μM, 1 U of Taq DNA polymerase (Amersham Biosciences, Freiburg), and 100 ng of genomic DNA used as a template. The oligonucleotide primers used included those reported previously for ermA, ermC, msrA/B, ereA, ereB, mphA, and mefA/E (43), ermB (15), the tet(M), tet(O), tet(S), and tet(W) ribosomal protection proteins, and the tet(K) and tet(L) tetracycline efflux proteins (1, 19), and PCR was performed as described before (1, 15, 19). In addition, custom-designed primers for the cat gene (Catfw1 [forward], 5′-TTA GGT TAT TGG GAT AAG TTA-3′, and Catrev [reverse], 5′-GCA TGR TAA CCA TCA CAW AC-3′), and the β-lactamase gene (bla) (Bla-forward, 5′-CAT ART TCC GAT AAT ASM GCC-3′; Bla-reverse, 5′-CGT STT TAA CTA AGT ATS GY-3′) were used, which amplified PCR products of 300 and 297 bp, respectively.
PCR amplification was done as described previously (1, 15, 31, 43), or for 35 cycles at annealing temperatures of 48°C (cat) or 51°C (bla) for 1 min, and extension was done at 72°C for 45 s. A final polymerization step of 5 min at 72°C ended the PCR protocol. The PCR products were subjected to electrophoresis on 1.8% agarose gels, and the products were visualized by staining with ethidium bromide.
PCR amplification and DNA sequencing of parts of antibiotic resistance-associated genes.
To investigate whether observed fluoroquinolone resistances were due to mutations in the quinolone resistance-determining regions (QRDR) of the gyrA and parC genes, the QRDR encoding regions were PCR amplified. The custom-designed primers for the gyrA gene were GyrAfw (5′-CAM CGK CGK ATT CTT TAC GGA ATG-3′) and GyrArev (5′-TTR TTG ATA TCR CGB AGC ATT TC-3′), and the primers for the parC gene were ParCfw (5′-TAT TCY AAA TAY ATC ATT CAR GA-3′) and ParCrev (5′-GCY TCN GTA TAA CGC ATM GCC G-3′). The amplification conditions for both the gyrA and the parC genes consisted of 35 cycles of 94°C for 1 min, 50°C for 1 min, and 72°C for 30 s.
Part of the cat gene was amplified by using the primers Catfw2 (5′-AGA MAA TTG GRA GAG AAA AGA G-3′) and Catrev (see above). This 568-bp gene fragment was amplified in 35 cycles of 94°C for 1 min, 52°C for 1 min, and 72°C for 40 s. A partial 405-bp ermB fragment was amplified as described by Gevers et al. (15) and was also sequenced. The resulting PCR products were sequenced bidirectionally at GATC Biotech (Konstanz, Germany), and the deduced amino acid sequences were aligned with those retrieved from the GenBank database by using the DNAStar CLUSTAL W multiple alignment tool. The DNA sequences obtained for the partial cat genes from L. acidophilus BFE 7429, L. delbrueckii subsp. bulgaricus BFE 7430, and S. thermophilus BFE 7420 were submitted to GenBank and received the accession numbers EF070730, EF070729, and EF070728, respectively. The DNA sequence of the partial ermB gene sequence from L. salivarius BFE 7441 was also submitted to GenBank and received the accession number EF070727.
Southern hybridization.
Large-scale plasmid DNA isolation of the erythromycin-resistant L. salivarius BFE 7441 strain was done as described above. Restriction enzyme analysis of plasmid DNA was performed using the restriction enzymes EcoRI, XbaI, PstI, KpnI, BamHI, AvaII, MluI, NotI, SphI, and XmaI (New England Biolabs, Frankfurt am Main, Germany) in separate reactions according to the manufacturer's recommendations. Samples were run on a 1% agarose gel, stained with ethidium bromide, and visualized under UV light. DNA was transferred onto a nylon membrane (Hybond N+; Amersham Pharmacia) according to standard methods (38) and then hybridized with an ermB-specific probe labeled with digoxigenin (Boehringer, Mannheim, Germany). The probe was obtained by PCR of the ermB gene using the oligonucleotide primers and amplification conditions described by Gevers et al. (15) and the DIG dUTP labeling kit for PCR (Roche Diagnostics).
Isolation of total RNA and RNA expression studies.
Total RNA was isolated from LAB strains that possess cat genes but which were not resistant to chloramphenicol, as well as from positive control strains that were chloramphenicol resistant (Table 1). Total RNA was isolated by using an RNeasy minikit (QIAGEN, Hilden, Germany), with the use of RNA protect solution (QIAGEN) and a DNase (catalog no. 79254; QIAGEN) digest according to the manufacturer's instructions. For RNA isolation, cells were grown until the mid-logarithmic growth phase, and the cell numbers were adjusted to 109 CFU/ml, as suggested by the manufacturer. The RNA quality was visually assessed by using denaturing gel electrophoresis, and the RNA quantity was measured spectrophotometrically at 260 nm as described in Sambrook et al. (38). The total RNA was adjusted to 400 ng/μl using diethyl pyrocarbonate-treated water (Ambicon, Huntingdon, United Kingdom), and 600 ng of total RNA was used for reverse transcriptase-PCR (RT-PCR). For RT-PCR, the ready-to-go RT-PCR beads of Amersham Biosciences (Freiburg, Germany) were used. The RT-PCR contained reagents as described above for amplification of antibiotic resistance genes and DNA was amplified after RT of the mRNA at 42°C for 30 min. The cat gene was amplified by using primers Catfw1 and Catrev as described above. Housekeeping genes that were amplified included part of the l-lactate dehydrogenase (ldhL) gene, as well as the gyrA or the parC genes, as described above. The ldhL gene fragment was amplified by using the primer LLDHfw (5′-GTT GCY AAC CCA GTT GAT ATC-3′) and the primer LLDHrev (5′-GTA CCA ATG TAA ATG TCG TTC).
Filter-mating experiments.
The transferability of the erythromycin resistance of the strain L. salivarius BFE 7441 was examined by filter mating. E. faecalis JH2-2 (resistant to rifampin at 16 μg/ml), L. lactis LMG 19460 (resistant to rifampin at 16 μg/ml), and E. faecalis OG1X (resistant to streptomycin at 128 μg/ml) were used as recipients in mating experiments as described previously (19). Transconjugants were spread plated on MRS agar plates containing 128 μg of erythromycin/ml and 32 μg of rifampin/ml. The plates were incubated for 24 to 48 h at 37°C.
RESULTS
Antibiotic resistance phenotypes.
