Abstract
Stable isotope probing (SIP) can be used to analyze the active bacterial populations involved in a process by incorporating 13C-labeled substrate into cellular components such as DNA. Relatively long incubation times are often used with laboratory microcosms in order to incorporate sufficient 13C into the DNA of the target organisms. Addition of nutrients can be used to accelerate the processes. However, unnatural concentrations of nutrients may artificially change bacterial diversity and activity. In this study, methanotroph activity and diversity in soil was examined during the consumption of 13CH4 with three DNA-SIP experiments, using microcosms with natural field soil water conditions, the addition of water, and the addition of mineral salts solution. Methanotroph population diversity was studied by targeting 16S rRNA and pmoA genes. Clone library analyses, denaturing gradient gel electrophoresis fingerprinting, and pmoA microarray hybridization analyses were carried out. Most methanotroph diversity (type I and type II methanotrophs) was observed in nonamended SIP microcosms. Although this treatment probably best reflected the in situ environmental conditions, one major disadvantage of this incubation was that the incorporation of 13CH4 was slow and some cross-feeding of 13C occurred, thereby leading to labeling of nonmethanotroph microorganisms. Conversely, microcosms supplemented with mineral salts medium exhibited rapid consumption of 13CH4, resulting in the labeling of a less diverse population of only type I methanotrophs. DNA-SIP incubations using water-amended microcosms yielded faster incorporation of 13C into active methanotrophs while avoiding the cross-feeding of 13C.
Molecular techniques provide increasingly sophisticated ways to characterize the diversity of active microbial populations in the natural environment. Combining stable isotopes (13C) with advanced molecular techniques provides new ways to understand the processes and organisms that drive biogeochemical cycles. Stable-isotope probing (SIP) is a cultivation-independent technique that enables taxonomic identity to be linked with function in the environment. Recently, SIP techniques, utilizing 13C-labeled substrates, have been used to determine the active bacteria within the total bacterial community that contribute to particular metabolic pathways (44, 50, 53, 56). Different biomarkers, including phospholipid fatty acids, ergosterol, DNA, and RNA, have been used in SIP-based studies (5, 39, 40, 52, 53). Of these biomarkers, nucleic acids may be the most useful because they contain the most taxonomic information and have the greatest turnover rates (discussed in references 13 and 56).
DNA recovered after SIP incubations can be separated into heavy, labeled DNA (13C) and unlabeled DNA (12C) by cesium chloride (CsCl) gradient centrifugation (53). Amplification products obtained after PCR with heavy and light DNA fractions as templates can then be used for direct cloning, sequencing, and phylogenetic identification or community profiling analyses, such as terminal restriction fragment length polymorphism (T-RFLP) and denaturing gradient gel electrophoresis (DGGE). DNA-SIP has been used previously to characterize bacteria metabolizing C1-compounds such as methane, methanol, and methyl halides in various environments (4, 26, 36, 37, 38, 43, 44, 53, 55), as well as multicarbon compounds, including acetate, glucose, caffeine, naphthalene, phenol, salicylate, phenanthrene, and 2,2′-dichlorobiphenyl (8, 28, 50, 60, 63, 69). DNA-SIP shows considerable promise for estimating the diversity of active populations of microorganisms in the environment (13, 42), and the benefits and pitfalls of this technique, as well as the influence of incubation period, substrate concentration, and cross-feeding, have been reviewed (17, 38, 56, 65). Initial studies (55) used relatively long incubation times (>40 days) to generate DNA with sufficient incorporation of label to visualize [13C]DNA on a gradient, but more recent studies have successfully used shorter incubation times (18, 19, 26, 36, 64). RNA-SIP studies (20, 40-42, 49) generally require shorter incubation periods since RNA is labeled faster than DNA (20, 29, 30, 38, 41), but the recovery of pure RNA from soil is still difficult.
DNA-SIP is particularly appropriate for study of the diversity of methane-oxidizing bacteria (MOB), using 13CH4 as a substrate (5, 10, 26, 36, 44, 48, 55). MOB use CH4 as the sole carbon and energy source and are currently classified as type I and type II methanotrophs, which differ in intracytoplasmic membrane structure, carbon assimilation pathways, fatty acid composition, and phylogeny (7, 21). The first step in the microbial oxidation of CH4 to CO2 is the conversion to methanol by the enzyme methane monooxygenase (MMO), which exists in two forms: a particulate membrane-bound form (pMMO) and a soluble, cytoplasmic form (sMMO). The presence of pMMO has been reported in all methanotrophs except the genus Methylocella (11, 14), whereas sMMO appears to occur only in a restricted range of methanotrophs. pmoA, encoding the active site subunit of pMMO, is therefore a suitable functional marker gene for methanotrophs and has been used in cultivation-independent methods to characterize methane-oxidizing communities in different environments.
The aim of the present study was to determine the effects of different incubation regimens on the outcome of DNA-SIP incubations using a field soil sample and to test the hypothesis that nutrient-amended microcosms will impact the outcome of DNA-SIP experiments. The diversity of active methanotrophs in a field soil sample was analyzed for three different incubation conditions by cultivation-independent techniques. In order to provide robust datasets for methanotroph 16S rRNA genes, both DGGE fingerprinting and clone library analysis of 16S rRNA genes were carried out. The data from these analyses were complemented with studies of the functional gene pmoA from active methanotrophs by clone library and microarray analyses (3).
MATERIALS AND METHODS
Sample description and potential methane oxidation rates.
Sampling of Wytham soil, a gleyic soil (pH 7.25; particle size, 22% clay [<0.005 mm], 31% silt, 47% sand [>0.05 mm]) was carried out in August 2004 at the University of Oxford farm, Wytham, Oxford, United Kingdom. At the time of sampling, the moisture content of the soil was 23.2% (wt/wt), assessed by determining the loss in mass after incubation of the soil at 105°C for 24 h. The soil did not produce methane, and it did not oxidize atmospheric concentrations of methane. The Wytham soil was air dried and sieved (2-mm-diameter pore size) to homogenize the soil. Roughly 20% of the initial mass was removed since it was >2.0 mm in diameter. Potential methane oxidation rates were measured on both crude and sieved soil (with adjusted water content) to determine the effects of sieving on potential activity. Activities at 20°C were determined in triplicate 1-g soil (equivalent to dry soil weight) samples in 120 ml, crimp-top serum vials, into which 0.6 ml of CH4 (0.5% [vol/vol]) had been injected with a gas-tight syringe. Samples were incubated in the dark at 20°C with gentle agitation. Headspace CH4 concentrations were measured daily by gas chromatography-flame ionization detection, and concentrations were calculated by comparison to a standard containing methane (0.5% CH4 in ultrapure He; Linde Gases). Measurements in triplicate for each time point gave errors of <5%.
Incubations for stable isotope probing.
DNA-SIP was carried out by using the methods of Radajewski and Murrell (54) and Morris et al. (44). Three soil incubations were established with 5 g of dried and sieved Wytham soil placed in a 120-ml crimp-top serum vial. Two were made into slurries using 5 ml of either 1/10th strength nitrate mineral salts (NMS) medium (66) or distilled water, and the third was adjusted to natural moisture content by the addition of 1.16 ml of distilled water. Vials were then sealed with a butyl rubber stopper and injected with 5 ml of 13CH4 (>99% pure; Linde Gases). Soil slurries were incubated in the dark at 20°C with gentle agitation, and headspace 13CH4 concentrations were measured every few days (depending on the rate of methane consumption) by gas chromatography. After >90% of the 13CH4 was consumed, the vials were flushed with air to remove any 13CO2 and to ensure that the slurries remained aerobic, and another 5 ml of 13CH4 was then added. The vials were incubated until 0.6 mmol of 13CH4 (three injections of 5 ml of CH4) for the NMS and water slurries and 0.2 mmol of 13CH4 (one injection of 5 ml of CH4) for the natural moisture content microcosm had been consumed. Slurries were then centrifuged (25,000 × g, 15 min) and, together with the soil pellet, were stored at −80°C until DNA extraction was carried out.
