Abstract
While looking for new means to limit the dissemination of antibiotic resistance, we evaluated the role of potentially probiotic bifidobacteria on the transfer of resistance genes between enterobacteria. Transfers of bla genes encoding extended-spectrum β-lactamases (SHV-5 and CTX-M-15) were studied in the absence or presence of bifidobacteria. In vitro, transfer frequencies of these bla genes decreased significantly in the presence of three of five tested strains, i.e., Bifidobacterium longum CUETM-89-215, Bifidobacterium bifidum CIP-56.7T, and Bifidobacterium pseudocatenulatum CIP-104168T. Four transfer experiments were conducted in the digestive tract of gnotobiotic mice, the first three observing the effect of B. longum CUETM-89-215, B. bifidum CIP-56.7T, and B. pseudocatenulatum CIP-104168T on blaSHV-5 transfer and the fourth experiment studying the effect of B. bifidum CIP-56.7T on blaCTX-M-15 transfer. These experiments revealed significant decreases in the transconjugant levels (up to 3 logs) in mice having received B. bifidum CIP-56.7T or B. pseudocatenulatum CIP-104168T compared to control mice. Bifidobacteria appear to have an inhibitory impact on the transfer of antibiotic resistance genes. The inhibitory effect is associated to specific bifidobacterial strains and may be related to the production of thermostable metabolites by these strains.
Antimicrobial resistance spreads rapidly among pathogens constituting nowadays an important public health problem. In hospital settings, multidrug-resistant bacteria are being isolated at an increasing rate and are having a significant impact on clinical practice and overall treatment cost (7, 15). Furthermore, in the community, a pool of antibiotic-resistant bacteria exists that either originates from hospital reservoirs or is a consequence of the use of antimicrobial substances in animals (14).
The evolution of bacterial strains toward multiresistance depends on the ecosystem in which they live. The digestive tract, colonized by a complex microbial flora up to 1012/g (wet weight) of gut contents, is a privileged site of horizontal transfer of plasmids carrying antibiotic resistance genes and contributes to the maintaining and the dissemination of resistance (23). Likewise, extended-spectrum β-lactamases (ESBLs), enzymes capable of efficiently hydrolyzing extended-spectrum cephalosporins and associated with several treatment failures, disseminate through plasmid transmission among Enterobacteriaceae, a main genera of the aero-anaerobic flora of the digestive tract (28). Under normal physiological conditions, resistant bacteria are repressed by the dominant digestive flora (33). However, the ingestion of antibiotics, even at low dosages, may disturb this equilibrium, modify the barrier effect of the dominant flora, and allow the proliferation of resistant strains. In addition, antibiotic therapy can exert selective pressure on plasmid-carrying strains, and certain studies have reported that antibiotics can even induce plasmid transfer under certain circumstances (4, 16, 19).
The concept of modulating the gut microflora by probiotics has emerged these last two decades. Probiotics are defined as live microorganisms that exert beneficial effects on the host physiology when administered at an adequate dosage (6). Probiotic bacteria are mainly part of two genera, i.e., Lactobacillus and Bifidobacterium, and act either by modifying the intestinal microbiota, increasing the number of beneficial bacteria, or direct contact on mucosal cells (5). A number of health effects have been associated with the use of probiotics. For instance, there is a strong evidence of the interest of probiotic administration in the reduction of the duration and the severity of diarrhea associated with rotavirus or in the prevention of antibiotic-associated diarrhea (30, 31). New studies support other beneficial effects for a broad set of disorders, including atopic disease, irritable bowel disease, and inflammatory bowel disease, but these studies need to be confirmed by additional clinical trials (5, 24, 26). However, the potential use of probiotics to limit the emergence of bacterial resistance and the dissemination of resistance genes has been briefly investigated. One team observed the beneficial effect of a Lactobacillus supplementation during treatment with penicillin and quinolone antibiotics (29), and another team revealed the inhibitory effect of the yogurt symbiosis on horizontal antibiotic resistance gene transfers (17, 18).
In this context, we sought to determine the role of bifidobacteria in the transfer of antibiotic resistance genes in vitro and in vivo and to evaluate the impact of different bifidobacterial species on the establishment of transconjugants in the digestive tracts of gnotobiotic mice.