The incidence of resistance of the starter and probiotic strains to some antibiotics varied considerably depending on the breakpoints used for determining the MICs (Tables 2 and 3). Generally, more strains were resistant to the antibiotics gentamicin, streptomycin, and ciprofloxacin when the MIC breakpoints suggested by the European Scientific Committee and Panel are used, i.e., SCAN (10) and FEEDAP (41), compared to the number of resistant strains based on the breakpoints proposed by Danielsen and Wind (8), reflecting the fact that the MIC breakpoints indicated by SCAN and FEEDAP are lower for these antibiotics. Interestingly, the incidences of resistance to these three antibiotics was generally very high (>71.1%) as indicated by the SCAN and FEEDAP (10, 41) breakpoint criteria and still noticeably high even by the Danielsen and Wind (8) criteria (Table 4). In contrast, the incidence of resistance to the antibiotics ampicillin, penicillin G, erythromycin, tetracycline, and chloramphenicol was generally low (between 0 and 6.7% of strains; Table 4) using the MIC breakpoint values as proposed by SCAN/FEEDAP (10, 41) or Danielsen and Wind (8). Resistance to Penicillin G, erythromycin, ampicillin, and tetracycline generally did not occur among S. thermophilus or Lactobacillus strains used as starters in yogurt fermentation or among L. lactis or the two Leuconostoc strains used as starters for cheese production (Table 2). In contrast, the same strains generally possessed ciprofloxacin, gentamicin, and streptomycin resistances according to the SCAN/FEEDAP (10, 41) breakpoints. When the Danielsen and Wind (8) breakpoints were applied to these latter antibiotics, however, they were found to be mostly ciprofloxacin resistant, whereas some strains were also streptomycin resistant but generally sensitive to gentamicin (Table 2). A different situation was encountered for the starter strains (mostly lactobacilli and two Pediococcus strains) used for meat fermentations, as well as for the probiotic strains. All of the ampicillin-, chloramphenicol-, penicillin G-, tetracycline-, and erythromycin-resistant strains, although few in number, occurred among the meat starter and probiotic strains (Table 3). In addition, like the yogurt and cheese starters, numerous strains used as meat starters possessed ciprofloxacin, gentamicin, and streptomycin resistances. In this case, however, resistances to ciprofloxacin, gentamicin, and streptomycin occurred according to both the breakpoint values proposed by SCAN/FEEDAP (10, 41) and the criteria of Danielsen and Wind (8). This indicated that these resistances among the lactobacilli were generally higher than those observed for the most S. thermophilus and L. lactis strains. However, a relatively low number of species were investigated here, and it would be interesting to determine whether other workers in the field obtain similar data in the future. Even in light of the relatively high MIC breakpoints proposed by Danielsen and Wind (8), some strains, especially the two Pediococcus strains, L. pentosus BFE 7437, and L. plantarum BFE 7440, still showed multiple (four or five different) antibiotic resistances (Table 3).
TABLE 2.
Antibiotic resistances of lactic acid bacteria starter strains used for the manufacture of yogurt and cheesea
| Substrate and starter strain | EUC antibiotic resistance (MIC [μg/ml])
|
(Multiple) resistances
|
|||||
|---|---|---|---|---|---|---|---|
| Am, PG, Em, Cl, Te | Ci (DW = >32) | Gm (DW = >128) | Sm (DW = >256) | EUC | DW | Genotype | |
| Yogurt | |||||||
| S. thermophilus BFE 7418 | ≤1 | 0.75 | 3 | 3 | |||
| S. thermophilus BFE 7419 | ≤1 | >32 | 16 | 8 | Ci, Gm | Ci | |
| S. thermophilus BFE 7420 | ≤1 | >32 | 32 | >256 | Ci, Gm, Sm | Ci, Sm | cat |
| S. thermophilus BFE 7421 | ≤1 | 0.5 | 3 | 128 | Sm | cat | |
| S. thermophilus BFE 7422 | ≤1 | >32 | 6 | 16 | Ci, Sm | Ci | cat |
| S. thermophilus BFE 7423 | ≤1 | >32 | 32 | 48 | Ci, Gm, Sm | Ci, Sm | cat |
| S. thermophilus BFE 7424 | ≤1 | >32 | 12 | 4 | Ci, Gm | Ci | cat |
| S. thermophilus BFE 7425 | ≤1 | 0.5 | 4 | 6 | cat | ||
| S. thermophilus BFE 7426 | ≤1 | >32 | 48 | 64 | Ci, Gm, Sm | Ci | cat |
| S. thermophilus BFE 7427 | ≤1 | >32 | 12 | 6 | Ci, Gm | Ci | cat |
| S. thermophilus BFE 7428 | ≤1 | 1 | 8 | 48 | Gm, Sm | cat | |
| L. acidophilus BFE 7429 | ≤1.5 | >32 | 48 | 12 | Ci, Gm | Ci | cat |
| L. delbrueckii subsp. bulgaricus BFE 7430 | ≤1.5 | >32 | 24 | >256 | Ci, Gm, Sm | Ci, Sm | cat |
| L. delbrueckii subsp. bulgaricus BFE 7431 | ≤1.5 | >32 | 48 | 128 | Ci, Gm, Sm | Ci | cat |
| L. helveticus BFE 7432 | ≤1.5 | >32 | 16 | 3 | Ci, Gm | Ci | |
| Cheese | |||||||
| L. lactis subsp. lactis BFE 7400 | ≤1.5 | 3 | 4 | 2 | |||
| L. lactis subsp. lactis BFE 7401 | ≤1.5 | 3 | 6 | 32 | Sm | ||
| L. lactis subsp. lactis BFE 7402 | ≤1.5 | 4 | 16 | 48 | Ci, Gm, Sm | ||
| L. lactis subsp. lactis BFE 7403 | ≤1.5 | >32 | 6 | 24 | Ci, Sm | Ci | |
| L. lactis subsp. lactis BFE 7404 | ≤1.5 | 8 | 12 | >256 | Ci, Gm, Sm | Sm | |
| L. lactis subsp. lactis BFE 7405 | ≤1.5 | 6 | 12 | >256 | Ci, Gm, Sm | Sm | |
| L. lactis subsp. lactis BFE 7406 | ≤1.5 | >32 | 8 | 48 | Ci, Gm, Sm | Ci | |
| L. lactis subsp. lactis BFE 7407 | ≤1.5 | >32 | 24 | >256 | Ci, Gm, Sm | Ci, Sm | |
| L. lactis subsp. lactis BFE 7408 | ≤1.5 | 8 | 24 | 6 | Ci, Gm | ||
| L. lactis subsp. lactis BFE 7409 | ≤1.5 | >32 | 6 | 32 | Ci, Sm | Ci | |
| L. lactis subsp. lactis BFE 7410 | ≤1.5 | >32 | 1 | 12 | Ci | Ci | |
| L. lactis subsp. diacetylactis BFE 7411 | ≤1.5 | 1.5 | 12 | 16 | Gm, Sm | ||
| L. lactis subsp. diacetylactis BFE 7412 | ≤1.5 | 0.5 | 6 | 12 | |||
| L. lactis subsp. diacetylactis BFE 7413 | ≤1.5 | 3 | 6 | 12 | |||
| L. lactis subsp. diacetylactis BFE 7414 | ≤1.5 | 3 | 12 | >256 | Gm, Sm | Sm | |
| L. lactis subsp. diacetylactis BFE 7415 | ≤1 (Cl = 4) | 6 | 6 | 16 | Ci, Sm | ||
| L. mesenteroides BFE 7416 | ≤1.5 | >32 | 1 | 8 | Ci, Sm | Ci | cat |
| L. pseudomesenteroides BFE 7417 | ≤1.5 | 16 | 8 | 192 | Ci, Gm, Sm | cat | |
The EU Commission (EUC) breakpoint values as suggested by SCAN (10) and FEEDAP (41) or the values Danielsen and Wind (8) (DW) are given. All strains in Table 2 were not resistant to ampicillin (Am), penicillin G (PG), tetracycline (Te), erythromycin (Em), and chloramphenicol (Cl) based on either the SCAN/FEEDAP or the DW MIC breakpoints; the breakpoints for these antibiotics, together with the breakpoints for ciprofloxacin (Ci), streptomycin (Sm), and gentamicin (Gm), are given in Table 3.
TABLE 3.