DNA extraction and separation of 13C- and 12C-labeled DNA.
DNA was extracted from each microcosm soil (5 g of soil plus liquid phase) using a CO2-cooled bead beating method (68) combined with a Bio 101 FastDNA spin kit for soil (QBiogene). This method was adapted from that described by Radajewski and Murrell (53) and Morris et al. (44). DNA was mixed with an equal weight of CsCl and transferred to 5.1-ml Beckman quick-seal centrifuge tubes. Ethidium bromide (100 μl of a 10-mg ml−1 solution) and CsCl solution (1 g ml−1) were used to top up the tubes, which were then centrifuged (265,000 × g, 20°C for 15 h) to separate the [12C]DNA and [13C]DNA fractions. The DNA from the NMS slurry produced two visible bands, separated by 1 cm, which were collected with syringes and 20-gauge needles (58). Even if a single DNA band ([12C]DNA band) was observed for the biomass from the water slurry and the natural moisture content microcosm, the second heavier fraction was collected from these tubes at the exact position of the [13C]DNA band (1 cm below the [12C]DNA band) observed in the identical CsCl gradient containing DNA extracted from the NMS slurry microcosm. Ethidium bromide was extracted from DNA by butanol extraction, and CsCl was removed from the DNA by dialysis. DNA was then precipitated and resuspended in 100 μl of Tris-EDTA buffer and stored at −20°C.
Molecular analyses.
The 13C- and 12C-labeled DNA fractions were used as a template for PCR. All reactions (50-μl volume) contained 1 μl of template DNA (1:10 dilution corresponding to 20 ng μl−1 and 1:100 dilution corresponding to a concentration of 2 ng μl−1) and were performed as described for each primer set.
Cloning.
Active methanotrophs from the three SIP microcosms were characterized at the domain level with primers that amplify the bacterial (27F and 1492R [34]) 16S rRNA genes (1,450 bp). The 12C- and 13C-labeled DNA fractions from the three SIP microcosms were also characterized at a functional level with primers that amplify the particulate methane monooxygenase gene (pmoA, A189/mb661 [9]), following the PCR conditions described by Bourne et al. (6). After PCR amplification with these two sets of primers, purified (QIAquick PCR purification kit; QIAGEN) amplicons were ligated into the pCR 2.1 vector supplied with a TOPO TA cloning kit (Invitrogen, San Diego, CA) to generate clone libraries according to the manufacturer's instructions. Individual colonies containing inserts were suspended in 3 ml of nutrient broth containing ampicillin (50 mg ml−1) and grown overnight at 37°C. Then, 50 16S rRNA gene clones and 20 pmoA gene clones were analyzed from each of the respective clone libraries. Small-scale preparations of plasmids were performed as described by Saunders and Burke (59), and plasmids were digested with the restriction enzymes EcoRI and RsaI. DNA fragments were resolved on 2% (wt/vol) agarose gels. Clones were grouped into operational taxonomic units (OTUs) according to RFLP patterns. Clones from each OTU were partially sequenced with the primers 357f (34) and M13F (58) for the 16S rRNA gene and pmoA gene clone libraries, respectively.
DGGE.
Fragments of 16S rRNA genes, 550 bp in length, were obtained with the primer combination 341f, with a GC clamp, and 907r (46). The concentration of PCR products was estimated by comparison with a molecular mass standard by electrophoresis in 1% (wt/vol) agarose gels, and 400 ng of PCR product from each sample was analyzed on 6% (wt/vol) polyacrylamide gels containing gradients of 30 to 70% denaturant (a 100% solution consists of 7 M urea and 40% [vol/vol] formamide). Gels were run at a constant voltage of 85 V for 16 h at 60°C in 1× TAE buffer (40 mM Tris, 20 mM acetic acid, 1 mM EDTA; pH 8.3). After electrophoresis, the gels were stained for 30 min in a SYBR green (Invitrogen) solution (1/10,000 in 1× TAE buffer), rinsed in 1× TAE buffer, and scanned. The number of bands per DGGE profile was determined, and individual bands of interest were excised, left overnight in 25 μl of MilliQ water, reamplified by PCR, and examined again by DGGE to ensure purity and correct electrophoretic mobility within the gels. PCR products yielding pure bands were purified by using the QiaQuick PCR purification system (QIAGEN) and sequenced.
Microarrays.
pmoA genes were amplified using the primer set A189/T7-mb661; the reverse primers contained the T7 promoter at the 5′ end, which enabled T7 RNA polymerase mediated in vitro transcription using the PCR products as templates to generate RNA template for the hybridization on microarrays as described by Bodrossy et al. (3) and modified by Stralis-Pavese et al. (62).
DNA sequencing and phylogenetic analysis.
DNA sequencing was performed in the University of Warwick Central Molecular Biology Services Laboratory by cycle sequencing with a BigDye dyedeoxy terminator ready reaction kit (Applied Biosystems, Warrington, United Kingdom) and ABI3100 capillary DNA sequencers. Phylogenetic analysis was carried out on the 16S rRNA gene and pmoA genes from clone libraries and also from PCR products generated from DGGE bands. Sequences were aligned with the related environmental 16S rRNA gene and pmoA gene sequences, obtained by using the Blastn program in BLAST version 2.1 using the CLUSTAL X program (www.ncbi.nlm.nih.gov/BLAST). All of the 16S rRNA gene sequences were edited to 450 bp, and pmoA gene sequences were edited to 500 bp for alignment analysis. Dendrograms were constructed by using the programs DNADIST, NEIGHBOR, SEQBOOT, and CONSENSE from PHYLIP version 3.65 (16), and phylogenetic tree files were analyzed by using TreeView (51).
Nucleotide sequence accession numbers.
The pmoA gene and 16S rRNA gene sequences retrieved from Wytham soil are available from GenBank under the respective accession numbers EF015784 to EF015807 and EF015808 to EF015857.
RESULTS
Potential methane oxidation rates.