MATERIALS AND METHODS
Bacterial strains.
Transfer of plasmidic bla genes encoding the ESBLs SHV-5 and CTX-M-15 was studied from two donor strains of clinical origin, Klebsiella pneumoniae CIP-104771 (9) and Salmonella enterica serovar Typhimurium CAM18 (20), to the recipient Escherichia coli K-12 strain resistant to nalidixic acid. The blaSHV-5 resides on a 140-kb plasmid that harbors also a kanamycin resistance gene and the blaCTX-M-15 on a 60-kb plasmid carrying a tetracycline resistance gene. The impact on the gene transfer of the following five bifidobacterial strains isolated from human fecal samples was investigated: Bifidobacterium longum CUETM-89-215, Bifidobacterium bifidum CIP-56.7T, B. bifidum BS119, Bifidobacterium pseudocatenulatum CIP-104168T, and B. pseudocatenulatum BS4. These bifidobacterial strains have been demonstrated previously to be sensitive to β-lactam antibiotics (21).
Media.
Culture of bifidobacteria was performed in TGYH broth containing tryptone peptone (30 g/liter), glucose (5 g/liter), yeast extract (20 g/liter), and hemin (5 mg/liter) for 48 h at 37°C in an anaerobic chamber (N2:CO2:H2; 80:10:10). Enterobacterial strains were grown in the same media for 18 h at 37°C in aerobiosis. After the mating step, enterobacterial populations were enumerated on eosin methylene blue medium (BioMerieux, Marcy l'Etoile, France) containing ceftazidime (4 mg/liter) for the donors, nalidixic acid (50 mg/liter) for the recipients, and nalidixic acid and ceftazidime for the transconjugants. Bifidobacteria were enumerated on Wilkins-Chalgren modified for bifidobacteria (WCB) medium (2) containing polymyxin B (10 mg/liter) and on WCB containing polymyxin B and ceftazidime (4 mg/liter) for those that could have acquired a bla gene. Eosin methylene blue plates were incubated aerobically for 24 h at 37°C, and WCB plates were incubated for 48 h at 37°C in an anaerobic chamber.
In vitro broth mating.
Conjugation experiments using K. pneumoniae (SHV-5) and E. coli or serovar Typhimurium (CTX-M-15) and E. coli were performed according to the broth mating procedure as follows. After culture in TGYH broth, 2 ml of the donor, 2 ml of the recipient, and 2 ml of a bifidobacterial culture were mixed in a sterile tube. A control tube was prepared by replacing the bifidobacterial culture with 2 ml of TGYH broth. After 5 h of anaerobic incubation at 37°C, the cells were diluted in regenerated sterile water supplemented with peptone in 10-fold series, and 100 μl of each dilution was plated on selective medium. Transfer frequencies were calculated by dividing the number of transconjugants by the number of donor colonies. Each experiment was repeated three times.
In order to investigate the mechanism of action of bifidobacteria on gene transfer, experiments were performed by replacing the bifidobacterial culture by either a heat-killed culture of bifidobacterial strain (culture in TGYH broth treated 20 min at 100°C), the supernatant of a culture in TGYH broth, or the bacterial pellet of a culture in TGYH broth, washed and suspended in TGYH broth.
In vivo mating experiments. (i) Mice.
Protocol for in vivo experiments was approved by the animal care and use committee of the Faculté des Sciences Pharmaceutiques et Biologiques (Paris, France). Germfree consanguineous C3H mice were maintained in separate isolators (J.C.E. Biotechnology, Vichy, France) and fed ad libitum with a commercial diet sterilized by gamma irradiation (4 Mrad) and supplied with autoclaved drinking water. The germfree status of mice was ensured before the beginning of the experiment by testing fecal samples for aerobic and anaerobic growth of bacteria and yeast.
(ii) Experimental design.
