Abstract
In the ocean's most extreme depths, pressures of 70 to 110 megapascals prevent the growth of all but the most hyperpiezophilic (pressure-loving) organisms. The physiological adaptations required for growth under these conditions are considered to be substantial. Efforts to determine specific adaptations permitting growth at extreme pressures have thus far focused on relatively few γ-proteobacteria, in part due to the technical difficulties of obtaining piezophilic bacteria in pure culture. Here, we present the molecular phylogenies of several new piezophiles of widely differing geographic origins. Included are results from an analysis of the first deep-trench bacterial isolates recovered from the southern hemisphere (9.9-km depth) and of the first gram-positive piezophilic strains. These new data allowed both phylogenetic and structural 16S rRNA comparisons among deep-ocean trench piezophiles and closely related strains not adapted to high pressure. Our results suggest that (i) the Circumpolar Deep Water acts as repository for hyperpiezophiles and drives their dissemination to deep trenches in the Pacific Ocean and (ii) the occurrence of elongated helices in the 16S rRNA genes increases with the extent of adaptation to growth at elevated pressure. These helix changes are believed to improve ribosome function under deep-sea conditions.
Low temperature and high hydrostatic pressure structure deep-sea communities outside of hydrothermal vents. Tight selection by these and other environmental parameters is considered the cause of the conspicuous absence of many deep-sea taxonomic groups from the deepest ocean environments (8, 44).
Both temperature and pressure exert their effects at many levels of bacterial physiology, from the structure of macromolecules to the rate of metabolic reactions. Adaptations to low temperature include alterations of membrane phospholipids, such as increased fatty acid unsaturation (43), enzymes characterized by high catalytic efficiency and reduced activation enthalpy (16, 20, 37, 45), and high levels of cold shock proteins, RNA helicases (9), and posttranscriptional modification of tRNA molecules (15), all of which may reduce the formation of unfavorable nucleic acid secondary structures at low temperature. In contrast with enthalpy-based temperature effects, the underlying cause of pressure effects arises from the promotion of reduced system volumes, in accordance with Le Chatelier's principle (5). Despite these thermodynamic differences, low temperature and high pressure share a surprising number of influences on biological processes. For example, membrane fluidity, permeability, and phase are similarly altered by both parameters.
As with psychrophiles, piezophiles (“high-pressure-loving” microbes) contain lipids with highly unsaturated fatty acids (6, 7). Indeed, the presence of unsaturated fatty acids is critical to growth ability at high pressure (3, 4, 19). Both low temperature and high pressure also alter protein quaternary structure (46) and nucleic acid secondary structure (50), and at the cellular level both parameters exert a pronounced influence on DNA replication and protein synthesis (27, 59).
While considerable insight has been garnered concerning the biological adaptation to extremes of temperature (13, 20), the lack of large numbers of well-characterized deep-sea organisms has limited analyses of the molecular details of pressure adaptation. This is in part a result of the technical difficulties of culturing piezophiles, including the need for pressurization systems and low-temperature incubators and for precautions to limit light exposure (57).
The recent genome analysis of the piezophilic model organism Photobacterium profundum strain SS9 (54) has suggested that adaptation to the deep sea involves a combination of modifications of gene structure and regulation. However, no piezospecific genes are yet known. Therefore, discerning general molecular trends associated with piezophily requires comparisons with other piezophilic extremophiles.
Here we present the isolation and characterization of the first hyperpiezophiles (strains for which optimal growth pressure is >60 MPa) obtained from the southern hemisphere and, from another trench environment, the first isolation of piezophilic gram-positive bacteria. The 16S rRNA-based phylogenetic assessments of these novel isolates and other geographically disseminated piezophiles reveal that (i) low-temperature piezophiles appear to be the descendants of psychrophiles present in the polar regions and (ii) stem elongation of helices 10 and 11 within the 16S rRNA molecule strongly correlates with the extent of piezophily.
MATERIALS AND METHODS
Collection of trench material for bacterial enrichment.
On 17 October 2001, scavenging amphipods of the species Hirondellea dubia were collected with insulated, baited, free-vehicle traps deployed to a depth of 9,856 m in the Kermadec Trench (32°01′.07S, 177°20′.99W). The temperature at this depth was recorded to be 1.8°C. The trap was estimated to be on the sea floor for 14 ± 0.5 h with an ascent time of 4 h 36 min to reach the sea surface, at which time the trap contents were at atmospheric pressure with a temperature of less than 5°C. The trap was recovered within 15 min of surfacing, and contents were immediately transferred to ice-chilled buckets. In a cold room (maintained at 4°C), several amphipods were placed into Kapak bags (Ampac Packaging, Cincinnati, OH) containing 5 ml of sterile, ice-cold filtered seawater. Bags were then sealed and subsequently pressurized to 99 MPa in pressure vessels which were maintained at 0°C for the duration of the cruise. Samples were at atmospheric pressure for less than 1 h. Once back at Scripps Institution of Oceanography, samples were maintained at 2°C (in pressure vessels) except for periods of less than 30 min to allow inoculation of enrichment cultures which were in turn incubated at high pressure.