Antibiotic resistances of lactic acid bacteria starter strains used for the manufacture of fermented sausages and of probiotic strains
| Product and starter strain | EUC antibiotic resistance (MIC [μg/ml])a
|
(Multiple) resistances
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Am (DW = 4)b | PG (DW = 4) | Em (DW = 1, 2, or 4) | Cl (DW = 16) | Te (DW 4, 8, or 64) | Ci (DW = >32) | Gm (DW = >128) | Sm (DW = >256) | EUC | DW | Genotype | |
| Sausage | |||||||||||
| L. plantarum BFE 7433 | 0.38 | 32 | 1 | 2 | 4 | >32 | >256 | >256 | Ci, Gm, Sm, PG | PG, Ci, Gm, Sm | |
| P. acidilactici BFE 7434 | 2 | 0.25 | 1 | 2 | 16 | >32 | 128 | >256 | Te, Ci, Gm, Sm | Te, Ci, Gm, Sm | cat |
| L. curvatus BFE 7435 | 0.38 | 0.19 | 1 | 1 | 1 | >32 | >256 | >256 | Ci, Gm, Sm | Ci, Gm, Sm, Em | |
| P. pentosaceus BFE 7436 | 4 | 0.75 | 1 | 2 | 16 | >32 | >256 | >256 | Am, Te, Ci, Gm, Sm | Am, Te, Ci, Gm, Sm | |
| L. pentosus BFE 7437 | 2 | 1 | 1.5 | 4 | 12 | >32 | >256 | >256 | Ci, Cl, Gm, Sm, Te | Ci, Gm, Sm | |
| Lactococcus lactis subsp. lactis BFE 7439 | 0.38 | 0.25 | 0.38 | 1 | 0.38 | >32 | 24 | >256 | Ci, Gm, Sm | Ci, Sm | |
| L. plantarum BFE 7440 | 0.38 | 8 | 1.5 | 12 | 16 | >32 | >256 | >256 | PG, Cl, Ci, Gm, Sm | PG, Te, Ci, Gm, Sm | |
| Probiotic strains | |||||||||||
| L. farciminis BFE 7438 | 0.38 | 0.25 | 0.38 | 1.5 | 1.5 | >32 | 128 | >256 | Ci, Gm, Sm | Ci, Gm, Sm | |
| L. salivarius BFE 7441 | 0.38 | 0.19 | >256 | 0.5 | 1.5 | >32 | 64 | >256 | Em, Ci, Gm, Sm | Em, Ci, Sm | ermB |
| L. rhamnosus BFE 7442 | 0.25 | 0.19 | 0.38 | 2 | 0.38 | 6 | 96 | 64 | Ci, Gm, Sm | - | |
| L. acidophilus BFE 7444 | 1 | 0.19 | 1.5 | 2 | 0.38 | 16 | 128 | >256 | Ci, Gm, Sm | Gm, Sm | |
| L. casei BFE 7445 | 1 | 0.25 | 0.5 | 1.5 | 0.38 | >32 | >256 | >256 | Ci, Gm, Sm | Ci, Gm, Sm | |
The EU Commission (EUC) breakpoint values as suggested by SCAN (10) and FEEDAP (41) or according to Danielsen and Wind (8) (DW) are given. For ciprofloxacin (Ci) the SCAN breakpoint value of 4 μg/ml was used for all strains except pediococci, for which 16 μg/ml was used. Breakpoints for ciprofloxacin or penicillin G (PG) were not suggested by FEEDAP. The FEEDAP breakpoints (superseding previous SCAN breakpoints) were as follows: ampicillin (Am) and erythromycin (Em), 4 μg/ml for all strains in this study; gentamicin (Gm), 8 μg/ml for obligately homofermentative lactobacilli (HFLB), L. lactis, and S. thermophilus strains, 4 μg/ml for Pediococcus and Leuconostoc spp., and 64 μg/ml for L. plantarum strains; streptomycin (Sm), 4 μg/ml for Pediococcus spp., 8 μg/ml for Leuconostoc spp., 16 μg/ml for L. lactis and S. thermophilus strains and for HFLB, and 64 μg/ml for L. plantarum strains; tetracycline (Te), 4 μg/ml for Pediococcus and Leuconostoc spp., as well as for L. lactis and S. thermophilus strains, 8 μg/ml for HFLB and 32 μg/ml for L. plantarum strains; chloramphenicol (Cl), 4 μg/ml for Pediococcus and Leuconostoc spp. and for HFLB and 8 μg/ml for L. lactis, S. thermophilus, and L. plantarum strains. Species-specific MIC breakpoints were defined by Danielsen and Wind (8) as follows: for erythromycin, 1 μg/ml for L. acidophilus, L. sakei, and L. curvatus, 2 μg/ml for L. paracasei and L. rhamnosus, and 4 μg/ml for L. plantarum and L. pentosus; for tetracycline, 4 μg/ml for L. paracasei, L. acidophilus, and L. rhamnosus, 8 μg/ml for L. sakei and L. curvatus, and 64 μg/ml for L. plantarum and L. pentosus.
For pediococci the SCAN (10) MIC breakpoint was stipulated to be 32 μg/ml for streptomycin, 4 μg/ml for gentamicin, and 16 μg/ml for ciprofloxacin.
TABLE 4.
Incidence of antibiotic resistance according to MIC breakpoint values of SCAN (10) and Danielsen and Wind (8)
| Antibiotic | % Incidence of resistance phenotype according to the MIC breakpoint value from:
|
|
|---|---|---|
| EU Scientific Commissiona | Danielsen and Wind (8) | |
| Gentamicin | 71.1 | 20.0 |
| Streptomycin | 73.3 | 40.0 |
| Ciprofloxacin | 77.8 | 60.0 |
| Tetracycline | 6.7 | 6.7 |
| Ampicillin | 2.2 | 4.4 |
| Penicillin G | NDb | 4.4 |
| Erythromycin | 2.2 | 2.2 |
| Chloramphenicol | 4.4 | 0 |
Detection and characterization of cat resistance genes.
In an attempt to relate these observed resistances to the presence of a resistance gene, we used PCR amplification or gene probing to detect known resistance genes. From the genomic DNA of both of the two strains showing chloramphenicol resistance (L. pentosus BFE 7437 and L. plantarum BFE 7440), the cat gene could not be amplified. Interestingly, 15 strains (33.3%) possessed the cat gene, even though these strains were not chloramphenicol resistant (Tables 2 and 3). Gene fragments were PCR amplified from the genomic DNA of three representative cat+ species—L. delbrueckii subsp. bulgaricus BFE 7430, L. acidophilus BFE 7429, and S. thermophilus BFE 7420—and sequenced to confirm their identity as part of the cat gene. These fragments encoded 171 amino acids, which were identical for L. delbrueckii subsp. bulgaricus BFE 7430 and S. thermophilus BFE 7420. The 171-amino-acid sequence of these latter two strains differed by three amino acids from that of L. acidophilus BFE 7429 (results not shown).
The amino acid sequence of the L. acidophilus BFE 7429 cat gene fragment showed 100% identity to the corresponding region from amino acids 23 to 193 of the 215-amino-acid cat gene from plasmid pIP501 of S. agalactiae (47) or plasmid pRE25 of E. faecalis RE25 (40). The corresponding sequences of L. delbrueckii subsp. bulgaricus BFE 7430 and S. thermophilus BFE 7420 showed 100% identity to the region from amino acids 23 to 193 of the 215-amino-acid cat gene from plasmid pC221 of S. aureus (42) or plasmid pTZ12 of B. subtilis (2).