In vivo transfer was studied in the absence (control groups) or presence of bifidobacteria. In the control groups, the recipient strain E. coli K-12 was inoculated first (8 log10 CFU) and then, 8 days later, after checking the implantation of the recipient strain, the mice were inoculated with the donor strain (8 log10 CFU) K. pneumoniae (referred to as blaSHV-5 mice) or serovar Typhimurium (referred to as blaCTX-M-15 mice). In the following experiments, germfree mice received, in addition to the donor and the recipient strains, one bifidobacterial strain. Every germfree mouse received first intragastrically 8 log10 CFU of the recipient E. coli K-12. Three days later, after verification of the implantation of the recipient strain, the mice were inoculated with 8 log10 CFU of the bifidobacterial strain, and 5 days later, after we checked the implantation of the recipient and the bifidobacterial strains, the mice were inoculated with 8 log10 CFU of the donor strain. Four experiments were carried out, the first three using the donor strain K. pneumoniae and the bifidobacterial strains B. longum CUETM-89-215, B. bifidum CIP-56.7T, and B. pseudocatenulatum CIP-104168T. The fourth experiment used the donor strain serovar Typhimurium and the bifidobacterial strain B. bifidum CIP-56.7T. Each experiment lasted 2 months and included six mice.
(iii) Analysis of fecal samples.
For each model, the evolution of the various bacterial populations in mice feces was studied since this reflects the number of bacteria in the cecum (10). Fecal samples (one to two pellets) collected five times per week by provoked defecation were diluted in 10-fold series and spread onto the appropriate plates supplemented with antibiotics by using the automated WASP spiral system (AES Laboratoire, Bruz, France). Bacterial counts were expressed per gram of feces and as log10 values.
Antibiotic susceptibility testing and ESBL production detection.
Ten transconjugants from each in vitro experiment and ten others from each fecal sample collected from mice were selected randomly for antibiotic susceptibility determination using the disk diffusion method according to the recommendations of the Comité de l'Antibiogramme de la Société Française de Microbiologie (3). The following commercial disks (Bio-Rad, Marne-la-Coquette, France) were tested: ceftazidime (30 μg), amoxicillin (20 μg)-clavulanic acid (10 μg), cefotaxime (30 μg), kanamycin (30 μg), tetracycline (30 IU), and nalidixic acid (30 μg). ESBL production was detected by the double-disk synergy test according to the method of Jarlier et al. (13). The presence of blaSHV and blaCTX-M genes in donor strains and in transconjugants selected during in vitro and in vivo experiments was confirmed by using PCR as described by Perilli et al. (25) and Mulvey et al. (22), respectively.
Statistical analysis.
Transfer frequencies with or without bifidobacteria were compared for in vitro experiments by using the Student test and for in vivo experiments by using the Mann-Whitney U test for small samples.
RESULTS
In vitro transfer.
In the absence of bifidobacteria, the transfer of blaSHV-5 and blaCTX-M-15 from K. pneumoniae CIP-104771 and serovar Typhimurium CAM18 to E. coli K-12 occurred at high frequencies equivalent to (42.02 ± 2.04) × 10−6 and (44.04 ± 1.82) × 10−4, respectively. Moreover, resistance genes carried by the studied plasmids were transferred at the same frequencies, i.e., the resistance gene to kanamycin was transferred along with blaSHV-5, and the resistance gene to tetracycline was transferred along with blaCTX-M-15. In the presence of bifidobacteria, a significant decrease in the transfer frequencies of resistance genes was observed with three of the five tested bifidobacterial strains, i.e., B. longum CUETM 89-215, B. bifidum CIP-56.7T, and B. pseudocatenulatum CIP-104168T (P ≤ 0.01) (Table 1) . The highest decreases in transconjugant counts were observed in conjugations including B. pseudocatenulatum CIP-104168T and were equivalent to 2.6 and 2.1 logs for blaSHV-5 and blaCTX-M-15 transfers, respectively. In contrast, insignificant changes in the transfer frequencies were noticed with B. bifidum BS119 and B. pseudocatenulatum BS4. In all experiments, the presence of bifidobacteria did not modify the levels of donors and recipients.
TABLE 1.