Strains AT7 and AT12 are in a collection of deep-sea piezophilic bacteria at the Scripps Institution of Oceanography. These two strains originated from a water sample taken by John Burke from the Aleutian Trench at 52°53′.1N, 163°0′.0W. The sample was from a depth of 2,500 m, where the temperature was 1.8°C. The sample was collected on 22 June 1981 and put in a narrow-mouth sterile plastic bottle, kept on wet ice, and returned to the Scripps Institution of Oceanography. Strains AT7 and AT12 are from enrichments incubated at 40 MPa and begun on 14 August 1981.
Isolation of piezophiles and growth characterization.
The decomposing amphipods and surrounding medium were briefly depressurized and immediately inoculated into a beaker containing full-strength 2216 Marine Broth (Difco Laboratories, Detroit, MI). From this slurry, approximately 15 ml was loaded into sterile Samco transfer pipettes (Samco Scientific Corp., San Fernando, CA), which were heat sealed and quickly pressurized to 99 MPa in pressure vessels (57, 60). The first enrichment, performed on 19 December 2001, was incubated in a rocking water bath at 2°C. A separate enrichment was performed on 30 April 2004, and cultures were grown in rocking water baths maintained at either 2°C or 8°C. Enrichments were microscopically examined after 3 weeks for evidence of bacterial growth. Once these cultures were established, the following procedure was followed to obtain clonal isolates. Enrichments were serially diluted into fresh 2216 Marine Broth with 4% gelatin. This mixture is liquid at room temperature (∼22°C) but hardens when cooled below 15°C, thus making it a suitable medium for growing and isolating colonies. The inoculated mixture was then loaded into fresh transfer pipettes and quickly pressurized to 99 MPa. These new cultures were incubated at the same temperature (either 2°C or 8°C) as the initial enrichment and periodically examined until colonies were observed within the gelatin matrix. Colonies were extracted directly from the transfer pipette bulbs by piercing the walls with a sterile needle (21 gauge) and syringe. Isolates were extruded into ice-cold 2216 Marine Broth, reloaded into transfer pipettes, and incubated at the appropriate temperature and pressure for growth. Established cultures were maintained by transferring them into fresh medium every 2 to 3 months. During one of these transfers Colwellia sp. strain KT27 was lost.
For growth-rate determinations as a function of pressure, cultures were inoculated into multiple heat-sealable bulbs and pressurized as described above. At prescribed time intervals one bulb was removed and the optical density (at 600 nm) of the culture was recorded.
16S characterization of isolates.
Genomic DNA was extracted from pure cultures using a DNeasy kit (QIAGEN, Valencia, CA). PCR amplification was performed using the general bacterial primers 27F and 1492R (17). Automated DNA sequencing was performed on an Amersham MEGABace 500 system (Amersham Pharmacia Biotech) using general eubacterial primers 27F (5′-AGAGTTTGATCCTGGCTCAG-3′), 518R (5′-GTATTACCGCGGCTGCTG-3′), 530F (5′-GTGCCAGCAGCCGCGG-3′), 907R (5′-CCGTCAATTCATTTGAGT-3′), 926F (5′-ACTCAAAGGAATTGACGG-3′), and 1492R (5′-GGTTACCTTGTTACGACTT-3′). The resulting chromatograms were assembled into contigs by use of Sequencher software (Gene Codes Corp., Ann Arbor, MI).
Phylogenetic and structural analyses.
Phylogenetic dendrograms were reconstructed with MEGA 2.1 (31) for neighbor-joining and minimum-evolution analysis and Treefinder (29) for maximum-likelihood analysis with an HKY (Hasegawa, Kishino, Yano) model of evolution (25). Since conservation of the topology of the trees was independent of the method used, only the maximum-likelihood trees are shown. Bootstrap support was from 1,000 bootstrap replicates. Structure analysis of the 16S rRNA variable regions was performed by dynamic programming energy minimization as implemented in the VIENNA RNA package (26). A folding temperature of 15°C was used, and GU pairing was allowed.
Semiquantitative PCR and RT-PCR.
Bacterial cells were grown in 2216 Marine Broth amended with glucose (20 mM) and HEPES (pH 7.5) (100 mM). For reverse transcription-PCR (RT-PCR), P. profundum SS9 was grown to an optical density at 600 nm of 0.1 to 0.2 and harvested, and total RNA was extracted using RNABee (Tel-Test, Friendswood, TX) and further purified with RNeasy columns (QIAGEN, Valencia, CA). Genomic DNA was removed using in-column digestion with DNase (QIAGEN, Valencia, CA).
RT-PCR was performed using a OneStep RT-PCR kit (QIAGEN, Valencia, CA) with partially degenerate primers PIEZOLOOPF (5′-GAAACGAYDGCTAATACCGCAT-3′) and PIEZOLOOPR (5′-TTACCYYACCAACWAGCTAATC-3′), which amplify the region containing helices 10 and 11 in most γ-proteobacteria.
Semiquantitative PCR was performed with Taq polymerase (Invitrogen, Carlsbad, CA) on genomic DNA with the same primers but omitting the reverse-transcriptase step in the PCR cycle.
For the quantification, PCR amplification products were removed at cycles 20, 25, and 30 and analyzed in 10% Tris-borate-EDTA acrylamide gels stained with ethidium bromide. The PCR program for amplification was 94°C for 5 min, 94°C for 25 s, 56°C for 25 s, and 72°C for 35 s followed by a final extension for 5 min. The gel images were analyzed for peak intensity using a ChemiImager 5500 system (Alpha Innotech Corp., San Leandro, CA), and the ratio between the intensities of the higher bands and the lowermost band was calculated.
