Abstract
A classification system based on Fourier transform infrared (FTIR) spectroscopy combined with artificial neural network analysis was designed to differentiate 12 serovars of Listeria monocytogenes using a reference database of 106 well-defined strains. External validation was performed using a test set of another 166 L. monocytogenes strains. The O antigens (serogroup) of 164 strains (98.8%) could be identified correctly, and H antigens were correctly determined in 152 (91.6%) of the test strains. Importantly, 40 out of 41 potentially epidemic serovar 4b strains were unambiguously identified. FTIR analysis is superior to PCR-based systems for serovar differentiation and has potential for the rapid, simultaneous identification of both species and serovar of an unknown Listeria isolate by simply measuring a whole-cell infrared spectrum.
Human listeriosis, caused by the pathogen Listeria monocytogenes, mainly is a consequence of the ingestion of contaminated food products and remains a significant public health problem (8, 14, 30). Since many different contamination routes for this ubiquitous bacterium have been reported (19, 27, 34), the development of rapid and accurate typing methods is of particular importance. A substantial number of sensitive, discriminatory, and reproducible typing technologies have therefore been developed (20).
L. monocytogenes expresses O-somatic and H-flagellar antigens which, by their unique combination, determine the serovar (serotype) of individual strains. Thirteen serovars have been found in this species by using specific and standardized sera (32). However, traditional serotyping presents a number of limitations such as the commercial availability and high cost of sera, as well as limited reproducibility. Palumbo et al. (28) therefore proposed a low-cost enzyme-linked immunosorbent assay combined with commercial antisera for the differentiation of L. monocytogenes serovars. This method, nevertheless, is of limited potential for application in routine laboratories due to the complex and laborious protocol. Consequently, very few diagnostic laboratories offer conventional serotyping of L. monocytogenes.
Most clinical isolates belong to serovars 1/2a, 1/2b, and 4b; among these, the majority of strains which have caused large outbreaks are serovar 4b (15). In contrast, most food strains belong to serovar 1/2c (12, 36). There is now considerable evidence that epidemic serovar 4b clones express specific virulence biomarkers (11) and carry distinct genetic markers (21, 36). So far, only a few well-known strains of serovars 1/2a and 4b have been used to study the virulence of L. monocytogenes. Virulence attributes specific to other clinical or food-related serovars cannot yet effectively be addressed. Although there are no specific legal specifications for serotyping in terms of food safety or risk assessment at this time, serovar differentiation may be helpful in the future when additional knowledge concerning the relation between the serovar and pathogenic potential of Listeria monocytogenes becomes available (14).
Infrared spectra of microorganisms reflect the overall structure of the molecular constituents of the cell (24, 25), and Fourier transform infrared spectroscopical (FTIR) analysis of intact cells has been used to distinguish bacteria at different taxonomic levels (10). Extensive reference libraries containing thousands of spectra of well-characterized microorganisms can be used for the rapid identification of unknown isolates at the species level (9, 17, 23, 26, 35). A powerful method for data processing to interpret these complex spectral patterns is an important key for a successful identification. Advanced multivariate methods such as artificial neural networks (ANN) have been shown to be especially advantageous for the analysis of subtle differences at, and partially below, the species level (7, 31, 33). Recently, Rebuffo et al. (29) described a superior ANN-based FTIR method for the identification of Listeria monocytogenes and related Listeria species.
FTIR studies on Escherichia coli and Salmonella enterica for differentiation of serotypes based on variations in their lipopolysaccharide have been undertaken (10, 16). However, neither study used a sufficient number of strains to be statistically relevant or resolved the differentiation of all serovars. Therefore, it remains unclear whether FTIR spectroscopy potentially may differentiate the serovars of pathogens, in particular if the technique is applied to a large number of strains covering a significant part of the intraspecific biodiversity. In this study, we therefore applied FTIR combined with ANN to a large and diverse collection of strains representing 12 serovars of L. monocytogenes in order to evaluate whether this method is suitable to discriminate specific spectral patterns which differentiate between L. monocytogenes serovars.
FTIR spectra of Listeria monocytogenes reflect serogroup- and serovar-specific markers.
