Abstract
Medicinal plants are valuable resources of natural antimicrobial materials. A novel small protein with antimicrobial activities, designated LJAMP1, was purified from the seeds of a medicinal herb, motherwort (Leonurus japonicus Houtt). LJAMP1 is a heat-stable protein with a molecular mass of 7.8 kDa and a determined isoelectric point of 8.2. In vitro assays showed that LJAMP1 inhibits the growth of an array of fungi and bacteria. The hyphal growth inhibition by LJAMP1 was more evident against hyphomycete fungi, such as Alternaria alternata, Cercospora personata, and Aspergillus niger. The N-terminal amino acid sequence of LJAMP1 was determined, and its coding gene was consequently cloned by the rapid amplification of cDNA ends. The gene LJAMP1 has no intron and encodes a polypeptide of 95 amino acids, in which the first 27 residues was deduced as a signal peptide. The mature LJAMP1 shows relatively low identity to plant napin-like storage proteins. Northern blot assays revealed that LJAMP1 is expressed preferentially in seeds. Bioassays in transgenic tobacco demonstrated that that overexpression of LJAMP1 significantly enhanced the resistance of tobacco against not only the fungal pathogen A. alternata but also the bacterial pathogen Ralstonia solanacearum, while no visible alteration in plant growth and development was observed.
Microorganisms are capable of causing disease in a myriad of plant hosts and responsible for great losses in economical crops, preventing effective food distribution worldwide (11, 14). Although a great amount of research has been performed to overcome the damage caused by phytopathogens, a major difficulty encountered is the lack of effective control against some phytopathogenic bacteria, e.g., Erwinia amylovora and Xanthomonas vesicatoria (14), and some phytopathogenic fungi, e.g., Verticillium dahliae (47). The search for novel molecules with a wide spectrum of antimicrobial activity against phytopathogens is of challenging interest in plant pathology.
Plant seeds are usually scattered in environments rich in microorganisms. To protect their seeds from the invasion of pathogens, plants have developed various defense systems during their evolution (26). The physical barriers of seeds, e.g., lower water content and hard seed shell, against bacterial and fungal invasion are destroyed during seed germination because the seed coat is ruptured by imbibition. The breakage of these physical barriers can allow the invasion of pathogens into seeds (44). Therefore, seed germination is a period vulnerable to pathogen attack in a plant's life cycle (26, 45). It is widely accepted that antimicrobial peptides or proteins participate in the protection of seeds against potential microbial invaders during their germination (7, 26, 43-45). Until now, in plant seeds many proteins with antimicrobial activities have been detected, including chitinases and β-1,3-glucanases (24, 28), defensins (6, 46), thionins (15), lipid transfer proteins (7, 8), 2S albumins (1, 44), and ribosome-inactivating proteins (3, 10, 35). These antimicrobial proteins may be employed to create pathogenic resistance in transgenic plants (1, 21).
Motherwort (Leonurus japonicus Houtt) is a medicinal herb used in China as a traditional emmenagogue (an agent that promotes menstrual discharge) and an antipyretic and is now known to produce valuable products which have potential application in the treatment of various diseases (9, 29, 30). However, so far the antimicrobial ingredients in motherwort have been poorly investigated. In screening for potent antimicrobial proteins from various plant seeds, we found that the extract from motherwort seeds exhibits strong inhibition of growth of the fungal pathogens tested. We purified an antimicrobial protein from the extract and found that the protein is small in molecular mass (7.8 kDa) and heat stable. Furthermore, this seed protein has not only antifungal but also antibacterial activities, indicating that this is a novel seed protein. The expression of the gene encoding this protein enhanced resistance to fungal and bacterial pathogens in transformed tobacco.
MATERIALS AND METHODS
Plant materials.
Motherwort (Leonurus japonicus Houtt) seeds were obtained from a local medicinal herb market, and plants were grown in a field. Tobacco plants (Nicotiana tabacum cv. Xanthi) were grown in a greenhouse at 26 to 28°C under natural and additional artificial light (14-h/10-h photoperiod at 150 μE/s/m2).
Antimicrobial assays.
All bacterial and fungal strains used in this study were kindly provided by the Department of Plant Pathology, Southwest University. For antifungal activity assays, 17 fungal species were used as listed in Table 1. All fungi were grown in potato dextrose broth (Difco), except Saccharomyces cerevisiae was cultivated in YPD (2% peptone, 1% yeast extract, 2% glucose and 2% agar, pH 5.8). The antifungal assay in vitro was performed as previously described (50). The determination of the concentration required for a 50% inhibition of fungal growth (IC50) was performed by liquid growth inhibition assays (41). For the antibacterial assay, the four bacterial species in Table 1, grown in Luria-Bertani medium at 28°C, were employed as previously described (42). The antifungal activity to inhibit conidial germination of fungi was measured by placing 20-μl conidial suspensions (1.5 × 106 conidia/ml) in 40 μl potato dextrose broth containing the sample on a sterile glass sheet. Conidial germination was directly examined on the glass sheet after 8 h of incubation in a moist chamber at 28°C.
TABLE 1.
Antimicrobial activity of LJAMP1a
| Tested pathogens | Conc. of inhibition (μM) |
|---|---|
| Bacteria | |
| Ralstonia solanacearum | 7.5-15 |
| Agrobacterium radiobacter | No |
| Bacillus subtilis | >15 |
| Escherichia coli | No |
| Fungi | |
| Alternaria alternata | <7.5 |
| Alternaria brassicae | <7.5 |
| Aspergillus niger | <7.5 |
| Botrytis maydis | <7.5 |
| Botrytis cinerea | 7.5-15 |
| Cercosporapersonata | <7.5 |
| Colletotrichumgloeosporiodes | >15 |
| Fusarium oxysporum | 7.515 |
| Penicillium digitatum | >15 |
| Pyriculariagrisea | 7.5-15 |
| Rhizoctonia solani | 7.5-15 |
| Rhizoctonia cerealis | <7.5 |
| Rhizopus nigrieans | >15 |
| Saccharomyces cerevisiae | >15 |
| Sclerotiniasclerotiorum | >15 |
| Trichoderma harzianum | <7.5 |
| Verticillium dahliae | 7.5-15 |
Antifungal activity of LJAMP1 against various bacterial and fungal species is shown. Purified LJAMP1 dissolved in sterile 20 mM sodium phosphate buffer, pH 6.2, was used for determination of antibacterial and antifungal activity. The inhibition of fungal and bacterial growth is expressed by the LJAMP1 concentration. No, no inhibition at 25 μM LJAMP1.
Protein extraction and isolation.