In order to determine why the strains in the present study did not show a resistance phenotype, the expression of the cat gene was studied at the mRNA level for three representative starter strains (P. acidilactici BFE 7434, L. acidophilus BFE 7429, and S. thermophilus BFE 7420), in addition to an E. faecium FAIR-E 151 positive control that showed a resistance phenotype in previous studies (19). P. pentosaceus BFE 7436, which did not contain a cat gene, was used as a negative control. Our RT-PCR results with specific primers for the cat gene showed that none of these starter strains investigated expressed the cat gene (Fig. 1) under both inducing (culture grown in MRS broth containing 0.015 μg of chloramphenicol/ml at 37°C) and noninducing (culture grown in MRS broth without chloramphenicol at 37°C) conditions. In contrast, the cat gene was expressed by the E. faecium FAIR-E 151 positive control under such inducing and noninducing conditions. Using our methodology, the expression of two housekeeping genes (either gyrA or ldhL) could be determined (Fig. 1), indicating the successful RNA isolation and RT of mRNA. Two weak bands of the wrong size were obtained after RT-PCR with the L. acidophilus BFE 7429 mRNA and cat primers. These bands were sequenced, and a BLAST search indicated homology to a fragment of the thioredoxin reductase gene of L. acidophilus NCFM (GenBank accession no. CP000033) but no homology to cat genes (results not shown).
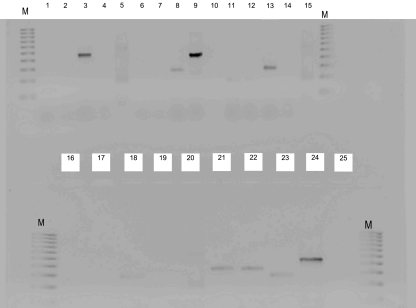
FIG. 1.
Results of RT-PCR amplification of chloramphenicol acetyltransferase (cat) genes under inducing or noninducing conditions, and selected housekeeping genes. Lanes 1 to 4, PCR of the P. acidilactici BFE 7434 (cat+, chloramphenicol-sensitive [CLs]) cat gene under noninduced (lane 1) and induced (lane 2) conditions and the l-lactate dehydrogenase (ldhL) gene (lane 3) and the ldhL gene (negative control, lane 4) after RNA digestion. Lanes 5 and 15, QIAGEN kit RT-PCR positive control. Lanes 6 to 10, PCR of the P. pentosaceus BFE 7436 (cat−, CLs) cat gene under noninduced (lane 6) and induced (lane 7) conditions and the gyrA gene (lane 8), ldhL gene (lane 9), and ldhL gene (lane 10) after RNA digestion. Lanes 11 to 20, PCR of the L. acidophilus BFE 7429 (cat+, CLs) cat gene under noninduced (lane 11) and induced (lane 12) conditions, the gyrA gene (lane 13) and the ldhL gene after RNA digestion (lane 14), the S. thermophilus BFE 7420 (cat+, CLs) cat gene under noninduced (lane 16) and induced (lane 17) conditions, and the gyrA gene (lane 18) and gyrA gene (lane 19) after RNA digestion. Lane 20, QIAGEN kit RT-PCR positive control. Lanes 21 to 25, PCR of E. faecium FAIR-E 151 (cat+, CLr) cat gene under noninduced (lane 21) and induced (lane 22) conditions and the gyrA gene (lane 23), the ldhL gene (lane 24), and the ldhL gene (lane 25) after RNA digestion.
Detection of tetracycline and ampicillin genes.
The two Pediococcus strains, P. acidilactici BFE 7434 and P. pentosaceus BFE 7436, as well as the L. pentosus strain BFE 7437, showed low resistance to tetracycline at 12 to 16 μg/ml. However, when investigated for the presence of resistance genes, neither of the genes encoding ribosomal protection proteins [tet(M), tet(O), tet(S), or tet(W)] nor genes encoding the tetracycline efflux pumps [tet(K) or tet(L)] could be amplified. A probe was developed to search for tet(K) and tet(L) efflux pump homologue sequences in the total DNA of these strains. Using Southern transfer, followed by hybridization with the digoxigenin-labeled tet(K/L) probe, no homologous sequences could be determined in the genomic DNAs of P. acidilactici BFE 7434, P. pentosaceus BFE 7436, and L. pentosus BFE 7437, whereas a positive signal was obtained with a tet(K)-positive E. faecalis FAIR-E 63 strain (19) (results not shown). In addition, a probe was developed based on an alignment of the tet(M), tet(O), and tet(S) ribosomal protection protein genes of different LAB species. In this case, however, use of the probe led to false-positive hybridization signals in all cases (also with the DNAs of the tetracycline-sensitive, negative control strains E. faecium FAIR-E 210 and E. faecalis FAIR-E 69), indicating that the probe probably cross-reacted with the sequence of the elongation factor EF-Tu and EF-G genes, which were reported to have considerable sequence homology to ribosomal protection protein genes (7).
As shown in Table 3, only one strain, P. pentosaceus BFE 7436 was resistant to ampicillin, and two strains, L. plantarum BFE 7433 and BFE 7440, were resistant to penicillin G. In both the ampicillin and the penicillin G resistance cases, the resistance phenotype was low, i.e., equal to or slightly higher than the breakpoint value of the respective antibiotic (Table 3). A β-lactamase gene could not be detected in any of these resistant strains using PCR with specific bla gene primers. As a positive control, the bla gene could be amplified using the same primers and amplification conditions using plasmid pUC19 (New England Biolabs) DNA as a template, which contains a β-lactamase gene.
Genetic characterization of ciprofloxacin resistance.
Ciprofloxacin resistance is known to be associated with mutations in the QRDR of the gyrA or the parC genes in various gram-positive or gram-negative bacteria, which lead to amino acid substitutions and result in the quinolone resistance phenotype (23, 36). In gram-positive bacteria, different fluoroquinolones have different levels of inhibitory activity against these two enzymes (18), and the findings of several studies suggest that the topoisomerase IV is the primary target of ciprofloxacin in staphylococci, streptococci, and enterococci (4, 17, 39). After PCR amplification and DNA sequencing, the amino acid sequences of the QRDR of selected starter strains with either resistant or sensitive phenotypes were deduced, and these are shown in Table 5. In the case of the ParC subunit of topoisomerase IV, in which the Ser 80 is typically substituted with Leu or Ile, such a substitution could not be observed for any one of the resistant L. lactis, S. thermophilus, L. acidophilus, P. pentosaceus. L. plantarum, or L. curvatus strains investigated (Table 5). Similarly, a Ser83-to-Arg substitution within the QRDR of the GyrA subunit of DNA gyrase could also not be observed for the resistant strains described above. Moreover, a further possible amino acid substitution in the QRDR of the GyrA subunit associated with quinolone resistance is the Glu87 substitution with either Gly or Lys (36). However, such a substitution also did not occur among the investigated ciprofloxacin-resistant starters in the present study (Table 5). A Glu87-to-Leu substitution was, however, noted for the L. acidophilus BFE 7429 strain. Interestingly, for this strain, the parC QRDR also showed some amino acid substitutions at positions 74, 84, and 88 (Table 5). However, such substitutions have not yet been reported to be associated with increases in quinolone resistance.
TABLE 5.