Nucleotide sequence accession numbers.
All the 16S rRNA gene sequences determined in this work were deposited in GenBank under accession numbers DQ027051 to DQ027062.
RESULTS AND DISCUSSION
Isolation of additional piezophiles.
Four new piezophiles were isolated, two from the Aleutian Trench and two from the Kermadec Trench. Strains AT7 and AT12, isolated from the Aleutian Trench, were identified as members of the genus Carnobacterium. These strains are closely related to the recently isolated Carnobacterium pleistocenium (42) and to Antarctic isolates of the family Carnobacteriaceae (22). Members of this family have been previously detected in rRNA surveys of deep-ocean sediments (38), but this is the first report of a piezophilic isolate of this species as well as the first gram-positive piezophile ever identified. The pressure range for growth of AT7 and AT12 was 0.1 to 60 MPa, with an optimum at 15 MPa (58). Interestingly the sister taxon Marinilactibacillus contains only two marine isolates, one psychrophilic (28) and the other psychropiezotolerant (51).
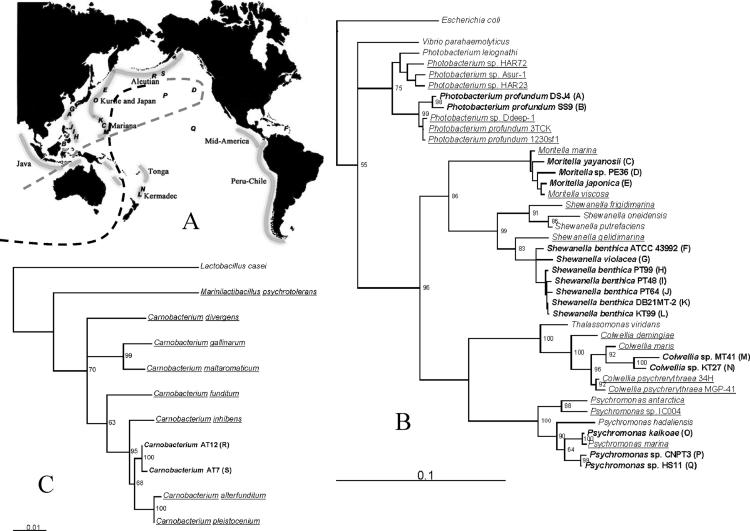
Strains KT27 and KT99 were acquired from a depth of 9,856 m (ambient temperature, 1.8°C) in the Kermadec Trench, located in the southwest Pacific Ocean. Both isolates are piezophilic and grow well at the in situ pressure of 98 MPa. They represent the first psychropiezophiles from the southern hemisphere. Phylogenetic analysis revealed that the closest cultured relatives of KT27 and KT99 are the isolates Colwellia sp. strain MT41 (56) and Shewanella sp. strain PT99 (18, 56), respectively (Fig. 1). Both MT41 and PT99 originated from extreme depths of deep-ocean trenches within the northern hemisphere of the Pacific Ocean.
FIG. 1.
Phylogenetic relationship between cultured deep-sea isolates and relatives from shallow waters. Maximum-likelihood trees were computed as described in Materials and Methods. Piezophilic and piezotolerant isolates are indicated in bold, with a corresponding letter referring to the approximate site of isolation on the world map. Psychrophilic and psychrotolerant strains are underlined. Reference and accession numbers for each sequence are given in Table S1 in the supplemental material. (A) Approximate world locations of the site of collection for each strain. The dashed line is the approximate path of the global conveyor belt in the Pacific Ocean; the black portion of the line represents the deep current, and the gray portion represents the surface current. (B) Phylogenetic tree depicting the relationships of the γ-proteobacteria strains. (C) Phylogenetic tree depicting the relationships of the Carnobacteriaceae strains.
Origin and dispersal of psychropiezophiles.
In order to obtain a broader perspective on the evolutionary history of existing psychropiezophiles, phylogenetic tree reconstruction was performed for a total of 20 psychropiezophiles and their closest gram-positive or gram-negative relatives (Fig. 1; also see Table S1 in the supplementary material). Interestingly, the 16S sequences of all the piezophiles share high similarity with those of nonpiezophilic isolates from Antarctica. Moreover, some genera containing piezophiles (Colwellia, Psychromonas, and Moritella) appear to be largely restricted to cold waters of various depths and are underrepresented in rRNA surveys (1) and absent from environmental shotgun sequences (53) obtained from temperate waters.
These facts argue against the idea of piezophily arising from shallow-water mesophilic sources, an evolutionary path that would also seem unlikely on the grounds that it would require concurrent adaptations to both high pressure and low temperature. Rather, the most parsimonious explanation of the data is that psychrophiles can give rise to psychropiezophiles. Pressure and temperature exert overlapping effects on many microbial processes (18). In most microorganisms increasing pressure results in a shift to higher cardinal growth temperatures (11, 30, 35, 56), suggesting a partial compensation between the two physical factors. Thus, a logical hypothesis is that initial acclimation to high pressure is facilitated by preexisting adaptations to low temperature. Genome comparisons within the well-studied γ subdivision of the phylum Proteobacteria might be very instructive in this context, since it includes two orders that contain psychropiezophiles: the Vibrionales and the Alteromonadales (18, 40, 41). The genome sequence of the piezophile Photobacterium profundum has already been noted for the similarity of its gene content to that of the cold-adapted Antarctic bacterium Pseudoalteromonas haloplanktis TAC125 (36).