In vitro transfer of bla genes between enterobacteria in the absence or presence of bifidobacteriaa
| Strain | Transfer of blaSHV-5 from K. pneumoniae CIP-104771 to E. coli K-12 |
Transfer of blaCTX-M-15 from serovar Typhimurium CAM18 to E. coli K-12 |
||||
|---|---|---|---|---|---|---|
| Transfer frequency | Transconjugant count (log10) | Decrease in transconjugant count (log10) | Transfer frequency | Transconjugant count (log10) | Decrease in transconjugant count (log10) | |
| No bifidobacteria | (42.02 ± 2.04) × 10−6 | 4.5 ± 0.3 | (44.04 ± 1.82) × 10−4 | 6.1 ± 0.1 | ||
| B. pseudocatenulatum BS4 | (41.23 ± 1.03) × 10−6 | 4.3 ± 0.1 | 0.2 | (70.15 ± 9.26) × 10−4 | 5.9 ± 0.3 | 0.2 |
| B. bifidum BS119 | (40.61 ± 6.50) × 10−6 | 4.3 ± 0.1 | 0.2 | (20.38 ± 1.12) × 10−4 | 5.8 ± 0.1 | 0.3 |
| B. longum CUETM-89-215 | (0.06 ± 0.02) × 10−6* | 2.1 ± 0.3* | 2.4 | (0.38 ± 0.24) × 10−4* | 4.7 ± 0.1* | 1.4 |
| B. bifidum CIP-56.7T | (0.07 ± 0.01) × 10−6* | 2.3 ± 0.2* | 2.2 | (0.18 ± 0.04) × 10−4* | 3.9 ± 0.2* | 2.2 |
| B. pseudocatenulatum CIP104168T | (0.07 ± 0.01) × 10−6* | 1.9 ± 0.4* | 2.6 | (0.25 ± 0.17) × 10−4* | 4.0 ± 0.1* | 2.1 |
Values represent the averages from a minimum of three experiments. The transfer frequency is the transconjugant counts relative to the donor counts after the mating period. *, P ≤ 1% in the Student test in comparison to values in the conjugations without bifidobacteria. Significant decreases in transconjugant counts are indicated in boldface.
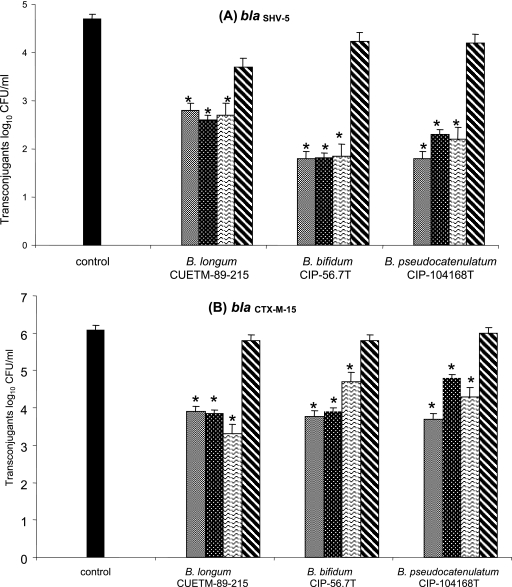
In order to explore the mechanism of action of the three bifidobacterial strains with an inhibitory impact on gene transfers in vitro (i.e., B. longum CUETM 89-215, B. bifidum CIP-56.7T, and B. pseudocatenulatum CIP-104168T), transfers of blaSHV-5 and blaCTX-M-15 were performed in the presence of heat-killed bifidobacteria, culture supernatants, or bifidobacterial pellets and compared to transfers in the presence of viable bifidobacteria. Transconjugant counts after conjugations in the presence of bifidobacterial pellets were similar to the ones found in conjugations performed in the absence of bifidobacteria (Fig. 1). However, assays in presence of killed bifidobacterial strains, and culture supernatants revealed much lower levels, close to the ones of conjugations including bifidobacterial cultures (Fig. 1).
FIG. 1.
Effects of different bifidobacterial preparations of B. longum CUETM-89-215, B. bifidum CIP-56.7T, and B. pseudocatenulatum CIP-104168T on transconjugant counts in the conjugation medium. (A) Transfer of blaSHV-5; (B) transfer of blaCTX-M-15. Data, expressed as means ± the standard errors, are from three independent experiments. Symbols: ▪, control; ░⃞, bifidobacterial culture; , killed bifidobacteria; , culture supernatant; ▧, bifidobacterial pellet. *, significantly different from frequencies obtained in absence of bifidobacteria (P ≤ 1% [Student test]).
In vivo transfer.