CLUSTAL W amino acid alignment of the QRDR of parC and gyrA of resistant and nonresistant strains published in the literature, as well as lactic acid bacteria strains investigated in this study
| Sequence type and strain | Resistance phenotypea | Partial ParC or GyrA (QRDR) amino acid sequenceb | Source or reference |
|---|---|---|---|
| ParC | |||
| E. faecalis ATCC 19433 | R | YHPHGDSSIYEAMVRLSQD | 36 |
| E. faecalis E52 | R | YHPHGDISIYEAMVRLSQD | 36 |
| E. faecium ATCC 19434 | S | YHPHGDSSIYEAMVRMSQD | 36 |
| E. faecium E138 | R | YHPHGDISIYEAMVRMSQD | 36 |
| S. agalactiae GTC1234 | R | FHPHGDFSIYDAMVRMSQD | 23 |
| L. lactis subsp. lactis BFE 7403 | R | FHPHGDSSIYEAMIRMSQD | This study |
| L. lactis subsp. lactis BFE 7406 | R | FHPHGDSSIYEAMIRMSQD | This study |
| L. lactis subsp. lactis BFE 7409 | R | FHPHGDSSIYEAMIRMSQD | This study |
| L. lactis subsp. diacetylactis BFE 7413 | S | FHPHGDSSIYEAMIRMSQD | This study |
| L. lactis subsp. diacetylactis BFE 7414 | S | FHPHGDSSIYEAMIRMSQD | This study |
| S. thermophilus BFE 7418 | S | FHPHGDSSIYDAMVRMSQD | This study |
| S. thermophilus BFE 7419 | R | FHPHGDSSIYDAMVRMSQD | This study |
| L. acidophilus BFE 7429 | R | YHPHGDSSIYGALVHLSQD | This study |
| P. pentosaceus BFE 7436 | R | FHPHGDSSIYEALVRMSQD | This study |
| L. plantarum BFE 7433 | R | FHPHGDSSIYEAMVRLSQD | This study |
| L. plantarum BFE 7440 | R | FHPHGDSSIYEAMVRLSQD | This study |
| L. curvatus BFE 7435 | R | FHPHGDSSIYEAMVRLSQD | This study |
| GyrA | |||
| E. faecalis ATCC 19433 | S | VMGKYHP HGDSAIYE | 36 |
| E. faecalis E52 | R | VMGKYHP HGDSAIYG | 36 |
| E. faecium ATCC 19434 | S | VMGKYHP HGDSAIYE | 36 |
| E. faecium E138 | R | VMGKYHP HGDRAIYE | 36 |
| E. faecium FE6/NE43 | R | VMGKYHP HGDSAIYG/K | 36 |
| L. lactis subsp. lactis BFE 7400 | R | VMGKYHP HGDSSIYE | This study |
| L. lactis subsp. lactis BFE 7406 | R | VMGKYHP HGDSSIYE | This study |
| L. lactis subsp. diacetylactis BFE 7413 | S | VMGKYHP HGDSSIYE | This study |
| L. lactis subsp. diacetylactis BFE 7414 | S | VMGKYHP HGDSSIYE | This study |
| S. thermophilus BFE 7418 | S | VMGKYHP HGDSSIYE | This study |
| S. thermophilus BFE 7419 | R | VMGKYHP HGDSFRKE | This study |
| L. curvatus BFE 7435 | R | MGKYHP HGDSAIYE | This study |
| L. plantarum BFE 7433 | R | KYHP HGDSAIYE | This study |
| L. plantarum BFE 7440 | R | VMGKYHP HGDSAIYE | This study |
| L. acidophilus BFE 7429 | R | VMGKFHP HGDSSIYL | This study |
R, resistant; S, sensitive.
Substituted amino acids that result in a resistant phenotype are indicated in boldface.
Detection and transfer of ermB gene.
Only one strain, L. salivarius BFE 7441 was resistant (32 μg/ml) to erythromycin (Table 3). This strain was shown to possess the ermB gene after PCR amplification with ermB-specific primers, but no ermB or ermC gene could be detected (result not shown). The ermB PCR product was sequenced, and the deduced 136-amino-acid sequence showed 100% similarity to amino acids 25 to 160 of the 219-amino-acid ermB gene of a group G Streptococcus sp. (49) and also 100% similarity to amino acids 45 to 180 of a 237-amino-acid ermB gene of E. faecium (16). The possibility that the ermB gene was located in the plasmid was investigated, and plasmids isolations were attempted on both the small and large scales. Plasmid DNA could not be detected by either of these methods. The possibility of the transfer of the ermB gene, for example, by a conjugative transposon, was investigated in filter-mating studies with L. lactis LMG 19460, E. faecalis JH2-2, and E. faecalis OG1X. No transconjugants could be obtained with either of these recipient LAB strains in at least triplicate filter-mating experiments for each strain. In a previous study, this methodology was used successfully to transfer tetracycline resistance genes from a tetracycline-resistant E. faecalis strain to the E. faecalis OG1X recipient (19).
DISCUSSION
Knowledge on the antibiotic resistance of LAB is still limited, possibly because of the large numbers of genera and species encountered in this group, as well as variances in their resistance spectra. What is becoming apparent, also from the results of the present study, is that the LAB starter or probiotic strains belonging to the genera Lactobacillus, Pediococcus, Leuconostoc, and Streptococcus are generally quite sensitive to clinically relevant antibiotics such as penicillin G, ampicillin, tetracycline, erythromycin, and chloramphenicol. Such resistances were lower than 10% of the isolates studied (6, 8, 21, 22), although Zarazaga et al. (52) reported an unusually high incidence of 26.2% of penicillin-resistant Lactobacillus isolates. In contrast, some resistances appear to be intrinsic for lactobacilli. These include resistance to aminoglycosides, quinolones, and glycopeptides (8, 25, 45, 51). In the present study, more than 70% of the isolates were resistant to gentamicin, streptomycin, and ciprofloxacin (Table 4) based on the MIC breakpoint values of SCAN and FEEDAP (10, 41). Similarly, more than 80% of the LAB starter and probiotic strains were previously reported to be aminoglycoside resistant (6, 8), while ciprofloxacin resistance was reported for more than 60% of the LAB strains examined by Zarazaga et al. (52).
The EFSA considers antibiotic resistances, especially transferable resistances, a safety concern and a decision criterion for determining a strain's QPS status (11). Although this step toward a safety evaluation is commendable, we foresee some problems with LAB antibiotic resistance determinations, which can lead to difficulties in safety evaluation. First, there are no approved standards for the phenotypic or genotypic evaluation of antibiotic resistances in food isolates (8, 21). Thus, already the choice of media is problematic, and in a previous study (13) we showed that MIC breakpoints vary considerably depending on the medium and the antibiotic used. Since MRS has been used in most studies and seems to be generally suited for the growth of many LAB and their antibiotic susceptibility determinations, it was used also here. However, MRS agar could not support the growth of streptococci, and thus M17 agar was used. Recently, Klare et al. (24) reported on a “general” broth medium for determining LAB antibiotic susceptibilities. These authors showed that this medium, consisting of Iso-Sensitest (90%) and MRS (10%) broth, optimally supported the growth of six Lactobacillus, two Pediococcus, and two Lactococcus strains, as well as various Bifidobacterium species (24). However, that study did not indicate whether the medium is suitable for the growth of the majority of Lactobacillus species (the genus currently consists of >80 species), in addition to Streptococcus, Leuconostoc, and Weissella spp., which were not tested by Klare et al. (24).
