Abstract
Escherichia coli O157:H7, a zoonotic human pathogen for which domestic cattle are a reservoir host, produces a Shiga toxin(s) (Stx) encoded by bacteriophages. Chromosomal insertion sites of these bacteriophages define three principal genotypes (clusters 1 to 3) among clinical isolates of E. coli O157:H7. Stx-encoding bacteriophage insertion site genotypes of 282 clinical and 80 bovine isolates were evaluated. A total of 268 (95.0%) of the clinical isolates, but only 41 (51.3%) of the bovine isolates, belonged to cluster 1, 2, or 3 (P < 0.001). Thirteen additional genotypes were identified in isolates from both cattle and humans (four genotypes), from only cattle (seven genotypes), or from only humans (two genotypes). Two other markers previously associated with isolates from cattle or with clinical isolates showed similar associations with genotype groups within bovine isolates; the tir allele sp-1 and the Q933W allele were under- and overrepresented, respectively, among cluster 1 to 3 genotypes. Stx-encoding bacteriophage insertion site typing demonstrated that there is broad genetic diversity of E. coli O157:H7 in the bovine reservoir and that numerous genotypes are significantly underrepresented among clinical isolates, consistent with the possibility that there is reduced virulence or transmissibility to humans of some bovine E. coli O157:H7 genotypes.
Escherichia coli O157:H7 is an important food- and waterborne zoonotic pathogen that expresses two cardinal virulence factors: the ability to produce one or more Shiga toxins (Stx) encoded by genes located in lambdoid bacteriophages (8, 33) and the ability to attach intimately to epithelial cells via expression of a pathogenicity island termed the locus of enterocyte effacement (LEE) (31).
Feng et al. described a plausible series of evolutionary events leading to the evolution of the dominant non-sorbitol-fermenting, β-glucuronidase-negative E. coli O157:H7 clade from a nontoxigenic progenitor, E. coli O55:H7 (5). Shaikh and Tarr (27) assayed clinical isolates of this dominant E. coli O157:H7 clade for the Stx-encoding bacteriophage insertion sites defined in the strains that have been sequenced (7, 24). Three principal groups of isolates sharing Stx bacteriophage insertion site genotypes were identified: isolates with an Stx2-encoding bacteriophage inserted at a location other than wrbA and with yehV occupied by a centrally truncated bacteriophage (cluster 1), isolates with an Stx2-encoding bacteriophage inserted into wrbA and with yehV occupied by a truncated bacteriophage as in cluster 1 (cluster 2), and isolates with a complete Stx1-encoding bacteriophage inserted into yehV and with an Stx2-encoding bacteriophage inserted into wrbA (cluster 3).
Association of the genetic diversity within the sorbitol-negative, β-glucuronidase-negative E. coli O157:H7 clade with prophage insertions has been demonstrated previously. Kim et al. (14) identified two major lineages within this clade using octamer-based genomic scanning and also found prophage sequences in some of the polymorphic bands detected by this method. Ohnishi et al. (22) used whole-genome PCR scanning and concluded that variation among bacteriophages is a major factor in generating genomic diversity within the O157 lineage. Comparative genomic microarray hybridization demonstrated that prophage or prophage-related elements accounted for >85% of the variably present genes in 12 E. coli isolates representing the O157:H7 evolutionary lineage (34).
Cattle are a major reservoir of E. coli O157:H7, but colonized animals typically exhibit no disease (6). E. coli O157 is ubiquitous on cattle farms, and the prevalence of cattle shedding this agent frequently is greater than 10% and can approach 100% (6). The high prevalence of this agent in cattle contrasts with the comparative rarity of human infection, despite the reportedly low infectious dose (13, 28, 32).
In this study, we characterized the bacteriophage insertion site genotypes of E. coli O157:H7 isolates from cattle and from humans and found increased genotype diversity among bovine isolates and differential representation of genotypes in these host species.
MATERIALS AND METHODS
Bacterial isolates.
Eighty E. coli O157:H7 isolates from cattle were randomly selected from isolates obtained during various Washington State University surveys between 1991 and 2004 (Table 1). All studies involving animals were performed under appropriate animal use guidelines with the approval of the WSU Institutional Animal Care and Use Committee. The random selection was stratified so that the group included 20 isolates each from four time periods (1991 to 1994, 1995 and 1996, 1997 to 2000, and 2001 to 2004), limited to single isolates from individual farms within each period. An additional set of stx1-negative, stx2-positive bovine isolates was selected to search for an unusual genotype, as described below.
TABLE 1.
Year and state or province of origin for isolates used in this study
| Year of isolation | Host | Study(ies) | No. of isolates obtained at the following locations:
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| WA | ID | OR | MT | WY | MO | KS | NE | TX | PA | AB | Total | |||
| 1984 | Human | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| 1986 | Human | 1 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 |
| 1987 | Human | 1 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4 |
| 1991 | Bovine | Va | 10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 10 |
| 1993 | Bovine | V | 5 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 7 |
| 1994 | Bovine | V | 2 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 |
| 1995 | Bovine | V | 5 | 3 | 3 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 12 |
| 1996 | Bovine | V | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 8 |
| 1997 | Human | 2 | 18 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 18 |
| Bovine | V | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | |
| 1998 | Human | 2, 3, 4 | 16 | 1 | 1 | 6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 24 |
| Bovine | V | 2 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 4 | |
| 1999 | Human | 2, 3, 4 | 38 | 1 | 1 | 25 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 66 |
| Bovine | V | 6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 6 | |
| 2000 | Human | 2, 3 | 42 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 44 |
| Bovine | V | 7 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 7 | |
| 2001 | Human | 3 | 27 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 29 |
| Bovine | V | 8 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 10 | |
| 2002 | Human | 2, 5 | 16 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 18 |
| Bovine | V | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | |
| 2003 | Human | 2, 5 | 18 | 0 | 2 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 23 |
| 2004 | Human | 2, 5 | 29 | 7 | 1 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 40 |
| Bovine | V | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 9 | 9 | |
| 2005 | Human | 2, 5 | 5 | 0 | 0 | 0 | 0 | 7 | 0 | 0 | 0 | 0 | 0 | 12 |
| Total | Human | All | 217 | 11 | 9 | 31 | 1 | 13 | 0 | 0 | 0 | 0 | 0 | 282 |
| Bovine | V | 51 | 7 | 4 | 0 | 0 | 0 | 1 | 2 | 1 | 1 | 13 | 80 | |
V indicates that isolates were obtained in various cattle studies at Washington State University.