In all experiments, recipient (i.e., E. coli) and donor (i.e., K. pneumoniae or serovar Typhimurium) strains colonized the gastrointestinal tracts efficiently, at stable levels ranging between 9 and 10 log10 CFU per g of feces.
Experiments in the absence of bifidobacteria (control groups).
Transconjugants were detected in the first samples collected 1 day after donor inoculation, revealing the ease with which transfers of blaSHV-5 or blaCTX-M-15 gene occurred in vivo. The transfer of both genes was confirmed by phenotype and genotype analysis: both ESBL production and either blaSHV-5 or blaCTX-M-15 genes were detected in all transconjugants resistant to nalidixic acid and ceftazidime. Transconjugants established at a mean level of 5 log10 CFU per g of feces in blaSHV-5 mice and 8 log10 CFU per g of feces in blaCTX-M-15 mice.
Experiments in the presence of bifidobacteria.
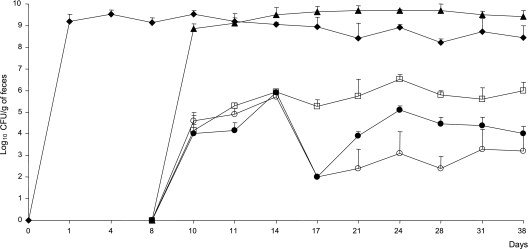
On days 10, 11, and 14 the level of transconjugants in blaSHV-5 mice was the same in all three groups treated with bifidobacteria, i.e., B. bifidum CIP-56.7T, B. pseudocatenulatum CIP-104168T, or B. longum CUETM-89-215. Starting at day 14, a significant decrease in transconjugant levels was observed when mice were associated with B. bifidum CIP-56.7T or B. pseudocatenulatum CIP-104168T (P ≤ 0.05). Indeed, B. bifidum CIP-56.7T and B. pseudocatenulatum CIP-104168T both led to 3.3-log decreases at day 17 and to 1.4- and 2.8-log decreases, respectively, in the last 3 weeks of the experiments (Fig. 2) . In contrast, B. longum CUETM-89-215 was not associated with a significant decrease in the transconjugant level in gnotobiotic mice. In blaCTX-M-15 mice, the bifidobacterial strain B. bifidum CIP-56.7T led to a 2.1-log decrease in transconjugant levels throughout the experiment (data not shown).
FIG. 2.
Bacterial counts in homogenized feces of gnotobiotic mice. On day 0, 18 germfree mice were inoculated with 108 E. coli K-12 recipients. On day 3, six of these mice were inoculated with 108 B. bifidum CIP-56.7T and six others were inoculated with 108 B. pseudocatenulatum CIP-104168T. The remaining six mice corresponded to our control group. On day 8, all mice were inoculated with 108 K. pneumoniae CIP-104771::blaSHV-5 donors. The figure shows the geometric averages of the recipient and donor counts in the 18 mice and the geometric averages of transconjugant counts in each group of six mice. The error bars indicate the standard deviations. Symbols: ⧫, E. coli K-12; ▴, K. pneumoniae CIP-104771::blaSHV-5; □, transconjugants in the absence of bifidobacteria; ○, transconjugants in the presence of B. bifidum CIP-56.7T; •, transconjugants in the presence of B. pseudocatenulatum CIP-104168T.
Moreover, in all experiments, and whatever the effect of bifidobacterial strains on gene transfer, none of the bifidobacterial strains acquired the bla antibiotic resistance genes.
DISCUSSION
Probiotics, mainly lactobacilli and bifidobacteria, are of increasing interest due to the growing evidence of health benefits associated with their use. In the present study, we revealed for the first time the beneficial role that can be played by bifidobacteria on antibiotic resistance gene transfers. Indeed, bifidobacterial strains led to a significant in vitro and in vivo decrease in the transfer frequency of resistance to β-lactams, kanamycin, and tetracycline antibiotics among enterobacteria. However, this effect was strain specific, since only three of the five bifidobacterial strains tested had an inhibitory effect in vitro.