A further problem with LAB antibiotic susceptibility determination is the specification of MIC breakpoint values. This is important, since it may affect decisions on whether resistances can be considered to be intrinsic. Again, there are no standards, and the National Committee for Clinical Laboratory Standards (35) does not stipulate MIC breakpoints for LAB, with the exception of Enterococcus spp. One problem is the large species variation and the possible resulting variation in MIC values between species and genera (8). Thus, SCAN (10) differentiated between lactobacilli and pediococci with regard to MIC breakpoints for the different antibiotics. To add to the confusion and demonstrating the complexity of the problem, FEEDAP (41) superseded these MIC breakpoints in 2005, now assigning breakpoint values for different LAB groups, species, and strains, i.e., for homofermentative and heterofermentative lactobacilli, enterococci, Pediococcus spp., Leuconostoc spp., L. plantarum strains, and L. lactis strains. Danielsen and Wind (8) suggested up to three different breakpoint values based on differences in the resistance of only 14 Lactobacillus species. Thus, if the antibiotic resistances of the other (>60) Lactobacillus species and of other LAB genera, for which breakpoints were not investigated or specified, are found to differ considerably in future, we may end up with a range of different species- or genus-specific breakpoint values that may increase further the current complexity.
Furthermore, the actual concentration of antibiotic that can be reached in human blood serum was not taken into consideration in LAB resistance determinations. For example, the serum antibiotic concentrations for penicillin, erythromycin, tetracycline, and chloramphenicol that can be reached in humans are 2.5, 2 to 3, 8, and 10 to 15 μg/ml, respectively (51). In this case, determinations of resistances greater than these values would be of academic interest but not of practical relevance.
Resistance to aminoglycoside antibiotics is considered to be intrinsic in LAB (6, 8, 22) and is attributed to the absence of cytochrome-mediated electron transport, which mediates drug uptake (6). In addition, it was shown that when lactobacilli were grown in medium containing bile, they became more sensitive to aminoglycosides, suggesting that membrane impermeability plays an important role in this intrinsic aminoglycoside resistance (9). Our results also showed that the LAB are intrinsically resistant when the SCAN or FEEDAP MIC breakpoints are used (Table 4). However, ≤40% of the strains (Table 4) showed either streptomycin or gentamicin resistance, respectively, when the MIC breakpoints of Danielsen and Wind are used (8). This indicates that these may be set too high. A total of 60 or 77.8% of strains in the present study were resistant to ciprofloxacin according to the MIC breakpoint values of Danielsen and Wind (8) or SCAN (10), respectively (Table 4). This also indicated an intrinsic resistance. However, the basis for this resistance is not clear. For some organisms, e.g., gram-positive cocci, resistance to quinolones has been described as a result of mutation in either gyrA of parC genes (10, 18, 33, 39). However, no such point mutations in the QRDR of the gyrA or parC genes could be determined. Although one L. acidophilus BFE 7429 strain could be shown to have amino acid substitutions, possibly as a result of point mutations in the QRDR of the parC and gyrA genes, these substitutions were not the typical ones previously associated with this kind of resistance. Furthermore, only one L. acidophilus strain was sequenced in the present study, and therefore it is not clear whether such different amino acid sequences in the QRDR are typical for this species. The present study was the first to investigate whether point mutations in the gyrA or parC genes may be responsible for fluoroquinolone resistance in LAB other than enterococci and streptococci. However, since this did not appear to be the case, the basis for this resistance could not be established.
In our study, we screened all strains by PCR for known resistance genes and thus were able to determine the presence of cat genes in 15 of 46 strains that phenotypically were not resistant to chloramphenicol (Tables 2 and 3). Furthermore, we could show that the cat gene was not expressed at the RNA level under both inducing and noninducing conditions (Fig. 1) and that the reason for the chloramphenicol sensitivity was therefore probably not the result of a mutation in the cat gene. Thus, speculatively, a mutation in the regulatory region may have resulted in the open reading frame not being expressed. Our study thus warns against the use of only genetic methods, such as PCR amplification or microarray screening, to determine LAB resistances, since this could lead to false assumptions of resistance. However, in many cases such investigations are done on both the phenotypic and the genetic level, which in this case is obviously preferable. Furthermore, the present study is the first to point out that such inactive cat genes occur among different LAB genera and species (i.e., the strains of L. delbrueckii subsp. bulgaricus, L. acidophilus, S. thermophilus, P. acidilactici, L. mesenteroides, and L. pseudomesenteroides used in the present study).
Zarazaga et al. (52) reported a quite high incidence (26.2% of investigated strains) of penicillin-resistant lactobacilli, but the genetic basis for this antibiotic resistance was not elucidated. Gevers et al. (15) isolated tetracycline-resistant lactobacilli (L. plantarum, L. sakei, L. curvatus, and L. alimentarius strains) from fermented sausages and found tet(M) to be the only resistance genotype. In our study, we found a few strains with low-level resistance to tetracycline and chloramphenicol, as well as to ampicillin and penicillin G. In all cases, the MIC values were equal or close to the MIC breakpoint values, and comparably high resistances as noted by Zarazaga et al. (52) and Gevers et al. (15) were not observed. A close investigation of underlying resistance genes, using either PCR or hybridization with a gene probe, could not determine the presence of any ribosomal protection proteins or efflux genes in the case of tetracycline resistance or β-lactamase genes in the case of β-lactam antibiotic resistance. This could mean that there are underlying resistance mechanisms or genes that have not been described thus far, as may also be the case for the quinolone resistance described above. Kastner et al. (21), using an antibiotic resistance gene-specific microarray, also noticed that some antibiotic resistances could not be traced back to specific genes and hypothesized that this may be the result of possible unknown resistance genes. The existence of such unknown resistance genes clearly would make verification of the observed phenotypic resistance at the genetic level difficult.
Alternatively, and in our eyes more likely, these MIC breakpoints may be set just too low at 4 μg/ml (8) for the β-lactam antibiotics and 4 to 8 μg/μl (41) for tetracycline. Thus, such breakpoints may allow the determination of some “borderline” resistant strains, which may be resistant as a result of some complex intrinsic features such as cell wall structure or metabolic properties (21). Thus, none of the typically associated resistance genes would be discernible. Such cases can only be critically evaluated when both the phenotypic and the genotypic resistance profiles of LAB starters and probiotic strains are investigated, as was done in the present study. Many of the earlier studies only concentrated on the resistance phenotypes and thus may have had problems in interpreting borderline resistance cases. Again, this may complicate safety determinations and present regulatory drawbacks if such a “borderline” antibiotic resistance has been determined for a particular strain, but none of the typical resistance genes could be identified, leading to confusion as to whether this resistance is acquired and/or transferable.
Only in one L. salivarius strain (BFE 7441) in the present study could a typical antibiotic resistance gene, ermB involved in erythromycin resistance, be determined. Although plasmid DNA was detected in this strain, it did not hybridize with an ermB gene probe. Instead, the gene probe hybridized with genomic DNA (result not shown). This strain showed a very high resistance profile (MIC > 256 μg/ml). Similar to our study, Kastner et al. (21) studied 161 LAB isolates for antibiotic resistance, and only one L. reuteri strain SD 2112 showed a high tetracycline resistance phenotype that could be correlated with a tet(W) resistance gene (21). Erythromycin resistance genes have been reported to occur on conjugative plasmids in lactobacilli such as plasmid pGT633 from L. reuteri strain 100-63 (44) or pLEM3 from L. fermentum LEM89 (12). However, the ermB gene in L. salivarius BFE 7441 appeared to be chromosomally encoded. To investigate the possibility of whether the ermB gene is located on a conjugative, integrated plasmid or possibly a transposon, as has been reported for the E. faecium strain 160-1 to which the ermB gene from our strain showed high homology, we used filter-mating experiments with various sensitive recipient strains. However, conjugative transposition to E. faecalis JH2-2, E. faecalis OG1X, and L. lactis LMG 19460 could not be observed, so that the possibility of transferability by transposon could not be confirmed. However, filter-mating studies that show the involvement of a transpositional event are hampered by many experimental factors and thus are variable in outcome (19). Furthermore, appropriate positive control strains for conjugation and/or transposition experiments and standard protocols for gene transfer are sorely lacking. Thus, another problem associated with safety determinations of starter strains is that once a resistance phenotype and an associated resistance determinant have been identified, it becomes difficult to show that this determinant is not transferable, especially if the resistance gene is not located on a plasmid and no standard protocols for showing genetic transfer are available.