Cold Antarctic waters and their associated shallow-water psychrophiles are a likely source of the piezophiles populating the deepest depths of the Pacific Ocean. Antarctic Bottom Water, mixing with North Atlantic Deep Water, forms the lower Circumpolar Deep Water, some of which escapes to the north, first encountering the Kermadec Trench and the Tonga Trench (49, 52). There, strains acquiring mutations enhancing their fitness at that depth could reproduce, and their descendants could then hitchhike on the “global conveyor belt” (49, 52) of deepwater circulation to other trenches. This dispersal mechanism could also explain the apparent paradox of the similarity between Arctic and Antarctic psychrophilic bacterial communities (48). Since the survival of a psychrophile crossing the equator in the warm surface waters is unlikely, the two communities should be geographically isolated and should therefore be evolving independently. Deep-sea currents provide a plausible mechanism for the mixing of Arctic and Antarctic psychrophilic communities. Alternatively, this community similarity could be a result of the current level of resolution in the available molecular data, as suggested by Whitaker et al. (55). In-depth comparisons between the genomes of psychrophiles from the two hemispheres, along with those of related piezophiles, should be undertaken to better evaluate this hypothesis.
Convergent evolution of rRNA structure among piezophilic γ-proteobacteria.
In the genera Photobacterium, Colwellia, and Shewanella, the majority of the 16S rRNA alignment disparities among sister strains from different depths comes from a few short insertions (Fig. 2).
FIG. 2.
Alignment of the relevant portion of the 16S rRNA gene, highlighting the elongated loops in the deep-sea strains.
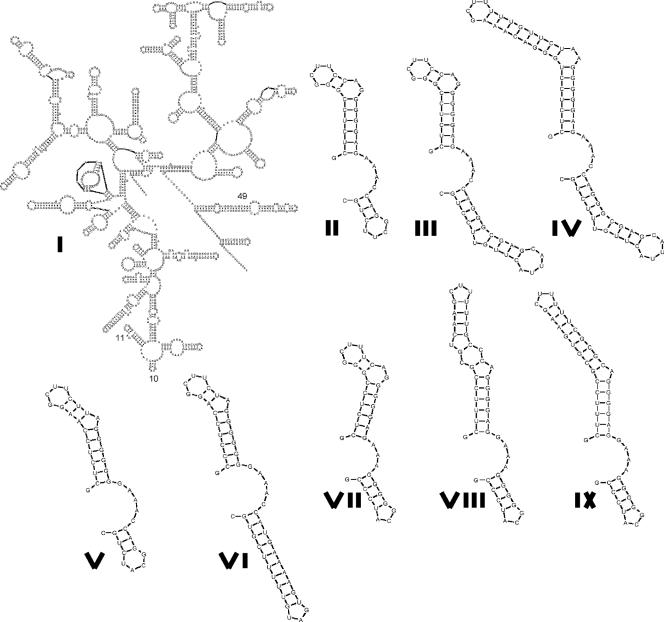
Folding models show that these insertions result in elongations to the stems of helices 10 (Escherichia coli positions 184 to 193) and 11 (Escherichia coli positions 198 to 219), while the loop sequence is conserved (Fig. 3). Although these two helices are hypervariable regions of the 16S rRNA (12), analysis of the alignment of 800 γ-proteobacterial sequences from the CRW website (12) (http://www.rna.icmb.utexas.edu) reveals that the longer stems are an almost exclusive feature of piezophiles and of hyperpiezophiles in particular. The only exceptions are Pasteurella testudinis and Salinivibrio costicola, which also possessed similarly structured longer loops.
FIG. 3.
Structural comparison of the relevant portions of the 16S rRNAs (helices 10 and 11) of piezophilic and nonpiezophilic strains. Alignments of the same regions are shown in Fig. 2. I, 16S structure of Escherichia coli, with the locations of helices 10, 11, and 49 indicated; II to IV, Photobacterium profundum SS9 helices 10 and 11 from different ribotypes; V, Colwellia psychrerythraea 34H; VI, Colwellia sp. strain MT41; VII, Shewanella oneidensis MR1; VIII, Shewanella benthica PT99; IX, Shewanella benthica KT99.
This unique feature of the piezophilic ribosome is consistent with the well-documented sensitivity of mesophile ribosomes to elevated hydrostatic pressure (24, 32). E. coli ribosome piezosensitivity has been linked to ribosome dissociation, with the 30S subunit being the most sensitive (33, 39). In fact, hybrid ribosomes containing the 30S subunit from the piezotolerant Pseudomonas bathycetes coupled to the 50S subunit of E. coli showed remarkable pressure tolerance in vitro (33).
Functionally, the stems encompassing E. coli residues 122 to 239 have been implicated in interactions with protein S20 of the ribosome (14), which is essential for the assembly of a functional ribosome. Mutants defective in S20 synthesis are impaired in their capability to associate 30S and 50S subunits and in translation initiation (23).