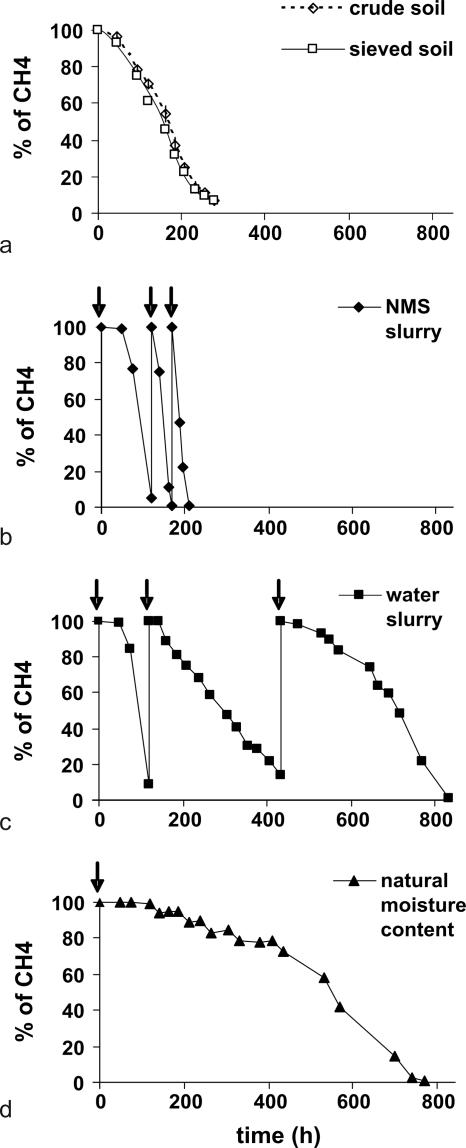
Potential methane oxidation rates were similar in the unprocessed Wytham soil and in soil that had been air dried and sieved (Fig. 1a), with values of 0.105 μmol of CH4 oxidized h−1 g of dry soil−1. Air drying and sieving therefore did not have a detectable influence on the capacity of the bacterial community to oxidize methane, and so soil that had been air dried and sieved was used in SIP incubations. Since the methane oxidation rates did not vary >5% between triplicates (Fig. 1a) used for each soil treatment, only one incubation was used in subsequent experiments.
FIG. 1.
(a) Methane consumption during measurement of the potential methane oxidation rates for unprocessed Wytham soil and the same soil after air drying, sieving, and adjustment to the original moisture content. (b to d) SIP was performed under three different incubation conditions: with NMS (b), with water (c), and at the natural moisture content (d). Arrows show the times for the injection of 5 ml of 13CH4. Methane oxidation activity was determined in triplicate, and the data presented are mean values (the standard errors were less than ±5%).
SIP incubations.
Stable isotope probing of the Wytham soil was carried out with 13CH4 in three microcosms under different conditions: in NMS slurry, in distilled water slurry, and in soil at its original moisture content. Figure 1b, c, and d show the methane consumption rates for the three microcosms. The soil in the NMS and water slurry microcosms consumed 15 ml of 13CH4 in 8.75 and 35 days, respectively. The rate of methane oxidation increased for each new methane injection in the NMS slurry microcosm with values of 0.558, 0.833, and 0.939 μmol of CH4 oxidized h−1 g of dry soil−1. These could be explained by the growth of the methane-oxidizing bacterial population due to the availability of nutrients in NMS medium. In contrast, in the water slurry microcosm, the rates of methane oxidation decreased after each methane injection with consecutive values of 0.557, 0.127, and then 0.100 μmol of CH4 oxidized h−1 g of dry soil−1, possibly through the rapid consumption of available nutrients in the early stages of incubation. Another possibility was that a shift in the population of the methanotrophs occurred, although this is less likely given the time scale and availability of nutrients. Incubation of the first two microcosms was terminated after the consumption of 15 ml of 13CH4. The soil in the third microcosm, which had natural moisture content, exhibited a very low methane oxidation rate (0.052 μmol of CH4 oxidized h−1 g of dry soil−1) and consumed the equivalent of only one 13CH4 aliquot (5 ml) in 32 days. Incubation was terminated at this time because the incorporation of a total of 15 ml of 13CH4 would have taken too long and such a period would almost certainly have modified the diversity of the total bacterial community.
Total DNA was extracted from microcosm samples and subjected to ultracentrifugation. Light DNA fractions ([12C]DNA) were visible under UV illumination in each tube, but the heavy DNA band ([13C]DNA) was only visible to the naked eye in the tube containing DNA from the NMS slurry microcosm. However, the gradient fractions corresponding in each tube to the light bands and the heavy bands were sampled: the [13C]DNA fraction was recovered 1 cm below the [12C]DNA fraction, the position of the [13C]DNA band observed in the NMS slurry tube. After butanol extraction and dialysis of the DNA, all fractions contained significant amounts of DNA for amplification of the 16S rRNA and functional genes (pmoA) by PCR.
Diversity of pmoA genes in 12C- and 13C-labeled DNA fractions.
The diversity of methane oxidizers was investigated by targeting the “functional gene” encoding the particulate methane mono-oxygenase (pmoA). In order to avoid bias potentially arising from one technique and to compare the results obtained by different methods, pmoA diversity was investigated by microarray analysis and by clone library analysis. These were done on 12C- and 13C-labeled DNA fractions from the three SIP incubations to compare the active MOB population with the total methanotroph community and to compare the effects of different incubation conditions during SIP experiments on the activation, stimulation, or repression of specific groups of methane oxidizers.
Microarray analyses yielded data on the diversity and semiquantitative information on the proportion of phylogenetically distinct pmoA sequences belonging to different phylogenetic groups or species of methanotrophs. The probes used on the pmoA microarray are comprehensive and cover all pmoA genes from methanotrophs that were known in 2004 (3, 62). The microarray experiments therefore give information on the general distribution of pmoA clones arising from SIP experiments between the type I and type II methanotrophs and on their affiliation with all genera of methanotrophs.
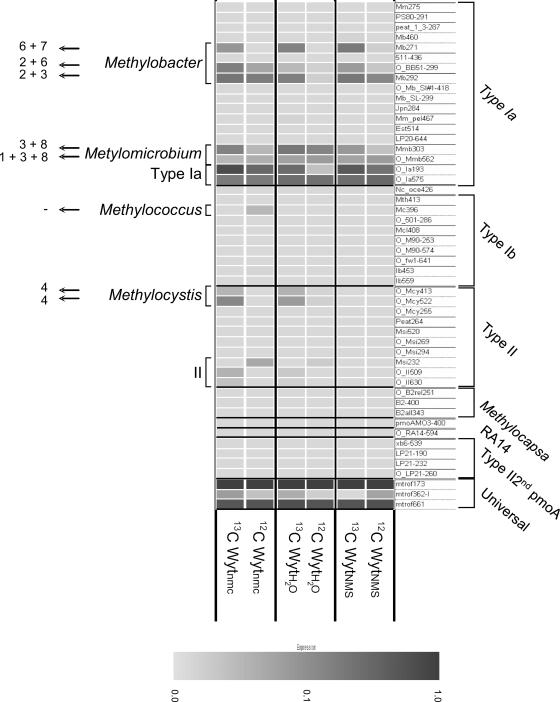
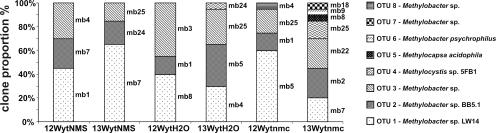
Figure 2 shows the microarray results obtained with the six DNA samples. In general, the diversity of pmoA sequences recovered from the [12C]DNA was less than the diversity of pmoA sequences in the [13C]DNA fraction in each of the three SIP experiments. The majority of methanotrophs were type I methanotrophs, represented mainly by Methylobacter-like clones and not Methylomonas. The diversity of active Methylobacter spp., irrespective of the SIP incubation conditions, was shared mainly between four subclusters represented by hybridization to the probes Mb271, O_BB51-299, and Mb292 (for a description of the probes, see reference 3). The main difference between the three SIP incubations was the activation of type II methanotrophs closely related to Methylocystis. Indeed, Methylocystis-like sequences that hybridized with the two specific probes—O_Mcy413 and O_Mcy522—were recovered from water and the natural moisture content SIP microcosms DNA. Analysis of the same DNA samples by cloning, RFLP analyses, and sequencing of representative clones from each OTU showed results remarkably consistent with those obtained by analysis with the microarrays (Fig. 3 and see Fig. S1 in the supplemental material). The same primer set (A189/mb661) was used in the two methods to enable comparison of results. The pmoA diversity found in the [12C]DNA fractions for the three different SIP treatments was mainly represented by three OTUs (Fig. 3). These three OTUs (OTUs 1, 2, and 3) are represented by members of the type I methanotrophs related to Methylobacter species (see Fig. S1 in the supplemental material). The same three OTUs were also found in the active fraction ([13C]DNA derived sequences) of the population. In the NMS slurry, methanotrophs containing pmoA from OTU 1 (closely related to Methylobacter sp. strain LW14 isolated from Lake Washington sediment) seem to be the most active bacteria. The proportion of the sequences increased from 45 to 65% between the 12C- and 13C-labeled DNA fractions. In contrast, the same OTU 1 seems to be less active during the water slurry and the natural moisture content SIP incubations. The water slurry treatment induced the activity of the methanotrophs from which the sequences belong to the OTU 2 (closely related to the uncultivated clone LOPB13.2 (AF358052) found in peat soil (44) representing 35% of the clones compared to the 15% recovered from the [12C]DNA fraction.