Mature dry seeds (200 g) were frozen in liquid nitrogen and homogenized to a fine powder in a laboratory blender. The resulting flour was suspended in 1 liter of buffer A (20 mM sodium phosphate buffer, pH 6.2) containing 50 mM KCl, 5 mM EDTA, 1 mM aprotinin, and 20 mM thiourea. After stirring for 12 h, the extract was centrifuged for 35 min at 32,000 × g. The supernatant was precipitated with solid ammonium sulfate at 75% saturation for 4 h, and then the pellet obtained by centrifugation at 32,000 × g for 35 min was dissolved in 100 ml buffer A and heated at 80°C for 10 min. After centrifugation to remove the heat-denatured protein precipitates, the supernatant was dialyzed against buffer A. The dialyzed solution was passed through a HiPrep 16/10 CM FF column on an ÄKTA Prime system (Amersham Biosciences, Sweden) preequilibrated with buffer A, and then the column was washed with the same buffer to remove unadsorbed proteins. The adsorbed proteins were eluted with an NaCl gradient (0 to 1 M) in buffer A. Antifungal activity against Trichoderma harzianum was monitored in each fraction. The antifungal fractions were pooled and centrifuged at 18,000 × g for 10 min, and the supernatant was then applied to reversed-phase high-performance liquid chromatography (HPLC) using a Hypersil C8 column (8 by 100 mm; 15 μm; Yilite Co., China) equilibrated with 0.05% trifluoroacetic acid (TFA) on a Gold HPLC system (Beckman). Separation of the fraction was performed with 0.05% TFA over 5 min and a linear gradient of 0 to 60% acetonitrile in 0.05% TFA over 45 min and 60% acetonitrile in 0.05% TFA over 10 min at a flow rate of 1.0 ml/min. An active fraction was further purified on an analytical C18 reversed-phase column (Aquapore OD-300, 4.6 by 250 mm, 7 μm; PerkinElmer) with a linear biphasic gradient of acetonitrile in 0.05% TFA from 0 to 25% over 5 min at a flow rate of 1.0 ml/min and from 25 to 30% over 35 min with a flow rate of 0.5 ml/min. Individual peak fractions were collected and then condensed in 2-kDa cutoff dialysis tubing (Sigma) with polyethylene glycol of 20 kDa. After dialysis against ultrapurified water, the samples were used for further analysis. All purification steps were performed at room temperature, and the column effluent was monitored by absorbance at 280 nm.
Electrophoresis and amino acid sequence determination.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was carried out on 0.75-mm-thick slab gels containing a 15.5% polyacrylamide separating gel with a 4% stacking gel using an electrophoresis cell (Amersham Biosciences, Sweden) according to the manufacturer's instructions. Polypeptide molecular mass markers and silver staining reagents were from Amersham Biosciences (Sweden). Protein concentration was measured by the Bradford method (5) using bovine serum albumin as a standard. The purified sample was subjected to SDS-PAGE as above and then electroblotted to a polyvinylidene difluoride membrane (Bio-Rad). Protein bands were visualized with Coomassie blue R and excised from the membrane. The N-terminal amino acid sequence determination was performed by the Laboratory for Protein Chemistry of Hunan Normal University (Changsha, China) on a protein sequencer (model no. 491; Applied Biosystems).
Gene cloning and RNA analyses.
Based on the N-terminal amino acid sequence, a degenerate primer, 5′-CCG CAG CAG CAG AC(ATCG) GT(ATCG) GA(AG)-3′, was designed. 3′ rapid amplification of cDNA ends (RACE) was performed with a 3′ full RACE kit (Takara, Japan) according to the manufacturer's instructions. The amplified product was purified and cloned into pGEM-T (Promega, WI) for sequencing. To amplify the 5′ flanking sequence of the gene, the Y-shaped adaptor-dependent extension (YADE) method (49) was employed. Based on the nucleotide sequence of the 3′ RACE products, two nested primers (A, 5′-GCTGTGACCTTAATCCGCAA-3′, and B, 5′-GCATCCTAGCCTTTCGATAC-3′) were designed. The Y-shaped adaptor with a blunt terminus was produced by annealing equal molarities of oligonucleotides 5′-CGGTAGGATCCCGCAGAACGACGGCCAG-3′and 5′-pCTGGCCGTCCAAGACGC-3′. Around 10 ng DraI-/SmaI-digested genomic DNA of motherwort was ligated to 4 μl of adaptor (8 μM), and then 1 μl of ligation products containing about 0.5 ng of genomic DNA was used as a template to sequentially perform linear amplification and exponential amplification. The reaction of linear amplification contained 1 μl of ligation products, 1× PCR buffer, 200 μM deoxynucleoside triphosphates, 1.5 mM MgCl2, 200 μM primer A, and 1 U Taq DNA polymerase (added when the reaction temperature rose to 94°C). The reaction started at 94°C for 5 min, and the following cycle was repeated 40 times: 94°C for 30 s, 56°C for 30 s, 72°C for 2 min, and a final extension at 72°C for 5 min. Finally, linear amplification products (1 μl) were used as the template of the exponential amplification with the adaptor primer (5′-CGGTAGGATCCCGCAGAAC-3′) and nested primer B. The reactions were amplified using a step-down PCR protocol (18), with one step for 5 min at 94°C, followed by 12 cycles at 94°C for 30 s, 58°C (decreasing 0.5°C per cycle) for 30 s, and 72°C for 2 min, then 30 cycles at 94°C for 30 s, 52°C for 30 s, and 72°C for 2 min, followed by 72°C for 10 min. The YADE product was purified and cloned into pGEM-T (Promega, Madison,WI) for sequencing. The products of YADE and the 3′ RACE sequence were overlapped with the SeqMan program of DNASTAR (DNASTAR, Madison, Wisconsin), and the contig sequence was further used to perform a similarity search with the BLASTX program to determine the putative initiator ATG. The coding region of the LJAMP1 gene was further amplified from the genomic DNA using the primers 5′-GCAGTCAAATGCTGCAGGGT-3′ and 5′-GCCGATAGGCCTTAATCATC-3′.
Total RNAs were extracted from various motherwort tissues and tobacco leaves using the guanidinium thiocyanate method (27). Northern blot hybridizations were carried out with 32P-labeled LJAMP1 cDNA probes using the Ready-To-Go random primer labeling kit (Amersham Biosciences, Sweden) as previously described (40).
Vector construction and plant transformation.