In conclusion, in Europe the adoption of the QPS system for safety evaluation must accommodate such problems in LAB antibiotic resistance determinations and allow flexible interpretation of results and not strict adherence to nonstandardized protocols or breakpoint values. The QPS system should allow leeway for interpretations of results, especially when these relate to the methodology for resistance phenotype determinations; determinations of MIC breakpoints for certain genera, species, or strains; the nondeterminability of a genetic basis of a resistance phenotype; and the transferability of resistance genes.
Acknowledgments
We thank Viva Diagnostika and particularly W. Wegst for supporting this study.
We thank I. Specht for excellent technical assistance.
Footnotes
Published ahead of print on 22 November 2006.
REFERENCES
- 1.Aminov, R. I., J. N. Garrigues-Jean, and R. J. Mackie. 2001. Molecular ecology of tetracycline resistance: development and validation of primers for detection of tetracycline resistance genes encoding ribosomal protection proteins. Appl. Environ. Microbiol. 67:22-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aoki, T., N. Noguchi, M. Sasatsu, and M. Kono. 1987. Complete nucleotide sequence of pTZ12, a chloramphenicol-resistance plasmid of Bacillus subtilis. Gene 51:107-111. [DOI] [PubMed] [Google Scholar]
- 3.Björkroth, J., and H. Korkeala. 1996. Evaluation of Lactobacillus sake contamination in vacuum-packaged sliced cooked meat products by ribotyping. J. Food Prot. 59:398-401. [DOI] [PubMed] [Google Scholar]
- 4.Brisse, S., A. C. Fluit, U. Wagner, P. Heisig, D. Milatovic, J. Verhoef, S. Scheuring, K. Kohrer, and F. J. Schmitz. 1999. Association of alterations in ParC and GyrA proteins with resistance of clinical isolates of Enterococcus faecium to nine different fluoroquinolones. Antimicrob. Agents Chemother. 43:2513-2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caplice, E., and G. F. Fitzgerald. 1999. Food fermentations: role of microorganisms in food production and preservation. Int. J. Food Microbiol. 50:131-149. [DOI] [PubMed] [Google Scholar]
- 6.Charteris, W. P., P. M. Kelley, L. Morelli, and J. K. Collins. 2001. Gradient diffusion antibiotic susceptibility testing of potentially probiotic lactobacilli. J. Food Prot. 64:2007-2014. [DOI] [PubMed] [Google Scholar]
- 7.Chopra, I., and M. Roberts. 2001. Tetracycline antibiotics: mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol. Mol. Biol. Rev. 65:232-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Danielsen, M., and A. A. Wind. 2003. Susceptibility of Lactobacillus ssp. to antimicrobial agents. Int. J. Food Microbiol. 82:1-11. [DOI] [PubMed] [Google Scholar]
- 9.Elkins, C. A., and L. B. Mullis. 2004. Bile-mediated sensitivity in Lactobacillus species likely results from increased membrane permeability attributable to cholic acid. Appl. Environ. Microbiol. 70:7200-7209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.European Commission. 2002. Opinion of the Scientific Committee on Animal Nutrition on the criteria for assessing the safety of micro-organisms resistant to antibiotics of human clinical and veterinary importance. European Commission, Health and Consumer Protection Directorate General, Directorate C, Scientific Opinions, Brussels, Belgium.
- 11.European Food Safety Authority. 2004. EFSA Scientific Colloquium Summary Report. QPS: qualified presumption of safety of microorganisms in food and feed. European Food Safety Authority, Brussels, Belgium.
- 12.Fons, M., T. Hégé, M. Ladiré, P. Raibaud, R. Ducluzeau, and E. Maguin. 1997. Isolation and characterization of a plasmid from Lactobacillus fermentum conferring erythromycin resistance. Plasmid 37:199-203. [DOI] [PubMed] [Google Scholar]
- 13.Franz, C. M. A. P., A. Hummel, and W. H. Holzapfel. 2005. Problems related to the safety assessment of lactic acid bacteria starter cultures and probiotics. Mitt. Lebensm. Hyg. 96:39-65. [Google Scholar]
- 14.Gevers, D., G. Huys, and J. Swings. 2003. In vitro conjugal transfer of tetracycline resistance from Lactobacillus isolates to other gram-positive bacteria. FEMS Microbiol. Lett. 225:125-130. [DOI] [PubMed] [Google Scholar]
- 15.Gevers, D., M. Danielsen, G. Huys, and J. Swings. 2003. Molecular characterisation of tet(M) genes in Lactobacillus isolates from different types of fermented dry sausage. Appl. Environ. Microbiol. 69:1270-1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hammerum, A. M., S. E. Flannagan, D. B. Clewell, and L. B. Jensen. 2001. Indication of transposition of the motile DNA element containing the vat(D) and ermB genes in Enterococcus faecium. Antimicrob. Agents Chemother. 45:3223-3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hooper, D. C. 2000. Mechanisms of action and resistance of older and newer fluoroquinolones. Clin. Infect. Dis. 31:4-28. [DOI] [PubMed] [Google Scholar]
- 18.Hooper, D. C. 2002. Fluoroquinolone resistance among gram-positive cocci. Infect. Dis. 2:530-538. [DOI] [PubMed] [Google Scholar]
- 19.Hummel, A., W. H. Holzapfel, and C. M. A. P. Franz. 2006. Characterisation and transfer of antibiotic resistance genes from enterococci isolated from food. Syst. Appl. Microbiol. [Epub ahead of print.] [DOI] [PubMed]
- 20.Huys G., K. D'Haene, and J. Swings. 2002. Influence of the culture medium on antibiotic susceptibility testing of food-associated lactic acid bacteria with the agar overlay disc diffusion method. Lett. Appl. Microbiol. 34:402-406. [DOI] [PubMed] [Google Scholar]
- 21.Kastner, S., V. Perreten, H. Bleuler, G. Hugenschmidt, C. Lacroix, and L. Meile. 2006. Antibiotic susceptibility patterns and resistance genes of starter cultures and probiotic bacteria used in food. Syst. Appl. Microbiol. 29:145-155. [DOI] [PubMed] [Google Scholar]
- 22.Katla, A.-K., H. Kruse, H. Johnsen, and H. Herikstad. 2001. Antimicrobial susceptibility of starter culture bacteria used in Norwegian dairy products. Int. J. Food Microbiol. 67:147-152. [DOI] [PubMed] [Google Scholar]
- 23.Kawamura, Y., H. Fujiwara, N. Mishima, Y. Tanaka, A. Tanimoto, S. Ikawa, Y. Itoh, and T. Ezaki. 2003. First Streptococcus agalactiae isolates highly resistant to quinolones, with point mutations in gyrA and parC. Antimicrob. Agents Chemother. 47:3605-3609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klare, I., C. Konstabel, S. Müller-Bertling, R. Reissbrodt, G. Huys, M. Vancanneyt, J. Swings, H. Goossens, and W. Witte. 2005. Evaluation of new broth media for microdilution antibiotic susceptibility testing of lactobacilli, ediococci, lactococci, and bifidobacteria. Appl. Environ. Microbiol. 71:8982-8986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klein, G., C. Hallmann, I. A. Casas, J. Abad, J. Louwers, and G. Reuter. 2000. Exclusion of vanA, vanB, and vanC type of glycopeptide resistance in strains of Lactobacillus reuteri and Lactobacillus rhamnosus used as probiotics by polymerase chain reaction and hybridisation methods. J. Appl. Microbiol. 89:815-824. [DOI] [PubMed] [Google Scholar]