Clinical E. coli O157:H7 isolates were obtained from five different studies (Table 1). Study 1 isolates were obtained from Seattle area patients of all ages (30). Study 2 isolates originated from children up to 10 years old in a four-state region (4, 12). Study 3 isolates originated from patients up to 21 years old at the Seattle Children's Hospital and Regional Medical Center (CHRMC) (16). Study 4 isolates originated from Montana patients of all ages (12). Study 5 isolates originated from CHRMC patients up to 21 years old. Only a single isolate from each recognized cluster of infections was included in this analysis to maximize the independence of the isolates. Outcomes (hemolytic-uremic syndrome [HUS] versus no HUS) were available for 248 patients, 51 of whom developed HUS. Each of these studies was performed in accordance with the human experimentation guidelines of the U.S. Department of Health and Human Services and with approval from one or more of the Institutional Review Boards of the CHRMC, the University of Washington, and Washington University.
The control strains used in this study included E. coli O157:H7 strains 87-14 (cluster 1) and 86-24 (cluster 2) (30), and EDL933 (= ATCC 43895; ATCC, Manassas, VA) (cluster 3), and E. coli K-12 strain INVαF′ (Invitrogen Corp., Carlsbad, CA).
Bovine isolate characterization.
Bovine isolates were tested for sorbitol fermentation and β-glucuronidase expression as described previously (18). O157 antigen expression was detected by latex agglutination (E. coli PRO O157; PL.070HD; Hardy Diagnostics, Santa Maria, CA). Multiplex PCR was used to detect eae, stx1, and stx2 in all bovine isolates (23).
For multilocus sequence typing, seven loci (aspC, clpX, fadD, icdA, lysP, mdh, and uidA) were sequenced and allele profiles were used to assign a sequence type (ST) to each bovine isolate (11, 25) using protocols available on the STEC Center website (http://www.shigatox.net/mlst).
Stx1 and Stx2 expression was detected by a gold-labeled immunosorbent assay (DuoPath Verotoxins, EMD Inc., Darmstadt, Germany) with broth culture supernatants, using carbadox (25 ng/ml; Sigma-Aldrich, St. Louis, MO) to induce Stx production. The verocytotoxicity of bovine isolates was confirmed by direct microscopic observation of Vero cell monolayers in 96-well plates 24 and 48 h after addition of 1:10 dilutions of filter-sterilized (0.45 μm; Millipore, Billerica, MA) overnight carbadox-induced broth culture supernatants in minimal essential medium (MEM) containing 5% fetal bovine serum (FBS). Total Stx production by bovine isolates was quantified by an enzyme immunoassay (Premier EHEC assay; Meridian Bioscience, Cincinnati, OH) with overnight carbadox-induced broth culture supernatants. E. coli DH5α (stx negative) was used as a negative control and E. coli O157:H7 strain EDL933 was used as a positive control for these Stx assays.
Stx-encoding bacteriophage insertion sites.
Study isolates were interrogated for Stx genes, for Stx-encoding bacteriophage-chromosome junctions, and for intact bacteriophage insertion site regions by PCR (27) (Table 2). The presence of intact chromosomal insertion site regions (yehV and wrbA) was confirmed in all isolates with negative reactions for Stx-encoding bacteriophage-chromosome junctions. Selected bovine isolates with variant patterns of bacteriophage insertion in yehV (right junction negative and left junction positive) were subjected to amplification of a longer segment of the variant right junction by long PCR (Platinum Taq High Fidelity DNA polymerase [Invitrogen]; 35 cycles of 94°C for 30 s, 58°C for 30 s, and 68°C for 5 min). Cloning (TA cloning kit; Invitrogen) and partial DNA sequencing (Amplicon Express, Pullman, WA) of the resulting product allowed us to design a new reverse primer (E2) (Table 2) specific for the variant right yehV-bacteriophage junction. An additional group of 128 stx2-positive, stx1-negative bovine isolates was subjected to PCR analysis of Stx-encoding bacteriophage insertion sites to screen for the cluster 2 genotype, which occurs in a minority of human clinical isolates (27) and was absent from the initial group of 80 randomly selected bovine isolates.
TABLE 2.
Nucleotide sequences of primers used for PCR analyses in this study
| Primer | Sequence (5′-3′) | Target | Reference(s) |
|---|---|---|---|
| A | AAGTGGCGTTGCTTTGTGAT | yehV insertion site region | 27 |
| B | AACAGATGTGTGGTGAGTGTCTG | ||
| A | AAGTGGCGTTGCTTTGTGAT | Bacteriophage-yehV right junction | 27 |
| E | GATGCACAATAGGCACTACGC | ||
| F | CACCGGAAGGACAATTCATC | Bacteriophage-yehV left junction | 27 |
| B | AACAGATGTGTGGTGAGTGTCTG | ||
| C | AGGAAGGTACGCATTTGACC | wrbA insertion site region | 27 |
| D | CGAATCGCTACGGAATAGAGA | ||
| C | AGGAAGGTACGCATTTGACC | Bacteriophage-wrbA right junction | 27 |
| G | ATCGTTCGCAAGAATCACAA | ||
| H | CCGACCTTTGTACGGATGTAA | Bacteriophage-wrbA left junction | 27 |
| D | CGAATCGCTACGGAATAGAGA | ||
| A | AAGTGGCGTTGCTTTGTGAT | Variant bacteriophage-yehV right junction | 27; this study |
| E2 | CAGGGAATCAATCGCAGTTT | ||
| 595 | CCGAAGAAAAACCCAGTAACAG | stx2-antiterminator Q gene junction | 33, 17 |
| Q933 | CGGAGGGGATTGTTGAAGGC | ||
| TirF | TCATGTTCAGCCGTTTATCG | tir | This study |
| TirR | AAGCTCAAGAGTTGCCCATC | ||
| A | AAGTGGCGTTGCTTTGTGAT | Bacteriophage-yehV right junction (∼8 kb) | 27; this study |
| R1 | GCAGATGATGAGCGTGATTG | ||
| FimH460F | CTTATGGCGGCGTGTTATCTA | FimH mannose-binding pocket flanking N135K | Shaikh et al., submitted |
| FimH460R | GCCAGTAGGCACTACCACATC | ||
| stx2c-F919 | CTGAACAGAAAGTCACAGTYTTTA | stx2c-specific region | This study |
| stx2c-R1100 | GGCCACTTTTACTGTGAATGTATC | ||
| O-F163 | ATGCGCAAGACATACGGATTC | Stx2c bacteriophage O gene | This study |
| O-R695 | TGCACAAACGCCCTGACATA | ||
| Q-F69 | GGGCGCATGGGTTTATTCA | Stx2c bacteriophage Q gene | This study |
| Q-R389 | ACTTCCCGTCGGCAGGTTG | ||
| mdh-F41 | AGGCGCTTGCACTACTGTTA | mdh | This study |
| mdh-R875 | AGCGCGTTCTGTTCAAATG |
Detection of additional genetic markers in bovine isolates.