Additional insight into the function of these helices comes from the genome analysis of Photobacterium profundum strain SS9 (54). This eurypiezophilic bacterium has a record number of ribosomal operons (16), with high intragenomic variability within the operons (4% nucleotide divergence). Most of the sequence variation within the 16S is due to indels in helices 10, 11, and 49 (E. coli nucleotide positions 1409 to 1491), allowing for a total of five ribotypes (Fig. 4). Helix 49 has also been implicated in the interaction with protein S20 and in interfacing the 30S and 50S subunits (21). Semiquantitative RT-PCR performed on total RNA extracted from P. profundum SS9 cells showed that all ribotypes are constitutively expressed regardless of the pressure conditions (data not shown).
FIG. 4.
Ribotype (A), location (B), and alignment (C) of the relevant portions for the 15 16S rRNA genes of P. profundum strain SS9. The data are derived from reference 54, using the genome assembly data from February 2004.
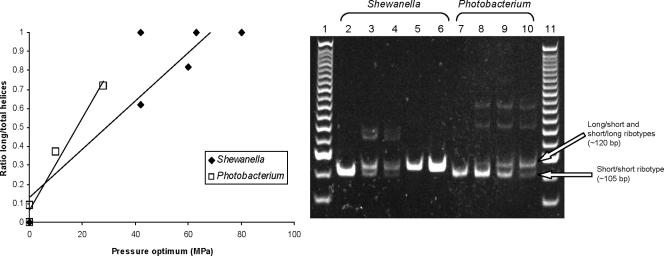
P. profundum contains both piezophilic and nonpiezophilic strains (10, 47). Among the different P. profundum strains, semiquantitative PCR revealed that the proportion of ribotypes with longer stems is directly correlated to the optimal growth pressure (r2 = 0.97) (Fig. 5), with the piezophilic strains of P. profundum (SS9, DSJ4) having a higher proportion of the longer stems. A similar trend is observed within Shewanella strains (r2 = 0.79) (Fig. 5), another cosmopolitan oceanic genus with members adapted to various depths. It is possible that the longer loops could have been acquired through lateral gene transfer of ribosomal operons, a process thought to contribute to the high intragenomic variability in genomes with multicopy 16S rRNA genes (2). In this scenario, shallow-water strains such as those belonging to the genera Photobacterium and Shewanella could acquire deep-sea piezophile ribosomal 16S rRNA genes and homologous recombination and gene conversion would then generate the various intragenomic ribotypes (34). Alternatively, the conditions in the deep sea might alter the selection pressure on loops 10, 11, and 49, generating the longer ribotypes. Interestingly, the sequence of the elongated loops appears to be genus specific in strains isolated thousand of miles apart (Fig. 2). Notably, not all piezophilic γ-proteobacterial 16S rRNA gene sequences available in GenBank exhibit the long loops. However, if only some of the rRNA operons within a given piezophile were to have the long loops then they might not be detected during 16S cloning and sequencing. Among the Shewanella strains examined, only F1A and KT99 have the longer ribotype exclusively (PCR analysis, Fig. 5). Alternatively, other ribosome modifications might compensate for the lack of these long loops at high pressure in other bacterial genera. Additional studies are currently under way to assess the physiological importance of the piezospecific loops.
FIG. 5.
(Left panel) Relationship between optimal growth pressure and loop elongation in the genera Shewanella and Photobacterium. (Right panel) Detection of ribotype composition in various strains by semiquantitative PCR. The assay was performed for 25 cycles as described in Materials and Methods (lane 1, 25-bp ladder; lane 2, PE36; lane 3, PT48; lane 4, PT64; lane 5, F1A; lane 6, KT99; lane 7, 3TCK; lane 8, 1230; lane 9, DSJ4; lane 10, SS9; lane 11, 25-bp ladder). Because of chimera formation, in Photobacterium strains only the short-short, the short-long, and the long-short ribotypes could be detected with the general primers. The long-long ribotype had to be amplified with specific primers.
Supplementary Material
Acknowledgments
We thank the staff and crew of R/V Melville for technical assistance. We are indebted to Lynne Talley for insightful discussion. We acknowledge Jean Brenchley and Jennifer Biddle for providing strains.
We are grateful to the National Science Foundation (grants OCE 99-07651, MCB02-37059, and MCB05-44524) for financial support. Additional support to F.M.L. was from the Ermanno Bellio award from the University of Padova.
This article is dedicated to the memory of Ermanno Bellio (1974-1998).
Footnotes
Published ahead of print on 8 December 2006.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Acinas, S. G., V. Klepac-Ceraj, D. E. Hunt, C. Pharino, I. Ceraj, D. L. Distel, and M. F. Polz. 2004. Fine-scale phylogenetic architecture of a complex bacterial community. Nature 430:551-554. [DOI] [PubMed] [Google Scholar]
- 2.Acinas, S. G., L. A. Marcelino, V. Klepac-Ceraj, and M. F. Polz. 2004. Divergence and redundancy of 16S rRNA sequences in genomes with multiple rrn operons. J. Bacteriol. 186:2629-2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allen, E. E., and D. H. Bartlett. 2000. FabF is required for piezoregulation of cis-vaccenic acid levels and piezophilic growth of the deep-sea bacterium Photobacterium profundum strain SS9. J. Bacteriol. 182:1264-1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Allen, E. E., D. Facciotti, and D. H. Bartlett. 1999. Monounsaturated but not polyunsaturated fatty acids are required for growth of the deep-sea bacterium Photobacterium profundum SS9 at high pressure and low temperature. Appl. Environ. Microbiol. 65:1710-1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Balny, C., P. Masson, and K. Heremans. 2002. Frontiers in high pressure biochemistry and biophysics. Elsevier, Amsterdam, The Netherlands.