The open reading frame region encoding LJAMP1 was cloned to downstream of the CaMV 35S promoter. The nos transcription terminator was placed downstream. Then the expression cassette with the CaMV 35S promoter-LJAMP1-nos terminator and CaMV 35S promoter-gus-nos terminator and Nos promoter-npt II (neomycin phosphotransferase II gene)-nos terminator were cloned in the binary vector pBIN19 (16). The construct was delivered into Agrobacterium tumefaciens LBA4404 (19) by the freeze-thawing method (2), and the resulting Agrobacterium strain was used for the transformation of tobacco leaf disk by cocultivation (25). The transgenic plants were regenerated under kanamycin selection (100 mg/liter) as previously described (20).
Analyses on transgenic tobacco.
For protein analysis, tobacco seeds were incubated at 26°C in a rotary shaker (170 rpm) for 3 days and then the germinating seeds were grown in sterile soil in multiwell plastic containers that were kept in a growth chamber at 80% relative humidity, from 25°C to 28°C, under a 14-h/10-h photoperiod at 150 μE/s/m2). Fresh leaf tissues (10 g) of tobacco were homogenized with 10 ml of buffer A containing 50 mM KCl, 5 mM EDTA, 1 mM aprotinin, and 20 mM thiourea. To identify the transgene products, the samples were analyzed by HPLC on a SOURCE 5RPC reversed-phase column (4.6 by 150 mm, 5 μm; Amersham Biosciences, Sweden) equilibrated with 0.05% TFA using an ÄKTA explorer 10S (Amersham Biosciences, Sweden), with 0.05% TFA over 5 min and a linear gradient of 0 to 45% acetonitrile in 0.05% TFA over 35 min at a flow rate of 1.0 ml/min.
To test the resistance of transgenic tobacco to infection by Alternaria alternata, the bioassay was performed. A. alternata strain was grown on potato dextrose agar at room temperature for 2 to 3 weeks. Spores were harvested in sterile tap water, counted with the hemocytometer, and then diluted to 1 × 106 spores/ml. Phytopathogenic fungus inoculation of tobacco was carried out by spraying the A. alternata spore suspension (1 × 106 spores/ml in sterile water) on leaves of 4-week-old plants. The inoculated plants were placed in a growth chamber at 90% relative humidity and 28°C for 3 days and further incubated at 80% relative humidity and 28°C for 7 days. Then the numbers and sizes of lesions on leaves were scored, and the variation of lesions was reflected by a five-class disease severity scale (DS) defined as follows: DS class 0, no lesion; DS class 1, mean size of lesions less than 1 mm; DS class 2, mean size of lesions from 1 to 2 mm; DS class 3, mean size of lesions from 2 to 5 mm; and DS class 4, mean size of lesions more than 5 mm. Disease resistance was expressed using a disease index (DI). The DI was calculated using the following formula: DI (%) = (∑I × j/4 × n) × 100, where i is the DS class, j is the number of disease leaves in each class, and n is the total number of leaves. Each treatment was replicated three times, and each replicate contained 18 plants.
To determine whether the overexpression of LJAMP1 in tobacco can provide resistance to the bacterial pathogen, we infected the tobacco plants with a Ralstonia solanacearum isolate, which causes bacterial wilt disease on tobacco. The bacterium was grown for 48 h at 30°C on Luria-Bertani medium. The bacterial suspension for inoculation was prepared by washing the medium surface with sterile tap water, and the bacterial population was adjusted to 107 cells/ml. Tobacco plants (4 weeks old) were inoculated by dipping the roots into the bacterial suspensions. The plants were kept in a growth chamber at 95% relative humidity and 28°C for 2 days and then at 80% relative humidity and 28°C for 6 days. The number of wilting leaves was recorded for each plant daily, and a five-class disease severity scale was calculated based on the previously described method (22). Each treatment was replicated three times, and each replicate contained 20 plants.
All data were analyzed by t test (P ≤ 0.05) using the software Origin v6 (OriginLab Co.).
Nucleotide sequence accession number.
The nucleotide sequence data reported for the LJAMP1 gene were deposited in GenBank Nucleotide Sequence Databases under the GenBank accession number AAW66631.
RESULTS
Purification of the antifungal protein, LJAMP1, from motherwort seeds.
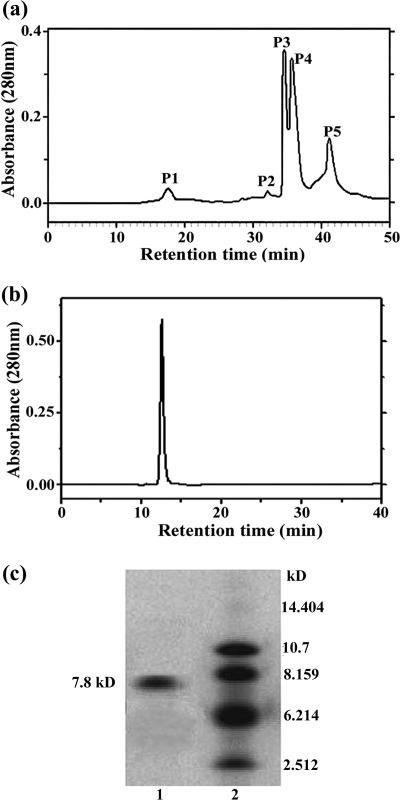
Medicinal plants often produce natural antimicrobial materials. In searching for antifungal peptides or proteins from plant seeds, we found that the extract from motherwort seeds shows strong activity against the growth of T. harzianum and that this antifungal activity is thermostable. To enrich for the antifungal proteins, we first heated the dissolved solution from ammonium sulfate precipitation and then applied the soluble fraction to cation-exchange chromatography. The eluted fractions were assayed for growth inhibition on the test fungus, T. harzianum. No antifungal activity was detected in the unbound fraction, but activity was found in a peak resulting from a linear NaCl gradient elution (data not shown). Then the antifungal fractions were subjected to the reversed-phase HPLC on a C8 semipreparation column. Five peaks were obtained (Fig. 1a), and then each peak was assayed for the antifungal activity. We found that the third and the fourth peaks (P3 and P4) (Fig. 1a) showed antifungal activities. Further purification for P3 was conducted through the reversed-phase HPLC on a C18 analytical column (P4 is currently under investigation), and only one protein peak was detected (Fig. 1b). This antifungal protein, designated LJAMP1 (Leonurus japonicus antimicrobial protein), was purified to homogeneity as determined by SDS-PAGE (Fig. 1c). We then confirmed that the antifungal activity is heat stable and resistant to high-temperature (80°C for 15 min) treatment. The molecular mass of LJAMP1 was estimated as approximately 7.8 kDa, calibrated with polypeptide markers on SDS-PAGE (Fig. 1c). Isoelectric focusing analysis showed that the isoelectric point of LJAMP1 is about 8.2 (data not shown). The N-terminal amino acid sequence of LJAMP1 was determined to be NH2-P-Q-S-Q-T-V-E-E-C-C-E-S-L-K-D-I.