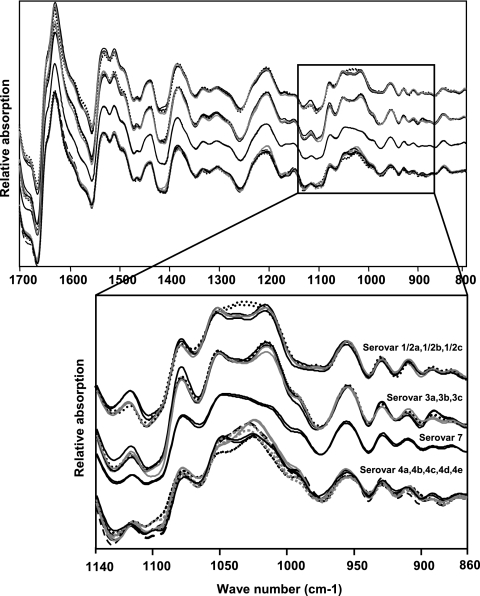
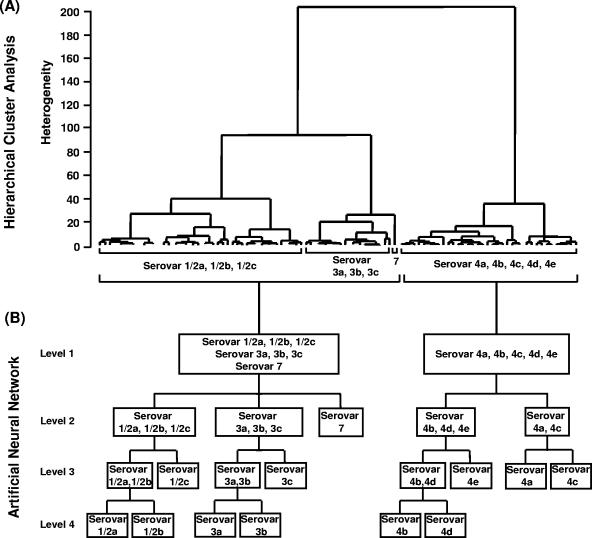
The Listeria monocytogenes strains used in this work were grown under standardized conditions on tryptone soy agar plates, and spectra were measured as described elsewhere (29). Figure 1 shows typical first derivatives of infrared spectra of 12 Listeria monocytogenes serovars. At a first glance, the polysaccharide region between 900 and 1,200 cm−1 displays the most prominent spectral differences among the four serogroups (1/2, 3, 7, and 4). This indicates that carbohydrate-containing structures are involved in serogroup discrimination. Moreover, subtle spectral differences within each serogroup due to serovar-specific markers were observed. FTIR spectra of 106 L. monocytogenes strains, including the 69 reference strains of L. monocytogenes studied by Rebuffo et al. (29) and comprising all known serovars (see Table S1 in the supplemental material) were used for hierarchical cluster analysis (HCA) (Fig. 2A). This HCA was performed as described by Rebuffo et al. (29) using the first derivative of the original spectra covering the regions from 900 to 1,200 and 1,400 to 1,800 cm−1. One major cluster corresponded to serogroups 1/2, 3, and 7, while another cluster corresponded to serogroup 4. This observation is in accordance with two distinct structural types of teichoic acids found previously for serogroups 1/2, 3, and 7 versus serogroup 4 (6). A serogroup 4-specific gene cassette which is absent in serovar 1/2b strains has been found (18), and DNA array studies of 13 genes involved in cell wall biosynthesis revealed the same grouping of L. monocytogenes serovars (4). These two major serovar groups were used to establish level 1 of a four-layer neural net (Fig. 2B). The construction and optimization of the ANN are described in more detail in the supplemental material.
FIG. 1.
Typical first derivatives of infrared spectra of 12 Listeria monocytogenes serovars. Black dotted line, WSLC 1427, 1/2a; black solid line, WSLC 1030, 1/2b; gray solid line, WSLC 1377, 1/2c; black dotted line, WSLC 1485, 3a; black solid line, WSLC 1444, 3b; gray solid line, WSLC 11082, 3c; black dotted line, WSLC 1939, 4a; black solid line, WSLC 1634, 4b; gray solid line, WSLC 11094, 4c; gray dotted line, WSLC 1045, 4d; black dashed line, WSLC 1761, 4e; black solid line, WSLC 1932, 7. Each strain is represented by two independent spectra to show the reproducibility of the measurements. Spectra are stacked to clearly show the spectral differences among the four serogroups (1/2, 3, 7, and 4). The polysaccharide region (1,200 to 900 cm−1) has been expanded to show the subtle differences between and within the serogroups. WSLC, Weihenstephan Listeria Collection.