- 6.Bartlett, D. H. 2006. Extremophilic Vibrionaceae, p. 156-171. In F. L. Thompson, B. Austin, and J. Swings (ed.), The biology of vibrios. ASM Press, Washington, DC.
- 7.Bartlett, D. H. 2002. Pressure effects on in vivo microbial processes. Biochim. Biophys. Acta 1595:367-381. [DOI] [PubMed] [Google Scholar]
- 8.Belyaev, G. M. 1989. Deep-sea ocean trenches and their fauna. Nauka Publishing House, Moscow, Russia. (In Russian.)
- 9.Berger, F., P. Normand, and P. Potier. 1997. capA, a cspA-like gene that encodes a cold acclimation protein in the psychrotrophic bacterium Arthrobacter globiformis SI55. J. Bacteriol. 179:5670-5676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Campanaro, S., A. Vezzi, N. Vitulo, F. M. Lauro, M. D'Angelo, F. Simonato, A. Cestaro, G. Malacrida, G. Bertoloni, G. Valle, and D. H. Bartlett. 2005. Laterally transferred elements and high pressure adaptation in Photobacterium profundum strains. BMC Genomics 6:122. http://www.biomedcentral.com/1471-2164/6/122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Canganella, F., J. M. Gonzalez, M. Yanagibayashi, C. Kato, and K. Horikoshi. 1997. Pressure and temperature effects on growth and viability of the hyperthermophilic archaeon Thermococcus peptonophilus. Arch. Microbiol. 168:1-7. [DOI] [PubMed] [Google Scholar]
- 12.Cannone, J. J., S. Subramanian, M. N. Schnare, J. R. Collett, L. M. D'Souza, Y. S. Du, B. Feng, N. Lin, L. V. Madabusi, K. M. Muller, N. Pande, Z. D. Shang, N. Yu, and R. R. Gutell. 2002. The comparative RNA web (CRW) site: an online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics 3:15. http://www.biomedcentral.com/1471-2105/3/2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Charlier, D., and L. Droogmans. 2005. Microbial life at high temperature, the challenges, the strategies. Cell. Mol. Life Sci. 62:2974-2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cormack, R. S., and G. A. Mackie. 1991. Mapping ribosomal protein S20-16S ribosomal-RNA interactions by mutagenesis. J. Biol. Chem. 266:18525-18529. [PubMed] [Google Scholar]
- 15.Dalluge, J. J., T. Hamamoto, K. Horikoshi, R. Y. Morita, K. O. Stetter, and J. A. McCloskey. 1997. Posttranscriptional modification of tRNA in psychrophilic bacteria. J. Bacteriol. 179:1918-1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.D'Amico, S., T. Collins, J. C. Marx, G. Feller, and C. Gerday. 2006. Psychrophilic microorganisms: challenges for life. EMBO Rep. 7:385-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.DeLong, E. F. 1992. Archaea in coastal marine environments. Proc. Natl. Acad. Sci. USA 89:5685-5689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.DeLong, E. F., D. G. Franks, and A. A. Yayanos. 1997. Evolutionary relationships of cultivated psychrophilic and barophilic deep-sea bacteria. Appl. Environ. Microbiol. 63:2105-2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.DeLong, E. F., and A. A. Yayanos. 1985. Adaptation of the membrane lipids of a deep-sea bacterium to changes in hydrostatic pressure. Science 228:1101-1102. [DOI] [PubMed] [Google Scholar]
- 20.Feller, G., and C. Gerday. 2003. Psychrophilic enzymes: hot topics in cold adaptation. Nat. Rev. Microbiol. 1:200-208. [DOI] [PubMed] [Google Scholar]
- 21.Firpo, M. A., and A. E. Dahlberg. 1998. The importance of base pairing in the penultimate stem of Escherichia coli 16S rRNA for ribosomal subunit association. Nucleic Acids Res. 26:2156-2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Franzmann, P. D., P. Hopfl, N. Weiss, and B. J. Tindall. 1991. Psychotrophic, lactic acid-producing bacteria from anoxic waters in Ace Lake, Antarctica; Carnobacterium funditum sp. nov. and Carnobacterium alterfunditum sp. nov. Arch. Microbiol. 156:255-262. [DOI] [PubMed] [Google Scholar]
- 23.Götz, F., E. R. Dabbs, and C. O. Gualerzi. 1990. Escherichia coli 30S mutants lacking protein S20 are defective in translation initiation. Biochim. Biophys. Acta 1050:93-97. [DOI] [PubMed] [Google Scholar]
- 24.Gross, M., and R. Jaenicke. 1990. Pressure-induced dissociation of tight couple ribosomes. FEBS Lett. 267:239-241. [DOI] [PubMed] [Google Scholar]
- 25.Hasegawa, M., H. Kishino, and T. A. Yano. 1985. Dating of the human ape splitting by a molecular clock of mitochondrial DNA. J. Mol. Evol. 22:160-174. [DOI] [PubMed] [Google Scholar]