FIG. 1.
Purification of LJAMP1. (a) The adsorbed fraction from a carboxymethylcellulose cation-exchange column was subjected to reversed-phase chromatography using HPLC on a C8 semipreparation column. (b) The P3 fraction shown in panel a was purified on an Aquapore OD-300 analytical C18 reversed-phase column. (c) Analysis by SDS-PAGE of the peak shown in panel b. Lane 1, 2.6 μg protein from the peak in panel b; lane 2, polypeptide molecular mass markers.
Antimicrobial activity of LJAMP1.
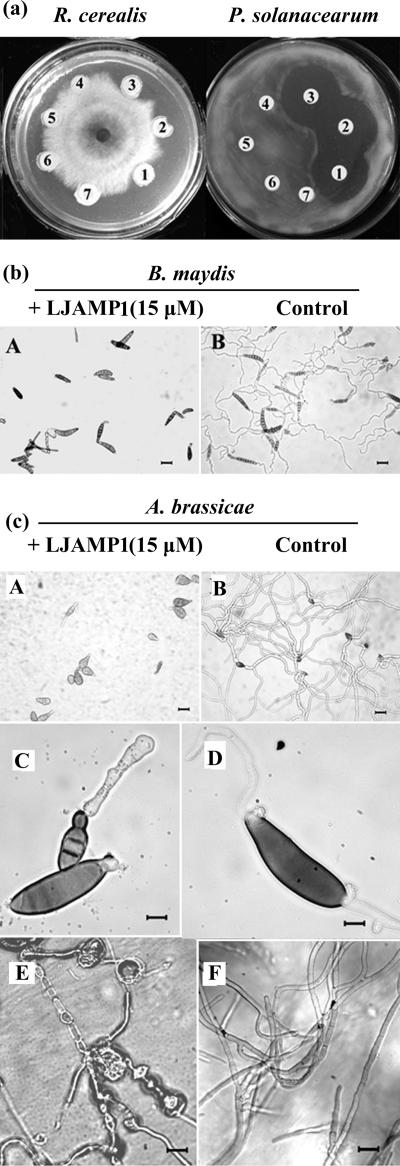
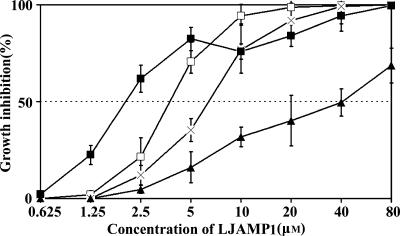
Antimicrobial activity of LJAMP1 was tested against an array of fungi and several bacteria in vitro. For fungi, the hyphal growth inhibition by LJAMP1 was more evident against hyphomycete fungi, such as A. alternata, Alternaria brassicae, Aspergillus niger, Botrytis maydis, Cercospora personata, and Rhizoctonia cerealis (Table 1 and Fig. 2). For bacteria, LJAMP1 showed inhibition of the growth of R. solanacearum and Bacillus subtilis but no action against Agrobacterium radiobacter and Escherichia coli (Table 1 and Fig. 2). To analyze the antifungal effect in detail, we evaluated the inhibition by LJAMP1 against B. maydis and A. brassicae microscopically. The spore germination of B. maydis and A. brassicae was inhibited by LJAMP1 at a concentration of 15 μM (Fig. 2b and c, panels A and B). The hyphae of A. brassicae, treated with LJAMP1, produced enlarged hyphal tips and swelling and extensive gnarl (Fig. 2c, panels C to F). In order to examine the protein IC50, dose-response curves were measured for four fungi. As showed in Fig. 3, the IC50s of LJAMP1 for A. alternata, C. personata, A. niger, and S. cerevisiae were 2.2, 3.9, 6.7, and 41.6 μM, respectively.
FIG. 2.
Antimicrobial activity of LJAMP1 in vitro. (a) Inhibition of growth on plate for the fungus R. cerealis and the bacterium R. solanacearum. LJAMP1 was added at concentrations of 7.5 μM (1), 15 μM (2), and 25 μM (3). Bovine serum albumin was added at concentrations of 7.5 μM (4), 15 μM (5), and 25 μM (6). Buffer A is also shown (7). (b) Effect on germination of spores for phytopathogenic fungus B. maydis. (c) Effect on germination of spores and growth of hyphae for phytopathogenic fungus A. brassicae. Spores and hyphae were incubated in potato dextrose broth medium with LJAMP1 at a final concentration of 15 μM and without LJAMP1 (control). Photomicrographs were taken after 6 h of incubation of B. maydis (b) and A. brassicae (c). In panels b and c, parts A and B, the scale bar indicates 100 μm; in panel c, parts C to F, the scale bar indicates 20 μm.
FIG. 3.
Dose-response effect of LJAMP1 in in vitro antifungal assays. Dose-response curves were derived from in vitro microtiter plate assays with LJAMP1 after incubation for 48 h at 26°C. Fungi tested were A. alternata (▪), C. personata (□), A. niger (×), and S. cerevisiae (▴). Each data point represents the mean of experiments (n = 4 each) performed three times.
Cloning and characterization of the gene encoding LJAMP1.
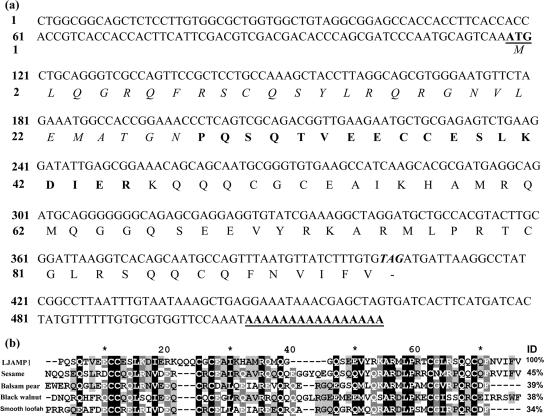
To clone the gene encoding LJAMP1, the RACE approach was employed. Based on the N-terminal amino acid sequence, a degenerate primer was designed for the amplification of the 3′ cDNA. A 350-bp cDNA fragment was amplified and sequenced. The N terminus of the deduced amino acid sequence of the cDNA matched perfectly with the N-terminal amino acid sequence of LJAMP1. To obtain the 5′ part of the cDNA, the Y-shaped-adaptor-dependent extension method (49) was used, which yielded another 500-bp fragment. The full-length cDNA of the LJAMP1 gene was constructed from the nucleotide sequence of the 5′ and 3′ parts, containing a 285-bp open reading frame encoding 95 amino acids. According to the N-terminal sequence of the purified LJAMP1, the mature protein is composed of 68 residues. The remaining 27 residues in the N-terminal region have characteristic features of a signal peptide found in excreted proteins (31). The polyadenylation site was found 117 bp downstream from the stop codon (Fig. 4a). Based on the cDNA sequence, a genome DNA fragment was amplified by PCR. By comparing the PCR product sequence with the cDNA sequence, we found that the gene encoding LJAMP1, named LJAMP1, is an intronless gene. In the BLAST search (www.ncbi.nlm.nih.gov/BLAST), the amino acid sequence of mature LJAMP1 was found to share low identity with a series of napin proteins (Fig. 4b). A 2S albumin from sesame (Sesamum indicum L.) showed 45% identity with LJAMP1, followed by a napin protein (39%) from balsam pear (Momordica charantia L.), a 2S albumin (38%) from black walnut (Juglans nigra L.), and a ribosome-inactivating protein (RIP; 34%) from smooth loofah (Luffa aegyptiaca L.).