The emergence of multiresistant bacteria is now an increasing problem that could be handled by applying antibiotic policies, such as the administration of shorter courses of antibiotics and/or the cycling of antibiotics (12). The use of probiotics can also be an alternative to the latter policies as suggested by Levy (16). In fact, probiotics, acting as a competitive flora, could avoid or suppress the gut colonization by resistant bacteria through their barrier effects. Sullivan et al. examined whether Lactobacillus sp. strain F19, in conjunction with antibiotics, prevents the establishment of resistant bacteria in the gastrointestinal tract and reported that the overall difference between the placebo and supplemented groups was limited (29). However, our study revealed that probiotics might interfere by reducing antibiotic resistance genes transfers.
Such an inhibitory effect was never described for bifidobacteria. A previous study demonstrated that the daily consumption of yogurt symbiosis, i.e., Lactobacillus bulgaricus and Streptococcus thermophilus, decreased plasmid transfer between two Escherichia coli strains in the digestive tract of mice colonized with human fecal flora (17). A later study involving the same model evaluated the effect of yogurt, heat-treated fermented milk, milk, and lactose solution intake on plasmid transfer and the establishment of the resulting transconjugants in the digestive tracts of mice. It was proposed, in that study, that the lactose, available at the same concentration in the above supplementations, was the main agent responsible for the inhibiting effects (18).
In our attempt to understand the inhibitory effect observed in our study, we monitored the evolution of bacterial populations at the end of the experiments. In fact, bifidobacterial strains can exert inhibitory effects on the growth of other bacterial species (11). However, in our experiments the level of donor and recipient populations remained stable in vitro and in vivo. The decrease in transfer efficiency was not due therefore to an inhibitory impact of bifidobacterial strains on either the donor or the recipient strains. This inhibitory effect could be related to an impact of bifidobacteria on the conjugation mechanism itself, altering its efficiency. To investigate this hypothesis, in vitro experiments were performed with heat-killed bacteria, culture supernatant, and bacterial pellet. Since no inhibition was observed with the bacterial pellet, the decrease in the transfer was not due to a problem of steric hindrance affecting the contact between donor and recipient. The threefold decrease in transconjugant levels observed when the conjugation medium included either heat-killed bifidobacterial strains or bacterium-free supernatants suggests that the inhibitory action is more likely due to thermostable metabolites produced by bifidobacteria. Bacterial conjugation is a complex phenomenon that involves a mating-pair formation system belonging to the donor strain (27). The components of the mating-pair formation system, comprising a minimal set of 10 proteins, form a membrane-spanning protein complex and a surface-exposed sex pilus, which both serve to establish intimate physical contact with a recipient bacterium. Thus, these bifidobacterial metabolites might act on the conjugation process by affecting the tip of the donor pili or altering the recipient cell surface (8). Inhibition of conjugation was previously described with agents that affect the formation of sex pili or allow plasmid curing of the donor strains, such as sodium dodecyl sulfate, organic solvents, proteases, high temperature, and acridine dyes (32).
To conclude, little is known about the environmental factors capable of limiting the spread of resistance genes, whereas the evolution of bacteria toward resistance to antibiotics, and even multidrug resistance, has become a major cause of concern worldwide (1). We demonstrated here the antagonistic impact of bifidobacteria on the transfer of antibiotic resistance genes. Further investigations are in progress in order to identify the strains associated with the highest inhibitory impact and to elucidate the mechanism of this effect. Nevertheless, the use of probiotics might provide a new means to limit the dissemination of antibiotic genes.
Acknowledgments
We thank Regis Dantier for technical assistance.
C.M. was supported by a research grant from the Chancellerie des Universités de Paris.
Footnotes
Published ahead of print on 22 November 2006.
REFERENCES
- 1.Alanis, A. J. 2005. Resistance to antibiotics: are we in the post-antibiotic era? Arch. Med. Res. 36:697-705. [DOI] [PubMed] [Google Scholar]
- 2.Butel, M. J., N. Roland, A. Hibert, F. Popot, A. Favre, A. C. Tessèdre, M. Bensaada, A. Rimbault, and O. Szylit. 1998. Clostridial pathogenicity in experimental necrotizing enterocolitis in gnotobiotic quails and protective role of bifidobacteria. J. Med. Microbiol. 47:391-399. [DOI] [PubMed] [Google Scholar]
- 3.Comité de l'Antibiogramme de la Société Française de Microbiologie. 2004. Communiqué 2004. Société Française de Microbiologie. http://www.sfm.asso.fr.