FIG. 2.
Microarray results showing hybridization efficiency obtained with the microarray probe set described in Stralis-Pavese et al. (62) on light and heavy DNA (12C and 13C) extracted from three SIP treatments: NMS-supplemented slurry, water-supplemented slurry, and under natural moisture content (nmc). The relative signal intensities are indicated by gray shading, as shown on the color bar (a value of 1 corresponding to the highest signal intensity obtained for the given probe during probe set validation). OTUs (Fig. 3 and see Fig. S1 in the supplemental material) corresponding to positive signals on specific probes are indicated by numbers. Methylobacter-, Methylomicrobium-, Methylococcus-, and Methylocystis-specific probes are as indicated. Ia, type Ia universal probes; II, type II universal probes. The sensitivity of the microarray allows the detection of MOB present at 5% or more of the total methanotroph community (i.e., the community targeted by the PCR).
FIG. 3.
Proportion of clones found in the different OTUs obtained from pmoA clone libraries constructed from both 12C- and 13C-labeled DNA from all three SIP incubations (20 pmoA clones per library). These clone libraries were designated WytNMS, WytH2O, and Wytnmc for the NMS-supplemented slurry, the water-supplemented slurry, and the natural moisture content microcosm, respectively. The number of representative clones per OTU is shown, with “mb” indicating that the clone library was constructed using PCR products derived from the mb661 primer set. The phylogenetic affiliation of the pmoA sequences indicated by mb4, mb7, mb1, etc., is shown in Fig. S1 in the supplemental material.
During the water slurry and the natural moisture content incubations, a fourth OTU (OTU 4) appeared in the [13C]DNA fraction (Fig. 3); the clones from this group belonged to type II methanotrophs closely related to Methylocystis sp. strain 5FB1 (see Fig. S1 in the supplemental material), an oligotrophic strain isolate from upland soil under a low methane mixing ratio (32). As shown by the microarray result, these two incubation treatments induced the activity of type II methanotrophs and not just that of type I methanotrophs. Finally, the main difference in the diversity of the active populations is shown in the natural moisture content SIP experiment, where seven OTUs were found. Two new OTUs (OTUs 6 and 7) containing pmoA belonging to type I methanotrophs were identified: one was closely related to the phylogenetic branch represented by pmoA from Methylobacter sp. strain LW12 and Methylobacter psychrophilus, and the second was closely related to some pmoA clones from OTU 3. The third new methanotroph group (OTU 5), which was detected in the [13C]DNA (active fraction) from the natural moisture content SIP incubation, belonged to type II methanotrophs, and its pmoA is closely related to Methylocapsa acidophila in a cluster represented by uncultivated clones isolated mainly from upland and forest soils, in a clade close to the upland soil cluster USCα (see Fig. S1 in the supplemental material).
The advantage of using SIP incubation was that the labeled active population was entirely separated from all of the other microorganisms that did not use the 13CH4. As a consequence, a 16S rRNA gene specific primer set can be used for PCR, which could detect both known (extant) and unknown (new) methanotrophs that have been labeled with 13C.
Total bacterial diversity in the [13C]DNA fraction.
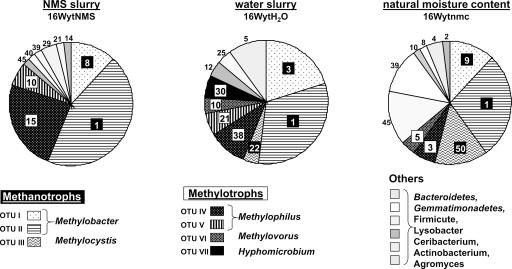
Again, two different methods were used to assess the diversity of 16S rRNA genes in light and heavy DNA fractions from the three SIP experiments. 16S rRNA gene clone libraries were constructed using the three [13C]DNA fractions, recovered from incubations with NMS, water, and original moisture content, as a template. One 16S rRNA gene clone library was constructed using the [12C]DNA fraction recovered from the SIP incubation with NMS. Of the 50 clones analyzed, none of the RFLP patterns was similar to those found for the [13C]DNA clone libraries. None of the clones from the library constructed using the [12C]DNA fraction was sequenced. For the three clone libraries derived from the “heavy” [13C]DNA fractions, 10 to 11 OTUs were found, 6 of which were detected at least in two libraries (Fig. 4). Generally, three OTUs (OTUs I, II, and III) corresponded to 16S rRNA genes from methanotrophs closely related to Methylobacter and Methylocystis spp., and four were affiliated with the 16S rRNA genes from methylotrophs (OTUs IV, V, VI, and VII) closely related to Methylophilus, Methylovorus, and Hyphomicrobium spp. All of the other OTUs represented by one or two clones from each library were affiliated with phylogenetic groups not considered to be methanotrophs or methylotrophs and belonging to Bacteroidetes; Firmicutes; Actinobacterium; and β, δ-, and γ-Proteobacteria (Fig. 4 and see Fig. S2 in the supplemental material). There is still of course the formal possibility that members of these genera might have the ability to oxidize methane, and this should be tested in the future.
FIG. 4.
Proportion of 16S rRNA genes in clone libraries constructed using [13C]DNA from the three SIP incubations (50 16S rRNA gene clones per library). The names of the representative groups of clones within each OTU correspond to the initial Blastn surveys of these sequences. The number of representative clones per OTU is indicated and their phylogenetic affiliations are shown in Fig. S2 in the supplemental material.