FIG. 4.
Nucleotide sequence and deduced amino acid sequence of the full-length LJAMP1 cDNA. (a) The cDNA gene sequence encoding LJAMP1. The deduced amino acid sequence is shown in one-letter code under the DNA sequence. The initiation codon and poly(A) are underlined. The deduced signal peptide sequence is italic, and the determined N-terminal amino acid sequence of mature LJAMP1 is bold.(b) Amino acid sequence alignment of LJAMP1 (accession no. AAW66631) with napin-like proteins from sesame (Sesamum indicum, accession no. AAK15088), balsam pear (Momordica charantia, accession no. CAD32398), and black walnut (Juglans nigra, accession no. AY102930). Protein sequences were aligned with the ClustalW algorithm. The ID column shows the identity of the protein sequences.
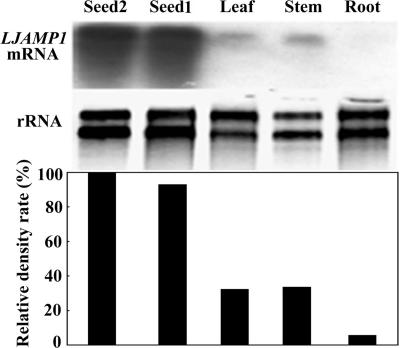
To determine LJAMP1 transcript accumulations in various organs of motherwort, Northern blot analysis was performed by using the cDNA probe and total RNA isolated from roots, stems, leaves, young seeds, and mature seeds. The hybridization signal was strong in young seeds and mature seeds, relatively weak in stems and leaves (Fig. 5), and undetected in roots. The density rates of the hybridization signal relative (LJAMP1 mRNA) to rRNA are 100, 93, 32.4, 33.7, and 5.7% for young seeds, mature seeds, stems, leaves, and roots, respectively. This expression pattern suggests that LJAMP1 is preferentially expressed in seeds.
FIG. 5.
RNA gel blot analysis of LJAMP1 transcripts in different tissues of motherwort. Total RNAs were extracted from leaves, stems, and roots of 5-week-old plants and from the seeds at the 30th (seed 1) and 50th (seed 2) days after anthesis. RNAs electrophoresed in a 1% agarose gel containing 6% formaldehyde were hybridized with a 32P-labeled LJAMP1 cDNA probe. Hybridizing signals were visualized by exposing the membrane to X-ray film (top). The gel was stained with ethidium bromide to detect rRNAs as control (bottom). The relative density rate (%) represents the relative percentages of LJAMP1 mRNA/rRNA in different tissues.
Overexpression of LJAMP1 in tobacco.
To evaluate the antimicrobial potential of LJAMP1, transgenic tobacco plants overexpressing the gene under the control of the 35S promoter were generated (see Fig. S1 in the supplemental material). As a control, tobacco was transformed with the empty vector. The regenerated plants were screened by kanamycin resistance and then by ß-glucuronidase (GUS) histochemical analysis (23). GUS-positive and kanamycin-resistant plantlets were obtained and grown in a greenhouse. The transgene insert numbers in the independent transformants (T0) were estimated by the segregation pattern for gus expression in the progeny resulting from self-fertilization (T1). Fourteen transgenic lines, showing a Mendelian inheritance ratio of 3:1 (positive/negative for gus expression) in the progenies were determined to have a single insert of transgenes. Then the isolation of homozygous transgenic plants with a single insert of the transgene was conducted in the progeny self-fertilized from T1 detected by GUS staining.
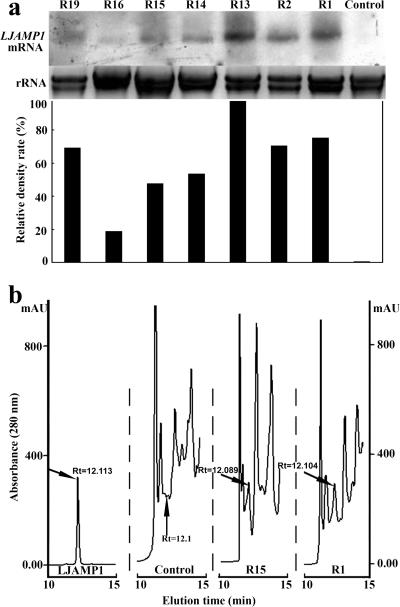
We randomly selected seven homozygous transgenic T2 lines with a single insert of the transgene for subsequent molecular analyses. Hybridization with LJAMP1 cDNA revealed different levels of accumulation of LJAMP1 transcripts in transgenic plants (Fig. 6a). Strong hybridization signals were seen in the transgenic lines R1, R2, and R13, moderate signals in lines R14, R15, and R19, and a weak signal in line R16. To detect the gene products of LJAMP1 in transgenic tobacco plants, the protein extracts from the leaves of all transgenic lines were subjected to HPLC and the HPLC profiles of the samples were compared with that of the purified LJAMP1. The LJAMP1-specific peak, which showed inhibition activity against R. solanacearum (data not shown), was detected in the transgenic lines, while no such peak was found in the control (Fig. 6b), indicating that LJAMP1 had been expressed stably in the transgenic tobacco plants.
FIG. 6.
The expression analysis of LJAMP1 in transgenic tobacco seedlings. (a) RNA gel blot analysis of LJAMP1 transcripts in transgenic tobacco seedlings. As a control, tobacco was transformed with an empty vector. The relative density rate (%) represents the relative percentages of LJAMP1 mRNA/rRNA in different lines. (b) HPLC profiles of proteins from transgenic lines control, R1 and R15, and the purified LJAMP1. Arrows indicate LJAMP1 peaks.