A multiplex PCR was designed to specifically detect the Stx2c-encoding bacteriophage (29). In the multiplex reaction we used 0.5 μl of 10 μM primers stx2c-F, stx2c-R, O-F, O-R, Q-F, and Q-R (Table 2) to amplify stx2c and the O and Q genes of the Stx2c-encoding bacteriophage, as well as primers to amplify the malate dehydrogenase (mdh) gene as a positive PCR control. Each 25-μl reaction mixture contained 2.5 μl of 10× buffer II (Applied Biosystems, Foster City, CA), 2.5 μl of 2 mM deoxynucleoside triphosphates, 2.0 μl of 25 mM MgCl2, the six Stx2c-encoding bacteriophage-specific primers listed above, 1 μl of each of the two mdh primers, 0.3 μl of 1.5 U AmpliTaq Gold (Applied Biosystems), 1 μl of a 25-ng/μl genomic DNA template, and 11.7 μl of double-distilled H2O. The amplification cycle included an initial denaturation step (94°C for 10 min), 35 amplification cycles (92°C for 1 min, 55°C for 1 min, and 72°C for 30 s), and a final extension step (72°C for 5 min). PCR products were visualized on ethidium bromide-stained 1.5% agarose gels by illumination with UV light. The presence of the Stx2c-encoding bacteriophage was inferred in isolates that produced the following four amplicons: mdh (835 bp), stx2c (182 bp), O (533 bp), and Q (321 bp).
The sp-1 polymorphism (D78E) in tir was detected by a real-time PCR TaqMan assay (J. L. Bono, J. E. Keen, M. L. Clawson, L. M. Durso, M. P. Heaton, and W. W. Laegreid, Abstr. 104th Gen. Meet. Am. Soc. Microbiol., abstr. C-388, 2005) or by sequencing a tir PCR product (Table 2). The junction between the antiterminator Q of bacteriophage 933W and stx2 was detected by PCR (Table 2) (17). An N135K polymorphism in fimH was detected by sequencing PCR products (Table 2) (N. Shaikh, N. J. Holt, J. R. Johnson, and P. I. Tarr, submitted for publication).
Adherence to MDBK cells was quantified as described by Monteville et al. (21). A three-color immunofluorescence analysis of the E. coli O157:H7 interaction with MDBK cells was performed using strains transformed with a green fluorescent protein (GFP) gene-containing plasmid, pMEK91 (20). MDBK cells (5 × 104 cells/well) were cultured on glass coverslips for 18 h at 37°C in a humidified incubator containing 5% CO2. The cells were infected with a 10:l dilution of a log-phase culture of GFP-transformed E. coli in 1 ml of Tris (10 mM, pH 7.4)-buffered minimal essential medium supplemented with 1% (vol/vol) FBS. Infected cells were incubated (3 h), rinsed with MEM, and reincubated in MEM containing FBS (4 h). Cells were washed three times with phosphate-buffered saline, fixed (2% paraformaldehyde), and permeabilized (0.1% Triton X-100). Actin was stained using tetramethylrhodamine isothiocyanate-labeled phalloidin (0.2 μg/ml; Sigma, St. Louis, MO). Coverslips were mounted using Vectashield (Vector Laboratories Inc., Burlingame, CA) with 4′,6-diamidino-2-phenylindole (DAPI) and were visualized using epifluorescence (Nikon Eclipse TE2000). Images were captured with the MetaMorph version 5 imaging software and were formatted using Adobe Photoshop 3.0.4.
Statistical analysis.
The distribution of Stx-encoding bacteriophage insertion site genotypes in human and cattle isolates and the associations between the fimH, tir sp-1, Q933, and stx2c markers and the Stx-encoding bacteriophage insertion site genotypes in isolates from cattle were evaluated by χ2 tests (SigmaStat; SPSS, Inc., Chicago, IL). Differences in Stx production and differences in adherence of selected E. coli O157:H7 isolates to MDBK cells were evaluated by one-way analysis of variance (SigmaStat), and where differences were detected, pairwise multiple comparisons were performed using the Tukey test.
RESULTS
Identification and characterization of bovine isolates.
All bovine isolates expressed the O157 antigen, were positive for eae and tir, and fermented lactose but not sorbitol; none expressed β-glucuronidase. All but one isolate were positive for stx1 or stx2 or both (Table 3). Multilocus sequence typing assigned 79 of the 80 isolates to ST-66, the most common ST of the β-glucuronidase-negative, non-sorbitol-fermenting clade of E. coli O157:H7 in the United States, consistent with their membership in this clade. The one remaining isolate was assigned to ST-350, which differs from ST-66 by a single nucleotide in fadD.
TABLE 3.
Genotyping data, isolate sources, and further characterization of bovine isolates of E. coli O157:H7
| Genotypea | Genotyping data
|
No. (%) of isolates from:
|
Additional markers (bovine isolates only)
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| stxb | yehVc | wrbAd | Cattle | Clinical sources | fim (N135K)e | Q933W-stx2f | stx2cg | tir sp-1 (D78E)h | |
| Cluster 1 | 2 | Occupied | Intact | 8 (10) | 85 (30.1) | N (5) | 7 | 1 | D (7), E (1) |
| Cluster 2 | 2 | Occupied | Occupied | 0 (0) | 7 (2.5) | ||||
| Cluster 3 | 1 + 2 | Occupied | Occupied | 33 (41.3) | 176 (62.4) | K (5) | 33 | 1 | D (32), E (1) |
| Genotype 4 | 2 | Intact | Occupied | 1 (1.3) | 5 (1.8) | K (1) | 1 | 0 | D (1) |
| Genotype 5 | 2 | Variant-R | Intact | 9 (11.3) | 4 (1.4) | N (5) | 1 | 8 | D (3), E (6) |
| Genotype 6 | 1 + 2 | Variant-R | Intact | 15 (18.8) | 2 (0.7) | N (5) | 10 | 13 | D (10), E (5) |
| Genotype 7 | 1 + 2 | Variant-R | Occupied | 5 (6.3) | 1 (0.3) | K (5) | 5 | 0 | D (5) |
| Genotype 8 | 1 | Occupied | Intact | 0 (0) | 1 (0.3) | ||||
| Genotype 9 | None | Intact | Intact | 0 (0) | 1 (0.3) | ||||
| Genotype 10 | None | Occupied | Intact | 1 (1.3) | 0 (0) | N (1) | 0 | 0 | D (1) |
| Genotype 11 | 2 | Intact | Intact | 2 (2.5) | 0 (0) | N (2) | 1 | 0 | D (1) |
| Genotype 12 | 2 | Variant-R | Occupied | 1 (1.3) | 0 (0) | K (1) | 1 | 0 | D (1) |
| Genotype 13 | 1 | Variant-R | Intact | 1 (1.3) | 0 (0) | K (1) | 1 | 0 | D (1) |
| Genotype 14 | 1 | Variant-R | Occupied | 2 (2.5) | 0 (0) | K (2) | 0 | 0 | D (2) |
| Genotype 15 | 1 + 2 | Intact | Intact | 1 (1.3) | 0 (0) | N (1) | 1 | 1 | D (1) |
| Genotype 16 | 1 + 2 | Occupied | Intact | 1 (1.3) | 0 (0) | N (1) | 1 | 0 | D (1) |
Stx-encoding bacteriophage insertion site genotypes. The cluster genotypes are genotypes identified in multiple clinical isolates in a previous study (27). The genotypes are based on patterns of Stx genes and Stx-encoding bacteriophage insertion sites described in this study.