- 26.Klein, G., A. Pack, and G. Reuter. 1998. Antibiotic resistance patterns of enterococci and occurrence of vancomycin-resistant enterococci in raw minced beef and pork in Germany. Appl. Environ. Microbiol. 64:1825-1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Landman, D., and J. M. Quale. 1997. Management of infections due to resistant enterococci: a review of therapeutic options. J. Antimicrob. Chemother. 40:161-170. [DOI] [PubMed] [Google Scholar]
- 28.Leclercq, R. 1997. Enterococci acquire new kinds of resistance. Clin. Infect. Dis. 24(Suppl. 1):S80-S84. [DOI] [PubMed] [Google Scholar]
- 29.Leroy, F., and L. De Vuyst. 2004. Lactic acid bacteria as functional starter cultures for the food fermentation industry. Trends Food Sci. Technol. 15:67-78. [Google Scholar]
- 30.Mogensen, G., S. Salminen, J. O'Brien, A. Ouwehand, W. H. Holzapfel, C. Shortt, R. Fondén, G. D. Miller, D. Donohue, M. Playne, R. Crittenden, B. Bianchi, B. Salvadori, and R. Zink. 2002. Food microorganisms—health benefits, safety evaluation and strains with documented history of use in foods. Bull. Int. Dairy Fed. 377:4-9. [Google Scholar]
- 31.Moore, J. E., B. C. Millar, X. Yongmin, N. Woodford, S. Vincent, C. E. Goldsmith, R. B. McClurg, M. Crowe, R. Hone, and P. G. Murphy. 2001. A rapid molecular assay for the detection of antibiotic resistance determinants in causal agents of infective endocarditis. J. Appl. Microbiol. 90:719-726. [DOI] [PubMed] [Google Scholar]
- 32.Morrison, D., N. Woodford, and B. Cookson. 1997. Enterococci as emerging pathogens of humans. J. Appl. Microbiol. Symp. Suppl. 83:89S-99S. [DOI] [PubMed] [Google Scholar]
- 33.Munoz, R., and A. G. De La Campa. 1996. ParC subunit of DNA topoisomerase IV of Streptococcus pneumoniae is a primary target of fluoroquinolones and co-operates with DNA gyrase A subunit in forming resistance phenotype. Antimicrob. Agents Chemother. 40:2252-2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Murray, B. E. 1990. The life and times of the Enterococcus. Clin. Microbiol. Rev. 3:46-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.NCCLS. 1997. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 4th ed., vol. 17, no. 2. National Committee for Clinical Laboratory Standards, Wayne, PA.
- 36.Petersen, A., and L. B. Jensen. 2004. Analysis of gyrA and parC mutations in enterococci from environmental samples with reduced susceptibility to ciprofloxacin. FEMS Microbiol. Lett. 231:73-76. [DOI] [PubMed] [Google Scholar]
- 37.Pitcher, D. G., N. A. Saunders, and R. J. Owen. 1989. Rapid extraction of bacterial genomic DNA with guanidium thiocyanate. Lett. Appl. Microbiol. 8:151-156. [Google Scholar]
- 38.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 39.Schmitz, F. J., M. E. Jones, B. Hofmann, B. Hansen, S. Scheuring, M. Luckefahr, A. Fluit, J. Verhoef, U. Hadding, H. P. Heinz, and K. Kohrer. 1998. Characterisation of grlA, grlB, gyrA, and gyrB mutations in 116 unrelated isolates of Staphylococcus aureus and effects of mutations on ciprofloxacin MIC. Antimicrob. Agents Chemother. 42:1249-1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schwarz, F. V., V. Perreten, and M. Teuber. 2001. Sequence of the 50-kb conjugative multiresistance plasmid pRE25 from Enterococcus faecalis RE25. Plasmid 46:170-187. [DOI] [PubMed] [Google Scholar]
- 41.Scientific Panel on Additives and Products or Substances Used in Animal Feed. 2005. Opinion of the scientific panel on additives and products or substances used in animal feed on the updating of the criteria used in the assessment of bacteria for resistance to antibiotics of human or veterinary importance. EFSA J. 223:1-12. [Google Scholar]
- 42.Shaw, W. V., D. G. Brenner, S. F. LeGrice, S. E. Skinner, and A. R. Hawkins. 1985. Chloramphenicol acetyltransferase gene of staphylococcal plasmid pC221. Nucleotide sequence analysis and expression studies. FEBS Lett. 179:101-106. [DOI] [PubMed] [Google Scholar]
- 43.Sutcliffe, J., T. Grebe, A. Tait-Kamradt, and L. Wondrack. 1996. Detection of erythromycin-resistant determinants by PCR. Antimicrob. Agents Chemother. 40:2562-2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tannock, G. W., J. B. Luchansky, L. Miller, H. Connell, S. Thode-Andersen, A. A. Mercer, and T. R. Klaenhammer. 1994. Molecular characterization of a plasmid-borne (pGT633) erythromycin resistance determinant (ermGT) from Lactobacillus reuteri 100-63. Plasmid 31:60-71. [DOI] [PubMed] [Google Scholar]
- 45.Temmerman, R., B. Pot, G. Huys, and J. Swings. 2002. Identification and antibiotic susceptibility of bacterial isolates from probiotic products. Int. J. Food Microbiol. 81:1-10. [DOI] [PubMed] [Google Scholar]
- 46.Teuber, M., L. Meile, and F. Schwarz. 1999. Acquired antibiotic resistance in lactic acid bacteria from food. Antonie Leeuwenhoek 76:135-155. [PubMed] [Google Scholar]
- 47.Trieu-Cuot, P., G. de Cespedes, and T. Horaud. 1992. Nucleotide sequence of the chloramphenicol resistance determinant of the streptococcal plasmid pIP501. Plasmid 28:272-276. [DOI] [PubMed] [Google Scholar]
- 48.van Belkum, M., and M. E. Stiles. 1995. Molecular characterization of genes involved in the production of the bacteriocin leucocin A from Leuconostoc gelidum. Appl. Environ. Microbiol. 61:3573-3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Woo, P. C., A. P. To, H. Tse, S. K. Lau, and K. Y. Yuen. 2003. Clinical and molecular epidemiology of erythromycin-resistant beta-hemolytic Lancefield group G-streptococci causing bacteremia. J. Clin. Microbiol. 41:51885191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wood, B. J. B., and W. H. Holzapfel. 1995. The genera of lactic acid bacteria. Blackie Academic and Professional, London, United Kingdom.
- 51.Yao, J. D. C., and R. C. Moellering. 1995. Antibacterial agents, p. 1281-1307 In P. R. Murray, E. J. Baron, M. A. Pfaller, F. C. Tenover, and R. H. Yolken (ed.), Manual of clinical microbiology, 7th ed. ASM Press, Washington, DC.
- 52.Zarazaga, M., Y. Sáenz, A. Portillo, C. Tenorio, F. Ruiz-Larrea, R. Del Campo, F. Baquero, and C. Torres. 1999. In vitro activities of ketolide HMR3647, macrolides, and other antibiotics against Lactobacillus, Leuconostoc, and Pediococcus isolates. Antimicrob. Agents Chemother. 43:3039-3041. [DOI] [PMC free article] [PubMed] [Google Scholar]