- 26.Hofacker, I. L., W. Fontana, P. F. Stadler, L. S. Bonhoeffer, M. Tacker, and P. Schuster. 1994. Fast folding and comparison of RNA secondary structures. Monatsh. Chem. 125:167-188. (In German.) [Google Scholar]
- 27.Ishii, A., T. Oshima, T. Sato, K. Nakasone, H. Mori, and C. Kato. 2005. Analysis of hydrostatic pressure effects on transcription in Escherichia coli by DNA microarray procedure. Extremophiles 9:65-73. [DOI] [PubMed] [Google Scholar]
- 28.Ishikawa, M., K. Nakajima, M. Yanagi, Y. Yamamoto, and K. Yamasato. 2003. Marinilactibacillus psychrotolerans gen. nov., sp. nov., a halophilic and alkaliphilic marine lactic acid bacterium isolated from marine organisms in temperate and subtropical areas of Japan. Int. J. Syst. Evol. Microbiol. 53:711-720. [DOI] [PubMed] [Google Scholar]
- 29.Jobb, G., A. von Haeseler, and K. Strimmer. 2004. TREEFINDER: a powerful graphical analysis environment for molecular phylogenetics. BMC Evol. Biol. 4:18. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 30.Kaye, J. Z., and J. A. Baross. 2004. Synchronous effects of temperature, hydrostatic pressure, and salinity on growth, phospholipid profiles, and protein patterns of four Halomonas species isolated from deep-sea hydrothermal-vent and sea surface environments. Appl. Environ. Microbiol. 70:6220-6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kumar, S., K. Tamura, I. B. Jakobsen, and M. Nei. 2001. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics 17:1244-1245. [DOI] [PubMed] [Google Scholar]
- 32.Landau, J. V. 1967. Induction transcription and translation in Escherichia coli: a hydrostatic pressure study. Biochim. Biophys. Acta 149:506-512. [DOI] [PubMed] [Google Scholar]
- 33.Landau, J. V., W. P. Smith, and D. H. Pope. 1977. Role of 30S ribosomal subunit, initiation factors, and specific ion concentration in barotolerant protein synthesis in Pseudomonas bathycetes. J. Bacteriol. 130:154-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liao, D. Q. 2000. Gene conversion drives within genic sequences: concerted evolution of ribosomal RNA genes in bacteria and archaea. J. Mol. Evol. 51:305-317. [DOI] [PubMed] [Google Scholar]
- 35.Marteinsson, V. T., P. Moulin, J. L. Birrien, A. Gambacorta, M. Vernet, and D. Prieur. 1997. Physiological responses to stress conditions and barophilic behavior of the hyperthermophilic vent archaeon Pyrococcus abyssi. Appl. Environ. Microbiol. 63:1230-1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Médigue, C., E. Krin, G. Pascal, V. Barbe, A. Bernsel, P. N. Bertin, F. Cheung, S. Cruveiller, S. D'Amico, A. Duilio, G. Fang, G. Feller, C. Ho, S. Mangenot, G. Marino, J. Nilsson, E. Parrilli, E. P. C. Rocha, Z. Rouy, A. Sekowska, M. L. Tutino, D. Vallenet, G. von Heijne, and A. Danchin. 2005. Coping with cold: the genome of the versatile marine Antarctica bacterium Pseudoalteromonas haloplanktis TAC125. Genome Res. 15:1325-1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Methé, B. A., K. E. Nelson, J. W. Deming, B. Momen, E. Melamud, X. J. Zhang, J. Moult, R. Madupu, W. C. Nelson, R. J. Dodson, L. M. Brinkac, S. C. Daugherty, A. S. Durkin, R. T. DeBoy, J. F. Kolonay, S. A. Sullivan, L. W. Zhou, T. M. Davidsen, M. Wu, A. L. Huston, M. Lewis, B. Weaver, J. F. Weidman, H. Khouri, T. R. Utterback, T. V. Feldblyum, and C. M. Fraser. 2005. The psychrophilic lifestyle as revealed by the genome sequence of Colwellia psychrerythraea 34H through genomic and proteomic analyses. Proc. Natl. Acad. Sci. USA 102:10913-10918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Newberry, C. J., G. Webster, B. A. Cragg, R. J. Parkes, A. J. Weightman, and J. C. Fry. 2004. Diversity of prokaryotes and methanogenesis in deep subsurface sediments from the Nankai Trough, Ocean Drilling Program Leg 190. Environ. Microbiol. 6:274-287. [DOI] [PubMed] [Google Scholar]
- 39.Niven, G. W., C. A. Miles, and B. M. Mackey. 1999. The effects of hydrostatic pressure on ribosome conformation in Escherichia coli: an in vivo study using differential scanning calorimetry. Microbiology 145:419-425. [DOI] [PubMed] [Google Scholar]
- 40.Nogi, Y., C. Kato, and K. Horikoshi. 2002. Psychromonas kaikoae sp. nov., a novel piezophilic bacterium from the deepest cold-seep sediments in the Japan Trench. Int. J. Syst. Evol. Microbiol. 52:1527-1532. [DOI] [PubMed] [Google Scholar]