PCR gene detection of stx1 (1) and/or stx2 (2) (23).
“Occupied” indicates bilateral detection of the yehV-Stx-1-encoding bacteriophage junctions (with the A-E and B-F primer pairs, respectively [Table 2]). “Intact” indicates detection of neither junction. “Variant-R” indicates that the left junction between the bacteriophage and the chromosome was detected but the right junction was not detected. Most yehV-Variant-R isolates share a different right junction sequence, as described in Results.
“Occupied” indicates bilateral detection of the wrbA-Stx-2-encoding bacteriophage junctions (with the C-G and D-H primer pairs [Table 2]). “Intact” indicates detection of neither junction.
N and K indicate the status of the N135K polymorphism (N. Shaikh et al., submitted for publication) in the FimH mannose-binding pocket in bovine isolates. All isolates were tested for genotype groups containing less than six isolates; five isolates were randomly selected for testing from each larger genotype group.
The numbers indicate the numbers of bovine isolates that were PCR positive for the Q-stx2 junction of phage 933W.
The numbers indicate the numbers of bovine isolates positive as determined by multiplex PCR for the Stx2c-encoding bacteriophage.
The numbers in parentheses indicate the numbers of bovine isolates in which the alleles of the D78E (T238A) single-nucleotide polymorphism in tir were detected.
Stx-encoding bacteriophage insertion sites.
Clinical isolates of E. coli O157:H7 (Table 1) were analyzed to determine their Stx-encoding bacteriophage insertion genotypes. Clusters 1 to 3, as defined previously, accounted for 268 (95%) of the 282 isolates (Table 3). Six additional, less frequent genotypes were identified among the clinical isolates. Genotype 4 (n = 5; 1.8%) was characterized by the presence of stx2, the absence of stx1, an intact yehV site, and a bacteriophage-occupied wrbA site; genotype 5 (n = 4; 1.4%) was characterized by the presence of stx2, the absence of stx1, amplification of the left but not the right yehV-bacteriophage junction (designated yehV-Variant-R), and an intact wrbA site; genotype 6 (n = 2; 0.7%) was characterized by the presence of both stx1 and stx2, yehV-Variant-R, and an intact wrbA site; genotype 7 (n = 1; 0.4%) was characterized by the presence of both stx1 and stx2, yehV-Variant-R, and a bacteriophage-occupied wrbA site; genotype 8 (n = 1; 0.4%) was characterized by the presence of stx1, the absence of stx2, a bacteriophage-occupied yehV site, and an intact wrbA site; and genotype 9 (n = 1; 0.4%) was characterized by the absence of stx1 and stx2 and intact yehV and wrbA sites (Table 3).
In contrast, the cluster 1 to 3 genotypes were found much less frequently among the bovine isolates (n = 41; 51.3%) (Table 3). The distribution of clusters 1 to 3 and “other” genotypes (all remaining genotypes) differed significantly in the clinical and bovine isolates (χ2 = 98.2, 3 df, P < 0.001; including no more than a single isolate per farm with the same genotype). One bovine isolate had genotype 4. Genotypes 5 (n = 9; 11.3%), 6 (n = 15; 18.8%), and 7 (n = 5; 6.3%) were each overrepresented among cattle isolates compared to the clinical isolates (in each case, χ2 > 9.8, 1 df, and P < 0.01). The remaining nine bovine isolates had seven additional Stx-encoding bacteriophage insertion site genotypes that were not detected among the clinical isolates. These genotypes, designated genotypes 10 to 16, each were found in only one or two strains; the specific results are shown in Table 3. Because the cluster 2 genotype was not detected among the randomly chosen set of 80 bovine isolates, 124 additional bovine isolates previously determined to have the appropriate stx1-negative, stx2-positive profile were screened for the cluster 2 genotype, and four of these strains (3.2%) had cluster 2 genotypes, confirming the presence of cluster 2 isolates in the bovine reservoir, albeit at a low frequency.
The yehV-Variant-R bacteriophage junction was characterized further in view of the relatively high frequency of genotypes with this reaction, particularly among bovine isolates. Considering the possibility that the region complementary to primer E (Table 2) had been deleted or disrupted, we attempted to amplify a larger segment of the right bacteriophage-yehV junction using yehV primer A (Table 2) paired with a primer designed to amplify approximately 7.5 kb of the right yehV-stx1 bacteriophage junction. This long PCR amplified a 7.5-kb product from the template of the positive control strain (EDL933; cluster 3); in contrast, it amplified 8-kb products from the templates of two yehV-Variant-R isolates (genotype groups 5 and 6). This PCR product was cloned, and a partial 5′ sequence was obtained. The initial 362 bp of this sequence was 100% identical to the yehV-stx1 prophage junction of enterohemorrhagic E. coli Sakai, but the next 738 bp was most similar (>96% identity) to E. coli O157:H7 strain Sakai genes ECs2153 and ECs2154, located adjacent to another cryptic prophage in the Sakai genome (GenBank accession no. EF081034). This sequence was used to design a new reverse primer, E2 (Table 1), which, combined with yehV forward primer A, specifically amplified the yehV-Variant-R junction sequence from 23 of 30 (76.7%) bovine yehV-Variant-R isolates. All bovine isolates in which yehV sites were intact (n = 13) or occupied (n = 10) were negative as determined by this PCR.
Stx and LEE effector expression by bovine E. coli O157:H7 isolates.
All bovine isolates in which one or more stx genes were detected produced verocytotoxigenic culture supernatants; the single stx-negative isolate (genotype 10) was nontoxic. The Stx expression detected by the gold-labeled immunosorbent assay agreed exactly with the stx detected by PCR for 76 of the 80 bovine isolates. The four exceptions included one genotype 5 isolate and two genotype 6 isolates that did not express detectable Stx2 and one genotype 16 isolate that did not express detectable Stx1. Five randomly selected bovine isolates from genotype groups with six or more isolates and all isolates from genotype groups with fewer members were evaluated for Stx production by an enzyme immunoassay (Table 4); significant differences in Stx production in different genotype groups were detected (P = 0.019, as determined by one-way analysis of variance). The Tukey multiple-comparison procedure indicated that the cluster 3 isolates tested produced significantly more Stx than the genotype 5 isolates tested produced; no other statistically significant differences in Stx production in different genotype groups were identified.