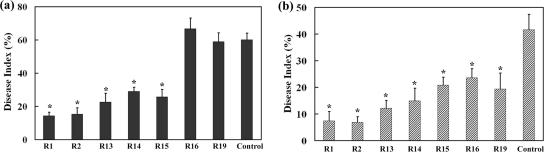
To evaluate disease resistance of the transgenic tobacco, the standard leaf assay was performed. The transgenic lines overexpressing LJAMP1 and the control transformed with the empty vector were challenged with the fungal pathogen A. alternata. From the disease index, R1 and R2 showed the highest resistance against A. alternata. R13, R14, and R15 also exhibited a significant increase of disease resistance compared with that of the control (Fig. 7a). We further challenged the tobacco plants with the bacterial pathogen R. solanacearum and found that the transgenic lines R1 and R2 also showed the highest resistance against the pathogen and R13, R14, R15, R16 and R19 showed significantly increased resistance (Fig. 7b). Comparing Fig. 6 and 7, the resistance levels were consistent with the expression levels of LJAMP1 in transgenic lines, indicating that the LJAMP1 expression contributes the enhancement of resistance against A. alternata and R. solanacearum in transgenic tobacco plants. Meanwhile, in our experiments, no visible changes were found in transgenic tobacco, suggesting that the overexpression of LJAMP1 does not alter plant growth and development.
FIG. 7.
Resistance of transgenic T2 tobacco plants inoculated with the fungus A. alternata and the bacterium R. solanacearum. (a) Fungal infection assays of tobacco plants holding LJAMP1. Mean values of disease indexes (%) estimated from four independent infection assays (nine plants per line) are shown. (b) Bacterial infection assays of tobacco transformed with LJAMP1. Mean values of disease indexes (%) estimated from three independent infection assays (12 plants per line) are shown. Asterisks indicate that the DI in transgenic lines was significantly different (P values was ≤0.05 by t test) compared with that of the DI in the control. Error bars indicate standard deviations.
DISCUSSION
In searching for antimicrobial proteins from plant seeds, we found that the extracts from motherwort seeds exerted strong antimicrobial activity. We purified an antimicrobial protein, named LJAMP1, from motherwort seeds and then cloned its cDNA. The BLAST search revealed that LJAMP1 is partially similar to a series of napin-like proteins from plants.
Napins, a member of the 2S albumin class of proteins, which are water-extractable storage proteins in seeds, are encoded by a gene family (4) and usually composed of a 4.5-kDa small subunit and a 10-kDa large subunit (17). They are derived from proteolytic cleavage of a precursor of about 180 amino acid residues (12, 33). However, some napin-like proteins were reported to be single-chain proteins. For instance, charantin, an RIP isolated from balsam pear, is a single-chain peptide and structurally related to the large chain of the napin-like proteins (36). Although the major function of napins is devoted to storage (4), some napins or napin-like proteins possess significant antifungal or antibacterial properties (4, 32). Our bioassay showed that LJAMP1 inhibits the growth of an array of both fungi and bacteria. Furthermore, we also assayed the efficacy of LJAMP1 in inhibiting the mycelial growth and spore germination of B. maydis and A. brassicae (Fig. 2b and c). In a comparison of the antimicrobial potencies of different antimicrobial proteins from plants, LJAMP1 displays a satisfied inhibitory effect (IC50 values from 17 to 52 μg/ml) against A. alternata, C. personata, and A. niger. For example, the growth inhibition of TRIP, a protein from tobacco, was about 50 μg/ml against a number of fungi and bacteria, and ME, a protein from Mirabilis expansa, is active against Rhizoctonia solani growth at a dose of 10 μg/ml (34).
Genes coding for defense proteins have already been employed to enhance plant resistance against fungal and bacterial phytopathogens (38, 39, 51). For instance, transgenic orange plants expressing a tomato thaumatin-like protein exhibited increased tolerance toward Phytophtora citrophthora (13); constitutive overexpression of an antimicrobial protein gene Ace-AMP1 from Allium cepa in Oryza sativa subsp. indica rice enhanced resistance against three major rice pathogens, Magnaporthe grisea, R. solani, and Xanthomonas oryzae(37); and the expressions of defense-related gene ch5B coding for a chitinase caused the reduction of disease symptoms in strawberry (cultivar Pájaro) infected with Botrytis cinerea (48). To confirm the antifungal and antibacterial function of LJAMP1, we constitutively expressed the LJAMP1 gene in transgenic tobacco. Infection assays for fungal and bacterial pathogens indicated that transgenic tobacco plants were characterized by resistance against R. solanacearum and A. alternata (see Fig. S2 in the supplemental material). The resistance of LJAMP1 in transgenic tobacco against fungal and bacterial diseases confirmed the antifungal and antibacterial effects of LJAMP1 in planta.
Taken together, the results of our study indicated that LJAMP1 is a novel, heat-stable, small single-chain antimicrobial protein with a wide range of antifungal and antibacterial activities. Because there were no visible negative effects on the growth of transgenic plants, LJAMP1 has potential application in plant protection by gene engineering.
Supplementary Material
Acknowledgments
We are grateful to Xiaojiu Wang for her technical assistance in bioassay and to Masahiro Sugiura for the critical reading of the manuscript.
This research was supported with funds from the National Natural Science Foundation of China (grant no. 30270147 and 30370916 to X. Yang).
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 8 December 2006.