FIG. 2.
(A) Hierarchical cluster analysis of the first derivatives of 106 L. monocytogenes strains belonging to 12 serovars and included in the reference data set. Spectral regions used were 900 to 1,200 cm−1 and 1,400 to 1,800 cm−1 (correlation with scaling to first range according to the OPUS FT-IR program [Bruker, Ettlingen, Germany]). Cluster analysis was performed using Ward's algorithm. (B) Artificial neural network classification scheme for the discrimination of serogroups and serovars.
Validation of FTIR-based serovar differentiation.
The identification potential of this ANN was evaluated by an internal validation (compare reference 29). This internal validation (Table 1) resulted in a correct identification of 100% of the somatic antigens (serogroup level). However, only 94.3% of the flagellar antigens (serovar level) could be identified. Then, an external validation was performed using a test set of 166 Listeria monocytogenes strains isolated from food, the environment, animals, and humans. These represented 12 serovars and included the 130 L. monocytogenes strains used for external validation by Rebuffo et al. (29) (see Table S2 in the supplemental material). These strains were either identified at the species level as described by Rebuffo et al. (29) and serotyped based on agglutination reactions with antisera for L. monocytogenes (Denka Seiken Co., Japan) according to the instructions of the manufacturer or had been previously serotyped by a reference laboratory. The external validation (Table 2) resulted in the correct typing rate of 98.8% at the serogroup level and 91.6% at the serovar level. Importantly, 40 out of 41 potentially outbreak-causing serovar 4b strains were identified correctly. From 87 serovar 1/2a plus 1/2b strains, 84 were assigned to either one of these two serovars. Five serovar 1/2b strains were misidentified as 1/2a. In total, only 3.1% of the potentially pathogenic serovars 1/2a, 1/2b, and 4b were grouped with a potentially nonpathogenic serovar. However, both internal and external validations were somewhat biased since only a few strains of serovars 3c, 4a, 4c, 4e, and 7 were available, which is due to the fact that these are rarely isolated from food and never from patients.
TABLE 1.
Internal validation of the infrared spectral reference database
| L. monocytogenes serovar | No. of strains:
|
|||
|---|---|---|---|---|
| Tested | For which O-somatic antigen (serogroup) correctly identified | For which H-flagellar antigens (serovar)
|
||
| Correctly identified | Misidentified | |||
| 1/2a | 23 | 23 | 21 | 2a |
| 1/2b | 7 | 7 | 7 | |
| 1/2c | 11 | 11 | 10 | 1b |
| 3a | 8 | 8 | 8 | |
| 3b | 12 | 12 | 12 | |
| 3c | 4 | 4 | 4 | |
| 4a | 3 | 3 | 3 | |
| 4b | 24 | 24 | 22 | 2c |
| 4c | 2 | 2 | 1 | 1c |
| 4d | 9 | 9 | 9 | |
| 4e | 2 | 2 | 2 | |
| 7 | 1 | 1 | 1 | |
| Total | 106 | 106 (100%) | 100 (94.3%) | 5.7 (6%) |
Strains misidentified as serovar 1/2c.
Strain misidentified as serovar 1/2a.
Strain(s) misidentified as serovar 4d.
TABLE 2.
External validation of the infrared spectral reference database
| L. monocytogenes serovar | No. of strains:
|
||||
|---|---|---|---|---|---|
| Tested | For which O-somatic antigens (serogroup)
|
For which H-flagellar antigens (serovar)
|
|||
| Correctly identified | Misidentified | Correctly identified | Misidentified | ||
| 1/2a | 59 | 57 | 2a | 56 | 3b |
| 1/2b | 36 | 36 | 31 | 5c | |
| 1/2c | 11 | 11 | 10 | 1c | |
| 3a | 3 | 3 | 3 | ||
| 3b | 5 | 5 | 5 | ||
| 3c | 2 | 2 | 1 | 1d | |
| 4a | 1 | 1 | 1 | ||
| 4b | 41 | 41 | 40 | 1e | |
| 4c | 3 | 3 | 2 | 1f | |
| 4d | 2 | 2 | 1 | 1f | |
| 4e | 2 | 2 | 1 | 1f | |
| 7 | 1 | 1 | 1 | ||
| Total | 166 | 164 (98.8%) | 2 (1.2%) | 152 (91.6%) | 14 (8.4%) |
Strains misidentified as serogroup 3.