TABLE 4.
Stx production by and cytoadherence of E. coli O157:H7 genotypes isolated from cattle
| Genotype | Stx productiona
|
Cytoadherenceb
|
||
|---|---|---|---|---|
| No. of isolates | Mean optical density (SD) | No. of isolates | Mean adherence (SD) | |
| Cluster 1 | 5 | 1.61 (0.88) | 2 | 39.7 (31.7) |
| Cluster 3 | 5 | 1.73 (0.93) | 2 | 9.8 (7.7) |
| Genotype 4 | 1 | 1.57 | NDc | |
| Genotype 5 | 5 | 0.24 (0.27) | 3 | 27.7 (19.6) |
| Genotype 6 | 5 | 0.57 (0.54) | 3 | 55.3 (82.7) |
| Genotype 7 | 5 | 1.53 (1.02) | 2 | 11.3 (6.4) |
| Genotype 8 | 1 | 0.08 | ND | |
| Genotype 9 | 2 | 1.87 (0.10) | ND | |
| Genotype 10 | 1 | 1.66 | ND | |
| Genotype 11 | 1 | 0.18 | ND | |
| Genotype 12 | 2 | 0.27 (0.04) | ND | |
| Genotype 13 | 1 | 0.31 | ND | |
| Genotype 14 | 1 | 1.00 | ND | |
Stx production as measured by an enzyme immunoassay using overnight broth cultures of E. coli O157:H7 isolates with bacteriophage induction by carbadox (25 ng/ml).
Cytoadherence to MDBK cells. The results are fold increases in the number of adherent CFU per well compared to the E. coli INVαF′ control strain.
ND, not determined.
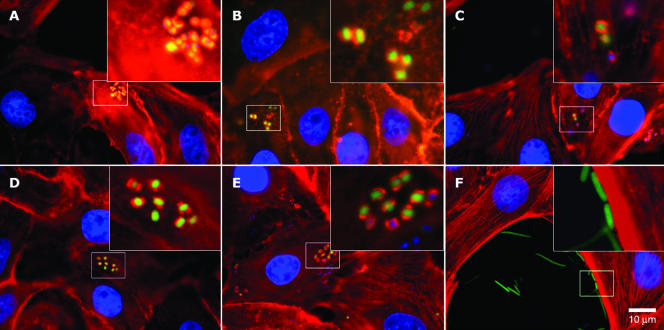
Randomly selected bovine isolates with the cluster 1 or cluster 3 genotype or genotype 5, 6, or 7 all exhibited comparable quantitative adherence to MDBK cells (Table 4). Similarly, randomly selected bovine isolates with the cluster 1, cluster 2, or cluster 3 genotype or genotype 5 or 6 all produced qualitatively similar adherent E. coli O157:H7 microcolonies and focal actin accumulation on MDBK cells (Fig. 1).
FIG. 1.
Actin polymerization associated with E. coli O157:H7 microcolonies adhering to MDBK cells: three-color immunofluorescence analysis of E. coli O157:H7 interaction with MDBK cells. Blue, DAPI staining of nuclear DNA; red, tetramethylrhodamine isothiocyanate-conjugated phalloidin staining of actin; green, GFP plasmid-transformed E. coli cells. (A) E. coli O157:H7 strain EDL933 (cluster 3; positive control); (B) E. coli O157:H7 strain 86-24 (cluster 2; positive control); (C) E. coli O157:H7 bovine isolate (cluster 1); (D) E. coli O157:H7 genotype 5 bovine isolate; (E) E. coli O157:H7 genotype 6 bovine isolate; (F) E. coli K-12 strain InvαF′ (negative control).
Geographic and temporal distributions of E. coli O157:H7 genotypes.
The bovine isolates originated from 69 different cattle farms in seven states in the United States and one Canadian province (Table 1). Neither secular nor geographic trends were observed in the distribution of Stx-encoding bacteriophage insertion site genotypes (data not shown). Twenty-five of the bovine isolates were obtained from predominantly beef breeds and 55 were obtained from dairy breeds, and there were not significant differences in genotype distribution between the breeds. Eight farms contributed more than one study isolate. Multiple isolates obtained during different time periods from single farms tended to share Stx-encoding bacteriophage insertion site genotypes; six farms contributed isolates with the same genotype on two (four farms) or three (two farms) occasions separated by 1 to 8 years. The genotypes repeatedly isolated from these single farms included cluster 1 (one farm) and 3 (three farms) genotypes and genotypes 5 and 11 (one farm each). Two isolates obtained 2 years apart from a seventh farm had genotype 6, while a third isolate from that farm obtained 4 years later was a cluster 3 strain. Finally, two isolates obtained 2 years apart on the eighth farm had the cluster 3 genotype and genotype 6.
Human isolates were recovered from patients in six states in the United States in two time periods, 1984 to 1987 and 1997 to 2005. Neither secular nor geographic trends were observed in the distribution of the Stx-encoding bacteriophage insertion site genotypes (data not shown).
Stx2c bacteriophage.
The Stx2c-encoding bacteriophage, as inferred by PCR detection of stx2c and specific O and Q gene sequences, was detected in 24 of the 80 bovine isolates, including cluster 1 and 3 genotype and genotype 5, 6, and 15 isolates (Table 3). stx2c occurred less frequently among bovine isolates with cluster 1 and 3 genotypes (2 of 41 isolates) than in bovine isolates with other genotypes (22 of 39 isolates; χ2 = 22.9, 1 df, P < 0.001) (Table 3). In the context of human disease, while E. coli O157:H7 with genes encoding Stx2c is unusual, it may be associated with more severe illness (12).
fimH mannose binding pocket polymorphism.
Representative bovine isolates (all isolates in genotype groups with less than five isolates and five randomly chosen isolates each for all other genotype groups) were examined for the presence of a single-nucleotide polymorphism in fimH resulting in an N135K amino acid sequence change in the mature FimH protein. Fourteen isolates with wrbA occupied by bacteriophage all carried the K allele, while 20 of 21 isolates with wrbA intact carried the N allele (χ2 = 27.3, 1 df, P < 0.001) (Table 3). The segment of wrbA amplified by primers C and D (Table 2) in the single wrbA-intact, K allele-positive isolate was identical to the segment of the same region in E. coli K-12 wrbA.
Q933W and tir polymorphic markers.
The bacteriophage Q933W-stx2 junction was detected by PCR in 40 of 41 cattle isolates with the cluster 1 or 3 genotype but in only 21 of 39 isolates having other genotypes (χ2 = 18.7, 1 df, P < 0.001). The T238A (D78E) tir polymorphism designated sp-1 was detected in only 2 of 41 cattle isolates with the cluster 1 or 3 genotype but in 11 of 38 isolates with other genotypes, particularly genotypes 5 (6 of 9 isolates) and 6 (5 of 15 isolates) (χ2 = 10.7, 1 df, P < 0.001).