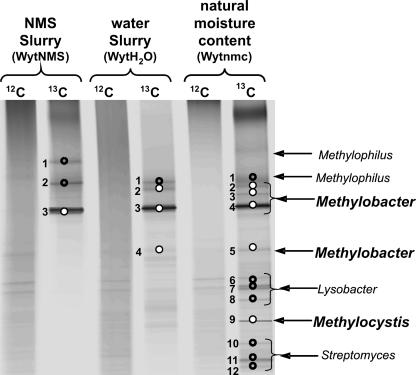
DGGE analysis was performed on light and heavy DNA fractions recovered from the three SIP experiments to compare the results with those of clone libraries (Fig. 5). There were very many bands observed in DGGE gels obtained with [12C]DNA since 16S rRNA genes from the total soil bacterial population were amplified. In contrast, well-defined bands were visualized for the DGGE pattern of 16S rRNA genes amplified from [13C]DNA fractions. The pattern obtained with heavy DNA from the SIP experiment carried out in soil at the original moisture content, presented more bands than that obtained with heavy DNA from the SIP experiment set up with water and with NMS. The bands were reamplified and sequenced, and a phylogenetic analysis was done combining 16S rRNA gene sequences from DGGE bands and clone libraries (see Fig. S2 in the supplemental material). One major DGGE band sequence (WytNMS3-DGGE) belonging to Methylobacter spp. was recovered from the heavy DNA fraction of the SIP experiment with NMS (Fig. 5). This band corresponded to sequences (16WytNMS1 was the representative clone) belonging to OTU II that grouped 44% of the 16S rRNA clones. Irrespective of the SIP treatments, this major Methylobacter-like group (OTU II), closely related to Methylobacter sp. strain LW14, was dominant in clone libraries and DGGE analyses. The OTU I (16WytNMS8, 16WytH2O3, and 16Wytnmc9) identified in the three clone libraries was composed of 16S rRNA gene sequences closely related to that of Methylobacter luteus. These 16S rRNA clone sequences grouped with two DGGE bands (WytH2O4- and Wytnmc5-DGGE), which were only visible in heavy DNA fractions from SIP experiments carried out with water and under natural moisture content. Finally, the third group (OTU III) of methanotroph 16S rRNA clones (represented by 16WytH2O22 and 16Wytnmc50) representing 4 and 14% of the clone libraries, for SIP experiments with water and under natural moisture content, respectively, belonged to the α-Proteobacteria closely related to Methylocystis. These clones grouped with one DGGE band (Wytnmc9-DGGE) recovered from heavy DNA from the SIP experiment done under natural moisture content.
FIG. 5.
Fingerprints from DGGE experiments targeting the 16S rRNA genes from 12C- and 13C-labeled DNA fractions from the three SIP incubations (NMS slurry, water slurry, and natural moisture content microcosms). Bands that were reamplified, purified, and sequenced are labeled with circles, and the phylogenetic affiliations of these 16S rRNA gene sequences, according to Blastn, are indicated.
As found with the 16S rRNA gene clone library analyses, the last two bands (WytNMS1- and WytNMS2-DGGE) visualized by DGGE from the SIP experiment with NMS were methylotrophs closely related to Methylophilus spp. These bacteria were probably labeled by the incorporation of methanol or other oxidation products arising from the metabolism of 13CH4. For the other [13C]DNA fractions from the two SIP experiments done with water and at the original moisture content, similar 16S rRNA gene DGGE bands (WytH2O1- and Wytnmc1-DGGE) corresponding to the same Methylophilus-like group were recovered. As might be expected, more diverse 16S rRNA gene sequences of methylotrophs were recovered with the clone libraries (OTU IV, V, VI, and VII) than with DGGE analysis. Finally, the two methods used confirmed that during the SIP experiment undertaken in natural moisture content conditions, ca. 35% of the bacterial population that incorporated 13C compounds were not methanotrophs but methylotrophs belonging to the various phylogenetic groups cited above.
DISCUSSION
Previous studies using SIP have used laboratory systems in which the incorporation of 13C-labeled substrate is optimized by the application of relatively high concentrations of substrates, repeated applications, minimization of losses of labeled substrates, and the addition of a relatively high nutrient concentration. A long incubation period can select for bacteria that are well adapted to laboratory enrichment conditions, leading to misinterpretation of results. In the present study, three SIP incubation conditions, with different nutrient regimens, were tested in order to determine the possible enrichment effects on the active MOB and subsequent DNA-SIP results obtained. The Wytham soil studied here was well suited to assessing the effect of the SIP incubation or treatment on MOB because the diversity of this functional group was quite low, facilitating identification of the differences in populations of the different types of methanotrophs. Consistency in results obtained by using different sensitive and complementary molecular techniques to assess the diversity of methanotrophs allowed confidence in the results obtained.
The SIP methodology facilitated the separation of DNA from active MOB from DNA of the whole microbial community. Diversity analyses of bacteria involved in this process could then be studied by fingerprinting methods such as DGGE and cloning and sequencing. Targeting the functional gene pmoA provides information on the functionality of bacteria in this environment. However, one drawback of this method is that the PCR primers used for amplification of pmoA gene have been designed using known pmoA sequences from extant methane oxidizers; hence, widely divergent pmoA sequences from unknown methanotrophs might not be detected by this method. Nevertheless, pmoA clone library and microarray analyses (3) combined with SIP technology, providing 13C-labeled DNA from active bacteria, allowed a detailed study of methanotroph diversity and activity. This was supported by DNA-SIP and 16S rRNA gene-based phylogenetic analyses using DGGE and clone libraries.
pmoA analyses revealed that 100% of the active methanotrophs detected by pmoA clone library and microarray analyses were type I methanotrophs closely related to Methylobacter and/or Methylomicrobium species in the NMS-enriched SIP incubation. However, a significant proportion of type II methane oxidizers closely related to Methylocystis were detected in the DNA-SIP incubations using water and natural moisture content. For the three SIP experiments, the type I methanotrophs detected were closely related to Methylobacter sp. strain LW14, a strain isolated from Lake Washington sediment (1) that has recently been renamed Methylosarcina lacus. Methanotrophs related to LW14 were active in the three different SIP incubation microcosms, suggesting that it played a major role in methane oxidation in situ in the Wytham soil. The two other groups of type I methanotrophs were closely related to a branch represented by Methylobacter sp. strain LW1 (9) and Methylomicrobium album (23) and to a cluster closely related to Methylobacter sp. strain BB5.1 (61). Type I methanotrophs are thought to prevail in systems with high nitrogen concentrations (2) and might outcompete type II methanotrophs under NMS-supplemented SIP incubation that can favor the enrichment of fast-growing bacteria (53, 56, 67). Many type II methanotrophs can fix N2 (45), and it is interesting that sequences from these groups of methanotrophs were recovered from water-supplemented and natural moisture content microcosms.
One interesting clone (13Wytnmc_mb8) belonging to OTU 5 (see Fig. S1 in the supplemental material), which accounted for 5% of the pmoA clone library from the natural moisture content SIP microcosm, is closely related to Methylocapsa acidiphila strain B2 (AJ278727) and to some pmoA sequences (DGGE band AC-A2, AJ868283) described by Knief and Dunfield (33) belonging to a group only detected with the mb661 primer, which was closely related to but clearly distinct from previously detected USCα (upland soil cluster) sequences. These unique pmoA gene sequences (for which no isolates are known) have also been identified in a number of culture-independent studies of environmental samples and are believed to belong to specialized MOB adapted to atmosphere trace levels of methane (22, 24, 25, 31, 32). It is interesting to speculate that if SIP microcosm incubation had been done with a methane concentration closer to atmospheric values, this cluster might have represented a larger proportion of the active methanotrophs.
The methanotroph diversity observed in 16S rRNA gene clone libraries was lower than that observed with pmoA, and one explanation for this could be that two species of Methylobacter can possess two different pmoA genes but similar 16S rRNA gene sequences which would not be discriminated by RFLP analyses of the clones. The results from a sequencing analyses of 16S rRNA gene clones and 16S rRNA DGGE bands were also congruent according to their phylogeny (see Fig. S2 in the supplemental material). The two techniques gave complementary information about 16S rRNA gene sequences from methanotrophs, and the major band observed on DGGE (WytNMS3-, WytH2O3-, and Wytnmc4-DGGE) grouped with clones from OTU II represented by 28 to 44% of the clones and was closely related to Methylobacter sp. strain LW14. This confirmed results from pmoA analyses that led to the identification of the same major Methylobacter-related group. Globally, the results from pmoA and 16S rRNA gene analyses are congruent, with both indicating that 100% of active methanotrophs were type I during the NMS-enriched SIP incubation. This may have been due to an enrichment effect due to the extra mineral salts provided. The results also showed the appearance of active type II Methylocystis strains during water SIP incubation and in SIP-natural moisture content microcosms, where these type II bacteria represent up to 25% of the active methanotrophs detected.