Strains misidentified as serovar 1/2c.
Strain(s) misidentified as serovar 1/2a.
Strain misidentified as serovar 3b.
Strain misidentified as serovar 4d.
Strain misidentified as serovar 4b.
Comparison of FTIR- and PCR-based serovar differentiation.
Due to the importance of serotyping L. monocytogenes, a few PCR-based methods have been proposed. However, some are limited to differentiation of strains into only two or three serovar groups (2, 13, 22) or to differentiation of only two serovars from the others (37). Others are more complicated since two or even three independent PCRs are needed (1, 13). In contrast, the PCR system developed by Doumith et al. (3), in a one-step multiplex PCR, allows the differentiation of L. monocytogenes strains into four “serovar groups”. Group 1 comprises serovars 1/2a and 3a; group 2 comprises serovars 1/2c and 3c; group 3 contains serovars 1/2b, 3b, and 7; and group 4 comprises serovars 4b, 4d, and 4e. However, individual serovars cannot be separated. We applied this PCR system to the 166 strains of our external-validation strain set. The PCR-based method correctly differentiated 159 (4 not typeable and 3 incorrectly typed [see Table S2 in the supplemental material]) out of the 166 strains (95.8%) to the serovar group level, which is comparable to its previous validation (5). FTIR-based serotyping was able to correctly discriminate 164 out of the 166 strains (98.8%) at the serogroup level. In addition, FTIR is much more discriminatory since 91.6% of the individual serovars can be determined (Table 2).
Conclusion.
Rebuffo et al. (29) previously described an ANN-based FTIR method for the reliable identification of all Listeria species in only 25 h. Here, we report on the development of an L. monocytogenes ANN subnet in order to additionally identify serogroups and serovars. The integration of both classification systems now offers the possibility to simultaneously identify Listeria at the species level, L. monocytogenes at the serogroup level, and most L. monocytogenes strains at the serovar level in a single step by simply measuring an infrared spectrum of a pure Listeria culture. Our data indicate that this method is superior to molecular approaches for L. monocytogenes serovar determination. We suggest that FTIR identification and serotyping constitute a rapid and inexpensive tool which may be suitable for diagnostic laboratories. This tool may be used routinely in food control to gain additional information on the pathogenic potential of strains isolated from the food-processing chain.
Supplementary Material
Acknowledgments
This work was supported in part by the Forschungskreis der Ernährungsindustrie e. V. (Bonn), the Arbeitskreis für industrielle Forschung, and the Ministry of Economics and Technology, project no. 14126N.
We thank H. Hof, Mannheim, Germany, C. Montel, Aurillac, France, and K. Pellicer, La Plata, Argentina, for kindly providing Listeria monocytogenes strains.
Footnotes
Published ahead of print on 1 December 2006.
Supplemental material for this article may be found at http://aem.mcb.org/.
REFERENCES
- 1.Borucki, M. K., and D. R. Call. 2003. Listeria monocytogenes serotype identification by PCR. J. Clin. Microbiol. 41:5537-5540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Comi, G., L. Cocolin, C. Cantoni, and M. Manzano. 1997. A RE-PCR method to distinguish Listeria monocytogenes serovars. FEMS Immunol. Med. Microbiol. 18:99-104. [DOI] [PubMed] [Google Scholar]
- 3.Doumith, M., C. Buchrieser, P. Glaser, C. Jacquet, and P. Martin. 2004. Differentiation of the major Listeria monocytogenes serovars by multiplex PCR. J. Clin. Microbiol. 42:3819-3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Doumith, M., C. Cazalet, N. Simoes, L. Frangeul, C. Jacquet, F. Kunst, P. Martin, P. Cossart, P. Glaser, and C. Buchrieser. 2004. New aspects regarding evolution and virulence of Listeria monocytogenes revealed by comparative genomics and DNA arrays. Infect. Immun. 72:1072-1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Doumith, M., C. Jacquet, P. Gerner-Smidt, L. M. Graves, S. Loncarevic, T. Mathisen, A. Morvan, C. Salcedo, M. Torpdahl, J. A. Vazquez, and P. Martin. 2005. Multicenter validation of a multiplex PCR assay for differentiating the major Listeria monocytogenes serovars 1/2a, 1/2b, 1/2c, and 4b: toward an international standard. J. Food Prot. 68:2648-2650. [DOI] [PubMed] [Google Scholar]
- 6.Fiedler, F., J. Seger, A. Schrettenbrunner, and H. P. R. Seeliger. 1984. The biochemistry of murein and cell wall teichoic acids in the genus Listeria. Syst. Appl. Microbiol. 5:360-376. [Google Scholar]