DISCUSSION
Our typing system revealed a diversity of Stx-encoding bacteriophage insertion site genotypes among E. coli O157:H7 from the bovine reservoir that was considerably broader than that expected on the basis of analysis of human isolates. Some of the genotypes detected among isolates from both species were markedly overrepresented in clinical isolates (clusters 1 and 3) or bovine isolates (genotypes 5 to 7). All genotypes represented by two or more clinical isolates (cluster 1 to 3 genotypes and genotypes 4 to 6) were identified in isolates from cattle (although discovery of a bovine cluster 2 strain required screening of additional isolates), consistent with the hypothesis that cattle are the reservoir for E. coli O157:H7 resulting in human disease. Most (10/13) of the genotype groups identified for the first time in this study had only one or two members, indicating that identification of additional diversity is likely to occur when more isolates, particularly isolates from the bovine reservoir, are analyzed.
We considered the possibility that lysogenization and excision events in the short term, such as during a single human or animal gastrointestinal passage, could result in changes in the Stx-encoding bacteriophage insertion site genotypes of E. coli O157:H7 characterized in this study. However, the available evidence generally does not support this possibility. First, previous studies of non-sorbitol-fermenting, β-glucuronidase-negative E. coli O157:H7 suggested that the acquisition of Stx-encoding phages was a key event in the evolution of this pathogenic clade from precursor strains and thus predated the divergence detected on the basis of these genotypes within the clade (5, 34). Consistent with this view, we demonstrated here the correlation between wrbA site occupancy and fimH (a nonprophage gene) single-nucleotide polymorphism and also the correlation between stx2c carriage and genotypes 5 and 6 (defined by insertion sites of other phages). Both of these correlations are inconsistent with highly dynamic phage movement. Second, we observed stability of cluster 1 and cluster 3 genotypes among multiple isolates from several farms over multiple years, which is inconsistent with the possibility that the phage exposure in bovine environments results in rapid conversion to “bovine” phage insertion site genotypes. More generally, pulsed-field gel electrophoresis profiles, which would be expected to be highly sensitive to genetic changes at the scale of bacteriophage insertions or excisions, have been empirically observed to be well conserved among epidemiologically related isolates of β-glucuronidase-negative E. coli O157:H7 (1, 2, 32). In contrast, in a different lineage of enterohemorrhagic E. coli O157, sorbitol-fermenting E. coli O157:NM, the pulsed-field gel electrophoresis patterns are less stable because of rapid bacteriophage recombination activity (3, 19). It will be important to specifically evaluate the stability of diverse Stx-encoding bacteriophage insertion site genotypes in future studies.
The heterogeneity of bovine E. coli O157:H7 genotypes reported here has implications for pathophysiology, evolutionary genetics, and food safety. First, two key virulence factors, the production of Stx and the ability to colonize the colonic surface through the functions of LEE-encoded products, are believed to be central to the pathophysiology of E. coli O157:H7-induced disease. Most isolates, including those with genotypes underrepresented among clinical isolates, nevertheless possessed both of these factors. Thus, it is possible either that these isolates differ in the level of expression of Stx or LEE-encoded products (as was demonstrated here with reduced Stx production for genotype 5 compared to the cluster 3 genotype) or that they lack another, currently unrecognized virulence factor(s). Therefore, identification of the genetic basis underlying the nonrandom distribution of these genotypes could expand our understanding of the pathophysiology of this important human pathogen. This could apply even more broadly to Stx-producing E. coli isolates belonging to other serotypes. Of the more than 250 serotypes of Stx-producing E. coli detected in the bovine reservoir, fewer than 100 have been associated with human illnesses (9, 10), and only a few serotypes cause most human infections. It is possible that some of the same genetic differences that account for the distribution of nonclinical genotypes of E. coli O157:H7 also account for the distribution of nonclinical Stx-producing E. coli serotypes.
Second, these findings shed additional light on the recent evolution of E. coli O157:H7. At any given time, the vast majority of E. coli O157:H7 isolates reside in the animal reservoirs and not in humans, but studies of the evolution of E. coli O157:H7 have focused on clinical isolates. Shaikh and Tarr proposed an evolutionary lineage in which evolving E. coli O157:H7 sequentially acquired Stx-encoding bacteriophages in specific chromosomal locations (27). It is possible that some of the genotypes identified here represent additional steps in this lineage. For example, genotype 9 has the fimH sequence, yehV and wrbA occupancy, and Stx profile expected of an immediate ancestor of the cluster 1 genotype. However, most of the new genotypes reported here appear to represent evolutionary offshoots within the β-glucuronidase-negative E. coli O157:H7 clade. For example, genotype 8 could represent a single excision of the Stx2-encoding bacteriophages from a cluster 3 E. coli O157:H7 strain. Additional genetic information is required in order to place such offshoots into a plausible phylogeny.
Third, these findings should prompt additional epidemiologic studies of E. coli O157:H7 in the animal reservoir, in the environment, and in contaminated food products in order to determine whether the cluster 1 to 3 genotypes differ from other genotypes in terms of the distribution among potential sources of human infection. It is also possible that regulatory attention should be focused more on the subset of E. coli O157:H7 genotypes overrepresented among clinical isolates.
Kim et al. (14) first reported differential distribution of E. coli O157:H7 lineages among isolates from cattle and humans that was consistent with differences in human infectivity or pathogenicity in certain isolates. These investigators used octamer-based scanning to identify two E. coli O157:H7 lineages, one of which more frequently contained human clinical isolates and the other of which more frequently contained bovine isolates. Interestingly, much of the variation seen with octamer-based scanning was associated with bacteriophage-related sequences. The results of subsequent investigations of a larger and more diverse panel of isolates using simplified techniques supported the hypothesis that there is nonrandom distribution of these lineages in cattle and human hosts but that the degree is considerably lower than that originally proposed (15, 35). The correlation, if any, between the lineages reported in these previous studies and the genotypes reported here is not known.
Roe et al. (26) reported differences between human clinical and bovine isolates in the proportion of cells expressing LEE-encoded EspA filaments, with human isolates having higher proportions of filament-expressing cells. The possibility that EspA expression varies between genotypes found in both bovine and clinical isolates (cluster 1 to 3 genotypes and genotype 4) and genotypes found only in bovine isolates has not been evaluated to date. However, data for in vitro cytoadherence and actin accumulation (Fig. 1 and Table 4) do not indicate that there are large intrinsic differences between these genotype groups in terms of LEE expression.