When assessing the bacterial diversity in the heavy DNA fractions, we recovered not only methanotrophs but also some known methylotrophic bacteria and some other phylogenetic groups (Gemmatimonadetes, Bacteroidetes, Xanthomonadaceae as Lysobacter, Actinobacteria, and Firmicutes) not previously known to be involved in methylotrophic processes. This could be due to contamination of the heavy DNA gradient fraction by [12C]DNA, in particular from bacteria with high GC contents (56) and/or cross-feeding mechanisms. In fact, the long incubation time of 35 days required for the consumption of methane in water and natural moisture content slurries probably increased the potential for the isotopic label to be passed down the food chain into the biomass of nontarget organisms. For example, it has previously been shown in elegant studies by Lueders et al. (37) using 13C-labeled methanol that trophic interactions with methylotrophs and fungi and protozoa can occur in prolonged SIP incubations. Methanol (35), CO2, and other 13C-labeled metabolites, formed from 13CH4 oxidation, could potentially accumulate, and these compounds could be substrates for nontarget microorganisms during too long an incubation time (44, 53). Furthermore, microbial scavenging of labeled biomass can rapidly occur (12, 41). This could largely explain the fact that methylotrophic bacteria such as Methylophilus, Methylovorus, and Hyphomicrobium were 13C labeled, because of their ability to oxidize methanol. Moreover, the other genera of soil bacterial identified in the heavy DNA fraction could be implicated in 13C recycling, but we cannot make any conclusions as to their role in the food chain, although it is interesting that Nercessian et al. (47) postulated in their study of the deep sea that the Gemmatimonadetes and Xanthomonadaceae could be candidates for the utilization of formaldehyde and formate, which are likely products of methane oxidation. Other potential problems with long incubation times are possible shifts in the populations of primary assimilators of methane; this might be reflected in the changing oxidation rates of methane (Fig. 1d) after about 500 h, which again highlights the need to carry out SIP incubations in as short a time as possible to incorporate sufficient 13C label for subsequent analysis.
In conclusion, the active MOB diversity observed for the water slurry and natural moisture content with 80% of type I methanotrophs and 20% of type II methanotrophs should better reflect the in situ diversity of methanotrophs. Both types of methanotrophs have been detected in various kinds of soils, including forest, rice, landfill, and agricultural soils (6, 15, 27, 57, 67). The Wytham soil is a field soil, which is not exposed to high concentrations of methane and, as for upland and forest soils (6, 33), significant proportions of type I methanotrophs closely related to Methylobacter and type II methanotrophs such as Methylocystis are present. In the future, it would be prudent to use conditions as close as possible to the natural environment and avoid unnecessarily long incubation periods when DNA-SIP is used, but often there will be a tradeoff between the length of time necessary to achieve sufficient 13C labeling of cells and the possibilities of cross-feeding of the 13C label. Sampling throughout the time course of SIP incubations will also enable labeled cells to be detected at the earliest opportunity, thus ensuring that the primary consumers of the 13C-labeled substrate are detected.
Supplementary Material
Acknowledgments
We thank NERC for funding this study through grant NE/B505389/1 to I.P.T, J.C.M, and J.I.P.
Footnotes
Published ahead of print on 12 November 2006.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Auman, A. J., S. Stolyar, A. M. Costello, and M. E. Lidstrom. 2000. Molecular characterization of methanotrophic isolates from freshwater lake sediment. Appl. Environ. Microbiol. 66:5259-5266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bodelier, P. L., P. Roslev, T. Henckel, and P. Frenzel. 2000. Stimulation by ammonium-based fertilizers of methane oxidation in soil around rice roots. Nature 403:421-424. [DOI] [PubMed] [Google Scholar]
- 3.Bodrossy, L., N. Stralis-Pavese, J. C. Murrell, S. Radajewski, A. Weilharter, and A. Sessitsch. 2003. Development and validation of a diagnostic microbial microarray for methanotrophs. Environ. Microbiol. 5:566-582. [DOI] [PubMed] [Google Scholar]
- 4.Borodina, E., M. J. Cox, I. R. McDonald, and J. C. Murrell. 2005. Use of DNA-stable isotope probing and functional gene probes to investigate the diversity of methyl chloride-utilizing bacteria in soil. Environ. Microbiol. 7:1318-1328. [DOI] [PubMed] [Google Scholar]
- 5.Boschker, H. T. S., S. C. Nold, P. Wellsbury, D. Bos, W. de Graaf, R. Pel, R. J. Parkes, and T. E. Cappenberg. 1998. Direct linking of microbial populations to specific biogeochemical processes by 13C-labeling of biomarkers. Nature 392:801-805. [Google Scholar]
- 6.Bourne, D. G., I. R. McDonald, and J. C. Murrell. 2001. Comparison of pmoA PCR primer sets as tools for investigating methanotroph diversity in three Danish soils. Appl. Environ. Microbiol. 67:3802-3809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bowman, J. P. 1999. The obligate methanotrophic bacteria: Methylococcus, Methylomonas, and Methylosinus. In M. Dworkin (ed.), The prokaryotes. Springer Verlag, New York, NY. http://link.springer-ny.com.