- 7.Goodacre, R., E. M. Timmins, P. J. Rooney, J. J. Rowland, and D. B. Kell. 1996. Rapid identification of Streptococcus and Enterococcus species using diffuse reflectance-absorbance Fourier transform infrared spectroscopy and artificial neural networks. FEMS Microbiol. Lett. 140:233-239. [DOI] [PubMed] [Google Scholar]
- 8.Gray, M. J., N. E. Freitag, and K. J. Boor. 2006. How the bacterial pathogen Listeria monocytogenes mediates the switch from environmental Dr. Jekyll to pathogenic Mr. Hyde. Infect. Immun. 74:2505-2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Helm, D., H. Labischinski, and D. Naumann. 1991b. Elaboration of a procedure for identification of bacteria using Fourier-transform IR spectral libraries: a stepwise correlation approach. J. Microbiol. Methods 14:127-142. [Google Scholar]
- 10.Helm, D., H. Labischinski, G. Schallehn, and D. Naumann. 1991. Classification and identification of bacteria by Fourier-transform infrared spectroscopy. J. Gen. Microbiol. 137:69-79. [DOI] [PubMed] [Google Scholar]
- 11.Jacquet, C., M. Doumith, J. I. Gordon, P. M. Martin, P. Cossart, and M. Lecuit. 2004. A molecular marker for evaluating the pathogenic potential of foodborne Listeria monocytogenes. J. Infect. Dis. 189:2094-2100. [DOI] [PubMed] [Google Scholar]
- 12.Jacquet, C., E. Gouin, D. Jeannel, P. Cossart, and J. Rocourt. 2002. Expression of ActA, Ami, InlB, and listeriolysin O in Listeria monocytogenes of human and food origin. Appl. Environ. Microbiol. 68:616-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jinneman, K. C., and W. E. Hill. 2001. Listeria monocytogenes lineage group classification by MAMA-PCR of the listeriolysin gene. Curr. Microbiol. 43:129-133. [DOI] [PubMed] [Google Scholar]
- 14.Kathariou, S. 2002. Listeria monocytogenes virulence and pathogenicity, a food safety perspective. J. Food Prot. 65:1811-1829. [DOI] [PubMed] [Google Scholar]
- 15.Kathariou, S. 2000. Pathogenesis determinants of Listeria monocytogenes, p. 295-314. In J. W. Cary, J. E. Linz, and D. Bhatnagar (ed.), Microbial foodborne diseases. Technomics Publishing Co., Inc., Lancaster, PA.
- 16.Kim, S., B. L. Reuhs, and L. J. Mauer. 2005. Use of Fourier transform infrared spectra of crude bacterial lipopolysaccharides and chemometrics for differentiation of Salmonella enterica serotypes. J. Appl. Microbiol. 99:411-417. [DOI] [PubMed] [Google Scholar]
- 17.Kümmerle, M., S. Scherer, and H. Seiler. 1998. Rapid and reliable identification of food-borne yeasts by Fourier-transform infrared spectroscopy. Appl. Environ. Microbiol. 64:2207-2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lei, X. H., F. Fiedler, Z. Lan, and S. Kathariou. 2001. A novel serotype-specific gene cassette (gltA-gltB) is required for expression of teichoic acid-associated surface antigens in Listeria monocytogenes of serotype 4b. J. Bacteriol. 183:1133-1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lemunier, M., C. Francou, S. Rousseaux, S. Houot, P. Dantigny, P. Piveteau, and J. Guzzo. 2005. Long-term survival of pathogenic and sanitation indicator bacteria in experimental biowaste composts. Appl. Environ. Microbiol. 71:5779-5786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu, D. 2006. Identification, subtyping and virulence determination of Listeria monocytogenes, an important foodborne pathogen. J. Med. Microbiol. 55:645-659. [DOI] [PubMed] [Google Scholar]
- 21.Liu, D., M. L. Lawrence, L. Gorski, R. E. Mandrell, A. J. Ainsworth, and F. W. Austin. 2006. Listeria monocytogenes serotype 4b strains belonging to lineages I and III possess distinct molecular features. J. Clin. Microbiol. 44:214-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Manzano, M., L. Cocolin, C. Cantoni, and G. Comi. 1998. A rapid method for the identification and partial serotyping of Listeria monocytogenes in food by PCR and restriction enzyme analysis. Int. J. Food Microbiol. 42:207-212. [DOI] [PubMed] [Google Scholar]
- 23.Maquelin, K., C. Kirschner, L. P. Choo-Smith, N. A. Ngo-Thi, T. van Vreeswijk, M. Stammler, H. P. Endtz, H. A. Bruining, D. Naumann, and G. J. Puppels. 2003. Prospective study of the performance of vibrational spectroscopies for rapid identification of bacterial and fungal pathogens recovered from blood cultures. J. Clin. Microbiol. 41:324-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Naumann, D., D. Helm, and H. Labischinski. 1991. Microbiological characterizations by FT-IR spectroscopy. Nature 351:81-82. [DOI] [PubMed] [Google Scholar]
- 25.Naumann, D., C. P. Schultz, and D. Helm. 1996. What can infrared spectroscopy tell us about the structure and composition of intact bacterial cell? p. 279-310. In H. Mantsch, and D. Chapmann (ed.), Infrared spectroscopy of biomolecules. Wiley-Liss, New York, NY.