- 4.Doucet-Populaire, F., P. Trieu-Cuot, A. Dosbaa, A. Andremont, and P. Courvalin. 1991. Inducible transfer of conjugative transposon Tn1545 from Enterococcus faecalis to Listeria monocytogenes in the digestive tracts of gnotobiotic mice. Antimicrob. Agents Chemother. 35:185-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.FAO/WHO. 2001. Health and nutritional properties of probiotics in food including powder milk with live lactic acid bacteria. Expert consultation. FAO/WHO, Cordoba, Argentina.
- 6.FAO/WHO. 2002. Guidelines for the evaluation of probiotics in food. FAO/WHO, London, Ontario, Canada. [Online.] http//www.who.int/foodsafety/fs_management/en/probiotic_guidelines.pdf.
- 7.Farr, B. M., C. D. Salgado, T. B. Karchmer, and R. J. Sherertz. 2001. Can. antibiotic-resistant nosocomial infections be controlled? Lancet Infect. Dis. 1:38-45. [DOI] [PubMed] [Google Scholar]
- 8.Firth, N., and K. Ippen-Ihler. 1996. Structure and function of the F factor and mechanism of conjugation, p. 2377-2401. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, DC.
- 9.French, G. L., K. P. Shannon, and N. Simmons. 1996. Hospital outbreak of Klebsiella pneumoniae resistant to broad-spectrum cephalosporins and beta-lactam-beta-lactamase inhibitor combinations by hyperproduction of SHV-5 beta-lactamase. J. Clin. Microbiol. 34:358-363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Freter, R., H. Brickner, J. Fekete, M. M. Vickerman, and K. E. Carey. 1983. Survival and implantation of Escherichia coli in the intestinal tract. Infect. Immun. 39:686-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gibson, G. R., and X. Wang. 1994. Regulatory effects of bifidobacteria on the growth of other colonic bacteria. J. Appl. Bacteriol. 77:412-420. [DOI] [PubMed] [Google Scholar]
- 12.Gould, I. M. 2005. Antibiotic policies in European hospitals. Med. Mal. Infect. 35(Suppl. 2):S123-S124. [DOI] [PubMed] [Google Scholar]
- 13.Jarlier, V., M. H. Nicolas, G. Fournier, and A. Philippon. 1988. Extended broad-spectrum beta-lactamases conferring transferable resistance to newer beta-lactam agents in Enterobacteriaceae: hospital prevalence and susceptibility patterns. Rev. Infect. Dis. 10:867-878. [DOI] [PubMed] [Google Scholar]
- 14.Kuhn, I., A. Iversen, M. Finn, C. Greko, L. G. Burman, A. R. Blanch, X. Vilanova, A. Manero, H. Taylor, J. Caplin, L. Dominguez, I. A. Herrero, M. A. Moreno, and R. Mollby. 2005. Occurrence and relatedness of vancomycin-resistant enterococci in animals, humans, and the environment in different European regions. Appl. Environ. Microbiol. 71:5383-5390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leverstein-van Hall, M. A., A. C. Fluit, H. E. Blok, A. T. Box, E. D. Peters, A. J. Weersink, and J. Verhoef. 2001. Control of nosocomial multiresistant Enterobacteriaceae using a temporary restrictive antibiotic agent policy. Eur. J. Clin. Microbiol. Infect. Dis. 20:785-791. [DOI] [PubMed] [Google Scholar]
- 16.Levy, S. B. 2002. The 2000 Garrod lecture: factors impacting on the problem of antibiotic resistance. J. Antimicrob. Chemother. 49:25-30. [DOI] [PubMed] [Google Scholar]
- 17.Maisonneuve, S., M. Ouriet, and Y. Duval-Iflah. 2000. Effects of yogurt intake on plasmid transfer and colonization with transconjugants in the digestive tract of mice associated with human faecal flora. FEMS Microbiol. Ecol. 31:241-248. [DOI] [PubMed] [Google Scholar]
- 18.Maisonneuve, S., M. F. Ouriet, and Y. Duval-Iflah. 2001. Comparison of yoghurt, heat treated yoghurt, milk and lactose effects on plasmid dissemination in gnotobiotic mice. Antonie Leeuwenhoek 79:199-207. [DOI] [PubMed] [Google Scholar]