- 8.Chang, Y. J., P. Long, R. Geyer, A. Peacock, C. Resch, K. Sublette, S. Pfiffner, A. Smithgall, R. T. Anderson, H. A. Vrionis, J. R. Stephen, R. Dayvault, I. Ortiz-Bernad, and D. C. White. 2005. Microbial incorporation of 13C-labeled acetate at the field scale: detection of microbes responsible for reduction of U(VI). Environ. Sci. Technol. 39:9039-9048. [DOI] [PubMed] [Google Scholar]
- 9.Costello, A. M., and M. E. Lidstrom. 1999. Molecular characterization of functional and phylogenetic genes from natural populations of methanotrophs in lake sediments. Appl. Environ. Microbiol. 65:5066-5074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crossman, Z. M., P. Ineson, and R. P. Evershed. 2005. The use of 13C labeling of bacterial lipids in the characterisation of ambient methane-oxidizing bacteria in soils. Organic Geochem. 36:769-778. [Google Scholar]
- 11.Dedysh, S. N., W. Liesack, V. N. Khmelenina, N. E. Suzina, Y. A. Trotsenko, J. D. Semrau, A. M. Bares, N. S. Panikov, and J. M. Tiedje. 2000. Methylocella palustris gen. nov., sp. nov., a new methane-oxidizing acidophilic bacterium from peat bogs, representing a novel subtype of serine-pathway methanotrophs. Int. J. Syst. Evol. Microbiol. 3:955-969. [DOI] [PubMed] [Google Scholar]
- 12.DeRito, C. M., G. M. Pumphrey, and E. L. Madsen. 2005. Use of field-based stable isotope probing to identify adapted populations and track carbon flow through a phenol-degrading soil microbial community. Appl. Environ. Microbiol. 71:7858-7865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dumont, C. G., and J. C. Murrell. 2005. Stable isotope probing: linking microbial identity to function. Nat. Rev. Microbiol. 3:499-504. [DOI] [PubMed] [Google Scholar]
- 14.Dunfield, P. F., V. N. Khmelenina, N. E. Suzina, Y. A. Trotsenko, and S. N. Dedysh. 2003. Methylocella silvestris sp. nov., a novel methanotroph isolated from an acidic forest cambisol. Int. J. Syst. Evol. Microbiol. 53:1231-1239. [DOI] [PubMed] [Google Scholar]
- 15.Eller, G., and P. Frenzel. 2001. Changes in activity and community structure of methane-oxidizing bacteria over the growth period of rice. Appl. Environ. Microbiol. 67:2395-2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feselstein, J. 1989. PHYLIP-phylogeny inference package (version 3.2). Cladistics 5:164-166. [Google Scholar]
- 17.Friedrich, M. W. 2006. Stable isotope probing of DNA: insights into the function of uncultivated microorganisms from isotopically labeled metagenomes. Curr. Opin. Biotechnol. 17:59-66. [DOI] [PubMed] [Google Scholar]
- 18.Gallagher, E., L. McGuinness, C. Phelps, L. Y. Young, and L. J. Kerkhof. 2005. 13C-carrier DNA shortens the incubation time needed to detect benzoate-utilizing denitrifying bacteria by stable-isotope probing. Appl. Environ. Microbiol. 71:5192-5196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ginige, M. P., P. Hugenholtz, H. Daims, M. Wagner, J. Keller, and L. L. Blackall. 2004. Use of stable-isotope probing, full-cycle rRNA analysis, and fluorescence in situ hybridization-microautoradiography to study a methanol-fed denitrifying microbial community. Appl. Environ. Microbiol. 70:588-596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Griffiths, R. I., M. Manefield, N. Ostle, N. McNamara, A. G. O'Donnell, M. J. Bailey, and A. S. Whiteley. 2004. 13CO2 pulse-labeling of plants in tandem with stable-isotope probing: methodological considerations for examining microbial function in the rhizosphere. J. Microbiol. Methods 58:119-129. [DOI] [PubMed] [Google Scholar]
- 21.Hanson, R. S., and T. E. Hanson. 1996. Methanotrophic bacteria. Microbiol. Rev. 60:439-471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Henckel, T., U. Jackel, S. Schnell, and R. Conrad. 2000. Molecular analyses of novel methanotrophic communities in forest soil that oxidize atmospheric methane. Appl. Environ. Microbiol. 66:1801-1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Holmes, A. J., N. J. P. Owens, and J. C. Murrell. 1995. Detection of novel marine methanotrophs using phylogenetic and functional gene probes after methane enrichment. Microbiology 141:1947-1955. [DOI] [PubMed] [Google Scholar]
- 24.Holmes, A. J., P. Roslev, I. R. McDonald, N. Iversen, K. Henriksen, and J. C. Murrell. 1999. Characterization of methanotrophic bacterial populations in soils showing atmospheric methane uptake. Appl. Environ. Microbiol. 65:3312-3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Horz, H. P., V. Rich, S. Avrahami, and B. J. Bohannan. 2005. Methane-oxidizing bacteria in a California upland grassland soil: diversity and response to simulated global change. Appl. Environ. Microbiol. 71:2642-2652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hutchens, E., S. Radajewski, M. G. Dumont, I. R. McDonald, and J. C. Murrell. 2004. Analysis of methanotrophic bacteria in Movile Cave by stable isotope probing. Environ. Microbiol. 6:111-120. [DOI] [PubMed] [Google Scholar]
- 27.Jensen, S., L. Øvreås, F. L. Daae, and V. Torsvik. 1998. Diversity in methane enrichments from agricultural soil revealed by DGGE separation of PCR amplified 16S rDNA fragments. FEMS Microbiol. Ecol. 26:17-26. [Google Scholar]
- 28.Jeon, C. O., W. Park, P. Padmanabhan, C. DeRito, J. R. Snape, and E. L. Madsen. 2003. Discovery of a bacterium, with distinctive dioxygenase, that is responsible for in situ biodegradation in contaminated sediment. Proc. Natl. Acad. Sci. USA 100:13591-13596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kemp, P. F. 1995. Can we estimate bacterial growth rates from rRNA content?, p. 279-302. In I. Joint (ed.), Molecular ecology of aquatic microbes. NATO ASI series, vol. G38. Springer-Verlag, Berlin, Germany. [Google Scholar]
- 30.Kerkhof, L., and B. Ward. 1993. Comparison of nucleic acid hybridization and fluorometry for measurement of RNA/DNA relationship with growth rate in a marine bacterium. Appl. Environ. Microbiol. 59:1303-1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Knief, C., A. Lipski, and P. F. Dunfield. 2003. Diversity and activity of methanotrophic bacteria in different upland soils. Appl. Environ. Microbiol. 69:6703-6714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Knief, C., and P. F. Dunfield. 2005. Response and adaptation of different methanotrophic bacteria to low methane mixing ratios. Environ. Microbiol. 7:1307-1317. [DOI] [PubMed] [Google Scholar]
- 33.Knief, C., S. Vanitchung, N. W. Harvey, R. Conrad, P. F. Dunfield, and A. Chidthaisong. 2005. Diversity of methanotrophic bacteria in tropical upland soils under different land uses. Appl. Environ. Microbiol. 71:3826-3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-175. In M. Goodfellow and E. Stackebrandt (ed.), Nucleic acid techniques in bacterial systematics. John Wiley & Sons, Inc., New York, NY.