- 26.Oberreuter, H., H. Seiler, and S. Scherer. 2002. Identification of coryneform bacteria and related taxa by Fourier-transform infrared (FT-IR) spectroscopy. Int. J. Syst. Evol. Microbiol. 52:91-100. [DOI] [PubMed] [Google Scholar]
- 27.Paillard, D., V. Dubois, R. Thiebaut, F. Nathier, E. Hoogland, P. Caumette, and C. Quentin. 2005. Occurrence of Listeria spp. in effluents of French urban wastewater treatment plants. Appl. Environ. Microbiol. 71:7562-7566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Palumbo, J. D., M. K. Borucki, R. E. Mandrell, and L. Gorski. 2003. Serotyping of Listeria monocytogenes by enzyme-linked immunosorbent assay and identification of mixed-serotype cultures by colony immunoblotting. J. Clin. Microbiol. 41:564-571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rebuffo, C. A., J. Schmitt, M. Wenning, F. von Stetten, and S. Scherer. 2006. Reliable and rapid identification of Listeria monocytogenes and Listeria species by artificial neural network-based Fourier transform infrared spectroscopy. Appl. Environ. Microbiol. 72:994-1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rudolf, M., and S. Scherer. 2001. High incidence of Listeria monocytogenes in European red smear cheese. Int. J. Food Microbiol. 63:91-98. [DOI] [PubMed] [Google Scholar]
- 31.Schmitt, J., and T. Udelhoven. 2001. Use of artificial neural networks in biomedical diagnosis, p. 379-419. In B. Yan (ed.), Infrared and Raman spectroscopy of biological materials. Marcel Dekker, New York, NY.
- 32.Seeliger, H. P. R., and B. Langer. 1979. Serotyping of Listeria monocytogenes and related species. Methods Microbiol. 13:31-49. [Google Scholar]
- 33.Udelhoven, T., D. Naumann, and J. Schmitt. 2000. Development of a hierarchical classification system with artificial neural networks and FT-IR spectra for the identification of bacteria. Appl. Spectrosc. 54:1471-1479. [Google Scholar]
- 34.Wagner, M., D. Melzner, Z. Bago, P. Winter, M. Egerbacher, F. Schilcher, A. Zangana, and D. Schoder. 2005. Outbreak of clinical listeriosis in sheep: evaluation from possible contamination routes from feed to raw produce and humans. J. Vet. Med. Ser. B 52:278-283. [DOI] [PubMed] [Google Scholar]
- 35.Wenning, M., H. Seiler, and S. Scherer. 2002. Fourier-transform infrared microspectroscopy, a novel and rapid tool for identification of yeasts. Appl. Environ. Microbiol. 68:4717-4721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yildirim, S., W. Lin, A. D. Hitchins, L. A. Jaykus, E. Altermann, T. R. Klaenhammer, and S. Kathariou. 2004. Epidemic clone I-specific genetic markers in strains of Listeria monocytogenes serotype 4b from foods. Appl. Environ. Microbiol. 70:4158-4164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang, W., and S. J. Knabel. 2005. Multiplex PCR assay simplifies serotyping and sequence typing of Listeria monocytogenes associated with human outbreaks. J. Food Prot. 68:1907-1910. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.