- 41.Nogi, Y., C. Kato, and K. Horikoshi. 1998. Taxonomic studies of deep-sea barophilic Shewanella strains and description of Shewanella violacea sp. nov. Arch. Microbiol. 170:331-338. [DOI] [PubMed] [Google Scholar]
- 42.Pikuta, E. V., D. Marsic, A. Bej, J. Tang, P. Krader, and R. B. Hoover. 2005. Carnobacterium pleistocenium sp. nov., a novel psychrotolerant, facultative anaerobe isolated from permafrost of the Fox Tunnel in Alaska. Int. J. Syst. Evol. Microbiol. 55:473-478. [DOI] [PubMed] [Google Scholar]
- 43.Russell, N. J. 1997. Psychrophilic bacteria—molecular adaptations of membrane lipids. Comp. Biochem. Physiol. 118:489-493. [DOI] [PubMed] [Google Scholar]
- 44.Russell, N. J., and D. S. Nichols. 1999. Polyunsaturated fatty acids in marine bacteria—a dogma rewritten. Microbiology 145:767-779. [DOI] [PubMed] [Google Scholar]
- 45.Siddiqui, K. S., and R. Cavicchioli. 2006. Cold-adapted enzymes. Annu. Rev. Biochem. 75:403-433. [DOI] [PubMed] [Google Scholar]
- 46.Silva, J. L., and G. Weber. 1993. Pressure stability of proteins. Annu. Rev. Phys. Chem. 44:89-113. [DOI] [PubMed] [Google Scholar]
- 47.Simonato, F., S. Campanaro, F. M. Lauro, A. Vezzi, M. D'Angelo, N. Vitulo, G. Valle, and D. H. Bartlett. 2006. Piezophilic adaptation: a genomic point of view. J. Biotechnol. 126:11-25. [DOI] [PubMed] [Google Scholar]
- 48.Staley, J. T., and J. J. Gosink. 1999. Poles apart: biodiversity and biogeography of sea ice bacteria. Annu. Rev. Microbiol. 53:189-215. [DOI] [PubMed] [Google Scholar]
- 49.Stommel, H. 1958. The abyssal circulation. Deep-Sea Res. 5:80-82. [Google Scholar]
- 50.Tang, G. Q., N. Tanaka, and S. Kunugi. 1998. In vitro increases in plasmid DNA supercoiling by hydrostatic pressure. Biochim. Biophys. Acta 1443:364-368. [DOI] [PubMed] [Google Scholar]
- 51.Toffin, L., K. Zink, C. Kato, P. Pignet, A. Bidault, N. Bienvenu, J. L. Birrien, and D. Prieur. 2005. Marinilactibacillus piezotolerans sp. nov., a novel marine lactic acid bacterium isolated from deep sub-seafloor sediment of the Nankai Trough. Int. J. Syst. Evol. Microbiol. 55:345-351. [DOI] [PubMed] [Google Scholar]
- 52.Tsujino, H., H. Hasumi, and N. Suginohara. 2000. Deep Pacific circulation controlled by vertical diffusivity at the lower thermocline depths. J. Phys. Oceanogr. 30:2853-2865. [Google Scholar]
- 53.Venter, J. C., K. Remington, J. F. Heidelberg, A. L. Halpern, D. Rusch, J. A. Eisen, D. Y. Wu, I. Paulsen, K. E. Nelson, W. Nelson, D. E. Fouts, S. Levy, A. H. Knap, M. W. Lomas, K. Nealson, O. White, J. Peterson, J. Hoffman, R. Parsons, H. Baden-Tillson, C. Pfannkoch, Y. H. Rogers, and H. O. Smith. 2004. Environmental genome shotgun sequencing of the Sargasso Sea. Science 304:66-74. [DOI] [PubMed] [Google Scholar]
- 54.Vezzi, A., S. Campanaro, M. D'Angelo, F. Simonato, N. Vitulo, F. M. Lauro, A. Cestaro, G. Malacrida, B. Simionati, N. Cannata, C. Romualdi, D. H. Bartlett, and G. Valle. 2005. Life at depth: Photobacterium profundum genome sequence and expression analysis. Science 307:1459-1461. [DOI] [PubMed] [Google Scholar]
- 55.Whitaker, R. J., D. W. Grogan, and J. W. Taylor. 2003. Geographic barriers isolate endemic populations of hyperthermophilic archaea. Science 301:976-978. [DOI] [PubMed] [Google Scholar]
- 56.Yayanos, A. A. 1986. Evolutional and ecological implications of the properties of deep-sea barophilic bacteria. Proc. Natl. Acad. Sci. USA 83:9542-9546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yayanos, A. A. 1995. Microbiology to 10,500 meters in the deep sea. Annu. Rev. Microbiol. 49:777-805. [DOI] [PubMed] [Google Scholar]
- 58.Yayanos, A. A., and E. F. DeLong. 1987. Deep-sea bacterial fitness to environmental temperatures and pressure, p. 17-32. In H. W. Jannasch, R. E. Marquis, and A. M. Zimmerman (ed.), Current perspectives in high pressure biology. Academic Press, Toronto, Ontario, Canada.
- 59.Yayanos, A. A., and E. C. Pollard. 1969. A study of effects of hydrostatic pressure on macromolecular synthesis in Escherichia coli. Biophys. J. 9:1464-1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yayanos, A. A., and R. Vanboxtel. 1982. Coupling device for quick high-pressure connections to 100 MPa. Rev. Sci. Instrum. 53:704-705. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.