- 35.Lieberman, R. L., and A. C. Rosenzweig. 2004. Biological methane oxidation: regulation, biochemistry, and active site structure of particulate methane monooxygenase. Crit. Rev. Biochem. Mol. Biol. 39:147-164. [DOI] [PubMed] [Google Scholar]
- 36.Lin, J. L., S. Radajewski, B. T. Eshinimaev, Y. A. Trotsenko, I. R. McDonald, and J. C. Murrell. 2004. Molecular diversity of methanotrophs in Transbaikal soda lake sediments and identification of potentially active populations by stable isotope probing. Environ. Microbiol. 6:1049-1060. [DOI] [PubMed] [Google Scholar]
- 37.Lueders, T., B. Wagner, P. Claus, and M. W. Friedrich. 2004. Stable isotope probing of rRNA and DNA reveals a dynamic methylotroph community and trophic interactions with fungi and protozoa in oxic rice field soil. Environ. Microbiol. 6:60. [DOI] [PubMed] [Google Scholar]
- 38.Madsen, E. L. 2006. The use of stable isotope probing techniques in bioreactor and field studies on bioremediation. Curr. Opin. Biotechnol. 17:92-97. [DOI] [PubMed] [Google Scholar]
- 39.Malosso, E., L. English, D. W. Hopkins, and A. G. O'Donnell. 2004. Use of 13C-labeled plant materials and ergosterool, PLFA, and NLFA analyses to investigate organic matter decomposition in Antarctic soil. Soil Biol. Biochem. 36:165-175. [Google Scholar]
- 40.Manefield, M., A. S. Whiteley, N. Ostle, P. Ineson, and M. J. Bailey. 2002. Technical considerations for RNA-based stable isotope probing: an approach to associating microbial diversity with microbial community function. Rapid Commun. Mass Spectrom. 16:2179-2183. [DOI] [PubMed] [Google Scholar]
- 41.Manefield, M., A. S. Whiteley, R. I. Griffiths, and M. J. Bailey. 2002. RNA stable isotope probing, a novel means of linking microbial community function to phylogeny. Appl. Environ. Microbiol. 68:5367-5373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Manefield, M., A. S. Whiteley, and M. J. Bailey. 2004. What can stable isotope probing do for bioremediation? Int. Biodeterior. Biodegrad. 54:163-166. [Google Scholar]
- 43.Miller, L. G., K. L. Warner, S. M. Baesman, R. S. Oremland, I. R. McDonald, S. Radajewski, and J. C. Murrell. 2004. Degradation of methyl bromine and methyl chloride in soil microcosms: use of stable C isotope fractionation and stable isotope probing to identify reactions and the responsible microorganisms. Geochim. Cosmochim. Acta 68:3271-3283. [Google Scholar]
- 44.Morris, S. A., S. Radajewski, T. W. Willison, and J. C. Murrell. 2002. Identification of the functionally active methanotroph population in a peat soil microcosm by stable-isotope probing. Appl. Environ. Microbiol. 68:1446-1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Murrell, J. C., and H. Dalton. 1983. Nitrogen fixation in obligate methanotrophs. J. Gen. Microbiol. 129:3481-3486. [Google Scholar]
- 46.Muyzer, G., T. Brinkhoff, U. Nübel, C. Santegoeds, H. Schäfer, and C. Wawer. 1998. Denaturing gradient gel electrophoresis (DGGE) in microbial ecology, p. 1-27. In A. D. L. Akkermans, J. D. van Elsas, and F. J. de Bruijn (ed.), Molecular microbial ecology manual, vol. 3.4.4. Kluwer Academic Publishers, Dordrecht, The Netherlands. [Google Scholar]
- 47.Nercessian, O., N. Bienvenu, D. Moreira, D. Prieur, and C. Jeanthon. 2005. Diversity of functional genes of methanogens, methanotrophs and sulfate reducers in deep-sea hydrothermal environments. Environ. Microbiol. 7:118-132. [DOI] [PubMed] [Google Scholar]
- 48.Nold, S. C., H. T. S. Boschker, R. Pel, and H. J. Laanbroek. 1999. Ammonium addition inhibits 13C-methane incorporation into methanotroph membrane lipids in freshwater sediment. FEMS Microbiol. Ecol. 29:81-89. [Google Scholar]
- 49.Ostle, N., A. S. Whiteley, M. J. Bailey, D. Sleep, P. Ineson, and M. Manefield. 2003. Active microbial RNA turnover in a grassland soil estimated using a 13CO2 spike. Soil Biol. Biochem. 35:877-888. [Google Scholar]
- 50.Padmanabhan, P., S. Padmanabhan, C. DeRito, A. Gray, D. Gannon, J. R. Spape, C. S. Tsai, W. Park, C. Jeon, and E. L. Madsen. 2003. Respiration of 13C-labeled substrates added to soil in the field and subsequent 16S rRNA gene analysis of 13C-labeled soil DNA. Appl. Environ. Microbiol. 69:1614-1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Page, R. D. M. 1996. TREEVIEW: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12:357-358. [DOI] [PubMed] [Google Scholar]
- 52.Pombo, S. A., J. Kleikemper, M. H. Schroth, and J. Zeyer. 2005. Field-scale isotopic labeling of phospholipid fatty acids from acetate-degrading sulfate-reducing bacteria. FEMS Microbiol. Ecol. 51:197-207. [DOI] [PubMed] [Google Scholar]
- 53.Radajewski, S., P. Ineson, N. R. Parekh, and J. C. Murrell. 2000. Stable-isotope probing as a tool in microbial ecology. Nature 403:646-649. [DOI] [PubMed] [Google Scholar]
- 54.Radajewski, S., and J. C. Murrell. 2001. Stable isotope probing for detection of methanotrophs after enrichment with 13CH4. Methods Mol. Biol. 179:149-157. [DOI] [PubMed] [Google Scholar]
- 55.Radajewski, S., G. Webster, D. S. Reay, S. A. Morris, P. Ineson, D. B., Nedwell, J. L. Prosser, and J. C. Murrell. 2002. Identification of active methylotroph populations in an acidic forest soil by stable-isotope probing. Microbiology 148:2331-2342. [DOI] [PubMed] [Google Scholar]
- 56.Radajewski, S., I. R. McDonald, and J. C. Murrell. 2003. Stable-isotope probing of nucleic acids: a window to the function of uncultured microorganisms. Curr. Opin. Biotechnol. 14:296-302. [DOI] [PubMed] [Google Scholar]
- 57.Reay, D. S., S. Radajewski, J. C. Murrell, N. P. McNamara, and D. B. Nedwell. 2001. Impact of land-use on the activity and diversity of methane oxidizing bacteria in forest soils. Soil Biol. Biochem. 33:1613-1623. [Google Scholar]
- 58.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 59.Saunders, S. E., and J. F. Burke. 1990. Rapid isolation of miniprep DNA for double strand sequencing. Nucleic Acids Res. 18:4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Singleton, D. R., S. N. Powell, R. Sangaiah, A. Gold, L. M. Ball, and M. D. Aitken. 2005. Stable-isotope probing of bacteria capable of degrading salicylate, naphthalene, or phenanthrene in a bioreactor treating contaminated soil. Appl. Environ. Microbiol. 71:1202-1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Smith, K. S., A. M. Costello, and M. E. Lidstrom. 1997. Methane and trichloroethylene oxidation by an estuarine methanotroph, Methylobacter sp. strain BB5.1. Appl. Environ. Microbiol. 63:4617-4620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stralis-Pavese, N., A. Sessitsch, A. Weilharter, T. Reichenauer, J. Riesing, J. Csontos, J. C. Murrell, and L. Bodossy. 2004. Optimisation of diagnostic microarray for application in analysing landfill methanotroph communities under different plant covers. Environ. Microbiol. 6:347-363. [DOI] [PubMed] [Google Scholar]
- 63.Tillmann, S., C. Strömpl, K. N. Timmis, and W. R. Abraham. 2005. Stable isotope probing reveals the dominant role of Burkholderia species in aerobic degradation of PCBs. FEMS Microbiol. Ecol. 52:207-217. [DOI] [PubMed] [Google Scholar]
- 64.Treonis, A. M., N. J. Ostle, A. W. Stott, R. Primrose, S. J. Grayston, and P. Ineson. 2004. Identification of groups of metabolically active rhizosphere microorganisms by stable-isotope probing of PLFAs. Soil Biol. Biochem. 36:533-537. [Google Scholar]
- 65.Whitby, C., M. J. Bailey, K. Killham, J. C. Murrell, J. I. Prosser, A. Whiteley, and H. M. Lappin-Scott. 2005. Stable isotope probing links taxonomy with function in microbial communities. ASM News 71:169-173. [Google Scholar]
- 66.Whittenbury, R., K. C. Phillips, and J. F. Wilkinson. 1970. Enrichment, isolation and some properties of methane-utilizing bacteria. J. Gen. Microbiol. 61:205-218. [DOI] [PubMed] [Google Scholar]
- 67.Wise, M. G., J. V. McArthur, and L. J. Shimkets. 1999. Methanotroph diversity in landfill soil: isolation of novel type I and type II methanotrophs whose presence was suggested by culture-independent 16S ribosomal DNA analysis. Appl. Environ. Microbiol. 65:4887-4897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yeates, C., and M. R. Gillings. 1998. Rapid purification of DNA from soil for molecular biodiversity analysis. Lett. Appl. Microbiol. 27:49-53. [Google Scholar]
- 69.Yu, C. P., and K. H. Chu. 2005. A quantitative assay for linking microbial community function and structure of a naphthalene-degrading microbial consortium. Environ. Sci. Technol. 39:9611-9619. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.