Our Stx-encoding bacteriophage insertion site data correlate with two other markers previously reported to be nonrandomly distributed among E. coli O157:H7 strains from cattle and humans. The stx2-Q933W antiterminator gene junction was more common in human isolates than in cattle isolates (17). The D78E tir polymorphism was more common in cattle isolates than in human isolates (Bono et al., Abstr. 104th Gen. Meet. Am. Soc. Microbiol.). Here, we show that these markers are nonrandomly distributed among E. coli O157:H7 genotypes in our set of cattle isolates and that their distribution is consistent with the frequency at which these genotypes occur in human disease isolates. Q933W occurred in all but one bovine cluster 1 to 3 genotype or genotype 4 isolate but significantly less frequently in isolates with other genotypes. The tir polymorphism sp-1 was significantly less common in bovine cluster 1 to 3 genotype or genotype 4 isolates than in isolates with other genotypes. These observations suggest that genotypes with diverse potentials for human disease distinguishable by their Stx-encoding bacteriophage insertion site genotypes coexist in bovine populations.
In summary, the diversity within the non-sorbitol-fermenting, β-glucuronidase-negative E. coli O157:H7 clade is underestimated by evaluation of clinical isolates alone. This diversity is more fully demonstrated by animal (bovine) isolates, and it appears that no single linear phylogeny could encompass this diversity. Studies of broader panels of isolates from cattle and perhaps other animal species are necessary to confirm the distribution of E. coli O157:H7 Stx-encoding bacteriophage insertion site genotypes in cattle and to understand the basis for their different occurrence in clinical isolates.
Acknowledgments
This project was funded in part by the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, under contracts N01-AI-30055 (WSU) and N01-AI-30058 (MSU) from USDA-NRICGP 0102147 and from the Agriculture Animal Health Program, College of Veterinary Medicine, Washington State University, Pullman, WA. Work in the lab of M.E.K. was supported by grant DK58911 from NIH. Some bovine isolates resulted from previous studies funded by the Washington State Beef Commission and by the International Life Sciences Institute Food Safety Committee. Collection of clinical strains in study 2 was supported by grant DK52081 from NIH, and collection of strains in studies 3 and 4 was supported by CDC cooperative agreements U50/CCU015040 and U50/CCU814408, respectively., Study 5 was supported by grant 2002-35200-02238 from the National Research Initiative of the USDA Cooperative State Research, Education and Extension Service.
We thank David Lacher (MSU) for designing and testing the Stx2c phage-specific multiplex.
Footnotes
Published ahead of print on 1 December 2006.
REFERENCES
- 1.Besser, T. E., D. D. Hancock, L. C. Pritchett, E. M. McRae, D. H. Rice, and P. I. Tarr. 1997. Duration of detection of fecal excretion of Escherichia coli O157:H7 in cattle. J. Infect. Dis. 175:726-729. [DOI] [PubMed] [Google Scholar]
- 2.Besser, T. E., B. L. Richards, D. H. Rice, and D. D. Hancock. 2001. Escherichia coli O157:H7 infection of calves: infectious dose and direct contact transmission. Epidemiol. Infect. 127:555-560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bielaszewska, M., R. Prager, W. Zhang, A. W. Friedrich, A. Mellmann, H. Tschape, and H. Karch. 2006. Chromosomal dynamism in progeny of outbreak-related sorbitol-fermenting enterohemorrhagic Escherichia coli O157:NM. Appl. Environ. Microbiol. 72:1900-1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cornick, N. A., S. Jelacic, M. A. Ciol, and P. I. Tarr. 2002. Escherichia coli O157:H7 infections: discordance between filterable fecal Shiga toxin and disease outcome. J. Infect. Dis. 186:57-63. [DOI] [PubMed] [Google Scholar]
- 5.Feng, P., K. A. Lampel, H. Karch, and T. S. Whittam. 1998. Genotypic and phenotypic changes in the emergence of Escherichia coli O157:H7. J. Infect. Dis. 177:1750-1753. [DOI] [PubMed] [Google Scholar]
- 6.Hancock, D. D., T. E. Besser, J. T. Lejeune, M. A. Davis, and D. H. Rice. 2001. The control of VTEC in the animal reservoir. Int. J. Food Microbiol. 66:71-78. [DOI] [PubMed] [Google Scholar]
- 7.Hayashi, T., K. Makino, M. Ohnishi, K. Kurokawa, K. Ishii, K. Yokoyama, C. G. Han, E. Ohtsubo, K. Nakayama, T. Murata, M. Tanaka, T. Tobe, T. Iida, H. Takami, T. Honda, C. Sasakawa, N. Ogasawara, T. Yasunaga, S. Kuhara, T. Shiba, M. Hattori, and H. Shinagawa. 2001. Complete genome sequence of enterohemorrhagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res. 8:11-22. [DOI] [PubMed] [Google Scholar]
- 8.Huang, A., J. Friesen, and J. L. Brunton. 1987. Characterization of a bacteriophage that carries the genes for production of Shiga-like toxin 1 in Escherichia coli. J. Bacteriol. 169:4308-4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hussein, H. S., and L. M. Bollinger. 2005. Prevalence of Shiga toxin-producing Escherichia coli in beef cattle. J. Food Prot. 68:2224-2241. [DOI] [PubMed] [Google Scholar]
- 10.Hussein, H. S., and T. Sakuma. 2005. Prevalence of Shiga toxin-producing Escherichia coli in dairy cattle and their products. J. Dairy Sci. 88:450-465. [DOI] [PubMed] [Google Scholar]
- 11.Hyma, K. E., D. W. Lacher, A. M. Nelson, A. C. Bumbaugh, J. M. Janda, N. A. Strockbine, V. B. Young, and T. S. Whittam. 2005. Evolutionary genetics of a new pathogenic Escherichia species: Escherichia albertii and related Shigella boydii strains. J. Bacteriol. 187:619-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jelacic, J. K., T. Damrow, G. S. Chen, S. Jelacic, M. Bielaszewska, M. Ciol, H. M. Carvalho, A. R. Melton-Celsa, A. D. O'Brien, and P. I. Tarr. 2003. Shiga toxin-producing Escherichia coli in Montana: bacterial genotypes and clinical profiles. J. Infect. Dis. 188:719-729. [DOI] [PubMed] [Google Scholar]
- 13.Keene, W. E., J. M. McAnulty, F. C. Hoesly, L. P. Williams, Jr., K. Hedberg, G. L. Oxman, T. J. Barrett, M. A. Pfaller, and D. W. Fleming. 1994. A swimming-associated outbreak of hemorrhagic colitis caused by Escherichia coli O157:H7 and Shigella sonnei. N. Engl. J. Med. 331:579-584. [DOI] [PubMed] [Google Scholar]