- 19.Moubareck, C., N. Bourgeois, P. Courvalin, and F. Doucet-Populaire. 2003. Multiple antibiotic resistance gene transfer from animal to human enterococci in the digestive tract of gnotobiotic mice. Antimicrob. Agents Chemother. 47:2993-2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moubareck, C., F. Doucet-Populaire, M. Hamze, Z. Daoud, and F. X. Weill. 2005. First extended-spectrum-beta-lactamase (CTX-M-15)-producing Salmonella enterica serotype Typhimurium isolate identified in Lebanon. Antimicrob. Agents Chemother. 49:864-865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moubareck, C., F. Gavini, L. Vaugien, M. J. Butel, and F. Doucet-Populaire. 2005. Antimicrobial susceptibility of bifidobacteria. J. Antimicrob. Chemother. 55:38-44. [DOI] [PubMed] [Google Scholar]
- 22.Mulvey, M. R., G. Soule, D. Boyd, W. Demczuk, and R. Ahmed. 2003. Characterization of the first extended-spectrum beta-lactamase-producing Salmonella isolate identified in Canada. J. Clin. Microbiol. 41:460-462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Netherwood, T., R. Bowden, P. Harrison, A. G. O'Donnell, D. S. Parker, and H. J. Gilbert. 1999. Gene transfer in the gastrointestinal tract. Appl. Environ. Microbiol. 65:5139-5141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parvez, S., K. A. Malik, K. S. Ah, and H. Y. Kim. 2006. Probiotics and their fermented food products are beneficial for health. J. Appl. Microbiol. 100:1171-1185. [DOI] [PubMed] [Google Scholar]
- 25.Perilli, M., E. Dell'Amico, B. Segatore, M. R. De Massis, C. Bianchi, F. Luzzaro, G. M. Rossolini, A. Toniolo, G. Nicoletti, and G. Amicosante. 2002. Molecular characterization of extended-spectrum beta-lactamases produced by nosocomial isolates of Enterobacteriaceae from an Italian nationwide survey. J. Clin. Microbiol. 40:611-614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Santosa, S., E. Farnworth, and P. J. Jones. 2006. Probiotics and their potential health claims. Nutr. Rev. 64:265-274. [DOI] [PubMed] [Google Scholar]
- 27.Schroder, G., S. Krause, E. L. Zechner, B. Traxler, H. J. Yeo, R. Lurz, G. Waksman, and E. Lanka. 2002. TraG-like proteins of DNA transfer systems and of the Helicobacter pylori type IV secretion system: inner membrane gate for exported substrates? J. Bacteriol. 184:2767-2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shah, A. A., F. Hasan, S. Ahmed, and A. Hameed. 2004. Characteristics, epidemiology and clinical importance of emerging strains of gram-negative bacilli producing extended-spectrum beta-lactamases. Res. Microbiol. 155:409-421. [DOI] [PubMed] [Google Scholar]
- 29.Sullivan, A., A. Johansson, B. Svenungsson, and C. E. Nord. 2004. Effect of Lactobacillus F19 on the emergence of antibiotic-resistant microorganisms in the intestinal microflora. J. Antimicrob. Chemother. 54:791-797. [DOI] [PubMed] [Google Scholar]
- 30.Sullivan, A., and C. E. Nord. 2005. Probiotics and gastrointestinal diseases. J. Intern. Med. 257:78-92. [DOI] [PubMed] [Google Scholar]
- 31.Szajewska, H., M. Ruszczynski, and A. Radzikowski. 2006. Probiotics in the prevention of antibiotic-associated diarrhea in children: a meta-analysis of randomized controlled trials. J. Pediatr. 149:367-372. [DOI] [PubMed] [Google Scholar]
- 32.Viljanen, P., and J. Boratynski. 1991. The susceptibility of conjugative resistance transfer in gram-negative bacteria to physicochemical and biochemical agents. FEMS Microbiol. Rev. 8:43-54. [DOI] [PubMed] [Google Scholar]
- 33.Vollaard, E. J., and H. A. Clasener. 1994. Colonization resistance. Antimicrob. Agents Chemother. 38:409-414. [DOI] [PMC free article] [PubMed] [Google Scholar]