REFERENCES
- 1.Agizzio, A. P., A. O. Carvalho, S. F. F. Ribeiro, O. L. T. Machado, E. W. Alves, L. A. Okorokov, S. S. Samarao, J. C. Bloch, M. V. Prates, and V. M. Gomes. 2003. A 2S albumin-homologous protein from passion fruit seeds inhibits the fungal growth and acidification of the medium by Fusarium oxysporum. Arch. Biochem. Biophys. 416:188-195. [DOI] [PubMed] [Google Scholar]
- 2.An, G. 1987. Binary Ti vectors for plant transformation and promoter analysis. Methods Enzymol. 153:292-305. [Google Scholar]
- 3.Barbieri, L., M. G. Battelli, and F. Stirpe. 1993. Ribosome-inactivating proteins from plants. Biochim. Biophys. Acta Rev. Biomembr. 1154:237-282. [DOI] [PubMed] [Google Scholar]
- 4.Barciszewski, J., M. Szymanski, and T. Haertle. 2000. Minireview: analysis of rape seed napin structure and potential roles of the storage protein. J. Protein Chem. 19:249-254. [DOI] [PubMed] [Google Scholar]
- 5.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 6.Broekaert, W. F., F. R. G. Terras, B. P. A. Cammue, and R. W. Osborn. 1995. Plant defensins: novel antimicrobial peptides as components of the host defense system. Plant Physiol. 108:1353-1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cammue, B. P. A., K. Thevissen, M. Hendriks, K. Eggermont, I. J. Goderis, P. Proost, J. Van Damme, R. W. Osborn, F. Guerbette, J. C. Kader, and W. F. Broekaert. 1995. A potent antimicrobial protein from onion seeds showing sequence homology to plant lipid transfer proteins. Plant Physiol. 109:445-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheng, C. S., D. Samuel, Y. J. Liu, J. C. Shyu, S. M. Lai, K. F. Lin, and P. C. Lyu. 2004. Binding mechanism of nonspecific lipid transfer proteins and their role in plant defense. Biochemistry 43:13628-13636. [DOI] [PubMed] [Google Scholar]
- 9.Chinwala, M. G., M. Gao, J. Dai, and J. Shao. 2003. In vitro anticancer activities of Leonurus heterophyllus sweet (Chinese motherwort herb). J. Altern. Complement. Med. 9:511-518. [DOI] [PubMed] [Google Scholar]
- 10.Dong, T. X., T. B. Ng, H. W. Yeung, and R. N. S. Wong. 1994. Isolation and characterization of a novel ribosome-inactivating protein, β-kirilowin, from the seeds of Trichosanthes kirilowii. Biochem. Biophys. Res. Commun. 199:387-393. [DOI] [PubMed] [Google Scholar]
- 11.Donini, M., C. Lico, S. Baschieri, S. Conti, W. Magliani, L. Polonelli, and E. Benvenuto. 2005. Production of an engineered killer peptide in Nicotiana benthamiana by using a Potato virus X expression system. Appl. Environ. Microbiol. 71:6360-6367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ericson, M. L., J. Rodin, M. Lenman, K. Glimelius, L. G. Josefsson, and L. Rask. 1986. Structure of the rapeseed 1.7 S storage protein, napin, and its precursor. J. Biol. Chem. 261:14576-14581. [PubMed] [Google Scholar]
- 13.Fagoaga, C., I. Rodrigo, V. Conejero, C. Hinarejos, J. J. Tuset, J. Arnau, J. A. Pina, L. Navarro, and L. Pena. 2001. Increased tolerance to Phytophthora citrophthora in transgenic orange plants constitutively expressing a tomato pathogenesis related protein PR-5. Mol. Breed. 7:175-185. [Google Scholar]
- 14.Ferre, R., E. Badosa, L. Feliu, M. Planas, E. Montesinos, and E. Bardaji. 2006. Inhibition of plant-pathogenic bacteria by short synthetic cecropin A-melittin hybrid peptides. Appl. Environ. Microbiol. 72:3302-3308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Florack, D. E. A., and W. J. Stiekema. 1994. Thionins—properties, possible biological roles and mechanisms of action. Plant Mol. Biol. 26:25-37. [DOI] [PubMed] [Google Scholar]
- 16.Frisch, D. A., L. W. Harris-Haller, N. T. Yokubaitis, T. L. Thomas, S. H. Hardin, and T. C. Hall. 1995. Complete sequence of the binary vector Bin 19. Plant Mol. Biol. 27:405-409. [DOI] [PubMed] [Google Scholar]
- 17.Gehrig, P. M., and K. Biemann. 1996. Assignment of the disulfide bonds in napin, a seed storage protein from Brassica napus, using matrix-assisted laser desorption ionization mass spectrometry. Pept. Res. 9:308-314. [PubMed] [Google Scholar]
- 18.Hecker, K. H., and K. H. Roux. 1996. High and low annealing temperatures increase both specificity and yield in touchdown and stepdown PCR. BioTechniques 20:478-485. [DOI] [PubMed] [Google Scholar]
- 19.Hoekema, A., P. R. Hirsch, P. J. J. Hooykaas, and R. A. Schilperoort. 1983. A binary plant vector strategy based on separation of vir- and T-region of the Agrobacterium tumefaciens Ti-plasmid. Nature 303:179-180. [Google Scholar]
- 20.Horsch, R. B., J. E. Fry, N. L. Hoffman, D. Eichholtz, S. G. Rogers, and R. T. Fraley. 1985. A simple method for transferring gene into plants. Science 227:1229-1231. [DOI] [PubMed] [Google Scholar]
- 21.Jach, G., B. Gornhardt, J. Mundy, J. Logemann, P. Pinsdorf, R. Leah, J. Schell, and C. Maas. 1995. Enhanced quantitative resistance against fungal disease by combinatorial expression of different barley antifungal proteins in transgenic tobacco. Plant J. 8:97-109. [DOI] [PubMed] [Google Scholar]
- 22.Jaynes, J. M., P. Nagpala, L. Destefano-Beltran, J. Hong Huang, J. Kim, T. Denny, and S. Cetiner. 1993. Expression of a cecropin B lytic peptide analog in transgenic tobacco confers enhanced resistance to bacterial wilt caused by Pseudomonas solanacearum. Plant Sci. 89:43-53. [Google Scholar]
- 23.Jefferson, R. A., T. A. Kavanagh, and M. W. Bevan. 1987. GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6:3901-3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leah, R., H. Tommerup, I. Svendsen, and J. Mundy. 1991. Biochemical and molecular characterization of three barley seed proteins with antifungal properties. J. Biol. Chem. 266:1564-1573. [PubMed] [Google Scholar]
- 25.Li, Y., G. Hagen, and T. J. Guilfoyle. 1992. Altered morphology in transgenic tobacco plants that overproduce cytokinins in specific tissues and organs. Dev. Biol. 153:386-395. [DOI] [PubMed] [Google Scholar]