- 14.Kim, J., J. Nietfeldt, and A. K. Benson. 1999. Octamer-based genome scanning distinguishes a unique subpopulation of Escherichia coli O157:H7 strains in cattle. Proc. Natl. Acad. Sci. USA 96:13288-13293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim, J., J. Nietfeldt, J. Ju, J. Wise, N. Fegan, P. Desmarchelier, and A. K. Benson. 2001. Ancestral divergence, genome diversification, and phylogeographic variation in subpopulations of sorbitol-negative, beta-glucuronidase-negative enterohemorrhagic Escherichia coli O157. J. Bacteriol. 183:6885-6897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klein, E. J., J. R. Stapp, C. R. Clausen, D. R. Boster, J. G. Wells, X. Qin, D. L. Swerdlow, and P. I. Tarr. 2002. Shiga toxin-producing Escherichia coli in children with diarrhea: a prospective point-of-care study. J. Pediatr. 141:172-177. [DOI] [PubMed] [Google Scholar]
- 17.Lejeune, J. T., S. T. Abedon, K. Takemura, N. P. Christie, and S. Sreevatsan. 2004. Human Escherichia coli O157:H7 genetic marker in isolates of bovine origin. Emerg. Infect. Dis. 10:1482-1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.LeJeune, J. T., T. E. Besser, D. H. Rice, J. L. Berg, R. P. Stilborn, and D. D. Hancock. 2004. Longitudinal study of fecal shedding of Escherichia coli O157:H7 in feedlot cattle: predominance and persistence of specific clonal types despite massive cattle population turnover. Appl. Environ Microbiol. 70:377-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mellmann, A., M. Bielaszewska, L. B. Zimmerhackl, R. Prager, D. Harmsen, H. Tschape, and H. Karch. 2005. Enterohemorrhagic Escherichia coli in human infection: in vivo evolution of a bacterial pathogen. Clin. Infect. Dis. 41:785-792. [DOI] [PubMed] [Google Scholar]
- 20.Mixter, P. F., J. D. Klena, G. A. Flom, A. M. Siegesmund, and M. E. Konkel. 2003. In vivo tracking of Campylobacter jejuni by using a novel recombinant expressing green fluorescent protein. Appl. Environ Microbiol. 69:2864-2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Monteville, M. R., J. E. Yoon, and M. E. Konkel. 2003. Maximal adherence and invasion of INT 407 cells by Campylobacter jejuni requires the CadF outer-membrane protein and microfilament reorganization. Microbiology 149:153-165. [DOI] [PubMed] [Google Scholar]
- 22.Ohnishi, M., J. Terajima, K. Kurokawa, K. Nakayama, T. Murata, K. Tamura, Y. Ogura, H. Watanabe, and T. Hayashi. 2002. Genomic diversity of enterohemorrhagic Escherichia coli O157 revealed by whole genome PCR scanning. Proc. Natl. Acad. Sci. USA 99:17043-17048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paton, A. W., and J. C. Paton. 1998. Detection and characterization of Shiga toxigenic Escherichia coli by using multiplex PCR assays for stx1, stx2, eaeA, enterohemorrhagic E. coli hlyA, rfb0111 and rfb0157. J. Clin. Microbiol. 36:598-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Perna, N. T., G. Plunkett III, V. Burland, B. Mau, J. D. Glasner, D. J. Rose, G. F. Mayhew, P. S. Evans, J. Gregor, H. A. Kirkpatrick, G. Posfai, J. Hackett, S. Klink, A. Boutin, Y. Shao, L. Miller, E. J. Grotbeck, N. W. Davis, A. Lim, E. T. Dimalanta, K. D. Potamousis, J. Apodaca, T. S. Anantharaman, J. Lin, G. Yen, D. C. Schwartz, R. A. Welch, and F. R. Blattner. 2001. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409:529-533. [DOI] [PubMed] [Google Scholar]
- 25.Reid, S. D., C. J. Herbelin, A. C. Bumbaugh, R. K. Selander, and T. S. Whittam. 2000. Parallel evolution of virulence in pathogenic Escherichia coli. Nature 406:64-67. [DOI] [PubMed] [Google Scholar]
- 26.Roe, A. J., S. W. Naylor, K. J. Spears, H. M. Yull, T. A. Dransfield, M. Oxford, I. J. McKendrick, M. Porter, M. J. Woodward, D. G. Smith, and D. L. Gally. 2004. Co-ordinate single-cell expression of LEE4-and LEE5-encoded proteins of Escherichia coli O157:H7. Mol. Microbiol. 54:337-352. [DOI] [PubMed] [Google Scholar]
- 27.Shaikh, N., and P. I. Tarr. 2003. Escherichia coli O157:H7 Shiga toxin-encoding bacteriophages: integrations, excisions, truncations, and evolutionary implications. J. Bacteriol. 185:3596-3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Strachan, N. J., D. R. Fenlon, and I. D. Ogden. 2001. Modelling the vector pathway and infection of humans in an environmental outbreak of Escherichia coli O157. FEMS Microbiol. Lett. 203:69-73. [DOI] [PubMed] [Google Scholar]
- 29.Strauch, E., C. Schaudinn, and L. Beutin. 2004. First-time isolation and characterization of a bacteriophage encoding the Shiga toxin 2c variant, which is globally spread in strains of Escherichia coli O157. Infect. Immun. 72:7030-7039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tarr, P. I., M. A. Neill, C. R. Clausen, J. W. Newland, R. J. Neill, and S. L. Moseley. 1989. Genotypic variation in pathogenic Escherichia coli O157:H7 isolated from patients in Washington, 1984-1987. J. Infect. Dis. 159:344-347. [DOI] [PubMed] [Google Scholar]
- 31.Torres, A. G., X. Zhou, and J. B. Kaper. 2005. Adherence of diarrheagenic Escherichia coli strains to epithelial cells. Infect. Immun. 73:18-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tuttle, J., T. Gomez, M. P. Doyle, J. G. Wells, T. Zhao, R. V. Tauxe, and P. M. Griffin. 1999. Lessons from a large outbreak of Escherichia coli O157:H7 infections: insights into the infectious dose and method of widespread contamination of hamburger patties. Epidemiol. Infect. 122:185-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Unkmeir, A., and H. Schmidt. 2000. Structural analysis of phage-borne stx genes and their flanking sequences in Shiga toxin-producing Escherichia coli and Shigella dysenteriae type 1 strains. Infect. Immun. 68:4856-4864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wick, L. M., W. Qi, D. W. Lacher, and T. S. Whittam. 2005. Evolution of genomic content in the stepwise emergence of Escherichia coli O157:H7. J. Bacteriol. 187:1783-1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang, Z., J. Kovar, J. Kim, J. Nietfeldt, D. R. Smith, R. A. Moxley, M. E. Olson, P. D. Fey, and A. K. Benson. 2004. Identification of common subpopulations of non-sorbitol-fermenting, beta-glucuronidase-negative Escherichia coli O157:H7 from bovine production environments and human clinical samples. Appl. Environ. Microbiol. 70:6846-6854. [DOI] [PMC free article] [PubMed] [Google Scholar]