- 26.Liu, Y., J. Luo, C. Xu, F. Ren, C. Peng, G. Wu, and J. Zhao. 2000. Purification, characterization, and molecular cloning of the gene of a seed-specific antimicrobial protein from pokeweed. Plant Physiol. 122:1015-1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Logemann, J., J. Schell, and L. Willmitzer. 1987. Improved method for the isolation of RNA from plant tissues. Anal. Biochem. 163:16-20. [DOI] [PubMed] [Google Scholar]
- 28.Melchers, L. S., M. Apotheker-de Groot, J. A. Knaap, A. S. Ponstein, M. B. Sela-Buurlage, J. F. Bol, B. J. C. Cornelissen, P. J. M. Elzen, and H. J. M. Linthorst. 1994. A new class of tobacco chitinases homologous to bacterial exo-chitinases displays antifungal activity. Plant J. 5:469-480. [DOI] [PubMed] [Google Scholar]
- 29.Milkowska-Leyck, K., B. Filipek, and H. Strzelecka. 2002. Pharmacological effects of lavandulifolioside from Leonurus cardiaca. J. Ethnopharmacol. 80:85-90. [DOI] [PubMed] [Google Scholar]
- 30.Nagasawa, H., H. Inatomi, M. Suzuki, and T. Mori. 1992. Further study on the effects of motherwort (Leonurus sibiricus L) on preneoplastic and neoplastic mammary gland growth in multiparous GR/A mice. Anti-cancer Res. 12:141-143. [PubMed] [Google Scholar]
- 31.Nakai, K., and M. Kanehisa. 1991. Expert system for predicting protein localization sites in gram-negative bacteria. Proteins 11:95-110. [DOI] [PubMed] [Google Scholar]
- 32.Neumann, G. M., R. Condron, I. Thomas, and G. M. Polya. 1996. Purification and sequencing of multiple forms of Brassica napus seed napin large chains that are calmodulin antagonists and substrates for plant calcium-dependent protein kinase. Biochim. Biophys. Acta Protein Struct. Mol. Enzymol. 1295:34-43. [DOI] [PubMed] [Google Scholar]
- 33.Ngai, P. H. K., and T. B. Ng. 2003. Isolation of a napin-like polypeptide with potent translation-inhibitory activity from Chinese cabbage (Brassica parachinensis cv. green-stalked) seeds. J. Pept. Sci. 9:442-449. [DOI] [PubMed] [Google Scholar]
- 34.Park, S. W., N. M. Stevens, and J. M. Vivanco. 2002. Enzymatic specificity of three ribosome-inactivating proteins against fungal ribosomes, and correlation with antifungal activity. Planta 216:227-234. [DOI] [PubMed] [Google Scholar]
- 35.Park, S. W., R. Vepachedu, N. Sharma, and J. M. Vivanco. 2004. Ribosome-inactivating proteins in plant biology. Planta 219:1093-1096. [DOI] [PubMed] [Google Scholar]
- 36.Parkash, A., T. B. Ng, and W. W. Tso. 2002. Purification and characterization of charantin, a napin-like ribosome-inactivating peptide from bitter gourd (Momordica charantia) seeds. J. Pept. Res. 59:197-202. [DOI] [PubMed] [Google Scholar]
- 37.Patkar, R., and B. Chattoo. 2006. Transgenic indica rice expressing ns-LTP-like protein shows enhanced resistance to both fungal and bacterial pathogens. Mol. Breed. 17:159-171. [Google Scholar]
- 38.Punja, Z. K. 2001. Genetic engineering of plants to enhance resistance to fungal pathogens—a review of progress and future prospects. Can. J. Plant Pathol. 23:216-235. [Google Scholar]
- 39.Punja, Z. K. 2006. Recent developments toward achieving fungal disease resistance in transgenic plants. Can. J. Plant Pathol. 28:S298-S308. [Google Scholar]
- 40.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 41.Serna, A., M. Maitz, T. O'Connell, G. Santandrea, K. Thevissen, K. Tienens, G. Hueros, C. Faleri, G. Cai, F. Lottspeich, and R. D. Thompson. 2001. Maize endosperm secretes a novel antifungal protein into adjacent maternal tissue. Plant J. 25:687-698. [DOI] [PubMed] [Google Scholar]
- 42.Sharma, N., S.-W. Park, R. Vepachedu, L. Barbieri, M. Ciani, F. Stirpe, B. J. Savary, and J. M. Vivanco. 2004. Isolation and characterization of an RIP (ribosome-inactivating protein)-like protein from tobacco with dual enzymatic activity. Plant Physiol. 134:171-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tassin, S., W. F. Broekaert, D. Marion, D. P. Acland, M. Ptak, F. Vovelle, and P. Sodano. 1998. Solution structure of Ace-AMP1, a potent antimicrobial protein extracted from onion seeds. Structural analogies with plant nonspecific lipid transfer proteins. Biochemistry 37:3623-3637. [DOI] [PubMed] [Google Scholar]
- 44.Terras, F. R., H. M. Schoofs, M. F. De Bolle, F. Van Leuven, S. B. Rees, J. Vanderleyden, B. P. Cammue, and W. F. Broekaert. 1992. Analysis of two novel classes of plant antifungal proteins from radish (Raphanus sativus L.) seeds. J. Biol. Chem. 267:15301-15309. [PubMed] [Google Scholar]
- 45.Terras, F. R. G., K. Eggermont, V. Kovaleva, N. V. Raikhel, R. W. Osborn, A. Kester, S. B. Rees, S. Torrekens, F. V. Leuven, J. Vanderleyden, B. P. A. Cammue, and W. F. Broekaert. 1995. Small cysteine-rich antifungal proteins from radish: their role in host defense. Plant Cell 7:573-588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thomma, B., B. P. A. Cammue, and K. Thevissen. 2002. Plant defensins. Planta 216:193-202. [DOI] [PubMed] [Google Scholar]
- 47.Tjamos, S. E., E. Flemetakis, E. J. Paplomatas, and P. Katinakis. 2005. Induction of resistance to Verticillium dahliae in Arabidopsis thaliana by the biocontrol agent K-165 and pathogenesis-related proteins gene expression. Mol. Plant-Microbe Interact. 18:555-561. [DOI] [PubMed] [Google Scholar]
- 48.Vellicce, G., J. Ricci, L. Hernández, and A. Castagnaro. 2006. Enhanced resistance to Botrytis cinerea mediated by the transgenic expression of the chitinase gene ch5B in strawberry. Transgenic Res. 15:57-68. [DOI] [PubMed] [Google Scholar]
- 49.Xiao, Y. H., M. Luo, W. G. Fang, K. M. Luo, L. Hou, X. Y. Luo, and Y. Pei. 2002. PCR walking in cotton genome using YADE method. Acta Genetic Sin. 29:62-66. [PubMed] [Google Scholar]
- 50.Yang, X., J. Li, X. Wang, W. Fang, M. J. Bidochka, R. She, Y. Xiao, and Y. Pei. 2006. Psc-AFP, an antifungal protein with trypsin inhibitor activity from Psoralea corylifolia seeds. Peptides 27:1726-1731. [DOI] [PubMed] [Google Scholar]
- 51.Zoubenko, O., F. Uckun, Y. Hur, I. Chet, and N. Tumer. 1997. Plant resistance to fungal infection induced by nontoxic pokeweed antiviral protein mutants. Nat. Biotech. 15:992-996. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.